Abstract
Women carrying germline mutations in BRCA1 are at a substantially elevated risk of breast cancer and their tumors typically have distinctive morphological features. We hypothesised that constitutional methylation of the BRCA1 promoter region could give rise to such breast cancers in women. We selected 255 women diagnosed with breast cancer before the age of 40 years for whom BRCA1 germline mutations had not been identified. 52 had five or more of nine BRCA1 mutation-associated morphological features (group 1), 39 had four (group 2), and 164 had three or less (group 3). The prevalence of detectable BRCA1 promoter methylation in peripheral blood DNA decreased from 31% to 10% to 5% across groups 1–3, respectively (p=0.000002) and was significantly greater than the 4% frequency in unaffected controls (p=0.004). Peripheral blood methylation was associated with a 3.5-fold (95% CI 1.4 – 10.5) increased risk of having early onset breast cancer. Methylation was consistently mosaic in the peripheral blood where the estimated allelic frequency of BRCA1 promoter methylation ranged from 0.1% to 17%. Group 1 women but not group 3 women with detectable methylation of peripheral blood DNA had high levels of BRCA1 promoter methylation of their tumor DNA, indicating that constitutional BRCA1 methylation strongly predisposes towards the development of BRCA1 methylated tumors that then have features resembling BRCA1 mutated tumors. Screening peripheral blood for BRCA1 promoter methylation might thus predict early-onset breast cancers. This raises the possibility of chemoprevention or other intervention to diminish the risk of developing breast cancer in these women.
Keywords: predisposition, cancer epigenetics, epimutation, prevention, biomarker
Introduction
Inactivating germline mutations in BRCA1 act as an initial driver of breast carcinogenesis (1). Carriers of BRCA1 germline mutations typically develop tumors that have characteristic morphological features that include a high mitotic index, a trabecular growth pattern, little or no tubule formation, malignant nuclear grade, necrosis, nuclear pleomorphism, circumscribed growth pattern, pushing margins and a moderate to intense lymphocytic infiltrate (2–3) (Southey et al., submitted).
Germline mutations in BRCA1 currently explain less than 5% of all breast cancers, even for those women diagnosed before the age of 40 years (4–7). However, a substantial proportion of breast cancers diagnosed in women who have been found not to carry BRCA1 mutations have the characteristic morphological features of the tumors arising in women with germline BRCA1 mutations. Indeed, only 25% of breast cancers diagnosed before age of 40 years with a BRCA1 mutation-associated morphology - defined by five or more of the above nine features - occur in women with a detectable BRCA1 germline mutation (Southey et al., submitted).
Consequently, alternative mechanisms that could inactivate BRCA1 are of interest. Constitutional methylation (methylation of specific genes detectable in normal tissues either in complete or in mosaic form) is increasingly being considered as a mechanism for cancer predisposition (8). Notably, constitutional methylation of the MLH1 gene has been reported in a proportion of colorectal cancer patients whose tumors phenocopy those arising in individuals with germline mutations of the MLH1 gene (9–11). Peripheral blood is a convenient tissue to assay for constitutional methylation as its collection is considered non-invasive.
It is likely that constitutional methylation of BRCA1 could also be associated with increased risk of breast carcinogenesis. We previously published a pilot study of familial breast cancer patients in which low to moderate levels of BRCA1 methylation were detected in the peripheral blood-derived DNA of three of seven mutation negative women that had tumors with typical BRCA1-type morphology. Given that documented cases of transmission of germline transmission of altered methylation patterns are rare (12), we considered it desirable to analyse constitutional methylation of BRCA1 independently of family history.
In this study, we have used a large population-based study of early-onset breast cancer in which the tumors have been carefully analysed with regard to their pathology, to determine the relationship of BRCA1 promoter methylation in readily accessible normal tissue (peripheral blood) to breast cancer susceptibility and tumor morphology.
Materials and Methods
Individuals and study samples
Investigations were performed after approval by Human Research Ethics Committees of the Peter MacCallum Cancer Institute and The University of Melbourne. Individuals in the study were enrolled in the Australian site (ABCFR) of the Breast Cancer Family Registry (BCFR). The ABCFR includes a population-based case-control-family study of early-onset breast cancer diagnosed before the age of 40 years (13–15). Written informed consent was obtained from each subject. DNA was extracted from stored buffy-coats as previously described (6) for those recruited before 1995 and from whole peripheral blood by the use of spin columns (Mini blood spin columns; Qiagen, Hilden, Germany) for those recruited from 1995 onward. BRCA1 mutation screening of these subjects included a combination of protein truncation testing, Sanger sequencing of exonic and flanking intronic regions, screening for large genomic alterations, specific testing for founder mutations, 2-dimensional gel electrophoresis and via BRCAnalysis at Myriad Genetics (6, 13–14, 16–18).
Pathology review
The first primary invasive breast tumor for each woman was retrieved for 457 of 856 tumors (54%) arising and reviewed as of December 2007. The reasons for not being able to retrieve tissue included: lack of participant consent, loss of material at the diagnostic centre, the diagnostic laboratory not being willing or able to provide the material, and inability to locate material in centres that had moved or had undergone amalgamation. These are unlikely to be related to age at diagnosis, family history or other factors potentially confounding the analyses of this study.
Retrieved cases were systematically reviewed and scored for morphology features. The pathology scoring system used defines BRCA1 mutation-associated morphology when the tumor has at least five of the following nine features: mitotic index of 50 per 10 high power fields or greater, malignant nuclear grade, little or no (<10%) tubule formation, a trabecular growth pattern (primary or secondary), a syncytial growth pattern, pushing margins (>50%), circumscribed, necrosis, moderate or intense lymphocytic infiltrate (Southey et al., submitted). Estrogen receptor (ER) and progesterone receptor (PR) status was collected for cancers from the diagnostic centres or obtained via standard immunohistochemistry (IHC) staining as previously described (19).
Selection of study groups
Of the cases without a germline BRCA1 mutation (or mutations in the breast cancer susceptibility genes BRCA2, TP53, CHEK2, ATM and PALB2), three groups were selected based on having high, intermediate and low numbers of the nine BRCA1 mutation-associated morphological features. Group 1 consisted of cases who were considered to have BRCA1 mutation-associated pathology as their tumors had five or more features (n=52), Group 2 cases had four features (n=39) and Group 3 cases had three or less features (n=164). Controls were women (n=169) who were unaffected at the time of recruitment into the ABCFR using the electoral rolls and frequency-matched for age.
Tumor-enriched DNA samples
Macrodissection to enrich for tumor tissue was performed using a 21-gauge syringe needle following pathological identification of the tumor rich areas. DNA was prepared using the DNeasy Tissue Kit (Qiagen) as per the manufacturer’s protocol except that proteinase K incubation at 56°C was carried out for 72 hours with supplementation of the proteinase K at 24 and 48 hours.
Sodium Bisulfite DNA modification
Where 1µg DNA was available, DNA was sodium bisulfite modified using the MethylEasy™ DNA Bisulfite Modification Kit (Human Genetic Signatures, Sydney, Australia) as per manufacturer’s protocol. Where less than 1µg DNA was obtained (predominantly when using formalin-fixed paraffin-embedded samples), DNA was sodium bisulfite modified using the Epitect Bisulfite Kit (Qiagen). CpGenome™ Universal Methylated DNA (Chemicon, Billerica, MA) was used as fully methylated control DNA (100%). For each modification experiment, similar amounts of fully methylated DNA and DNA extracted from a panel of normal individuals (unmethylated DNA) were also modified.
Quantification of bisulfite modified DNA
All bisulfite modified DNA samples were assayed for relative amounts of DNA using MethyLight analysis of a region free of CpG dinucleotides within the hydroxymethylbilane synthase gene (HMBS; Genbank: NC_000011.8) on the Rotor-Gene 3000 (Corbett Life Sciences, Sydney, Australia). The primers used were 5’-ggTTtgaTTTtTtgTttTagggttatT and 5’-tAccaccaAtcaacactcctcaAA with the dual labelled probe (5’-TtgTTTtaggTtTTaTTaTtgaagtagaggTagggg). Capital letters denote bases corresponding to converted cytosines. A combined pool of bisulfite modified DNA from normal individuals was diluted with PCR-grade water to obtain a standard curve of relative DNA amounts of 100%, 50%, 10%, 5%, 1% and 0%.
BRCA1 methylation analysis
The MethyLight primers encompassed CpG dinucleotides at −37 and −29 (forward primer; 5’-ggtagTTTTttggtttTcgtggTaac) and +27 and +44 (reverse primer; 5’-cccgtccaAAaaAtctcaAcg) with the MGB labelled probe (ActcacgccgcgcaA) spanning CpG dinucleotides at +14, +16 and +19. The relative methylation level (percentage methylated reference) was determined using Ct values relative to the HMBS assay. A standard curve including fully methylated DNA and unmethylated DNA samples was included in each experiment. Amplification efficiency (E) and take-off (T) for each sample were obtained using the RotorGene software (21).
MS-HRM primers assessed either 9 CpG dinucleotides (−37, −29, −21, −19, +8, +14, +16, +19, +27) in a 122 base pair (bp) amplicon for the peripheral blood analysis (forward primer: 5’-TtgTtgTttagcggtagTTTTttggt; reverse primer: 5’-aAcctAtcccccgtccaAAaa) or 4 CpG dinucleotides (−37, −29, −21 and −19) in a 81 bp amplicon for the FFPE tumor analysis (forward primer as above; reverse primer: 5’-caAtcgcaAttttaatttatctAtaattccc).
Bisulfite modified fully methylated DNA was diluted in pooled bisulfite modified DNA from normal individuals to obtain a standard curve comprising 100%, 10%, 5%, 1%, 0.1% and 0% methylated alleles. Each bisulfite modified peripheral blood DNA sample was run in triplicate on the RotorGene 6000 (Corbett, now available as the RotorGeneQ, Qiagen). When assaying bisulfite modified tumor-enriched DNA samples, each sample was run in duplicate. The methylation level of each DNA sample was determined visually by comparing it against the standard curve.
Digital MS-HRM and bisulfite sequencing of amplified products
Digital MS-HRM was performed by limiting dilution (20, 22). Selected amplified products were bisulfite sequenced using Big Dye Terminator Chemistry v3.1 (Applied Biosystems, Foster City, CA). Amplified MS-HRM products were diluted 1 in 20 times with sterile water and subjected to standard PCR sequencing protocols followed by ethanol precipitation. Clean sequencing products were analysed on the 3730 Genetic Analyzer (Applied Biosystems).
SNP Genotyping analysis using High Resolution Melt (HRM) analysis
Primers were designed to flank four BRCA1 promoter region single nucleotide polymorphisms (rs8176072, rs8176073, rs11655505 and rs799906). Each reaction mix consisted of 1X GeneAmp PCR Buffer II, 3.0mM MgCl2, 100µM dNTPs, 200nM each of forward and reverse primers, 450nM Syto9 green-fluorescent nucleic acid stain, 0.375U AmpliTaq Gold DNA polymerase, and approximately 50ng DNA extracted from blood spots on Guthrie cards. Cycling conditions on the RotorGene 6000 were as follows: initial polymerase activation at 95°C for 15 minutes followed by 50 cycles of denaturation at 95°C for 15 seconds and annealing at 60°C for 60 seconds. Fluorescent data was acquired to the green channel at the end of the annealing step and melting was analysed after rapid cooling from 95°C to 70°C.
Statistical analysis
The cancer family history, pathology and molecular features listed in Table 3 were assessed for association with BRCA1 promoter methylation in the tumor (some methylation versus none) using simple and multiple unconditional logistic regression. The best fitting multivariate model was defined to be the one with the lowest Bayesian information criterion (23). Unconditional logistic regression was also used to estimate the association of methylation with disease and to assess evidence for differences in prevalences across case groups. All p-values were based on the likelihood ratio test and all calculations were performed using R version 2.7.2 (24).
Table 3. The association of BRCA1 mutation-associated and other pathological features with BRCA1 promoter region methylation in the peripheral blood.
For each feature potentially predictive of BRCA1 promoter methylation status, the number (proportion in parentheses) of women with the feature by BRCA1 methylation status, the odds ratio (OR) with 95% confidence interval and corresponding statistical significance (p), the negative predictive value (NPV) and positive predictive value (PPV). The overall number (N) of women with for whom the data was available are given in the headings of each category of predictive feature.
| FEATURE |
BRCA1 promoter methylated |
BRCA1 promoter non-methylated |
OR (95% CI) | p | NPV | PPV |
|---|---|---|---|---|---|---|
| FAMILY HISTORY | N = 26 | N = 224 | ||||
| One or more 1st degree relative with breast cancer < 60 yrs | 4 (15%) | 19 (8%) | 2 (0.53–5.8) | 0.3 | 0.9 | 0.17 |
| One or more 1st or 2nd degree relative with ovarian cancer | 1 (4%) | 2 (1%) | 4.4 (0.2–48) | 0.3 | 0.9 | 0.33 |
| Strong family history | 5 (19%) | 25 (11%) | 1.9 (0.59–5.1) | 0.3 | 0.9 | 0.17 |
| One or more 1st degree relative with breast cancer ≥ 60 yrs | 1 (4%) | 8 (4%) | 1.1 (0.057–6.2) | 0.9 | 0.9 | 0.11 |
| One or more 2nd degree relative with breast cancer | 6 (23%) | 67 (30%) | 0.7 (0.25–1.7) | 0.5 | 0.89 | 0.08 |
| MORPHOLOGICAL FEATURES | N = 28 | N = 226 | ||||
| Trabecular growth pattern | 15 (54%) | 34 (15%) | 6.5 (2.9–15) | <0.0001 | 0.94 | 0.31 |
| High mitotic index | 14 (50%) | 22 (10%) | 9.3 (3.9–22) | <0.0001 | 0.94 | 0.39 |
| Necrosis | 16 (57%) | 68 (30%) | 3.1 (1.4–7) | 0.005 | 0.93 | 0.19 |
| Circumscribed growth pattern | 14 (50%) | 31 (14%) | 6.3 (2.7–15) | <0.0001 | 0.93 | 0.31 |
| Moderate or intense lymphocytic infiltrate | 26 (93%) | 182 (81%) | 3.1 (0.89–20) | 0.08 | 0.96 | 0.12 |
| Syncytial growth pattern | 6 (21%) | 8 (4%) | 7.4 (2.3–23) | 0.002 | 0.91 | 0.43 |
| Malignant nuclear grade | 24 (86%) | 191 (85%) | 1.1 (0.39–3.9) | 0.9 | 0.9 | 0.11 |
| Pushing margins (> 50%) | 2 (7%) | 5 (2%) | 3.4 (0.47–17) | 0.2 | 0.89 | 0.29 |
| Little or no tubule formation | 21 (75%) | 168 (74%) | 1 (0.44–2.7) | 0.9 | 0.89 | 0.11 |
| ER and PR STATUS | N = 24 | N = 212 | ||||
| ER negative | 15 (62%) | 71 (33%) | 3.3 (1.4–8.2) | 0.006 | 0.94 | 0.17 |
| PR negative | 12 (50%) | 59 (28%) | 2.6 (1.1–6.2) | 0.03 | 0.93 | 0.17 |
Results
Peripheral blood methylation analysis
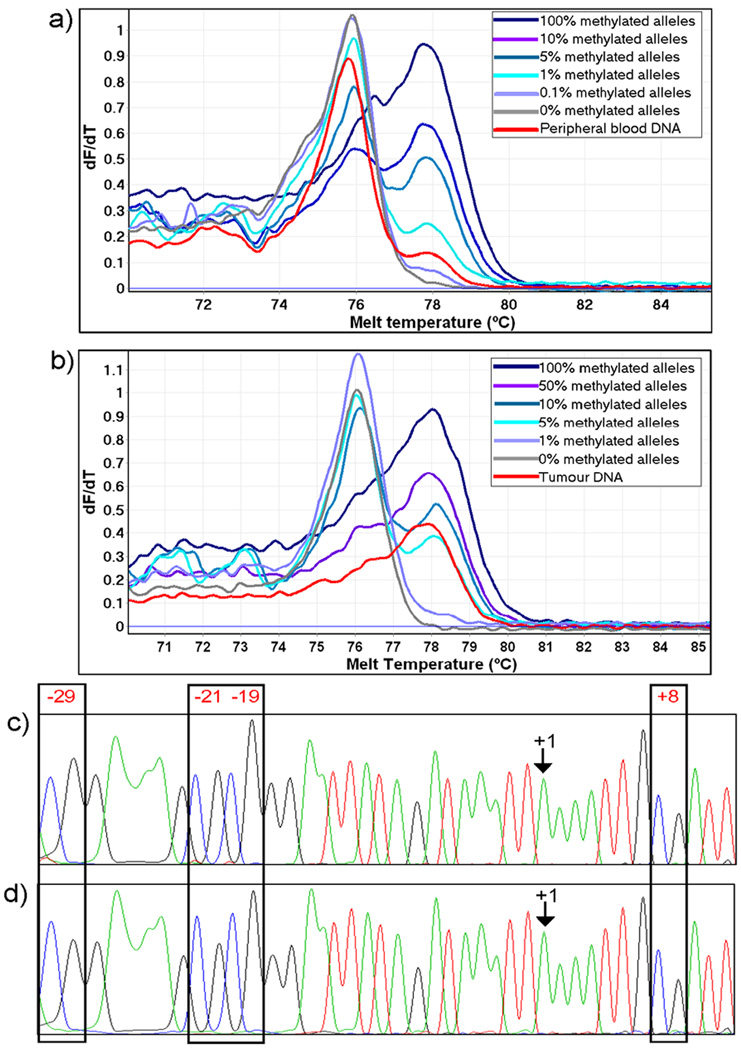
We used two independent assays to determine the proportion of methylated BRCA1 alleles: MethyLight and Methylation Sensitive-High Resolution Melting (MS-HRM). The regions covered by each assay were overlapping but not identical (20), due to the different requirements of primer design for these two assays (25). MS-HRM, which provides a semi-quantitative measurement of methylation was used to confirm the levels of methylation as estimated by MethyLight. MS-HRM gave well-defined peaks (Figure 1) indicating that BRCA1 promoter region methylation was homogeneous within the region of the 9 CpGs analysed by the 124 bp MS-HRM assay.
Figure 1. BRCA1 promoter methylation detected in the peripheral blood and corresponding tumor.
MS-HRM Tm curves are shown for the peripheral blood DNA (panel a) and corresponding tumor-enriched DNA (panel b) of individual P1.16. The DNA is shown compared to a set of dilution standards. The blood was analysed using the 122 bp fragment whereas the tumor was analysed using the 81bp fragment. Forward sequences for the same individual are shown for a methylated clone from the peripheral blood DNA (panel c) and corresponding tumor-enriched DNA (panel d).
Sixteen of the 52 (30.8%) peripheral blood DNA samples from group 1 women (whose breast tumors had five or more BRCA1 mutation-associated morphological features) were found to be methylated in the BRCA1 promoter using MethyLight (Table 1). Calculation of relative methylation levels using relative Ct values showed that seven had low level methylation (≤1% of DNA methylated), six had intermediate level methylation (more than 1% but less than 5% of DNA methylated), and three had moderate level promoter region methylation (ranging from more than 5% to 17%) (Table 1; Supplementary Table S1).
Table 1. Peripheral blood DNA methylation detected in the BRCA1 promoter for the study groups.
The final methylation value for each individual was determined by assessing the relative methylation levels from MethyLight and MS-HRM experiments. Group 1 are women with five or more of nine BRCA1 mutation-associated morphological features, group 2 have four, and group 3 have three or less. The full data is in supplementary tables S1–S4.
| Group 1 (n=52) | Group 2 (n=39) | Group 3 (n=164) | Controls (n=169) | |
|---|---|---|---|---|
| ≤ 1% methylated alleles | 7 | 2 | 4 | 4 |
| > 1% methylated alleles | 6 | 1 | 4 | 2 |
| ≥ 5% methylated alleles | 3 | 1 | 0 | 0 |
| Not methylated | 36 | 35 | 156 | 163 |
| Proportion with BRCA1 promoter methylation | 16/52 (30.8%) | 4/39 (10.3%) | 8/164 (4.9%) | 6/169 (3.6%) |
Four of the women in the intermediate group (group 2) had BRCA1 promoter methylation in their peripheral blood (4/39; 10.3%) (Table 1). Two women had moderate levels of methylation while one had intermediate and one had low levels of methylation, (Table 1; Supplementary Table S2).
The frequency and level of measurable methylation in peripheral blood DNA samples for group 3 women and for unaffected controls were strikingly similar to each other. Of the 164 affected women in group 3, only eight (4.9%) had detectable BRCA1 promoter methylation in their peripheral blood DNA (Table 1; Supplementary Table S3). Four had low-level methylation and four had intermediate levels of methylation (Table 1). Interestingly, two of those peripheral blood DNAs scored as intermediate level by MethyLight were negative and weakly positive by MS-HRM indicating possible heterogeneous methylation. BRCA1 promoter methylation was detected in only six (3.6%) of the 169 peripheral blood DNAs from controls. Three had low-level methylation and three had intermediate level methylation (Table 1; Supplementary Table S4).
Therefore, the prevalence of BRCA1 promoter region methylation in DNA from peripheral blood differed between cases and controls (p=0.004) as calculated by logistic regression. Within cases, it differed by group (p = 2×10−6) consistent with increasing prevalence with greater number of BRCA1 mutation-associated morphology features.
Tumor methylation and histopathology
Tumor tissue was available for 20 women in group 1 and 32 women in group 3. Nine (45%) of the tumor-enriched DNA specimens from the 20 available group 1 tumors had methylation in the BRCA1 promoter region. All were highly methylated (50–100% methylated alleles). With the exception of two samples, all tumor-enriched DNAs that displayed BRCA1 promoter region methylation were from women who also displayed BRCA1 promoter methylation in their corresponding peripheral blood DNA (Table 2). Significantly, there were no instances in group 1 where peripheral blood with detectable methylation was not associated with a tumor-enriched DNA sample with markedly increased methylation (Supplementary Table S5).
Table 2. Comparison of methylation values in all the BRCA1 methylated tumor DNAs with the corresponding peripheral blood DNA from Group 1 and Group 3 women.
Both MethyLight and MS-HRM methylation are shown. Twenty tumors were available from group 1 women with five or more of nine BRCA1 mutation-associated morphological features and 32 tumors were available from group 3 women with three or less features. Only the methylated tumors are shown below. Consensus methylation values for the corresponding peripheral blood DNA are shown. The full data is in supplementary tables S5 and S6.
| Tumor ID | Tumor content (%) | MethyLight (%) | MS-HRM (%) | Corresponding peripheral blood DNA (%) |
|---|---|---|---|---|
| Tumor-enriched DNA samples from group 1 women | ||||
| T1.1 | 92 | 150 | 100 | <1 |
| T1.2 | 95 | 203.31 | 100 | <1 |
| T1.3 | 95 | 168.78 | 100 | 1–5 |
| T1.4 | 90 | 95.23 | 50 | 1–5 |
| T1.5 | 80 | 78.93 | 50–100 | >5 |
| T1.6 | 80 | 72.73 | 50 | Not methylated |
| T1.7 | 70 | 56.51 | 100 | Not methylated |
| T1.8 | 90 | 44.45 | 100 | <1 |
| T1.9 | 90 | 22.89 | 100 | 1–5 |
| Tumor-enriched DNA samples from group 3 women | ||||
| T3.1 | 80 | 36.08 | 50 | Not methylated |
| T3.2 | 60 | 150.19 | 50–100 | Not methylated |
| T3.3 | 60 | Not methylated | 1 | 1–5 |
Three of the available 32 tumor-enriched DNA samples from group 3 women had detectable methylation at the BRCA1 promoter region. For one of the samples, there was moderate methylation (more than 1% and less than 5% methylated alleles) in the corresponding peripheral blood DNA. In contrast to group 1, this case showed no increase of methylation in its tumor-enriched DNA (Table 2), indicating that the cancer did not arise from a BRCA1 methylated clone and that the level of methylation seen represented a background level. The remaining two group 3 tumor-enriched DNA specimens were strongly methylated but there was no detectable methylation in the corresponding peripheral blood DNA (Supplementary Table S6).
Simple logistic regression (Table 3) showed that the following pathology features were individually predictive of BRCA1 promoter methylation: syncytial, trabecular and circumscribed growth patterns; high mitotic index; and necrosis. ER and PR status were also predictive of BRCA1 promoter methylation but no family history variables were. Multivariate logistic regressions identified high mitotic index and circumscribed growth patterns as the best predictors of methylation, with estimated odds ratios (95% confidence interval (CI)) of 5.8 (2.3 – 15.0) and 3.4 (1.3 – 8.7) respectively. The odds of methylation were estimated to increase by a factor of 1.63 (95% CI 1.33 – 2.03) for each additional pathology feature present (p-trend=0.0001). From an analysis which included the case and the control groups, BRCA1 promoter region methylation was estimated to be associated with a 3.5-fold (95% CI 1.4 – 10.5) increased risk of breast cancer before age 40 years. (p=0.004).
Family members of cases
We screened peripheral blood DNA from the family members of nine women (eight from group 1 and one from group 3) who had moderate to high-levels of methylation (1% or greater) at the BRCA1 promoter region in their peripheral blood DNA (Table 4a). Neither MethyLight nor MS-HRM analysis detected methylation at any level in any of the family members. We also identified methylation negative individuals with family members who had been diagnosed with either breast and/or ovarian cancer (Table 4b). Neither MS-HRM nor MethyLight detected methylation at any level in the peripheral blood DNA of any of these affected family members.
Table 4. Lack of detectable peripheral blood BRCA1 methylation in family members of women in this study.
4a) Peripheral blood BRCA1 promoter methylation in family members of women with moderate levels of peripheral blood BRCA1 promoter methylation. 4b) BRCA1 promoter methylation in peripheral blood DNA of affected family members (breast and/or ovarian cancer) of group 1 and group 3 women without peripheral blood methylation.
| Individual ID | Peripheral blood methylation (%) of the proband | Relationship to the proband | Cancer status (age of diagnosis) | Peripheral blood methylation |
|---|---|---|---|---|
| 4a) Relatives of probands with detectable peripheral blood methylation | ||||
| Group 1 | ||||
| P1.1 | >5 | Sister | Not affected | Not methylated |
| P1.2 | >5 | Sister | Not affected | Not methylated |
| Sister | Not affected | Not methylated | ||
| P1.5 | 1–5 | Paternal Aunt | Breast (69) | Not methylated |
| P1.6 | 1–5 | Maternal Aunt | Ovary (65) | Not methylated |
| P1.7 | 1 | Sister | Breast (37) Overlapping lesion of the breast (51) | Not methylated |
| P1.8 | 1 | Mother | Breast (45) | Not methylated |
| P1.13 | 1–5 | Father | Not affected | Not methylated |
| Mother | Tongue (46 years); Breast (51 years) | Not methylated | ||
| Maternal Uncle | Not affected | Not methylated | ||
| Sister | Not affected | Not methylated | ||
| Brother | Not affected | Not methylated | ||
| P1.14 | >5 | Mother | Not affected | Not methylated |
| Group 3 | ||||
| P3.8 | 3.5–5 | Father | Not affected | Not methylated |
| 4b) Other relatives with breast or ovarian cancer | ||||
| Group 1 | ||||
| P1.9 | Not methylated | Mother | Breast (47) | Not methylated |
| Group 3 | ||||
| P3.2 | Not methylated | Mother | Breast (68) | Not methylated |
| P3.3 | Not methylated | Mother | Breast (74) | Not methylated |
| Paternal Aunt | Breast (76) | Not methylated | ||
| P3.4 | Not methylated | Mother | Breast (72) | Not methylated |
| P2.5 | Not methylated | Mother | Ovary (68) | Not methylated |
Genotyping analysis for BRCA1 promoter single nucleotide polymorphisms
All group 1 women were genotyped for four SNPs (rs8176072: A>T, rs8176073: A>G, rs11655505: A>G and rs799906: C>T) in the BRCA1 promoter region to assess if there was an association with detectable BRCA1 promoter methylation in the peripheral blood (Table 5). There was a trend (significant at the 5% level in the case of rs799906 and rs11655505) for detectable methylation to be associated with the presence of the minor allele. All three peripheral blood DNA displaying moderate levels of methylation (at least 5% methylated alleles) were heterozygous for both rs799906 and rs11655505.
Table 5. Association between BRCA1 promoter region genetic variants and BRCA1 promoter methylation detected in group 1 women.
| rs number | Genotype | Methylated (n=16) |
Not methylated (n=36) |
Allelic test P-value |
OR (95% CI) |
|---|---|---|---|---|---|
| rs8176072 | TT | 16 | 32 | 0.31 | 0 |
| AT | 0 | 4 | |||
| AA | 0 | 0 | |||
| rs8176073 | AA | 16 | 35 | 1 | 0 |
| AG | 0 | 1 | |||
| GG | 0 | 0 | |||
| rs11655505 | CC | 4 | 22 | 0.035 | 2.7 (1.11–2.67) |
| CT | 10 | 12 | |||
| TT | 2 | 2 | |||
| rs799906 | AA | 4 | 23 | 0.017 | 3.11 (1.25 to 7.74) |
| AG | 10 | 10 | |||
| GG | 2 | 2 |
Discussion
It is reasonable to consider that constitutional epimutations, when present, can act as the first step in carcinogenesis in a manner analogous to germline mutations. It is also reasonable to consider that cancers with a driver lesion in a given gene should have a similar pathology, regardless of whether the driver lesion is genetic or epigenetic.
This study sought to determine whether detectable constitutional epimutations in the BRCA1 gene were frequent in the peripheral blood using a population based study of women with early-onset breast cancer who did not have BRCA1 germline mutations, but differed according to the extent to which their tumors exhibited BRCA1 mutation-associated pathology features. This would have implications for not only our understanding of breast cancer pathogenesis but also would raise the possibility of using BRCA1 directed prevention measures for unaffected women with constitutional epimutations.
The BRCA1 exon 1a promoter comprises 11 CpG sites between BRCA1 exon 1a and the first exon of NBR2 (26). Dobrovic and Simpfendorfer first showed that the BRCA1 promoter region was methylated in about 20% of breast tumors (27). This finding was confirmed and BRCA1 promoter region methylation was also shown for cases of ovarian cancer (26–33). The identification of BRCA1 methylation in two cancer types that are driven by germline BRCA1 mutations, and the lack of BRCA1 methylation in other cancers such as colorectal cancer and leukaemia that were not associated with germline BRCA1 mutations, argued that BRCA1 methylation could play a pathogenic role in breast and ovarian cancer (29–30). Thus, constitutional methylation of BRCA1, in which the promoter is methylated throughout the somatic tissues, could be a predisposing factor if present.
Peripheral blood is a convenient tissue to assay for constitutional methylation. We previously reported a pilot study of BRCA1 methylation in the peripheral blood of a small group of BRCA1 mutation negative women from a familial cancer registry (20). Three out of seven women whose tumors showed BRCA1 mutation-associated pathology had detectable methylation of the BRCA1 exon 1a promoter (at levels between 1 and 12%) in their peripheral blood. All had an early age of onset of breast cancer. The corresponding tumors from these women were heavily methylated at the BRCA1 promoter, suggesting that BRCA1 epimutations could act as an alternative mechanism leading to breast cancer predisposition and that they might be particularly important in early-onset disease (20). We therefore set to test these initial observations by assessing BRCA1 promoter methylation in a much larger set of 255 women with early-onset breast cancer from a population-based study (13, 15).
The prevalence of BRCA1 promoter region methylation in DNA from peripheral blood for group 1 individuals with five or more BRCA1 mutation-associated morphology features was 31%, considerably higher than for the group 3 individuals with few (three or less) features (5%; p=2×10−6), and for the intermediate group with four features (10%; p=0.02), and for the controls (4%; p=2×10−7). The prevalence of BRCA1 promoter methylation did not significantly differ between the group 3 women and the unaffected controls (p=0.5).
The amount of BRCA1 methylation in the peripheral blood DNA (when present) was not at the expected 50% level expected of a germline epimutation indicating that mosaicism was present. It varied markedly between the samples examined (0.1% to 17% of BRCA1 alleles). There was good concordance between the MethyLight and the MS-HRM results (Supplementary Tables S1–S4). The two methodologies take very different approaches to the quantification of methylation, giving confidence in the accuracy of the results.
It could be argued that our findings were due to circulating breast cancer cells which were methylated for BRCA1. This is unlikely as circulating tumor cells are present at very low levels and can only be isolated from the blood after enrichment, as studies from our laboratory and other laboratories have shown (34). The sensitivity of our methylation assays is 0.1%. It is unlikely that cancer cells are present in the peripheral circulation at levels equal to or higher than this in any breast cancer patient let alone any normal control woman.
Nine of the 20 tumors examined from women in group 1 had high level (defined as more than 50%) BRCA1 methylation whereas none of the 32 tumors examined from women in group 3 had high or even moderate level methylation. Importantly, the tumors were strongly methylated for each of the seven group 1 women with detectable peripheral blood methylation for whom a tumor could be obtained. This contrasted dramatically with the one woman from group 3 with detectable peripheral blood methylation for whom we were also able to test a tumor. Her tumor showed no increase of methylation relative to that seen in peripheral blood. These results are consistent with BRCA1 methylation driving the development of tumours with BRCA1-associated pathology.
Reports of the relationship between BRCA1 promoter region methylation and clinicopathological features have been conflicting. However, most studies show a clear tendency for BRCA1 methylated tumors to occur in young women with high-grade, estrogen receptor–negative, progesterone receptor–negative tumors (33, 35). Several studies pointed out the resemblance between BRCA1 methylated and BRCA1 mutated tumors. Esteller et al. (2000) reported that BRCA1 methylation was associated with the less common medullary (67% methylated) and mucinous (55% methylated) subtypes, which are over-represented in families carrying BRCA1 mutations (30). Turner et al. (2007) reported that BRCA1 methylation occurred in 63% of metaplastic breast cancers (36).
We hypothesise that there are likely to be two groups of BRCA1 promoter region methylated tumors. In “first hit” tumors, BRCA1 methylation predisposes to and initiates tumorigenesis and these tumors accordingly resemble BRCA1 mutation-positive tumors morphologically. In tumors in which BRCA1 methylation occurs later during tumor progression, the loss of BRCA1 would not be expected play a major role in determining the tumor’s morphology. This may be more common in post-menopausal tumors. Interestingly, population-based studies of women with BRCA1 mutations indicate that they are primarily at risk of early-onset breast cancer, with risk falling as the women age (13).
Two tumors from women in group 3 showed high level without corresponding peripheral blood methylation. It is likely that these are examples of BRCA1 methylation that occurred during a later stage of tumor evolution and thus did not drive the breast cancer.
The origin of constitutional methylation of the BRCA1 promoter remains obscure. There is very little evidence of transmission of an epimutation in human heredity. In most cases, what is transmitted is genetic variation, both in cis and in trans sequences which alter the propensity of a locus to methylate (8). Cis-acting sequences that affect methylation in normal tissues have been identified for the RIL and MGMT loci among others (37). For the MGMT gene, we have previously shown that a promoter SNP plays a major role in determining the probability of the methylation of the MGMT promoter CpG island in normal tissues (38). The same SNP affects the probability of MGMT methylation in colorectal cancer and adjacent colonic tissues (39).
We thus assessed whether genetic variants in the promoter region of the BRCA1 gene bore any relation to the probability of BRCA1 promoter region methylation in the women with tumors showing BRCA1 mutation-associated features (Table 5). Moderate associations were observed for the rs11655505: A>G and rs799906: C>T SNPs. Women with the minor alleles (T allele of rs11655505 /G allele of rs799906) were more likely to be methylated. The association of methylation with both SNPs was significant at the 5% level. These two genetic variants have been reported to alter promoter activity (40). Surprisingly, carriers of the T allele of rs11655505 were reported as having a reduced risk of breast cancer in a Chinese population, although in that study the protective effect was most strongly seen for women aged more than 45 years (40). However, a recent genotyping study of Caucasian populations did not find any association between rs11655505 and breast cancer risk (41). The study included the ABCFS data set from which our cases derived, and although there was a lack of significant association, there was a weak trend for the T allele to have an increased risk of breast cancer, concordant with our results.
The absence of strongly predisposing genetic variants is consistent with the lack of somatic methylation in family members. We analysed two groups of immediate family members of the affected individuals in this study (Table 4a and 4b). In the first group, we were interested in determining if family members (irrespective of affected status) of cases with moderate to high levels of germline methylation were also methylated in BRCA1. In the second group, we were interested in evaluating BRCA1 promoter methylation levels for cancer affected family members where proband methylation levels were not detected. We were unable to detect any methylation in either group. The results indicate that the BRCA1 promoter methylation detected in the index cases was not inherited but rather arose de novo in the women.
This study has shown that the BRCA1 peripheral blood methylation identifies a small group of women in the population (our best estimate for Australian women is 4%) who have about a 3.5-fold increased risk of breast cancer before age 40 years due to epigenetic loss of BRCA1. This hypothesis is further supported when BRCA1 promoter methylation of both tumor-enriched DNA samples and tumor pathology were considered. If this methylation were causal, then our study predicts that about 11% of early-onset breast cancers would be attributable to this phenomenon. Given that less than 10% of early-onset reast cancers arise in women with germline mutations in BRCA1, constitutional epimutations of BRCA1 might explain as much, if not more, of early-onset breast cancer as inherited germline defects.
In conclusion, we have described a group of women with early-onset breast cancer who have detectable methylation of the BRCA1 promoter region in the peripheral blood and have tumors that are strongly methylated at the BRCA1 promoter and possess morphological features consistent with carrying BRCA1 germline mutations. We postulate that BRCA1 epimutations are also present in the breast tissue of these young women and drive breast carcinogenesis in a manner analogous to BRCA1 germline mutations.
BRCA1 methylation thus may be an important new biomarker for breast cancer predisposition. Our findings, if replicated by further studies, could have major implications for prevention by screening women for epigenetic changes that increase risk of early-onset breast cancer. It also raises the possibility for cancer prevention in that women with epimutations might have their risk of developing breast cancer reduced by appropriate chemoprevention (42) or even by dietary manipulation.
Supplementary Material
The individuals are numbered P1.1 to P1.52. Both MethyLight and MS-HRM estimates are shown along with a consensus methylation score based on both assays. In two cases, where there was a very low MethyLight value and no positivity for MS-HRM, the DNAs were scored as negative.
The individuals are numbered P2.1 to P2.39. Both MethyLight and MS-HRM estimates are shown along with a consensus methylation score based on both assays. In one case where there was a low MS-HRM value and no positivity for MethyLight, the DNA was scored as negative.
The individuals are numbered P3.1 to P3.164. Both MethyLight and MS-HRM estimates are shown along with a consensus methylation score based on both assays. In one case, where there was a very low MethyLight value and no positivity for MS-HRM, the DNA was scored as negative. In two cases, where there was a moderate MethyLight value and no or very low positivity for MS-HRM, the DNAs were scored as positive.
The individuals are numbered PU.1 to PU.169. Both MethyLight and MS-HRM estimates are shown along with a consensus methylation score based on both assays. In one case where there was a very low MethyLight value and no positivity for MS-HRM, the DNA was scored as negative.
Both MethyLight and MS-HRM estimates are shown. The tumor pathology score is the number of BRCA1 mutation-associated pathological features seen in the tumor.
Both MethyLight and MS-HRM estimates are shown. The tumor pathology score is the number of BRCA1 mutation-associated pathological features seen in the tumor.
Acknowledgements
We thank the participants in this study. We also thank Elena Takano for assistance in the collection of tumor blocks and slides, Mukta Rayoo and Max Yan for assisting the pathology review of the tumor sections for microdissection, Minh Bui for assistance with the statistical analysis, and Andrea Tesoriero and Letitia Smith for technical input.
Financial support, including the source and number of grants:
This study was supported by a grant to AD from the US Department of Defense Breast Cancer Research Program under award number W81XWH-06-1-0670 and by grants to AD and MCS from the Cancer Council of Victoria. The Australian Breast Cancer Family Registry (ABCFR) was supported by the National Health and Medical Research Council of Australia (NHMRC) [145604], the United States National Institutes of Health (NIH) [CA102740-01A2], and by the United States National Cancer Institute, National Institutes of Health [CA-06-503] through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and principal investigators Cancer Care Ontario [CA69467], Columbia University [CA69398], Fox Chase Cancer Center [CA69631], Huntsman Cancer Institute [CA69446], Northern California Cancer Center [CA69417], University of Melbourne [CA69638]. The ABCFR was initially supported by the NHMRC, the New South Wales Cancer Council and the Victorian Health Promotion Foundation. The ABCFR has also received support from the Victorian Breast Cancer Research Consortium of which MCS and JH are group leaders. JLH is an Australia Fellow of the NHMRC. MCS is a Senior Research Fellow of the NHMRC. EMW was supported by the Dora Lush postgraduate biomedical scholarship from the NHMRC. Views and opinions of, and endorsements by the authors do not reflect those of the US Army or the US Department of Defense. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of collaborating centers in the BCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR.
Footnotes
Disclosure of Potential Conflicts of Interest
There are no potential conflicts of interest involved.
References
- 1.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.Armes JE, Egan AJ, Southey MC, et al. The histologic phenotypes of breast carcinoma occurring before age 40 years in women with and without BRCA1 or BRCA2 germline mutations: a population-based study. Cancer. 1998;83:2335–2345. [PubMed] [Google Scholar]
- 3.Lakhani SR, Gusterson BA, Jacquemier J, et al. The pathology of familial breast cancer: histological features of cancers in families not attributable to mutations in BRCA1 or BRCA2. Clin Cancer Res. 2000;6:782–789. [PubMed] [Google Scholar]
- 4.Langston AA, Malone KE, Thompson JD, Daling JR, Ostrander EA. BRCA1 mutations in a population-based sample of young women with breast cancer. N Engl J Med. 1996;334:137–142. doi: 10.1056/NEJM199601183340301. [DOI] [PubMed] [Google Scholar]
- 5.Newman B, Mu H, Butler LM, et al. Frequency of breast cancer attributable to BRCA1 in a population-based series of American women. JAMA. 1998;279:915–921. doi: 10.1001/jama.279.12.915. [DOI] [PubMed] [Google Scholar]
- 6.Southey MC, Tesoriero AA, Andersen CR, et al. BRCA1 mutations and other sequence variants in a population-based sample of Australian women with breast cancer. Br J Cancer. 1999;79:34–39. doi: 10.1038/sj.bjc.6690008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whittemore AS, Gong G, Itnyre J. Prevalence and contribution of BRCA1 mutations in breast cancer and ovarian cancer: results from three U.S. population-based case-control studies of ovarian cancer. Am J Hum Genet. 1997;60:496–504. [PMC free article] [PubMed] [Google Scholar]
- 8.Dobrovic A, Kristensen LS. DNA methylation, epimutations and cancer predisposition. Int J Biochem Cell Biol. 2009;41:34–39. doi: 10.1016/j.biocel.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Gazzoli I, Loda M, Garber J, Syngal S, Kolodner RD. A hereditary nonpolyposis colorectal carcinoma case associated with hypermethylation of the MLH1 gene in normal tissue and loss of heterozygosity of the unmethylated allele in the resulting microsatellite instability-high tumor. Cancer Res. 2002;62:3925–3928. [PubMed] [Google Scholar]
- 10.Hitchins M, Williams R, Cheong K, et al. MLH1 germline epimutations as a factor in hereditary nonpolyposis colorectal cancer. Gastroenterology. 2005;129:1392–1399. doi: 10.1053/j.gastro.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 12.Hitchins MP, Wong JJ, Suthers G, et al. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356:697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- 13.Hopper JL, Southey MC, Dite GS, et al. Population-based estimate of the average age-specific cumulative risk of breast cancer for a defined set of protein-truncating mutations in BRCA1 and BRCA2. Australian Breast Cancer Family Study. Cancer Epidemiol Biomarkers Prev. 1999;8:741–747. [PubMed] [Google Scholar]
- 14.Dite GS, Jenkins MA, Southey MC, et al. Familial risks, early-onset breast cancer, and BRCA1 and BRCA2 germline mutations. J Natl Cancer Inst. 2003;95:448–457. doi: 10.1093/jnci/95.6.448. [DOI] [PubMed] [Google Scholar]
- 15.John EM, Hopper JL, Beck JC, et al. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:R375–R389. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith LD, Tesoriero AA, Ramus SJ, et al. BRCA1 promoter deletions in young women with breast cancer and a strong family history: a population-based study. Eur J Cancer. 2007;43:823–827. doi: 10.1016/j.ejca.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leong T, Whitty J, Keilar M, et al. Mutation analysis of BRCA1 and BRCA2 cancer predisposition genes in radiation hypersensitive cancer patients. Int J Radiat Oncol Biol Phys. 2000;48:959–965. doi: 10.1016/s0360-3016(00)00728-8. [DOI] [PubMed] [Google Scholar]
- 18.Neuhausen SL, Ozcelik H, Southey MC, et al. BRCA1 and BRCA2 mutation carriers in the Breast Cancer Family Registry: an open resource for collaborative research. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-0153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCredie MR, Dite GS, Giles GG, Hopper JL. Breast cancer in Australian women under the age of 40. Cancer Causes Control. 1998;9:189–198. doi: 10.1023/a:1008886328352. [DOI] [PubMed] [Google Scholar]
- 20.Snell C, Krypuy M, Wong EM, Loughrey MB, Dobrovic A. BRCA1 promoter methylation in peripheral blood DNA of mutation negative familial breast cancer patients with a BRCA1 tumour phenotype. Breast Cancer Res. 2008;10:R12. doi: 10.1186/bcr1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Candiloro IL, Mikeska T, Hokland P, Dobrovic A. Rapid analysis of heterogeneously methylated DNA using digital methylation-sensitive high resolution melting: application to the CDKN2B (p15) gene. Epigenetics Chromatin. 2008;1:7. doi: 10.1186/1756-8935-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwarz G. Estimating the dimension of a model. Annals of Statistics. 1978;6:461–464. [Google Scholar]
- 24.R: A language and environment for statistical computing. Vienne, Austria: R Foundation for Statistical Computing; [Google Scholar]
- 25.Wojdacz TK, Hansen LL, Dobrovic A. A new approach to primer design for the control of PCR bias in methylation studies. BMC Res Notes. 2008;1:54. doi: 10.1186/1756-0500-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu CF, Chambers JA, Solomon E. Complex regulation of the BRCA1 gene. J Biol Chem. 1997;272:20994–20997. doi: 10.1074/jbc.272.34.20994. [DOI] [PubMed] [Google Scholar]
- 27.Dobrovic A, Simpfendorfer D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 1997;57:3347–3350. [PubMed] [Google Scholar]
- 28.Rice JC, Massey-Brown KS, Futscher BW. Aberrant methylation of the BRCA1 CpG island promoter is associated with decreased BRCA1 mRNA in sporadic breast cancer cells. Oncogene. 1998;17:1807–1812. doi: 10.1038/sj.onc.1202086. [DOI] [PubMed] [Google Scholar]
- 29.Bianco T, Chenevix-Trench G, Walsh DC, Cooper JE, Dobrovic A. Tumour-specific distribution of BRCA1 promoter region methylation supports a pathogenetic role in breast and ovarian cancer. Carcinogenesis. 2000;21:147–151. doi: 10.1093/carcin/21.2.147. [DOI] [PubMed] [Google Scholar]
- 30.Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 31.Baldwin RL, Nemeth E, Tran H, et al. BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer Res. 2000;60:5329–5333. [PubMed] [Google Scholar]
- 32.Niwa Y, Oyama T, Nakajima T. BRCA1 expression status in relation to DNA methylation of the BRCA1 promoter region in sporadic breast cancers. Jpn J Cancer Res. 2000;91:519–526. doi: 10.1111/j.1349-7006.2000.tb00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Catteau A, Morris JR. BRCA1 methylation: a significant role in tumour development? Semin Cancer Biol. 2002;12:359–371. doi: 10.1016/s1044-579x(02)00056-1. [DOI] [PubMed] [Google Scholar]
- 34.Raynor MP, Stephenson SA, Pittman KB, et al. Identification of circulating tumour cells in early stage breast cancer patients using multi marker immunobead RT-PCR. J Hematol Oncol. 2009;2:24. doi: 10.1186/1756-8722-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei M, Xu J, Dignam J, et al. Estrogen receptor alpha, BRCA1, and FANCF promoter methylation occur in distinct subsets of sporadic breast cancers. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9766-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner NC, Reis-Filho JS, Russell AM, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–2132. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- 37.Boumber YA, Kondo Y, Chen X, et al. An Sp1/Sp3 binding polymorphism confers methylation protection. PLoS Genet. 2008;4:e1000162. doi: 10.1371/journal.pgen.1000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Candiloro IL, Dobrovic A. Detection of MGMT promoter methylation in normal individuals is strongly associated with the T allele of the rs16906252 MGMT promoter single nucleotide polymorphism. Cancer Prev Res (Phila Pa) 2009;2:862–867. doi: 10.1158/1940-6207.CAPR-09-0056. [DOI] [PubMed] [Google Scholar]
- 39.Ogino S, Hazra A, Tranah GJ, et al. MGMT germline polymorphism is associated with somatic MGMT promoter methylation and gene silencing in colorectal cancer. Carcinogenesis. 2007;28:1985–1990. doi: 10.1093/carcin/bgm160. [DOI] [PubMed] [Google Scholar]
- 40.Chan KY, Liu W, Long JR, et al. Functional polymorphisms in the BRCA1 promoter influence transcription and are associated with decreased risk for breast cancer in Chinese women. J Med Genet. 2009;46:32–39. doi: 10.1136/jmg.2007.057174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verderio P, Pizzamiglio S, Southey MC, et al. A BRCA1 promoter variant (rs11655505) and breast cancer risk. J Med Genet. 47:268–270. doi: 10.1136/jmg.2009.073544. [DOI] [PubMed] [Google Scholar]
- 42.Rubinstein WS. Hereditary breast cancer: pathobiology, clinical translation, and potential for targeted cancer therapeutics. Fam Cancer. 2008;7:83–89. doi: 10.1007/s10689-007-9147-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The individuals are numbered P1.1 to P1.52. Both MethyLight and MS-HRM estimates are shown along with a consensus methylation score based on both assays. In two cases, where there was a very low MethyLight value and no positivity for MS-HRM, the DNAs were scored as negative.
The individuals are numbered P2.1 to P2.39. Both MethyLight and MS-HRM estimates are shown along with a consensus methylation score based on both assays. In one case where there was a low MS-HRM value and no positivity for MethyLight, the DNA was scored as negative.
The individuals are numbered P3.1 to P3.164. Both MethyLight and MS-HRM estimates are shown along with a consensus methylation score based on both assays. In one case, where there was a very low MethyLight value and no positivity for MS-HRM, the DNA was scored as negative. In two cases, where there was a moderate MethyLight value and no or very low positivity for MS-HRM, the DNAs were scored as positive.
The individuals are numbered PU.1 to PU.169. Both MethyLight and MS-HRM estimates are shown along with a consensus methylation score based on both assays. In one case where there was a very low MethyLight value and no positivity for MS-HRM, the DNA was scored as negative.
Both MethyLight and MS-HRM estimates are shown. The tumor pathology score is the number of BRCA1 mutation-associated pathological features seen in the tumor.
Both MethyLight and MS-HRM estimates are shown. The tumor pathology score is the number of BRCA1 mutation-associated pathological features seen in the tumor.