Abstract
Many organic pollutants are readily degradable by microorganisms in soil, but the importance of soil organic matter for their transformation by specific microbial taxa is unknown. In this study, sorption and microbial degradation of phenol and 2,4-dichlorophenol (DCP) were characterized in three soil variants, generated by different long-term fertilization regimes. Compared with a non-fertilized control (NIL), a mineral-fertilized NPK variant showed 19% and a farmyard manure treated FYM variant 46% more soil organic carbon (SOC). Phenol sorption declined with overall increasing SOC because of altered affinities to the clay fraction (soil particles <2 mm in diameter). In contrast, DCP sorption correlated positively with particulate soil organic matter (present in the soil particle fractions of 63–2000 μm). Stable isotope probing identified Rhodococcus, Arthrobacter (both Actinobacteria) and Cryptococcus (Basidiomycota) as the main degraders of phenol. Rhodococcus and Cryptococcus were not affected by SOC, but the participation of Arthrobacter declined in NPK and even more in FYM. 14C-DCP was hardly metabolized in the NIL variant, more efficiently in FYM and most in NPK. In NPK, Burkholderia was the main degrader and in FYM Variovorax. This study demonstrates a strong effect of SOC on the partitioning of organic pollutants to soil particle size fractions and indicates the profound consequences that this process could have for the diversity of bacteria involved in their degradation.
Keywords: dichlorophenol, microbial diversity, phenol, soil organic matter, stable isotope probing
Introduction
Most organic compounds entering the soil, including many man-made pollutants, are degradable by indigenous microorganisms. The compounds' molecular structure defines not only their potential uptake and metabolic transformation by the microbial cells, but also their sorption and desorption behavior, which have a strong effect on their mass transfer within the soil matrix (Bosma et al., 1997). Sorption and desorption processes in soils can occur at mineral, organo-mineral or organic particle surfaces, and, considering the complex composition and architecture of a soil, the overall sorption of an organic compound is, in fact, a composite of many particular surface interactions occurring in parallel (Wu and Gschwend, 1986). Although sorption and desorption to particular soil materials, for example, pure clay minerals, can be well established and mathematically characterized, the soil complexity and its natural variability make it very hard to predict mass transfer rates of organic compounds in real soils (Karickhoff, 1981; Haggerty and Gorelick, 1998).
Soil organic matter is considered to be the single most important constituent influencing sorption of compounds in soil (Farenhorst, 2006). It originates from plant, animal and microbial residues, and, it is composed of many different organic molecules. The organic material may be highly degraded and stabilized, especially when forming complexes with mineral particles; or it may be potentially more accessible to microbial degradation as particulate organic matter (POM), the latter typically with characteristic features of plant residues (Schulten et al., 1993). Depending on the soil particle size fractions, which can be distinguished as clay (<2 μm), fine silt (2–20 μm), coarse silt (20–63 μm) and sand (63–2000 μm), the composition of their associated organic matter is also different (Christensen, 1992; Guggenberger et al., 1994; Kögel-Knabner et al., 2008). This heterogeneity of soil organic matter also has a tremendous effect on their sorption potential (Xing et al., 1996; Huang et al., 2003), and it can thus be expected that partitioning of organic pollutants is strongly affected by soil particle size fractions. Recently, we could demonstrate that these particle size fractions within one soil are also colonized by distinct microbial communities (Neumann et al., 2013).
The objective of this study was to analyze whether the different sorbent qualities of soil organic matter of soil particle size fractions would affect the partitioning of organic pollutants and thereby possibly the diversity of microorganisms involved in their degradation. In order to compare the importance of soil organic matter for the particular size fractions, three soil variants from the ‘Static fertilization experiment' from Bad Lauchstädt, Germany, were analyzed and compared. Different fertilization regimes over a period of >100 years had generated soil variants, which substantially differed in their amount of soil organic carbon (SOC) and microbial biomass (Böhme et al., 2005).
Phenol and 2,4-dichlorophenol (DCP) were selected as environmentally relevant model compounds for organic pollutants. Both are listed among the priority pollutants of the EPA (United States Environmental Protection Agency), and they are readily degradable in soil (Ghisalba, 1983; Benoit et al., 1999; Lillis et al., 2009). Soil bacteria and fungi that transform these compounds under laboratory conditions have been described (Haider et al., 1974; Middelhoven, 1993). However, as for most organic pollutants, the implications of their partitioning to soil particle size fractions and the importance of soil organic matter therein for their microbial degradation are not understood.
In this study, we assessed (1) the sorption of phenol and DCP to three soil organic matter variants and their particle size fractions; (2) the degradation of phenol and DCP by following the release of 14CO2 from 14C-labeled compounds during soil incubations and (3) the diversity of bacteria and fungi degrading the compounds. DNA stable isotope probing (SIP) was used to achieve the third goal. SIP, as introduced by Radajewski et al. (2000), allows to access the nucleic acids of microorganisms participating in the degradation of 13C-labeled compounds without culturing them in growth media. The method was suitable to detect and identify microorganisms involved in the degradation of different organic pollutants (Mahmood et al., 2005; Cupples and Sims, 2007; Martin et al., 2012; Uhlik et al., 2012), and, thus, it was selected to identify the actively phenol and DCP-degrading microbial community and compare their diversity from the three SOC variants selected for this study.
Materials and methods
Soil, soil variants and soil sampling
The soil of this study originated from the ‘Static fertilization experiment' in Bad Lauchstädt, Germany (51°24′N, 11°53′E). It is a Haplic Chernozem derived from loess (Altermann et al., 2005); a silt loam consisting of 21% (wt/wt) clay, 67.8% silt and 11.2% sand (Blair et al., 2006). The ‘Static fertilization experiment' started in the year 1902 and is described in more detail elsewhere (Körschens et al., 1998; Böhme and Böhme, 2006). Three fertilization variants were chosen for this study: the NIL variant with no addition of agricultural fertilizers, the NPK variant with mineral fertilizer on an annual basis and the FYM variant with mineral fertilizer on an annual basis and farmyard manure at 20 t ha−1 every second year (Blair et al., 2006).
The soils of this study were collected in October 2008, 2 years after the last farmyard manure application. Samples were taken from the plough layer (0–30 cm depth). For each variant, combined samples originated from four different subplots of 25 m2 of which two pairs were located next to each other, and the others in a distance of 16.5 m. The soils were sieved (mesh size 2 mm), adjusted to 50–55% of their total water holding capacity and stored at 4 °C. Before experiments, they were adapted to room temperature for 48 h.
Physicochemical soil analyses
Total organic carbon was determined by dry combustion (Element Analyzer; TruSpec CN, Leco, St Joseph, MI, USA). The pH was determined in soil suspensions (10 g in 20 ml of 0.01 M CaCl2 solution).
Soil microbial biomass was quantified by chloroform–fumigation incubation (Jenkinson and Powlson, 1976) for characterization of field collected soils, and by chloroform–fumigation extraction (Vance et al., 1987) for analyzing the partitioning of 14C-label from 14C-phenol and 14C-DCP after incubations in soil microcosms.
Soil fractionation was based on the ultrasonication wet-sieving centrifugation protocol described by Amelung et al. (1998), with modifications described elsewhere (Neumann et al., 2013). POM was separated by density fractionation and quantified as described before (Neumann et al., 2013).
Organic pollutants and adsorption studies
Phenol (>99% purity; Sigma Aldrich Chemie, Steinheim, Germany) and DCP (99%, Sigma Aldrich Chemie) were chosen for this study. [UL-14C] phenol (14C-phenol) and [UL-14C] 2,4-dichlorophenol (14C-DCP) were both obtained from GE Healthcare (Chalfont St Giles, UK). [UL-13C] phenol (13C-phenol; 99.2 atom % 13C) was obtained from IsoSciences (King of Prussia, PA, USA) and [UL-13C] 2,4-dichlorophenol (13C-DCP;>99 atom % 13C) from Campro-Scientific (Veenendaal, The Netherlands).
Sorption isotherms to soils were established using the OECD protocol 106 (OECD, 2000). Accordingly, microbial activity was inhibited by addition of HgCl2 (Wolf et al., 1989). 14C-labeled compounds (15 kBq for phenol; 20.6 kBq for DCP) were added as tracers to the soil suspensions. After equilibration under shaking at 250 r.p.m. for 48 h and 20 °C in the dark, the suspensions were centrifuged for 10 min at 2450 × g and 500 μl-aliquots of the supernatants were analyzed for 14C-activity by liquid scintillation counting in 18 ml of a scintillation cocktail (Rotiszint eco-plus, Roth, Karlsruhe, Germany; Liquid scintillation counter LS1801, Beckman Coulter, La Brea, CA, USA). The centrifuged sediments were frozen at −20 °C. For soil fractionation, the thawed sediments were suspended in 50 ml of distilled water. The particle size fractions were dried at 40 °C and the adsorbed 14C-activity was determined after combustion (Biological Oxidizer Ox 300, Harvey Instrument Corp., Hillsdale, NJ, USA), captured as 14CO2 in a scintillation cocktail (Oxysolve C-400, Zinsser-Analytic, Frankfurt, Germany), by liquid scintillation counting.
Soil incubations with14C- and13C-labeled phenol and DCP
Degradation experiments were performed in soil microcosms, each consisting of a 250-ml flask with 50 g soil (dry weight) at 50–55% water saturation, under continuous flow of water-saturated CO2-free air (Martens, 1985). At the onset of the incubations, phenol or DCP solutions, equivalent to an end concentration of 5 mM in the soil aqueous phase and containing 14C-labeled compounds (10.83 kBq for phenol; 11.56 kBq for DCP) were added to the soils. Microcosms were incubated for 15 days with phenol, or 21 days with DCP, and liberated 14CO2, captured in 2 N NaOH, was analyzed by liquid scintillation counting, as described above. Four independent replicated microcosms were analyzed for each soil and experimental variant.
For SIP (Neufeld et al., 2007), soil incubations were conducted under the same conditions, but with 5 g of soil in 100-ml flasks (initial concentration in soil water 5 mM; 50–55% water saturation) with 13C-phenol and 13C-DCP, respectively. Corresponding controls with non-labeled compounds were also set up and analyzed. Samplings were conducted according to the respective degradation activity seen with the formation of 14CO2 after 1, 2, 4, 7, and 14 days for phenol, and after 9, 14 or 21 days for DCP, respectively. For each sampling, soil incubations were terminated by freezing the samples to −20 °C.
DNA extraction
DNA was extracted from thawed soil samples with the Bio101 FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH, USA). Cell lysis was achieved by two subsequent steps of bead beating, each at 6.5 m s−1 in a FastPrep-24 apparatus (MP Biomedicals). The DNA solutions were purified by Geneclean columns (MP Biomedicals) and quantified by PicoGreen (MoBiTec, Göttingen, Germany). For each microcosm, DNA was extracted from two 0.5 g soil aliquots and combined.
DNA analyses of CsCl-gradient fractions
Equal amounts of DNA from three independent microcosms were pooled and 600 ng of DNA were added to a CsCl solution with a density of 1.72 g ml−1. The density was analyzed with an AR200 digital refractometer (Reichert, Depew, NY, USA). Density centrifugations were performed in 4.9 ml OptiSeal tubes (Beckman Coulter) in an Optima L-100 XP ultracentrifuge (vertical rotor VTI 65.1, Beckman Coulter) for ⩾ 40 h at maximum relative centrifugation force (RCFmax) of 180 000 × g and 20 °C. A total of 15 fractions, each approximately 350 μl, were obtained by displacing the gradient solution with distilled water using an LK B pump (Produkar AB, Sweden) and the DNA was precipitated with polyethylene glycol.
Partial bacterial 16S ribosomal RNA (rRNA) genes were amplified from DNA using primers Com1 and Com2 (Schwieger and Tebbe, 1998), and NSI1 and 58A2R for fungal 18S-ITS1-5.8S rRNA sequences (Martin and Rygiewicz, 2005) under conditions described therein. The reverse primers were phosphorylated to be suitable for subsequent genetic profiling using single-strand conformation polymorphism (SSCP; Schwieger and Tebbe, 1998). Triplicate amplifications, each in a 50 μl volume, were performed for each sample and then pooled. After digestion of the phosphorylated DNA-strand with lambda exoncuclease (New England Biolabs, Frankfurt, Germany), DNA was analyzed by SSCP as described before (Dohrmann and Tebbe, 2004).
Cloning of PCR products and DNA sequence analyses
The identification of DNA sequences recovered from bands of SSCP profiles followed the protocol described elsewhere, with three clones selected to identify each band of interest (Dohrmann and Tebbe, 2005).
DNA sequences were also identified from clone libraries of rRNA gene fragments amplified from DNA of SIP gradient fractions with a density of 1.72 g ml−1 and denser, when there were no indications of PCR products from the corresponding fractions obtained from DNA originating from microcosms amended with unlabeled (12C) compounds. For bacteria, DNA was amplified with primers Com1 and Com2 as described above. For fungi primers ITS1f (Gardes and Bruns, 1993) and ITS4r (White et al., 1990), targeted an 18S-ITS1-5.8S-ITS2-28S rRNA gene fragment. Fungi-specific amplifications were performed in a total volume of 50 μl containing template DNA, 0.025 U μl−1 of HotStarTaq polymerase (Qiagen, Hilden, Germany), 1 × buffer, 0.2 mM dNTP, 0.5 μM of each primer, a final concentration of MgCl2 of 2.75 mM, and bovine serum albumin (Roche, Mannheim, Germany) in a final concentration of 0.4 μg μl−1. Thermal cycling conditions: 15-min initial denaturation at 95 °C, 30 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s and extension at 72 °C for 60 s, and a final primer extension at 72 °C for 5 min. Triplicate PCR products were pooled, purified with the QIAquick PCR purification system (Qiagen) and cloned in Escherichia coli JM109 cells using the pGEM-T Vector System II (Promega, Madison, WI, USA). For each soil variant, 50–60 clones were randomly selected and sent for DNA sequencing to GATC-Biotech (Konstanz, Germany).
Nucleotide sequences were checked with the GATC Viewer (GATC-Biotech) and Mega 4 (Tamura et al., 2007). The Pintail program was used to screen for chimera (Ashelford et al., 2005) which, in case of detection, were removed. Phylogenetic assignment and further processing of the sequences was done using the ARB software (database ssu_jan05, Ludwig et al., 2004). Corrected sequences were compared with DNA sequences from public databases (NCBI: http://www.ncbi.nlm.nih.gov; RDP: http://rdp.cme.msu.edu; Cole et al., 2009) using the BLAST function (Altschul et al., 1990).
Mathematical analyses
The degree of proton dissociation of phenol and DCP under different pH values was determined by using the Henderson–Hasselbalch equation. Phylogenetic trees were calculated with the tree builder function of the ARB-software (Ludwig et al., 2004), using the AxML+ algorithm (Stamatakis et al., 2004). Results displayed as bar plots, line plots or line-scatter plots are expressed as mean and s.d. For statistical analyses, one-way analysis of variance and successive Bonferroni t-tests were conducted using Sigma-Plot (11.0; Systat Software, Chicago, IL, USA).
DNA sequence accession
All DNA sequences obtained in this study have been submitted to the EMBL database. Their accession numbers are HG422058–HG422170.
Results
Selected characteristics of the three soil variants NIL, NPK and FYM
The long-term treatment with mineral and organic fertilizer lowered the pH by 0.9 and 1.0 in comparison with NIL with no significant differences between NPK and FYM. Fertilization increased the amounts of SOC and total microbial biomass, with FYM having a stronger effect than NPK (Table 1). The application of FYM caused an increase in the proportion of the sand-sized particle size fractions because of more POM.
Table 1. Characterization of the three soil variants selected for this study from the ‘static fertilization experiment' in Bad Lauchstädt, Germany.
| Long-term fertilization regime* |
Soil variant |
||
|---|---|---|---|
| NIL | NPK | FYM | |
| |
No fertilizer |
Mineral fertilizer |
Mineral fertilizer plus farmyard manure |
| Soil organic carbon (g kg−1) | 17.7a | 21.1b | 25.9c |
| Soil microbial biomass (mg kg−1)** | 129a | 164b | 264c |
| pH value | 7.4a | 6.4b | 6.5b |
| Soil particle size fractions (%)*** | |||
| Sand and POM (63–2000 μm) | 5.8 | 6.3 | 7.8 |
| Coarse silt (20–63 μm) | 40.1 | 39.6 | 39.0 |
| Fine silt (2–20 μm) | 31.1 | 34.1 | 30.1 |
| Clay (<2 μm) | 23.2 | 20.4 | 23.2 |
Abbreviation: POM, particulate organic matter.
Values followed by different alphabets indicate differences between soil variants.
Fertilizations were conducted on an annual basis according to good agricultural practice since 1902; farmyard manure was only applied every second year—sampling was conducted in October 2008, 24 months after the last application of farmyard manure.
Determined by chloroform–fumigation incubation, according to Jenkinson and Powlson (1976).
Achieved using the ultrasonication wet-sieving centrifugation method according to Amelung et al. (1998), modified by Neumann et al. (2013).
Most soil organic matter, measured as SOC, was associated with the clay fraction and its content was not significantly affected by the different long-term fertilization regimes (Table 2). In contrast, the SOC levels of the largest particle size fraction (sand plus POM) showed a strong response, with an increase of 59% in the NPK and 314% in FYM.
Table 2. Amount and partitioning of SOC to soil particle size fractions.
| Particle size fractionsa |
Amount of soil organic carbon per g particle size fraction (μg g−1) (contribution to total Corg
in non-fractionated soil (%)) |
||
|---|---|---|---|
|
Soil variantsb |
|||
| NIL | NPK | FYM | |
| Sand and POM (63–2000 μm) | 9.2±1.1 (3) | 14.6±0.6 (5) | 38.1±4.7 (10) |
| Coarse silt (20–63 μm) | 4.0±0.2 (9) | 3.4±0.1 (8) | 11.1±0.5 (18) |
| Fine silt (2–20 μm) | 14.6±0.3 (28) | 15.9±0.9 (30) | 18.8±1.3 (26) |
| Clay (<2 μm) | 43.4±1.6 (60) | 44.5±0.9 (58) | 46.8±3.1 (46) |
Abbreviations: POM, particulate organic matter; SOC, soil organic carbon.
Achieved using the ultrasonication wet-sieving centrifugation method according to Amelung et al. (1998), modified by Neumann et al. (2013).
For more details, see Table 1 and Materials and methods section.
Sorption of phenol and DCP
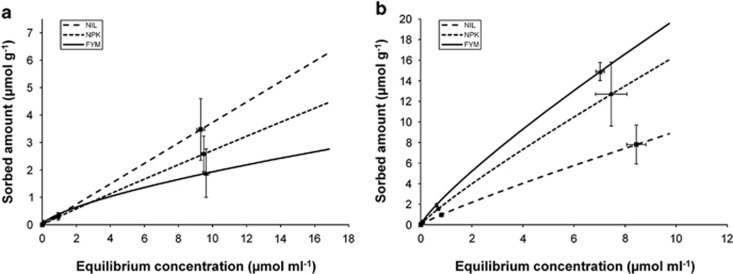
Sorption of both organic pollutants (phenol, DCP) occurred for all initial concentrations (0.001–10 mM) tested (Figure 1). Sorption of DCP was always stronger than that of phenol. Although the absolute quantity of sorbed compounds increased with their higher initial concentrations, the relative amount tended to decline, except for phenol in the NIL variant. For phenol, long-term fertilization caused a decline in the amount of sorbed compound, which correlated negatively with the total amount of SOC. The opposite was seen with DCP: sorption increased with SOC.
Figure 1.
Freundlich isotherms of phenol (a) and DCP (b) sorbed by soil variants generated by long-term fertilization receiving no fertilization (NIL), mineral fertilization (NPK) and mineral and organic fertilization (FYM) for a period of >100 years; for more details, see Materials and methods section. The initial concentrations of phenol and DCP in the soil aqueous phase were 0.001, 0.01, 0.1, 1 and 10 mM, respectively.
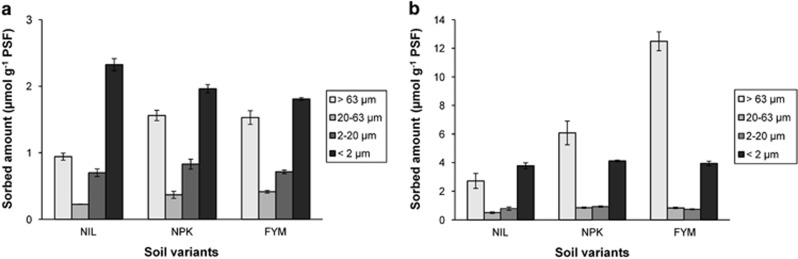
Particle size fractionations of soils incubated with 10 mM initial concentrations of the respective 14C-labeled organic compounds revealed that the largest proportion of phenol was adsorbed to clay, whereas most of the DCP was found with the sand-sized particle size fraction containing POM (Figure 2). Apparently, these dominant fractions also caused the overall contrasting effects of soil organic matter seen in the non-fractionated soils: although the negative correlation between SOC and sorbed phenol was due to processes at the clay fraction, the positive correlation of DCP with SOC was caused by the sand–POM fraction. A separate experiment confirmed that sorption of DCP (as well as for phenol) in the sand-sized fraction was caused to a great extent by POM and not by the mineral sand particles (data not shown). The overall negative correlation between SOC and phenol sorption masked a positive correlation with SOC in the sand-sized fraction and in coarse silt.
Figure 2.
Sorption of phenol (a) and DCP (b) to soil particle size fractions of the three soil variants NIL, NPK and FYM, respectively. The initial concentration in the soil aqueous phase was 10 mM.
Degradation of phenol and DCP
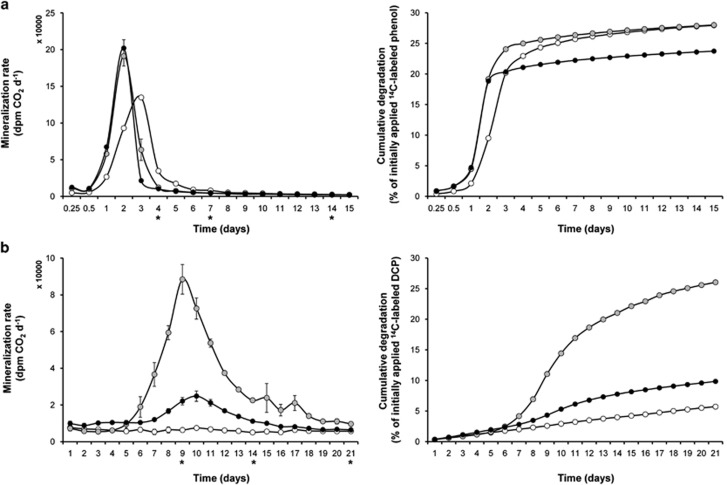
14CO2 was released in NPK and FYM soil variants from 14C-phenol (initial concentration 5 mM) at the same initial rates (Figure 3a) with a maximum after 2 days. The maximum activity of NIL was 24-h delayed. The cumulative curves showed that the mineralization of phenol during the 15-day period was equally efficient in NIL and NPK and 14% less with FYM. Mass balances after 15 days indicated a total recovery of 14C-labeled compounds (originating from phenol) of 91–92%, including 14CO2 and 14C-residues in soil. A total of 2.1–2.3% of this carbon allocation were detected in the microbial biomass (Supplementary Table S1).
Figure 3.
Mineralization of 14C-labeled phenol (a) and DCP (b) in soil microcosms, including the three soil variants NIL (empty circles), NPK (gray-filled circles), FYM (filled circles), respectively. Left panels show mineralization rates, right panels show the cumulative fractions collected as 14CO2. Stars at the x axis in the left panels indicate the time points of sampling for SIP (for results, see Figures 4 and 5).
The rates of 14C-DCP mineralization differed highly between the soil variants. The highest degradation rates and strongest cumulative degradation was found with NPK: a significant production of 14CO2 started after 6 days, reached a maximum after 9 days and then declined until 21 days (Figure 3b). Somewhat similar kinetics, however, with much lower activities were detected for the FYM variant. When the microcosms were sacrificed after 21 days, mass balances indicated a total 14C-recovery of 93–100%. A total of 0.15% (NIL variant) to 0.64% (NPK) of this carbon allocation were detected in the microbial biomass (Supplementary Table S1).
In a separate experiment, it was tested whether the addition of nitrogen, phosphorous or potassium (NPK) would stimulate DCP degradation by the NIL and FYM soil variants. However, this addition had no effect on the degradation rates of DCP or the total recovery of added 14C (Supplementary Table S2). In another experiment, it was tested whether the presence of 5 mM DCP was toxic for soil microorganisms by measuring basal respiration in presence and absence of DCP for the three soil variants. There was no adverse effect observed (data not shown).
Identification of microorganisms involved in the degradation of phenol
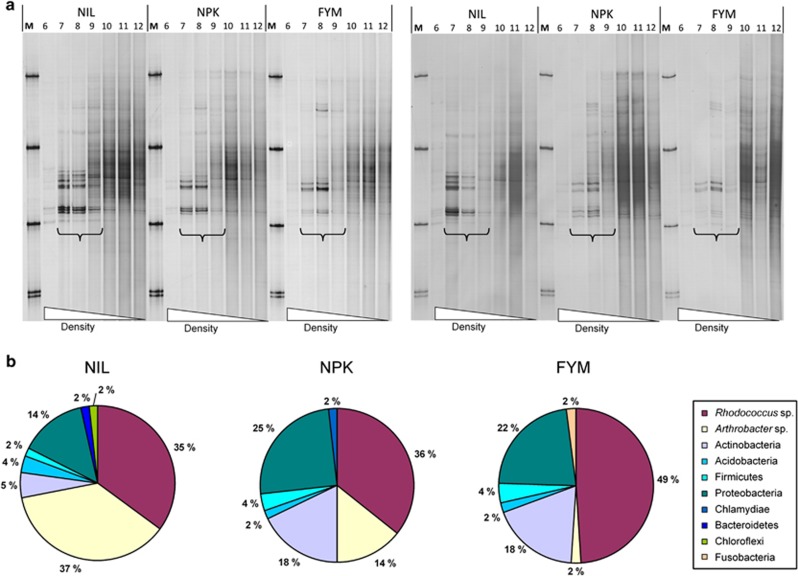
Density centrifugations with DNA extracted from microcosms incubated for 4 and 7 days, respectively, yielded no heavy (13C-enriched) DNA extracted from control soils incubated with 12C-phenol, as indicated by a lack of PCR products targeting the bacterial or the fungal rRNA genes (Supplementary Figures S1 and S2). Similarly, there were no visible PCR products for both targeted domains with DNA extracted from soils amended with 13C-phenol immediately after its addition to soil (data not shown). After 24-h soil incubation with 13C-phenol, several faint bands became detectable (no figure), and their intensities increased after 4 days of incubation (Figure 4a, left panel). The pattern of these bands was repeated in 7-day (Figure 4a) and 14-day (no figure) samples. Two groups of bands appeared equally strongly in the DNA extracted from the NIL variant. Although the intensities of the upper group of bands was similar with DNA from the NPK and NIL variant, the lower group of bands apparently declined in intensities in NPK, and even more in FYM (Figure 4a). DNA sequencing of upper bands indicated that they mainly originated from Rhodococcus sp. (sequence identity ⩾99%), whereas the lower bands were caused by Arthrobacter sp. (⩾99%) (Supplementary Figure S3 and Supplementary Table S3). Independent clone libraries from density fractions with 1.72 g ml−1 confirmed the dominant occurrence of these two Actinobacteria and the decline of Arthrobacter with increasing amounts of SOC in the fertilized variants (Figure 4b). The three clone libraries also revealed the presence of other Actinobacteria and several members of the phyla Proteobacteria (including Alpha-, Beta-, Gamma- and Deltaproteobacteria), Firmicutes, as well as singletons from other phylogenetic groups (Supplementary Table S4). Overall, 64 different bacterial taxa (phylotypes) were detected.
Figure 4.
(a) SSCP profiles of partial 16S rRNA gene products PCR amplified from density gradient fractions of soil DNA from microcosms incubated with 13C-phenol. Comparisons of results obtained from soil variants NIL, NPK and FYM, incubated over a period of 4 days (left panel) and 7 days (right panel), respectively. Brackets indicate lanes with density fractions in the range of 1.71–1.73 g ml−1. Each band represents a specific PCR product. No specific PCR products were detected in the corresponding density fractions with DNA from soil incubated with unlabeled (12C) phenol as a control (shown on SSCP gels in the Supplementary Figure S1). For more details of the DNA sequences from these bands, see Supplementary Figure S3 and Supplementary Table S3. (b). Phylogenetic assignment of partial 16S rRNA genes from clone libraries of the same soil DNA solutions after 4 days of incubation (for more details, see Supplementary Table S4).
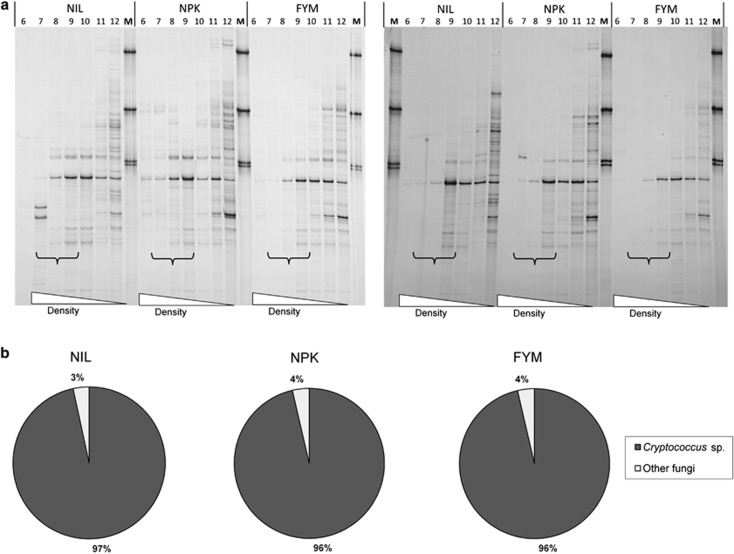
With fungi-specific PCR, one dominant and six less dominant bands appeared in SSCP profiles of the heavy DNA fractions after 4 and 7 days (Figure 5a). No differences were seen between band intensities obtained from the three soil variants. Sequencing of the dominant band indicated that it originated from a member of the Basidiomycota (99% identity to the yeast Cryptococcus sp.), whereas the less dominant bands came from Ascomycota and Zygomycota, respectively (Supplementary Figure S4 and Supplementary Table S5). Independent clone libraries based on larger DNA fragments confirmed the dominance of Cryptococcus sp. (sequence identity ⩾99%) independent of the soil variant (Figure 5b; Supplementary Table S6).
Figure 5.
(a) SSCP profiles of PCR amplified fungal 18S-ITS1-5.8S rRNA gene products from density gradient fractions of soil DNA from microcosms incubated with 13C-phenol. Results are shown for samples obtained from soil variants NIL, NPK and FYM, incubated over a period of 4 days (left panel) and 7 days (right panel), respectively. Brackets indicate lanes with density fractions in the range of 1.71–1.73 g ml−1. Each band indicates a specific PCR product. No specific PCR products were detected in the corresponding fractions with DNA from control soil incubated with unlabeled phenol (see Supplementary Figure S2). More details on the DNA sequences from the bands detected from the fractions with 13C-enriched DNA are shown in the Supplementary Figure S4 and Supplementary Table S5. (b) Phylogenetic assignment of the 18S-ITS1-5.85S-ITS2-28S rRNA sequences from clone libraries of the same soil DNA solutions after 4 days of incubation (for more details, see Supplementary Table S6).
Identification of microorganisms involved in the degradation of DCP
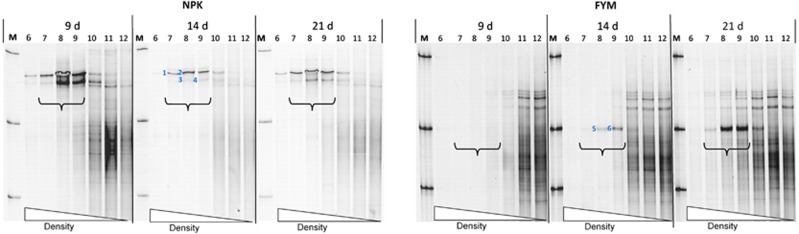
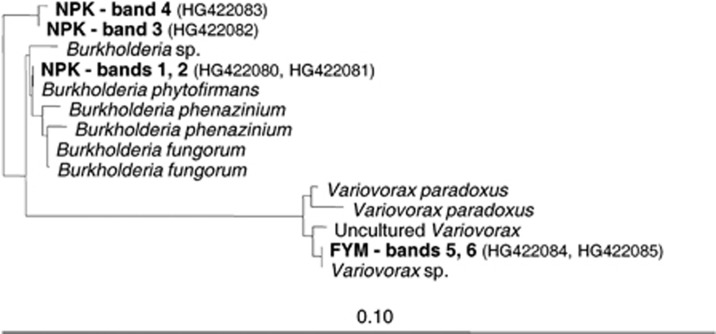
Owing to small degradation rates of DCP in the NIL variant, SIP was only performed with DNA from NPK and FYM. Controls from microcosms incubated with 12C-DCP were analyzed after incubations for 9 and 21 days, respectively. At the first sampling, after 9 days, bacterial PCR products were detected in the heavy DNA fractions of the NPK variant incubated with 13C-DCP, but not with DNA from the 13C-DCP FYM variant, or both variants incubated with 12C-DCP (Figure 6; Supplementary Figure S5). The two different SSCP bands, which were detected with 13C-DCP, persisted in samples taken after 14 and 21 days, respectively. For the FYM variant incubated with 13C-DCP, one dominant band was detected in the heavy DNA fractions after 14 days, and the same band also after 21 days. Faint bands were also detected in the corresponding fractions after 21 days of incubation with 12C-DCP (Supplementary Figure S5). However, those bands were in the same position in the SSCP gels as the bands from the lighter fraction, whereas the bands from incubations with 13C-DCP were clearly in a different position, demonstrating that those were caused by DNA enriched in 13C. DNA sequencing revealed that the two upper bands from the NPK variant incubated with 13C-DCP consisted of nearly identical sequences with full identity to 16S rRNA genes of Burkholderia sp. (Figure 7). The 13C-enriched band detected from DNA of the FYM variant could be assigned with 100% sequence identity to a Variovorax sp. No PCR products were detected with fungi-specific PCR from any of the CsCl-gradient fractions, indicating that fungi did not utilize significant amounts of DCP for growth.
Figure 6.
SSCP profiles of partial 16S rRNA gene products PCR amplified from density gradient fractions of soil DNA from microcosms incubated with 13C-DCP. Comparisons of results obtained from soil variants NIL, NPK and FYM, incubated over a period of 9, 14 and 21 days, respectively. Brackets indicate lanes with density fractions in the range of 1.71–1.73 g ml−1. Each band represents a specific PCR product. No specific PCR products were detected in the corresponding density fractions with DNA from control soil incubated with unlabeled (12C) DCP (Supplementary Figure S5). Bands (DNA) indicated by numbers 1–6 were sequenced and characterized for the phylogenetic assignment (see Figure 7).
Figure 7.
Phylogenetic tree (maximum likelihood) of 16S rRNA gene sequences cloned from six different bands extracted from SSCP gels (see Figure 6). Bands were obtained from heavy DNA fractions (enriched with 13C-labeled DNA) by PCR amplification with DNA extracted from soil microcosms incubated with 13C-DCP.
Discussion
The three soil variants of this study differed in their total amount of soil organic matter, measured as SOC, and in their pH. Although there was a gradual increase of SOC in response to the intensity of fertilization (NIL<NPK<FYM), the pH of both fertilized variants was 0.9–1.0 units below the pH of NIL (7.4), and not significantly different from each other. Similarly for phenol and DCP, the three key parameters selected for this study, that is, (1) sorption, (2) mineralization rates and (3) diversity of the microbial communities participating in their degradation, the dissimilarity of these parameters to NIL increased with the amount of SOC, but did not correlate with differences in pH. Considering their different pKa values (phenol: 9.99; DCP: 7.89), >99.7% of the phenol molecules in NIL and the fertilized controls must have been non-dissociated. However, with DCP, almost 25% would be negatively charged in the NIL, but only 3–4% in the fertilized variants. The minor importance of these pronounced charge differences on our results can be explained by the heterogeneous sorption potential of organic matter (Xing and Pignatello, 1996) for both ionic and non-ionic compounds (Ahmad et al., 2001; Tulp et al., 2009).
Opposing patterns of sorption were found for phenol and DCP. For phenol, the overall sorption to soil declined with increasing amounts of SOC, and for DCP sorption it increased. Particle size fractionation revealed that the smallest and largest size fractions, that is, clay and sand-sized particles, were the most important sorbents; and they also explained these contrasting results: the importance of clay as a sorbent is linked to its large surface area, which, for these particular soil variants, represented 73–76% of the total (Neumann et al., 2013). On the other hand, the sorptive capacity of the largest fraction can be explained by presence of POM. The 1.5- and 4-fold higher amounts of SOC in this fraction from NPK and FYM, respectively, clearly explains the increased sorption of DCP; and it also demonstrates a relatively low affinity of POM for phenol. Considering that the long-term fertilization did not affect the amounts of SOC detected in the clay fraction, it is not as straightforward for phenol to explain the effect of clay on its sorption. Recently, however, advanced spectroscopic methods revealed qualitative differences in the composition of soil organic matter at the clay fractions from these soil variants (Demyan et al., 2012). It was suggested that soil organic matter of clay consists of at least two different fractions: one responsive to environmental change, like induced by long-term fertilization, and the other not (Leifeld and Kögel-Knabner, 2005). Thus, phenol sorption by clay may have been dominated by interactions with the responsive constituents, whereas DCP sorption would have been under a stronger influence of interactions with the more recalcitrant fraction.
The degradation of phenol proceeded faster than the degradation of DCP. In contrast to DCP, phenol is a common natural constituent of soil organic material, as it exists in lignin-related fractions, and thus, a higher abundance of microorganisms capable of degrading this compound can be expected. Therefore, it was not surprising that the two fertilized variants with generally more microbial biomass and cells (Böhme et al., 2005; Neumann et al., 2013) degraded the compound more quickly than the NIL soil. After 6 days of incubation, however, the mineralization of phenol in NIL and NPK soils stabilized at a higher rate than FYM, suggesting a lower bioavailability of phenol. While the POM fraction of NIL and NPK originated exclusively from previously cultivated crops, FYM also contained organic material from former farmyard manure applications. As the nature of the organic material is important for sorption of phenol (Xing et al., 1994), binding to organic residues from farmyard manure might have been stronger than to organic matter of the NIL and NPK variants.
For 14C-DCP degradation, there was a small and continuous release of CO2 in soils from the NIL variant, suggesting that there was no enrichment of microorganisms utilizing DCP as a carbon or energy source. Several soil bacteria and fungi have shown the capacity to cometabolically transform DCP under laboratory conditions (Haider et al., 1974; Hofrichter et al., 1994). Our detection of 14C incorporated into the soil microbial biomass, however, indicates the use of DCP as a carbon source. In FYM and NPK, the activity peaks in mineralization of DCP almost coincided 9–10 days after its application, and the quantification of the 14C-label in the microbial biomass indicated more efficient DCP degradation in NPK (0.64% label in biomass) than in FYM (0.33%). Considering the strong correlation between DCP sorption and presence of POM, this indicates that the mass transfer of DCP to microbial cells with a potential to degrade the compound was lower in FYM, thereby limiting its degradation (Bosma et al., 1997).
Among a consortium of bacteria and fungi enriched by DNA-SIP with 13C-phenol, two different Actinobacteria (Arthrobacter sp. and Rhodococcus sp.) and one yeast (Cryptococcus) were identified as the main degraders. Other Cryptococcus isolates degraded phenol in laboratory media (Middelhoven, 1993; Bergauer et al., 2005) and, in one DNA-SIP study, a member of the same order (Tramellales) was detected as a phenol degrader in soil (DeRito and Madsen, 2009). Equally, Arthrobacter and Rhodococcus isolates can degrade phenol (Margesin et al., 2005; Karigar et al., 2006; Martinkova et al., 2009), and members of both genera were detected by DNA-SIP as phenanthrene degraders in soil (Cebron et al., 2011). Another DNA-SIP study with phenol retrieved mainly Gammaproteobacteria from a silt loam soil (Padmanabhan et al., 2003). The diverging results suggest that soil and environmental properties have a strong effect on the diversity of bacteria involved in phenol degradation.
The diversity of bacteria enriched with 13C-DCP as a substrate was lower than with 13C-phenol and no fungi were detected. The differences between the two soil variants NPK and FYM, however, were striking: a Burkholderia member metaboled DCP in NPK soil and a Variovorax in FYM. Isolates of both genera are known for their capacity to degrade phenolic compounds (Rapp and Timmis, 1999) and 13C-phospholipid analyses, as well as DNA-SIP indicated that Burkholderia frequently participate in the metabolism of aromatic compounds in soil (Tillmann et al., 2005; Uhlik et al., 2012).
Soil organic matter, the key variable of this study, had apparently a huge effect on the diversity of microorganisms involved in the degradation of both phenol and DCP. For phenol, there was a gradual decline of Arthrobacter with increasing amounts of SOC, whereas the other two degraders, Rhodococcus and Cryptococcus were not affected. Although it cannot be excluded that differences in the microorganisms contributing to the degradation of phenol or DCP were also affected by structurally different microbial communities as they may have evolved during the long-term fertilizations, we hypothesize that the observed differences in phenol and DCP degraders could be linked to different spatial preferences of the respective microorganisms within the soil matrix: as the differences in the availability of phenol were mainly caused by sorption at the clay fractions, cells of Arthrobacter could be more closely attached to clay, experiencing the qualitative changes of the SOC in its microhabitat surfaces. In contrast, cells of Rhodococcus or Cryptococcus would preferentially be located in less affected microsites, as it was the case for POM, sand or silt. Niche preference for clay within a soil matrix has, in fact, been demonstrated for a Mycobacterium degrading polycyclic aromatic hydrocarbons (Uyttebroek et al., 2006).
The dominance of Burkholderia in the more actively DCP mineralizing NPK soil does not necessarily mean that this organism is generally a better degrader than the Variovorax detected in FYM soil. Further information about the spatial preference of both DCP degraders for specific soil microsites could answer whether the lower degradation in FYM was mainly caused by the limited mass transfer of DCP described above, or in fact by eco-physiological adaptations of both degraders.
In conclusion, this study demonstrates, with the example of two phenolic compounds of different sorptive behavior, the strong effect of soil organic matter on the partitioning of organic pollutants to soil microsites, as represented by particle size fractions, and it indicates the profound consequences that this process could have for the diversity of bacteria involved in their degradation.
Acknowledgments
We thank Ines Merbach (UFZ, Experimental Station Bad Lauchstädt) and Kai Totsche (University of Jena) for supporting this work, Anja B Dohrmann for discussion, Denis Finke for help with the adsorption studies, and Karin Trescher and Katja Krause for excellent technical assistance. This study contributes to SPP1315 (Biogeochemical Interfaces in Soils), financially support by the German Research Foundation, DFG (project numbers Te383/3-1 and Te383/3-2).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ahmad R, Kookana RS, Alston AM, Skjemstad JO. The nature of soil organic matter affects sorption of pesticides. 1. Relationships with carbon chemistry as determined by 13C CPMAS NMR spectroscopy. Environ Sci Technol. 2001;35:878–884. doi: 10.1021/es001446i. [DOI] [PubMed] [Google Scholar]
- Altermann M, Rinklebe J, Merbach I, Körschens M, Langer U, Hofmann B. Chernozem - soil of the year 2005. J Plant Nutr Soil Sc. 2005;168:725–740. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment serach tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amelung W, Zech W, Zhang X, Follett RF, Tiessen H, Knox E, et al. Carbon, nitrogen, and sulfur pools in particle-size fractions as influenced by climate. Soil Sci Soc Am J. 1998;62:172–181. [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol. 2005;71:7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit P, Barriuso E, Soulas G. Degradation of 2,4-D, 2,4-dichlorophenol, and 4-chlorophenol in soil after sorption on humified and nonhumified organic matter. J Environ Qual. 1999;28:1127–1135. [Google Scholar]
- Bergauer P, Fonteyne PA, Nolard N, Schinner F, Margesin R. Biodegradation of phenol and phenol-related compounds by psychrophilic and cold-tolerant alpine yeasts. Chemosphere. 2005;59:909–918. doi: 10.1016/j.chemosphere.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Blair N, Faulkner RD, Till AR, Korschens M, Schulz E. Long-term management impacts on soil C, N and physical fertility - Part II: Bad Lauchstadt static and extreme FYM experiments. Soil Till Res. 2006;91:39–47. [Google Scholar]
- Böhme L, Böhme F. Soil microbiological and biochemical properties affected by plant growth and different long-term fertilisation. Eur J Soil Biol. 2006;42:1–12. [Google Scholar]
- Böhme L, Langer U, Böhme F. Microbial biomass, enzyme activities and microbial community structure in two European long-term field experiments. Agr Ecosyst Environ. 2005;109:141–152. [Google Scholar]
- Bosma TNP, Middeldorp PJM, Schraa G, Zehnder AJB. Mass transfer limitation of biotransformation: quantifying bioavailability. Environ SciTechnol. 1997;31:248–252. [Google Scholar]
- Cebron A, Louvel B, Faure P, France-Lanord C, Chen Y, Murrell JC, et al. Root exudates modify bacterial diversity of phenanthrene degraders in PAH-polluted soil but not phenanthrene degradation rates. Environ Microbiol. 2011;13:722–736. doi: 10.1111/j.1462-2920.2010.02376.x. [DOI] [PubMed] [Google Scholar]
- Christensen BT.1992Physical fractionation of soil and organic matter in primary particle size and density separatesIn: Stewart BA (ed). Advances in Soil Science Springer Verlag: New York; 1–90. [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupples AM, Sims GK. Identification of in situ 2,4-dichlorophenoxyacetic acid-degrading soil microorganisms using DNA-stable isotope probing. Soil Biol Biochem. 2007;39:232–238. [Google Scholar]
- Demyan MS, Rasche F, Schulz E, Breulmann M, Mueller T, Cadisch G. Use of specific peaks obtained by diffuse reflectance Fourier transform mid-infrared spectroscopy to study the composition of organic matter in a Haplic Chernozem. Eur J Soil Sci. 2012;63:189–199. [Google Scholar]
- DeRito CM, Madsen EL. Stable isotope probing reveals Trichosporon yeast to be active in situ in soil phenol metabolism. ISME J. 2009;3:477–485. doi: 10.1038/ismej.2008.122. [DOI] [PubMed] [Google Scholar]
- Dohrmann AB, Tebbe CC.2004Microbial community analysis by PCR-single-strand conformation polymorphism (PCR-SSCP)In: Kowalchuk GA, de Bruijn F, Head IM, Akkermans ADL, van Elsas JD (eds). Molecular Microbial Ecology Manual Kluwer Academic Publishers: Dordrecht, The Netherlands; 809–838. [Google Scholar]
- Dohrmann AB, Tebbe CC. Effect of elevated tropospheric ozone on the structure of bacterial communities inhabiting the rhizosphere of herbaceous plants native to Germany. Appl Environ Microbiol. 2005;71:7750–7758. doi: 10.1128/AEM.71.12.7750-7758.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farenhorst A. Importance of soil organic matter fractions in soil-landscape and regional assessments of pesticide sorption and leaching in soil. Soil Sci Soc Am J. 2006;70:1005–1012. [Google Scholar]
- Gardes M, Bruns TD. ITS primers with enhanced specificity for Basidiomycetes - Applications to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Ghisalba O. Microbial degradation of chemical waste, an alternative to physical methiods of waste-disposal - chemical wastes and their biodegradation - an overview. Experientia. 1983;39:1247–1257. doi: 10.1007/BF01990362. [DOI] [PubMed] [Google Scholar]
- Guggenberger G, Christensen BT, Zech W. Land-use effects on the composition of organic matter in particle size separates of soil. 1. Lignin and carbohydrate signature. Eur J Soil Sci. 1994;45:449–458. [Google Scholar]
- Haggerty R, Gorelick SM. Modeling mass transfer processes in soil columns with pore-scale heterogeneity. Soil Sci Soc Am J. 1998;62:62–74. [Google Scholar]
- Haider K, Jagnow G, Kohnen R, Lim SU. Degradation of chlorinated benzenes, phenols and cyclohexane derivatives by benzene and phenol utilizing soil bacteria under aerobic conditions. Arch Microbiol. 1974;96:183–200. doi: 10.1007/BF00590175. [DOI] [PubMed] [Google Scholar]
- Hofrichter M, Bublitz F, Fritsche W. Unspecific degradation of halogenated phenols by the soil fungus Penicillium frequentans BI-7/2. J Basic Microbiol. 1994;34:163–172. doi: 10.1002/jobm.3620340306. [DOI] [PubMed] [Google Scholar]
- Huang WL, Ping PA, Yu ZQ, Fu HM. Effects of organic matter heterogeneity on sorption and desorption of organic contaminants by soils and sediments. Appl Geochem. 2003;18:955–972. [Google Scholar]
- Jenkinson DS, Powlson DS. Effects of biocidal treatments on metabolism in soil. 5. Methods for measuring soil microbial biomass. Soil Biol Biochem. 1976;8:209–213. [Google Scholar]
- Karickhoff SW. Semiempirical estimation of sorption of hydrophobic pollutants on natural sediments and soils. Chemosphere. 1981;10:833–846. [Google Scholar]
- Karigar C, Mahesh A, Nagenahalli M, Yun DJ. Phenol degradation by immobilized cells of Arthrobacter citreus. Biodegradation. 2006;17:47–55. doi: 10.1007/s10532-005-3048-y. [DOI] [PubMed] [Google Scholar]
- Kögel-Knabner I, Ekschmitt K, Flessa H, Guggenberger G, Matzner E, Marschner B, et al. An integrative approach of organic matter stabilization in temperate soils: linking chemistry, physics, and biology. J Plant Nutr Soil Sc. 2008;171:5–13. [Google Scholar]
- Körschens M, Weigel A, Schulz E. Turnover of soil organic matter (SOM) and long-term balances - Tools for evaluating sustainable productivity of soils. Z Pflanz Bodenkunde. 1998;161:409–424. [Google Scholar]
- Leifeld J, Kögel-Knabner I. Soil organic matter fractions as early indicators for carbon stock changes under different land-use. Geoderma. 2005;124:143–155. [Google Scholar]
- Lillis L, Doyle E, Clipson N. Comparison of DNA- and RNA-based bacterial community structures in soil exposed to 2,4-dichlorophenol. J Appl Microbiol. 2009;107:1883–1893. doi: 10.1111/j.1365-2672.2009.04369.x. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood S, Paton GI, Prosser JI. Cultivation-independent in situ molecular analysis of bacteria involved in degradation of pentachlorophenol in soil. Environ Microbiol. 2005;7:1349–1360. doi: 10.1111/j.1462-2920.2005.00822.x. [DOI] [PubMed] [Google Scholar]
- Margesin R, Fonteyne PA, Redl B. Low-temperature biodegradation of high amounts of phenol by Rhodococcus spp. and basidiomycetous yeasts. Res Microbiol. 2005;156:68–75. doi: 10.1016/j.resmic.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Martens R. Limitations of the application of the fumigation technique for biomass estimations in amended soils. Soil Biol Biochem. 1985;17:57–63. [Google Scholar]
- Martin F, Torelli S, Le Paslier D, Barbance A, Martin-Laurent F, Bru D, et al. Betaproteobacteria dominance and diversity shifts in the bacterial community of a PAH-contaminated soil exposed to phenanthrene. Environ Pollut. 2012;162:345–353. doi: 10.1016/j.envpol.2011.11.032. [DOI] [PubMed] [Google Scholar]
- Martin KJ, Rygiewicz PT. Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 2005;5:28. doi: 10.1186/1471-2180-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinkova L, Uhnakova B, Patek M, Nesvera J, Kren. V. Biodegradation potential of the genus Rhodococcus. Environ Int. 2009;35:162–177. doi: 10.1016/j.envint.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Middelhoven WJ. Catabolism of benzene compounds by ascomycetous and basidiomycetous yeasts and yeast-like fungi - a literature review and expeirmental approach. A Van Leeuw J Microb. 1993;63:125–144. doi: 10.1007/BF00872388. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW, et al. DNA stable-isotope probing. Nat Protoc. 2007;2:860–866. doi: 10.1038/nprot.2007.109. [DOI] [PubMed] [Google Scholar]
- Neumann D, Heuer A, Hemkemeyer M, Martens R, Tebbe CC. Response of microbial communities to long-term fertilization depends on their microhabitat. FEMS Microbiol Ecol. 2013;86:71–84. doi: 10.1111/1574-6941.12092. [DOI] [PubMed] [Google Scholar]
- OECD . OECD Guidelines for Testing of Chemicals Section 1: Physico-Chemical Properties. OECD: Paris; 2000. Adsorption-desorption using a batch equilibrium method. Test 106. Organisation for Economic Co-Operation and Development (ed) [Google Scholar]
- Padmanabhan P, Padmanabhan S, DeRito C, Gray A, Gannon D, Snape JR, et al. Respiration of 13C-labeled substrates added to soil in the field and subsequent 16S rRNA gene analysis of 13C-labeled soil DNA. Appl Environ Microbiol. 2003;69:1614–1622. doi: 10.1128/AEM.69.3.1614-1622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radajewski S, Ineson P, Parekh NR, Murrell JC. Stable-isotope probing as a tool in microbial ecology. Nature. 2000;403:646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- Rapp P, Timmis KN. Degradation of chlorobenzenes at nanomolar concentrations by Burkholderia sp. strain PS14 in liquid cultures and in soil. Appl Environ Microbiol. 1999;65:2547–2552. doi: 10.1128/aem.65.6.2547-2552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten HR, Leinweber P, Sorge C. Composition of organic matter in particle size fractions of an agricultural soil. J Soil Sci. 1993;44:677–691. [Google Scholar]
- Schwieger F, Tebbe CC. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl Environ Microbiol. 1998;64:4870–4876. doi: 10.1128/aem.64.12.4870-4876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis AP, Ludwig T, Meier H. The AxML program family for maximum likelihood-based phylogenetic tree inference. Concurr Comp-Pract E. 2004;16:975–988. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tillmann S, Strompl C, Timmis KN, Abraham WR. Stable isotope probing reveals the dominant role of Burkholderia species in aerobic degradation of PCBs. FEMS Microbiol Ecol. 2005;52:207–217. doi: 10.1016/j.femsec.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Tulp HC, Fenner K, Schwarzenbach RP, Goss KU. pH-Dependent sorption of acidic organic chemicals to soil organic matter. Environ Sci Technol. 2009;43:9189–9195. doi: 10.1021/es902272j. [DOI] [PubMed] [Google Scholar]
- Uhlik O, Wald J, Strejcek M, Musilova L, Ridl J, Hroudova M, et al. Identification of bacteria utilizing biphenyl, benzoate, and naphthalene in long-term contaminated soil. PLoS One. 2012;7:e490653. doi: 10.1371/journal.pone.0040653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyttebroek M, Breugelmans P, Janssen M, Wattiau P, Joffe B, Karlson U, et al. Distribution of the Mycobacterium community and polycyclic aromatic hydrocarbons (PAHs) among different size fractions of a long-term PAH-contaminated soil. Environ Microbiol. 2006;8:836–847. doi: 10.1111/j.1462-2920.2005.00970.x. [DOI] [PubMed] [Google Scholar]
- Vance ED, Brookes PC, Jenkinson DS. Microbial biomass measurements in forest soils - the use of chloroform fumigation incubation method in strongly acidic soils. Soil Biol Biochem. 1987;19:697–702. [Google Scholar]
- White TJ, Burns T, Lee S, Taylor JW.1990Analysis of phylogenetic relationship by amplification and direct sequencing of ribosomal RNA genesIn: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds). PCR Protocols: A Guide to Methods and Applications Academic Press: San Diego; 315–322. [Google Scholar]
- Wolf DC, Dao TH, Scott HD, Lavy TL. Influence of sterilization methods on selected soil microbiological, physical, and chemical properties. J Environ Qual. 1989;18:39–44. [Google Scholar]
- Wu SC, Gschwend PM. Sorption kinetics of hydrophobic organic compounds to natural sediments and soils. Environ Sci Technol. 1986;20:717–725. doi: 10.1021/es00149a011. [DOI] [PubMed] [Google Scholar]
- Xing BS, McGill WB, Dudas MJ, Maham Y, Hepler L. Sorption of phenol by selected biopolymers - Isotherms, energetics, and polarity. Environ Sci Technol. 1994;28:466–473. doi: 10.1021/es00052a019. [DOI] [PubMed] [Google Scholar]
- Xing BS, Pignatello JJ. Time-dependent isotherm shape of organic compounds in soil organic matter: implications for sorption mechanism. Environ Toxicol Chem. 1996;15:1282–1288. [Google Scholar]
- Xing BS, Pignatello JJ, Gigliotti B. Competitive sorption between atrazine and other organic compounds in soils and model sorbents. Environ Sci Technol. 1996;30:2432–2440. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.