Abstract
BACKGROUND
Upper gastrointestinal adenocarcinomas are a common cause of cancer-related deaths. In this study, the authors investigated the prevalence and biological significance of Aurora Kinase A (AURKA) overexpression in upper gastrointestinal adenocarcinomas.
METHODS
Quantitative real-time polymerase chain reaction (qRT-PCR) and immunohistochemical staining on tumor tissue microarrays (TMA) were used to study the expression of AURKA in upper gastrointestinal adenocarcinomas. To investigate the biological and signaling impact of AURKA, the authors used multiple in vitro assays that included 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), TUNEL (terminal deoxynucleotidyl transferase–mediated nick-end labeling), cytochrome C release, flow cytometry, luciferase reporter, and Western blot analysis.
RESULTS
Frequent overexpression of AURKA transcript in upper gastrointestinal adenocarcinomas was detected compared with normal samples (47%; P = .001). The immunohistochemical analysis of 130 tumors demonstrated moderate-to-strong immunostaining of AURKA in >50% of upper gastrointestinal adenocarcinomas. By using camptothecin as a drug-induced apoptosis in vitro model, the authors demonstrated that the expression of AURKA provided protection against apoptosis to gastrointestinal cancer cells (AGS and RKO) (P =.006) and RIE-1 primary intestinal epithelial cells (P =.001). The AURKA overexpression mediated an increase in phosphorylation of AKTSer473 with an increase in HDM2 level. The shRNA-knockdown of AKT in AURKA-overexpressing cells reversed this effect and showed a significant increase in the p53 protein level, indicating a possible nexus of AURKA/AKT/p53. Indeed, overexpression of AURKA led to a remarkable reduction in the transcription activity of p53, with subsequent reductions in transcript and protein levels of its downstream proapoptotic transcription targets (p21, BAX, NOXA, and PUMA).
CONCLUSIONS
Study results indicated that AURKA provides potent antiapoptotic properties to gastrointestinal cells by regulating levels of p53 through the AKT/HDM2 axis.
Keywords: AURKA, gastric, Barrett, cancer, survival, apoptosis, p53
Upper gastrointestinal adenocarcinomas of the stomach and esophagus are poorly responsive to therapy and have an unfavorable outcome.1 Upper gastrointestinal adenocarcinomas are the second most common cause of cancer-related death in the world and are characterized by complex molecular changes. Several epidemiological studies have indicated that the incidence of proximal adenocarcinomas of the gastroesophageal junction and lower esophagus is rising faster than ever before in the Western world.2,3
Aurora Kinase A (AURKA) is 1 of the 3 serine/threonine kinases (A, B, and C) that are evolutionarily conserved and regulate mitotic progression in various organisms.4 It controls centrosome maturation and separation, mitotic entry, spindle formation, and chromosome alignment.5,6 The human AURKA gene is located in chromosome band 20q13, a locus that is frequently amplified in breast, bladder, ovarian, pancreatic, and gastrointestinal cancers.7–10 In normal cells, the AURKA protein level increases periodically during the cell cycle from G2 to M phase and is enriched at the centrosome and mitotic spindle.5,11 It is noteworthy that overexpression of AURKA correlates with and is predisposing to chromosomal instability in several tumors.12 Cytological analysis has revealed that overexpression of AURKA results in centrosome amplification and cytokinesis failure and, thus, production of aneuploid cells.8 Several inhibitors of Aurora kinases, such as hesperadin,13 ZM447439,14 and VX-68015 have been developed and are in different phases of clinical trials.10,15 These evolving drugs hold a promise of gene-targeted therapy in cancer.
Recent studies have indicated that AURKA overexpression increases resistance to taxol-induced apoptosis in HeLa cells.16 AURKA interacts at multiple levels within the p53 pathways, suggesting that these proteins form part of an integrated functional network. 17 In this study, we investigated the prevalence and biological significance of AURKA overexpression in upper gastrointestinal adenocarcinomas and demonstrated the activation of the AURKA/AKT/HDM2 axis as a potential paradigm for regulating p53 and cancer-cell survival.
MATERIALS AND METHODS
Tissue Samples
Paraffin-embedded tissue blocks from 130 gastric and/or esophageal resections due to upper gastrointestinal adenocarcinomas, performed between 1995 and 2005, were obtained from Vanderbilt University Medical Center (Nashville, Tenn) for immunohistochemical analysis. They included 44 lower esophageal, 43 gastroesophageal junction, 8 cardia, and 30 distal gastric (antrum and body) tumors. Tumor grading was performed according to World Health Organization (WHO) standards. DNA and mRNA purification were performed by using Qiagen purification kits (Qiagen, Hilden, Germany). Single-strand cDNA was synthesized by using the Advantage real-time polymerase chain reaction (RT-PCR Kit; Clontech, Palo Alto, Calif).
Quantitative real-time polymerase chain reaction (qRT-PCR)
mRNA was isolated from 25 normal gastric cardia samples and 45 primary upper gastrointestinal adenocarcinomas. Gene-specific primers for AURKA, PUMA, NOXA, and HPRT1 were designed, and the results were normalized to HPRT1 as described earlier. 18 All primers were purchased from GeneLink (Hawthorne, NY), and their sequences are available from the authors upon request. qRT-PCR was performed by using an iCycler (Bio-Rad, Hercules, Calif) with a threshold cycle number determined with the use of iCycler software version 3.0. The reactions were performed in triplicate, and threshold cycle numbers were averaged. The fold change in all samples was calculated according to the formula 2(Rt–Et)/2(Rn–En), as described previously.7
Tissue microarrays and immunohistochemistry of AURKA protein
All tumor and normal gastric and esophageal mucosal epithelial tissues were histologically verified, and representative regions were selected for inclusion in a tissue microarray (TMA). The tumors were classified into intestinal and diffuse types19 and ranged from well differentiated (WD) to poorly differentiated (PD). Clinical staging was performed according to American Joint Committee on Cancer (AJCC) criteria.20 Tissue cores with a diameter of 0.5 mm were retrieved from the selected regions of the donor blocks and punched to the recipient block by using a manual tissue-array instrument (Beecher, Silver Spring, Md). Sections (5 µm) were transferred to polylysine-coated slides (SuperFrostPlus; Menzel-Glaser, Brunschwig, Germany) and incubated at 37°C for 2 hours. The resulting TMA was used for immunohistochemical analysis. An avidin-biotin immunoperoxidase assay was performed after pretreatment in a microwave with a citrate buffer for 20 minutes, and rabbit anti-AURKA (KR051; 1:100 dilutions; TransGenic, Kobe, Japan) was applied at room temperature. Immunohistochemical results were evaluated for intensity and staining frequency of nuclear and cytoplasmic components. The intensity of staining was graded 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). The frequency was graded from 0 to 4 according to the percentage of positive cells as follows: 0, ≤3%; 1, ≤4% to 25%; 2, ≤26% to 50%; 3, ≤51% to 75%; 4, ≥75%. The products of multiplication of the intensity and frequency grades were then classified into an index core on a scale of 0 to 3: index score 0 = product of 0 (negative), index score 1 = products of 1 and 2 (weak), index score 2 = products of 3 and 4 (moderate), index score 3 = products of 6 through 12 (strong).
Cell culture, vectors, siRNA, and transfection
AGS, RKO, and RIE-1 cells were used in this study. Cells were cultured in F-12 (HAM) or Dulbecco modified eagle medium (DMEM) together with 10% fetal bovine serum (Invitrogen, Carlsbad, Calif) at 37°C in an atmosphere containing 5% CO2. AGS and RKO cells were obtained from American Tissue Culture Collection (ATCC, Manassas, Va). RIE-1 cells are spontaneously immortalized, nontransformed, and from an epidermal growth factor-responsive cell line (A generous gift from Dr. Robert. J. Coffey, Jr. at Vanderbilt University). The expression plasmid for AURKA was generated by polymerase chain reaction (PCR) amplification of the full-length coding sequence of AURKA and cloned in-frame into pcDNA3.1. A synthetic Flag tag sequence was added at the N-terminus of AURKA. Retroviral expression constructs, pBabe-puro, containing either the full-length of the AURKA coding sequence (pBabe puro-AURKA) or a kinase-dead AURKA mutant (D274A) were purchased from Addgene (Cambridge, Mass). AURKA-specific shRNA sequence (GATCCCC ATGCCCTGTCTTACTGTCATTCAAGAGATGACAGTAAGACAGGGCATTTTTTA) was cloned in retroviral expression construct pMSCV-siRNA-GFP by using Bgl II and Hind III restriction enzymes. A green fluorescent protein (GFP)-specific shRNA was cloned in the same vector and used as a negative control. In addition, we obtained validated small-interfering RNA (siRNA) oligonucleotides specific for AKT (#42,811) and a negative control siRNA (#4611) (Ambion, Austin, Tex). Transient transfections were performed by using Fugene 6 (Roche, Indianapolis, Ind) and Lipofectamine (Invitrogen, Carlsbad, Calif) following the manufacturers’ protocols.
Retroviral transduction
Ecopack 293 cells (5 × 105 per well) (Clontech, Palo Alto, Calif) were plated in a 6-well plate. Cells were transfected with retroviral vectors; empty pBabe-puro (control) and pBabe-puro-AURKA, respectively. After 48 hours of transfection, viruses were collected, filtered by using 0.45 µM filters, and added to RIE-1 cells in the presence of 8 µg/mL polybrene. The culture medium was replaced with fresh medium after 24 hours of transduction.
MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay
AURKA-overexpressing AGS cells (5 × 103 per well) were plated in a 96-well plate. The MTT Cell Assay kit from American Type Culture Collection (ATCC, Manassas, Va) was used to perform this assay as recommended by the manufacturer. Cells were plated in triplicate, and the assay carried out measurements for 3 days.
TUNEL (terminal deoxynucleotidyl transferase–mediated nick-end labeling) apoptosis assay
RKO cells were seeded into 8-well chamber slides and transfected with 400 ng of pcDNA3-flag-AURKA and pcDNA3 empty vector (control) per well. Cells were then treated either with camptothecin (5 µM) or dimethyl sulfoxide (DMSO; vehicle, control) for 24 hours. After 24 hours, DMSO-treated and camptothecin-treated cells were stained by terminal deoxynucleotidyl transferase–mediated nick-end labeling (TUNEL) according to the manufacturer’s instructions (Roche, Indianapolis, Ind), and then observed by tetramethylrhodamine isothiocyanate (TRITC) (red fluorescence). AURKA overexpression was determined by immunofluorescence staining in duplicate wells with the Flag antibody (Cell Signaling Technology, Boston, Mass) and fluorescein isothiocyanate (FITC)-goat anti-rabbit immunoglobulin G (IgG) (heavy + light [H + L]) conjugate (green; Jackson Immuno Research, West Grove, Pa). TUNEL-positive cells and AURKA-expressing cells (20 random fields at magnification of 40×, >400 cells) were then counted.
Cytochrome C release
AGS cells that stably expressed AURKA or pcDNA3 empty vector (control) were cultured into 8-well slide chambers (12 × 103 cell per well). Cells were then treated with DMSO (vehicle, control) or 20 µM camptothecin for 4 hours at 37°C. Cytochrome C release was detected by immunofluorescence by using mouse monoclonal anticytochrome-C antibody and by following the manufacturer’s recommendations (Pharmingen, San Diego, Calif).
Fluorescence assorted cell sorting
Cells were trypsinized, washed twice with 1× ice-cool phosphate-buffered saline (PBS), and then resuspended in 0.2 mL ice-cold PBS, then fixed in 1mL icecool 70% ethanol, and incubated for 1 hour at −20°C. The cells were then centrifuged, resuspended in 1 mL PBS and treated with propidium iodide (50 µg/mL) and RNase (1 µg/mL) for 30 minutes at 37°C, and then analyzed by BD LSR II flow cytometer (Becton Dickinson, Franklin Lakes, NJ). The data were analyzed by Becton Dickinson’s BD FACS Diva software.
Western blot analysis
Cell lysates were prepared in a phosphate saline buffer (PBS) containing 1× protease cocktail inhibitor (Pierce, Rockford, Ill) and centrifuged at 3500 rpm for 10 minutes at 4°C. The protein concentration was measured by using a Bio-Rad protein assay (Bio-Rad, Hercules, Calif). Protein (10–15 µg) from each sample was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto a nitrocellulose membrane. Target proteins were detected by using specific antibodies as follows: p53, HDM2, and p21Waf1 (Oncogene, San Diego, Calif); ACTIN, AKT, pAKTSer473, BAX, Anti-Flag, PUMA (Cell Signaling Technology, Boston, Mass); AURKA (TransGenic, Kobe, Japan); and NOXA (Imgenex, San Diego, Calif).
Luciferase assays
Luciferase activity assays were performed by using the pG13-Luc reporter plasmid, which contains 13 tandem repeats of the p53 consensus DNA-binding sites and measures the p53 transcription activity.21 The pcDNA3-p53 and pcDNA3-ΔNp73 are described elsewhere.22 PUMA-Luc luciferase vector was used to investigate the PUMA promoter activity. We determined the luciferase activity by using a Dual-Luciferase Reporter Assay kit (Promega, Madison, Wis). Results were normalized by using the Renilla luciferase activity (Renilla Luciferase Assay Lysis Buffer; Promega, Madison, Wis) or protein normalization as indicated. Results were averaged from 3 independent experiments and expressed as mean values with their individual data points.
Statistical Analysis
For qRT-PCR data, mRNA expression levels between groups were assessed by using Kruskal-Wallis tests. When the overall effect was present, group-by-group comparisons were made while adjusting the significance level with the Scheffe method.23 Statistical comparisons were also performed by using the Student t test. P < .05 was considered to represent a statistically significant difference. For the immunohistochemical analysis results, patient characteristics and clinical variables were tabulated, and Fisher exact tests were used to compare these characteristics between the normal and the AURKA-overexpressed groups.
RESULTS
Frequent Overexpression of AURKA in Upper Gastrointestinal Adenocarcinomas
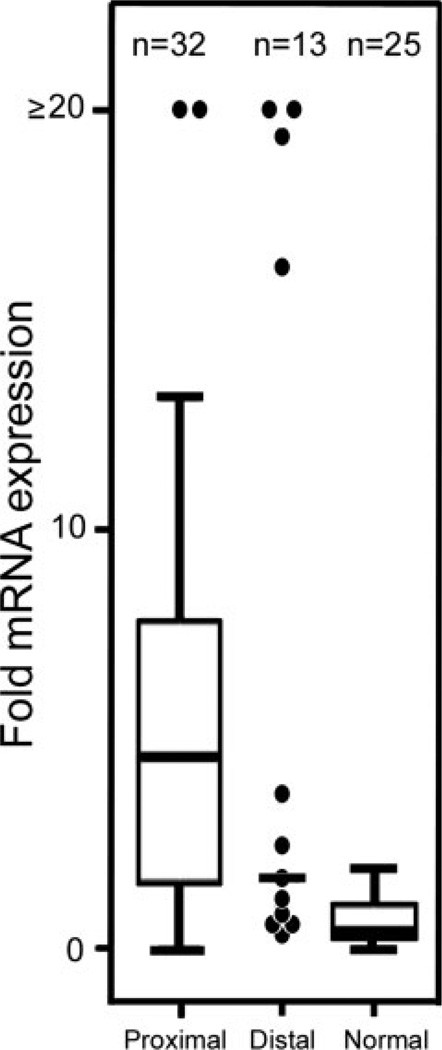
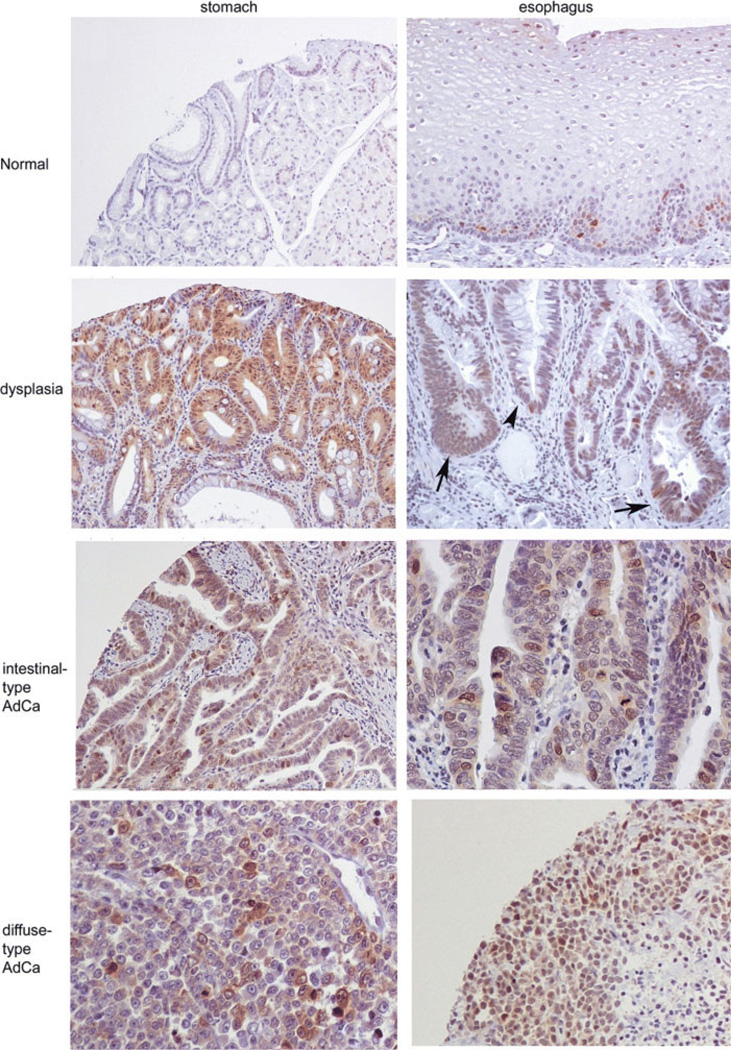
We observed the overexpression of AURKA at the mRNA level in 23 of 32 (71.9%; P =.001) proximal (gastroesophageal junction and lower esophagus) and in 5 of 13 (38.5%; P =.013) distal (body and antrum) tumors, whereas 0 of 25 normal gastric cardia samples showed overexpression (Fig. 1). The expression levels of AURKA mRNA in 32 proximal (P < .0001; 95% CI, 2.2–5.4) and in 13 distal (P < .004; 95% CI, 0.2–14.4) tumors were significantly higher than in normal gastric cardia samples (n = 25). Proximal tumors showed more overexpression of AURKA (≥2.5-fold) than distal tumors (P =.036) (Fig. 1). However, the difference in expression levels in proximal and distal samples was not statistically significant (P =.48). The immunohistochemical analysis of AURKA on TMA showed moderate-to-strong immunostaining in 15 of 30 (50%) body and antrum, in 6 of 8 (75%) cardia, and in 53 of 87 (61%) gastroesophageal and lower esophageal adenocarcinomas (Fig. 2). The immunostaining was predominantly cytoplasmic with a variable degree of nuclear staining. Interestingly, when available, adjacent tissues from intestinal metaplasia and gastric dysplasia demonstrated overexpression of AURKA, suggesting that this could be an early event in tumorigenesis (Fig. 2). On logistic regression, there were no statistically significant associations between AURKA expressions and the tumor site, histologic type, grade, or tumor, node, metastasis (TNM) classification.
FIGURE 1.
Frequent overexpression of AURKA at the mRNA level in upper gastrointestinal adenocarcinomas is shown. Forty-five upper gastrointestinal adenocarcinomas and 25 normal gastric cardia samples were studied for mRNA expression by using an iCycler (Bio-Rad, Hercules, Calif). The average threshold cycle number for all normal samples was used as a reference value. All results were normalized to the expression of HPRT1 in the same sample. Each tumor sample was compared with 25 normal gastric cardia samples and normalized to HPRT1 expression. Overexpression (≥2.5-fold) of AURKA mRNA was detected in 28 of 45 (62.2%) tumors. In proximal samples (esophageal and gastroesophageal junction), 23 of 32 (71.9%) showed overexpression, whereas only 5 of 13 (46.2%) distal samples (antrum and body of the stomach) showed overexpression.
FIGURE 2.
Immunohistochemical analysis reveals overexpression of AURKA in upper gastrointestinal adenocarcinomas. This panel contains representative images of AURKA immunohistochemistry on tumor tissue microarray that contained 130 upper gastrointestinal adenocarcinomas. Normal gastric glandular epithelia demonstrate no immunoreactivity (original magnification, ×100). Normal esophageal squamous epithelium shows absence of immunostaining in the superficial layer and weak-to-moderate immunostaining in the basal layer. Dysplasia of the antrum and esophagus (Barrett-related) demonstrates moderate immunostaining for AURKA (original magnification, ×100). Intestinal-type and diffuse-type adenocarcinomas of the stomach and esophagus show moderate to strong immunostaining (original magnification, ×100).
AURKA Overexpression Enhances Cell Survival and Counteracts Drug-induced Apoptosis
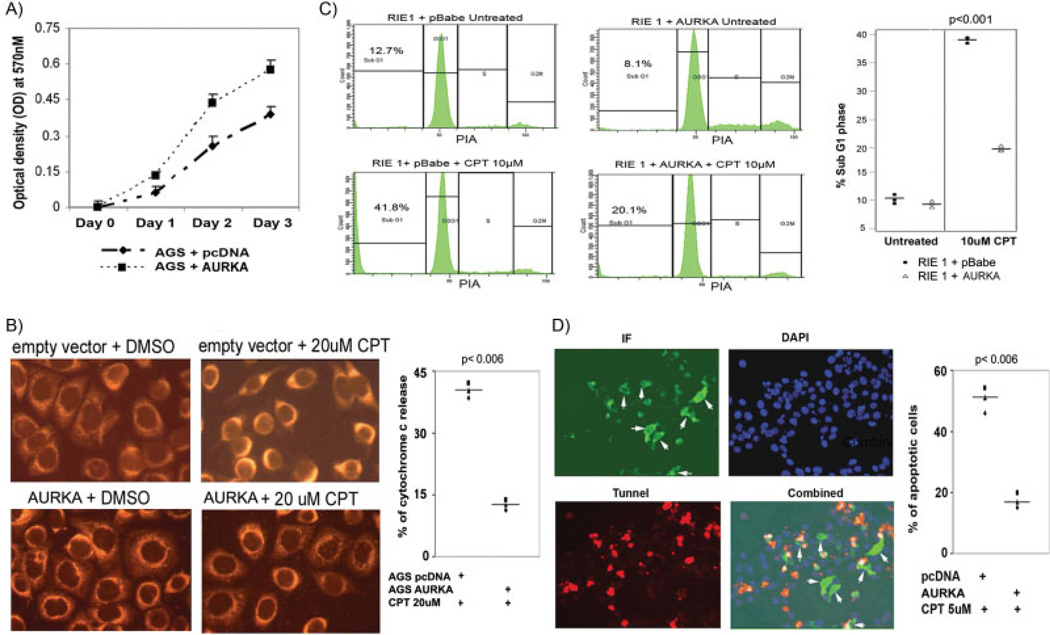
We observed a steep increase in the survival of AGS cells that overexpressed AURKA compared with cells that expressed control vector on MTT assay (Fig. 3A). This finding suggested that AURKA overexpression provided cells with a proproliferative or survival advantage. Therefore, we performed additional experiments to study the role of AURKA in cancer-cell survival after drug-induced apoptosis. The treatment of AGS cells that stably expressed AURKA, with 20 µM camptothecin for 4 hours, confirmed our hypothesis and did not induce the release of cytochrome C, as depicted by mitochondrial punctuate staining. Conversely, cells that expressed empty vector (control) demonstrated a diffuse cytosolic staining that indicated a release of cytochrome C and, thus, progression of these cells toward apoptosis (Fig. 3B). This finding in cancer cells led us to investigate whether a similar effect could be observed in primary gastrointestinal epithelial cells (RIE-1), which could indicate a role for AURKA in early tumorigenesis. Flow cytometry analysis of RIE-1 cells, transduced with AURKA-expressing retroviral construct, showed a significant decrease in the SubG1 phase compared with cells that expressed control vector. A 50% reduction in the SubG1 phase was observed in AURKA-overexpressing RIE-1 cells when they were treated with 10 µM camptothecin for 24 hours (Fig. 3C). This experiment was repeated 3 times; results are summarized in a bar graph (Fig. 3C). For further confirmation, we analyzed the antiapoptotic potential of AURKA overexpression in gastrointestinal cancer cells by using TUNEL assay on RKO cells after they were treated with camptothecin. RKO cells harbor a wild-type p53 and are considered to be a sensitive measure for p53-dependent apoptosis after DNA damage.22,24 Indeed, the AURKA-expressing RKO cells showed a considerable reduction (≥3-fold) in apoptosis compared with cells that expressed empty vector (control) (Fig. 3D). Taken together, these results suggested that AURKA expression protected cells from drug-induced toxicity.
FIGURE 3.
AURKA overexpression enhances cell survival and counteracts apoptosis. A) AGS cells that overexpressed AURKA or empty vector control were plated in a 96-well plate (5 × 103 cells per well). Cell survival was determined by MTT assay by measuring optical density (OD) at 570 nM for 3 days. AGS cells that overexpressed AURKA showed a steep increase in cell survival compared with control (empty vector). B) AGS cells that overexpressed AURKA prevented mitochondrial release of cytochrome C. AGS cells that overexpressed AURKA showed a punctuate mitochondrial staining of cytochrome C after they were treated with 20 µM camptothecin (CPT) for 4 hours, indicating that these cells did not release mitochondrial cytochrome C and suggesting their viability. Conversely, AGS cells that expressed empty pcDNA vector (control) demonstrated a bright, diffuse, cytoplasmic staining, indicating a release of cytochrome C and progression of these cells toward apoptosis. The bar graph demonstrates quantification of cells that showed a release of cytochrome C. C) RIE-1 cells were transduced with pBabe (control) and pBabe-AURKA vectors and treated with 10 µM camptothecin for 24 hours. Control cells had an increase in the percentage of cells in the SubG1 phase compared with cells that overexpressed AURKA (pBabe-AURKA); thus, cells that overexpress AURKA are resisting drug-induced apoptosis. The bar graph summarizes the percentage of cells in the SubG1 phase of 3 independent experiments in RIE-1 cells. Individual data points are displayed, and the horizontal bar indicates the mean value. D) Overexpression of AURKA conferred resistance to camptothecin. TUNEL assay was performed in RKO cells that harbored wild-type p53. RKO cells were transfected with AURKA or empty pcDNA vector (control) and treated with 5 µM camptothecin for 24 hours or left untreated. After fixation with 4% paraformaldehyde, each well underwent TUNEL staining (red) and immunofluorescence with COOH-terminus, AURKA-specific antibody (green fluorescence). DAPI (blue) was used as a nuclear-counter stain to count all cells. The combined picture (red, green, and blue) indicates that AURKA-expressing cells (white arrows) are virtually protected from camptothecin-induced apoptosis. The bar graph summarizes results and demonstrates the percentage of apoptosis in AURKA-expressing cells compared with empty pcDNA vector (control).
AURKA Regulates p53 Through an AKT-dependent Mechanism
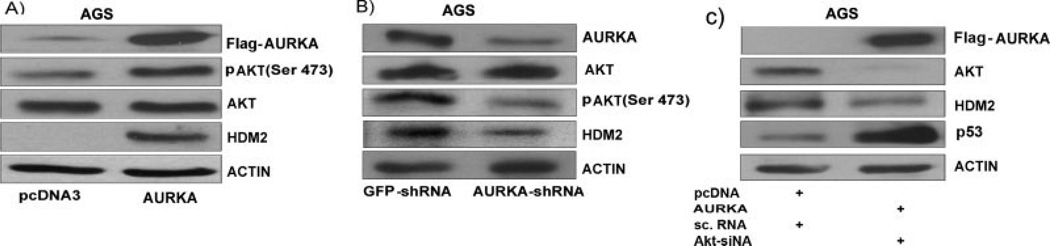
Activation of AKT signaling provides potent prosurvival and antiapoptotic signals to cancer cells.25,26 Therefore, we tested whether AURKA could regulate the level of phospho-AKT in upper gastrointestinal adenocarcinomas. Western blot analysis of AGS cells, after transient transfection with AURKA, demonstrated an increase in the phosphorylation level of AKTSer473 (Fig. 4A). AGS cells harbor a wild-type p53 and were protected from drug-induced apoptosis when AURKA was overexpressed (Fig. 3B). Therefore, we tested whether AURKA could regulate p53 activity through the AKT-dependent mechanism in upper gastrointestinal adenocarcinomas. As shown in Figure 4A, the increase in pAKT was associated with an increase in HDM2 protein level. HDM2 plays a critical role in negatively regulating levels of p53 through ubiquitination-dependent mechanisms. This finding indicated that the AURKA/AKT axis could be responsible for down-regulating p53 and protecting cancer cells from apoptosis. We confirmed that AURKA can mediate an increase in the AKTSer473 phosphorylation in MKN-45 and SEG1 cells (data not shown). We also validated results and performed the reverse experiment by knocking down the endogenous AURKA in AGS cells. The reduction in AURKA levels resulted in a decrease in AKTSer473 phosphorylation (Fig. 4B) and HDM2 levels. Moreover, the knockdown of AKT in AGS cells that overexpressed pcDNA3-Flag-AURKA, detected by Flag, led to a reduction in HDM2 levels with a notable increase in p53, thus confirming that the AURKA/AKT axis is playing a role in regulating p53 (Fig. 4C). These data demonstrate that AURKA may play a major role in regulating p53 through AKT-dependent mechanism(s) by affecting the levels of HDM2, a major regulator of p53. Therefore, our results provide an important additional mechanism for regulating p53 in cancer and suggest that this mechanism could be important in upper gastrointestinal adenocarcinomas.
FIGURE 4.
AURKA-AKT axis regulates p53 through HDM2. A) Western immunoblot analysis after AURKA overexpression in AGS cells demonstrated that AURKA mediates up-regulation in phosphorylation of AKTSer473. In addition to the increase in the phosphorylation level of AKT, there was an increase in the levels of HDM2 at the protein level. B) The knockdown of AURKA in AGS cells by using AURKA-specific shRNA led to a decrease in AURKA-mediated AKT phosphorylation and a subsequent decrease in HDM2 levels, indicating that the observed increase in AKT phosphorylation and HDM2 levels were mediated by AURKA overexpression. C) Knockdown of AKT in AGS cells that overexpressed AURKA led to a reduction in HDM2 levels with a notable increase in p53 levels. These data confirmed that AURKA can regulate p53 levels through AKT-dependent mechanism(s) by modulating levels of HDM2, a major regulator of p53.
AURKA Suppresses the Transcription Activity and Protein Level of p53 and Its Downstream Targets
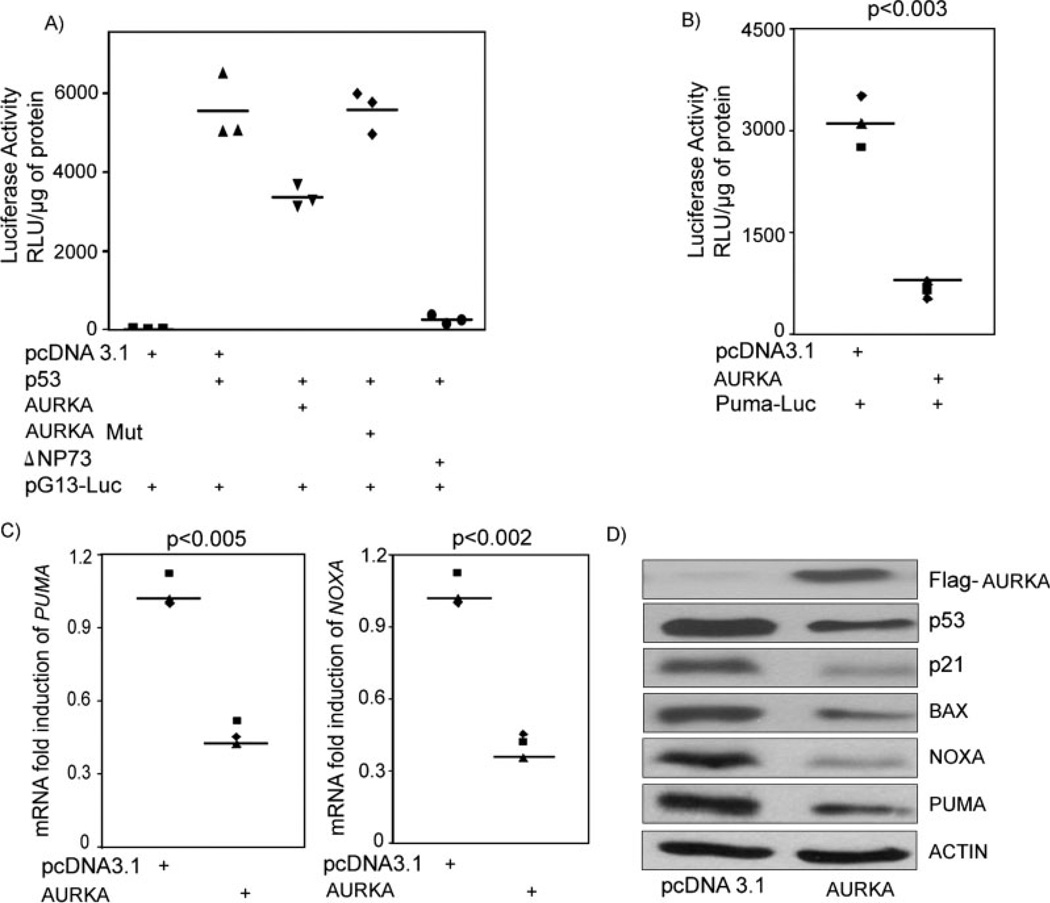
The aforementioned results suggested that regulation of p53 by AURKA is an important mechanism in upper gastrointestinal adenocarcinomas. Therefore, we investigated the impact of this regulation on p53 transcription activity and its down stream targets. We investigated the transcriptional activity of p53 in AGS cells by using pG13-Luc reporter plasmids. pG13-Luc contains 13 binding sites for p53 and is used as a measure of p53 transcription activity. Transfection of p53, as expected, led to a remarkable induction of pG13 luciferase activity. On the other hand, cotransfection of p53 together with AURKA showed a significant reduction in pG13 activity compared with p53 alone. In addition, cotransfection of p53 with AURKA-mutant D274A (kinase dead) did not suppress pG13 activity (Fig. 5A). As expected, cotransfection of ΔNp73 (positive control) with p53 suppressed pG13 activity. To confirm the reduction in p53 transcription activity, we investigated the effect of AURKA on known proapoptotic targets of p53: PUMA, NOXA, BAX, and p21. The luciferase assay that used the PUMA-Luc vector in AGS cells cotransfected with AURKA showed a 5-fold decrease in luciferase activity (Fig. 5B). Quantification by qRT-PCR of mRNA levels of PUMA and NOXA indicated a 53% and a 60% decrease, respectively, in their mRNA expression levels in AGS cells that overexpressed AURKA compared with controls (Fig. 5C). Western blot analysis confirmed down-regulation of the p53 targets, PUMA, NOXA, p21, and BAX (Fig. 5D), at the protein level. Therefore, our results confirm that AURKA down-regulates p53 and its proapoptotic transcription targets in upper gastrointestinal adenocarcinomas and indicate that the AURKA/AKT axis is a potential mechanism for providing potent antiapoptotic and prosurvival advantages to cancer cells in patients with upper gastrointestinal adenocarcinomas.
FIGURE 5.
AURKA regulates the p53 transcription activity and its downstream proapoptotic targets. A) Luciferase activity was seen by using pG13-Luc reporter vector in AGS cells. The pG13-Luc is a reporter plasmid containing 13 tandem repeats of the p53 consensus DNA-binding sites and measures p53 transcription activity. pG13-Luc was transfected with different combinations of expression vectors as shown in the x-axis. The transfection of p53 led to a remarkable induction of luciferase activity of the pG13 reporter. The transfection of ΔNp73, a known protein that inhibits p53, led to suppression of pG13 luciferase activity, as expected. A cotransfection of p53 with AURKA suppressed pG13 reporter activity. A cotransfection of p53 and ΔNp73 led to suppression of transcriptional activity of wild-type p53, as indicated by reduction in pG13 activity. B) AURKA overexpression resulted in a reduction of luciferase activity of Puma-Luc reporter, a p53 downstream target. C) Quantitative real-time RT-PCR (qRT-PCR) for PUMA and NOXA indicated a 53% and 60% decrease in their mRNA expression levels, respectively, in AGS cells that overexpressed AURKA. Individual data points are displayed, and straight bars indicate mean values. D) Western blot analysis with cell lysates from AGS cells transfected with pcDNA3-AURKA that were compared with cells transfected with empty pcDNA3 (control) confirmed down-regulation of p53 downstream targets: PUMA, NOXA, p21, and BAX.
DISCUSSION
Upper gastrointestinal adenocarcinomas are the second most common cause of cancer-related deaths in the world.27 The incidence of adenocarcinoma of the cardia, gastroesophageal junction, and lower esophagus has been rapidly rising, 5-fold to 6-fold in the past few decades, especially in patients younger than 50 years of age.3,28,29 Upper gastrointestinal adenocarcinomas are characterized by poor response to therapy and an unfavorable clinical outcome that reflect an inherent protective mechanism in these tumors against drug-induced apoptosis.30,31 We have demonstrated frequent overexpression of AURKA in upper gastrointestinal adenocarcinomas and identified the AURKA/AKT axis as an important mechanism that provides cancer cells with potent antiapoptotic properties through regulating p53-dependent apoptosis.
In this study, we detected the frequent overexpression of AURKA at the mRNA and protein levels in upper gastrointestinal adenocarcinomas, and interestingly, this overexpression was more prevalent in gastroesophageal junction adenocarcinomas and lower esophageal, Barrett-related adenocarcinomas (BAs) than in antrum and body gastric adenocarcinomas (P =.036). We could not identify an association between AURKA overexpression and histopathological parameters such as tumor grade, TNM classification, and lymph-node metastasis. Overexpression of AURKA has been reported in other tumors such as human breast, bladder, colon, ovarian, and pancreatic cancers.8,11 Therefore, our findings confirm that AURKA overexpression is an important mechanism for several cancer types. We observed AURKA overexpression in premalignant lesions including dysplasia. This is of particular interest because AURKA plays a major role in centrosome maturation and cell division, and its overexpression can induce tumor progression by promoting chromosomal instability, which leads to aneuploidy in bladder cancer cells.32 Chromosomal instability is the hallmark of adenocarcinomas of the stomach and esophagus; thereby, AURKA overexpression may play a central role in their early tumorigenesis cascade. However, further studies are needed to address this important mechanism in upper gastrointestinal adenocarcinomas.
Upper gastrointestinal adenocarcinomas are typically characterized by poor response to therapy and low survival rates.30,31 We detected a remarkable effect of AURKA overexpression on inhibition of drug-induced apoptosis in several gastrointestinal cells. Cells that overexpressed AURKA prevented release of cytochrome C from mitochondria. A decrease in cytochrome C release led to inactivation of caspases, thus protecting cells from apoptosis.33 This antiapoptotic effect was further confirmed in AGS and RIE-1 cells after they were treated with camptothecin. Overexpression of AURKA, therefore, could be a factor that contributes to poor clinical outcome in patients with adenocarcinomas of the stomach and esophagus.
We have demonstrated that AURKA regulates phospho-AKT levels. AKT regulates fundamental cellular processes linked to tumorigenesis such as cell-cycle progression, adhesion, and cell survival.25,26 In line with this, we found that AURKA overexpression regulated HDM2 and p53 protein levels in an AKT-dependent mechanism. HDM2 is a major regulator of p53 that regulates its degradation by promoting ubiquitination via E3 ubiquitination-ligase activity and by cytoplasmic 26S proteasome.10 The knockdown of AKT in AGS cells that overexpressed AURKA led to a significant reduction of HDM2 levels with a parallel induction of p53 protein levels. These results confirm that the AURKA-AKT axis is required for regulating HDM2-p53 levels in this model. Therefore, our findings provide new insight into the role of AURKA in regulating p53-dependent apoptosis and enrich existing knowledge of AURKA and p53, as previous reports have shown that AURKA regulates p53 protein levels through direct interaction and phosphorylation of p53 at phosphorylation at Ser31517 and Ser215.34 The p53 protein is a major regulator of apoptosis, and several cancer drugs work through induction of DNA damage and activation of p53-mediated apoptosis.35,36 The biological outcome of p53 depends on its ability to activate its downstream proapoptotic transcription targets.37,38 Although we have not studied the phosphorylation status of p53, the total protein level and the transcription activity of p53 were dramatically impaired by AURKA overexpression. This observation is supported by changes in transcript and protein levels of several p53 downstream targets, p21, PUMA, NOXA, and BAX. Taken together, our results indicate that AURKA overexpression regulates p53 through the AKT/HDM2 axis in upper gastrointestinal adenocarcinomas.
In summary, we demonstrated frequent overexpression of AURKA in upper gastrointestinal adenocarcinomas. Up-regulation of AURKA provided cancer cells with a drug-resistant phenotype and survival properties. The AURKA/AKT axis is a novel molecular mechanism that underlines the oncogenic potential of AURKA in upper gastrointestinal adenocarcinomas.
Acknowledgments
This study was supported by the National Cancer Institute grants R01CA106176 (WER) and the GI SPORE CA 95103.
We thank Mrs. Elvira Dzhura for her technical assistance.
The contents of this work are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute or Vanderbilt University.
REFERENCES
- 1.Hohenberger P, Gretschel S. Gastric cancer. Lancet. 2003;362:305–315. doi: 10.1016/s0140-6736(03)13975-x. [DOI] [PubMed] [Google Scholar]
- 2.Blot WJ, Devesa SS, Fraumeni JF., Jr Continuing climb in rates of esophageal adenocarcinoma: an update. JAMA. 1993;270:1320. [PubMed] [Google Scholar]
- 3.Devesa SS, Blot WJ, Fraumeni JF., Jr Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer. 1998;83:2049–2053. [PubMed] [Google Scholar]
- 4.Crane R, Kloepfer A, Ruderman JV. Requirements for the destruction of human Aurora-A. J Cell Sci. 2004;117(pt 25):5975–5983. doi: 10.1242/jcs.01418. [DOI] [PubMed] [Google Scholar]
- 5.Marumoto T, Hirota T, Morisaki T, et al. Roles of aurora-A kinase in mitotic entry and G2 checkpoint in mammalian cells. Genes Cells. 2002;7:1173–1182. doi: 10.1046/j.1365-2443.2002.00592.x. [DOI] [PubMed] [Google Scholar]
- 6.Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.el-Rifai W, Powell SM. Molecular and biologic basis of upper gastrointestinal malignancy. Gastric carcinoma. Surg Oncol Clin N Am. 2002;11:273–291. viii. doi: 10.1016/s1055-3207(02)00004-2. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, Kuang J, Zhong L, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 9.Sen S, Zhou H, Zhang RD, et al. Amplification/overexpression of a mitotic kinase gene in human bladder cancer. J Natl Cancer Inst. 2002;94:1320–1329. doi: 10.1093/jnci/94.17.1320. [DOI] [PubMed] [Google Scholar]
- 10.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 11.Bischoff JR, Anderson L, Zhu Y, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyoshi Y, Iwao K, Egawa C, Noguchi S. Association of centrosomal kinase STK15/BTAK mRNA expression with chromosomal instability in human breast cancers. Int J Cancer. 2001;92:370–373. doi: 10.1002/ijc.1200. [DOI] [PubMed] [Google Scholar]
- 13.Hauf S, Cole RW, LaTerra S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ditchfield C, Johnson VL, Tighe A, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrington EO, Newton J, Morin N, Rounds S. Barrier dysfunction and RhoA activation are blunted by homocysteine and adenosine in pulmonary endothelium. Am J Physiol Lung Cell Mol Physiol. 2004;287:L1091–L1097. doi: 10.1152/ajplung.00421.2003. [DOI] [PubMed] [Google Scholar]
- 16.Anand S, Penrhyn-Lowe S, Venkitaraman AR. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/s1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 17.Katayama H, Sasai K, Kawai H, et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz DR, Wu R, Kardia SL, et al. Novel candidate targets of beta-catenin/T-cell factor signaling identified by gene expression profiling of ovarian endometrioid adenocarcinomas. Cancer Res. 2003;63:2913–2922. [PubMed] [Google Scholar]
- 19.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 20.Greene FL, Stewart AK, Norton HJ. A new TNM staging strategy for node-positive (stage III) colon cancer: an analysis of 50,042 patients. Ann Surg. 2002;236:416–421. doi: 10.1097/00000658-200210000-00003. discussion 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaika A, Irwin M, Sansome C, Moll UM. Oncogenes induce and activate endogenous p73 protein. J Biol Chem. 2001;276:11310–11316. doi: 10.1074/jbc.M005737200. [DOI] [PubMed] [Google Scholar]
- 22.Zaika AI, Slade N, Erster SH, et al. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med. 2002;196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu JC. Multiple Comparisons: Theory and Methods. London: Chapman & Hall/CRC Press; 1996. [Google Scholar]
- 24.Belkhiri A, Zaika A, Pidkovka N, Knuutila S, Moskaluk C, el-Rifai W. Darpp-32: a novel antiapoptotic gene in upper gastrointestinal carcinomas. Cancer Res. 2005;65:6583–6592. doi: 10.1158/0008-5472.CAN-05-1433. [DOI] [PubMed] [Google Scholar]
- 25.Dudek H, Datta SR, Franke TF, et al. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 26.Kulik G, Klippel A, Weber MJ. Antiapoptotic signalling by the insulin-like growth factor I receptor, phosphatidylinositol 3-kinase, and Akt. Mol Cell Biol. 1997;17:1595–1606. doi: 10.1128/mcb.17.3.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roder DM. The epidemiology of gastric cancer. Gastric Cancer. 2002;(5) suppl 1:5–11. doi: 10.1007/s10120-002-0203-6. [DOI] [PubMed] [Google Scholar]
- 28.Drewitz DJ, Sampliner RE, Garewal HS. The incidence of adenocarcinoma in Barrett’s esophagus: a prospective study of 170 patients followed 4.8 years. Am J Gastroenterol. 1997;92:212–215. [PubMed] [Google Scholar]
- 29.Spechler SJ. Barrett’s esophagus and esophageal adenocarcinoma: pathogenesis, diagnosis, and therapy. Med Clin North Am. 2002;86:1423–1445. vii. doi: 10.1016/s0025-7125(02)00082-2. [DOI] [PubMed] [Google Scholar]
- 30.Earle CC, Maroun JA. Adjuvant chemotherapy after curative resection for gastric cancer in non-Asian patients: revisiting a meta-analysis of randomised trials. Eur J Cancer. 1999;35:1059–1064. doi: 10.1016/s0959-8049(99)00076-3. [DOI] [PubMed] [Google Scholar]
- 31.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 32.Fraizer GC, Diaz MF, Lee IL, Grossman HB, Sen S. Aurora-A/STK15/BTAK enhances chromosomal instability in bladder cancer cells. Int J Oncol. 2004;25:1631–1639. [PubMed] [Google Scholar]
- 33.Wang XX, Liu R, Jin SQ, Fan FY, Zhan QM. Overexpression of Aurora-A kinase promotes tumor cell proliferation and inhibits apoptosis in esophageal squamous cell carcinoma cell line. Cell Res. 2006;16:356–366. doi: 10.1038/sj.cr.7310046. [DOI] [PubMed] [Google Scholar]
- 34.Yan X, Cao L, Li Q, et al. Aurora C is directly associated with Survivin and required for cytokinesis. Genes Cells. 2005;10:617–626. doi: 10.1111/j.1365-2443.2005.00863.x. [DOI] [PubMed] [Google Scholar]
- 35.Bykov VJ, Issaeva N, Shilov A, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 36.Yang H, Burke T, Dempsey J, et al. Mitotic requirement for aurora A kinase is bypassed in the absence of aurora B kinase. FEBS Lett. 2005;579:3385–3391. doi: 10.1016/j.febslet.2005.04.080. [DOI] [PubMed] [Google Scholar]
- 37.Wong HK, Fricker M, Wyttenbach A, et al. Mutually exclusive subsets of BH3-only proteins are activated by the p53 and c-Jun N-terminal kinase/c-Jun signaling pathways during cortical neuron apoptosis induced by arsenite. Mol Cell Biol. 2005;25:8732–8747. doi: 10.1128/MCB.25.19.8732-8747.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yakovlev AG, Di Giovanni S, Wang G, Liu W, Stoica B, Faden AI. BOK and NOXA are essential mediators of p53-dependent apoptosis. J Biol Chem. 2004;279:28367–28374. doi: 10.1074/jbc.M313526200. [DOI] [PubMed] [Google Scholar]