Abstract
Propionate is produced in the human large intestine by microbial fermentation and may help maintain human health. We have examined the distribution of three different pathways used by bacteria for propionate formation using genomic and metagenomic analysis of the human gut microbiota and by designing degenerate primer sets for the detection of diagnostic genes for these pathways. Degenerate primers for the acrylate pathway (detecting the lcdA gene, encoding lactoyl-CoA dehydratase) together with metagenomic mining revealed that this pathway is restricted to only a few human colonic species within the Lachnospiraceae and Negativicutes. The operation of this pathway for lactate utilisation in Coprococcus catus (Lachnospiraceae) was confirmed using stable isotope labelling. The propanediol pathway that processes deoxy sugars such as fucose and rhamnose was more abundant within the Lachnospiraceae (based on the pduP gene, which encodes propionaldehyde dehydrogenase), occurring in relatives of Ruminococcus obeum and in Roseburia inulinivorans. The dominant source of propionate from hexose sugars, however, was concluded to be the succinate pathway, as indicated by the widespread distribution of the mmdA gene that encodes methylmalonyl-CoA decarboxylase in the Bacteroidetes and in many Negativicutes. In general, the capacity to produce propionate or butyrate from hexose sugars resided in different species, although two species of Lachnospiraceae (C. catus and R. inulinivorans) are now known to be able to switch from butyrate to propionate production on different substrates. A better understanding of the microbial ecology of short-chain fatty acid formation may allow modulation of propionate formation by the human gut microbiota.
Keywords: acrylate pathway, gut microbiota, propanediol pathway, propionate, succinate pathway
Introduction
The human large intestine is inhabited by a diverse microbial community that influences host health through a number of mechanisms, including the production of metabolites, a barrier function against pathogens, and interactions with the host's immune system and physiology (Flint et al., 2012b). The breakdown of non-digestible carbohydrates originating from the diet leads to the formation of fermentation acids, mainly the short-chain fatty acids (SCFAs) acetate, propionate and butyrate. SCFAs are absorbed by the host and used as an energy source but also have a variety of distinct physiological effects. Butyrate, in particular, is believed to counteract colorectal cancer and inflammation (Hamer et al., 2008; Berni Canani et al., 2012). Propionate also has potential health-promoting effects that include anti-lipogenic, cholesterol-lowering, anti-inflammatory and anti-carcinogenic action (Hosseini et al., 2011; Vinolo et al., 2011). Furthermore, the potential role of propionate in enhancing satiety (Arora et al., 2011) is of increasing interest given the rising incidence of obesity across the world. Recent proteomic work suggests that some of the effects of propionate at the cellular level differ from the action of butyrate (Kilner et al., 2012). The recently deorphanised SCFA receptors FFA2 and FFA3 are likely to mediate some of the actions of propionate; however, the exact mechanisms remain to be established (Ulven, 2012). Enhancing propionate production in the colon through dietary intervention with non-digestible carbohydrates is an attractive approach for increasing satiety and maintaining health; however, in-depth knowledge of propionate producers within the gut microbiota is required for the development of dietary strategies.
In order to understand and manipulate SCFA formation by the human colonic microbiota, we need to know which phylogenetic groups, substrates and pathways have major roles in the formation of each acid. Relevant information is now available for butyrate formation (Louis and Flint, 2009, Louis et al., 2010) but is currently lacking for propionate. Three different biochemical pathways for propionate production are known to be present in the microbiota (Figure 1). Bacteroidetes utilise the succinate pathway via methylmalonyl-CoA (Macy and Probst, 1979), which is also present in several Firmicutes bacteria belonging to the recently proposed new class of Negativicutes (formerly classed as Veillonellaceae or Clostridial cluster IX (Marchandin et al., 2010)). Bacteroidetes mainly utilise polysaccharides and peptides for growth (Macy and Probst, 1979, Flint et al., 2012a), whereas in Firmicutes propionate formation has been reported from organic acids as well (Seeliger et al., 2002; Watanabe et al., 2012). Veillonella parvula gains additional energy from succinate in the presence of lactate as the main growth substrate (Janssen, 1992), whereas Phascolarctobacterium succinatutens, isolated from human faeces, can grow on succinate alone (Watanabe et al., 2012). Selenomonas ruminantium strains isolated from the rumen produce lactate, acetate and propionate from carbohydrates, and some are also able to utilise lactate for growth (Bryant, 1956; Gilmour et al., 1994). This class of bacteria remains understudied in the human gut and it remains to be established which genera dominate in this ecosystem. The acrylate pathway for propionate formation has been characterised in detail in the soil bacterium Clostridium propionicum (Hetzel et al., 2003), and the corresponding genes have recently been described (Kandasamy et al., 2013). This pathway is also present in the rumen bacterium Megasphaera elsdenii within the Negativicutes (Hino and Kuroda, 1993), which produces butyrate during growth on glucose, but propionate during growth on lactate (Hino and Kuroda, 1993). It is possible to distinguish the succinate pathway from the acrylate pathway by incubation with stable isotope-labelled substrates (Bourriaud et al., 2005; Morrison et al., 2006). Finally, several different bacteria are known to produce 1,2-propanediol from deoxy sugars such as fucose and rhamnose, or via different pathways from dihydroxyacetonephosphate or lactate (Saxena et al., 2010). In some bacteria, including Salmonella enterica serovar Typhimurium, 1,2-propanediol can be further metabolised to propionate or propanol (Bobik et al., 1999). Propionate formation from fucose via propanediol has also been described in the human gut anaerobe Roseburia inulinivorans, which produces butyrate rather than propionate when grown on glucose (Scott et al., 2006).
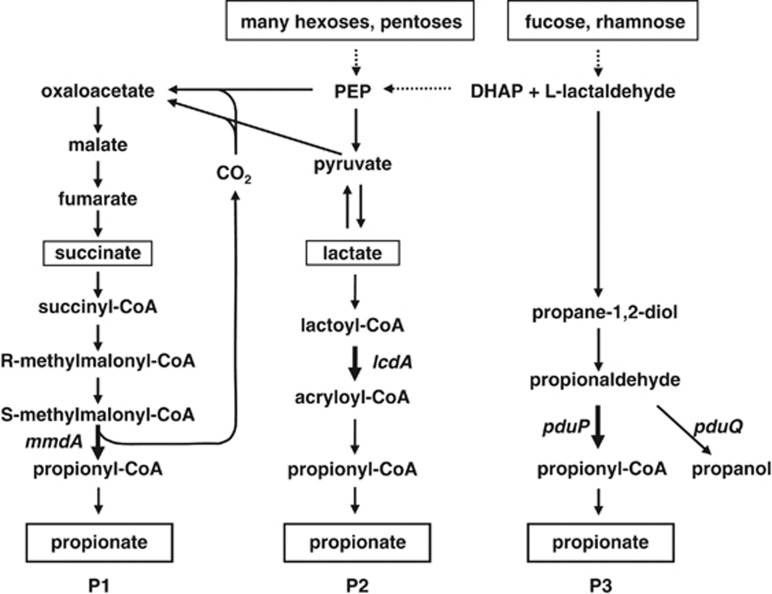
Figure 1.
Known pathways for propionate formation in human gut bacteria. (P1), Succinate pathway; (P2), acrylate pathway; (P3), propanediol pathway. Substrates utilised are shown in boxes. Genes targeted as molecular markers for the specific pathways are indicated. DHAP, dihydroxyacetonephosphate; PEP, phosphoenolpyruvate.
Our primary aim here was to explore the distribution of the three known pathways for propionate production within the human microbiota with genomic and metagenomic approaches. In addition, we demonstrate the operation of the acrylate pathway for propionate production for the first time in an isolated human colonic bacterium by means of stable isotope labelling.
Materials and methods
Bacterial isolation and cultivation
Strain GD/7 was isolated on yeast extract-casitone-fatty acid (YCFA) medium (Lopez-Siles et al., 2012) containing 25 mM DL lactate from a faecal sample of a 42-year-old man consuming an omnivorous diet as described previously (Louis et al., 2004). The 16S rRNA sequence of GD/7 was determined (accession number EU266552) as described before (Louis et al., 2004) and blastn analysis revealed that it was 97% identical to Coprococcus catus VPI-C6-61. A fluorescent in situ hybridisation (FISH) probe was designed for C. catus and was used to determine the abundance of this species in human volunteers (for details see Supplementary Information S1). All other bacterial strains used were described before (Dabek et al., 2008) or obtained from the German Collection of Microorganisms and Cell Cultures, the American Type Culture Collection, the National Collection of Industrial and Marine Bacteria (as indicated by DSM, ATCC and NCIMB numbers) or from the Rowett Institute strain collection (V. parvula L59). Bacteria were grown anaerobically in M2GSC (Miyazaki et al., 1997) or YCFAGSC medium (Lopez-Siles et al., 2012) apart from Akkermansia muciniphila DSM 22959 (M2GSC + 0.2% procine mucin, Sigma-Aldrich, Gillingham, UK) and V. parvula L59 (M2GSC medium with maltose replacing starch + 1% lactate). Growth experiments on specific substrates were performed in triplicate on basal YCFA medium supplemented with substrates as detailed in the results (at 0.5% substrate, unless specified otherwise). Growth experiments were performed by inoculating 0.1 ml of overnight culture into 7.5 ml of anaerobic medium prepared in Hungate tubes (in a 100% CO2 atmosphere) followed by incubation for 24 h at 37 °C (Lopez-Siles et al., 2012). The SCFA and lactate content of samples was determined by gas chromatography analysis following conversion to t-butylmethylsilyl derivatives (Richardson et al., 1989).
Determination of concentrations and 13C enrichments in propionate
Duplicate culture supernatant samples from YCFA and 25 mM L-lactate with additional universally 13C-labelled L-lactate at 10 molar % excess were prepared for estimation of concentrations by isotope dilution and for determination of enrichments of SCFA and lactate by gas chromatography coupled with mass spectrometry. They were measured by analysis of the tert-butyldimethylsilyl derivatives. Procedures were as described previously (Belenguer et al., 2007). The mass spectrometer was operated under electron impact ionisation conditions. For the concentration determinations, appropriate corrections were applied for the enrichments in the sample. Lactate flows to propionate, either through acrylate or through succinate, were obtained from the changes in labelled propionate (M+2 and M+3), assuming that the succinate route gives half M+2 and M+3.
Degenerate primer design, PCR and clone library analysis
Degenerate primers (Table 1) specific for genes lcdA (lactoyl-CoA dehydratase subunit alpha), pduP (CoA-dependent propionaldehyde dehydrogenase), pduQ (propanol dehydrogenase; for further details see Supplementary Information) and mmdA (methylmalonyl-CoA decarboxylase α-subunit of Negativicutes, equivalent to propionyl-CoA carboxylase β-subunit of Bacteroidetes, for details see Results) were designed by visual inspection of protein sequence alignments of genes detected by blastp analysis (Altschul et al., 1990) against the non-redundant database at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and using the CoGe database (http://genomevolution.org/CoGe/) (Lyons and Freeling, 2008). Query sequences: LcdA, C. propionicum AEM62994; PduP, R. inulinivorans ABC25528; PduQ, R. inulinivorans ABC25529; and MmdA, Bacteroides thetaiotaomicron NP_810363. Degenerate primer design was carried out as described previously (Louis et al., 2004). Primer specificity was validated with genomic DNA prepared with a DNeasy Blood and Tissue Kit (Qiagen, Manchester, UK) from pure cultures of 21 bacterial strains (Supplementary Table S1). To confirm specificity of the primers, they were used to amplify the respective genes from faecal DNA from a healthy human volunteer. Fresh faeces was homogenised for 3 × 1 min in a stomacher and 0.2 g was used to extract genomic DNA using the FastDNA spin kit for soil (MP Biomedicals, Cambridge, UK). PCRs contained 10 ng template DNA, 3 mM MgCl2, 10 mM dNTPs, 100 nM of each primer (Table 1) and 1 U Taq polymerase (Bioline, London, UK) per 50 μl reaction, and were performed at 95 °C for 3 min, followed by 30 cycles at 95 °C for 30 s, primer-specific annealing temperature (Table 1) for 30 s, 72 °C for 40 s (1 min for lcdA, 2 min for 16S rRNA gene) and a final extension at 72 °C for 5 min (10 min for 16S rRNA gene). Clone libraries specific for Bacteroidetes and Negativicutes were constructed by amplification of faecal DNA with primers targeting the 16S rRNA gene (Table 1). For primer design of Negativicutes primers see Supplementary Information. The PCR amplification protocol was the same as described above. PCR products were cloned and sequenced as described before (Louis et al., 2010). Sequence accession numbers were KF385507 (lcdA of Clostridium lactatifermentans DSM 14214) and KF385508-KF385859 (faecal clones).
Table 1. Primers used in this study.
| Target gene | Primer name | Primer sequence | Amplicon size (bp) | Ann. temp.a | Reference |
|---|---|---|---|---|---|
| Functional genes | |||||
| lcdA | lcdAF | 5′-CTGGTGTGCTGGWSIGCIWSIGTIGCNCC-3′ | 771 | 58 | This study |
| lcdAR | 5′-CAGATAGGTCCAIAYIGCDATNCCYTCCCA-3′ | This study | |||
| pduP | pduPF1 | 5′-GTGGATGARACIGGIATGGGNAAYGTNGG-3′ | 1088 | 57 | This study |
| pduPF2b | 5′-TGGCTGGAACATGGIAAYMRICAYWSIGCICA-3′ | This study | |||
| pduPR | 5′-CAATAGCCYTCICCICCRAAICCIADNGC-3′ | This study | |||
| pduQ | pduQF | 5′-GATCTGAAYAARMARAAYRTIGGIGGIGC-3′ | 584 | 57 | This study |
| pduQR1b | 5′-CGGATCATCTTTRAAIATCATRCAISWNAC-3′ | This study | |||
| pduQR2 | 5′-ATGCGGGTTAATNKKIGCRTCICCICCRTTDAT-3′ | This study | |||
| mmdA | mmdAF | 5′-AATGACTCGGGIGGIGCIMGNATHCARGA-3′ | 874 | 56 | This study |
| mmdAR | 5′-GATTGTTACYTTIGGIACNGTNGCYTC-3′ | This study | |||
| 16S rRNA gene | |||||
| Universal | 7-f | 5′-AGAGTTTGATYMTGGCTCAG-3′ | ∼1495 | 52 | Satokari et al. (2001) |
| 1510-r | 5′-ACGGCTACCTTGTTACGACTT-3′ | Satokari et al. (2001), modified | |||
| Bacteroidetes | Bac303F | 5′-GAAGGTCCCCCACATTG-3′ | 610 | 60 | Bartosch et al. (2004) |
| BacPre-rev | 5′-CTTTGAGTTTCACCGTTGCCGG-3′ | Wood et al. (1998), modified | |||
| Negativicutes | IX552Fc | 5′-GTTGTCCGGAATYATTGGGC-3′ | 321 | 63 | This study |
| IX854R2Ad | 5′-ATTGCGTTAACTCCGGCACA-3′ | Daly and Shirazi-Beechey (2003); modified | |||
| IX854R2Gd | 5′-ATTGCGTTAACTCCGGCACG-3′ | Daly and Shirazi-Beechey (2003); modified | |||
ann. temp.: annealing temperature.
Primers have not been used here for clone library analysis but can be used to amplify across pduP and pduQ.
Primer is not entirely specific for Negativicutes and will work specifically only in conjunction with reverse primers.
Both primers are used together at equimolar concentrations.
Metagenomic mining methods
The deduced protein sequences of mmdA from V. parvula (accession number ZP_06259922), lcdA from C. propionicum (AEM62994) and pduP and pduQ from R. inulinivorans (ABC25528 and ABC25529) were blasted (tblastn) against the metagenomic data set comprising 124 European individuals (Qin et al., 2010) on the in-house Rowett/BioSS Beowulf cluster as described before (McIntosh et al., 2012). Matches with at least 50% sequence identity for LcdA and 55% for PduP and PduQ were selected on the basis of the blastp results in the non-redundant (nr) database indicating which identity level is likely to represent a true hit (>60% for LcdA, as blastp results with query sequence AEM62994 had at least 60% identity for strains known to carry this pathway, followed by a sharp drop in sequence identity to ⩽43% for other matches; >65% for PduP and PduQ, as blastp results with query sequences ABC25528/9 had at least 73% identity for strains known to carry this pathway, followed by a sharp drop in sequence identity to ⩽58% for PduP (but not PduQ) two matches of 63% identity were also present in Thermoanaerobacterium spp.). Maximum likelihood phylogenetic trees (poisson model, 100 times bootstrap) were constructed using Mega5 (Tamura et al., 2011). Sequences with at least 95% identity were grouped into phylotypes. Individual bacterial genomes were obtained from multiple sources and included 20 strains of human colonic Firmicutes bacteria (listed in Table 2) isolated at the Rowett Institute that were sequenced at the Sanger Institute as part of the MetaHit EU project (http://www.sanger.ac.uk/pathogens/metahit/).
Table 2. Occurrence of genes diagnostic for different pathways of propionate and butyrate production in the genomes of dominant species of human colonic anaerobes.
| Phylum, family and species | Origin | % Protein sequence identitya | ||||
|---|---|---|---|---|---|---|
|
Fermentation pathwayb
and diagnostic genec |
||||||
| P1 | P2 | P3 | B1 | B2 | ||
| |
|
MmdAd |
LcdA |
PduP |
Buk |
BCoAT |
| Actinobacteria | ||||||
| Bifidobacterium adolescentis DSM 20083 | Adult intestine | 48 | No | No | No | No |
| Bacteroidetes | ||||||
| Bacteroides thetaiotaomicron VPI-5482, DSM 2079T | Human faeces | 60 | No | No | 49 | No |
| Bacteroides fragilis YCH46 | Clinical | 62 | No | No | 52 | No |
| Bacteroides vulgatus DSM 1447T | Human faeces | 60 | No | No | 50 | No |
| Firmicutes-Lachnospiraceae | ||||||
| Roseburia intestinalis M50/1 | Human faeces | 35 | No | No | No | 94 |
| Eubacterium rectale A1-86 | Human faeces | 38 | No | No | No | 81 |
| Roseburia inulinivorans A2-194 | Human faeces | 38 | No | 100 | No | 87 |
| Unknown sp SR1/5 | Human faeces | 44 | No | 78 | No | No |
| Ruminococcus obeum A2-162 | Human faeces | 30 | No | 77 | No | No |
| Ruminococcus gnavus ATCC 29149 | Human faeces | 39 | No | 78 | No | No |
| Ruminococcus torques L2-14 | Human faeces | 40 | No | 74 | No | No |
| Unknown sp SM4/1 | Human faeces | No | No | No | No | 82 |
| Clostridium sp. M62/1 | Human faeces | 36 | 39 | No | No | 76 |
| Clostridiales bacterium SS3/4 | Human faeces | 38 | 37 | No | No | 73 |
| Eubacterium hallii L2-7 | Human faeces | 38 | No | 53 | No | 77 |
| Anaerostipes hadrus SSC/2 | Human faeces | No | No | No | No | 74 |
| Coprococcus catus GD/7 | Human faeces | 39 | 67 | No | No | 75 |
| Coprococcus eutactus L2-50 | Human faeces | 40 | No | No | 100 | 51 |
| Coprococcus eutactus ART55/1 | Human faeces | 42 | No | No | 92 | 49 |
| Coprococcus comes ATCC 27758 | Human faeces | 39 | No | No | 76 | No |
| Clostridium symbiosum WAL-14163 | Human faeces | 54 | 37 | 33 | No | 76 |
| Clostridium asparagiforme DSM 15981 | Human faeces | 40 | 36 | 43 | No | No |
| Clostridium sp. MSTE9 (cluster XIVb) | Oral | 54 | 78 | 53 | No | 41 |
| Firmicutes-Ruminococcaceae | ||||||
| Faecalibacterium prausnitzii A2-165 | Human faeces | No | No | No | No | 74 |
| Faecalibacterium prausnitzii S3L/3 | Human faeces | No | No | No | No | 73 |
| Faecalibacterium prausnitzii L2-6 | Human faeces | No | No | No | No | 74 |
| Ruminococcus bromii L2-63 | Human faeces | 34 | No | 32 | No | No |
| Eubacterium siraeum 70/3 | Human faeces | 35 | No | 34 | No | No |
| Eubacterium siraeum V10Sc8a | Human faeces | 35 | No | 34 | No | No |
| Firmicutes–Negativicutes | ||||||
| Veillonella parvula DSM 2008 | Intestinal tract | 99 | No | No | No | No |
| Dialister succinatiphilus YIT 11850, DSM 21274T | Human faeces | 77 | No | No | No | No |
| Phascolarctobacterium succinatutens YIT 12067 | Human faeces | 81 | No | No | No | No |
| Selenomonas ruminantium subsp. lactilytica TAM 6421 | Bovine rumen | 81 | No | No | No | No |
| Megasphaera elsdenii LC1 DSM 20460T | Sheep rumen | 74 | 62 | No | No | 55 |
| Firmicutes–Peptostreptococcaceae | ||||||
| Clostridium difficile 630 | Clinical | No | 43 | 32 | 62 | 39 |
| Firmicutes–Clostridiaceae | ||||||
| Clostridium botulinum A2 Kyoto | Infant botulism | No | 42 | 35 | 65 | No |
| Clostridium botulinum C Eklund | No | 75 | No | No | No | |
| Clostridium novyi NT | Gas gangrene | No | 76 | 31 | No | No |
| Clostridium beijerickii NCIMB 8052 | Corn meal | No | No | 51 | 67 | 41 |
| Verrucomicrobia | ||||||
| Akkermansia muciniphila DSM 22959T | Human faeces | 52 | No | No | No | No |
On the basis of blastp and tblastn analysis of genomes in the NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and CoGe (http://genomevolution.org/CoGe/) databases. Per cent protein identity is given for all matches >30% with >80% query coverage. Shading from light grey to black: 40–49%, 50–59%, 60–69%, 70–100%.
Propionate pathways: P1, succinate pathway; P2, acrylate pathway; P3, propanediol pathway (see Figure 1). Butyrate pathways: B1, butyrate kinase pathway (as Bacteroides do not produce butyrate, Buk matches below 60% in the Bacteroidetes genomes are likely to have another function); B2, butyryl-CoA:acetate CoA-transferase pathway (Louis and Flint, 2009).
Genes used for blastp analyses are MmdA, ZP_06259922; LcdA, AEM62994; PduP, ABC25528; Buk, AAR19758; BCoAT, AAX19660.
Genes with high sequence similarity to MmdA may also be involved in other pathways as detailed in the main text.
Results
Succinate pathway
The succinate pathway of propionate formation has mainly been studied in bacteria belonging to the class Negativicutes. In V. parvula, the decarboxylation of methylmalonyl-CoA to propionyl-CoA is coupled with sodium transport across the membrane, which facilitates ATP generation via a sodium-translocating ATPase (Dimroth and von Ballmoos, 2008), and the respective genes for the sodium pump decarboxylase have been characterised (mmdA-E, (Huder and Dimroth, 1993)). The other pathway genes are encoded upstream of the mmd gene cluster, with the same gene configuration also being present in the human colonic anaerobes Dialister succinatiphilus YIT 11850 and P. succinatutens YIT 12067 (Supplementary Table S2). Genes with high similarity to V. parvula ATCC 17745 mmdA were also found in the genomes of the rumen species M. elsdenii DSM 20460 and S. ruminantium TAM 6421, although most of the remaining pathway genes were lacking in M. elsdenii (Supplementary Table S2).
Bacteroidetes strains also carry genes with similarity to mmdA (Table 2), and this gene was therefore chosen as a molecular target for this pathway, as it exhibited the highest level of sequence identity between Negativicutes and Bacteroidetes (Supplementary Table S2). Degenerate primers were designed and validated against pure bacterial DNA of 21 strains, which indicated that they worked specifically for bacteria carrying the mmdA gene, most of which are known to use the succinate pathway (Supplementary Table S1). A clone library was constructed from human faecal DNA to confirm specificity of the primers, and clone libraries of 16S rRNA genes specific for both phylogenetic groups of bacteria carrying this pathway were generated in parallel. For the functional gene primers, 64 of 65 analysed clone sequences belonged to Bacteroidetes, with the final sequence having 72% identity to Clostridium hathewayi DSM 13479 ZP_6112601 (the C. hathewayi gene exhibits 67% protein sequence identity to V. parvula MmdA; however, only one further gene (mmdB, 60% protein identity) with similarity to the V. parvula gene cluster ZP_06259927-18 was found by blastp analysis in this bacterium). The clone distribution between different species agreed well with a Bacteroidetes-specific clone library based on the 16S rRNA gene (Supplementary Table S3), whereas a 16S rRNA gene clone library targeted specifically against Negativicutes detected mainly Phascolarctobacterium faecium (32 clones; also 1 clone closely related to Veillonella dispar, out of 44 clones analysed, Supplementary Table S3). P. faecium is reported to convert succinate to propionate (Del Dot et al., 1993).
Acrylate pathway
Analysis of the acrylate pathway genes revealed the lactoyl-CoA dehydratase sequence to be most suitable as a marker gene for this pathway (Supplementary Table S4). The protein sequences of genes lcdA, lcdB and lcdC from the soil bacterium C. propionicum (accession numbers AEM62993-5), encoding the α- and β-subunits of the enzyme as well as the activator (Kandasamy et al., 2013), were blasted against non-redundant protein sequences at NCBI, and phylogenetic trees were generated from the blast results (data not shown). Genes belonging to a limited number of bacteria (C. propionicum, C. botulinum type C and D, C. novyi, Megasphaera spp., C. catus, Desulfosporosinus spp., Peptinophilus indolicus and Fusobacterium sp.) formed a separate cluster for all three genes, which was most clearly delineated from other sequences for LcdA. The presence of C. botulinum type C and D but not other C. botulinum types in this cluster is in line with those strains (together with C. novyi) being able to convert lactate to propionate, whereas other C. botulinum types cannot utilise lactate (Rainey et al., 2009). Furthermore, this cluster contained M. elsdenii, known to carry the pathway (Hino and Kuroda, 1993), and we therefore postulated that it represents true lactoyl-CoA dehydratase genes. Analysis of genome arrangements of lcdABC was in agreement with this, as all the bacteria within this cluster contained propionate CoA-transferase two genes upstream, whereas other bacteria carrying genes of lower identity to lcdABC did not (Supplementary Table S4).
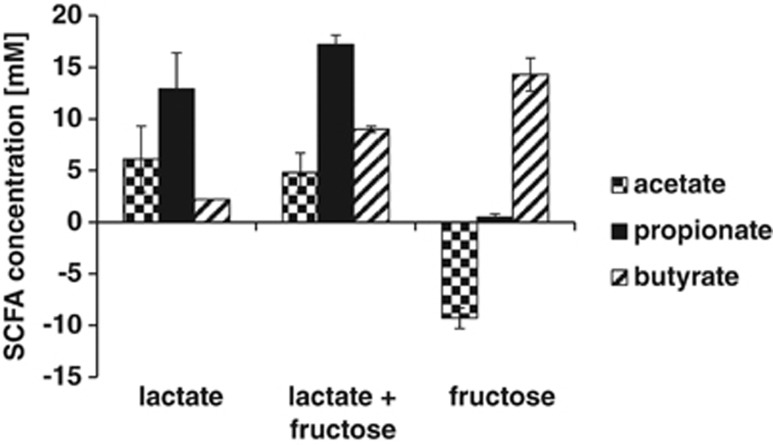
Interestingly, the gut bacterium C. catus GD/7 (Lachnospiraceae) also contained two homologues of the lactoyl-CoA dehydratase genes, one of which had a similar genome arrangement to that of C. propionicum and M. elsdenii (Supplementary Table S4). C. catus has been reported to produce butyrate and propionate, whereas Coprococcus eutactus and Coprococcus comes produce butyrate together with formate or lactate, respectively, as main fermentation products (Ezaki, 2009a). We tested different Coprococcus strains for their ability to utilise lactate for growth. C. eutactus ATCC 27759 and C. comes ATCC 27758 could not grow on lactate, but C. catus GD/7 was able to grow with 25 mM lactate on YCFA medium containing 30 mM acetate. Propionate became the main product and substantial lactate utilisation also occurred in the presence of 10 mM fructose, whereas growth on fructose in the absence of lactate led to the production of butyrate and net consumption of acetate (Figure 2). In order to determine the pathway used for lactate utilisation, 25 mM DL lactate with additional universally 13C-labelled lactate at 10 molar % excess was added to the batch culture medium. All three labelled carbons are expected to appear in propionate when the acrylate pathway is used to convert lactate to propionate, whereas 50% of the propionate formed should contain only two labelled carbons when the succinate pathway is used (Gottschalk, 1979; Counotte et al., 1981). On this basis, it was estimated that 94–96% of the 13C from lactate flowed to propionate via the acrylate pathway, regardless of whether growth was with lactate alone or with lactate and fructose. Probe Ccat62, specific for C. catus, was designed and employed to determine the abundance of the C. catus group in faecal samples from 10 healthy volunteers (5 females and 5 males) between 24 and 60 years of age. C. catus was detected in samples from 9 out of 10 volunteers at mean populations of 2.02 × 107 cells g−1, averaging 0.035% of total bacteria detected with the broad Eub338 probe.
Figure 2.
Fermentation acid profiles for C. catus GD/7 grown (24 h) on basal YCFA medium with 30 mM acetate, supplemented with either lactate (25 mM), fructose (10 mM) or both substrates (average and s.d. of triplicate experiments).
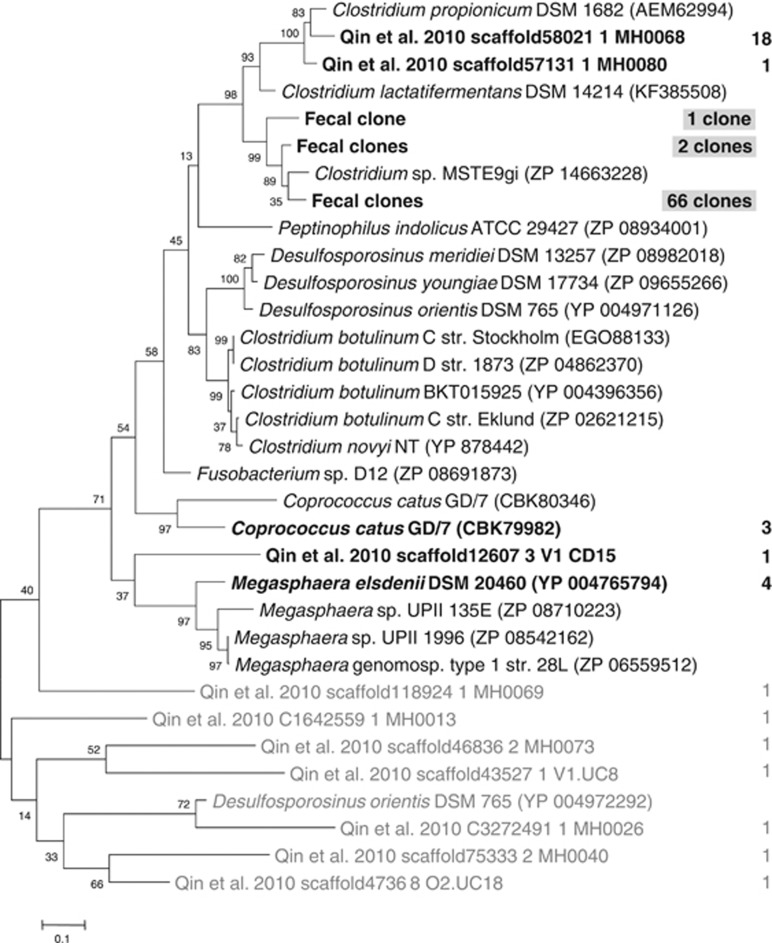
To establish how widespread the acrylate pathway is within the human gut microbiota, the human metagenomic data set from Qin et al. (2010), comprising data from 124 individuals (85 healthy, overweight and obese subjects and 39 IBD patients), was searched to identify genes with similarity to the lcdA gene from C. propionicum. Thirty-four hits with over 50% identity were found, with most of them falling within a cluster related to C. propionicum. Other sequences were closely related to M. elsdenii or C. catus, and one sequence (scaffold12607 3 V1 CD15) was not closely related to reference sequences (Figure 3, Supplementary Table S5). Thus, it appears that this pathway for propionate formation is present in a very restricted number of human gut bacteria. Degenerate primers designed against lcdA were employed to investigate whether they could specifically amplify the gene from a human faecal sample. Clone library analysis of 78 clones revealed that the primers showed limited cross-specificity to other genes (9 clones carried genes not closely related to lcdA). Three types of novel lcdA genes clustering with the oral isolate Clostridium sp. MSTE9gi and C. propionicum were found (Figure 3). C. lactatifermentans is a chicken strain phylogenetically closely related to C. propionicum that utilises lactate and produces propionate (Rainey et al., 2009). In order to investigate whether one of the unknown sequences represents C. lactatifermentans, we cloned and sequenced the lcdA gene fragment from C. lactatifermentans DSM 14214. The sequence clustered with C. propionicum-related sequences but did not correspond to any of the unknown sequences (Figure 3).
Figure 3.
Phylogenetic tree of deduced protein sequence of lactoyl-CoA dehydratase gene lcdA. Database matches to C. propionicum (AEM62994) of at least 60% as well as the top hit below this cutoff (Desulfosporosinus orientis YP_004972292, 41%) are shown. The lcdA gene fragment of C. lactatifermentans DSM 14214 was sequenced in this study. The number of hits within the metagenomic data set of Qin et al. (2010) with at least 50% identity is indicated to the right (sequences with at least 95% identity were grouped; for a list of all hits see Supplementary Table S5). Sequences from clone library analysis of a human faecal sample (⩾95% identity grouped) are shaded. Grey tree branches indicate genes with lower identity to C. propionicum assumed not to be bona fide lcdA genes.
Propanediol pathway
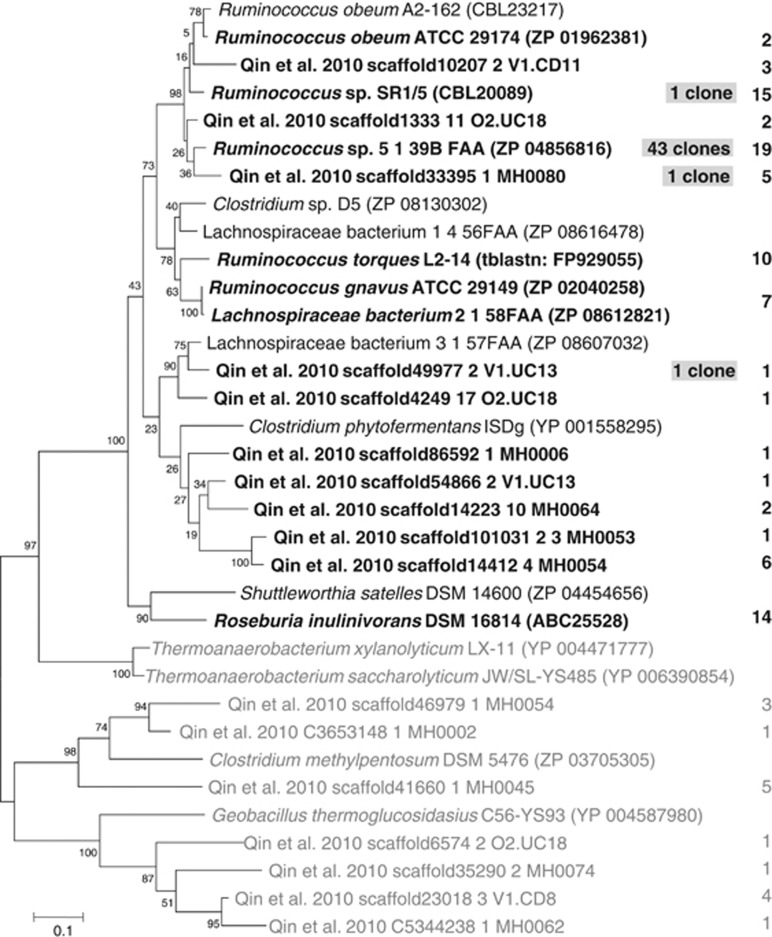
The propanediol pathway has been detected in the phylogenetically distant bacteria Salmonella enterica serovar Typhimurium (Bobik et al., 1999) and R. inulinivorans (Scott et al., 2006), suggesting that it might be present in other gut bacteria. The gene pduP, encoding a CoA-dependent propionaldehyde dehydrogenase that converts propionaldehyde to propionyl-CoA, was investigated for its suitability as a marker for this pathway. Blastp matches of R. inulinivorans PduP (ABC25528) of over 70% identity were found for several strains, most of which belonged to human gut Lachnospiraceae bacteria related to R. obeum (Figure 4). These bacteria are not reported to produce propionate (R. obeum produces acetate, R. torques produces lactate and acetate and R. gnavus produces acetate and formate on PYG medium, Ezaki, 2009b). However, propionate production via this pathway is dependent on the carbohydrate available for growth in other bacteria, with fucose and rhamnose reported as being propionigenic (Scott et al., 2006; Saxena et al., 2010). We therefore tested growth and fermentation product formation of R. obeum A2-162 in the presence of those sugars compared with glucose (OD650 at 24 h 0.38±0.02 on rhamnose and 0.96±0.06 on fucose vs 1.08±0.04 on glucose). Fermentation acids produced on glucose were mainly acetate, formate and lactate (20.9±4.8, 17.2±1.3 and 4.2 ±0.1 mM), with little propionate (0.2±0.1 mM) detected. On fucose, R. obeum A2-162 revealed a clear switch to propionate (19.9±0.8 mM) along with other fermentation products (26.5±1.0 mM acetate, 11.4±1.0 mM formate and 3.7±0.1 mM lactate). On rhamnose, R. obeum formed 10.6±1.4 mM propionate in addition to other fermentation products (18.8±2.9 mM acetate, 8.1±0.8 mM formate and 2.0±0.1 mM lactate), thus providing strong evidence that the propanediol pathway is indeed functional in this bacterium.
Figure 4.
Phylogenetic tree of deduced protein sequence of CoA-dependent propionaldehyde dehydrogenase pduP. Database matches to R. inulinivorans (ABC25528) of at least 60% as well as the top hits below this cutoff (Clostridium methylpentosum ZP_03705305, 57% Geobacillus thermoglucosidasius YP_004587980, 54%) are shown. The number of hits within the metagenomic data set of Qin et al. (2010) with at least 55% identity is indicated to the right (sequences with at least 95% identity were grouped; for a list of all hits see Supplementary Table S6). Numbers of sequences from clone library analysis of a human faecal sample (⩾95% identity grouped) are shaded. Grey tree branches indicate gene with lower identity to R. inulinivorans assumed not to be bona fide pduP genes.
The Qin et al. (2010) metagenomic database was searched for genes similar to PduP from R. inulinivorans (ABC25528) to investigate how widespread this pathway is within the gut microbial community. In total, 106 hits with at least 55% identity were found. Ninety metagenomic hits had over 70% identity to R. inulinivorans PduP, with 67 displaying over 95% identity to known bacteria (Figure 4, Supplementary Table S6). Twenty-three sequences distributed across 10 phylotypes did not correspond to known bacteria and thus represent novel strains carrying this pathway. The sequences with over 70% identity to the known PduP gene of R. inulinivorans formed a tight phylogenetic cluster (Figure 4), indicating that those genes represent bona fide pduP genes.
To enable a more targeted analysis of the pathway, degenerate primers were designed against pduP and used to amplify the gene from a human faecal sample. Forty-six clones were analysed, 43 of which were closely related to the sequence of Ruminococcus sp. 5_1_39B FAA, with three singletons closely related to Ruminococcus sp. SR1/5 and two sequences from the Qin et al. data set, respectively (Figure 4). As it was difficult to find highly specific sequence regions for degenerate primer design for this gene, primers were also designed for adjacent gene pduQ (encoding propanol dehydrogenase) to establish whether this gene could be used as an alternative marker gene. The Qin et al. (2010) data set was mined and a clone library was prepared from the same faecal sample as used for pduP. Comparison of the metagenomic hits revealed good agreement for both genes (Supplementary Figure S1, Supplementary Table S7). PduQ clone libraries were again dominated by Ruminococcus sp. 5_1_39B FAA, but 23 hits were also found for a metagenomic sequence only once detected in the pduP clone library (Supplementary Figure S1).
Discussion
We have employed a combination of genomic analysis and microbial physiology to investigate pathways of microbial propionate formation in the human gut microbiota. An overview of the pathway distribution in dominant human gut bacteria is given in Figure 5. The succinate pathway appears to be the most abundant route for propionate formation, as it is present in the abundant phylum Bacteroidetes as well as in several Negativicutes bacteria. Furthermore, tblastn analysis of the metagenomic data set of Qin et al. (2010) revealed 478 matches of at least 60% to the V. parvula MmdA gene (ZP_06259922, data not shown). This number likely underestimates the number of true hits, as the level of sequence identity between the Negativicutes and Bacteroidetes genes is around 60% (Table 2). Most of the pathway reactions are shared with central carbon metabolism pathways (conversions within the citrate cycle, amino acid degradation and so on) but the decarboxylation of methylmalonyl-CoA to propionyl-CoA appears to be specific to the succinate pathway and it was hoped that the corresponding gene could be taken as diagnostic for this pathway. Sequences amplified with the degenerate mmdA primers were almost exclusively related to Bacteroidetes and no mmdA genes were recovered from Negativicutes despite evidence that the genes were amplified well from purified DNA of representative Negativicutes (Supplementary Table S1). This is presumably due to the fact that Bacteroidetes are more abundant than Negativicutes in the human gut microbiota and also carry several copies of the gene (see below). Transcarboxylases, decarboxylases and carboxylases form part of a biotin-enzyme family (Lombard and Moreira, 2011) and the methylmalonyl-CoA decarboxylase alpha subunit (mmdA) from V. parvula shares similarity with the 12S subunit of methylmalonyl-CoA transcarboxylase of the dairy strain Propionibacterium freudenreichii (Kumar Bhat and Berger, 2007) and with the β-subunit of propionyl-CoA carboxylases. Many strains of Bacteroides carry several genes closely related to MmdA (for example, both B. thetaiotaomicron VPI-5482 and B. fragilis YCH46 have three genes with 58–62% identity to MmdA from V. parvula, Supplementary Table S2). Although there is no evidence for propionate carboxylation to methylmalonyl-CoA in Bacteroidetes, we did detect biotin carboxylase (BC) domains, which are considered to be present only in carboxylases, next to two Bacteroides mmdA genes (B. thetaiotaomicron VPI_5482 NP_810362 and NP_810828 and B. fragilis YCH46 YP_098894 and YP_100806). This is in contrast to an earlier report that concluded that BC domains were not associated with propionyl-CoA carboxylase genes in Bacteroidetes genomes (Lombard and Moreira, 2011). The presence of multiple mmdA gene copies, some of which might not encode true decarboxylases, therefore precluded assessment of the succinate pathway for propionate formation by degenerate PCR targeting the mmdA gene. As the phylogenetic distribution of this pathway is fairly coherent, targeting of Bacteroidetes and Negativicutes 16S rRNA genes offers a more satisfactory strategy for quantification of bacteria carrying this important pathway. Although M. elsdenii has an mmdA homologue, it lacks other pathway genes, suggesting that the mmdA-related gene product performs another function in this species. M. elsdenii is, however, able to produce propionate via the acrylate pathway.
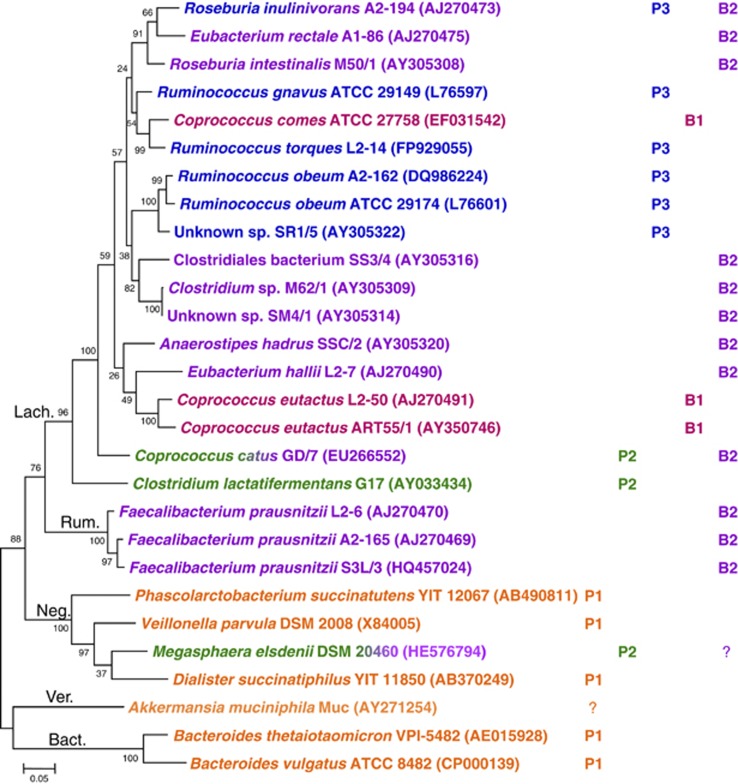
Figure 5.
Distribution of different pathways for propionate and butyrate formation in dominant human gut bacteria, based on genome searches of corresponding genes (Table 2; note that no sequence information is currently available for C. lactatifermentans), metagenomic mining (Figures 3 and 4) and validation of degenerate primers (Supplementary Table S1). C. lactatifermentans and M elsdenii originate from animal hosts (chicken and sheep, respectively). They are included as closely related bacteria are carriers of the acrylate pathway based on human metagenome mining (Figure 3). 16S rRNA sequences were obtained from the Ribosomal Database Project (Cole et al., 2009). Phylogenetic assignment—Lach.: Lachnospiraceae, Rum.: Ruminococcaceae, Neg.: Negativicutes, Ver.: Verrucomicrobia, Bact.: Bacteroidetes. P1–P3, B1–B2: Propionate and butyrate pathways as per Table 2. Question marks indicate the presence of genes with ⩾50% sequence identity (Table 2). As M. elsdenii is known to produce butyrate and A. munciniphila propionate, this has been taken into consideration for the assignment of pathways.
The acrylate pathway for propionate formation was found to have a very limited distribution within human gut bacteria, and the genera shown to carry it in this study belong to different Firmicutes families. The most abundant lactoyl-CoA dehydratase sequences were most closely related to the Lachnospiraceae (Clostridial cluster XIVb) bacterium C. propionicum. Only three further sequence types were found, related to C. catus (Lachnospiraceae), M. elsdenii (Negativicutes), or not related to a cultured isolate. The abundance of these bacteria appears to be quite low, based on both the limited number of metagenomic matches found and the enumeration of C. catus by FISH analysis. Although we have demonstrated the operation of the acrylate pathway here for the first time in a human colonic isolate, C. catus GD/7, this pathway seems unlikely to have a dominant role in propionate formation in the human gut. The propanediol pathway, however, appears to be more widespread within the gut community and is present in some relatively abundant bacteria. The phylogenetic analysis indicates that most, if not all, bacteria carrying this pathway belong to the Lachnospiraceae related to Ruminococcus spp. (several of the misclassified ruminococci within the Lachnospiraceae are being reclassified to the genus Blautia, Liu et al., 2008) and R. inulinivorans. This pathway is likely to have an important role in propionate fermentation from deoxy sugars. One of these, fucose, is a major component of host-derived glycans, suggesting that this pathway could be indicative of the ability to utilise host-derived substrates. A. muciniphila is a mucin-degrading bacterium originating from the human gut that produces propionate (Derrien et al., 2004). Its genome contains a gene with low-level (58%) protein sequence identity to PduQ (Supplementary Figure S1); however, the closest match to PduP only has 24% protein sequence identity with a coverage of 56% (data not shown); thus, it appears that this bacterium does not harbour the propanediol pathway. No matches to LcdA were found either, but a gene with 52% protein sequence identity to MmdA is present (Table 2). The mmdA degenerate primer binding sites in this gene are only partially conserved (data not shown) and it remains to be established whether A. muciniphila uses this pathway for propionate generation. Rhamnose is another deoxy sugar that may lead to propionate formation via the propanediol pathway, and increased serum propionate levels in response to rhamnose ingestion have been reported in humans (Vogt et al., 2004). In addition to the type of carbohydrate available to the microbiota, other factors also have to be considered with regard to propionate production in the gut. Thus, pH is an important determinant in the competition between Bacteroidetes and Firmicutes (Walker et al., 2005; Duncan et al., 2009), and the level of propionate production in Bacteroidetes is dependent on carbon dioxide levels (Macfarlane and Gibson, 1997).
The lcdA and pduP gene degenerate primers designed here for the acrylate and propanediol pathways should prove valuable in future for probing the phylogenetic diversity of propionate producers in different individuals—for example, by using signature-tagged versions of the primers in conjunction with pyrosequencing. It should be recognised, however, that the primers may not amplify genes from the different phylogenetic groups in a precisely quantitative manner, as sequence variations can be expected to lead to some variation in the efficiency of amplification. Thus, it has to be decided on a case-by-case basis, depending on the respective gene sequences, the phylogenetic distribution and how well the bacteria carrying the pathway are characterised, whether targeting the functional gene or specific phylogenetic groups via a phylogenetic marker gene (such as the 16S rRNA gene) is the better option for quantification of functional groups.
It is unusual for isolated anaerobic bacteria to produce both propionate and butyrate as major products from the fermentation of sugars (Chen et al., 1977). Table 2 and Figure 5 confirm that most human colonic bacteria that possess diagnostic genes for butyrate synthesis (either butyrate kinase or butyryl-CoA:acetate CoA-transferase) lack genes concerned with propionate formation, and vice versa. Thus, these two products represent alternative ‘hydrogen sinks' whose pathways have apparently evolved in different phylogenetic groups of anaerobic bacteria. Thus, there appears to be no evidence at present for the succinate pathway in the Lachnospiraceae or Actinobacteria, or for butyrate synthetic pathways in Bacteroidetes or Actinobacteria. Two exceptions were, however, found that are of some interest. C. catus was shown here to be able to utilise lactate via the acrylate pathway to produce propionate, although forming butyrate via the butyryl-CoA:acetate CoA-transferase route when grown on fructose. Meanwhile, R. inulinivorans was shown previously to utilise the propanediol pathway for propionate formation from fucose, although producing butyrate from glucose. Growth on fucose resulted in a dramatic change in the pattern of gene expression (Scott et al., 2006). In Salmonella, acquisition of the propanediol pathway is estimated to require many genes, encoding not only the pathway enzymes themselves but also the associated proteins that are involved in polyhedral body formation and synthesis of required cofactors (Bobik et al., 1999). Acquisition of additional fermentation pathways, presumably by horizontal gene transfer, may have arisen from selection for an expansion in the ability to utilise alternative substrates in the highly competitive environment of the colon.
In conclusion, the targeted analysis of three known biochemical pathways for propionate formation provides valuable information on their likely importance within the human colonic microbiota that will, in conjunction with improved knowledge of the underlying physiology of the respective bacteria and further studies on the level of gene expression, help to design strategies to boost the formation of this important metabolite in the human gut.
Acknowledgments
The Rowett Institute of Nutrition and Health receives financial support from the Scottish Government Rural and Environmental Sciences and Analytical Services. Alvaro Belenguer received financial support from the Spanish Ministry of Education and Science. Nicole Reichardt is funded by a Scottish Government Strategic Partnership on Food and Drink Science. We thank Wolfgang Buckel, Douglas Morrison, Tom Preston, Graeme Milligan, Lynda Williams and Janice Drew for helpful discussions, Freda Farquharson for help with bacterial cultures and supply of genomic DNA, Graham Calder and Gerald Lobley for help with stable isotope analysis and Tony Travis for help with bioinformatic analysis.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arora T, Sharma R, Frost G. Propionate. Anti-obesity and satiety enhancing factor. Appetite. 2011;56:511–515. doi: 10.1016/j.appet.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Bartosch S, Fite A, Macfarlane GT, McMurdo MET. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl Environ Microbiol. 2004;70:3575–3581. doi: 10.1128/AEM.70.6.3575-3581.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenguer A, Duncan SH, Holtrop G, Anderson SE, Lobley GE, Flint HJ. Impact of pH on lactate formation and utilization by human fecal microbial communities. Appl Environ Microbiol. 2007;73:6526–6533. doi: 10.1128/AEM.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berni Canani R, Di Costanzo M, Leone L. The epigenetic effects of butyrate: potential therapeutic implications for clinical practice. Clin Epigenetics. 2012;4:4. doi: 10.1186/1868-7083-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J Bacteriol. 1999;181:5967–5975. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourriaud C, Robins RJ, Martin L, Kozlowski F, Tenailleau E, Cherbut C, et al. Lactate is mainly fermented to butyrate by human intestinal microfloras but inter-individual variation is evident. J Appl Microbiol. 2005;99:201–212. doi: 10.1111/j.1365-2672.2005.02605.x. [DOI] [PubMed] [Google Scholar]
- Bryant MP. The characteristics of strains of Selenomonas isolated from bovine rumen contents. J Bacteriol. 1956;72:162–167. doi: 10.1128/jb.72.2.162-167.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Cummins CS, Johnson JL, Smibert RM, Smith LD, Wilkins TD. Anaerobe Laboratory Manual. V.P.I. Anaerobe Laboratory Virginia Polytechnic Institute and State University: Blacksburg, Virginia, USA; 1977. [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counotte GHM, Prins RA, RHAM Janssen, De Bie MJA. Role of Megasphaera elsdenii in the fermentation of DL-[2-13C]lactate in the rumen of dairy cattle. Appl Environ Microbiol. 1981;42:649–655. doi: 10.1128/aem.42.4.649-655.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabek M, McCrae SI, Stevens VJ, Duncan SH, Louis P. Distribution of β-glucosidase and β-glucuronidase activity and of β-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol Ecol. 2008;66:487–495. doi: 10.1111/j.1574-6941.2008.00520.x. [DOI] [PubMed] [Google Scholar]
- Daly K, Shirazi-Beechey SP. Design and evaluation of group-specific oligonucleotide probes for quantitative analysis of intestinal ecosystems: their application to assessment of equine colonic microflora. FEMS Microbiol Ecol. 2003;44:243–252. doi: 10.1016/S0168-6496(03)00032-1. [DOI] [PubMed] [Google Scholar]
- Del Dot T, Osawa R, Stackebrandt E. Phascolarctobacterium faecium gen. nov, spec. nov., a novel taxon of the Sporomusa group of bacteria. Syst Appl Microbiol. 1993;16:380–384. [Google Scholar]
- Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia municiphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- Dimroth P, von Ballmoos C. ATP synthesis by decarboxylation phosphorylation. Results Probl Cell Differ. 2008;45:153–184. doi: 10.1007/400_2007_045. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Thomson JM, Flint HJ. The role of pH in determining the species composition of the human colonic microbiota. Environ Microbiol. 2009;11:2112–2122. doi: 10.1111/j.1462-2920.2009.01931.x. [DOI] [PubMed] [Google Scholar]
- Ezaki T. Bergey's Manual of Systematic Bacteriology, Volume Three the Firmicutes. Springer: Dordrecht, Heidelberg, London, New York; 2009. Genus VII. Coprococcus Holdemann and Moore 1974, 260AL; pp. 940–941. [Google Scholar]
- Ezaki T. Bergey's Manual of Systematic Bacteriology, Volume Three the Firmicutes. Springer; 2009. Genus I. Ruminococcus Sijpesteijn 1948, 152AL; pp. 1016–1018. [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- Gilmour M, Flint HJ, Mitchell WJ. Multiple lactate dehydrogenase activities of the rumen bacterium Selenomonas ruminantium. Microbiology. 1994;140:2077–2084. doi: 10.1099/13500872-140-8-2077. [DOI] [PubMed] [Google Scholar]
- Gottschalk G. Bacterial Metabolism. Springer Verlag: New York, Heidelberg, Berlin; 1979. [Google Scholar]
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer R- Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- Hetzel M, Brock M, Selmer T, Pierik AJ, Golding BT, Buckel W. Acryloyl-CoA reductase from Clostridium propionicum: an enzyme complex of propionyl-CoA dehydrogenase and electron-transferring flavoprotein. Eur J Biochem. 2003;270:902–910. doi: 10.1046/j.1432-1033.2003.03450.x. [DOI] [PubMed] [Google Scholar]
- Hino T, Kuroda S. Presence of lactate dehydrogenase and lactate racemase in Megasphaera elsdenii grown on glucose or lactate. Appl Environ Microbiol. 1993;59:255–259. doi: 10.1128/aem.59.1.255-259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini E, Grootaert C, Verstraete W, Van de Wiele T. Propionate as a health-promoting microbial metabolite in the human gut. Nutr Rev. 2011;69:245–258. doi: 10.1111/j.1753-4887.2011.00388.x. [DOI] [PubMed] [Google Scholar]
- Huder JB, Dimroth P. Sequence of the sodium ion pump methylmalonyl-CoA decarboxylase from Veillonella parvula. J Biol Chem. 1993;268:24564–24571. [PubMed] [Google Scholar]
- Janssen PH. Growth yield increase and ATP formation linked to succinate decarboxylation in Veillonella parvula. Arch Microbiol. 1992;157:442–445. doi: 10.1007/BF00249102. [DOI] [PubMed] [Google Scholar]
- Kandasamy V, Vaidyanathan H, Djurdjevic I, Jayamani E, Ramachandran KB, Buckel W, et al. Engineering Escherichia coli with acrylate pathway genes for propionic acid synthesis and its impact on mixed-acid fermentation. Appl Microbiol Biotechnol. 2013;97:1191–1200. doi: 10.1007/s00253-012-4274-y. [DOI] [PubMed] [Google Scholar]
- Kilner J, Waby JS, Chowdry J, Khan AQ, Noirel J, Wright PC, et al. A proteomic analysis of differential cellular responses to the short-chain fatty acids butyrate, valerate and propionate in colon epithelial cancer cells. Mol BioSyst. 2012;8:1146–1156. doi: 10.1039/c1mb05219e. [DOI] [PubMed] [Google Scholar]
- Kumar Bhat R, Berger S. New and easy strategy for cloning, expression, purification, and characterization of the 5S subunit of transcarboxylase from Propionibacterium f. shermanii. Prep Biochem Biotechnol. 2007;37:13–26. doi: 10.1080/10826060601039394. [DOI] [PubMed] [Google Scholar]
- Liu C, Finegold SM, Song Y, Lawson PA. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae. Int J Syst Evol Microbiol. 2008;58:1896–1902. doi: 10.1099/ijs.0.65208-0. [DOI] [PubMed] [Google Scholar]
- Lombard J, Moreira D. Early evolution of the biotin-dependent carboxylase family. BMC Evol Biol. 2011;11:232. doi: 10.1186/1471-2148-11-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJM, Garcia-Gil LJ, Flint HJ. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol. 2012;78:420–428. doi: 10.1128/AEM.06858-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Duncan SH, McCrae SI, Millar J, Jackson MS, Flint HJ. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol. 2004;186:2099–2106. doi: 10.1128/JB.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ Microbiol. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- Lyons E, Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Gibson GR. Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine. Chapman and Hall: London; Gastrointestinal Microbiology. 1997;Vol. I:269–318. [Google Scholar]
- Macy JM, Probst I. The biology of gastrointestinal bacteroides. Annu Rev Microbiol. 1979;33:561–594. doi: 10.1146/annurev.mi.33.100179.003021. [DOI] [PubMed] [Google Scholar]
- Marchandin H, Teyssier C, Campos J, Jean-Pierre H, Roger F, Gay B, et al. Negativicoccus succinicivorans gen. nov., sp. nov., isolated from human clinical samples, emended description of the family Veillonellaceae and description of Negativicutes classis nov., Selenomonadales ord. nov. and Acidaminococcaceae fam. nov. in the bacterial phylum Firmicutes. Int J Syst Evol Microbiol. 2010;60:1271–1279. doi: 10.1099/ijs.0.013102-0. [DOI] [PubMed] [Google Scholar]
- McIntosh FM, Maison N, Holtrop G, Young P, Stevens VJ, Ince J, et al. Phylogenetic distribution of genes encoding β-glucuronidase activity in human colonic bacteria and the impact of diet on faecal glycosidase activities. Environ Microbiol. 2012;14:1876–1887. doi: 10.1111/j.1462-2920.2012.02711.x. [DOI] [PubMed] [Google Scholar]
- Miyazaki K, Martin JC, Marinsek-Logar R, Flint HJ. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B(1)4. Anaerobe. 1997;3:373–381. doi: 10.1006/anae.1997.0125. [DOI] [PubMed] [Google Scholar]
- Morrison DJ, Mackay WG, Edwards CA, Preston T, Dodson B, Weaver LT. Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate. Br J Nutr. 2006;96:570–577. [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey FA, Hollen BJ, Small A. Bergey's Manual of Systematic Bacteriology, Volume Three the Firmicutes. Springer: Dordrecht, Heidelberg, London, New York; 2009. Genus I. Clostridium Prazmowski 1880, 23AL; pp. 738–828. [Google Scholar]
- Richardson AJ, Calder AG, Stewart CS, Smith A. Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography. Lett Appl Microbiol. 1989;9:5–8. [Google Scholar]
- Satokari RM, Vaughan EE, Akkermans ADL, Saarela M, de Vos WM. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2001;67:504–513. doi: 10.1128/AEM.67.2.504-513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena RK, Anand P, Saran S, Isar J, Agarwal L. Microbial production and applications of 1,2-propanediol. Indian J Microbiol. 2010;50:2–11. doi: 10.1007/s12088-010-0017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KP, Martin JC, Campbell G, Mayer C-, Flint HJ. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium ‘Roseburia inulinivorans'. J Bacteriol. 2006;188:4340–4349. doi: 10.1128/JB.00137-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger S, Janssen PH, Schink B. Energetics and kinetics of lactate fermentation to acetate and propionate via methylmalonyl-CoA or acrylyl-CoA. FEMS Microbiol Lett. 2002;211:65–70. doi: 10.1111/j.1574-6968.2002.tb11204.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulven T. Short-chain free fatty acid receptors FFA2/GPR43 and FFA3/GPR41 as new potential therapeutic targets. Front Endocrinol (Lausanne) 2012;3:111. doi: 10.3389/fendo.2012.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinolo MAR, Rodrigues HG, Nachbar RT, Curi R. Regulation of inflammation by short chain fatty acids. Nutrients. 2011;3:858–876. doi: 10.3390/nu3100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt JA, Pencharz PB, Wolever TMS. L-rhamnose increases serum propionate in humans. Am J Clin Nutr. 2004;80:89–94. doi: 10.1093/ajcn/80.1.89. [DOI] [PubMed] [Google Scholar]
- Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ. pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol. 2005;71:3692–3700. doi: 10.1128/AEM.71.7.3692-3700.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Nagai F, Morotomi M. Characterization of Phascolarctobacterium succinatutens sp. nov., an asaccharolytic, succinate-utilizing bacterium isolated from human feces. Appl Environ Microbiol. 2012;78:511–518. doi: 10.1128/AEM.06035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J, Scott KP, Avguštin G, Newbold CJ, Flint HJ. Estimation of the relative abundance of different Bacteroides and Prevotella ribotypes in gut samples by restriction enzyme profiling of PCR- amplified 16S rRNA gene sequences. Appl Environ Microbiol. 1998;64:3683–3689. doi: 10.1128/aem.64.10.3683-3689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.