Fe uptake mechanisms and transcriptional control
Iron (Fe) is an essential micronutrient for almost all living organisms and represents one of the most versatile metals in biology, being involved in many ubiquitous metabolic processes such as respiration and photosynthesis, and required as a co-factor for numerous enzymes (Sperotto et al., 2010; Grillet et al., 2014a). In plants, Fe deficiency can cause severe chlorosis, growth arrest, and even plant death. Although highly abundant in the earth's crust, Fe phytoavailability is usually low, mainly because Fe3+ forms insoluble Fe oxides (Lemanceau et al., 2009). To circumvent this problem, plants developed mechanisms to acquire Fe from the rhizosphere (Sperotto et al., 2012; Grillet et al., 2014a).
Fe uptake mechanisms were classically separate into two strategies. Strategy I, or reduction strategy, is carried out by all plants except those from Poaceae family, and consists of: (a) H+ extrusion by P-type ATPases to acidify the rhizosphere and increase Fe3+ solubility (e.g., AtAHA2, Arabidopsis H+-pump ATPase); (b) reduction of Fe3+ by a plasma membrane (PM)-bound ferric chelate reductase to more soluble Fe2+ (e.g., AtFRO2, Ferric Reductase Oxidase); and (c) Fe2+ absorption into root epidermal cells by transmembrane transporters (e.g., AtIRT1, Iron-Regulated Transporter) (Hindt and Guerinot, 2012; Ivanov et al., 2012). All three components of this strategy increase their activities during Fe deficiency. Strategy II, or chelation strategy, is used by plants from Poaceae family, and involves: (a) synthesis and release of small molecular weight compounds of the mugineic acid family called phytosiderophores (PS) into the rhizosphere, which bind Fe3+ with high affinity, via TOM1/OsZIFL4 (Nozoye et al., 2011; Ricachenevsky et al., 2011); and (b) Fe(III)-PS complex uptake into root cells by a Yellow Stripe/Yellow Stripe-Like (YSL) transporters. Both processes (PS excretion and Fe(III)-PS transport) are increased in response to Fe deficiency.
Since both maize (Zea mays) ys1 mutant (defective for Fe(III)-PS transport) and Arabidopsis thaliana irt1 mutant (defective for Fe2+ transport) cannot survive under Fe deficiency conditions, it was first widely accepted that these two Fe uptake strategies were the main mechanisms for Fe acquisition in each plant group. However, later work on rice (Oryza sativa) showed that two functional Fe2+ transporters, OsIRT1 and OsIRT2, were expressed in roots upon Fe deficiency (Ishimaru et al., 2006; Walker and Connolly, 2008). It was proposed that rice uses a combined strategy, which has all features of a strategy II plant (PS release through TOM1/OsZIFL4 and Fe(III)-PS uptake through OsYSL15, the YS1 ortholog—Inoue et al., 2009; Lee et al., 2009) and some features of a strategy I plant (Fe2+ uptake using IRT transporters). The other two components of strategy I plants, proton extrusion, and Fe(III)-chelate reductase activity, were not detected in Fe-deficient rice roots (Ishimaru et al., 2006). Further evidence for combined strategy was provided by rice plants carrying a mutation in the NICOTIANAMINE AMINOTRANSFERASE (NAAT) gene, a key enzyme in PS synthesis. This mutant, which lacks PS, is able to grow if Fe2+ is supplied as Fe source (Cheng et al., 2007). Based on these findings, it was proposed that the ability to absorb Fe2+ evolved in rice as an adaptation to the soil conditions in flooded paddies, where Fe2+ is more abundant than Fe3+ (Ishimaru et al., 2006; Walker and Connolly, 2008; Hindt and Guerinot, 2012). So far, rice is the only plant described to use the combined strategy mechanism.
A number of studies described key players and major transcriptional networks that control Fe homeostasis in both grasses and non-grasses (Hindt and Guerinot, 2012; Ivanov et al., 2012). Interestingly, orthologous genes have been described in rice and Arabidopsis thaliana, showing similar roles. The bHLH transcription factor FIT (FER-like iron-deficiency-induced transcription factor) from A. thaliana interacts with bHLH038 and bHLH039 to regulate IRT1 and FRO2 under Fe deficiency (Yuan et al., 2008). FIT has no ortholog in rice, but bHLH38/39 are highly similar to OsIRO2 (Hindt and Guerinot, 2012), a known downstream regulator of Fe deficiency-responsive genes. OsIRO2 regulates the Fe(III)-PS transport-related genes, but not OsIRT1 (Ogo et al., 2007). OsIDEF1, acting upstream of OsIRO2, and OsIDEF2, are transcriptional regulators of distinct but partially overlapping branches of Fe deficiency response in rice (Ogo et al., 2008; Kobayashi et al., 2009) However, no ortholog for OsIDEF1 or OsIDEF2 was described in A. thaliana, although similar genes are found in the genomic sequence (Kobayashi and Nishizawa, 2012).
In A. thaliana, a second regulatory network is controlled by bHLH transcription factor named POPEYE (PYE), which targets distinct metal homeostasis genes. PYE seems to be regulated by interacting partners such as BRUTUS (BTS), an E3 ubiquitin ligase with metal and DNA binding domains that negatively regulates the response to Fe deficiency (Long et al., 2010; Kobayashi et al., 2013). In rice, Zheng et al. (2011) identified a negative regulator of the Fe deficiency response, OsIRO3, which could be the ortholog of PYE (Hindt and Guerinot, 2012). Interestingly, BTS orthologs OsHRZ1 and OsHRZ2 were recently characterized as negative regulators of Fe uptake and Fe utilization genes (Kobayashi et al., 2013). Thus, it seems that control of Fe deficiency response is partly conserved between A. thaliana and rice. Moreover, Urzica et al. (2012) performed a trans-system analysis looking for genes responsive to low Fe supply in Chlamydomonas reinhardtii, A. thaliana and rice, and observed that BTS/HRZs, IRT1, and IRT2 are conserved throughout the plant lineage.
Evolution of the Fe deficiency response
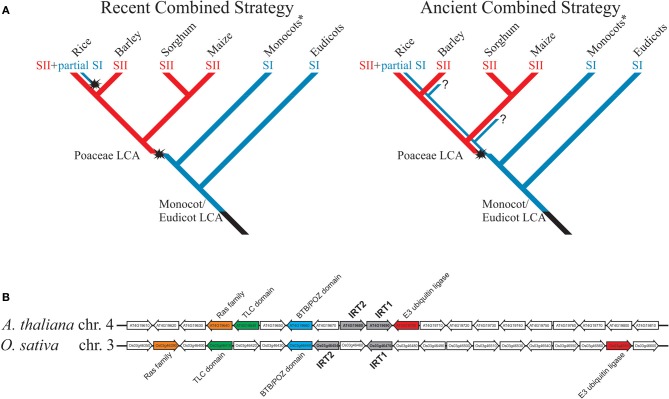
Currently there is no model for the evolution of Fe deficiency response in plants, especially in Poaceae, and few studies have focused on testing the hypothesis that rice is the only combined strategy species. Considering the available evidence, two models are possible (Figure 1A). In the first, named “recent combined strategy,” combined strategy is an evolutionary novelty restricted to rice or close ancestral species, not shared with other extant species from Poaceae, which all use strategy II, and rice has acquired ability to induce an IRT1-like transporter under Fe deficiency, partially resembling strategy I, as an adaptation to flooding. It implies that Poaceae last common ancestor (LCA) has lost strategy I-based Fe acquisition capacity and gained strategy II-based Fe uptake mechanism before diversification within the family (Figure 1A).
Figure 1.
(A) Models for evolution of rice combined strategy for Fe uptake. Blue lines represent plants using strategy I for Fe uptake; red lines represent plants using strategy II. Evolutionary novelties are highlighted, as the appearance of strategy II in Poaceae group and the putative re-appearance of IRT1-mediated Fe2+ uptake in rice, also known as the combined strategy (strategy II + partial strategy I). Question marks denote unclear points. Monocots* are all non-Poaceae monocotyledonous plants. SI = strategy I; SII = strategy II; LCA = last common ancestor. The selected monocot species are only to illustrate the divergence of Poaceae species, representing the subfamilies Ehrhartoideae (rice), Pooideae (barley), and Panicoideae (maize and sorghum). (B) Microsynteny between AtIRT1/AtIRT2 and OsIRT1/OsIRT2 loci. Conserved genes between rice and Arabidopsis were found using CoGe tool GEVo (http://genomevolution.org/CoGe/) and the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/). Homologs are shown with same colors; IRT1 and IRT2 in each genome are in gray.
In the second model, named “ancient combined strategy,” combined strategy is considered an evolutionary ancient trait (Figure 1A). Poaceae LCA has gained the ability to use strategy II for Fe uptake, but maintained strategy I. During Poaceae diversification and speciation, both strategy I and strategy II-specific genes were available for natural selection, and thus distinct groups could have adapted differently to respond to low Fe condition. As an example, the rice lineage has maintained IRT1-like up-regulation from the original strategy I response, while rhizosphere acidification and Fe reduction traits were lost. While all Poaceae would use the more efficient Fe acquisition mechanism strategy II (Curie and Briat, 2003), the model predicts that extant species could also show partial strategy I as rice does (i.e., IRT1-like Fe2+ uptake).
Indirect evidence favors the ancient combined strategy model. OsIRT1 ortholog in barley (Hordeum vulgare), HvIRT1, is also up-regulated by Fe deficiency and transports Fe, Mn, Zn, and Cd (Pedas et al., 2008). The maize ortholog, ZmIRT1, was both described as not Fe regulated by Nozoye et al. (2013) and as Fe regulated by Li et al. (2013). Either way, Li et al. (2013) observed strong up-regulation of ZmIRT1 under Fe deficiency and showed ZmIRT1 ability to complement Fe and Zn uptake-defective yeast strains. Interestingly, IRT1-like genes described in Poaceae clustered together with AtIRT1 in a phylogenetic analysis (Li et al., 2013). AtIRT2 and OsIRT2, two genes similar to AtIRT1 and OsIRT1 that code for Fe transporters (Ishimaru et al., 2006; Vert et al., 2009), are part of the same cluster. Strikingly, AtIRT1/AtIRT2 and OsIRT1/OsIRT2 gene pairs are localized in tandem in their respective genomic regions and, despite monocot/dicot divergence dates 120 to 200 million years ago (Salse et al., 2002), they still show some degree of microsynteny (Figure 1B). These data indicate that OsIRT1 (and probably OsIRT2) shares not only functional similarity but also common evolutionary origin with AtIRT1, as the ancient combined strategy model predicts. Moreover, phenolics were described as important for Fe deficiency response in A. thaliana and rice, indicating that less understood aspects of Fe deficiency response are conserved between strategy I and strategy II plants (Bashir et al., 2011; Rodríguez-Celma et al., 2013; Fourcroy et al., 2014; Schmid et al., 2014).
We should also consider the likeliness of each model. In recent combined strategy model, the LCA IRT1 gene would have lost Fe deficiency responsiveness, presumably through deleterious mutations related to promoter activity (i.e., hampering transcription factor binding, interaction with enhancers, etc.). For OsIRT1 to be able to respond again to low Fe concentration in combined strategy, such mutations (or any changes that rendered IRT1 non-regulated) would need to be reversed. That implies re-activation of non-functional regulatory sequences on the promoter of the same gene, which is part of the ZIP gene family of around ten members in Poaceae genomes, several of them encoding Fe transporters (Li et al., 2013), and re-insertion into intricate regulatory circuits (Kobayashi and Nishizawa, 2012). Although possible, the ancient combined strategy model is more parsimonious, predicting that IRT1 function was conserved through plant lineage evolution, and that preference for combined strategy or strategy II in Poaceae was a late adaptation (Figure 1A).
Conclusions and future perspectives
It has long been established that Poaceae species rely on strategy II mechanism, while all other plant groups use strategy I. Rice has been considered an exception to the Poaceae-strategy II rule (Ishimaru et al., 2006; Sperotto et al., 2012); however, although it is clear that strategy II is the main mechanism for Fe uptake in graminaceous plants, the presence of IRT1-based Fe2+ transport in roots might play a non-overlapping role, not only in rice but in other species (Ishimaru et al., 2006; Pedas et al., 2008; Li et al., 2013). The ancient combined strategy model proposed here states that AtIRT1 ortolog kept their ancient function, observed early in the plant lineage (Urzica et al., 2012), all the way from monocot/eudicot split to extant Oryza sativa species, rather than re-emerged with the same function in rice (Figure 1). Then, it is possible that distinct Poaceae subfamilies and species evolved independently to strategy II-exclusive or combined strategy-like strategies.
Many studies have focused on Fe acquisition genes and underlying signaling pathway controlling these strategies, but most available data is on model species A. thaliana and rice (Hindt and Guerinot, 2012; Ivanov et al., 2012; Kobayashi and Nishizawa, 2012). Besides these, studies in maize, barley and Brachypodium distachyon have indicated the role of YSL transporters in Fe(III)-PS acquisition (Curie et al., 2001; Murata et al., 2006; Yordem et al., 2011). However, the role of IRT1-like transporters is still poorly understood. Studies on the role of IRT1, as well as other strategy I-related genes such as Fe3+-reductase, in Poaceae species, should shed light into how exclusive rice combined strategy is.
We should also consider that other species might use variants of strategy I and strategy II, or even rely on distinct mechanisms. Recently, it was demonstrated that peanut (Arachis hypogaea), an eudicot, is able to absorb Fe(III)-PS complexes through AhYSL1 transporter. The complexes, however, are only present after intercropping with maize, which secretes PS in the soil, increasing Fe efficiency of peanut (Xiong et al., 2013). An exciting new study described a previously unknown Fe uptake mechanism, where Fe is delivered to embryos of pea (Pisum sativum) and A. thaliana in the form of Fe(III)-malate/citrate complexes, and is then chemically reduced to Fe2+ by ascorbate, which is effluxed from embryos, for subsequent uptake (Grillet et al., 2014b).
With lowering costs and increased access to technologies such as next-generation sequencing (Mardis, 2013) and genome editing (Gaj et al., 2013), it is becoming feasible to perform comparative genomics and transcriptomic studies with plants species for which genetic resources are not available, and eventually test key genes identified. These comparisons will allow testing the models discussed here, as well as uncovering new genes and strategies for Fe acquisition.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to authors whose relevant work could not be cited due to the lack of space. The authors are supported by CNPq and Centro Universitário UNIVATES.
References
- Bashir K., Ishimaru Y., Shimo H., Kakei Y., Senoura T., Takahashi R., et al. (2011). Rice phenolics efflux transporter 2 (PEZ2) plays an important role in solubilizing apoplasmic iron. Soil Sci. Plant Nutr. 57, 803–812 10.1080/00380768.2011.637305 [DOI] [Google Scholar]
- Cheng L., Wang F., Shou H., Huang F., Zheng L., He F., et al. (2007). Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiol. 145, 1647–1657 10.1104/pp.107.107912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C., Briat J. F. (2003). Iron transport and signaling in plants. Ann. Rev. Plant Biol. 54, 183–206 10.1146/annurev.arplant.54.031902.135018 [DOI] [PubMed] [Google Scholar]
- Curie C., Panaviene Z., Loulergue C., Dellaporta S. L., Briat J. F., Walker E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409, 346–349 10.1038/35053080 [DOI] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F., 3rd. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405 10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourcroy P., Sisó-Terraza P., Sudre D., Savirón M., Reyt G., Gaymard F., et al. (2014). Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol. 201, 155–167 10.1111/nph.12471 [DOI] [PubMed] [Google Scholar]
- Grillet L., Mari S., Schmidt W. (2014a). Iron in seeds - loading pathways and subcellular localization. Front. Plant Sci. 4:535 10.3389/fpls.2013.00535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet L., Ouerdane L., Flis P., Hoang M. T., Isaure M. P., Lobinski R., et al. (2014b). Ascorbate efflux as a new strategy for iron reduction and transport in plants. J. Biol. Chem. 289, 2515–2525 10.1074/jbc.M113.514828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindt M. N., Guerinot M. L. (2012). Getting a sense for signals: regulation of the plant iron deficiency response. Biochim. Biophys. Acta 1823, 1521–1530 10.1016/j.bbamcr.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Kobayashi T., Nozoye T., Takahashi M., Kakei Y., Suzuki K., et al. (2009). Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 284, 3470–3479 10.1074/jbc.M806042200 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y., Suzuki M., Tsukamoto T., Suzuki K., Nakazono M., Kobayashi T., et al. (2006). Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 45, 335–346 10.1111/j.1365-313X.2005.02624.x [DOI] [PubMed] [Google Scholar]
- Ivanov R., Brumbarova T., Bauer P. (2012). Fitting into the harsh reality: regulation of iron deficiency responses in dicotyledonous plants. Mol. Plant 5, 27–42 10.1093/mp/ssr065 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Itai R. N., Ogo Y., Kakei Y., Nakanishi H., Takahashi M., et al. (2009). The rice transcription factor IDEF1 is essential for the early response to iron deficiency, and induces vegetative expression of late embryogenesis abundant genes. Plant J. 60, 948–961 10.1111/j.1365-313X.2009.04015.x [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Nagasaka S., Senoura T., Itai R. N., Nakanishi H., Nishizawa N. K. (2013). Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 4, 2792 10.1038/ncomms3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Nishizawa N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152 10.1146/annurev-arplant-042811-105522 [DOI] [PubMed] [Google Scholar]
- Lee S., Chiecko J. C., Kim S. A., Walker E. L., Lee Y., Guerinot M. L., et al. (2009). Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol. 150, 786–800 10.1104/pp.109.135418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemanceau P., Bauer P., Kraemer S., Briat J. F. (2009). Iron dynamics in the rhizosphere as a case study for analyzing interactions between soils, plants and microbes. Plant Soil 321, 513–535 10.1007/s11104-009-0039-5 [DOI] [Google Scholar]
- Li S., Zhou X., Huang Y., Zhu L., Zhang S., Zhao Y., et al. (2013). Identification and characterization of the zinc-regulated transporters, iron-regulated transporter-like protein (ZIP) gene family in maize. BMC Plant Biol. 13:114 10.1186/1471-2229-13-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T. A., Tsukagoshi H., Busch W., Lahner B., Salt D. E., Benfey P. N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22, 2219–2236 10.1105/tpc.110.074096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis E. R. (2013). Next-generation sequencing platforms. Annu. Rev. Anal. Chem. 6, 287–303 10.1146/annurev-anchem-062012-092628 [DOI] [PubMed] [Google Scholar]
- Murata Y., Ma J. F., Yamaji N., Ueno D., Nomoto K., Iwashita T. (2006). A specific transporter for iron(III)-phytosiderophore in barley roots. Plant J. 46, 563–572 10.1111/j.1365-313X.2006.02714.x [DOI] [PubMed] [Google Scholar]
- Nozoye T., Nagasaka S., Kobayashi T., Takahashi M., Sato Y., Uozumi N., et al. (2011). Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 286, 5446–5454 10.1074/jbc.M110.180026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozoye T., Nakanishi H., Nishizawa N. K. (2013). Characterizing the crucial components of iron homeostasis in the maize mutants ys1 and ys3. PLoS ONE 8:e62567 10.1371/journal.pone.0062567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogo Y., Itai R. N., Nakanishi H., Kobayashi T., Takahashi M., Mori S., et al. (2007). The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 51, 366–377 10.1111/j.1365-313X.2007.03149.x [DOI] [PubMed] [Google Scholar]
- Ogo Y., Kobayashi T., Nakanishi I. R., Nakanishi H., Kakei Y., Takahashi M., et al. (2008). A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. J. Biol. Chem. 283, 13407–13417 10.1074/jbc.M708732200 [DOI] [PubMed] [Google Scholar]
- Pedas P., Ytting C. K., Fuglsang A. T., Jahn T. P., Schjoerring J. K., Husted S. (2008). Manganese efficiency in barley: identification and characterization of the metal ion transporter HvIRT1. Plant Physiol. 148, 455–466 10.1104/pp.108.118851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricachenevsky F. K., Sperotto R. A., Menguer P. K., Sperb E. R., Lopes K. L., Fett J. P. (2011). ZINC-INDUCED FACILITATOR-LIKE family in plants: lineage-specific expansion in monocotyledons and conserved genomic and expression features among rice (Oryza sativa) paralogs. BMC Plant Biol. 11:20 10.1186/1471-2229-11-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Celma J., Lin W. D., Fu G. M., Abadía J., López-Millán A. F., Schmidt W. (2013). Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiol. 162, 1473–1485 10.1104/pp.113.220426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J., Piégu B., Cooke R., Delseny M. (2002). Synteny between Arabidopsis thaliana and rice at the genome level: a tool to identify conservation in the ongoing rice genome sequencing project. Nucleic Acids Res. 30, 2316–2328 10.1093/nar/30.11.2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid N. B., Giehl R. F., Döll S., Mock H. P., Strehmel N., Scheel D., et al. (2014). Feruloyl-CoA 6′-Hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in arabidopsis. Plant Physiol. 164, 160–172 10.1104/pp.113.228544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperotto R. A., Ricachenevsky F. K., Stein R. J., Waldow V. A., Fett J. P. (2010). Iron stress in plants: dealing with deprivation and overload. Plant Stress 4, 57–69 [Google Scholar]
- Sperotto R. A., Ricachenevsky F. K., Waldow V. A., Fett J. P. (2012). Iron biofortification in rice: it's a long way to the top. Plant Sci. 190, 24–39 10.1016/j.plantsci.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Urzica E. I., Casero D., Yamasaki H., Hsieh S. I., Adler L. N., Karpowicz S. J., et al. (2012). Systems and trans-system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell 24, 3921–3948 10.1105/tpc.112.102491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G., Barberon M., Zelazny E., Séguéla M., Briat J. F., Curie C. (2009). Arabidopsis IRT2 cooperates with the high-affinity iron uptake system to maintain iron homeostasis in root epidermal cells. Planta 229, 1171–1179 10.1007/s00425-009-0904-8 [DOI] [PubMed] [Google Scholar]
- Walker E. L., Connolly E. L. (2008). Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 11, 530–535 10.1016/j.pbi.2008.06.013 [DOI] [PubMed] [Google Scholar]
- Xiong H., Kakei Y., Kobayashi T., Guo X., Nakazono M., Takahashi H., et al. (2013). Molecular evidence for phytosiderophore-induced improvement of iron nutrition of peanut intercropped with maize in calcareous soil. Plant Cell Environ. 36, 1888–1902 10.1111/pce.12097 [DOI] [PubMed] [Google Scholar]
- Yordem B. K., Conte S. S., Ma J. F., Yokosho K., Vasques K. A., Gopalsamy S. N., et al. (2011). Brachypodium distachyon as a new model system for understanding iron homeostasis in grasses: phylogenetic and expression analysis of Yellow Stripe-Like (YSL) transporters. Ann. Bot. 108, 821–833 10.1093/aob/mcr200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Wu H., Wang N., Li J., Zhao W., Du J., et al. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18, 385–397 10.1038/cr.2008.26 [DOI] [PubMed] [Google Scholar]
- Zheng L., Ying Y., Wang L., Wang F., Whelan J., Shou H. (2011). Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol. 10:166 10.1186/1471-2229-10-166 [DOI] [PMC free article] [PubMed] [Google Scholar]