ABSTRACT
The human oral cavity is home to a large and diverse community of viruses that have yet to be characterized in patients with periodontal disease. We recruited and sampled saliva and oral biofilm from a cohort of humans either periodontally healthy or with mild or significant periodontal disease to discern whether there are differences in viral communities that reflect their oral health status. We found communities of viruses inhabiting saliva and the subgingival and supragingival biofilms of each subject that were composed largely of bacteriophage. While there were homologous viruses common to different subjects and biogeographic sites, for most of the subjects, virome compositions were significantly associated with the oral sites from which they were derived. The largest distinctions between virome compositions were found when comparing the subgingival and supragingival biofilms to those of planktonic saliva. Differences in virome composition were significantly associated with oral health status for both subgingival and supragingival biofilm viruses but not for salivary viruses. Among the differences identified in virome compositions was a significant expansion of myoviruses in subgingival biofilm, suggesting that periodontal disease favors lytic phage. We also characterized the bacterial communities in each subject at each biogeographic site by using the V3 hypervariable segment of the 16S rRNA and did not identify distinctions between oral health and disease similar to those found in viral communities. The significantly altered ecology of viruses of oral biofilm in subjects with periodontal disease compared to that of relatively periodontally healthy ones suggests that viruses may serve as useful indicators of oral health status.
IMPORTANCE
Little is known about the role or the constituents of viruses as members of the human microbiome. We investigated the composition of human oral viral communities in a group of relatively periodontally healthy subjects or significant periodontitis to determine whether health status may be associated with differences in viruses. We found that most of the viruses present were predators of bacteria. The viruses inhabiting dental plaque were significantly different on the basis of oral health status, while those present in saliva were not. Dental plaque viruses in periodontitis were predicted to be significantly more likely to kill their bacterial hosts than those found in healthy mouths. Because oral diseases such as periodontitis have been shown to have altered bacterial communities, we believe that viruses and their role as drivers of ecosystem diversity are important contributors to the human oral microbiome in health and disease states.
INTRODUCTION
We are in the early stages of understanding the tremendous diversity harbored within the human microbiome and its significant role in human health. Viruses inhabiting human body surfaces may be key factors in shaping human microbial ecology (1–7), but the potential role of viral communities in human health and disease remains largely unexplored (2, 8–10). Microbial communities can now be studied in greater detail because of the increased accessibility of sequencing technology and improved analytical capabilities (11). There have been numerous studies of the bacterial communities inhabiting human body surfaces, such as the skin (12, 13) and the gastrointestinal (14, 15), respiratory (16, 17), and genitourinary tracts (18–20) but fewer studies of the viral communities inhabiting these sites. While studies of viral communities have shown that viruses on human body surfaces are diverse (5–7, 21), studies have yet to illuminate how viral community diversity and membership pertain to human health and disease.
Periodontitis is a highly prevalent oral disease among adults (22) that results from inflammation of the supporting structures of the teeth. Some have hypothesized that the disease is caused by the host immune response to the presence of specific pathogens (23–27). Historically, microbiological aspects of periodontal disease and dental caries have been studied by culture-based or PCR/hybridization-based methods to detect pathogens collected from subgingival plaque. Bacterial species such as Porphyromonas gingivalis (28), Tannerella forsythia (29), Aggregatibacter actinomycetemcomitans (30), Streptococcus mutans (31), and Treponema denticola (32) have been implicated as etiological agents of periodontal disease by using these or similar methods (33–35). Herpesviruses such as herpes simplex virus 1 (HSV-1), cytomegalovirus (CMV), and Epstein-Barr virus (EBV) have also been looked at in association with periodontitis (36–41). Certain studies have shown an increased presence of HSV-1, CMV, and/or EBV in subgingival plaque at sites of periodontitis (36, 39, 40), while others studies have shown no association (38, 42–44). Now, as our paradigms for understanding the interconnection between microbes and human health change, much of the study of microbes in periodontal disease has shifted toward studying communities rather than individual pathogens (45–50). Rather than verification of the presence of a few viruses present in periodontitis, we are interested in the broader dynamics of the communities of organisms present on and interacting with the gingiva and associated tissues.
The oral cavity contains multiple soft and hard tissue surfaces creating diverse niches that harbor a wide range of microbiota, including >1,000 different bacterial taxa (51). While herpesviruses may be present in the oral cavity, there is a much larger population of viruses present, the majority of which are bacteriophage (6, 21). Many of these phage belong to the Caudovirus families Siphoviridae (generally lysogenic with intermediate host ranges), Myoviridae (typically lytic with relatively broad host ranges), and Podoviridae (typically lytic with relatively narrow host ranges) (52, 53). Communities of oral viruses are highly personalized and vary according to host sex (6, 54). Unrelated household contacts share significant proportions of their viromes, which suggests that substantial environmental influences affect oral viral ecology (21). Because oral viruses have been shown to elicit host immune responses, they could potentially play a role in shaping oral immunity and disease pathogenesis (10, 54). In addition, we previously demonstrated that human oral viruses carry substantial gene functions that may be involved in the pathogenic functions of their host bacteria (6), which suggests a more subtle role for viruses as members of the human oral microbiome.
In this study, we investigated oral viral community membership in a cohort of periodontally healthy subjects and those with disease. We examined viruses from planktonic saliva and from subgingival and supragingival biofilms to determine whether there are significant differences in viral community membership by oral biogeographic site and to determine whether the ecology of human oral viral communities reflects oral health status.
RESULTS
Human subjects and isolation of viruses.
We recruited 16 human subjects and sampled their saliva, subgingival plaque, and supragingival plaque. Seven of the 16 subjects had periodontal disease, while the other 9 had good overall periodontal health (Table 1). Because of the relatively low biomass at each tooth, we pooled the subgingival or supragingival plaque to have sufficient biomass to examine each subject individually.
TABLE 1 .
Characteristics of study subjects
| Status and subject | Age (yr) |
Ethnicity | Sex | Diagnosis | Smoking | Comorbidity, other information |
|---|---|---|---|---|---|---|
| Significant periodontal disease | ||||||
| D1 | 73 | Caucasian | Male | Chronic severe generalized periodontitis |
Yes | None |
| D2 | 66 | Caucasian | Male | Chronic severe generalized periodontitis |
Yes | Hypertension |
| D3 | 62 | Caucasian | Female | Chronic moderate generalized periodontitis |
No | Hypothyroidism, vegetarian |
| D4 | 69 | Caucasian | Female | Chronic severe generalized periodontitis |
No | None |
| D5 | 48 | Caucasian | Female | Chronic moderate generalized periodontitis |
Yes | None |
| D6 | 73 | Caucasian | Male | Chronic severe generalized periodontitis |
Yes | None |
| D7 | 27 | Asian | Male | Generalized aggressive mild periodontitis |
No | Amoxicillin in prior 3 mo |
| Healthy or with mild periodontal disease |
||||||
| H1 | 25 | Asian | Male | Healthy | No | None |
| H2 | 25 | Caucasian | Male | Healthy | No | None |
| H3 | 51 | Hispanic | Female | Moderate gingivitisa | No | Diabetes |
| H4 | 34 | African American | Male | Healthy | No | None |
| H5 | 32 | Asian | Female | Healthy | No | Diabetes |
| H6 | 24 | Asian | Female | Healthy | No | None |
| H7 | 27 | Caucasian | Male | Healthy | No | None |
| H8 | 44 | Caucasian | Female | Chronic mild generalized periodontitisb |
No | None |
| H9 | 50 | Hispanic | Female | Chronic mild generalized periodontitisb |
Yes | None |
Has signs of gingival inflammation but no periodontal attachment loss or bone loss. Gingivitis affects about half of U.S. adults.
Has signs of gingival inflammation and, at most, 2-mm attachment loss. Nine percent of U.S. adults have this condition.
We isolated viruses from the saliva, subgingival plaque, and supragingival plaque of each subject for a total of 48 separate viromes. DNA viruses were enriched according to our previously described protocols (6) by CsCl density gradient ultracentrifugation, followed by extraction of DNA from intact virions. All viromes were sequenced by semiconductor sequencing (55) for a total of 29,076,203 reads after processing (10,386,122 from saliva, 8,659,122 from subgingival plaque, and 10,030,956 from supragingival plaque) with a mean length of 146 nucleotides (nt). We sequenced an average of 1,817,263 reads per subject and 605,754 per individual virome. Each virome was screened for the presence of contaminating cellular nucleic acids by BLASTN analysis (E score, <10−5) against a composite database of 16S rRNA and the Human Reference Genome database (ftp://ftp.ncbi.nlm.nih.gov/genomes/H_sapiens/). None of the supragingival or subgingival plaque viromes had any identifiable homologues to 16S rRNA, and only 3 of the 16 salivary viromes had single identifiable 16S rRNA homologues (see Table S1 in the supplemental material), indicating that each of the viromes was relatively free of contaminating bacterial nucleic acids. While we did identify homologues to human DNA in almost all viromes, the mean percentage was low (mean, 0.38%; range, 0.00 to 7.15%). All homologues to human DNA were removed prior to further analysis.
Examination of viruses in saliva and oral biofilm.
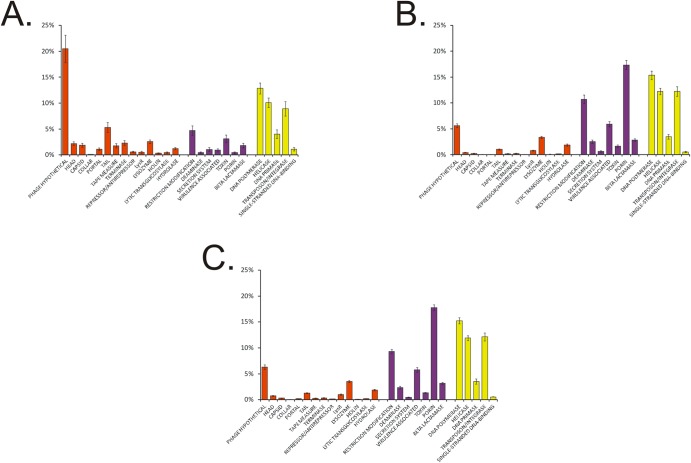
Because longer contigs are more likely to generate productive BLAST searches, we assembled the reads for all viromes in each subject. The mean number of contigs per subject was 3,468 (763 for saliva, 1,201 for subgingival plaque, and 1,504 for supragingival plaque), with a mean length of 1,041 nt (955 for saliva, 1,105 for subgingival plaque, and 1,061 for supragingival plaque). The median length of all contigs was 509 nt (491 for saliva, 516 for subgingival plaque, and 513 for supragingival plaque) (see Fig. S1A in the supplemental material), and the median GC content was 44% (44% for saliva, 43% for subgingival plaque, and 44% for supragingival plaque) (see Fig. S1B). We subjected all contigs to BLASTX analysis against the NCBI Nonredundant (NR) database to identify homologous sequences. Similar to prior studies, there was a limited number of contigs that had identifiable homologues. A significantly higher percentage of salivary contigs than contigs derived from plaque had identifiable viral homologues (40.09% of those from saliva, 27.87% of those from subgingival plaque, and 29.09% of those from supragingival plaque; P < 0.0001) (Fig. 1). The substantial differences in the structural genes identified in contigs from each biogeographic site (40.7% of the contigs from saliva, 12.4% of those from subgingival plaque, and 16.3% of those from supragingival plaque) account for much of the observed differences. These differences are likely explained by a lack of viruses present in the NR database that are homologous to subgingival and supragingival microbiota. Similar differences were not observed for homologues involved in viral replication and integration, where 37.0% of the contigs from saliva had these homologues, 43.8% of those from subgingival plaque had them, and 43.3% of those from supragingival plaque had them (Fig. 1).
FIG 1 .
Bar graphs of the mean percentages of contigs (± the standard errors) with viral homologues in the NR database from all of the subjects. Panels: A, saliva; B, subgingival plaque; C, supragingival plaque.
Personalized oral viruses by biogeographic site.
We compared the oral viruses in each subject by biogeographic site (saliva versus subgingival plaque versus supragingival plaque) to determine whether there were identifiable viruses specific to each subject by site. We used BLASTN analysis (E score, <10−10) to quantify the numbers and patterns of shared homologous contigs among all 16 subjects at each site. There were numerous homologous viruses among the saliva samples from many subjects, particularly among healthy subjects H5 to H8 (see Fig. S2A in the supplemental material), as well as among the subgingival and supragingival plaque samples from all of the subjects (see Fig. S2C and E, respectively). However, the pattern of homologous viruses observed in heat maps also suggested that despite some shared viruses, many oral viruses were specific to both subgingival and supragingival plaque samples from an individual. This pattern suggested that many viruses of oral biofilm were unique to each individual. We also performed global assemblies from the reads from all of the subjects by biogeographic site and found similar patterns in saliva (see Fig. S2B) and subgingival (see Fig. S2D) and supragingival plaque (see Fig. S2F) samples that suggested that many of the oral viruses were specific to individuals.
We performed permutation tests in which we randomly sampled the virome contigs to measure the proportions of intrasubject and intersubject homologous viruses to determine whether the viral ecology at each biogeographic site was significantly individual specific. For subgingival plaque, we found that viromes demonstrated significant levels of individual-specific contigs for 14 of the 16 subjects (P < 0.05) and in supragingival plaque for 13 of the 16 subjects (P < 0.05) (Table 2). In subgingival plaque from subjects with periodontal disease, 87.8% ± 4.3% of the contigs had intrasubject homologues and 69.0% ± 13.6% had intersubject homologues. Similar results were found for periodontally healthy subjects or those with mild disease, with 88.0% ± 2.8% of the contigs with intrasubject homologues and 77.0% ± 4.3% with intersubject homologues. In supragingival plaque from the significant periodontal disease group, 89.5% ± 1.9% of the viral contigs had intrasubject homologues and 71.2% ± 11.9% had intersubject homologues; in that from periodontally healthy subjects, 88.1% ± 3.1% of the viral contigs had intrasubject homologues and 75.0% ± 6.9% had intersubject homologues. Similar results were also found for globally assembled contigs from all of the subjects by biogeographic site, where significant levels of individual-specific contigs were identified for all 16 subjects in subgingival plaque (P < 0.05; see Table S2 in the supplemental material) and for 14 of the 16 subjects in supragingival plaque (P < 0.05). These data indicate that there is a significant association between individual subjects and their biofilm viral community membership, regardless of oral health status.
TABLE 2 .
Subgingival and supragingival virome homologues within and between subjects
| Subject | % Homology in subgingival plaque |
P valueb | % Homology in supragingival plaque |
P valueb | ||
|---|---|---|---|---|---|---|
| Intrasubjecta | Intersubjecta | Intrasubjecta | Intersubjecta | |||
| D1 | 91.46 ± 3.19 | 79.89 ± 4.97 | 0.0083 | 90.97 ± 3.07 | 84.39 ± 3.61 | 0.0479 |
| D2 | 92.06 ± 3.21 | 67.56 ± 6.67 | 0.0005 | 92.26 ± 2.43 | 43.76 ± 9.61 | <0.0001 |
| D3 | 87.58 ± 2.90 | 48.75 ± 7.15 | <0.0001 | 81.49 ± 3.36 | 63.97 ± 4.29 | 0.0009 |
| D4 | 87.11 ± 3.13 | 65.79 ± 5.22 | 0.0004 | 84.55 ± 2.34 | 62.48 ± 3.50 | <0.0001 |
| D5 | 88.53 ± 4.25 | 84.25 ± 3.81 | 0.1913 | 88.58 ± 4.23 | 73.53 ± 5.48 | 0.0114 |
| D6 | 90.60 ± 3.19 | 76.35 ± 4.80 | 0.0058 | 92.37 ± 3.35 | 79.36 ± 5.58 | 0.0260 |
| D7 | 88.84 ± 3.69 | 76.13 ± 4.81 | 0.0186 | 84.40 ± 3.24 | 75.19 ± 3.42 | 0.0255 |
| H1 | 87.23 ± 2.64 | 65.70 ± 5.62 | 0.0001 | 87.31 ± 2.66 | 68.98 ± 3.99 | 0.0005 |
| H2 | 84.52 ± 2.90 | 74.00 ± 3.66 | 0.0056 | 84.51 ± 3.42 | 79.77 ± 3.09 | 0.0024 |
| H3 | 84.80 ± 3.35 | 66.25 ± 4.43 | 0.0019 | 86.52 ± 3.72 | 82.41 ± 3.02 | 0.0043 |
| H4 | 89.29 ± 2.39 | 80.49 ± 3.19 | 0.0006 | 89.72 ± 2.11 | 77.16 ± 3.41 | 0.0125 |
| H5 | 86.81 ± 2.76 | 82.42 ± 2.98 | 0.0626 | 93.90 ± 1.91 | 80.54 ± 4.21 | 0.0001 |
| H6 | 92.62 ± 1.95 | 80.09 ± 4.16 | 0.0001 | 85.93 ± 2.74 | 78.82 ± 3.04 | <0.0001 |
| H7 | 91.42 ± 2.46 | 78.12 ± 4.39 | 0.0005 | 89.86 ± 2.64 | 78.52 ± 4.22 | 0.1747 |
| H8 | 91.04 ± 2.23 | 66.98 ± 5.06 | <0.0001 | 86.71 ± 3.05 | 73.74 ± 3.51 | 0.1381 |
| H9 | 84.94 ± 2.84 | 80.72 ± 2.62 | 0.1008 | 87.32 ± 2.61 | 72.52 ± 4.17 | <0.0001 |
Based on the mean of 10,000 iterations. One thousand random contigs were sampled per iteration.
Empirical P value based on the fraction of times the estimated percent homologous contigs for each subject exceeds that for different subjects. Values that indicate statistical significance are in bold.
Viromes from saliva from all of our periodontally healthy subjects or those with mild disease demonstrated significantly higher levels of intrasubject shared homologues than intersubject shared homologues (mean, 94.8% ± 2.6% compared to 78.3% ± 5.3% [P < 0.05] for all healthy subjects) (see Table S3 in the supplemental material). For subjects with significant periodontal disease, the proportions of intrasubject and intersubject shared homologues in saliva were not significantly different for four of the seven subjects (mean for all of the subjects, 94.2% ± 1.7% compared to 87.4% ± 6.8%). For globally constructed assemblies, significant levels of individual-specific contigs were found in saliva from all 16 subjects (P < 0.05; see Table S4 in the supplemental material).
Biogeographic differences among viral communities.
We compared the virome contigs across all of the subjects by biogeographic site to determine whether there were significant proportions of virome contigs specific to each site. We did not identify any significant associations with biogeographic sites among the viruses within saliva or among the viruses within biofilms (7.8% ± 12.0% versus 5.1% ± 7.8%; P = 0.258) (Table 3). In contrast, we found highly significant associations by biogeographic site among the viruses in subgingival plaque compared to other sites (37.5% ± 9.0% versus 10.6% ± 8.7%; P = 0.0122) and supragingival plaque compared to other sites (34.5% ± 7.0% versus 10.5% ± 8.6%; P = 0.0087). These data indicate that the biogeographic site may be an important determinant of oral viral ecology. We found similar trends in the data when examining globally constructed assemblies from all reads and biogeographic sites; however, these differences were not statistically significant (see Table S5 in the supplemental material).
TABLE 3 .
Viral homologues and shared 16S rRNA OTUs within and between subject groups
| Site and/or status | % Homology in virome |
P valueb | % Homology in 16S rRNA |
P valueb | ||
|---|---|---|---|---|---|---|
| Within groupa | Between groupsa | Within groupa | Between groupsa | |||
| Sites | ||||||
| Saliva | 7.81 ± 12.03 | 5.07 ± 7.78 | 0.2584 | 47.50 ± 34.17 | 11.58 ± 25.04 | 0.1671 |
| Subgingival plaque | 37.53 ± 8.98 | 10.60 ± 8.70 | 0.0122 | 73.01 ± 16.96 | 40.76 ± 35.83 | 0.2201 |
| Supragingival plaque | 34.45 ± 6.99 | 10.50 ± 8.62 | 0.0087 | 74.29 ± 16.43 | 40.30 ± 36.16 | 0.2143 |
| Health status | ||||||
| Saliva | ||||||
| Healthy | 7.06 ± 9.67 | 1.06 ± 2.68 | 0.1090 | 34.08 ± 37.82 | 36.01 ± 32.96 | 0.3416 |
| Periodontal disease | 12.43 ± 12.40 | 1.07 ± 2.61 | 0.1254 | 70.11 ± 12.74 | 34.02 ± 32.82 | 0.1464 |
| Subgingival plaque | ||||||
| Healthy | 38.63 ± 6.40 | 13.56 ± 5.64 | <0.0001 | 73.41 ± 15.39 | 71.15 ± 15.83 | 0.4382 |
| Periodontal disease | 37.16 ± 10.86 | 13.59 ± 5.81 | 0.0221 | 77.23 ± 11.93 | 70.67 ± 15.02 | 0.3474 |
| Supragingival plaque | ||||||
| Healthy | 31.25 ± 5.91 | 12.18 ± 4.11 | 0.0009 | 62.27 ± 34.50 | 65.23 ± 27.16 | 0.3862 |
| Periodontal disease | 34.66 ± 5.64 | 12.11 ± 4.05 | 0.0018 | 74.20 ± 13.26 | 64.44 ± 27.66 | 0.4181 |
Based on the mean of 10,000 iterations. One thousand random contigs were sampled per iteration.
Empirical P value based on the fraction of times the estimated percent homologous contigs or shared OTUs for each group exceeded that between groups. Values that indicate statistical significance are in bold.
Comparison of viruses in orally healthy persons with those in persons with periodontal disease.
We next compared the oral viruses of relatively periodontally healthy subjects with those of subjects with significant disease to determine whether viral community composition might be associated with oral health status. We performed principal-coordinate analysis (PCOA) to compare the patterns of variation in shared homologues across all of the subjects and biogeographic sites. Many of the salivary viromes were similarly clustered on the basis of host disease status (Fig. 2A). A similar trend was also identified in oral biofilm, where most of the subgingival and supragingival viromes from subjects with significant periodontal disease were similarly clustered (Fig. 2B). We next quantified the proportion of homologous contigs across viromes of relatively periodontally healthy subjects or disease to determine whether the patterns of variation observed in PCOA were supported statistically. For saliva, the proportion of homologous virome contigs was greater for comparisons among subjects with significant periodontal disease (12.4% ± 12.4%) than between subjects with different oral health status (1.1% ± 2.6%), but this difference was not statistically significant (Table 3). The proportion of shared virome homologues was much greater among subjects with significant periodontal disease than in comparisons between subjects with different oral health status for subgingival plaque (37.2 ± 10.9% versus 13.6 ± 5.8%; P = 0.022) and supragingival plaque (34.7 ± 5.6% versus 12.1 ± 4.1%; P = 0.002) (Table 3). These data indicate that human oral viral ecology in oral biofilm is significantly associated with oral health status. While similar trends were found in the data for globally constructed assemblies, the data were not significant for any biogeographic site (see Table S5 in the supplemental material).
FIG 2 .
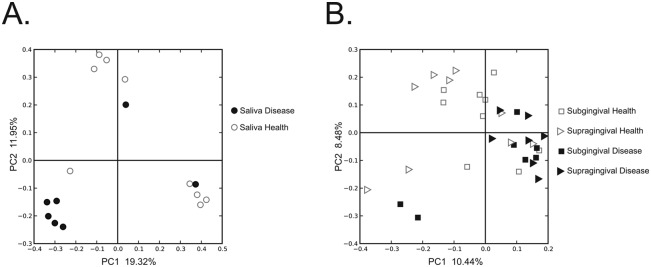
PCOA of beta diversity present in the viromes of each subject at each biogeographic site. Relatively periodontally healthy subjects are represented in white, and subjects with significant periodontal disease are represented in black. Circles represent saliva, squares represent subgingival plaque, and triangles represent supragingival plaque. Panels: A, saliva; B, subgingival and supragingival plaque.
We characterized the virus families in all of the subjects to determine whether significant differences existed in periodontally healthy subjects and those with disease by biogeographic site. In samples from each site in orally healthy subjects, members of the family Siphoviridae were the most abundant virus types (Fig. 3), and their generally lysogenic lifestyle suggests that lysogeny is the preferred state of viruses in orally healthy subjects. In addition, the abundance of podoviruses from each site was similar but the relative abundance of myoviruses varied considerably by site and disease state. Myoviruses were significantly more abundant in saliva from healthy individuals (Fig. 3A), though siphoviruses remained the most abundant viral type in healthy subjects and in those with disease. In subgingival plaque, however, we found myoviruses to be significantly more abundant in those with periodontal disease than in healthy subjects (Fig. 3B). Myoviruses are generally lytic, and their predominance in subjects with periodontal disease suggests a more active role for viruses in driving bacterial diversity in the subgingival crevice. The myoviruses in subgingival plaque from subjects with periodontal disease were even more abundant than siphoviruses, which was not observed in any other oral microenvironment studied. No significant differences were identified in the virus families found in supragingival plaque (Fig. 3C). Thus, virome membership was significantly altered in subjects with periodontal disease, predominantly as a result of the increased abundance of myoviruses in their subgingival plaque.
FIG 3 .
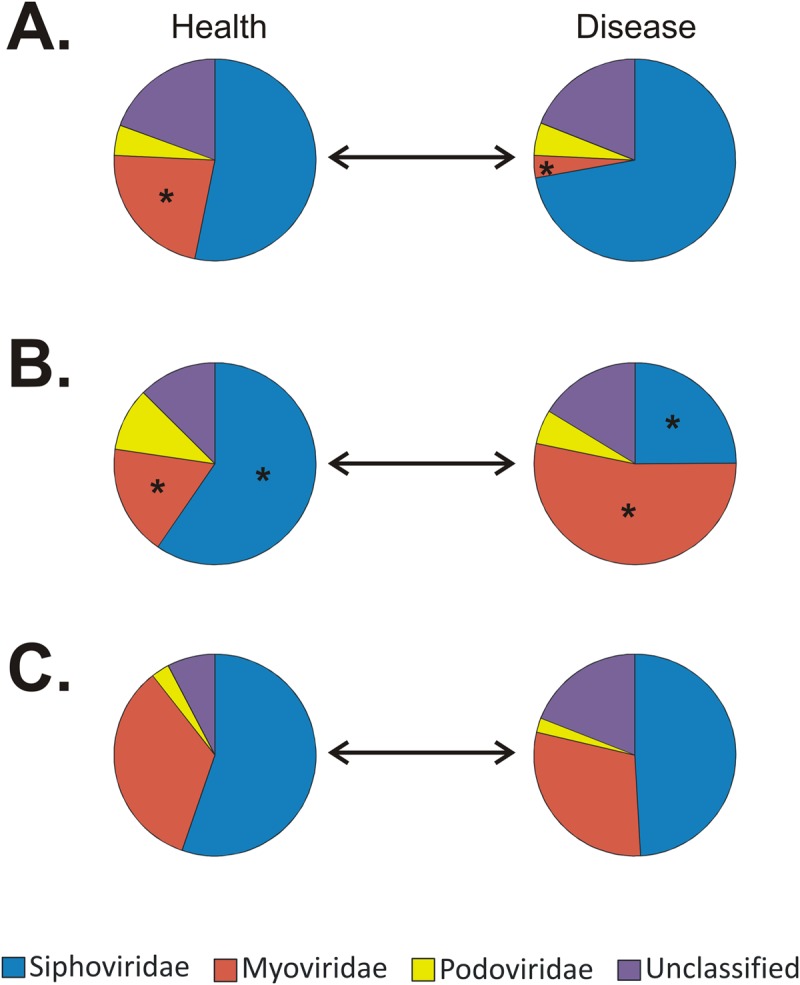
Pie charts of bacteriophage families present in the saliva (A) and subgingival (B) or supragingival plaque (C) samples of relatively periodontally healthy subjects (left) and those with disease (right). Asterisks indicate significant differences (P ≤ 0.05) between the proportions of virus families identified in periodontally healthy subjects and those with disease.
Characterization of bacterial communities.
On the basis of our findings that there were significant associations between oral viruses and individual subjects (see Fig. S2 in the supplemental material; Table 2), biogeographic sites (Table 3), and oral health status (Table 3), we investigated whether similar trends might also be identifiable for oral bacterial communities. We examined these communities on the basis of the V3 hypervariable region of the 16S rRNA. We sequenced 880,410 16S rRNA reads after processing, for a mean of 55,026 reads per subject and 18,342 reads per biogeographic site. We performed rarefaction analyses, which demonstrated that most of the diversity present in each subject had been adequately sampled for saliva (see Fig. S3A in the supplemental material) and subgingival (see Fig. S3B) and supragingival plaque (see Fig. S3C). Diversity of bacterial communities was estimated by using the Shannon diversity index (H′). In the subgingival and supragingival plaque samples from most of our subjects, the estimated species diversity was greater in relatively orally healthy subjects (H′, 6.5) than in those with significant periodontal disease (H′, 5.3). When the data for all of the subjects and biogeographic sites by oral health status were combined, the estimated species diversity in relatively orally healthy subjects exceeded that in subjects with periodontal disease (see Fig. S3D; H′, 7.0 in healthy subjects versus 6.4 in those with disease). Species richness in saliva exceeded that in both subgingival and supragingival plaque, regardless of oral health status (see Fig. S3E).
We used PCOA to examine the patterns of variation observed in oral bacterial communities by biogeographic site. We found variation differentiating the salivary microbiota from those of oral biofilm (Fig. 4A); however, there was little distinction between the subgingival and supragingival biota. We also quantified the proportion of shared bacterial operational taxonomic units (OTUs) by biogeographic site in all of our subjects to determine whether there were statistically significant differences in the biota at each site. The proportion of the biota shared within each biogeographic site was greater than that shared by different sites (47.5% versus 11.6% for saliva, 73.0% versus 40.8% for subgingival plaque, and 74.3% versus 40.3% for supragingival plaque), but none of these differences was statistically significant (Table 3). We found a distinct variation in the viromes of relatively orally healthy subjects and those with significant periodontal disease (Fig. 2), but similar significant trends could not be identified for the bacterial biota (Fig. 4B and Table 3). There was a significant association between virome constituents and oral biogeographic sites (Table 3), but no significant associations were identified in the bacterial biota (see Tables S6 and S7 in the supplemental material).
FIG 4 .
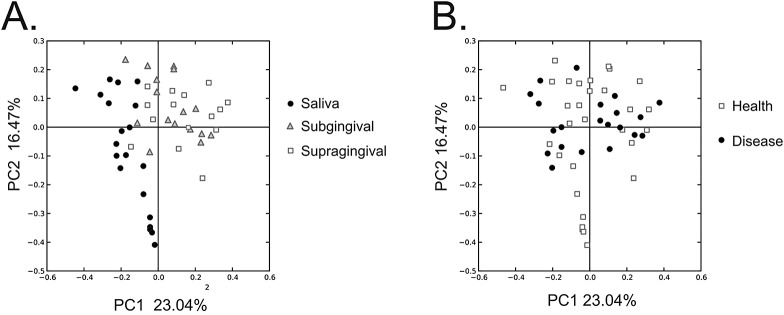
PCOA of beta diversity present in the bacterial communities of each subject at each biogeographic site. Panels: A, classified by biogeographic site; B, classified by oral health status.
Taxonomic compositions of viral and bacterial communities.
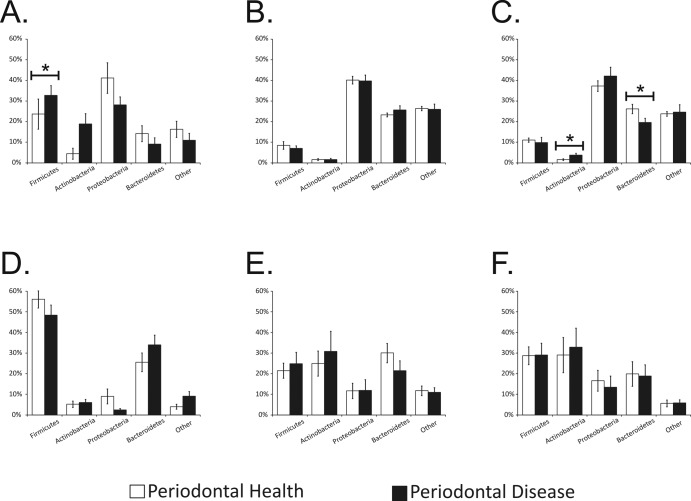
We compared the putative taxonomic compositions of viral communities across each biogeographic site to determine whether there were identifiable differences at a high taxonomic level. We found that viruses of Firmicutes and Proteobacteria were highly prevalent in saliva, followed by those of Bacteroidetes and Actinobacteria (Fig. 5A). In contrast, viruses of Proteobacteria and Bacteroidetes predominated in oral biofilm, with those of Firmicutes and Actinobacteria representing only a small minority of the viruses identified (Fig. 5B and C). The relative proportions of Firmicutes, Actinobacteria, and Bacteroidetes phage were all significantly different (P < 0.05) when the taxonomic compositions of viral communities in saliva and oral biofilm were compared. Many of these differences were also reflected in the taxonomies of the bacterial communities, where Firmicutes predominated in saliva, compared to a relatively even distribution of Firmicutes, Actinobacteria, and Bacteroidetes in oral biofilm (Fig. 5D to F).
FIG 5 .
Bar plots (means ± standard errors of the means) of putative viral host taxonomy (A and C) and bacterial taxonomy based on 16S rRNA (D to F) at the phylum level. Each phylum is shown on the x axis, and the percentage of contigs or OTUs identified belonging to the observed phyla is shown on the y axis. Panels A and D represent saliva, panels B and E represent subgingival plaque, and panels C and F represent supragingival plaque. White bars represent relatively periodontally healthy subjects, and black bars represents subjects with significant periodontal disease. Asterisks indicate significant differences (P ≤ 0.05) between phyla in periodontally healthy subjects and those with disease.
We also examined the taxonomic composition of the viral communities in relatively periodontally healthy subjects with that of the communities in subjects with significant disease. No significant differences were identified in subgingival plaque from periodontally healthy subjects and those with disease, despite the preponderance of myoviruses in subjects with disease. This suggests that the significant differences in viral communities may be more closely related to the virus families present than to their putative bacterial hosts (Fig. 3B). In saliva, there was a trend to lower levels of Firmicutes and Actinobacteria in orally healthy subjects than in those with disease, although only the difference in Firmicutes levels was statistically significant (Fig. 5A). In supragingival plaque, there was a significant decrease in the proportion of Bacteroidetes and an increase an Actinobacteria phage in subjects with periodontal disease (Fig. 5C), but the biological basis of these findings in saliva and supragingival plaque is unclear.
DISCUSSION
Human body surfaces are inhabited by endemic viral communities whose role on human body surfaces has not been thoroughly investigated. We previously demonstrated that there are robust communities of viruses that inhabit human saliva (6) and that many of these viruses are highly persistent over time (54). Because many of these persistent oral viruses are bacteriophage, they could play a role in determining oral bacterial community membership. We investigated the membership of the oral viral community in relatively periodontally healthy subjects and those with significant disease to determine whether there might exist ecological distinctions in the viral populations reflected in oral health status. We examined the viruses present in planktonic saliva and in subgingival and supragingival biofilms to provide a broad overview of different oral ecological niches. Our finding that membership in the biofilm virome is significantly associated with periodontal health and disease is the first statistically supported evidence that oral viral community membership is associated with a human disease condition.
Bacteriophage are significant drivers of bacterial diversity in a variety of ecosystems (1–7), and most of the viruses identified in saliva and dental plaque were phage (Fig. 3). In this study, many of the phage encountered at all oral sites were predicted to be siphoviruses, which generally have lysogenic lifestyles by integrating into their host genomes. Lysogenic oral viruses live in dynamic equilibrium with their cellular hosts and, as a result, are highly persistent members of the human oral microbiome (54). Myoviruses are often lytic and, because of their increased virulence for their host bacteria, may have a great impact upon local bacterial diversity. Myoviruses were highly prevalent in the subgingival crevice in subjects with periodontal disease (Fig. 3B), suggesting an expanded role in driving bacterial diversity in oral biofilm. In the development of periodontal disease, the surfaces of the gums and bones pull away from the teeth, forming pockets that are generally inhabited by different bacteria. While the profound differences in the subgingival virobiota may merely reflect changes in the bacterial biota that colonize these exposed pockets, the differences in the subgingival plaque virome in subjects with periodontal disease may also have other biological implications. The lytic phage in the subgingival crevice likely help to shape the local microbiota and contribute to the local microbial community structure and alter local biodiversity.
There were significant differences between the putative bacterial hosts of viruses in saliva and those of viruses in oral biofilm when examined at a high taxonomic level (Fig. 5). Similar taxonomic differences were not observed when periodontally healthy subjects were compared with those with disease, suggesting that much of the observed differences in our subjects may have been due to changes in the relative abundance of virus families rather than their bacterial hosts. Because of their substantial coevolution with their bacterial hosts, culture-independent methods based on patterns of nucleotide usage are relatively accurate in predicting the hosts of lysogenic viruses at the genus level (56). However, techniques used to predict bacterial hosts are generally less successful at predicting the hosts of lytic viruses; thus, we opted to characterize lytic viruses only at a high taxonomic level in this study to reduce the potential for inaccuracy.
Our data show a strong link between oral viruses and periodontal health (Table 3), biogeographic sites (Table 3), and individual subjects (Table 2) but likely only partially describe the robust communities of viruses that inhabit the human oral cavity. Our methods and sequencing depth were designed to characterize the most abundant oral viruses (6) and likely offer limited insight into the less-abundant members of the human oral virome. This analysis included only DNA viruses, and it remains unclear whether the mouth is also inhabited by RNA viral communities. Because of the limited starting volume of saliva and the relatively scarce quantities of DNA recovered, we used multiple-displacement amplification (MDA) before sequencing each virome. MDA may introduce biases into sequence data, particularly when relatively small quantities of starting nucleic acids are used (57). There are no available collections of non-MDA-treated oral viromes for comparison; however, substantial systematic and directional amplification biases would be necessary to reproduce the statistically significant trends found in these viromes. We used two separate techniques to assemble the sequence data, and they showed similar trends in individual-specific, biogeographic site-specific, and oral health status-specific viruses in each subject or subject group. We preferred to create the initial assemblies from each subject rather than use global assemblies from all of the subjects to reduce the potential for chimerism. Although the two methods showed similar trends, only the method that uses assemblies from individual subjects produced statistically significant results (Tables 2 and 3).
While this study demonstrated a significantly altered oral viral ecology in subjects with periodontal disease, other studies have investigated viral community constituents in other disease conditions. There are conserved viral genotypes in the respiratory tracts of human subjects with cystic fibrosis (2), which could potentially be attributable to shared bacterial ecology. Our data, however, indicate a high level of commonality among bacterial species representation between relatively periodontally healthy subjects and those with more advanced disease but still showed that viruses were significantly associated with oral health status. A study of gut microbial communities in persons with Crohn’s disease showed lower viral and bacterial diversity than in controls (58); however, both could potentially have been explained by antibiotic administration. Only one healthy subject in this study received any antibiotics (Table 1), and thus, antibiotics cannot explain the significant differences between the viral communities found in periodontally healthy subjects and those found in subjects with disease (Table 3). A recent study demonstrated a strong association between (i) host immune status and the use of antiviral therapies and (ii) viral community constituents in human plasma (8), indicating that viral communities respond to drug-mediated perturbations. The effects of such perturbations on human oral viral communities have yet to be reported on.
Biogeographic differences in human microbial ecology have been noted for cellular microbiota inhabiting different body surfaces. Our approach is the first to assess oral biogeographic differences in the viral microbiota of the human microbiome. Because of the substantial biomass necessary to evaluate DNA viruses by our techniques, we had to pool plaque samples from individual teeth in order to have sufficient biomass to examine subgingival and supragingival viral communities. As expected, the viral (Fig. 3B) and bacterial (Fig. 5A) biota of subgingival and supragingival plaque samples had similarities; however, each was quite distinct from that of saliva. The difference in viral ecology between planktonic saliva and biofilm communities is likely attributable to differences in the ecology of the bacteria and archaea that inhabit each individual niche and the various lifestyles observed (Fig. 3). Biogeographic differences in the cellular microbiota may also explain our relative inability to identify sequences homologous to many biofilm-derived viral structural genes (Fig. 1), as the biofilm had abundant bacteria belonging to Bacteroidetes and Actinobacteria (Fig. 5D to F) relative to Firmicutes (highly represented in saliva) and the representation of viruses for these bacteria may be heterogeneous in the NR database. There are several factors that likely contribute to our finding significant associations between viruses, individual subjects, and oral health status, while not identifying similar trends in the bacterial biota. First, the difference in the techniques used (metagenomics versus 16S rRNA amplicon sequencing) probably accounted for some of the differences observed, likely resulting in lower resolution for the detection of differences among the members of the bacterial biota. Second, our analysis of viral and bacterial community constituents was based on the relative abundance of community members rather than just their presence or absence. When taking into account only the presence or absence of taxa, both the virobiota and the bacterial biota showed significant subject specificity and associations with oral health status (data not shown). Lastly, many human subjects have bacterial species in common; however, the prophage in these genomes vary considerably (59, 60). The relative host specificity of these prophage, particularly when they are expressed as virions, may account for much of the subject specificity detailed in this report. Therefore, the relatively large proportion of lysogenic siphoviruses identified in each subject and at each biogeographic site would contribute substantially to the subject specificity observed.
The data produced in this study and those of other studies characterizing human viral community ecology together suggest that viral communities respond to perturbation (8) and environmental factors similar to their counterpart bacterial communities. We recently demonstrated that unrelated household contacts are significantly more likely to have oral viruses in common (61), suggesting that they may be exposed to viruses from the same environmental reservoir. Furthermore, oral viruses are significantly associated with the sex of their human host (54), suggesting that host factors such as hormones may play a formative role in human viral ecology. The reservoir of antibiotic resistance is expanded in the mouse gut virome in response to antibiotic perturbation (62). In gut viromes, diet plays an important formative role in the ecology of viral communities (3). While the nature of the perturbations that result in periodontal disease in humans may be variable, our data demonstrate that altered oral viral ecology is an associated feature of significant periodontitis. Viruses of bacteria have the potential to eradicate their hosts or to provide them with beneficial gene functions (6); thus, the predominantly lytic viruses in the subgingival crevice may have the capacity to shape the natural history of the oral microbiome in persons with periodontal disease. While the oral microbiome has been hypothesized to play a role in the development of periodontal disease (63–65), the role of viruses in these microbial communities has not been elucidated. Because of their potential to shape human microbial communities, as well as host immune responses, viruses likely also play an important role in human oral health status.
MATERIALS AND METHODS
Human subjects.
Subject recruitment and enrollment were approved by the University of California, San Diego, and the Western University Administrative Panels on Human Subjects in Medical Research. All of the subjects signed informed-consent forms indicating their willingness to participate in this study. Each subject underwent a baseline periodontal examination, including measurements of probing depths, clinical attachment loss, gingival index, plaque index, and gingival irritation (66), and their oral health status was recorded (Table 1). Dental plaque was collected from subgingival and supragingival biofilm samples from teeth 3, 9, 12, 19, 25, and 28 and placed into 200 µl of 0.02-µm-filtered phosphate-buffered saline (PBS; Fisher Scientific, Chico, CA). Approximately 3 ml of saliva was also collected from each subject without stimulation. All specimens were immediately frozen on dry ice and stored at −20°C until use in this study. All of the subjects completed a questionnaire detailing their dietary habits and comorbidities. Exclusion criteria included preexisting medical conditions that could result in substantial immunosuppression. All of the participating subjects were unrelated, and only one had received any antibiotics in the 3 months prior to the beginning of the study (Table 1).
Isolation and sequencing of oral viruses
Subgingival and supragingival plaque samples from each subject were pooled separately, washed twice in 0.02 micron filtered PBS, and spun at 6,000 × g for 10 min to pellet the biofilm. The biofilm was then incubated at 37°C for 30 min and vortexed vigorously for 10 min to separate out viruses. The biofilm was then spun at 6,000 × g for 10 min, and the supernatants were treated in the same manner as the saliva samples, by using previously described methods for enrichment and extraction of nucleic acids from viruses (6). Briefly, samples were filtered sequentially with 0.45- and 0.2-µm filters (VWR, Radnor, PA) to remove cellular and other debris and purified on a cesium chloride gradient. Only the fraction with a density corresponding to most known bacteriophage (67) was retained, further purified on Amicon YM-100 protein purification columns (Millipore, Inc., Billerica, MA), treated with DNase I, and subjected to lysis and DNA purification with the Qiagen UltraSens virus kit (Qiagen, Valencia, CA). The resulting DNA was amplified with the GenomiPhi V2 DNA amplification kit (GE Healthcare, Pittsburgh, PA), fragmented to roughly 200 to 400 bp with a Bioruptor (Diagenode, Denville, NJ), made into libraries with the Ion Plus Fragment Library kit (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions, and sequenced with 314 chips on an Ion Torrent Personal Genome Machine (PGM; Life Technologies, Grand Island, NY) (55).
Processing and analysis of virome sequences.
Sequencing reads were trimmed according to modified Phred scores of 0.5 with CLC Genomics Workbench 4.65 (CLC bio USA, Cambridge, MA), and low-complexity reads (where >25% of the length was due to homopolymer tracts) were removed prior to further analysis. After trimming and removal of low-complexity reads, any remaining reads with substantial length variation (<50 or >300 nt) or reads with ambiguous characters were removed prior to further analysis. Reads were screened for homology to a composite 16S rRNA database including the Ribosomal Database Project database (68), the Green Genes database (69) and the Silva database (70) by BLASTN analysis with an E score cutoff value of 10−5. Reads were also screened for homology to the Human Reference Database at ftp://ftp.ncbi.nlm.nih.gov/genomes/H_sapiens/ by BLASTN analysis with an E score cutoff value of 10−5. Any reads homologous to sequences in the human database were removed prior to further analysis. Reads then were assembled with CLC Genomics Workbench 4.65 (CLC bio, Cambridge, MA) to construct contigs based on 98% identity with a minimum of 50% read overlap, consistent with criteria developed to discriminate between highly related viruses (71). Because the shortest reads were 50 nt, the minimum tolerable overlap was 25 nt and the mean overlap was no less than 73 nt, depending on the characteristics of each virome. Any contigs of <200 nt or with ambiguous characters were removed prior to further analysis. Length and GC content variation among contigs were assessed by using box-and-whisker plots created with Microsoft Excel 2007 (Microsoft Corp., Redmond, WA).
We used BLASTX analysis against the NR database (E score cutoff, 10−5) to find viral homologues to contigs from each subject and biogeographic site. Homologous contigs were determined by parsing BLASTX results for known viral genes, including replication, structural, transposition, restriction/modification, hypothetical, and other genes previously found in viruses for which the E score was at least 10−5. Each individual virome contig was annotated by this technique (Fig. 1); however, if the best hit for any portion of the contig was to a gene with no known function, lower-level hits were used as long as they had known functions and still met the E score cutoff. The annotation data were compiled for all of the subjects and used to determine the relative proportion of assembled contigs that contained viral homologues. The phylum of the cellular hosts for each annotated contig was used to determine taxonomic distributions in each subject and at each biogeographic site. The relative abundances of virus families were determined by BLASTX analysis of the SEED database with MG-RAST (72). Analysis of shared homologues in each virome was performed by creating custom BLAST databases for each virome, comparing each database with all other viromes by BLASTN analysis (E score, <10−10). Heat maps were generated on the basis of shared homologues across all of the subjects and depicted with JAVA Treeview (73). Heat map data were normalized on the basis of the total number of viral contigs for each virome. PCOA was performed on homologous virome contigs by using binary Sorensen distances and Qiime (74). We also used a separate technique for assembly by constructing global assemblies from all of the reads from all of the subjects and all of the time points with 98% identity over a minimum of 50% read overlap. The contribution of each subject and time point to each assembly was assessed and used to construct heat maps with Java Treeview (73).
Analysis of 16S rRNA.
Genomic DNA was prepared from saliva or pooled subgingival or supragingival plaque from each subject with the Qiagen QIAamp DNA minikit (Qiagen, Valencia, CA). Each sample was subjected to a bead-beating step prior to nucleic acid extraction with Lysing Matrix-B (MP Bio, Santa Ana, CA). We amplified the bacterial 16S rRNA V3 hypervariable region with the forward primer 341F (CCTACGGGAGGCAGCAG) fused with the Ion Torrent Adaptor A sequence and one of 23 unique 10-bp bar codes and reverse primer 514R (ATTACCGCGGCTGCTGG) fused with the Ion Torrent Adaptor P1 from each subject and biogeographic site (75). PCRs were performed with Platinum PCR SuperMix (Invitrogen, Carlsbad, CA) with the following cycling parameters: 94°C for 10 min, followed by 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 30 s and a final elongation step of 72°C for 10 min. Resulting amplicons were purified on a 2% agarose gel stained with SYBR Safe (Invitrogen, Carlsbad, CA) with the MinElute PCR purification kit (Qiagen, Valencia, CA). Amplicons were further purified with AMPure beads (Beckman Coulter, Brea, CA), and molar equivalents were determined for each sample with a Bioanalyzer 2100 HS DNA kit (Agilent Technologies, Santa Clara, CA). Samples were pooled into equimolar proportions and sequenced on 314 chips with an Ion Torrent PGM according to the manufacturer’s instructions (Life Technologies, Grand Island, NY) (55). Resulting sequence reads were removed from the analysis if they were <130 nt, had any bar code or primer errors, contained any ambiguous characters, or contained any stretch of >6 homopolymers. Sequences were assigned to their respective samples on the basis of their 10-nt bar code sequences and analyzed further with Qiime (74). Briefly, representative OTUs from each set were chosen at a minimum sequence identity of 97% with UClust (76) and aligned with PyNast (77) against the GreenGenes database (69). Multiple alignments then were used to create phylogenies with FastTree (78), and taxonomy was assigned to each OTU with the RDP classifier (79, 80). PCOA was performed on the basis of beta diversity by using weighted UniFrac distances (81). Qiime was also used to calculate Shannon diversity indices.
Statistical analysis.
To assess whether viromes had significant overlap within or between subjects or subject groups, we performed a permutation test based on resampling (10,000 iterations). We simulated the distribution of the fraction of shared virome homologues from different subjects, biogeographic sites, or oral health statuses that were randomly chosen across all of the subjects and sites. For each set, we computed the summed fraction of shared homologues by using 1,000 random contigs between randomly chosen subjects or subject groups and from these computed an empirical null distribution of our statistic of interest (the fraction of shared homologues). The simulated statistics within each subject or group of subjects was referred to the null distribution of intersubject or intergroup comparisons, and the P value was computed as the fraction of times the simulated statistic for the each exceeded the observed statistic. An identical analysis was performed at the OTU level for the 16S rRNA taxonomic assignments. This technique was also used to assess the relative contributions of individual subjects and time points to global virome assemblies constructed from the reads of all of the subjects at all of the time points. We assessed whether any randomly selected contig had a higher proportion of intrasubject reads than intersubject reads recruited in its assembly.
Nucleotide sequence accession numbers.
The virome and 16S rRNA sequences obtained in this study are available for download in the MG-RAST database (http://metagenomics.anl.gov/) under the project Phage Biofilm Study or under consecutive individual accession numbers 4547358.3 to 4547405.3 for the viromes and 4547630.3 to 4547677.3 for the 16S rRNAs.
SUPPLEMENTAL MATERIAL
Box-and-whisker plots of the median viral contig length (A) and GC content (B) of each subject at each biogeographic site. The gray boxes represent the third quartile, the black boxes represent the first quartile, and the intersection of the gray and black boxes represents the second quartile (median). The error bars represent the minimum and maximum of all data points. The median contig length or GC content is shown on the y axis, and each subject from D1 to D7 and H1 to H9 from left to right is shown on the x axis. Download
Heat map of homologous contigs in the saliva and subgingival and supragingival plaque of all of the subjects (A, C, and E, respectively) and the contribution of reads to global assemblies in saliva from all of the subjects at all of the time points (B, D, and F, respectively). The heat maps are organized by subject and oral health status, where each column represents an individual subject. In panels A, C, and E, the rows represent contigs from each individual subject. The “matrix-like” appearance of the heat map is due to the high intensity of homologous contigs within each subject. In panels B, D, and F, the rows represent contigs assembled from all of the subjects at all of the time points, and each column represents the relative contribution of each subject to each individual contig. Download
Rarefaction analysis on the basis of chao1 estimates of bacterial OTUs present in each subject by biogeographic site. Panels: A, saliva; B, subgingival plaque; C, supragingival plaque; D, all biogeographic sites combined by oral health status; E, all relatively periodontally healthy subjects or those with periodontal disease combined by biogeographic site. Download
Virome reads from saliva (a) and subgingival (b) and supragingival plaque (c) samples.
Recruitment of reads to subgingival and supragingival plaque contigs within and between subjects and sexes.
Saliva virome homologues within and between subjects.
Recruitment of reads to salivary contigs within and between subjects and sexes.
Recruitment of reads to globally assembled virome contigs within and between subject groups.
Saliva shared 16S rRNA OTUs within and between subjects.
Subgingival and supragingival plaque sample shared 16S rRNA OTUs.
ACKNOWLEDGMENTS
This study was supported by the Robert Wood Johnson Foundation, the Burroughs Wellcome Fund, and NIH 1K08AI085028 to D.T.P.
D.T.P. conceived and designed the experiments. M.L., S.R.A., R.R.-S., and M.N. performed the experiments. D.T.P., T.S.R., and S.R.A. analyzed the data. T.K.B. contributed reagents and performed examinations. D.T.P. and S.R.A. wrote the manuscript.
Footnotes
Citation Ly M, Abeles SR, Boehm TK, Robles-Sikisaka R, Naidu M, Santiago-Rodriguez T, Pride DT. 2014. Altered oral viral ecology in association with periodontal disease. mBio 5(3):e01133-14. doi:10.1128/mBio.01133-14.
REFERENCES
- 1. Willner D, Furlan M, Schmieder R, Grasis JA, Pride DT, Relman DA, Angly FE, McDole T, Mariella RP, Jr, Rohwer F, Haynes M. 2011. Metagenomic detection of phage-encoded platelet-binding factors in the human oral cavity. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4547–4553. 10.1073/pnas.1000089107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F. 2009. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One 4:e7370. 10.1371/journal.pone.0007370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. 2011. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21:1616–1625. 10.1101/gr.122705.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466:334–338. 10.1038/nature09199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, Gouilh MA, Pariente K, Segondy M, Burguière A, Manuguerra JC, Caro V, Eloit M. 2012. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One 7:e38499. 10.1371/journal.pone.0038499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pride DT, Salzman J, Haynes M, Rohwer F, Davis-Long C, White RA, III, Loomer P, Armitage GC, Relman DA. 2012. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 6:915–926. 10.1038/ismej.2011.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. 2013. Rapid evolution of the human gut virome. Proc. Natl. Acad. Sci. U. S. A. 110:12450–12455. 10.1073/pnas.1300833110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, Okamoto J, Snyder TM, Cornfield DN, Nicolls MR, Weill D, Bernstein D, Valantine HA, Quake SR. 2013. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell 155:1178–1187. 10.1016/j.cell.2013.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lepage P, Colombet J, Marteau P, Sime-Ngando T, Doré J, Leclerc M. 2008. Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut 57:424–425. 10.1136/gut.2007.134668 [DOI] [PubMed] [Google Scholar]
- 10. Duerkop BA, Hooper LV. 2013. Resident viruses and their interactions with the immune system. Nat. Immunol. 14:654–659. 10.1038/nrm3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou Y, Gao H, Mihindukulasuriya KA, La Rosa PS, Wylie KM, Vishnivetskaya T, Podar M, Warner B, Tarr PI, Nelson DE, Fortenberry JD, Holland MJ, Burr SE, Shannon WD, Sodergren E, Weinstock GM. 2013. Biogeography of the ecosystems of the healthy human body. Genome Biol. 14:R1. 10.1186/gb-2013-14-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blaser MJ, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Estrada I, Gao Z, Clemente JC, Costello EK, Knight R. 2013. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. ISME J. 7:85–95. 10.1038/ismej.2012.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang L, Lu X, Nossa CW, Francois F, Peek RM, Pei Z. 2009. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology 137:588–597. 10.1053/j.gastro.2009.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang YJ, Charlson ES, Collman RG, Colombini-Hatch S, Martinez FD, Senior RM. 2013. The role of the lung microbiome in health and disease. A National Heart, Lung, and Blood Institute workshop report. Am. J. Respir. Crit. Care Med. 187:1382–1387. 10.1164/rccm.201303-0488WS [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, Flores SC, Fontenot AP, Ghedin E, Huang L, Jablonski K, Kleerup E, Lynch SV, Sodergren E, Twigg H, Young VB, Bassis CM, Venkataraman A, Schmidt TM, Weinstock GM, Microbiome Lung HIV Project 2013. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 187:1067–1075. 10.1164/rccm.201210-1913OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hummelen R, Macklaim JM, Bisanz JE, Hammond JA, McMillan A, Vongsa R, Koenig D, Gloor GB, Reid G. 2011. Vaginal microbiome and epithelial gene array in post-menopausal women with moderate to severe dryness. PLoS One 6:e26602. 10.1371/journal.pone.0026602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oakley BB, Fiedler TL, Marrazzo JM, Fredricks DN. 2008. Diversity of human vaginal bacterial communities and associations with clinically defined bacterial vaginosis. Appl. Environ. Microbiol. 74:4898–4909. 10.1128/AEM.02884-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stokholm J, Schjørring S, Eskildsen CE, Pedersen L, Bischoff AL, Følsgaard N, Carson CG, Chawes BL, Bønnelykke K, Mølgaard A, Jacobsson B, Krogfelt KA, Bisgaard H. 18 November 2013. Antibiotic use during pregnancy alters the commensal vaginal microbiota. Clin. Microbiol. Infect. (Epub ahead of print.) 10.1111/1469-0691.12411 [DOI] [PubMed] [Google Scholar]
- 21. Robles-Sikisaka R, Ly M, Boehm T, Naidu M, Salzman J, Pride DT. 2013. Association between living environment and human oral viral ecology. ISME J. 7:1710–1724. 10.1038/ismej.2013.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ, CDC Periodontal Disease Surveillance Workgroup 2012. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J. Dent. Res. 91:914–920. 10.1177/0022034512457373 [DOI] [PubMed] [Google Scholar]
- 23. Assuma R, Oates T, Cochran D, Amar S, Graves DT. 1998. IL-1 and TNF antagonists inhibit the inflammatory response and bone loss in experimental periodontitis. J. Immunol. 160:403–409 [PubMed] [Google Scholar]
- 24. Graves DT, Delima AJ, Assuma R, Amar S, Oates T, Cochran D. 1998. Interleukin-1 and tumor necrosis factor antagonists inhibit the progression of inflammatory cell infiltration toward alveolar bone in experimental periodontitis. J. Periodontol. 69:1419–1425. 10.1902/jop.1998.69.12.1419 [DOI] [PubMed] [Google Scholar]
- 25. Garlet GP, Cardoso CR, Campanelli AP, Ferreira BR, Avila-Campos MJ, Cunha FQ, Silva JS. 2007. The dual role of p55 tumour necrosis factor-alpha receptor in Actinobacillus actinomycetemcomitans-induced experimental periodontitis: host protection and tissue destruction. Clin. Exp. Immunol. 147:128–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Al-Rasheed A, Scheerens H, Rennick DM, Fletcher HM, Tatakis DN. 2003. Accelerated alveolar bone loss in mice lacking interleukin-10. J. Dent. Res. 82:632–635. 10.1177/154405910308200812 [DOI] [PubMed] [Google Scholar]
- 27. Jain S, Darveau RP. 2010. Contribution of Porphyromonas gingivalis lipopolysaccharide to periodontitis. Periodontol. 2000 54:53–70. 10.1111/j.1600-0757.2009.00333.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Darveau RP, Hajishengallis G, Curtis MA. 2012. Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res. 91:816–820. 10.1177/0022034512453589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gersdorf H, Meissner A, Pelz K, Krekeler G, Göbel UB. 1993. Identification of Bacteroides forsythus in subgingival plaque from patients with advanced periodontitis. J. Clin. Microbiol. 31:941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ximénez-Fyvie LA, Haffajee AD, Socransky SS. 2000. Microbial composition of supra- and subgingival plaque in subjects with adult periodontitis. J. Clin. Periodontol. 27:722–732. 10.1034/j.1600-051x.2000.027010722.x [DOI] [PubMed] [Google Scholar]
- 31. Loesche WJ. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sela MN. 2001. Role of Treponema denticola in periodontal diseases. Crit. Rev. Oral Biol. Med. 12:399–413. 10.1177/10454411010120050301 [DOI] [PubMed] [Google Scholar]
- 33. Ximénez-Fyvie LA, Haffajee AD, Socransky SS. 2000. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 27:648–657. 10.1034/j.1600-051x.2000.027009648.x [DOI] [PubMed] [Google Scholar]
- 34. Socransky SS, Haffajee AD, Teles R, Wennstrom JL, Lindhe J, Bogren A, Hasturk H, van Dyke T, Wang X, Goodson JM. 2013. Effect of periodontal therapy on the subgingival microbiota over a 2-year monitoring period. I. Overall effect and kinetics of change. J. Clin. Periodontol. 40:771–780. 10.1111/jcpe.12117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saygun I, Nizam N, Keskiner I, Bal V, Kubar A, Açıkel C, Serdar M, Slots J. 2011. Salivary infectious agents and periodontal disease status. J. Periodontal Res. 46:235–239. 10.1111/j.1600-0765.2010.01335.x [DOI] [PubMed] [Google Scholar]
- 36. Imbronito AV, Okuda OS, Maria de Freitas N, Moreira Lotufo RF, Nunes FD. 2008. Detection of herpesviruses and periodontal pathogens in subgingival plaque of patients with chronic periodontitis, generalized aggressive periodontitis, or gingivitis. J. Periodontol. 79:2313–2321. 10.1902/jop.2008.070388 [DOI] [PubMed] [Google Scholar]
- 37. Chalabi M, Rezaie F, Moghim S, Mogharehabed A, Rezaei M, Mehraban B. 2010. Periodontopathic bacteria and herpesviruses in chronic periodontitis. Mol. Oral Microbiol. 25:236–240. 10.1111/j.2041-1014.2010.00571.x [DOI] [PubMed] [Google Scholar]
- 38. Stein JM, Yekta S, Kleines M, Ok D, Kasaj A, Reichert S, Schulz S, Scheithauer S. 2013. Failure to detect an association between aggressive periodontitis and the prevalence of herpesviruses. J. Clin. Periodontol. 40:1–7. 10.1111/jcpe.12021 [DOI] [PubMed] [Google Scholar]
- 39. Sharma R, Padmalatha O, Kaarthikeyan G, Jayakumar ND, Varghese S, Sherif K. 2012. Comparative analysis of presence of cytomegalovirus (CMV) and Epstein-Barr virus-1 (EBV-1) in cases of chronic periodontitis and aggressive periodontitis with controls. Indian J. Dent. Res. 23:454–458. 10.4103/0970-9290.104948 [DOI] [PubMed] [Google Scholar]
- 40. Das S, Krithiga GS, Gopalakrishnan S. 2012. Detection of human herpes viruses in patients with chronic and aggressive periodontitis and relationship between viruses and clinical parameters. J. Oral Maxillofac. Pathol. 16:203–209. 10.4103/0973-029X.98502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cappuyns I, Gugerli P, Mombelli A. 2005. Viruses in periodontal disease—a review. Oral Dis. 11:219–229. 10.1111/j.1601-0825.2005.01123.x [DOI] [PubMed] [Google Scholar]
- 42. Dawson DR, III, Wang C, Danaher RJ, Lin Y, Kryscio RJ, Jacob RJ, Miller CS. 2009. Salivary levels of Epstein-Barr virus DNA correlate with subgingival levels, not severity of periodontitis. Oral Dis. 15:554–559. 10.1111/j.1601-0825.2009.01585.x [DOI] [PubMed] [Google Scholar]
- 43. Saygun I, Kubar A, Sahin S, Sener K, Slots J. 2008. Quantitative analysis of association between herpesviruses and bacterial pathogens in periodontitis. J. Periodontal Res. 43:352–359. 10.1111/j.1600-0765.2007.01043.x [DOI] [PubMed] [Google Scholar]
- 44. Nibali L, Atkinson C, Griffiths P, Darbar U, Rakmanee T, Suvan J, Donos N. 2009. Low prevalence of subgingival viruses in periodontitis patients. J. Clin. Periodontol. 36:928–932. 10.1111/j.1600-051X.2009.01476.x [DOI] [PubMed] [Google Scholar]
- 45. Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8:481–490. 10.1038/nrmicro2337 [DOI] [PubMed] [Google Scholar]
- 46. Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. 2006. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J. Clin. Microbiol. 44:3665–3673. 10.1128/JCM.00317-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lim YW, Evangelista JS, III, Schmieder R, Bailey B, Haynes M, Furlan M, Maughan H, Edwards R, Rohwer F, Conrad D. 2014. Clinical insights from metagenomic analysis of sputum samples from patients with cystic fibrosis. J. Clin. Microbiol. 52:425–437. 10.1128/JCM.02204-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Castelino M, Eyre S, Upton M, Ho P, Barton A. 2014. The bacterial skin microbiome in psoriatic arthritis, an unexplored link in pathogenesis: challenges and opportunities offered by recent technological advances. Rheumatology (Oxford) 53:777–784 [DOI] [PubMed] [Google Scholar]
- 49. Peterson SN, Snesrud E, Liu J, Ong AC, Kilian M, Schork NJ, Bretz W. 2013. The dental plaque microbiome in health and disease. PLoS One 8:e58487. 10.1371/journal.pone.0058487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ling Z, Kong J, Jia P, Wei C, Wang Y, Pan Z, Huang W, Li L, Chen H, Xiang C. 2010. Analysis of oral microbiota in children with dental caries by PCR-DGGE and barcoded pyrosequencing. Microb. Ecol. 60:677–690. 10.1007/s00248-010-9712-8 [DOI] [PubMed] [Google Scholar]
- 51. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J. Bacteriol. 192:5002–5017. 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wichels A, Biel SS, Gelderblom HR, Brinkhoff T, Muyzer G, Schütt C. 1998. Bacteriophage diversity in the North Sea. Appl. Environ. Microbiol. 64:4128–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sullivan MB, Waterbury JB, Chisholm SW. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047–1051. 10.1038/nature01929 [DOI] [PubMed] [Google Scholar]
- 54. Abeles SR, Robles-Sikisaka R, Ly M, Lum AG, Salzman J, Boehm TK, Pride DT. 20 March 2014. Human oral viruses are personal, persistent and gender-consistent. ISME J. (Epub ahead of print.) 10.1038/ismej.2014.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J. 2011. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352. 10.1038/nature10242 [DOI] [PubMed] [Google Scholar]
- 56. Pride DT, Wassenaar TM, Ghose C, Blaser MJ. 2006. Evidence of host-virus co-evolution in tetranucleotide usage patterns of bacteriophages and eukaryotic viruses. BMC Genomics 7:8. 10.1186/1471-2164-7-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim KH, Bae JW. 2011. Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl. Environ. Microbiol. 77:7663–7668. 10.1128/AEM.00289-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Perez-Brocal V, Garcia-Lopez R, Vazquez-Castellanos JF, Nos P, Beltran B, Latorre A, Moya A. 2013. Study of the viral and microbial communities associated with Crohn’s disease: a metagenomic approach. Clin. Transl. Gastroenterol. 4:e36. 10.1038/ctg.2013.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cooke FJ, Wain J, Fookes M, Ivens A, Thomson N, Brown DJ, Threlfall EJ, Gunn G, Foster G, Dougan G. 2007. Prophage sequences defining hot spots of genome variation in Salmonella enterica serovar Typhimurium can be used to discriminate between field isolates. J. Clin. Microbiol. 45:2590–2598. 10.1128/JCM.00729-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Green NM, Beres SB, Graviss EA, Allison JE, McGeer AJ, Vuopio-Varkila J, LeFebvre RB, Musser JM. 2005. Genetic diversity among type emm28 group A Streptococcus strains causing invasive infections and pharyngitis. J. Clin. Microbiol. 43:4083–4091. 10.1128/JCM.43.8.4083-4091.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Robles-Sikisaka R, Ly M, Boehm T, Naidu M, Salzman J, Pride DT. 2013. Association between living environment and human oral viral ecology. ISME J. 7:1710–1724. 10.1038/ismej.2013.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Modi SR, Lee HH, Spina CS, Collins JJ. 2013. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499:219–222. 10.1038/nature12212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ge X, Rodriguez R, Trinh M, Gunsolley J, Xu P. 2013. Oral microbiome of deep and shallow dental pockets in chronic periodontitis. PLoS One 8:e65520. 10.1371/journal.pone.0065520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Huang S, Yang F, Zeng X, Chen J, Li R, Wen T, Li C, Wei W, Liu J, Chen L, Davis C, Xu J. 2011. Preliminary characterization of the oral microbiota of Chinese adults with and without gingivitis. BMC Oral Health 11:33. 10.1186/1472-6831-11-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wade WG. 2013. The oral microbiome in health and disease. Pharmacol. Res. 69:137–143. 10.1016/j.phrs.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 66. Löe H. 1967. The Gingival Index, the Plaque Index and the Retention Index Systems. J. Periodontol. 38(Suppl):610–616. 10.1902/jop.1967.38.6.610 [DOI] [PubMed] [Google Scholar]
- 67. Murphy FA, Fauquet CM, Ghabrial BSA, Jarvis AW, Martelli GP, Mayo MA, Summers MD. 1995. Virus taxonomy: the sixth report of the International Committee on Taxonomy of Viruses. Springer Verlag, New York, NY. http://webcache.googleusercontent.com/search?q=cache:http:// jpkc.jluhp.edu.cn/zwkx/zwbl/improve/ref/APSNET/life.anu edu.au/viruses/report.htm [Google Scholar]
- 68. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkp353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. GreenGenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, Azam F, Rohwer F. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 99:14250–14255. 10.1073/pnas.202488399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. 10.1186/1471-2105-9-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Saldanha AJ. 2004. Java TreeView—extensible visualization of microarray data. Bioinformatics 20:3246–3248. 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- 74. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Whiteley AS, Jenkins S, Waite I, Kresoje N, Payne H, Mullan B, Allcock R, O’Donnell A. 2012. Microbial 16S rRNA Ion Tag and community metagenome sequencing using the Ion Torrent (PGM) platform. J. Microbiol. Methods 91:80–88. 10.1016/j.mimet.2012.07.008 [DOI] [PubMed] [Google Scholar]
- 76. Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 77. Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 79. Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650. 10.1093/molbev/msp077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lozupone C, Hamady M, Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. 10.1186/1471-2105-7-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Box-and-whisker plots of the median viral contig length (A) and GC content (B) of each subject at each biogeographic site. The gray boxes represent the third quartile, the black boxes represent the first quartile, and the intersection of the gray and black boxes represents the second quartile (median). The error bars represent the minimum and maximum of all data points. The median contig length or GC content is shown on the y axis, and each subject from D1 to D7 and H1 to H9 from left to right is shown on the x axis. Download
Heat map of homologous contigs in the saliva and subgingival and supragingival plaque of all of the subjects (A, C, and E, respectively) and the contribution of reads to global assemblies in saliva from all of the subjects at all of the time points (B, D, and F, respectively). The heat maps are organized by subject and oral health status, where each column represents an individual subject. In panels A, C, and E, the rows represent contigs from each individual subject. The “matrix-like” appearance of the heat map is due to the high intensity of homologous contigs within each subject. In panels B, D, and F, the rows represent contigs assembled from all of the subjects at all of the time points, and each column represents the relative contribution of each subject to each individual contig. Download
Rarefaction analysis on the basis of chao1 estimates of bacterial OTUs present in each subject by biogeographic site. Panels: A, saliva; B, subgingival plaque; C, supragingival plaque; D, all biogeographic sites combined by oral health status; E, all relatively periodontally healthy subjects or those with periodontal disease combined by biogeographic site. Download
Virome reads from saliva (a) and subgingival (b) and supragingival plaque (c) samples.
Recruitment of reads to subgingival and supragingival plaque contigs within and between subjects and sexes.
Saliva virome homologues within and between subjects.
Recruitment of reads to salivary contigs within and between subjects and sexes.
Recruitment of reads to globally assembled virome contigs within and between subject groups.
Saliva shared 16S rRNA OTUs within and between subjects.
Subgingival and supragingival plaque sample shared 16S rRNA OTUs.