Abstract
We describe initial steps for interrogating whole genome sequence (WGS) data to characterize the genetic architecture of a complex trait, such as high density lipoprotein cholesterol (HDL-C). We estimate that common variation contributes more to HDL-C heritability than rare variation, and screening for Mendelian dyslipidemia variants identified individuals with extreme HDL-C. WGS analyses highlight the value of regulatory and non-protein coding regions of the genome in addition to protein coding regions.
To date, WGS has been used to uncover mutations giving rise to rare Mendelian disorders1,2, but not to explore the genetic architecture of common complex traits. This study evaluates WGS in a population-based sample of well-phenotyped individuals to assess three major genotype-phenotype relationships in an unbiased, hypothesis-free, and coordinated approach to: (1) estimate the relative contribution of common and rare variation to the heritability of a quantitative trait relevant to health and disease; (2) identify individuals who carry variants causing Mendelian disease and analyze the effects of these variants on phenotypes in apparently asymptomatic individuals; and (3) determine the relative value of regulatory and non-protein coding regions of the genome compared with the protein coding portions.
We observed more than 25 million variants from sequencing 962 individuals (Supplementary Table 1) at 6-fold average depth and annotated genomic variation across regions and functional domains (see Supplementary Information). The largest proportion of observed variants was intergenic (58%), and 6% of variants were in annotated functional domains, with non-coding RNAs (ncRNAs) housing the largest fraction (Supplemental Table 2). Of the 252,764 exonic variants identified, 154,133 (61%) were either non-synonymous, stop-gains, or stop-losses. Common variation (MAF>1%) represented 35.7% of the ~25 million variants identified (Supplementary Table 3). The proportion of rare variation (MAF<1%) was 70.8% in exonic regions (UTR and coding), 65.3% in intronic regions, and 64.4% in intergenic regions.
To illustrate several analytic approaches for WGS analyses of a complex trait, we selected HDL-C, a health-related phenotype that is widely and accurately measured. High HDL-cholesterol is associated with a reduced risk of cardiovascular disease,3 but to date the role of HDL-raising therapies in lowering disease risk has not been established. Based on the correlation among twin and other relative pairs, the estimated heritability of HDL-C is 47% to 76% (Supplementary Table 4). We extended Yang et al.’s linear mixed models4,5 to estimate the contribution of variants with different MAF bins to the heritability. For each MAF bin used for estimating the genetic relationship matrix in the linear mixed model, we adopted an adjustment threshold, θ, specifically matching that MAF bin, to account for the distribution of unobserved causal variants. We validated our method by computer simulation (details of the estimation method and simulation validation can be found in the Online Methods). Of the whole genome sequence data reported here, we estimate that common variants (MAF > 1%) explain 61.8% (standard error 14.2) of the HDL-C variance and rare variants (MAF < 1%) explain an additional 7.8% (standard error 9.8) of the variance (Supplementary Table 5). In the estimate of variance explained by each chromosome (Supplementary Fig. 1), there was a positive correlation with both the chromosome length (r=0.37) and the total number of common variants observed on each chromosome (r=0.40).
We observed 1,243 variants previously reported to cause Mendelian disorders in the Human Gene Mutation Database (HGMD-DM).6 Altered HDL-C levels were a published characteristic (www.omim.org) for 10 of these variants in 4 genes (Supplementary Table 6), each of which was validated by an independent genotyping technology. For each of the 10 variants, a Wilcoxon rank sum test was conducted and none of these variants were significant based on a Bonferroni adjusted significance threshold of p = 0.005. Figure 1 highlights three examples representing diverse clinical presentation among individuals carrying known pathologic variants. One individual is heterozygous for a variant in ATP-binding cassette, sub-family A member 1 (ABCA1) that was first reported in a large kindred with Tangier disease7 whose HDL-C lies in the low extreme of the distribution (8.1 percentile, Wilcoxon rank sum p-value = 0.07). Variable penetrance is demonstrated in two individuals with an ABCA1 intronic variant first reported in a kindred with hypoalphalipoproteinemia and premature coronary disease.8 One of the individuals has very low HDL-C (4.6 percentile) while the other has less severely reduced levels (31.1 percentile, Wilcoxon rank sum p-value for both individuals = 0.06). Two individuals are heterozygous for the same mutation in apolipoprotein A-I (APOA1) that was first reported in a patient with severe hypoalphalipoproteinemia.9 Both present with low HDL-C levels (the 10.1 and 6.2 percentile, Wilcoxon rank sum p-value = 0.02).
Figure 1.

HDL-C distribution for carriers of identified Mendelian variation. HDL-C levels are adjusted for age, sex, body mass index, and cohort field center. Dotted lines indicate the 10th, 50th, and 90th percentiles.
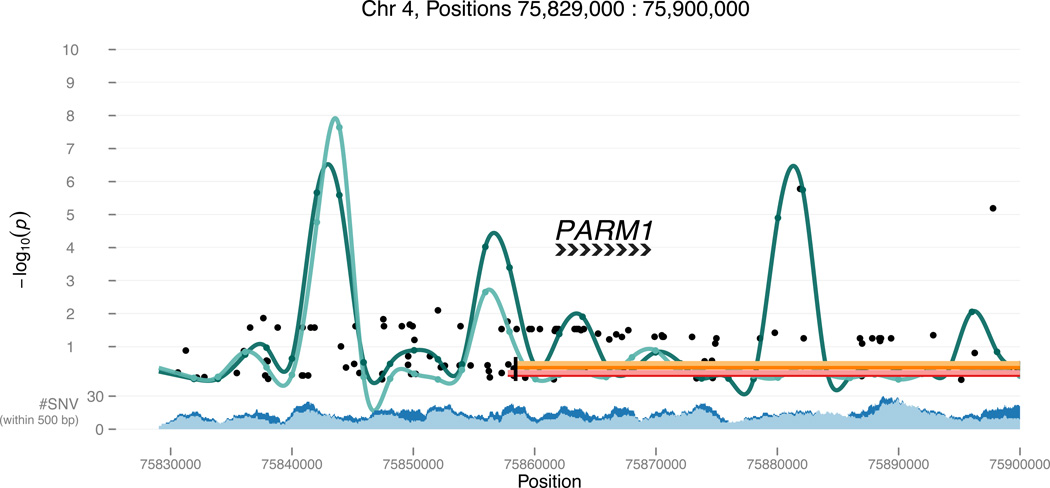
Our approach to evaluating phenotype-WGS association allows for a global survey of the entire genomic landscape, complemented by an annotation-based assessment of the genome. Results for two statistically significant regions of the genome are shown in Figure 2A and 2B, which respectively include the annotated regulatory unit (SKAT rare variant test p=1.09×10−7) and sliding window (T1 rare variant test p=2.69×10−8) with the strongest association. Results for the entire genome are available at http://www.chargeconsortium.com/main/lachesis along with a tool, Lachesis, to view any region of interest. Three types of results are shown: single SNP analyses (black dots), a sliding window aggregating the contribution of rare variants (green lines), and burden tests for a priori annotated regions (orange and red lines). Supplementary Table 7 summarizes the number of tests that were conducted for each type of analysis.
Figure 2.
Survey of the genomic landscape using Lachesis.
Global assessment of the genome is accomplished by interrogating common variation in a comprehensive genome-wide association study (super-GWAS)(black dots in Figure 2). Additionally, a sliding window of physical distance (4 kb) was used to evaluate the aggregate contribution of rare (T1, light green line) and all (SKAT, dark green line) variation across the entire genome. Annotation-based analyses at the gene level contrast the contribution to phenotypic variation from protein-coding variation (exon only, orange lines) and the addition of gene-based regulatory regions (exon +, red lines). Non-coding regions, such as miRNAs and lincRNAs, and annotated regulatory regions (ORegAnno15) throughout the genome were also evaluated for phenotypic association with the aggregate contribution of rare and common variation in these regulatory domains. Other annotations can be added to these analyses as they emerge.
As an illustration of these data, a snapshot of the genomic contribution to inter-individual variation in HDL-C is displayed in Figure 2A for a region on chromosome 16 encompassing the cholesteryl ester transfer protein (CETP) gene. WGS illustrates the contribution of variation beyond that previously identified from genome-wide association studies for HDL-C (e.g., index variant rs3764261 located 5’ of CETP10). Prior work indicates that CETP is likely subject to regulatory variation that affects both transcription initiation and inter-individual splicing of exon 9.11 WGS shows the full array of common variation contributing to HDL-C in the 5’ region of CETP, and highlights a CEBPβ transcription factor binding site, (OREG0001947). This domain at position 56995236 coincides with rs1800775, the −629 variant previously suggested as functional via allelic reporter gene and gel shift assays.12 A second set of regulatory elements (OREG0023819) reside in intron 2 and span a region from 56998238 to 56998689 encompassing a novel variant at 56998522 bp. The intron 2 regions are robustly supported by ENCODE data including histone markers, DNAse I hypersensitivity sites and TF ChIP-seq experiments.
A feature of the ability to scan the genome using the sliding window approach is the capability of agnostically identifying potentially informative genomic regions in the absence of prior biologic information. A region on chromosome 4 near PARM1 presented in Figure 2B was identified by a sliding window test that counts the number of alternate alleles across variants with MAF <1% (T1) and a Sequence Kernel Association Test (SKAT) that considers the contribution of all variation within the window. The association signals are near a transcript of uncertain coding potential (TCONS_00008503), and markers of histone and transcription factor activity. The presumed promoters of prostate androgen-regulated mucin-like protein 1 precursor (PARM1) and TCONS_00008503 appear to lie in head-to-head formation indicating potential co-regulation.
The results presented here document the tractable nature of whole genome sequence analysis for revealing the genetic architecture of common complex phenotypes in humans. We have opted to control costs while still obtaining a large sample size by targeting a depth of coverage that yields a high probability of identifying variants contributing to the genetic architecture of human disease. A sample size of 1,000 individuals at 6-fold average coverage yields a high probability (>98%) of detecting variants down to a frequency of 0.5%.13 We have taken a multifaceted analytical approach that is meant to establish a starting point for future investigation of other phenotypes, larger sample sizes, and updated models of genome function. By using whole genome sequencing instead of genome-wide association markers or candidate gene sequencing, we are able to obtain an unbiased glimpse of the relative contribution of rare and common variation to the heritability of a model trait. The results indicate that the majority (i.e. 61.8%) of the heritability of HDL-C can be attributable to common variation. Given the results of GWAS and targeted resequencing, these common variants likely represent true polygenic variation with small effects, which are of limited diagnostic utility but may be important for identifying novel biologic pathways. The results show that from the entire distribution of randomly sampled individuals, a portion of them in the tails of the phenotype distribution have rare variants with large effects14, and some of these rare variants are previously identified Mendelian conditions. The approach and the data also demonstrate the value added of whole genome sequencing beyond that afforded by whole exome sequencing. Clearly, there are emerging challenges and opportunities for annotating and integrating the functional and public health anatomies of the human genome.
ONLINE METHODS
Description of whole genome sequencing and calling
Library construction processes for the Illumina pipeline are fully automated at the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC). This automated pipeline uses the Biomek NX Span 8 liquid handler in tandem with Biomek FX or NX platforms. Established automated steps within the library construction process include DNA aliquoting, end-repair, 5’ adenylation, adaptor ligation, library amplification and sample pooling using the Biomek Span 8 platform. SPRI-bead purification associated with end-repair, nick translation and PCR amplification steps are all performed on Biomek FX and NX platforms. All processes within library construction are fully LIMS integrated for tracking sample identities on reaction plates to prevent sample swaps. LIMS interfacing allows downstream sequencing data to be deconvoluted, with reads being assigned to the appropriate barcodes/libraries/samples during analysis. To date, over 15,000 libraries have been completed using these automated methods for capture and WGS applications with up to 96 sample barcodes now employed for multiplexing. For this project, Illumina PE libraries were barcoded with standard Illumina multiplex adaptors and pooled for sequencing in sets of three samples to generate an average of 6-fold sequence coverage per sample.
Methods for WGS sequencing followed standard Illumina PairEnd library protocols with minor modifications. DNA concentration was determined by pico green assays while DNA integrity was determined through Agilent Bioanalyzer traces and agarose gels. Libraries were constructed using 1ug of genomic DNA in 100ul volume and sheared into fragments of approximately 300 base pairs in a Covaris plate with E210 system (Covaris, Inc. Woburn, MA). The setting was 10% Duty cycle, Intensity of 4, 200 Cycles per Burst, for 120 seconds. Fragment size was checked using a 2.2 % Flash Gel DNA Cassette (Lonza, Cat. No.57023). The fragmented DNA was end-repaired in 90ul total reaction volume containing sheared DNA, 9ul 10− buffer, 5ul END Repair Enzyme Mix and H2O (NEBNext End-Repair Module; Cat. No. E6050L) and then incubated at 20°C for 30 minutes. A-tailing was performed in a total reaction volume of 60ul containing end-repaired DNA, 6ul 10− uffer, 3ul Klenow Fragment (NEBNext dA-Tailing Module; Cat. No. E6053L) and H2O followed by incubation at 37°C for 30 minutes. Illumina multiplex adapter ligation (NEBNext Quick Ligation Module Cat. No. E6056L) was performed in a total reaction volume of 90ul containing 18ul 5− buffer, 5ul ligase, 0.5ul 100uM adaptor and H2O at room temperature for 30 minutes. After ligation, PCR was performed with Illumina PE 1.0 and modified barcode primers in 170µl reactions containing 85 2− Phusion High-Fidelity PCR master mix, adaptor ligated DNA, 1.75ul of 50uM each primer and H2O. The standard thermocycling for PCR was 5’ at 95°C for the initial denaturation followed by 6–10 cycles of 15 s at 95°C, 15 s at 60°C and 30 s at 72°C and a final extension for 5 min. at 72°C. Agencourt® XP® Beads (Beckman Coulter Genomics, Inc.; Cat. No. A63882) was used to purify DNA after each enzymatic reaction. After bead purification, PCR product quantification and size distribution was determined using the Caliper GX 1K/12K/High Sensitivity Assay Labchip (Hopkinton, MA, Cat. No. 760517). Mean depth of coverage was 6.2-fold, with minimum coverage of 4.0-fold and maximum coverage of 17.4-fold. Six-fold average coverage for each individual was accomplished through barcoding of each sample during library generation followed by a 3-plex sample pooling strategy for instrument loading. Each pool was then loaded into two HiSeq lanes and then data for individual samples merged to generate the 6-fold coverage (~24Gb/sample). Mapping was performed using the Burrows-Wheeler Aligner (BWA)16 with reference genome GRCh37 (hg19).
The SNPTools (http://www.hgsc.bcm.tmc.edu/cascade-tech-software_snp_tools-ti.hgsc) pipeline was developed at BCM-HGSC for single nucleotide variant (SNV) calling. SNPTools is an integrative pipeline which can achieve high quality for (1) variant site discovery, (2) genotype likelihood estimation, and (3) genotype/haplotype inference via imputation. The SNPTools algorithm includes a variance ratio statistic for the initial site discoveries. This statistic compares the difference in variation contributed from the variant read coverage (in case that they are true positives), and the variation that would be due to sequencing and mapping errors (in case that they are false positives). The larger the variance ratio statistic, the more likely the site is a true polymorphic site. In the CHARGE WGS dataset, we applied the default cutoff of 1.5 for the variance ratio statistic. SNPTools next estimates the genotype likelihoods (GLs) by clustering all candidate sites within each particular BAM to overcome data heterogeneity using a mixture model. This step is named the “BAM-specific Binomial Mixture Model (BBMM).” BBMM normalizes the heterogeneity within each sample BAM by estimating BAM and sample specific parameters. It takes into consideration all the variant sites identified from the sample collection (~25 million sites in the final sample of 962 individuals), and clusters across sites in one particular BAM to estimate the GLs. By clustering the scale read depth across millions of sites, it substantially reduces the variance and improves the accuracy in the GL estimation. The GLs are used by the imputation engine to refine the individual genotypes and phase haplotypes.
WGS quality control and quality assurance
Quality control and quality assurance checks were performed on the WGS using the VCF with imputed genotypes from SNPTools. The procedures described below were completed for all individuals from all cohorts. Pair-wise identity-by-state (IBS) was evaluated by PLINK17 to identify pairs of individuals with greater than expected allele sharing. Individuals with excess IBS were consistent across all chromosomes considered, and segregated from the rest of the population. A cutoff of 0.94 pair-wise IBS was used to identify pairs of individuals showing greater than expected allele sharing. This threshold identified 25 pairs of individuals from FHS with known familial relationship (i.e., 9 parent-offspring pairs, 4 sibling pairs, 7 avuncular pairs, 1 first cousin pair, and four unrelated pairs). Two FHS individuals are represented in two separate pairs. From each set of individuals, the sample with the lower WGS coverage was removed from further analysis. An additional three pairs that were identified were comprised of a set of two individuals from ARIC and two sets of ARIC-CHS pairs. From each pair of individuals, the sample with the lower WGS coverage was removed from further analysis. Based on evaluation of IBS, a total of 26 individuals (23 FHS, 2 ARIC, 1 CHS) were removed.
Principal components analysis (PCA) was used to identify possible population substructure and sample abnormalities. The set of variants for PCA was restricted to variants with MAF > 5% and linkage disequilibrium between variants of r2<0.30. Consistently, two individuals from FHS were identified as outliers by PC1 and were removed from further analyses. The two FHS individuals identified by PCA were also outliers for singleton counts and total number of heterozygous counts and were removed from analyses. Higher order PCs showed minor levels of population structure. Following sample-level QC, a total of 962 individuals (404 ARIC, 237 CHS, and 321 FHS) were available for the genotype-phenotype analyses reported here.
Variant-level quality assurance was achieved by a comparison of WGS with exome sequence available on 886 of the 962 individuals. Comparing SNPs identified in 886 overlapping samples having high coverage whole exome capture sequence (average coverage = 115− per sample), the overall genotype concordance rate was higher than 99% across the spectrum of MAF. The rediscovery probability in these data for sites with MAF>0.5% and a sample size of ≥500 is greater than 95%. Even at MAF=0.2–0.5%, the rediscovery probability remained as high as 80%, and ~45% at MAF=0.1%. The rate improves with increased sample size, and therefore our results provided a lower bound of rediscovery rate for the 962 samples. The concordance rate between the WGS and GWAS array data was 99%. When compared with the deep sequencing exome data from the same set of individuals, we estimated that SNPTools has effectively discovered more than 95% of the variants with MAF>0.5%, whereas the estimated discovery power for singletons and doubletons is limited to 45%.
Heritability Estimation
The genetic relationship matrix (GRM) of all individuals A* was estimated using a mixed linear model.4 Specifically, , the adjusted relationship between individual i and j, was calculated as
where b is an empirical adjustment parameter related to the number of SNVs used for estimation and the minor allele frequency (MAF) bins (to adjust the difference between the SNVs used and the unobserved distribution of true causal SNVs, see Yang et al.4 for details). And
is an unadjusted estimation of the relationship between individual i and j based on N SNVs, where xik is the counting (0, 1 or 2) of a specific allele (e.g. the minor allele) of the k-th SNV of the i-th individual, and pk is the frequency of that allele in the whole sample. Estimating b depends on a MAF threshold θ for unobserved causal variants. Throughout our analysis, we matched the θ(s) with the MAF bin(s) of the actual variants used for Aij estimation. For example, to adjust the Aij based on SNVs with 0.01 < MAF ≤ 0.05, 0.01 < θ ≤ 0.05 was used.
To estimate the heritability explained by common autosome SNVs (MAF>0.01), a mixed linear model y = Xβ + gc + ε was defined, where y is the phenotype vector, β is the vector of fixed effects (we used age, gender, BMI, and study center for our analysis) and gc is the vector of cumulated genetic effects explained by the common SNVs. The vector gc is assumed to follow where is the GRM estimated using common autosome SNVs. The heritability was estimated as using the restricted maximum likelihood (REML) method18, where is the phenotypic variance.
To partition into chromosomes, GRM ( for chromosome l) was estimated separately using selected SNVs (e.g. those with MAF>0.01) from each chromosome. Then REML was used to analyze the linear model, , where gl is the vector of cumulated genetic effects explained by the selected SNVs on chromosome l and .5 The contribution of each chromosome to the total heritability was estimated as . Pearson’s correlation coefficient between and chromosome length (or the number of total and common SNVs on the chromosome) was estimated and t-test was used to estimate the significance of the correlation.
The GRMs were estimated using the SNVs of different MAF bins. The corresponding linear model was defined as y = Xβ + Σigbi + ε, where gbi is the vector of cumulated genetic effects explained by the SNVs of the ith MAF bin, with . Finally, REML was used to estimate and its contribution to the total .
For each σ2 estimated above, the standard error of the estimation was calculated based on the Fisher information matrix, which however produced lower standard errors than the empirical standard deviations in simulations. Standard error of hc was approximated using equation (A1.19b) of Lynch and Walsh.19
Validation for estimates of genetic variance accounted for by rare variants using simulation
To validate our extension of Yang et al.’s4 method for estimating the genetic variance of rare variants, we conducted a simulation experiment. To reduce computational burden, we randomly selected 100,000 SNPs per chromosome from the observed 22 autosome genotypes of the 962 individuals (totally 2,200,000 SNPs) as the observed genotypes. We simulated the phenotypes of the 962 individuals with (i) high-coverage scenario: randomly selecting 5,000 SNPs as the causal SNPs; and (ii) low-coverage scenario: randomly selecting 5,000 SNPs as the causal SNPs and then randomly remove some of the SNPs mimicking the reduced discovery rate for the rare variants at 6-fold coverage. We used an empirical function for discovery rate: ,i = 1,2,…,20, where i is minor allele count (MAC) bin. For example, singletons (MAC=1) discovery rate is 0.34, i.e. 66% missing; doubletons (MAC=2) discovery rate is 0.48, i.e. 52% missing. There is no missing for variants with MAC>20. This empirical function is based on comparing high-coverage (average 115−) exome data of 886 individuals (from the 962 individuals) to the low-coverage whole genome sequencing data of the same region of the same individuals. We generated the effects of causal variants (u) from a uniform distribution (all effects are the same, e.g. ui=1 whatever the frequency of the variant) and calculated the genetic score of each individual by , where ur and uc are the effect sizes of rare and common variants respectively,nr and nc are the total number of rare and common variants respectively, and z is coded as 0, 1 or 2 for genotype qq, Qq or QQ. We generated residual non-genetic effects (e) from normal distribution with mean of 0 and variance of Vg(1−h2)/h2, where Vg is the empirical variance of genetic score, and h2 is the heritability. We calculated the phenotypic value of each individual by y = g + e. The total h2 was set at 50% and the effects of each rare variants as 1, 2, and 4. All the effects for common variants are set as 1.
Each simulation was repeated 10 times and the means and standard deviations of the true and estimated genetic variance due to rare variants are shown in Supplementary Figure 2. Across all scenarios, the averages of the estimated genetic variances due to rare variants were not significantly different to the corresponding true genetic variances simulated, although larger standard deviations were observed for the scenarios with larger genetic variances. This was also observed for genetic variances due to common variants (data not shown). Compared to high-coverage scenarios, low-coverage scenarios show no significant bias.
To investigate the impacts of SNP density, assumed effect sizes of causal loci and different MAC separation on our simulation, we conducted additional simulations with following assumptions: (1) We selected all the SNPs from chromosome 1 and 2 that passed quality control (totally 4,147,948 SNPs) as the observed genotypes, instead of sampling 100,000 SNPs per chromosome; (2) The jth causal locus has an effect size so that each causal locus has equal contribution to the heritability; (3) SNPs were separated into three MAC bins: rare (MAC≤ 0.01), low frequency (0.01<MAC≤ 0.05) and common (0.05<MAC); and (4) The total heritability was assumed to be 80%. Again ten replications were simulated. Supplementary Figure 3 shows the means and the standard deviations of the true and estimated genetic variances based on the ten replications. In general, the estimated genetic variances matched the true values for all three MAC bins and for both high-coverage and low-coverage scenarios, suggesting our estimating method is robust to SNP density, assumed effect size and separation of MAC bins.
Identification of Mendelian Variation
To identify known Mendelian variants influencing HDL-C, we used the strengths of the Human Gene Mutation Database (HGMD) and the Online Mendelian Inheritance in Man (OMIM20) catalogue. This approach had two stages: first identifying all Mendelian variants within our cohort, then applying a clinically-derived gene-based filter to the set of annotated sequences.
A complete list of HGMD (version 2011.4) single nucleotide substitutions were annotated using ANNOVAR.21 Among the 25,124,797 SNVs observed in the 962 individuals, a total of 3,920 SNVs matched HGMD, of which 1,688 were designated as disease-causing (DM) alleles. We then removed variants with only association or dubious evidence for pathogenicity as indicated in the HGMD disease field, reducing the observed set of HGMD-DM variants to 1,243.
Independently, we queried OMIM for HDL-C related terms (e.g., “HDL”, "Hyperalphalipoprotein”, and “Hypoalphalipoprotein") to identify genes causing Mendelian disorders influencing HDL-C levels. The intersection of this gene filter with the set of HGMD-DM variants, identified 14 variants in our cohort. After reviewing the OMIM clinical descriptions of the corresponding disorders, we removed four variants not directly related to HDL-C levels. Supplementary Table 6 describes the carriers of the 10 HGMD-DM variants related to HDL-C and their percentile within the HDL-C sample distribution adjusted for age, sex, body mass index, and cohort field center. All 10 variants were technically validated in the 21 individuals that carry them by either capture-based exome sequencing, Sequenom genotyping, or both.
Super-GWAS
All SNVs with MAF>1% were evaluated for association with HDL-C, assuming an additive genetic model. Age, gender, BMI, and study center were included as covariates. A total of 8,969,552 SNVs were tested. The Manhattan and Q-Q plot for the super-GWAS for HDL-C are shown in Supplementary Figure 4 and 5.
Genome scanning by sliding window
The sliding window across the genome considered the aggregate contribution of variants to the trait. Supplementary Figure 6 shows the composition of the 1.33 million windows, covering 2.7 billion autosomal base pairs. The median number of common (MAF > 1%) and rare (MAF < 1%) SNVs in a window were 12 and 28 respectively (IQR 8–17 and 19–28). Sliding windows were of size 4 kb, began at position 0 bp for each chromosome, and the skip length was 2 kb.
Primary considerations for the choice of tests was: (1) to represent a test with an ability to be statistically powered for the scenario where all variants in the defined window have effects in the same direction as well as a test that allows for omni-directional effects of the variants, (2) ease of calculation and implementation, including computational speed, (3) application to quantitative and qualitative outcomes. The T1 test generates a statistic for each person that counts the number of variant alleles across SNVs with MAF <1%. The summary statistic for each person is used in the linear regression model and a Wald test was used to assess significance. SKAT performs a score test for the model that includes all variants within the window.22 This allows for heterogeneous effect of the variants within a window, but has lower power for homogeneous effects relative to T1. For linear regression, the score test comes from the model:
where Xik = kth covariate for subjects i (i=1, …, n)
Gij = jth genotype for subject i, j=1 to p
α0,αk and βj are regression parameters
SKAT is a weighted sum of the score tests for each individual variant, where the weights are specified by the user. For testing the null hypothesis H0: β=0, SKAT takes a simple form. Let gij be the genotype of the jth variant for the ith person. For a given set of weights, wj for each variant j = 1 to p, the score test can be expressed as
μ̂i0 is the predicted value of yi from the model when there are no genotypes in the model
We chose wi = f(MAF(gi)), where f(x) is the density of a Beta(1,25) distribution, which gives more testing weight to rare variants relative to common variants.
Q-Q plots for the sliding window evaluation of the aggregate contribution of variants to HDL-C are shown in Supplementary Figure 7 for T1 (a) and SKAT (b).
Sources of regulatory and non-coding annotation
All annotations were obtained with positional mapping to the human genome build GRCh37 (hg19). The Open REGulatory ANNOtation database (ORegAnno) (http://www.oreganno.org/, accessed March 2012) provided a resource of 23,119 regulatory assays mapped to the human genome, with corresponding literature citations.23 Human long intergenic non-coding RNAs (lincRNAs) (n=8,195) were taken from a catalog based on RNA-sequencing of 24 human tissues and cell types24 and subject to additional padding as with RefSeq genes. Regulatory and non-coding annotation sources were evaluated with SKAT and T1 (Supplementary Figures 8 and 9) on the basis of variants from whole genome sequencing that intersected with the individual annotation entries.
Like the sliding window across the genome, annotation-based analyses considered the aggregate contribution of variants within a defined region. Gene-based methods contrasted the contribution to phenotypic variation from protein-coding variation and the addition of gene-based regulatory regions. Gene regions were annotated using the RefSeq database25, including definition of exon boundaries. A total of 216,362 exonic regions were captured across the genome in a total of 23,357 genes. The gene-based regulatory regions were identified using RefSeq as follows: 500 base pairs upstream (5-prime) from exon 1 and downstream (3-prime) from the last exon, the first 500 base pairs of intron 1, and 10 base pairs into each intron for the identification of splice sites. The aggregate contribution of variants to HDL-C was determined within exon-only (Supplementary Figure 10) and gene-based regulatory regions (Supplementary Figure 11) using T1 and SKAT.
Significance thresholds and false positive control
Correlation between tests will influence the joint behavior of p-values. However, we note that a significance threshold controls the expected number of false-positives regardless of the correlation between tests.26 For instance, the sliding window analysis was comprised of two tests on each of 1.3 million windows, for a total of 2.6 million tests. If one were to use a significance threshold of p=4.0×10−7, the number of expected false positives among all sliding window tests would be less than 1.05. We have refrained from specifying precise significance thresholds, as acceptable false positive rates will differ depending on the nature of follow-up studies. However, we recommend using different significance thresholds for each analysis to account for the differing number of tests used. In this way, the results from the gene-based tests are not hidden by the large number of super-GWAS tests. The study-wide false positive rate is then less than the sum of the expected number of false positives from each analysis. Supplementary Table 7 summarizes the number of tests that were conducted for each type of analysis.
Supplementary Material
ACKNOWLEDGMENTS
Atherosclerosis Risk in Communities (ARIC) Study: This ARIC study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C).
The authors thank the staff and participants of the ARIC study for their important contributions. Ancillary study support has been provided by National Heart, Lung, and Blood Institute sponsored project RC2HL102419-02.
Cardiovascular Health Study: This CHS research was supported by NHLBI contracts N01-HC-85239, N01-HC-85079 through N01-HC-85086; N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, HHSN268201200036C and NHLBI grants HL080295, HL087652, HL105756 with additional contribution from NINDS. Additional support was provided through AG-023629, AG-15928, AG-20098, and AG-027058 from the NIA. See also http://www.chs-nhlbi.org/pi.htm.
Framingham Heart Study of the National Heart, Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. This work was supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (contract N01-HC-25195).
AUTHOR DECLARATION
B.M.P. serves on the DSMB for a clinical trial of a device funded by the manufacturer (Zoll LifeCor) and on the Steering Committee of the Yale Open Data Access Project funded by Medtronic.
Footnotes
AUTHOR CONTIRBUTIONS
A.C.M., A.V., A.D.J. and X.L. all contributed equally to this work. J.Y., D.M., F.Y., E.B., and R.G. were responsible for design and implementation of the whole genome sequencing and variant calling. A.L. and K.R. contributed to the analysis of Mendelian variation. X.L. and C.Z. contributed to the estimation of heritability. A.C.M., A.V., and A.D.J. performed statistical analysis of the whole genome sequence and phenotype data. G.H., C.J.O, and B.M.P were involved in participant recruitment, consenting and examination. A.C.M., A.V., A.D.J., X.L., J.B., G.H., C.J.O., B.M.P., L.A.C., R.G., and E.B. jointly conceived the study and contributed to preparation and editing of the manuscript.
References
- 1.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lupski JR, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. New England Journal of Medicine. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castelli WP, et al. HDL cholesterol and other lipids in coronary heart disease. The cooperative lipoprotein phenotyping study. Circulation. 1977;55:767–772. doi: 10.1161/01.cir.55.5.767. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stenson PD, et al. Human Gene Mutation Database: towards a comprehensive central mutation database. J Med Genet. 2008;45:124–126. doi: 10.1136/jmg.2007.055210. [DOI] [PubMed] [Google Scholar]
- 7.Brousseau ME, et al. Novel mutations in the gene encoding ATP-binding cassette 1 in four tangier disease kindreds. J Lipid Res. 2000;41:433–441. [PubMed] [Google Scholar]
- 8.Rhyne J, Mantaring MM, Gardner DF, Miller M. Multiple splice defects in ABCA1 cause low HDL-C in a family with hypoalphalipoproteinemia and premature coronary disease. BMC Med Genet. 2009;10:1. doi: 10.1186/1471-2350-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weers PM, et al. Novel N-terminal mutation of human apolipoprotein A-I reduces self-association and impairs LCAT activation. J Lipid Res. 2011;52:35–44. doi: 10.1194/jlr.M007500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papp AC, et al. Cholesteryl Ester Transfer Protein (CETP) polymorphisms affect mRNA splicing, HDL levels, and sex-dependent cardiovascular risk. PLoS ONE. 2012;7:e31930. doi: 10.1371/journal.pone.0031930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dachet C, Poirier O, Cambien F, Chapman J, Rouis M. New functional promoter polymorphism, CETP/-629, in cholesteryl ester transfer protein (CETP) gene related to CETP mass and high density lipoprotein cholesterol levels: role of Sp1/Sp3 in transcriptional regulation. Arterioscler Thromb Vasc Biol. 2000;20:507–515. doi: 10.1161/01.atv.20.2.507. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21:940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romeo S, et al. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montgomery SB, et al. ORegAnno: an open access database and curation system for literature-derived promoters, transcription factor binding sites and regulatory variation. Bioinformatics. 2006;22:637–640. doi: 10.1093/bioinformatics/btk027. [DOI] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patterson H, Thompson R. Recovery of inter-block information when block sizes are unequal. Biometrika. 1971;58:545–554. [Google Scholar]
- 19.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: 1998. [Google Scholar]
- 20.McKusick-Nathans Institute of Genetic Medicine, editor. Baltimore, MD: Johns Hopkins University; [Accessed May 2012]. Online Mendelian Inheritance in Man, OMIM. www.omim.org. [Google Scholar]
- 21.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu MC, et al. Rare-variant association testing for sequencing data with the sequence kernel association test. American Journal of Human Genetics. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffith OL, et al. ORegAnno: an open-access community-driven resource for regulatory annotation. Nucleic Acids Res. 2008;36:D107–D113. doi: 10.1093/nar/gkm967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon A, Glazko G, Qiu X, Yakolev A. Control of the mean number of false discoveries, Bonferroni and stability of multiple testing. Annals of Applied Statistics. 2007;1:179–190. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.