Abstract
Microbial biogeography is influenced by the combined effects of passive dispersal and environmental selection, but the contribution of either factor can be difficult to discern. As thermophilic bacteria cannot grow in the cold seabed, their inactive spores are not subject to environmental selection. We therefore conducted a global experimental survey using thermophilic endospores that are passively deposited by sedimentation to the cold seafloor as tracers to study the effect of dispersal by ocean currents on the biogeography of marine microorganisms. Our analysis of 81 different marine sediments from around the world identified 146 species-level 16S rRNA phylotypes of endospore-forming, thermophilic Firmicutes. Phylotypes showed various patterns of spatial distribution in the world oceans and were dispersal-limited to different degrees. Co-occurrence of several phylotypes in locations separated by great distances (west of Svalbard, the Baltic Sea and the Gulf of California) demonstrated a widespread but not ubiquitous distribution. In contrast, Arctic regions with water masses that are relatively isolated from global ocean circulation (Baffin Bay and east of Svalbard) were characterized by low phylotype richness and different compositions of phylotypes. The observed distribution pattern of thermophilic endospores in marine sediments suggests that the impact of passive dispersal on marine microbial biogeography is controlled by the connectivity of local water masses to ocean circulation.
Keywords: biogeography, endospores, marine microorganisms, ocean currents, thermophiles
Introduction
Microorganisms display distinct biogeographic patterns (reviewed in Foissner, 2006; Green and Bohannan, 2006; Martiny et al., 2006; Ramette and Tiedje, 2007; Lindstrom and Langenheder, 2012), yet the mechanisms controlling their distribution in the environment are difficult to distinguish and thus not well understood. Four fundamental processes—selection, drift, dispersal and mutation—have been proposed for creating and maintaining microbial biogeographic patterns (Hanson et al., 2012), updating the classical concept of dispersal, speciation and extinction as the main factors determining biogeography. Regardless of theoretical framework, dispersal of microbial cells has a central role in shaping the spatial distribution of microbial biodiversity (Green and Bohannan, 2006; Fierer, 2008). In its strictest sense, microbial dispersal is defined as the physical movement of cells between two locations, but an extended definition additionally includes successful establishment—that is, physiological activity and growth of migrated cells—at the receiving location (Hanson et al., 2012). The existence of physical dispersal barriers for microorganisms has traditionally been questioned. Under the Baas Becking paradigm of microbial cosmopolitanism—‘everything is everywhere, but, the environment selects'—it is hypothesized that microorganisms possess unlimited dispersal capabilities due to their large population sizes and short generation times. In this paradigm, environmental factors are the sole determinants of observed microbial distribution patterns (Baas Becking, 1934; Finlay, 2002; Fenchel and Finlay, 2004). More recently, multiple studies have put forward evidence for dispersal limitation among microorganisms (Papke et al., 2003; Whitaker et al., 2003; Green et al., 2004; Reche et al., 2005; Martiny et al., 2006; Ghiglione et al., 2012; Sul et al., 2013). Controversy remains, however, partly due to differences in definitions of dispersal and the operational taxonomic units used to classify microorganisms (Hanson et al., 2012), but mostly because the biogeography of free-living microorganisms is the result of a combination of various evolutionary and ecological processes that make it difficult to unravel the relative influence of passive transport (Martiny et al., 2006; Ramette and Tiedje, 2007). We use the term dispersal here to describe physical movement by passive transport, but not colonization of the new location.
The presence of dormant endospores of thermophilic members of the bacterial phylum Firmicutes in cold marine sediments is the result of passive transport from warm environments and has been proposed as a natural model for selectively investigating the dispersal of microbial cells in the oceans (Bartholomew and Paik, 1966; Isaksen et al., 1994; Hubert et al., 2009; de Rezende et al., 2013). The inactivity of these model organisms enables their biogeography to be investigated largely without any confounding influence of environmental selection. Therefore, any observed non-random spatial distribution patterns of these organisms should be directly attributable to the influence of dispersal limitation. Ocean currents and eventual sedimentation have been invoked as dispersal vectors for supplying thermophilic Firmicutes spores to Arctic fjord sediments off the coast of Svalbard at an estimated rate exceeding 108 spores per square meter per year (Hubert et al., 2009). Once deposited in the cold sediment, these spores lie dormant as members of the rare biosphere and the ‘microbial seed bank' (Pedros-Alio, 2012; Gibbons et al., 2013), but they can be induced to germinate rapidly during sediment incubation experiments at high temperature (Hubert et al., 2009).
Enrichment of thermophilic anaerobes from dormant spores in high-temperature incubation experiments enables selective focus on a specific, culturable part of the rare biosphere that is of exceptional interest for studying long-term and long-distance dispersal. Bacterial endospore dispersal represents an upper boundary for the dispersal capabilities of vegetative cells, as bacterial endospores are metabolically inert, highly stress resistant and able to survive unfavorable conditions for long periods (Nicholson et al., 2000). For example, a half-life of up to 440 years was estimated for endospores of thermophilic sulfate-reducing bacteria deposited in Aarhus Bay in the Baltic Sea (de Rezende et al., 2013); these viable endospores decreased in abundance with depth but were still detected at 6.5 m below seafloor in 4500-year-old sediment, suggesting a life span that could allow the global dispersal of spores through thermohaline circulation that fully connects the world oceans on a time scale of ∼1000–2700 years (DeVries and Primeau, 2011). Diverse populations of dormant bacterial spores become active when sediment is heated (to 50–60 °C). Once germinated, the vegetative cells catalyze the mineralization of organic matter via extracellular enzymatic hydrolysis, fermentation and sulfate reduction (Hubert et al., 2010). So far, the phylogenetic composition of these dormant, thermophilic communities has only been studied in a few locations, that is, the sediments of West Svalbard fjords and Aarhus Bay. Even though these locations are ∼3000 km apart, they share at least two thermophilic Desulfotomaculum phylotypes with identical 16S rRNA and dissimilatory (bi)sulfite reductase (dsrAB) gene sequences (de Rezende et al., 2013). Although this is the first intriguing evidence for long-distance dispersal of these thermophilic endospores, their large-scale biogeography in the global ocean remains unexplored.
We thus analyzed the richness, phylogeny and distribution of endospores of thermophilic bacteria in marine sediments on global and regional scales to address the following questions: are thermophilic spores randomly distributed, which would be indicative of unlimited dispersal, or does the biogeography of thermophilic spores show signs of dispersal limitation? If these spores are not randomly distributed, how do local hydrography, sedimentation and major ocean currents impact their distribution? To investigate the biogeography of thermophilic spores, sediment samples from 81 locations around the world ocean, including regional Arctic samples from the Svalbard archipelago and the Baffin Bay that connect differently to global ocean circulation, were amended with organic substrates, pasteurized and incubated at 50 °C under anoxic conditions. Pasteurization kills the vegetative community of mesophilic or psychrophilic microorganisms. Shifts in microbial community activity and composition, owing to germination and growth of thermophilic endospores during the incubation, were then monitored by measuring sulfate reduction rates and by using multiplex pyrosequencing of bacterial 16S rRNA gene amplicons. Subsequently, the phylogeny and biogeography of enriched thermophilic phylotypes was analyzed.
Materials and methods
Marine sediment samples
The sample set comprised marine sediments from 81 locations around the world ocean, including two regional Arctic sample sets from Svalbard fjords and the Baffin Bay and samples from hydrothermally influenced sediments in the Guaymas Basin (43–150 °C, Gulf of California) that comprises potential source environments of endospore-forming thermophiles (Figure 1, Supplementary Table S1). Most sediment samples were collected from the seafloor surface (0–10 cm below seafloor) at coastal or deep sea, open ocean sites with in situ temperatures ranging from 0 to 30 °C. Samples were stored at 4 °C or frozen at −20 °C until germination experiments.
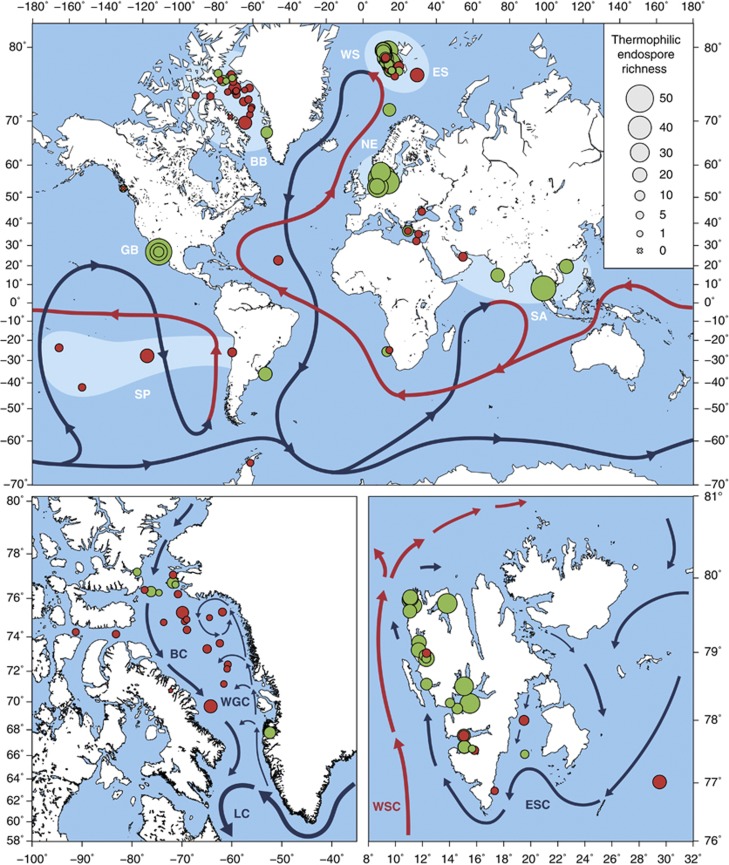
Figure 1.
Global and regional maps show the sediment-sampling sites and selected major ocean currents. Circle sizes represent richness of thermophilic Firmicutes endospore phylotypes (crosses indicate a richness of 0). Symbol color indicates whether thermophilic sulfate reduction was detected during the high temperature incubation (green=positive, red=negative). Global map (top) shows the global thermohaline circulation (warm/surface currents in red, cold/deep water currents in blue; adapted and simplified from Rahmstorf (2002)). Broad geographic regions studied are highlighted in lighter blue (WS, West Svalbard; ES, East Svalbard; BB, Baffin Bay; NE, Northern Europe; GB, Guaymas Basin; SA, South Asia; SP, South Pacific). Map of Baffin Bay (bottom left): local currents reproduced from Lloyd et al. (2005); BC, Baffin Island Current; WGC, West Greenland Current; LC, Labrador Current. Map of Svalbard (bottom right): local currents reproduced from Knies et al. (2007); WSC, West Spitsbergen Current; ESC, East Spitsbergen Current.
Sediment incubation and sulfate reduction measurements
Sediment was homogenized and mixed with sterile anoxic synthetic seawater medium at a 1:2 (w/w) ratio under constant flow of N2 gas. The sediment slurries were amended with organic substrates: formate, lactate, acetate, succinate, propionate, butyrate, ethanol (each to a final concentration of 0.5 mM) and/or with freeze-dried Spirulina cells (1.5 g l−1). Two 12-ml aliquots of slurried sediment were transferred to Hungate tubes under constant flow of N2 and pasteurized for 20 min at 80 °C and incubated in parallel at 50 °C. Before the incubation, one aliquot received ∼720 kBq 35S-labeled carrier-free sulfate tracer for sulfate reduction measurement. The incubations were sub-sampled by syringe-needle after 0, 56, 72 and/or 120 h. Aliquots of 3 ml from the 35S-sulfate slurries were mixed with 6 ml 20% zinc acetate solution and stored at −20 °C until sulfate reduction was determined using a single-step cold chromium distillation method (Kallmeyer et al., 2004). From the non-radioactive incubations, 2 ml subsamples were pelleted by centrifugation and frozen at −20 °C for subsequent DNA extraction.
Multiplex amplicon pyrosequencing
DNA was extracted from sediment slurries using the PowerSoil DNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA). Bacterial 16S rRNA gene amplicons were obtained and supplied with barcodes and pyrosequencing adaptors using a two-step PCR approach with low cycle numbers (20+5) and pooling of triplicate PCR reactions to reduce variability associated with barcoded pyrosequencing primers (Berry et al., 2011). The PCR primers targeting most bacteria (909F: 5′-ACTCAAAKGAATWGACGG-3′, 1492R: 5′-NTACCTTGTTACGACT-3′) and conditions were described previously (Berry et al., 2011). Final PCR products were purified using the Agencourt Ampure XP system (Beckman Coulter, Vienna, Austria) and the DNA concentration was determined using a Quant-iT PicoGreen dsDNA Assay (Invitrogen, Darmstadt, Germany). For better coverage of samples with higher expected bacterial diversity, amplicons were then pooled at a 2:1 ratio of 0 h and 120 h time points, respectively. Sequencing was performed on a GS FLX or GS FLX+ instrument using Titanium chemistry (Roche, Mannheim, Germany) by the Norwegian High-Throughput Sequencing Centre (Oslo, Norway) or by Eurofins MWG Operon (Ebersberg, Germany). In total, 1 493 577 reads with an average length of 567 nucleotides (nt) were received. Sequences were trimmed and erroneous sequencing reads removed using the PyroNoise implementation in mothur (Quince et al., 2009; Schloss et al., 2009) and sorted according to barcode using QIIME (Caporaso et al., 2010), yielding a total of 1 196 847 usable reads with an average length of 353 nt. A 97% identity threshold was used for clustering reads into phylotypes with UCLUST (Edgar, 2010). Representative sequences were aligned with mothur using the Needleman-Wunsch pairwise alignment method default settings (Schloss et al., 2009). Chimeras were detected using Chimera Slayer (Haas et al., 2011) and excluded from further analysis. Pyrosequencing data are archived at the NCBI Sequence Read Archive under accession SRP028774.
Identification of putative thermophilic endospore phylotypes
Two criteria were used to identify putative thermophilic endospore phylotypes. First, species-level phylotypes had to be significantly enriched in at least one sediment sample after incubation at high temperature. This criterion is based on the assumption that thermophilic endospores will survive the initial pasteurization and germinate and grow during the ensuing 50 °C incubation. Significant enrichment of phylotypes was determined using a two-proportion T-test and P-values were corrected for multiple comparisons using the false discovery rate method in R (Benjamini and Hochberg, 1995) to account for uncertainty due to sequence sampling depth. Corrected P-values⩽0.01 were considered significant. Second, phylotypes had to be affiliated with the phylum Firmicutes, to which all known endospore-forming bacteria belong (it was shown recently that endosporulation likely evolved only once at the base of the Firmicutes tree) (Abecasis et al., 2013). This approach does not survey all inactive cells in the sampled environments, but hones in on a physiological subset of the rare biosphere (Lennon and Jones, 2011). The clear advantage of this strategy is that taxa that were active and subject to environmental selection in situ are excluded from the analysis, allowing passive dispersal to be evaluated in isolation. Representative sequences of enriched Firmicutes phylotypes were then automatically aligned using the web-based SINA aligner (Pruesse et al., 2012) and imported into the ARB-SILVA database SSU Ref NR 111 (Quast et al., 2013) using the ARB software package (Ludwig et al., 2004). The alignment was manually curated and used to re-cluster the representative sequences into species-level phylotypes of ⩾97% sequence similarity using the average neighbor algorithm in mothur. A phylotype was called present at a location if at least one sequence of this phylotype was detected before and/or after the 120 h incubation of sediment from this location.
Phylogeny
A maximum likelihood (RAxML) tree was calculated with almost full-length 16S rRNA sequences (⩾1400 nt) of closely related reference bacteria or environmental clones based on 1222 alignment positions by using a 50% sequence conservation filter for bacteria. Using the ARB Parsimony Interactive tool, the short amplicon pyrosequences were subsequently added to this tree one at a time by using the 50% sequence conservation filter and alignment filters covering the individual length of each representative phylotype sequence, without changing the overall tree topology. Trees were visualized using iTOL (Letunic and Bork, 2006).
Network analysis
Two different kinds of networks were built based on the phylotype presence/absence matrix. First, a network of phylotype co-occurrence was produced for phylotypes present in at least five sites and with a minimum Spearman correlation coefficient of 0.6 (and P<0.0001 based on permutation testing) (Barberán et al., 2012). Focus on phylotypes that occurred at multiple sites reduced network complexity and facilitated identification of the core thermophilic endospore community. Second, a network of sites based on phylotype diversity was produced based on Bray-Curtis similarity of at least 0.6. Networks were plotted using the ‘network' package in R (Butts et al., 2012).
Distance-decay analysis
Jaccard similarities were calculated from the phylotype presence/absence matrix using the ‘vegan' package in R (Oksanen et al., 2012). Distances were calculated from latitude and longitude coordinates of sampling sites using the Vincenty inverse formula for ellipsoids and World Geodetic System (WGS) 84 ellipsoid parameters and calculated using R (R Development Core Team, 2008). A linear model was fit to the data and the statistical significance of the resulting regression parameters was evaluated by analysis of variance in R. To determine which phylotypes were dispersal-limited, a permutation-testing approach was used. The geographical dispersal of each phylotype was calculated as the mean distance between sites at which it was found. A null distribution of mean distances was produced by randomly selecting from all sites an equally sized subset (e.g. if a phylotype was found at seven sites, then seven sites would be randomly selected). The mean distance between the random subsets was calculated, and this process was repeated for 10 000 re-samplings. The probability that the observed mean distance was due to chance was calculated and corrected for multiple comparisons and P-values ⩽0.05 were considered significant.
Geographical maps were drawn using Generic Mapping Tools (Wessel and Smith, 1998).
Results and discussion
Non-uniform distribution of thermophilic Firmicutes endospores in marine sediments
We studied the biogeography of thermophilic endospores in Arctic and other permanently cold or temperate marine sediments, because longevity of endospores makes them ideal study objects for understanding the time-averaged impact of dispersal on marine microbial biogeography. While there might be differences in endospore survival between different bacteria (Nicholson et al., 2000; McKenney et al., 2013), results are interpreted under the assumption that differences in endospore distribution are dependent on dispersal and not on environmental selection.
Anoxic, high-temperature incubations of pasteurized marine sediment (mostly from the top few centimeters) (Supplementary Results and Discussion, Supplementary Figure S1) from 81 different locations (Supplementary Table S1) led generally to a reduction in bacterial alpha diversity due to germination and growth of thermophilic endospores combined with the death and DNA decay of vegetative cells (Supplementary Materials and Methods, Supplementary Table S2). Principle coordinate analysis of weighted UniFrac distances confirmed a shift in phylogenetic composition of bacterial 16S rRNA genes after the incubation of most, but not all, sediment samples (Supplementary Figure S2). In total, we identified 146 thermophilic endospore phylotypes (hereafter called ‘thermospore phylotypes') across all high-temperature incubations (Supplementary Table S3). Thermospore phylotypes were detected in samples from almost all of the investigated locations (n=79/81) (Figure 1). Most of the 146 thermospore phylotypes were affiliated with the orders Clostridiales (61.0%) and Bacillales (17.8%); the most represented families were Clostridiaceae (32.2%), Peptococcaceae (17.8%) and Bacillaceae (14.4%) (Supplementary Figure S3, Supplementary Table S3). The phylogenetic identity and potential physiology of thermospore phylotypes corroborate and greatly expand upon previous studies that investigated thermophilic endospores in marine sediments (Bartholomew and Paik, 1966; Isaksen et al., 1994; Vandieken et al., 2006; Hubert et al., 2009, 2010; Ji et al., 2012; de Rezende et al., 2013) (Supplementary Results and Discussion). A screening of environmental amplicon databases showed that 16S rRNA gene sequences of the identified thermospore phylotypes are very rarely detected by environmental surveys in the marine environment (Supplementary Results and Discussion, Supplementary Table S4), which is likely consistent with the widespread occurrence of endospore taxa in low relative abundance in many habitats.
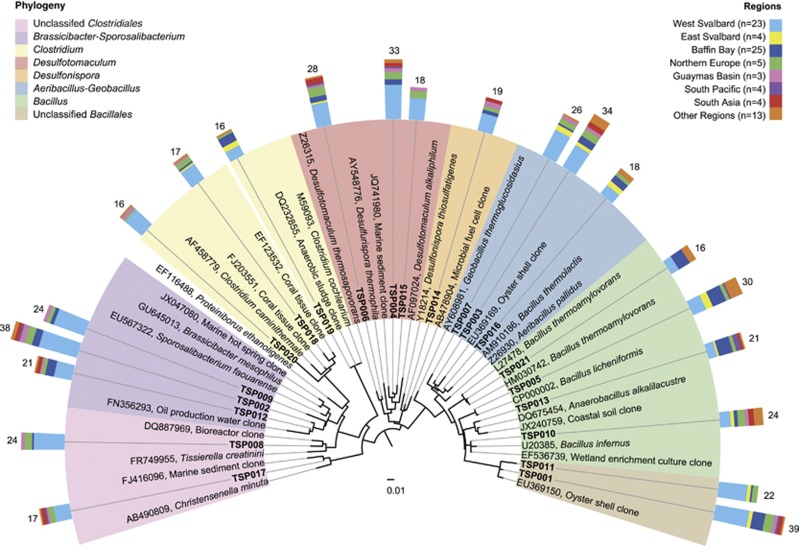
The different sampling locations showed considerable differences in thermospore phylotype richness, despite an absence of strict endemism of phylotypes that occurred at multiple locations (that is, exclusive presence in only one geographic region). The number of detected thermospore phylotypes per site ranged from 0 to 51 (on average, 13.3±11.6 (s.d.), n=81) (Supplementary Table S1). Thermospore phylotype richness was generally higher in Northern Europe (36.4±10.2 per site, n=5), the Guaymas Basin (28.7±18.6 per site, n=3), West Svalbard (17.9±8.3 per site, n=23) and South Asia (22.8±15.2 per site, n=4), whereas sites in the Baffin Bay (5.4±4.7 per site, n=25), the South Pacific (9.5±6.5 per site, n=4), East Svalbard (10.3±6.2 per site, n=4) and other regions (7.2±5.7 per site, n=13) showed comparatively lower phylotype richness (Figure 1). These differences in site occupancy indicate non-random variation in the distribution of thermospore phylotypes across the investigated sites. Out of the 146 thermospore phylotypes, 21 were detected at more than 15 locations and were widely distributed across different oceanic regions (‘cosmopolitan phylotypes', Figure 2, Supplementary Figures S4 and 5). This cosmopolitan distribution of thermospore phylotypes suggests that these taxa (i) are globally dispersed by ocean circulation and/or (ii) derive from multiple, globally widespread source environments that support growth of similar communities of thermophilic, endospore-forming bacteria. The cosmopolitan thermospore phylotypes were related to Bacillus (TSP005, TSP010, TSP013, TSP021), the Aeribacillus-Geobacillus lineage (TSP003, TSP007, TSP016), Desulfonispora (TSP014), Desulfotomaculum (TSP004, TSP006, TSP015), Clostridium (TSP018, TSP019, TSP020), the Brassicibacter-Sporosalibacterium lineage (TSP002, TSP009, TSP012), or were only assigned at the family or order level (Clostridiales Family XI. Incertae Sedis: TSP008; Christensenellaceae: TSP017; Bacillales: TSP001, TSP011) due to lack of close, cultivated relatives (Figure 2). In contrast, 82 thermospore phylotypes were found only at five or fewer locations (Supplementary Figures S3 and 4), indicating more restricted occurrence and/or low abundance below the detection limit of our approach.
Figure 2.
Phylogeny and geographic distribution of cosmopolitan thermophilic endospores. 16S rRNA-based phylogenetic tree of thermospore phylotypes (TSPs) that were detected at 15 or more sites. Scale bar indicates 1% sequence divergence as inferred from RAxML. Phylogenetic affiliations are highlighted in different colors. Colored bars indicate broad geographic regions where the thermospore phylotypes were present. Numbers indicate the number of sites at which a thermospore phylotype was detected, whereas the height of each bar color indicates the number of those phylotypes detected in each ocean region. TSP, thermospore phylotype. See Supplementary Figure S3 for an extended tree showing the phylogeny and geographic distribution of all 146 thermospore phylotypes.
Global biogeography patterns of thermophilic endospores show the influence of dispersal limitation
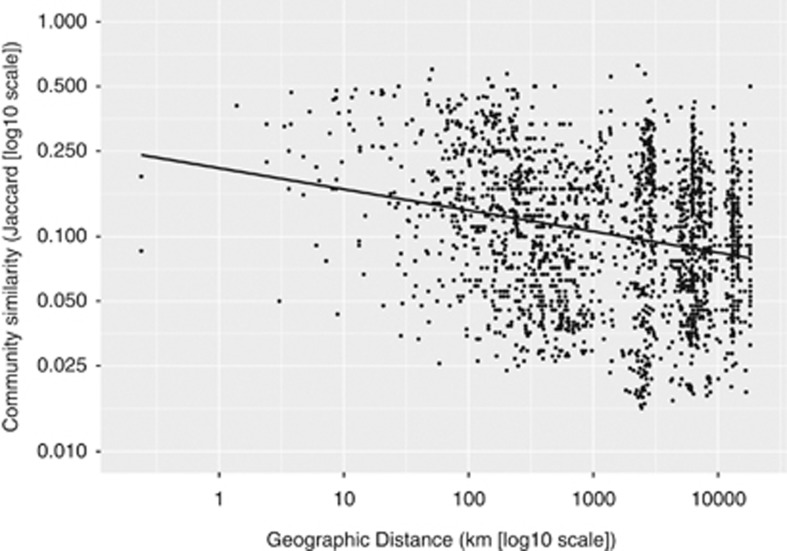
To analyze the relationship between geography and differences in endospore community structure, we plotted similarity between thermophilic endospore communities vs geographic distance for each location pair. This analysis revealed a significant, negative distance-decay curve (linear regression of log10-transformed variables with slope=−0.68177, intercept=−0.09818, P<0.001) (Figure 3). This result shows that thermophilic endospore communities are non-randomly distributed, which suggests that dispersal limitation is influencing the beta-diversity of thermospore phylotypes. Alternatively, the shape of the distance-decay curve may be influenced by differences in the ability of endospore phylotypes to resist decay during dispersal (Nicholson et al., 2000; McKenney et al., 2013). To account for this, we investigated if the distribution of individual phylotypes, which should be less biased by within-phylotype variances in endospore survival, is significantly different from a random geographic distribution. Twelve of 146 thermospore phylotypes (TSP001, TSP007, TSP008, TSP009, TSP011, TSP015, TSP020, TSP027, TSP060, TSP061, TSP096, TSP111) were seemingly dispersal-limited because they had significantly geographically clustered occurrence; that is, the average geographic distance between the locations of each phylotype was significantly less than would be expected from its random occurrence at the same number of locations.
Figure 3.
Distance decay of thermophilic endospore communities. The relationship between thermophilic endospore community similarity (Jaccard similarity, log10-transformed) and geographic distance (in kilometers, log10-transformed) is shown. The linear regression of log-transformed variables is significant (P<0.001) and has a negative slope (m=−0.68) and shows that communities of thermophilic endospores are non-randomly distributed.
Correlation network analyses reveal a widely distributed core of frequently co-occurring thermophilic endospores
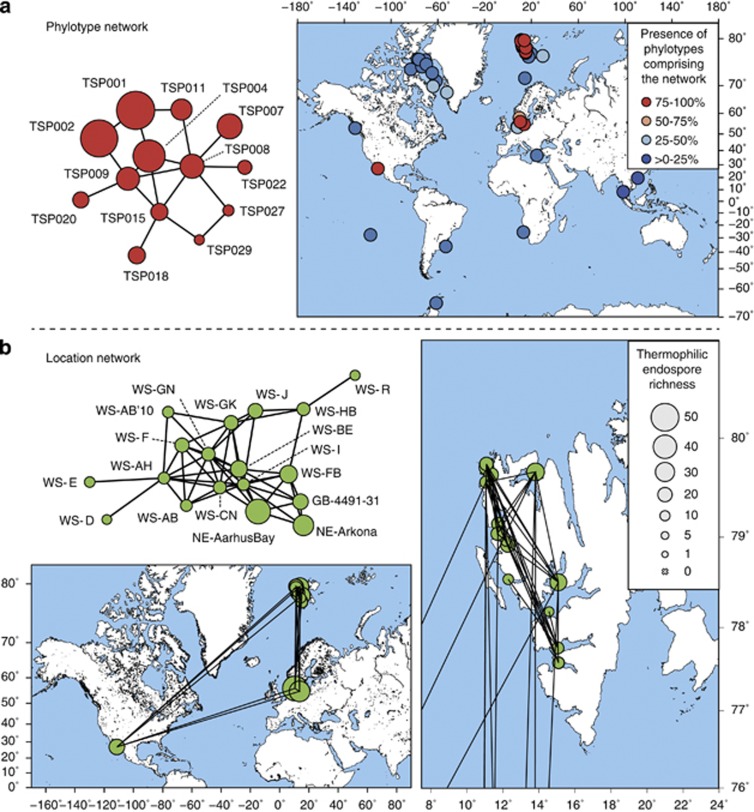
Network analysis was recently incorporated into microbial biogeography research to explore co-occurrence patterns of microbial taxa (Fuhrman, 2009; Barberán et al., 2012). We used correlation network analysis to better describe the biogeography patterns of thermophilic endospores identified by the distance-decay curve and investigated which phylotypes tend to co-occur; that is, which phylotypes are found together at some sites and are commonly absent from others. The co-occurrence of thermospore phylotypes was calculated using non-parametric Spearman correlations of thermospore phylotype presence/absence across all sampling sites and, to explore groups of co-occurring phylotypes, network analysis was used for visualization. We identified eight small networks of different complexity, that is, numbers of nodes and edges (connections) (Supplementary Figure S6A). The largest network was characterized by four highly connected, central phylotypes (⩾4 connections; TSP004, TSP008, TSP009, TSP015) and nine peripheral phylotypes that occurred at several locations (Figure 4a). Once we identified the most frequently co-occurring phylotypes, we explored similarities between thermospore phylotype communities by inferring association networks of sediment locations. The largest network contained 18 locations, of which many harbored several thermospore phylotypes (Figure 4b), whereas 10 other, much smaller networks were composed of ⩽4 locations (Supplementary Figure S6B). The 18-location network was composed of most West Svalbard samples, but also samples that were geographically very distant from the Svalbard archipelago, namely two samples from the Baltic Sea (Aarhus Bay, Arkona Basin) and one sample from the Gulf of California (Guaymas Basin) (Figure 4b). This network contains a phylogenetically diverse core of co-occurring thermophilic endospores that are widely but not ubiquitously distributed in the oceans (for example, one of the phylotypes, TSP001, was detected in 39 sediments). This raises questions about whether certain thermospore phylotypes share common source environments from where they are disseminated in a non-random manner over long distances and along similar, as yet unidentified travel routes.
Figure 4.
Network analysis of thermophilic endospore co-occurrence (a) and location (b). (a) Largest network of co-occurring thermospore phylotypes. Each node represents a thermospore phylotype (TSP). Presence of an edge between two nodes shows a strong correlation between these two phylotypes, which is indicative for co-occurrence. Circle size indicates site occupancy. Map shows locations where phylotypes of the network are present. The circle color indicates how many of the 13 phylotypes comprising the network are present at this site. (b) Largest location network. Each node represents a location. Presence of an edge between two nodes corresponds to a high Bray-Curtis similarity (⩾0.6) between the endospore communities at these two locations. Circle size indicates thermospore phylotype richness. Maps show the global locations and correlations of these sites. The map on the right shows a magnification of Svalbard for enhanced resolution.
Hydrothermal sediments of the Guaymas Basin as potential source environments for endospore-forming thermophiles
Based on the identity, physiology and sedimentation rates of thermophilic spores in Svalbard and Aarhus Bay, we have previously postulated biogeochemical and geological characteristics of hypothetical source environments for these bacteria (Hubert et al., 2009, 2010; de Rezende et al., 2013). First, such anoxic marine environments must be warm enough to allow vegetative growth of the diverse community of anaerobic, endospore-forming thermophiles. Second, sufficiently strong fluxes (e.g., fluid transport) from these environments into the water column must physically transport cells into circulating seawater. Consistent with this general description are, for example, pressurized gas or oil reservoirs in seabed sediments, and parts of oceanic spreading centers including hydrothermal vents and the sediments overlying them (Head et al., 2003; Cowen, 2004; Brazelton et al., 2006; Judd and Hovland, 2007; Hutnak et al., 2008; Hubert and Judd, 2010). Anaerobic thermophiles in these hot sediment and ocean crust habitats are connected with the overlying cold ocean water via advection of seabed fluids such as hydrocarbons and recirculating water discharged from the ocean crust (Judd and Hovland, 2007; Hutnak et al., 2008).
The hydrothermal surface sediments and the general geology and geography of Guaymas Basin satisfy these criteria (Supplementary Results and Discussion). Accordingly, a very large proportion (44.5%) of all 146 thermospore phylotypes were detected overall in the three Guaymas Basin sediments investigated. Although surveys with much finer genetic resolution than offered by the 16S rRNA gene approach are required to prove that identical microorganisms co-occur at distant locations, it is intriguing that the Guaymas Basin sediment community comprised 15 of 25 cosmopolitan thermospore phylotypes and 8 of the 13 co-occurring phylotypes (including all four highly connected phylotypes in the largest network) (Figure 4a). High thermospore phylotype richness despite restricted access due to the geographic isolation of the semi-enclosed Gulf of California supports the hypothesis that environments in the Guaymas Basin are sources of thermophilic spores to the overlying water column. Thermohaline water circulation in the Gulf of California is primarily influenced by wind, which alters directions and water transport between the gulf and the Pacific Ocean seasonally (Bray, 1988). It is conceivable that Guaymas Basin-derived spores embark on and survive long-distance travel to far away Svalbard and North Sea sediments via the global thermohaline circulation (Rahmstorf, 2000) (Figure 1). However, it may be more likely that these cold destinations are supplied with thermophilic spores from nearby warm environments that were not investigated in this study, and that dispersal barriers may prevent thermophilic endospores that escape the hot anoxic sediments in the Guaymas Basin from reaching far away water masses in the north Atlantic. Possible examples of potential source environments closer to Svalbard and the Baltic Sea could include systems such as the Lost City hydrothermal vent field associated with the mid-Atlantic ridge (Kelley et al., 2005; Brazelton et al., 2006) or the oil-bearing sediments in the North Sea (Gittel et al., 2009), which both support thermophilic sulfate-reducing Desulfotomaculum species that are closely related to the thermospore phylotypes identified in this study. It is conceivable that thermophilic bacteria get distributed along the mid-oceanic ridges throughout the world oceans, as exemplified by hydrothermal vent animals and their symbionts (Petersen et al., 2010; Vrijenhoek, 2010; Teixeira et al., 2011), creating an extended series of source habitats from where thermophilic endospores may be further dispersed to the cold sediments tested in this study. The location network shown in Figure 4b, thus likely represents a fraction of a much larger network of warm sources and warm or cold destinations for dispersing thermophilic endospores that are interconnected through ocean currents.
Anthropogenic sources may also contribute to the presence of anaerobic thermophiles in marine sediments. North Sea oil production systems have been proposed as possible sources for thermophilic Desulfotomaculum species in Aarhus Bay sediments (de Rezende et al., 2013) and close relatives to thermospore phylotypes detected in the present study were found in oil production water (Gittel et al., 2009). Close relatives were also detected in several anthropogenic, as well as natural terrestrial habitats such as bioreactors, activated sludge, compost and soil environments (Supplementary Tables S3 and 4). However, the phylotypes we have identified were enriched in artificial seawater, which is suggestive of a marine origin. Also, the presence of a widely distributed thermophilic Desulfotomaculum alkaliphilum phylotype (Figure 2) at depths corresponding to ∼4500 years of sedimentation in Aarhus Bay (de Rezende et al., 2013) indicates that there is a natural long-term dispersal of thermophilic endospores occurring in the marine environment that is independent of human activity.
Differences in thermophilic spore richness and phylogeny in Arctic regions correlate with connectivity of major water masses
The variable compositions of thermophilic endospore communities in Arctic sediments from the Baffin Bay and the east and west sides of the Svalbard archipelago suggest that thermospore phylotypes are not equally dispersed across these Arctic regions. Sediments from the Baffin Bay contained a significantly lower number of thermospore phylotypes than sediments from West Svalbard (Table 1). While sediments from only four locations on the East coast of Svalbard were available, we detected a similar yet not statistically significant trend of lower richness in East compared with West Svalbard sediments. Analysis of more samples from East Svalbard is required to confirm this trend. We additionally revealed a contrasting geographical distribution in the potential for thermophilic sulfate reduction that was generally consistent with the observed differences in thermospore phylotype richness (Figure 1). Thermophilic sulfate reduction was significantly more prevalent in West Svalbard than in East Svalbard and Baffin Bay sediments (Table 1).
Table 1. Comparison of thermospore phylotype richness, phylogenetic composition and prevalence of thermophilic sulfate reduction between Arctic regions.
| West Svalbard (n=23) | East Svalbard (n=4) | Baffin Bay (n=25) | West Svalbard vs Baffin Baya,b | West Svalbard vs East Svalbarda | East Svalbard vs Baffin Baya | |
|---|---|---|---|---|---|---|
| Thermospore phylotype richnessc | 17.9±8.3 | 10.3±6.2 | 5.4±4.7 | P<0.001 | Not significant | Not significant |
| Clostridiaceae richness | 6.0±2.8 | 2.0±1.8 | 1.5±2.1 | P<0.001 | P=0.02 | Not significant |
| Peptococcaceae richness | 3.4±2.9 | 2.0±1.6 | 0.6±0.9 | P<0.001 | Not significant | Not significant |
| Bacillaceae richness | 2.2±1.9 | 3.0±1.4 | 2.4±1.4 | Not significant | Not significant | Not significant |
| Thermophilic sulfate reduction (% of sites tested positive) | 87 | 25 | 24 | P<0.001 | P=0.02 | Not significant |
Significant differences between Arctic regions were determined by pairwise comparisons using the Mann–Whitney U-test for richness and Fisher's exact test for thermophilic sulfate reduction. P-values ⩽0.05 were considered significant.
All significant P-values in this column are also significant (P<0.01) when only surface (0–10 cm, n=13) or only deep (10–121 cm, n=12) sediment samples are analyzed.
All richness values are presented as mean±s.d.
The differences in thermophilic spore richness and metabolic potential were also reflected by the phylogenetic composition of thermophilic endospore communities in these Arctic regions (notwithstanding the aforementioned weak indications for strict endemism for any individual phylotypes). Of the three major families, West Svalbard sediments exhibit significantly more thermospore phylotypes belonging to the Clostridiaceae and the Peptococcaceae than Baffin Bay sediments, whereas Bacillaceae phylotypes were not differentially distributed between the Arctic regions (Table 1). Again, we observed a similar pattern between West Svalbard and East Svalbard, although differences were only significant for Clostridiaceae phylotypes (Table 1). No significant differences between East Svalbard and Baffin Bay sediments were found among these three taxonomic families (Table 1). Furthermore, frequently co-occurring thermospore phylotypes (e.g. TSP004, TSP008, TSP009, TSP015), as identified by network analysis (Figure 4), were almost exclusively limited to West Svalbard sediments. The observed distribution pattern is not impacted by the fact that 12 of 25 Baffin Bay sediment samples originate from a sediment depth below 10 cm (Supplementary Table S1). Restricting our analyses to only the shallow and deeper Baffin Bay sediments showed (i) no significant difference between them (e.g., thermospore phylotype richness: surface samples=4.8±4.7, deeper samples=6.1±4.9) and (ii) that both had the same significant differences compared with West Svalbard regarding richness, phylogenetic composition and prevalence of thermophilic sulfate reduction (Table 1).
One dispersal-limiting factor that might be responsible for the different thermophilic endospore communities in Svalbard and Baffin Bay sediments is the rate at which spores are deposited to the seafloor. Based on sediment thickness and age, sedimentation rates in the Baffin Bay and off the coast of Svalbard are estimated to be in the same order of magnitude (5–25 cm ka−1, Kallmeyer et al. (2012)). In contrast, reports for a few locations in these regions suggest that sedimentation rates are lower in the Baffin Bay (6.5 cm ka−1, Simon et al. (2012)) than in Svalbard fjords (180 cm ka−1, Hald et al. (2001); 190 cm ka−1, Hubert et al. (2009)). However, it is currently unknown how efficient endospores in the water column are deposited to the sediment, that is, how the specific rate of spore sedimentation relates to the overall sedimentation rate. It is noteworthy that for most locations our enrichment inoculum derives from homogenized sediment from 0 to 10 cm depth and therefore integrates endospore communities that accumulated over 50 to 1500 years of deposition. Given the longevity of thermophilic spores (de Rezende et al., 2013), our approach may diminish the impact of sedimentation rates on the recovered richness of thermophilic spore communities.
Besides the impact of local hydrography, sedimentation, and the possibility that unknown, local sources contribute to biogeography of thermophilic endospores, we hypothesized that dispersal-driven distribution also depends on how local currents in the sampled regions are connected to global ocean circulation. The observed West–East difference in thermophilic endospore community structure in Svalbard appears to correlate with the regional hydrography of the archipelago, although the difference is not statistically significant in all parameters (Table 1). Two major ocean currents define the hydrography of Svalbard. The warm West Spitsbergen Current, which is an extension of the Gulf Stream, flows from the Atlantic Ocean northwards, whereas the East Spitsbergen Current transports cold water from the Arctic Ocean southwards (Loeng, 1991) (Figure 1). Although the core of the West Spitsbergen Current that flows along the continental slope is separated from the West Spitsbergen shelf waters by the Arctic Front, extensive cross-front exchange takes place below 50 m depth (Saloranta and Svendsen, 2001) and it has been shown that the West Spitsbergen Current extends its influence deep into the fjord system of West Svalbard (MacLachlan et al., 2007).
The formation of deep and bottom water masses in the Baffin Bay are not fully understood (Tang et al., 2004), but they are generally influenced by cold Arctic Ocean water via the Baffin Island Current in the north and by the Atlantic Ocean via the West Greenland Current (Figure 1). The low thermospore phylotype richness in the Arctic water-influenced sediments in the Baffin Bay suggests a lower influx of thermophiles derived from the Arctic Ocean. Reduced thermospore phylotype richness in the Arctic Ocean is possibly due to limited thermophile sources and lower connectivity to water masses from the rest of the world oceans where thermophile sources may be far more abundant.
Recent studies of vegetative microbial communities in the global ocean have suggested that hydrography and geographic isolation of the Arctic Ocean represents an effective dispersal barrier for microorganisms (Galand et al., 2010; Ghiglione et al., 2012; Hamdan et al., 2013; Sul et al., 2013). By tracking thermophilic endospores whose Arctic biogeography is only controlled by passive dispersal, our results demonstrate this dispersal limitation to be true. Importantly, evidence presented here suggests that this dispersal limitation even holds true for marine bacteria with enhanced survival capacities that are less prone to death during long-term and long-distance dispersal under unfavorable environmental conditions, that is, perhaps the most likely candidates to not be dispersal-limited. Different phylotypes of globally widespread thermophilic endospores co-occur at multiple distantly separated locations and thus could be simultaneously dispersed from common sources and/or along similar paths according to non-random mechanisms. Global dispersal routes and frequencies of marine microbial taxa are largely dependent on the degree of connectivity between major oceanic water masses. Dispersal barriers of this kind exert greater control on the composition of marine microbial communities in water masses that participate minimally in global ocean circulation such as the Arctic Ocean.
Acknowledgments
We gratefully acknowledge the provision of sediment samples by Jan Amend, Gail Lee Arnold, Carol Arnosti, Elisa Bayraktarov, Antje Boetius, Gerhard Bohrmann, Ellen Damm, Steve D'Hondt, Tim Ferdelman, Moritz Holtappels, Lars Holmkvist, Sabine Kasten, Martin Krüger, Connie Lovejoy, Maren Nickel, Aude Picard, Roy Price, Nils Risgaard-Petersen, Alberto Robador, Søren Rysgaard, Joanna Sawicka, Verona Vandieken, and Thorsten Wilhelm. We thank China Hanson for her valuable comments on a draft version of this paper. This work was financially supported by the Austrian Science Fund (FWF, P20185-B17 and P25111-B22 to AL).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abecasis AB, Serrano M, Alves R, Quintais L, Pereira-Leal JB, Henriques AO. A genomic signature and the identification of new sporulation genes. J Bacteriol. 2013;195:2101–2115. doi: 10.1128/JB.02110-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas Becking LGM. Geobiologie of inleiding tot de milieukunde. W.P. Van Stockum & Zoon: The Hague, The Netherlands; 1934. [Google Scholar]
- Barberán A, Bates ST, Casamayor EO, Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012;6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew JW, Paik G. Isolation and identification of obligate thermophilic sporeforming bacilli from ocean basin cores. J Bacteriol. 1966;92:635–638. doi: 10.1128/jb.92.3.635-638.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
- Berry D, Mahfoudh KB, Wagner M, Loy A. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl Environ Microbiol. 2011;77:7846–7849. doi: 10.1128/AEM.05220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NA. Thermohaline circulation in the Gulf of California. J Geophys Res. 1988;93:4993–5020. [Google Scholar]
- Brazelton WJ, Schrenk MO, Kelley DS, Baross JA. Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl Environ Microbiol. 2006;72:6257–6270. doi: 10.1128/AEM.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butts CT, Handcock MS, Hunter DR.2012. Network: classes for relational data. R package version 1.7-1. http://statnet.org/ .
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen JP. The microbial biosphere of sediment-buried oceanic basement. Res Microbiol. 2004;155:497–506. doi: 10.1016/j.resmic.2004.03.008. [DOI] [PubMed] [Google Scholar]
- de Rezende JR, Kjeldsen KU, Hubert CR, Finster K, Loy A, Jorgensen BB. Dispersal of thermophilic Desulfotomaculum endospores into Baltic Sea sediments over thousands of years. ISME J. 2013;7:72–84. doi: 10.1038/ismej.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries T, Primeau F. Dynamically and observationally constrained estimates of water-mass distributions and ages in the global ocean. J Phys Oceanogr. 2011;41:2381–2401. [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Fenchel T, Finlay BJ. The ubiquity of small species: Patterns of local and global diversity. Bioscience. 2004;54:777–784. [Google Scholar]
- Fierer N.2008Microbial biogeography: patterns in microbial diversity across space and timeIn: Zengler K, (ed.)Accessing Uncultivated Microorganisms: From the Environment to Organisms and Genomes and Back ASM Press: Washington DC; 95–115. [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Foissner W. Biogeography and dispersal of micro-organisms: a review emphasizing protists. Acta Protozool. 2006;45:111–136. [Google Scholar]
- Fuhrman JA. Microbial community structure and its functional implications. Nature. 2009;459:193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- Galand PE, Potvin M, Casamayor EO, Lovejoy C. Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J. 2010;4:564–576. doi: 10.1038/ismej.2009.134. [DOI] [PubMed] [Google Scholar]
- Ghiglione JF, Galand PE, Pommier T, Pedros-Alio C, Maas EW, Bakker K, et al. Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc Natl Acad Sci USA. 2012;109:17633–17638. doi: 10.1073/pnas.1208160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons SM, Caporaso JG, Pirrung M, Field D, Knight R, Gilbert JA. Evidence for a persistent microbial seed bank throughout the global ocean. Proc Natl Acad Sci USA. 2013;110:4651–4655. doi: 10.1073/pnas.1217767110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittel A, Sørensen KB, Skovhus TL, Ingvorsen K, Schramm A. Prokaryotic community structure and sulfate reducer activity in water from high-temperature oil reservoirs with and without nitrate treatment. Appl Environ Microbiol. 2009;75:7086–7096. doi: 10.1128/AEM.01123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Bohannan BJ. Spatial scaling of microbial biodiversity. Trends Ecol Evol. 2006;21:501–507. doi: 10.1016/j.tree.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Green JL, Holmes AJ, Westoby M, Oliver I, Briscoe D, Dangerfield M, et al. Spatial scaling of microbial eukaryote diversity. Nature. 2004;432:747–750. doi: 10.1038/nature03034. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hald M, Dahlgren T, Olsen T-E, Lebesbye E. Late Holocene palaeoceanography in Van Mijenfjorden, Svalbard. Polar Res. 2001;20:23–35. [Google Scholar]
- Hamdan LJ, Coffin RB, Sikaroodi M, Greinert J, Treude T, Gillevet PM. Ocean currents shape the microbiome of Arctic marine sediments. ISME J. 2013;7:685–696. doi: 10.1038/ismej.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson CA, Fuhrman JA, Horner-Devine MC, Martiny JBH. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat Rev Microbiol. 2012;10:497–506. doi: 10.1038/nrmicro2795. [DOI] [PubMed] [Google Scholar]
- Head IM, Jones DM, Larter SR. Biological activity in the deep subsurface and the origin of heavy oil. Nature. 2003;426:344–352. doi: 10.1038/nature02134. [DOI] [PubMed] [Google Scholar]
- Hubert C, Loy A, Nickel M, Arnosti C, Baranyi C, Bruchert V, et al. A constant flux of diverse thermophilic bacteria into the cold Arctic seabed. Science. 2009;325:1541–1544. doi: 10.1126/science.1174012. [DOI] [PubMed] [Google Scholar]
- Hubert C, Arnosti C, Bruchert V, Loy A, Vandieken V, Jorgensen BB. Thermophilic anaerobes in Arctic marine sediments induced to mineralize complex organic matter at high temperature. Environ Microbiol. 2010;12:1089–1104. doi: 10.1111/j.1462-2920.2010.02161.x. [DOI] [PubMed] [Google Scholar]
- Hubert C, Judd A.2010Using microorganisms as prospecting agents in oil and gas explorationIn: Timmis KN, (ed.)Handbook of Hydrocarbon and Lipid Microbiology Springer-Verlag: Berlin, Heidelberg; 2711–2725. [Google Scholar]
- Hutnak M, Fisher AT, Harris R, Stein C, Wang K, Spinelli G, et al. Large heat and fluid fluxes driven through mid-plate outcrops on ocean crust. Nat Geosci. 2008;1:611–614. [Google Scholar]
- Isaksen MF, Bak F, Jorgensen BB. Thermophilic sulfate-reducing bacteria in cold marine sediment. FEMS Microbiol Ecol. 1994;14:1–8. [Google Scholar]
- Ji S, Wang S, Tan Y, Chen X, Schwarz W, Li F. An untapped bacterial cellulolytic community enriched from coastal marine sediment under anaerobic and thermophilic conditions. FEMS Microbiol Lett. 2012;335:39–46. doi: 10.1111/j.1574-6968.2012.02636.x. [DOI] [PubMed] [Google Scholar]
- Judd AG, Hovland M. Seabed fluid flow: the impact of geology, biology and the marine environment. Cambridge University Press: Cambridge; 2007. [Google Scholar]
- Kallmeyer J, Ferdelman TG, Weber A, Fossing H, Jorgensen BB. A cold chromium distillation procedure for radiolabeled sulfide applied to sulfate reduction measurements. Limnol Oceanogr-Meth. 2004;2:171–180. [Google Scholar]
- Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D'Hondt S. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci USA. 2012;109:16213–16216. doi: 10.1073/pnas.1203849109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DS, Karson JA, Früh-Green GL, Yoerger DR, Shank TM, Butterfield DA, et al. A serpentinite-hosted ecosystem: the Lost City hydrothermal field. Science. 2005;307:1428–1434. doi: 10.1126/science.1102556. [DOI] [PubMed] [Google Scholar]
- Knies J, Brookes S, Schubert CJ. Re-assessing the nitrogen signal in continental margin sediments: New insights from the high northern latitudes. Earth Planet Sc Lett. 2007;253:471–484. [Google Scholar]
- Lennon JT, Jones SE. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol. 2011;9:119–130. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2006;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Lindstrom ES, Langenheder S. Local and regional factors influencing bacterial community assembly. Env Microbiol Rep. 2012;4:1–9. doi: 10.1111/j.1758-2229.2011.00257.x. [DOI] [PubMed] [Google Scholar]
- Lloyd JM, Park LA, Kuijpers B, Moros M. Early holocene palaeoceanography and deglacial chronology of Disko Bugt, West Greenland. Quaternary Sci Rev. 2005;24:1741–1755. [Google Scholar]
- Loeng H. Features of the physical oceanographic conditions of the Barents Sea. Polar Res. 1991;10:5–18. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLachlan SE, Cottier FR, Austin WE, Howe JA. The salinity: δ18O water relationship in Kongsfjorden, western Spitsbergen. Polar Res. 2007;26:160–167. [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- McKenney PT, Driks A, Eichenberger P. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol. 2013;11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000;64:548–572. doi: 10.1128/mmbr.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. 2012. Vegan: community ecology package. R package version 2.0-4. http://CRAN.R-project.org/package=vegan .
- Papke RT, Ramsing NB, Bateson MM, Ward DM. Geographical isolation in hot spring cyanobacteria. Environ Microbiol. 2003;5:650–659. doi: 10.1046/j.1462-2920.2003.00460.x. [DOI] [PubMed] [Google Scholar]
- Pedros-Alio C. The rare bacterial biosphere. Ann Rev Mar Sci. 2012;4:449–466. doi: 10.1146/annurev-marine-120710-100948. [DOI] [PubMed] [Google Scholar]
- Petersen JM, Ramette A, Lott C, Cambon-Bonavita M-A, Zbinden M, Dubilier N. Dual symbiosis of the vent shrimp Rimicaris exoculata with filamentous gamma- and epsilonproteobacteria at four Mid-Atlantic Ridge hydrothermal vent fields. Environ Microbiol. 2010;12:2204–2218. doi: 10.1111/j.1462-2920.2009.02129.x. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glockner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria; 2008. [Google Scholar]
- Rahmstorf S. The thermohaline ocean circulation: A system with dangerous thresholds? An editorial comment. Climatic Change. 2000;46:247–256. [Google Scholar]
- Rahmstorf S. Ocean circulation and climate during the past 120,000 years. Nature. 2002;419:207–214. doi: 10.1038/nature01090. [DOI] [PubMed] [Google Scholar]
- Ramette A, Tiedje JM. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb Ecol. 2007;53:197–207. doi: 10.1007/s00248-005-5010-2. [DOI] [PubMed] [Google Scholar]
- Reche I, Pulido-Villena E, Morales-Baquero R, Casamayor EO. Does ecosystem size determine aquatic bacterial richness. Ecology. 2005;86:1715–1722. [Google Scholar]
- Saloranta TM, Svendsen H. Across the Arctic front west of Spitsbergen: high-resolution CTD sections from 1998-2000. Polar Res. 2001;20:177–184. [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon Q, St-Onge G, Hillaire-Marcel C. Late Quaternary chronostratigraphic framework of deep Baffin Bay glaciomarine sediments from high-resolution paleomagnetic data. Geochem Geophy Geosy. 2012;13:Q0AO03. [Google Scholar]
- Sul WJ, Oliver TA, Ducklow HW, Amaral-Zettler LA, Sogin ML. Marine bacteria exhibit a bipolar distribution. Proc Natl Acad Sci USA. 2013;110:2342–2347. doi: 10.1073/pnas.1212424110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CCL, Ross CK, Yao T, Petrie B, DeTracey BM, Dunlap E. The circulation, water masses and sea-ice of Baffin Bay. Prog Oceanogr. 2004;63:183–228. [Google Scholar]
- Teixeira S, Cambon-Bonavita M-A, Serrão EA, Desbruyéres D, Arnaud-Haond S. Recent population expansion and connectivity in the hydrothermal shrimp Rimicaris exoculata along the Mid-Atlantic Ridge. J Biogeogr. 2011;38:564–574. [Google Scholar]
- Vandieken V, Knoblauch C, Jørgensen BB. Desulfotomaculum arcticum sp. nov., a novel spore-forming, moderately thermophilic, sulfate-reducing bacterium isolated from a permanently cold fjord sediment of Svalbard. Int J Syst Evol Microbiol. 2006;56:687–690. doi: 10.1099/ijs.0.64058-0. [DOI] [PubMed] [Google Scholar]
- Vrijenhoek RC. Genetic diversity and connectivity of deep-sea hydrothermal vent metapopulations. Mol Ecol. 2010;19:4391–4411. doi: 10.1111/j.1365-294X.2010.04789.x. [DOI] [PubMed] [Google Scholar]
- Wessel P, Smith WHF. New, improved version of generic mapping tools released. Eos. 1998;79:579–579. [Google Scholar]
- Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.