Abstract
The growing need for medical diagnostics in resource limited settings is driving the development of simple, standalone immunoassay devices. A capillary flow device using polymerization based amplification is capable of blocking a microfluidic channel in response to target biomaterials, enabling multiple modes of detection that require little or no supplemental instrumentation.
Microfluidic biodetection platforms are advantageous over their macro-scale counterparts, owing to lower analyte volumes, decreased reagent cost, and short characteristic times for diffusion and adsorption.1 Microfluidic immunoassay systems are limited in their capacity for simple, standalone modes of detection. This shortcoming is especially challenging in biodetection in resource limited settings which serve most of the global population.2 In areas with minimal laboratory infrastructure, the low analyte volumes would drastically reduce the need for sterile collection materials and the subsequent disposal of biologically contaminated waste.3 Moreover, rapid diagnostic times and higher sensitivity are a necessity for these locations to provide immediate treatment on the first visit.2, 3 While electrochemical and spectroscopic approaches to detection are well suited to sensitive detection, their requirement for supplemental instrumentation and power sources4 limit their implementation outside of a developed medical setting. Additionally, fluorescent detection often enables improved signal to noise over visual detection schemes, but it is restricted by a requirement of specialized light sources and filters for detection.5 Here, we demonstrate the use of polymerization based amplification (PBA) as a means for simple detection in microfluidic systems. This technique enables multiple modes of detection, each allowing detection with a minimum of supplemental detection equipment (visual observation, pressure monitoring, or by redirection of fluid flow). PBA is an approach towards enhanced biodetection, which utilizes the specific, rapid and repetitive nature of radical polymerization to amplify the presence of a targeted biomolecule.6–9 In this work, we employ an enzyme-mediated redox polymerization scheme, where glucose oxidase (GOx) is used in conjunction with a monomer mixture to initiate a reaction cascade, ultimately yielding a readily observable polymer hydrogel.10 Specifically, GOx reacts with glucose and oxygen to generate gluconolactone and hydrogen peroxide. The hydrogen peroxide rapidly reacts with the Fe2+ to generate hydroxyl radicals which initiate propagating polymer chains and gelation within the microfluidic channel.
A glucose oxidase-avidin (GOx-Av) conjugate is used to achieve specific binding of the initiating enzyme (GOx) to a protein target through immunolabeling with biotinylated antibodies. The GOx-Av conjugate is bound to the surface of a microfluidic channel through conventional immunoassay techniques only in the presence of target (Figure 1A–C). After thorough rinsing, the channel is exposed to an aqueous monomer solution consisting of poly(ethylene glycol) diacrylate (PEGDA) as the reactive monomer, glucose, MES buffer, iron sulfate, and optional labeling moieties (Evans Blue or rhodamine b methacrylate). In channels exposed to the target, the enzyme is immobilized to the channel walls and will rapidly polymerize the monomer. In channels not exposed to the target, GOx-Av will be rinsed away, no polymerization will commence, and the monomer species are subsequently rinsed from the channel. Improved shelf stability of the amplification substrate materials (monomers) relative to traditional ELISA substrates is a major asset in settings with little laboratory infrastructure, as the monomer species do not require refrigerated storage.
Fig. 1.
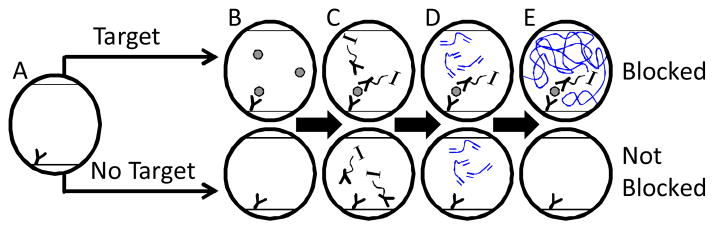
Schematic representation using polymerization to amplify the presence of a targeted biomaterial to completely block a microfluidic channel. A) Capture antibody coated channel; and B) binding of target; C) selective binding of initiator complex, denoted as “I,” through immunolabeling; D–E) selective polymerization and rinsing.
Where many PBA schemes result in a nano or micro scale polymer film, the GOx mediated PBA scheme utilizes a two-mode amplification system (enzymatic amplification AND polymerization amplification) which generates a macro-scale polymer from surface mediated reactions, capable of gelling monomer solutions in systems as large as standard 96 well plates.11–13 When confined to microfluidic environments, the system rapidly gels, and blocks the microfluidic channel (Figure 1 E). The microfluidic devices in this study were designed to be easily conjugated and are sufficiently large to permit rapid fluid flow using the capillary action of a paper towel or similar material. Devices were fabricated by placing a stoichiometric ratio of PEGDA and pentaerythritol tetra (3-mercaptopropionate) with Irgacure 184 between two acrylate-coated, glass microscope slides. A mask alignment system was used to specifically irradiate the monomer to form polymer walls. Unreacted monomer was removed with methanol. Device techniques and fabrication methods14 are found in detail in the electronic supplementary information.
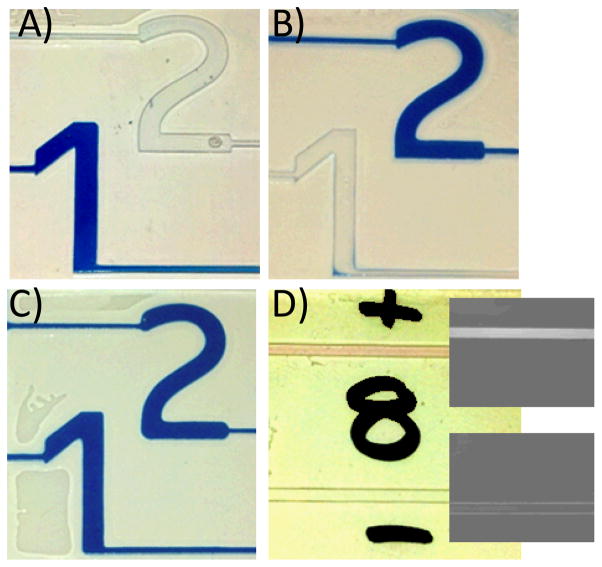
When labeling moieties are included, the presence of polymer can be readily observed in transparent microfluidic devices. For visual detection, Evans Blue is exceptionally effective at staining PEG hydrogels and does not significantly alter the limit of detection of the polymerization amplification. In Figure 2, a proof of concept multiplexed detection device discerns between two antibody systems. Thiolated15 mouse and rabbit antibodies were covalently conjugated to unreacted acrylate groups on the walls of the microfluidic channel via Michael addition in the presence of triethanol amine.16 The channels are shaped into numbers for easily interpreted results, where channel 1 was reacted with mouse IgG and channel 2 was reacted with rabbit IgG. The devices were then reacted with a 1:250 dilution of biotinylated secondary antibodies (2A: anti-mouse, 2B: anti-rabbit, 2C: anti-mouse and anti-rabbit) in phosphate buffered saline (PBS) with 0.1% bovine serum albumin (BSA). The devices were rinsed, exposed to 50 μg mL−1 GOx-Av, rinsed and then exposed to a monomer solution supplemented with 800 μg mL−1 Evans Blue. All reaction and rinse steps were for 5 minutes, and upon rinsing with deionized water, positive signals were clearly observed in appropriate microchannels (polymerization in 1, 2, and both microchannels for 2A, 2B, and 2C, respectively). See ESI for details and further discussion.
Fig. 2.
Visual detection and discrimination of antibody pairs. A–C) PBA is used to form a hydrogel in the presence of blue dye specifically in response to biodetection. Unreacted monomer and dye are then rinsed away with water. Channel 1 is coated with mouse IgG. Channel 2 is coated with rabbit IgG. A) Device exposed to analyte solution containing biotinylated antibodies against mouse. B) Device exposed to analyte solution containing biotinylated antibodies against rabbit. C) Device exposed to analyte solution containing biotinylated antibodies against both mouse and rabbit. D) “+” Channel: Specific formation of fluorescent polymer in response to avidin/biotin recognition. “−” Channel: No non-specific response from unconjugated glucose oxidase.
We also demonstrated fluorescent detection using a rhodamine-B methacrylate co-monomer added to the monomer formulation. The inclusion of 35 mM rhodamine B methacrylate does not alter the limit of detection of the polymerization system, but allows for either visual or fluorescent detection of the polymer gel (Figure 2D and insets).
Beyond visual and fluorescent detection, blockage of fluid flow in response to biodetection is an additional feature of the formation of a crosslinked polymer matrix within the channel. Biorecognition-responsive materials for microfluidic valves, where flow decisions occur independent of external action, hold the potential to drastically change the landscape of lab on a chip devices. We demonstrate PBA technology as a simple approach that is capable of readily altering the flow pattern of microfluidic devices in response to antigen detection without external detection equipment or external valve actuation. This approach could be of particular importance to the field of centrifugally-driven microfluidic devices,17 where external coupling is difficult. The lack of external detection and actuation vastly simplifies a biodetection device and its external dependencies.
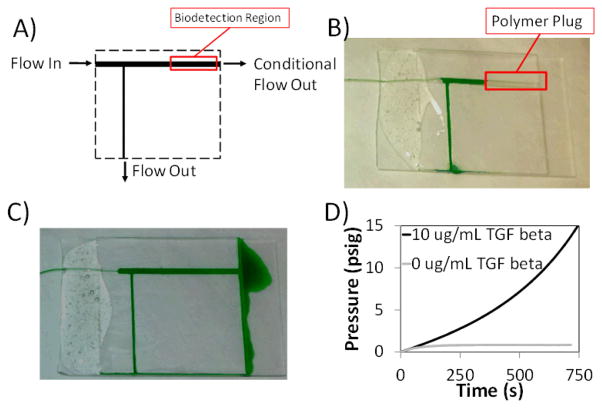
We designed a simple device (Figure 3A and ESI) to demonstrate the ability for the biodetection to alter the flow patterns in a device. When glucose oxidase is specifically bound to the channel through biotin/avidin interactions, the channel polymerizes, blocking subsequent fluid flow. In the absence of this biorecognition, there is no polymerization, and flow is permitted through the biodetection region of the channel. A main channel is coated with 2 mg mL−1 thiolated, biotinylated BSA via Michael addition in the presence of triethanol amine. The device is then blocked and exposed to 32 μg mL−1 of either GOx-Av (Figure 3B) or unconjugated GOx (Figure 3C) as a negative control against nonspecific binding. The devices were rinsed and then exposed to a monomer solution without additional labeling moieties. After a 5 minute polymerization detection period, a syringe was used to flush the devices with deionized water containing green food coloring. Figure 3B and 3C show the flow diverted by biorecognition and the unaltered flow pattern, respectively. With biorecognition and polymerization (Figure 3B), a polymer plug prevents flow across the main channel (flow from left to right), and flow of the food coloring is entirely diverted downward where it exits the device. Without biorecognition, the monomer solution does not gel, and does not impede the flow from left to right across the biodetection region. Experimental details are provided in the ESI.
Fig. 3.
A) Diagram representing devices shown in B and C. B) Diversion of flow through polymerization based amplification of the positive biotin/avidin recognition by the glucose oxidase-avidin complex. C) Lack of flow diversion when polymerization based amplification is performed on a system exposed to unconjugated glucose oxidase. D) Backpressure resulting from exposure to 0 or 10 μg mL−1 TGF-β and subsequent polymerization based amplification of a device built to detect TGF-β. A syringe pump generates pressure by depressing an air-filled syringe at a rate of 50 μL s−1.
We investigated the pressure tolerance of this microfluidic detection system in response to the detection of human transforming growth factor β1 (TGF-β) in a traditional sandwich assay format.18 Microchannel walls were coated with thiolated antibodies against TGF-β via Michael addition in the presence of triethanol amine.15, 16 Analyte solutions consisting of 0 or 10 μg mL−1 TGF-β were introduced for 5 minutes. The channels were then sequentially contacted for 5 minutes with 20 μg mL−1 biotinylated antibodies against TGF-β, 100 μg mL−1 GOx-Av, and the monomer solution, with rinses in PBS containing 0.1 % BSA between each step. Following the 5 minute polymerization, the devices were connected to a 60 mL syringe pump filled with room air, and pumped at a rate of 50 μLs−1, while the system back pressure was monitored. Representative pressure curves are presented in Figure 3D. The devices exposed to the analyte solution containing 10 μg mL−1 TGF-β generated a polymer plug capable of resisting pressures greater than 15.0 psig. The devices exposed to the negative control analyte solution were cleared of unreacted monomer at pressures below 1 psig. This dramatic distinction (>15 fold) in yield pressure between a positive and negative represents a simple approach to diagnostic readout based on pressure. A portable, rugged device interface with small bicycle pump connected to a pressure gauge would be adequate to enable biodetection in resource limited environments.
Conclusions
This work represents a critical first step in the development of PBA technology for equipment free diagnostics. PBA rapidly forms a stable polymer gel in response to the target biomaterial in a microfluidic environment. The resulting polymer can include labeling species for visual and fluorescent detection modes, or the polymer can be used as a barrier to flow for pressure and fluid flow modes of detection. The devices can be operated with capillary action, requiring no external power source, and the modes of detection are suitable for clear distinction between positive and negative results with little or no supplemental diagnostic equipment. The rapid, equipment-free nature of this approach is promising for potential deployment in low-infrastructure diagnostic sites. In addition to the application in resource limited settings, stimuli-responsive channel blockage represents a novel approach towards altering the flow characteristics of a device based on the recognition of the target species. Successful implementation of this approach would enable the design of more complex microfluidic architectures capable of performing distinct tasks based on the presence or absence of a given target.
Supplementary Material
Acknowledgments
This work was supported by NIH R21 CA 127884, NIH 5T32 HL007670-18, and NIH R21 EB012188.
Footnotes
Electronic Supplementary Information (ESI) available: A detailed description of materials and methods are in the ESI. See DOI: 10.1039/b000000x/
Notes and references
- 1.Ng AHC, Uddayasankar U, Wheeler AR. Analytical and Bioanalytical Chemistry. 397:991–1007. doi: 10.1007/s00216-010-3678-8. [DOI] [PubMed] [Google Scholar]
- 2.Urdea M, Penny LA, Olmsted SS, Giovanni MY, Kaspar P, Shepherd A, Wilson P, Dahl CA, Buchsbaum S, Moeller G, Hay Burgess DC. Nature. 2006;444:73–79. doi: 10.1038/nature05448. [DOI] [PubMed] [Google Scholar]
- 3.Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 4.Yi CQ, Zhang Q, Li CW, Yang J, Zhao JL, Yang MS. Analytical and Bioanalytical Chemistry. 2006;384:1259–1268. doi: 10.1007/s00216-005-0252-x. [DOI] [PubMed] [Google Scholar]
- 5.Avens HJ, Chang EL, May AM, Berron BJ, Seedorf GJ, Balasubramaniam V, Bowman CN. Journal of Nanoparticle Research. 2011;13:331–346. [Google Scholar]
- 6.Sikes HD, Jenison R, Bowman CN. Lab on a Chip. 2009;9:653–656. doi: 10.1039/b816198d. [DOI] [PubMed] [Google Scholar]
- 7.Avens HJ, Berron BJ, May AM, Voigt KR, Seedorf GJ, Balasubramaniam V, Bowman CN. Journal of Histochemistry & Cytochemistry. 2011;59:76–87. doi: 10.1369/jhc.2010.955948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sikes HD, Hansen RR, Johnson LM, Jenison R, Birks JW, Rowlen KL, Bowman CN. Nature Materials. 2008;7:52–56. doi: 10.1038/nmat2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson LM, Avens HJ, Hansen RR, Sewell HL, Bowman CN. Australian Journal of Chemistry. 2009;62:877–884. [Google Scholar]
- 10.Johnson LM, Fairbanks BD, Anseth KS, Bowman CN. Biomacromolecules. 2009;10:3114–3121. doi: 10.1021/bm900846m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berron BJ, Ba X, Johnson LM, Bowman CN. Biotechnology and Bioengineering. 2011;108:1521–1528. doi: 10.1002/bit.23101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwata H, Hata Y, Matsuda T, Ikada Y. Journal of Polymer Science Part a-Polymer Chemistry. 1991;29:1217–1218. [Google Scholar]
- 13.Iwata H, Hata Y, Matsuda T, Taki W, Yonekawa Y, Ikada Y. Biomaterials. 1992;13:891–896. doi: 10.1016/0142-9612(92)90111-z. [DOI] [PubMed] [Google Scholar]
- 14.Ashley JF, Cramer NB, Davis RH, Bowman CN. Lab on a Chip. 11:2772–2778. doi: 10.1039/c1lc20189a. [DOI] [PubMed] [Google Scholar]
- 15.Lin CC, Anseth KS. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:6380–6385. doi: 10.1073/pnas.1014026108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khire VS, Lee TY, Bowman CN. Macromolecules. 2007;40:5669–5677. [Google Scholar]
- 17.Cho YK, Lee JG, Park JM, Lee BS, Lee Y, Ko C. Lab on a Chip. 2007;7:565–573. doi: 10.1039/b616115d. [DOI] [PubMed] [Google Scholar]
- 18.McCall JD, Lin CC, Anseth KS. Biomacromolecules. 12:1051–1057. doi: 10.1021/bm101379v. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.