Abstract
Neurodevelopmental disorders such as intellectual disability, autism spectrum disorder and schizophrenia lack precise boundaries in their clinical definitions, epidemiology, genetics and protein–protein interactomes. This calls into question the appropriateness of current categorical disease concepts. Recently, there has been a rising tide to reformulate neurodevelopmental nosological entities from biology upward. To facilitate this developing trend, we propose that identification of unique proteomic signatures that can be strongly associated with patient's risk alleles and proteome-interactome-guided exploration of patient genomes could define biological mechanisms necessary to reformulate disorder definitions.
Keywords: AP-3, autism spectrum disorder, BLOC-1, DTNBP1, dysbindin, schizophrenia
Neurodevelopmental disorders (NDDs) are multifaceted conditions characterized by impairments in cognition, communication, behavior and/or motor skills resulting from abnormal brain development. Intellectual disability, communication disorders, autism spectrum disorder (ASD), attention deficit/hyperactivity disorder (ADHD) and schizophrenia fall under the umbrella of NDD.1, 2, 3 Currently, there are no biomarkers to diagnose NDD or to differentiate between them. Rather, these disorders are categorized into discrete disease entities, based on clinical presentation.1 This is problematic, as many symptoms are not unique to a single NDD, and several NDDs have clusters of symptoms in common. For example, impaired social cognition is common to ASD and schizophrenia,4, 5, 6, 7 and psychosis is observed not only in schizophrenia but also in those with bipolar disorder or major depressive disorder.8, 9 Thus, such overlap of clinical symptoms presents a challenge for nosology and course of treatment. This is in stark contrast to other disorders, such as cardiovascular diseases, where diagnosis is rooted in biological manifestations, biomarkers and pathophysiology. The diffuse clinical boundaries among NDD calls into question the appropriateness of current disease definitions.10, 11, 12, 13 Here, we advocate reformulating current nosological categories with novel disorder definitions rooted in the biology of processes that are awry in NDDs. We predict that biological disorder definitions will change the way we use symptomology for diagnosis.
Neurodevelopmental disorders: boundary definitions from genomes
The hypothesis that NDDs are distinct nosological entities predicts that genetic factors associated with risk for or causation of a given disorder should segregate with diagnostic categories; thus, in classical terms, there should be little or no overlap among the genetic factors implicated in each NDD. That is, the genes that operate in one disorder should not be involved in another. However, genetic epidemiology reveals substantive overlap between genes conferring risk for or causing NDDs.
Genetic defects associated with risk or causation of NDDs range from large chromosomal deletions to single-nucleotide polymorphisms (SNPs). Notably, among major genomic defects, a number of chromosomal deletions are associated with intellectual disability, ASD and schizophrenia.12, 14, 15, 16 Among the most frequent are 1q21.1, 16p11.2 and 22q11.2.12, 17 The large number of genes affected by these deletions should cause little surprise that they give rise to disorders with overlapping phenotypes. However, smaller genetic modifications, specifically SNPs in non-coding regions, are shared among diverse NDDs.18 Genetic overlap among NDDs extends to monogenic defects that affect the coding sequence and expression of a single polypeptide encoded by the gene (for example, SHANK3, NRXN1, DISC1, FMR1, MECP2, GPHN). Patients carrying these mutations are diagnosed either with intellectual disability, ASD, schizophrenia or combinations of thereof.19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33 Monogenic genetic defects affect subunits of obligated and stable protein complexes. For example, the adaptor complex AP-3 is an obligate heterotetramer that generates vesicles from early endosomes bound to lysosomes/synapses;34, 35 human mutations in a neuronal-specific AP-3 subunit (AP3B2) associate with ASD.36, 37 Thus, irrespective of the size of a genetic defect, there is a continuously expanding list of affected genes that do not respect categorical diagnostic boundaries among NDDs.
Neurodevelopmental disorders: boundary definitions from interactomes
Protein interaction networks (interactomes) to which NDD genes belong also overlap. Interactomes built from genes associated with intellectual disability, ASD, ADHD and schizophrenia converge on common molecular pathways.38 Genes associated with these NDDs intersect on one out of 700 genes catalogued as risk factors. However, the list of common proteins shared by these NDDs increases to 147 out of the 700 genes simply by expanding the gene catalog to include predicted first-degree interacting neighbors obtained from protein–protein interaction databases.38 These computational studies support the concept that the interactomes associated with NDDs overlap. However, the power of these types of studies is limited by the present quality of protein interaction databases, which are incomplete, are only moderately curated to accommodate newly published findings and are often populated by results not confirmed by alternative biochemical, genetic and/or functional approaches.39, 40, 41 Furthermore, protein interaction databases are biased by the experimental approach used in their generation; for example, most protein interaction databases poorly represent membrane proteins that are not amenable to exploration by traditional yeast two hybrid or pull-downs with recombinant proteins.42
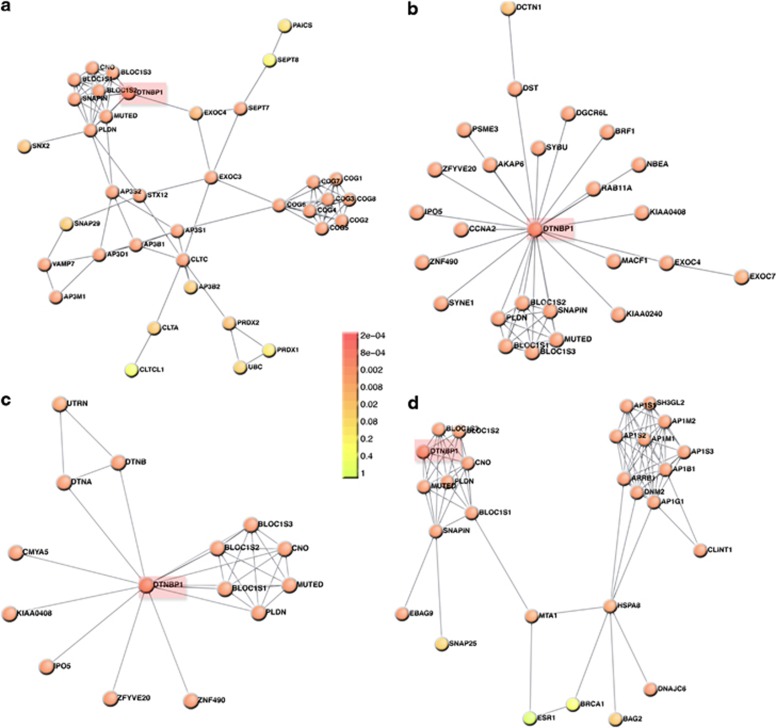
The interactome of the schizophrenia susceptibility gene DTNBP1 well illustrates several of these problems (Figure 1). DTNBP1 encodes dysbindin, a subunit of the BLOC-1 complex.43, 44, 45, 46, 47, 48, 49, 50, 51 This complex participates in membrane protein trafficking between endosomes and lysosomes, and between endosomes located in neuronal cell bodies and the synapse.35, 50, 51 Published in silico dysbindin interactomes52, 53 differ from biochemically and genetically tested protein interaction networks.37 However, discrepancies among interactomes expand beyond those published (Figures 1a and d). Three protein interaction databases report associations that differ from each other in interactor identities. Furthermore, feeding those associations into a rigorous algorithm for ‘de novo' generation of interactomes reveals different network topologies (Figures 1a and d).54 Only one of these four dysbindin interactomes links dysbindin with the adaptor complex AP-3, despite multiple biochemical, cell biology, and genetic evidence that these complexes interact in vivo and in vitro (Figure 1a).55, 56, 57, 58, 59, 60, 61, 62 This deficiency in existing databases has immediate ramifications, as mutations in AP3B2 associated with ASD cannot be linked to schizophrenia through the BLOC-1 subunit dysbindin.36, 37 AP3B2 is not an isolated instance. Rather, only the experimentally defined dysbindin interactome identifies SNAP29 and CLTCL1.37 SNAP29 has been identified as a de novo risk factor for schizophrenia, while both SNAP29 and CLTCL1 map to the chromosome interval affected in velocardiofacial (chromosome 22q11.2 deletion) syndrome.17, 63 This syndrome closely associates with schizophrenia, ASD and intellectual disability.17 Gaps in content and quality in relation to protein interaction databases are important, as these repositories are the foundation for molecular connectivity between genetic defects associated with a given disorder. These deficiencies are missed opportunities for establishing molecular mechanisms of disease and finding mechanistic commonalities among NDDs. Thus, we argue in favor of generating interactomes confirmed by biochemical, genetic and/or functional strategies. Epidemiological genomics offer the field a good selection of solid candidate genes with which to begin this quest.
Figure 1.
DTNBP1–dysbindin interactomes differ in their constituents and topology. Interactomes were assembled with the Dapple algorithm (http://www.broadinstitute.org/mpg/dapple/dapple.php)54 using as inputs the dysbindin associated proteins identified by affinity chromatography (a), and interactors reported in three protein–protein interaction databases: (b) Biogrid (http://thebiogrid.org/), (c) Genemania (http://www.genemania.org/) and (d) String 9.05 (http://string.embl.de/). Red boxes highlight DTNBP1. Note that the identity of interacting proteins differs among interactomes. Color code represents a Dapple estimated probability that a protein would be as connected to other proteins (directly or indirectly) by chance as is depicted. Only interactome A presents a biochemically and genetically confirmed interaction between the adaptor complex AP-3 and the dysbindin-containing BLOC-1 complex.
Neurodevelopmental disorders: ‘guilty by association' mechanisms of disease and their inclusion in interactomes
Loss of one protein function due to a genetic mutation can alter levels or activity of other proteins that interact either directly or indirectly with the mutant protein. These ‘guilty by association' proteins can be the actual culprits of disease phenotypes. The concept is illustrated readily by Marfan syndrome, a connective tissue disorder in which morbidity and mortality are chiefly associated with aortic aneurisms.64, 65 This disease is caused by mutations in the extracellular matrix protein fibrillin-1 (FBN1), which organizes into 10 nm fibers.64, 65 An old pathogenic hypothesis considered that loss of fibrillin fibers decreased blood vessel resilience to mechanical stress.64, 65 However, there is a new conceptualization of this syndrome that pinpoints abnormal TGFβ signal transduction as the main culprit in its vascular pathology. This unexpected shift can be understood from the fact that fibrillin binds and presents TGFβ to its receptor at the optimal concentration, time and location.64, 65, 66 Thus, TGFβ is a ‘guilty by association with fibrillin' protein.
Subunits of protein complexes are particularly susceptible to being ‘guilty by association' proteins. Genetic defects, or even non-pathogenic allelic variation affecting a single subunit of a protein complex, frequently lead to downregulation and/or covariation of other complex subunits.67, 68, 69, 70, 71, 72 DTNBP1 null mutations abrogating dysbindin expression downregulate most subunits of the BLOC-1 complex, despite the monogenic character of the mutation.44, 50, 69 Reciprocally, genetic defects on other BLOC-1 subunits decrease dysbindin cellular content.44, 50 ‘Guilty by association' proteins in the dysbindin interactome extend beyond intrinsic components of the BLOC-1 complex. These proteins include membrane protein cargoes such as VAMP7 (VAMP7), a synaptic vesicle fusogenic membrane protein (SNARE) implicated in spontaneous synaptic vesicle fusion and the Menkes disease copper transporter (ATP7A), the adaptor complex AP-3, RhoGEF1 (ARHGEF1) and BDNF (BDNF), a neurotrophin with a long history of association with several NDDs.57, 59, 73, 74, 75, 76 None of these proteins whose levels are affected by mutations in DTNBP1, or other BLOC-1 complex subunits, can be identified in current protein interaction databases that focus on physical protein–protein interactions. This problem prevents their inclusion in any analysis seeking to connect genetic defects found in genome-wide associations studies to relevant molecular pathology.
Creating a nosology from genome informed proteomes-interactomes
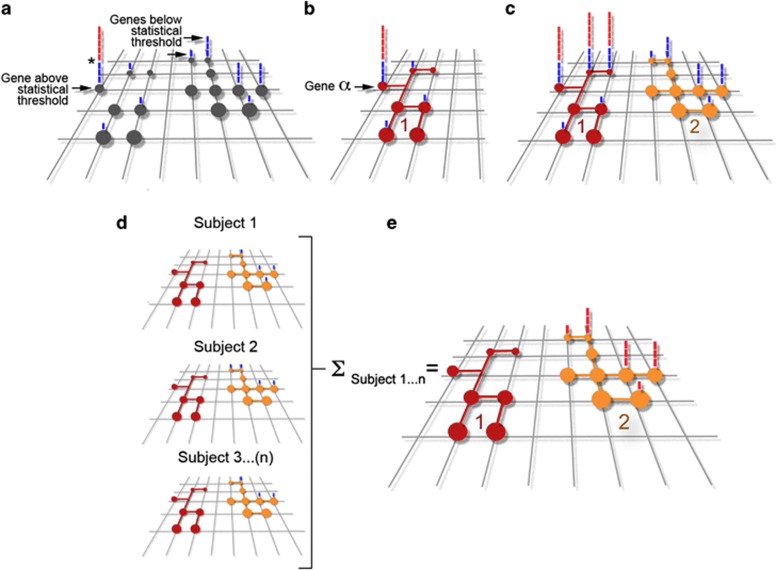
Genome-wide association studies (GWAS) search the genomes of clinically defined patient populations for genetic markers that reach a threshold of statistical significance to associate with disease risk (Figure 2a). This approach encounters the problem that these disorders are polygenic and that categorical NDD definitions are not linked to biological markers or molecular phenotypes.77, 78 Thus, it is likely that genetically heterogeneous patient cohorts in these studies gather multiple molecular mechanisms of disease. However, these studies offer powerful insight when a particular genetic marker reaches statistical significance, despite the ‘noise' introduced by the polygenic character of these disorders and the problems intrinsic to categorical NDD definitions. Genetic defects associated with one or multiple NDD should be seen as the tip of the iceberg to unravel biological mechanisms of disease. Interactomes of gene products consistently implicated in NDDs (‘tip of the iceberg genes') are a fertile ground to search for disease mechanisms.54, 79 This prediction stems from the hypothesis that genomes of patients affected by polygenic NDD should concentrate alleles that affect the expression or function of genes whose products belong to or modulate a relevant pathway.80 We illustrate this concept in Figure 2b where gene-α has reached statistical significance in a population GWAS. The product encoded by gene-α is a bait to ‘fish out' the red protein interaction network (Red interactome B1, Figure 2b). The biochemical definition of interactome 1 would occur irrespective of whether interactome 1 contains products encoded by genes carrying defects that do not cross a population statistical threshold (Figure 2b).
Figure 2.
Models of cross-fertilization between genomes, proteomes and interactomes. Grid in diagrams (a) to (e) depicts a polygenic genetic landscape associated with a NDD. Circles represent defined genes within the grid that when affected in different combinations trigger a NDD. Bars above each gene indicate a subject where a gene defect was found on a GWAS. Blue bars are those subjects that have a defect in a gene below statistical threshold, which is marked by the asterisk in (a). Red bars above a gene represent subjects that have a defect in a gene above statistical threshold. (b) Depicts a ‘tip of the iceberg gene α' and the network to which it belongs represented by the connected red circles (interactome 1). (c) Depicts three ‘tip of the iceberg genes' and the network to which they belong (interactome 1). The yellow interactome 2 is constituted by genes below statistical threshold as defined by gene-centric GWAS statistical analysis. (d) Represents genetic defects (blue bars) in two interactomes per patient (subjects 1–3). Note that in all patients there are no gene defects in the red interactome. E depicts hypothetical results of an interactome-centric GWAS that includes subjects 1–3 in (d). The yellow interactome 2 is now above statistical threshold as defined by an interactome-centric GWAS statistical analysis. See text for details.
This genome to proteome ‘reverse' approach is not foreign to current genomic studies, in which bioinformatics of protein–protein interaction databases are used to find connections between gene defects that associate with a disorder at a GWAS level36, 79 (Figure 2c). However, mapping GWAS results back to an interactome requires the availability of several network genes that cross a statistical threshold (Red interactome 1, Figure 2c) as well as pre-existing and reliable protein interaction databases. Genes below statistical threshold in the red network C1 would not contribute to the identification of the C1 interactome (Red interactome C1, Figure 2c). Moreover, current criteria to allocate GWAS results to an interactome would miss the yellow interactome C2 where genes encoding interactome products are all below statistical threshold (Figure 2c).
How can we obtain mechanistic insight from studying ‘omes'? We propose two non-exclusive approaches to define the biology of NDDs using protein–protein interaction networks and genomics. The first approach is through the definition of ‘tip of the iceberg gene' protein networks, such as those depicted by the red interactomes in Figures 2b and c. Second, reliable protein interactomes can be used as a query matrix to explore patient's genomes for genetic defects or variants targeting interactome-encoding loci. Different patients may carry defects in one or more genes encoding products belonging to an interactome. Each gene defect does not reach statistical significance in a ‘gene-centric' GWAS study (Subject 1–3, Figure 2d). However, collective analysis of the genomes in a cohort of patients (Subject 1–3, Figure 2d) shows significant enrichment of genetic defects clustered on a common pathway (compare red and yellow interactomes, Figure 2e). The association of a biological mechanism, defined by an already known and reliable interactome, with the genome of affected individuals would occur, although each gene in isolation would have not risen above statistical threshold. In this case, statistical significance is assigned to a collection of genes defining an interaction network rather than a single gene (Figure 2e).
These solutions depend on reliable protein interactions networks. As mentioned above, the quality of protein–protein interaction databases commonly used is substandard. This is due to a lack of thorough biochemical, functional and/or genetic confirmation of interactions. We posit that it is possible to extract more information about disease mechanisms and disorder boundaries from current GWAS studies if reliable protein interaction maps were to exist. As these are either not available or they are in construction, we propose to focus efforts on defining the interactomes of (a) NDDs ‘tip of the iceberg genes' as well as (b) ‘guilty by association' proteins detected in the proteomes of cells carrying genetic defects in ‘tip of the iceberg genes'. These and other experimentally confirmed interactomes (yellow interactome 2 in Figure 2e) would allow us to extract novel genetic information from existing and future GWAS.
Creating a genome-independent nosology from proteomes-interactomes
Human proteomes are hereditable molecular phenotypes72 and as such constitute valuable, yet untapped, resources to create disorder classifications rooted in molecules and their pathways. The study of proteomes shares with the analysis of genomes its quantitative and unbiased character. However, proteomes and interactomes offer the distinctive advantage of being executors of phenotypic programs in cells and tissues. Therefore, proteomes and interactomes are causally closer to the identity of disease mechanisms than genomes. Proteomes are already beginning to shed light on complex neurological disorders such as schizophrenia.81, 82 However, we should not limit ourselves to just exploring postmortem brains of subjects grouped solely by their clinical features. Instead, we advocate for the study of proteomes from cells isolated from individuals that are genetically related. Cell proteomes from affected probands compared with their unaffected first-degree relatives offer a great prospect for the identification of hereditable or de novo abnormalities in molecular phenotypes. Evidently, in the context of NDDs, human inducible pluripotent stem cells are a great resource, as they can be differentiated into neurons.83 However, it is likely that the molecular mechanisms affected in NDDs are common to many, if not all cells. For example, Fragile X syndrome or velocardiofacial syndrome, where multiple tissues are affected 12. Thus, fibroblasts or lymphoblasts from human pedigrees are likely to offer valuable insights into neuronal disorders. We predict that proteomes built from genetically related subjects' cells will bridge two camps. On one hand, proteomes will help us to interpret results from genome-wide analyses. On the other hand, they will guide us to define NDD mechanisms at levels of complexity higher than the traditional single genes or proteins. These would include, for instance, subcellular compartments, such as synapses or mitochondria, and deficits in tissue organization, such as those in neural circuits. Genomes, proteomes and interactomes give us vantage points, the inevitable next step is to dive deep into the biology emerging from and converging to them.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM077569 and NS42599) and CHOA Children's Center for Neuroscience to VF, and by Principal Investigator grant 07/IN.1/B960 from Science Foundation Ireland to JLW.
The authors declare no conflict of interest.
References
- American Psychiatric Association . DSM-5, Development. American Psychiatric Association; 2013. [Google Scholar]
- Reiss AL. Childhood developmental disorders: an academic and clinical convergence point for psychiatry, neurology, psychology and pediatrics. J Child Psychol Psychiatr Allied discipl. 2009;50:87–98. doi: 10.1111/j.1469-7610.2008.02046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport JL, Giedd JN, Gogtay N. Neurodevelopmental model of schizophrenia: update 2012. Mol Psychiatr. 2012;17:1228–1238. doi: 10.1038/mp.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz B. Theory of mind and neurodevelopmental disorders of childhood. Pediatr Res. 2011;69 (5 Pt 2:101R–108R. doi: 10.1203/PDR.0b013e318212c177. [DOI] [PubMed] [Google Scholar]
- Sugranyes G, Kyriakopoulos M, Corrigall R, Taylor E, Frangou S. Autism spectrum disorders and schizophrenia: meta-analysis of the neural correlates of social cognition. PloS ONE. 2011;6:e25322. doi: 10.1371/journal.pone.0025322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King BH, Lord C. Is schizophrenia on the autism spectrum. Brain Res. 2011;1380:34–41. doi: 10.1016/j.brainres.2010.11.031. [DOI] [PubMed] [Google Scholar]
- Lugnegard T, Unenge Hallerback M, Hjarthag F, Gillberg C. Social cognition impairments in Asperger syndrome and schizophrenia. Schizophrenia Res. 2013;143:277–284. doi: 10.1016/j.schres.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Owoeye O, Kingston T, Scully PJ, Baldwin P, Browne D, Kinsella A, et al. Epidemiological and Clinical Characterization Following a First Psychotic Episode in Major Depressive Disorder: Comparisons With Schizophrenia and Bipolar I Disorder in the Cavan-Monaghan First Episode Psychosis Study (CAMFEPS) Schizophrenia Bullet. 2013;39:756–765. doi: 10.1093/schbul/sbt075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domschke K. Clinical and molecular genetics of psychotic depression. Schizophrenia Bullet. 2013;39:766–775. doi: 10.1093/schbul/sbt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatr. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- Hyman SE. The diagnosis of mental disorders: the problem of reification. Ann Rev Clin Psychol. 2010;6:155–179. doi: 10.1146/annurev.clinpsy.3.022806.091532. [DOI] [PubMed] [Google Scholar]
- Moreno-De-Luca A, Myers SM, Challman TD, Moreno-De-Luca D, Evans DW, Ledbetter DH. Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol. 2013;12:406–414. doi: 10.1016/S1474-4422(13)70011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam D. Mental health: On the spectrum. Nature. 2013;496:416–418. doi: 10.1038/496416a. [DOI] [PubMed] [Google Scholar]
- Doherty JL, O'Donovan MC, Owen MJ. Recent genomic advances in schizophrenia. Clin Genet. 2012;81:103–109. doi: 10.1111/j.1399-0004.2011.01773.x. [DOI] [PubMed] [Google Scholar]
- Bassett AS, Scherer SW, Brzustowicz LM. Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. Am JPsychiatr. 2010;167:899–914. doi: 10.1176/appi.ajp.2009.09071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010;11:402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Craddock N, Kendler K, Lee PH, Neale BM, Nurnberger JI, et al. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionel AC, Vaags AK, Sato D, Gazzellone MJ, Mitchell EB, Chen HY, et al. Rare exonic deletions implicate the synaptic organizer Gephyrin (GPHN) in risk for autism, schizophrenia and seizures. Hum Mol Genet. 2013;22:2055–2066. doi: 10.1093/hmg/ddt056. [DOI] [PubMed] [Google Scholar]
- Iqbal Z, Vandeweyer G, van der Voet M, Waryah AM, Zahoor MY, Besseling JA, et al. Homozygous and heterozygous disruptions of ANK3: at the crossroads of neurodevelopmental and psychiatric disorders. Hum Mol Genet. 2013;22:1960–1970. doi: 10.1093/hmg/ddt043. [DOI] [PubMed] [Google Scholar]
- Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- Kirov G, Rujescu D, Ingason A, Collier DA, O'Donovan MC, Owen MJ. Neurexin 1 (NRXN1) deletions in schizophrenia. Schizophrenia Bullet. 2009;35:851–854. doi: 10.1093/schbul/sbp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching MS, Shen Y, Tan WH, Jeste SS, Morrow EM, Chen X, et al. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet. 2010;153B:937–947. doi: 10.1002/ajmg.b.31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf CP, Boone PM, Sampath S, Williams C, Bader PI, Mueller JM, et al. Phenotypic spectrum and genotype-phenotype correlations of NRXN1 exon deletions. Eur J Hum Genet. 2012;20:1240–1247. doi: 10.1038/ejhg.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Siddiqui TJ, Huashan P, Yokomaku D, Hamdan FF, Champagne N, et al. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum Genet. 2011;130:563–573. doi: 10.1007/s00439-011-0975-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Lam CW, Yeung WL, Ko CH, Poon PM, Tong SF, Chan KY, et al. Spectrum of mutations in the MECP2 gene in patients with infantile autism and Rett syndrome. J Med Genet. 2000;37:E41. doi: 10.1136/jmg.37.12.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Calfa G, Percy AK, Pozzo-Miller L. Experimental models of Rett syndrome based on Mecp2 dysfunction. Exp Biol Med. 2011;236:3–19. doi: 10.1258/ebm.2010.010261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Lazar G, Couvert P, Desportes V, Lippe D, Mazet P, et al. MECP2 mutation in a boy with language disorder and schizophrenia. Am J Psychiatr. 2002;159:148–149. doi: 10.1176/appi.ajp.159.1.148-a. [DOI] [PubMed] [Google Scholar]
- St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- Kilpinen H, Ylisaukko-Oja T, Hennah W, Palo OM, Varilo T, Vanhala R, et al. Association of DISC1 with autism and Asperger syndrome. Mol Psychiatr. 2008;13:187–196. doi: 10.1038/sj.mp.4002031. [DOI] [PubMed] [Google Scholar]
- Porteous DJ, Millar JK, Brandon NJ, Sawa A. DISC1 at 10: connecting psychiatric genetics and neuroscience. Trends Mol Med. 2011;17:699–706. doi: 10.1016/j.molmed.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Litwa K, Seong E, Burmeister M, Faundez V. Neuronal and non-neuronal functions of the AP-3 sorting machinery. J Cell Sci. 2007;120 (Pt 4:531–541. doi: 10.1242/jcs.03365. [DOI] [PubMed] [Google Scholar]
- Larimore J, Tornieri K, Ryder PV, Gokhale A, Zlatic SA, Craige B, et al. The schizophrenia susceptibility factor dysbindin and its associated complex sort cargoes from cell bodies to the synapse. Mol Biol Cell. 2011;22:4854–4867. doi: 10.1091/mbc.E11-07-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A, Larimore J, Werner E, So L, Moreno-De-Luca A, Lese-Martin C, et al. Quantitative proteomic and genetic analyses of the schizophrenia susceptibility factor dysbindin identify novel roles of the biogenesis of lysosome-related organelles complex 1. J Neurosci. 2012;32:3697–3711. doi: 10.1523/JNEUROSCI.5640-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristino AS, Williams SM, Hawi Z, An JY, Bellgrove MA, Schwartz CE, et al. Neurodevelopmental and neuropsychiatric disorders represent an interconnected molecular system Mol Psychiatradvance online publication, 26 February 2013; doi: 10.1038/mp.2013.16(e-pub ahead of print). [DOI] [PubMed]
- Gokhale A, Perez-Cornejo P, Duran C, Hartzell HC, Faundez V. A comprehensive strategy to identify stoichiometric membrane protein interactomes. Cell Logistics. 2012;2:189–196. doi: 10.4161/cl.22717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wallqvist A, Reifman J. Inferring high-confidence human protein-protein interactions. BMC Bioinformatics. 2012;13:79. doi: 10.1186/1471-2105-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimera R, Sales-Pardo M. Missing and spurious interactions and the reconstruction of complex networks. Proc Natl Acad Sci USA. 2009;106:22073–22078. doi: 10.1073/pnas.0908366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito GC, Andrews DW. Removing bias against membrane proteins in interaction networks. BMC Syst Biol. 2011;5:169. doi: 10.1186/1752-0509-5-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, et al. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol psychiatr. 2012;17:887–905. doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiani CA, Dell'angelica EC. Dysbindin-containing complexes and their proposed functions in brain: from zero to (too) many in a decade. ASN Neuro. 2011;3:e00058. doi: 10.1042/AN20110010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K, Ong WY, Blake DJ, Tang D, Louneva N, Carlson GC, et al. Dysbindin-1 and its protein family, with special attention to the potential role of dysbindin-1 in neuronal functions and the pathophysiology of schizophreniaIn: Kantrowitz Ja (ed).Handbook of Neurochemistry and Molecular Neurobiology vol. 27Springer Science: New York, NY, USA; 107–241.2009 [Google Scholar]
- Talbot K, Eidem WL, Tinsley CL, Benson MA, Thompson EW, Smith RJ, et al. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest. 2004;113:1353–1363. doi: 10.1172/JCI20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot K, Louneva N, Cohen JW, Kazi H, Blake DJ, Arnold SE. Synaptic dysbindin-1 reductions in schizophrenia occur in an isoform-specific manner indicating their subsynaptic location. PloS ONE. 2011;6:e16886. doi: 10.1371/journal.pone.0016886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, LeGros RP, Louneva N, Yeh L, Cohen JW, Hahn CG, et al. Dysbindin-1 in dorsolateral prefrontal cortex of schizophrenia cases is reduced in an isoform-specific manner unrelated to dysbindin-1 mRNA expression. Hum Mol Genet. 2009;18:3851–3863. doi: 10.1093/hmg/ddp329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weickert CS, Rothmond DA, Hyde TM, Kleinman JE, Straub RE. Reduced DTNBP1 (dysbindin-1) mRNA in the hippocampal formation of schizophrenia patients. Schizophrenia Res. 2008;98:105–110. doi: 10.1016/j.schres.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin AP, Gokhale A, Larimore J, Faundez V. Cell biology of the BLOC-1 complex subunit dysbindin, a schizophrenia susceptibility gene. Mol Neurobiol. 2011;44:53–64. doi: 10.1007/s12035-011-8183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder PV, Faundez V. Schizophrenia: the "BLOC" may be in the endosomes. Sci Signal. 2009;2:pe66. doi: 10.1126/scisignal.293pe66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Sun J, Riley BP, Thiselton DL, Kendler KS, Zhao Z. The dystrobrevin-binding protein 1 gene: features and networks. Molecular Psychiatr. 2009;14:18–29. doi: 10.1038/mp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Feng Y, Hao C, Guo X, Cui Y, He M, et al. The BLOC interactomes form a network in endosomal transport. J Genet Genomics. 2007;34:669–682. doi: 10.1016/S1673-8527(07)60076-9. [DOI] [PubMed] [Google Scholar]
- Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro SM, Falcon-Perez JM, Tenza D, Setty SR, Marks MS, Raposo G, et al. BLOC-1 Interacts with BLOC-2 and the AP-3 Complex to Facilitate Protein Trafficking on Endosomes. Mol Biol Cell. 2006;17:4027–4038. doi: 10.1091/mbc.E06-05-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto R, Ohi K, Okada T, Yasuda Y, Yamamori H, Hori H, et al. Association analysis between schizophrenia and the AP-3 complex genes. Neurosci Res. 2009;65:113–115. doi: 10.1016/j.neures.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Newell-Litwa K, Chintala S, Jenkins S, Pare JF, McGaha L, Smith Y, et al. Hermansky-Pudlak protein complexes, AP-3 and BLOC-1, differentially regulate presynaptic composition in the striatum and hippocampus. J Neurosci. 2010;30:820–831. doi: 10.1523/JNEUROSCI.3400-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell-Litwa K, Salazar G, Smith Y, Faundez V. Roles of BLOC-1 and AP-3 Complexes in Cargo Sorting to Synaptic Vesicles. Mol Biol Cell. 2009;20:1441–1453. doi: 10.1091/mbc.E08-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G, Craige B, Styers ML, Newell-Litwa KA, Doucette MM, Wainer BH, et al. BLOC-1 complex deficiency alters the targeting of adaptor protein complex-3 Cargoes. Mol Biol Cell. 2006;17:4014–4026. doi: 10.1091/mbc.E06-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneichi-Kuroda S, Taya S, Hikita T, Fujino Y, Kaibuchi K. Direct interaction of Dysbindin with the AP-3 complex via its mu subunit. Neurochem Int. 2009;54:431–438. doi: 10.1016/j.neuint.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Hikita T, Taya S, Fujino Y, Taneichi-Kuroda S, Ohta K, Tsuboi D, et al. Proteomic analysis reveals novel binding partners of dysbindin, a schizophrenia-related protein. J Neurochem. 2009;110:1567–1574. doi: 10.1111/j.1471-4159.2009.06257.x. [DOI] [PubMed] [Google Scholar]
- Oyama S, Yamakawa H, Sasagawa N, Hosoi Y, Futai E, Ishiura S. Dysbindin-1, a schizophrenia-related protein, functionally interacts with the DNA- dependent protein kinase complex in an isoform-dependent manner. PloS ONE. 2009;4:e4199. doi: 10.1371/journal.pone.0004199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, McCarthy S, Michaelson JJ, Vacic V, Burdick KE, Yoon S, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72:951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay ME, Dietz HC. Lessons on the pathogenesis of aneurysm from heritable conditions. Nature. 2011;473:308–316. doi: 10.1038/nature10145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge DP, Dietz HC. Therapy of Marfan syndrome. Ann Review Med. 2008;59:43–59. doi: 10.1146/annurev.med.59.103106.103801. [DOI] [PubMed] [Google Scholar]
- Munger JS, Sheppard D. Cross talk among TGF-beta signaling pathways, integrins, and the extracellular matrix. Cold Spring Harbor Perspect Biol. 2011;3:a005017. doi: 10.1101/cshperspect.a005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantheti P, Qiao X, Diaz ME, Peden AA, Meyer GE, Carskadon SL, et al. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron. 1998;21:111–122. doi: 10.1016/s0896-6273(00)80519-x. [DOI] [PubMed] [Google Scholar]
- Peden AA, Rudge RE, Lui WW, Robinson MS. Assembly and function of AP-3 complexes in cells expressing mutant subunits. J Cell Biol. 2002;156:327–336. doi: 10.1083/jcb.200107140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang Q, Oiso N, Novak EK, Gautam R, O'Brien EP, et al. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nature Genet. 2003;35:84–89. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic M, Dell'Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, et al. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc Natl Acad Sci USA. 2010;107:10442–10447. doi: 10.1073/pnas.0913293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Candille SI, Choi Y, Xie D, Jiang L, Li-Pook-Than J, et al. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Leal-Ortiz S, Foss SM, Waites CL, Garner CC, Voglmaier SM, et al. v-SNARE composition distinguishes synaptic vesicle pools. Neuron. 2011;71:474–487. doi: 10.1016/j.neuron.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez DM, Kavalali ET. The role of non-canonical SNAREs in synaptic vesicle recycling. Cell Logistics. 2012;2:20–27. doi: 10.4161/cl.20114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder PV, Vistein R, Gokhale A, Seaman MN, Puthenveedu M, Faundez V. The WASH Complex, an Endosomal Arp2/3 Activator, Interacts with the Hermansky-Pudlak Syndrome Complex BLOC-1 and its Cargo Phosphatidylinositol-4-kinase Type II Alpha. Mol Biol Cell. 2013;24:2269–2284. doi: 10.1091/mbc.E13-02-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autry AE, Monteggia LM. Brain-derived neurotrophic factor and neuropsychiatric disorders. Pharmacological Rev. 2012;64:238–258. doi: 10.1124/pr.111.005108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Shields J. A polygenic theory of schizophrenia. Proc Natl Acad Sci USA. 1967;58:199–205. doi: 10.1073/pnas.58.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Huang L, Jia P, Li M, Su B, Zhao Z, et al. Protein-protein interaction and pathway analyses of top schizophrenia genes reveal schizophrenia susceptibility genes converge on common molecular networks and enrichment of nucleosome (Chromatin) assembly genes in schizophrenia susceptibility loci Schizophrenia Bullet 2013(e-pub ahead of print). [DOI] [PMC free article] [PubMed]
- Sullivan PF. Puzzling over schizophrenia: schizophrenia as a pathway disease. Nature Med. 2012;18:210–211. doi: 10.1038/nm.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English JA, Pennington K, Dunn MJ, Cotter DR. The neuroproteomics of schizophrenia. Biol Psychiatr. 2011;69:163–172. doi: 10.1016/j.biopsych.2010.06.031. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D. Proteomics tackling schizophrenia as a pathway disorder. Schizophrenia Bullet. 2012;38:1107–1108. doi: 10.1093/schbul/sbs094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LS. New frontiers in human cell biology and medicine: can pluripotent stem cells deliver. J Cell Biol. 2012;199:577–581. doi: 10.1083/jcb.201209118. [DOI] [PMC free article] [PubMed] [Google Scholar]