ABSTRACT
The broad range and diversity of interferon-stimulated genes (ISGs) function to induce an antiviral state within the host, impeding viral pathogenesis. While successful respiratory viruses overcome individual ISG effectors, analysis of the global ISG response and subsequent viral antagonism has yet to be examined. Employing models of the human airway, transcriptomics and proteomics datasets were used to compare ISG response patterns following highly pathogenic H5N1 avian influenza (HPAI) A virus, 2009 pandemic H1N1, severe acute respiratory syndrome coronavirus (SARS-CoV), and Middle East respiratory syndrome CoV (MERS-CoV) infection. The results illustrated distinct approaches utilized by each virus to antagonize the global ISG response. In addition, the data revealed that highly virulent HPAI virus and MERS-CoV induce repressive histone modifications, which downregulate expression of ISG subsets. Notably, influenza A virus NS1 appears to play a central role in this histone-mediated downregulation in highly pathogenic influenza strains. Together, the work demonstrates the existence of unique and common viral strategies for controlling the global ISG response and provides a novel avenue for viral antagonism via altered histone modifications.
IMPORTANCE
This work combines systems biology and experimental validation to identify and confirm strategies used by viruses to control the immune response. Using a novel screening approach, specific comparison between highly pathogenic influenza viruses and coronaviruses revealed similarities and differences in strategies to control the interferon and innate immune response. These findings were subsequently confirmed and explored, revealing both a common pathway of antagonism via type I interferon (IFN) delay as well as a novel avenue for control by altered histone modification. Together, the data highlight how comparative systems biology analysis can be combined with experimental validation to derive novel insights into viral pathogenesis.
INTRODUCTION
Acute respiratory tract infections represent a considerable threat, causing significant morbidity and mortality globally (1). In particular, emerging pathogens, including influenza A virus and coronavirus, have caused minor to major outbreaks of viral pneumonia worldwide (2). In the current work, we compared the host response to four distinct emerging respiratory viruses: a highly pathogenic H5N1 avian influenza (HPAI) virus, influenza A/Vietnam/1203/2004 (H5N1; referred to as H5N1-VN1203); a 2009 pandemic influenza virus, A/California/04/2009 (H1N1; referred to as H1N1-09); severe acute respiratory syndrome coronavirus (SARS-CoV); and the recently emergent Middle East respiratory syndrome CoV (MERS-CoV). Each viral family causes severe disease, encodes several immune-modulatory elements, and remains a threat for future pandemics (2, 3). However, despite similar disease manifestations, influenza viruses and CoVs also exhibit sharp contrasts in terms of replication, immune stimulation, and overall lethality (4). Together, the similarities and differences offer an opportunity to identify both conserved and pathogen-specific host responses important during respiratory virus infection.
While previous studies have undertaken global analysis built on systems biology datasets (5–8), we took a new approach that focused on a parameter known to be important to viral infection: the interferon (IFN)-stimulated gene (ISG) response. Type I IFN induces a signaling cascade that provides the first line of defense against viral pathogens and initiates transcription of hundreds of ISGs that have antiviral, immune modulatory, and cell regulatory functions (9). Successful viral pathogens, including CoVs and influenza viruses, have evolved genetic functions that antagonize pathogen recognition as well as ISG effector functions (10, 11). Yet, as a whole, the ISG response has never been globally examined in the context of multiple viral pathogens in the same system; therefore, we sought to compare and contrast ISG control strategies employed by influenza A and pathogenic CoVs.
Using virologic and transcriptomic data, the results demonstrated distinct approaches used by each virus to interfere with the global ISG response. Differences and similarities were noted between strains within each virus family, and expression patterns were further validated by proteomic data. Finally, computational and empirical studies provided insights into conserved and novel mechanisms of ISG control. Whereas the HPAI virus actively manipulated the ISG response with up- and downregulation of ISG subsets, H1N1-09 produced strong, uniform induction. In contrast, SARS-CoV and MERS-CoV successfully delayed ISG expression until after peak viral titers were achieved. Notably, MERS-CoV also downregulated a subset of ISGs, overlapping part of the signature seen with HPAI virus. Mechanistic studies revealed that absent and delayed IFN induction was responsible for the ISG antagonism observed in the CoVs. In addition, ISG downregulation in both HPAI virus and MERS-CoV was not due to disruption of signaling but rather correlates with altered histone modification, a novel avenue to impede ISG transcription. Finally, varied antagonism of HPAI virus mutants suggested that NS1 contributes to ISG control via altered histone methylation and may impact virulence in other severe influenza virus infections. Together, the data highlight unique and conserved approaches used by disparate respiratory viral pathogens to manipulate and control the global ISG responses.
RESULTS
Type I IFN treatment and viral infections induce diverse ISG expression profiles.
ISG expression varies based on cell and tissue type. Therefore, we set out to define ISGs in Calu3 cells, a human respiratory cell line permissive for both CoV and influenza infection. Calu3 cells were treated with type I IFN (IFN-α or IFN-β), resulting in differential expression of >350 genes (log2 fold change [FC] of >1.5, adjusted P value of <0.05; Fig. 1A). Using these data, a consensus ISG list was developed from genes induced at more than one time point (see Table S1 in the supplemental material) and then used to examine ISG expression changes following infection. While cytopathic effect (CPE) in both H5N1-VN1203 and H1N1-09 required a slightly lower multiplicity of infection (MOI) to maximize comparable time points, infection of the cultures was uniform (>95% infection), as measured by CPE or fluorescent virus infection. In addition, each infection maintained similar kinetics in terms of both replication and genomic RNA production despite differing endpoint titers (Fig. 1B and C).
FIG 1 .
Type I IFN treatment and viral infections induce diverse ISG expression profiles. (A) Total number of genes induced by type I IFN treatment with a log2 FC of >1.5 at indicated time points in Calu3 cells. Red numbers indicate genes included in the consensus ISG list (see Table S1 in the supplemental material). All ISG and analysis available online as outlined in data dissemination (supplement). Viral titers (B) and viral genomic RNA (C) following infection of Calu3 cells with H5N1-VN1203 (MOI of 1; blue), H1N1-09 (MOI of 3; red), SARS-CoV (MOI of 5; green), or MERS-CoV (MOI of 5; orange). (D) Global ISG transcriptional response to IFN-α treatment or infection. Genes ordered by MERS-CoV or H5N1-VN1203 expression levels 24 hpi and grouped into subsets based on fold change: downregulated in both MERS-CoV and H5N1-VN1203 (purple; <−0.75), downregulated in H5N1-VN1203 (blue; <−0.75), minimal stimulation (green; 0.75 to −0.75), and upregulated (orange; >0.75). Values represent log2 FC compared to time-matched mocks.
Following IFN-α treatment or infection, the ISG RNA expression patterns showed both similarities and stark contrasts (Fig. 1D). H1N1-09 quickly and robustly induced the majority (97%) of ISGs, mimicking type I IFN treatment. However, H5N1-VN1203 induced robust induction in only 35% of the consensus-annotated ISGs (log2 FC of >0.75 at 24 h postinfection [hpi]) (Fig. 1D, orange panel); in contrast, a significant subset of ISGs (26%) had only minimal induction (green panel). Finally, the largest percentage (39%; log2 FC of <−0.75 at 24 hpi, purple and blue panels) was downregulated relative to mock following H5N1-VN1203 infection. Based on these groupings, the same ISGs were almost uniformly upregulated in response to H1N1-09 infection (Fig. 1D). In contrast, ISG expression was neither strongly up- nor downregulated following the first 12 h of SARS-CoV infection; only after 24 to 36 hpi are the majority (88% at 72 hpi) of ISGs induced. However, several ISGs showed only minimal, if any, stimulation (e.g., TLR3, SERPIN1), and ACE2, the receptor for SARS-CoV, was downregulated, corresponding with previous reports (12). For MERS-CoV, the first 12 h also found no significant ISG induction, after which expression mirrored H5N1-VN1203 with upregulation (42%), no stimulation (39%), and downregulation (19%) of ISG subsets (see Table S1). Together, these results demonstrate contrasting and overlapping approaches used by H5N1-VN1203, H1N1-09, SARS-CoV, and MERS-CoV in their regulation of the global ISG response.
Proteomic analysis validated ISG antagonism following CoV and influenza virus infection.
In parallel to RNA expression experiments, infected Calu3 cells were examined by global proteomics analysis. Numerous factors affect proteomics coverage, including a protein’s solubility, hydrophobicity, and location within the cell, thus preventing analysis of major protein subsets, including secreted and membrane-bound peptides. Despite these limitations, significant changes in protein levels provided independent validation of the RNA expression trends. For H1N1-09, the limited control of ISG RNA expression also manifested in robust ISG protein production; 27 ISG proteins were among 530 significantly upregulated host proteins (P or G test value of <0.05 at any time point; see Fig. S1A in the supplemental material), with production detected as early as 12 hpi and the majority of ISG protein (63%) produced by 24 hpi. In contrast, only 3 of the 382 proteins significantly upregulated by H5N1-VN1203 belonged to the consensus ISG list, validating the RNA expression analysis. While SARS-CoV induces 20 ISG proteins over its 72-h time course, none of these ISG proteins are detected prior to 30 hpi; similarly, MERS-CoV induces five ISG proteins, with only one (STAT1) detected prior to 18 hpi, thus confirming delayed ISG induction in both CoVs.
Examination of individual protein expression patterns also confirmed these trends. Although most ISGs are absent in mock samples, two detectable proteins were expressed at basal levels: STAT1 and PKR. H5N1-VN1203 infection had reduced protein expression of both STAT1 (27.4% decrease at 24 hpi) and PKR (28.5% decrease at 18 hpi) relative to that of mock; similarly, MERS-CoV reduced the protein level of STAT1 (15.8% decrease at 18 to 24 hpi) and PKR (20.2% decrease at 18 to 24 hpi) (see Fig. S1B). In contrast, both proteins were significantly increased following both H1N1-09 and SARS-CoV infection; however, the increase occurred only at late times (>30 hpi) for SARS-CoV, contrasting that for H1N1-09. Further examination of individual ISGs found a consistent linkage between mRNA expression (see Fig. S1B, solid line) and protein abundance (dotted line). For H1N1-09, mRNA upregulation by 3 to 7 hpi corresponded to parallel protein production 7 to 12 h later for ISGs; representative kinetics for MX1, OAS2, and IFIT family members are shown (see Fig. S1C). A similar mRNA/protein kinetic relationship was observed for SARS-CoV, showing delayed RNA induction, which allowed viral replication to peak prior to ISG protein production (see Fig. S1D). For both H5N1-VN1203 and MERS-CoV, this analysis was excluded due to insufficient detectable data to produce a representative curve. Together, the proteomics data validated the RNA expression results and confirmed unique strategies to control the ISG response between different viral strains and families.
Delayed IFN induction mediates ISG antagonism by CoVs.
Having established differential ISG regulation at both the RNA and protein levels, we next sought to determine the means of control. One possible global mechanism is to prevent, disrupt, or delay initial induction of either type I or type III IFN, another important antiviral cytokine. While several IFN-independent ISGs exist, the vast majority of genes are augmented by IFN production (9); naturally, both CoV and influenza family members have developed approaches to prevent this induction by disturbing the sensing pathways (10, 11). Yet, despite the presence of antagonists, both H5N1-VN1203 and H1N1-09 infection resulted in robust transcription of type I and type III IFN molecules (Fig. 2A); IFN-β1, IFN-α5, and IFN-λ1 were each strongly induced in both influenza strains. These results are consistent with previous reports for both viruses (13, 14) and indicated host recognition via intact sensing pathways despite the presence of IFN antagonists like NS1. In contrast, SARS-CoV infection resulted in IFN-β induction only after 12 h (Fig. 2); IFN-α5 and IFN-λ1 were even further delayed and dampened compared to those in the influenza infections. Similarly, MERS-CoV failed to induce IFN genes prior to 12 h but also induced more robust IFN-α5 relative to that of SARS-CoV, possibly implicating signaling differences in antagonism between the two CoVs. Together, the IFN induction data indicated that unlike the influenza strains, CoV infections more tightly block recognition and/or disrupt IFN induction. Coupled with previous reports of the sensitivity of SARS- and MERS-CoV to type I IFN (15, 16), these data argued that delayed ISG expression enhances CoV infection.
FIG 2 .
Delayed IFN induction mediates ISG antagonism by CoVs. Individual gene RNA expression of IFNB1, IFNL1, and IFNA5 molecules derived from microarray following H5N1-VN1203 (blue), H1N1-09 (red), SARS-CoV (green), or MERS-CoV (orange) infection.
Transcriptional factor binding preference corresponds with differential ISG expression.
For H1N1-09 infection, robust IFN expression syncs with observed global ISG induction (Fig. 1D). In contrast, while the strong induction of type I and III IFN corresponds to previous reports for H5N1-VN1203 (14), the up- and downregulation of ISG subsets indicate that the global IFN response is altered by the more virulent influenza strain. Notably, while blocking recognition pathways and IFN signaling have been described as means to impair ISG induction (10, 17), neither approach would directly result in ISG downregulation relative to mock. In fact, the results suggest that H5N1-VN1203 ISG antagonism occurs after pathway activation, possibly through interfering with transcriptional factor binding. However, ingenuity pathway analysis (IPA) of IRF7-, STAT1-, and NF-κB-dependent genes demonstrated no uniform pattern of either specific induction or repression 12 h after H5N1-VN1203 infection (see Fig. S2A in the supplemental material; also, data not shown); in addition, IPA upstream Z scoring, a value based on downstream expression of connected genes, revealed lower activation scores for these transcriptional factors in H5N1-VN1203 than in H1N1-09 but no evidence of repression (Z score < −2.0), reflecting both increased and decreased downstream ISG expression (see Fig. S2B to D). The analysis suggested that H5N1-VN1203 does not achieve downregulation via blockade of transcriptional factors; instead, these data and IFN expression values indicate that upstream ISG signaling remains intact.
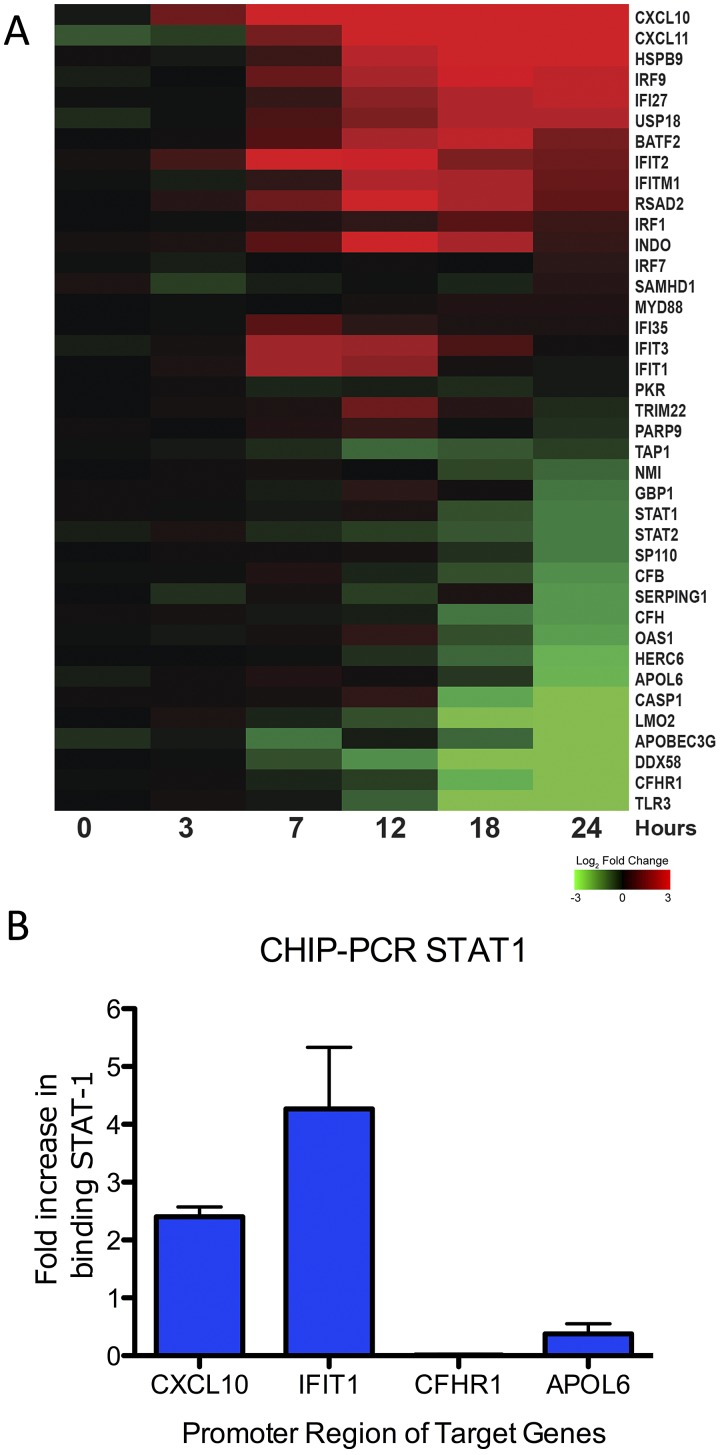
While unable to completely block activation, H5N1-VN1203 may dampen the total amount of activated transcriptional factor via antagonists like NS1. With limited availability, transcriptional factor binding preference may drive differential expression. To explore this possibility, we focused on STAT1-dependent/augmented ISGs as determined by the IPA knowledge base; the results identified broad variation in ISG expression despite their similar dependence on STAT1 (Fig. 3A). To verify that transcriptional factor binding contributed to this observation, we performed chromatin immunoprecipitations (ChIP) targeting STAT1, followed by quantitative real-time PCR (qPCR) to examine target promoter binding. The results indicated that CXCL10 and IFIT1, highly expressed ISGs following H5N1-VN1203 infection (Fig. 3A), had a corresponding increase in STAT1 binding in their 5′ promoter region at 12 hpi (Fig. 3B). In contrast, CFHR1 and APOL6, ISGs with decreased expression, had no increase in STAT1 binding despite the presence of activated STAT1 in those cells. For each of these genes, we had identified consensus STAT1 GAS motifs (TTC[N 2–4]GAA) within 1 kb of the translational start site and 200 bp downstream (18); the number of these motifs varied by gene (CXCL10 [2], IFIT1 [1], CFHR1 [1], APOL6 [1]) but did not dictate expression of STAT1. Notably, similar variation despite STAT1 dependence was observed following MERS-CoV infection as well as with STAT1 ChIP-PCR results (see Fig. S3 in the supplemental material). Together, the results indicated that differential promoter binding exists between genes activated by STAT1 and possibly contributes to differential ISG expression in H5N1-VN1203 and MERS-CoV infections.
FIG 3 .
STAT1 binding corresponds with differential ISG expression. (A) RNA expression of consensus ISGs linked to STAT1 following H5N1-VN1203 infection. Values represent log2 FC relative to time-matched mocks. (B) Chromatin immunoprecipitation with antibodies against phospho-STAT1 (Santa Cruz) followed by qPCR of the 5′UTR of upregulated genes CXCL10 and IFIT1 or downregulated genes CFHR1 and APOL6 in the context of H5N1-VN1203 infection. Values represent fold increase binding compared to mock on a linear scale.
Altered histone modification plays a significant role in both H5N1 and MERS-CoV ISG antagonism.
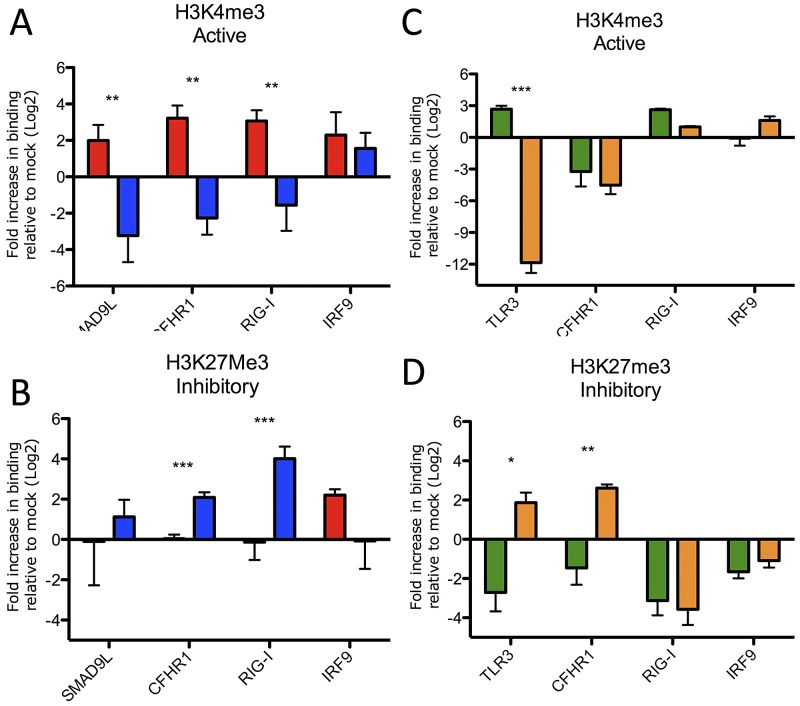
A number of factors can impact transcriptional factor binding preference, including the promoter binding sequence, site of phosphorylation, and underlying chromatin structure (19, 20). We initially chose to focus on chromatin structure based on a number of studies that implicated a role for chromatin remodeling during influenza virus infection (21–23) and identification of histone mimic (24). Histone modification can mediate a variety of diverse biological processes, including gene regulation (25, 26). These changes can result in rapid chromatin remodeling, including activation (opening) or repression (closing) depending on the location, type, and number of modifications. While the NS1 histone mimic motif is absent in both H1N1-09 and H5N1-VN1203, other findings suggest that histone remodeling strategies may be conserved across influenza strains (21–23). Coupled with substantial downregulation of ISG subsets during H5N1 infection, these facts encouraged us to investigate histone changes. We performed ChIP using antibodies for either an active form of histone H3 (H3K4me3) or a repressive form (H3K27me3) in the context of H5N1-VN1203 or H1N1-09 infection (25). We then targeted the 5′ untranslated region (UTR) of three ISGs with differential expression between the influenza strains (CFHR1, DDX58 [encodes RIG-I], and SMAD9L) and one with similar expression (IRF9) by quantitative real-time PCR. For H1N1-09, H3K4me3 was strongly associated with the 5′UTR of upregulated ISGs compared to mock (Fig. 4A). In contrast, these same genes, with downregulated RNA expression in H5N1-VN1203 infection, were also found to have significantly reduced association with H3K4me3, suggesting the absence of the active H3 motif. Examining the repressive H3K27me3 revealed enhanced association with CFHR1, DDX58, and SMAD9L during H5N1-VN1203 infection (Fig. 4B); no significant H3K27me3 association was observed for these genes following H1N1-09 infection. IRF9, an ISG upregulated following both H1N1-09 and H5N1-VN1203 infection, demonstrated similar trends for both H3K4me3 and H3K27me3, with no significant differences observed between the strains (Fig. 4A and B). Together, the results indicate that altered histone modification occurs in the context of H5N1-VN1203 infection and these patterns of change correlate with the observed differences in ISG antagonism.
FIG 4 .
Altered histone modifications play a significant role in both H5N1 and MERS-CoV ISG antagonism. Chromatin immunoprecipitation with antibodies against H3K4me3 (A and C) or H3K27me3 (B and D) followed by qPCR of the 5′UTR of the identified genes 12 h postinfection with H1N1-09 (red), H5N1-VN1203 (blue), SARS-CoV (green), or MERS-CoV (orange). Values represent fold increase binding compared to mock on a log2 scale. P values based on Student’s t test and are marked as indicated: *, <0.05; **, <0.01; ***, <0.001.
Having demonstrated significant differences in histone modification patterns that correlate with expression outcomes between influenza strains, we returned to the CoVs. While both MERS-CoV and SARS-CoV maintain delayed IFN induction (Fig. 2), MERS-CoV also downregulated a subset of ISGs with kinetics similar to those of H5N1-VN1203 (Fig. 1D); no similar subset was observed during SARS-CoV infection. Given its increased sensitivity to type I IFN (15, 16), one possibility is that MERS-CoV employed an additional layer of ISG antagonism via altered histone modification. To test this idea, ChIP-PCR was again employed following both MERS-CoV and SARS-CoV infection. The results demonstrated that two genes (TLR3 and CFHR1) downregulated in both H5N1-VN1203 and MERS-CoV, but not SARS-CoV infection, had decreased association with activating H3K4me3 (Fig. 4C) and increased association with the repressive H3K27me3 motif (Fig. 4D). In contrast, DDX58, a gene downregulated by H5N1-VN1203 but not MERS-CoV, actually had reduced H3K27me3 association following MERS-CoV infection. Similar to previous reports (27), the activating histone mark was variable in uninduced SARS-CoV-infected samples at 12 hpi; however, a late time point (36 hpi) that corresponded with robust ISG expression demonstrated increased H3K4me3 and reduced H3K27me3 modifications relative to mock (see Fig. S4 in the supplemental material). Together, the data suggested that while the activating motif was less predictable, the dominant H3K27me3 repressive histone mark was associated only with ISGs downregulated by both MERS-CoV and H5N1-VN1203 (28).
NS1 mediates ISG manipulation in virulent influenza virus strains.
To explore how H5N1-VN1203 mediates ISG control, we next focused on viral proteins linked to H5N1-VN1203 virulence and IFN manipulation: NS1, PB1-F2, and PB2 (29–32). To test the contributions of these viral proteins, Calu3 cells were infected with H5N1-VN1203 mutants containing a C-terminal truncation of NS1 (NS1trunc124), a deletion of PB1-F2 (PB1-F2del), or PB2 encoding an amino acid substitution (PB2-K627E) that significantly reduces polymerase activity. For each, viral replication and ISG induction was compared to those of the wild type (WT) and demonstrated attenuated replication, noting that each mutant virus replicated to equivalent titers at 18 to 24 h (see Fig. S5A in the supplemental material). However, ISG induction kinetics varied between both the WT virus and each mutant (Fig. 5A). For the PB1-F2del and PB2-K627E mutants, a significant subset of ISGs (48.5% for PB1-F2del and 38.8% for PB2-K627E; data not shown) had augmented expression (>1.5 log2 FC) compared to the WT at 24 hpi; however, 95% of genes downregulated in WT infections also remained downregulated (24%, <−0.75 log2 FC expression) or uninduced (74%, <1.5 log2 FC expression) by both of these mutants. In contrast, compared to WT H5N1-VN1203, the NS1trunc124 mutant induced a larger subset and augmented expression (71%) of ISGs (>1.5 log2 FC) (data not shown). Importantly, of the 41 consensus ISGs downregulated during H5N1 infection, 38 were no longer downregulated and over half (51%) were upregulated (>1.5 log2 FC expression) following infection with the mutant NS1 virus. Proteomics analysis validated these results, demonstrating increased protein expression of 11 ISGs in H5N1-NS1trunc124 infection, including MDA5, MX1, and IFIT1, which were not detectable in WT infection (see Fig. S3B). Together, these results suggest that the NS1 C terminus plays a role in selective ISG downregulation and makes a strong contribution to global ISG control following H5N1-VN1203 infection.
FIG 5 .
H5N1-VN1203 mediates control over the global ISG response primarily through the activity of NS1. (A) RNA expression of downregulated ISGs following each infection condition. Downregulated gene list was derived from Fig. 1C, and values represent fold change (log2 FC) compared to time-matched mocks. (B and C) ChIP against H3K4me3 (B) and H3K27me3 (C) followed by qPCR of the 5′UTR of the identified genes within the context of H5N1-VN1203 (blue) or H5N1 NS1trunc124 (white) infection. Values represent fold increases in binding compared to mock on a log2 scale. (D) Global transcription of consensus ISG from A549 cells infected with WT 1918 H1N1 or 1918 H1N1 encoding TX/91 NS1 (MOI of 2). RNA expression values represent fold change (log2 FC) compared to time-matched mock.
Having established NS1 as a required component for strong ISG manipulation and downregulation, we next sought to determine if histone modifications of specific ISGs were modified in H5N1-NS1trunc124 infections. For this, we repeated the ChIP-PCR experiment with WT and NS1trunc124 viruses (Fig. 5B and C). Similar to what was previously observed, ISGs that were transcriptionally downregulated in WT infections (i.e., SMAD9L, CFHR1, and DDX58) were strongly associated with repressed histones (H3K27me3) and only weakly associated with active histones (H3K4me3). In contrast, the absence of full-length NS1 resulted in augmented association of the same transcripts with the active H3K4me3 and reduced binding to the inhibitory H3K27me3 mark. Together, the data demonstrated that histone modifications were altered in the absence of full-length NS1 and may result in the augmented ISG RNA and protein expression following NS1trunc124 mutant virus infection.
We next sought to determine if NS1 from another highly pathogenic virus mediated a similar global ISG response. With this in mind, a meta-analysis was conducted on mRNA expression data from A549 cells infected with pandemic 1918 H1N1 and a 1918 H1N1 mutant expressing NS1 from a seasonal H1N1 strain (A/Texas/36/91 [TX]) (33). Each of these viruses maintained a full-length NS1 but lacked the histone mimic motif found in H3N2. Confirming the original findings, several ISGs had a substantial increase in expression after infection with recombinant virus encoding the TX NS1 compared to the WT 1918 NS1 (Fig. 5D). However, expanding the examination to the consensus ISG list derived from Calu3 cells to the A549 cell metadata revealed that expression of ISGs was not uniformly downregulated with 1918 NS1. Instead, many ISGs had equivalent expression levels, and several (IFITM1, ZBP1, HSH2D, etc.) had greater expression with 1918 NS1 than TX NS1. These results suggest that NS1 substitution from a more virulent strain does not always result in uniform ISG augmentation but rather targets ISG manipulation based on strain specific NS1 activity. Notably, ISG downregulation was absent in the less virulent TX NS1 (0 genes) compared to in the 1918 NS1 (21% <−0.25 log2 FC at 24 hpi); coupled with data from H5N1-VN1203 and MERS-CoV infections, these results suggest that pathogenic respiratory viruses may downregulate a subset of ISGs via histone modification and contribute to effective infection and enhanced virulence.
DISCUSSION
This study demonstrates contrasting, similar, and novel avenues used by virulent respiratory viruses to dampen and impair the global ISG response. HPAI utilizes rapid manipulation resulting in both strong up- and downregulation of ISG subsets rather than a binary ISG response. In contrast, the less virulent H1N1-09 virus fails to modulate either ISG transcripts or protein, producing a robust antiviral state which may impact peak titers. Similarly, SARS-CoV infection stimulates robust ISG transcription and protein production; however, ISGs are delayed 24 to 48 h, permitting SARS-CoV to achieve peak titers prior to ISG production. Finally, MERS-CoV not only delays ISG induction but also manipulates a subset of ISGs, similar to H5N1-VN1203. Together, the data highlight methods that pathogenic viruses utilize to control the host antiviral state. In each case, the strategy contributes to successful infection and may explain differences in virulence seen between viral families and strains.
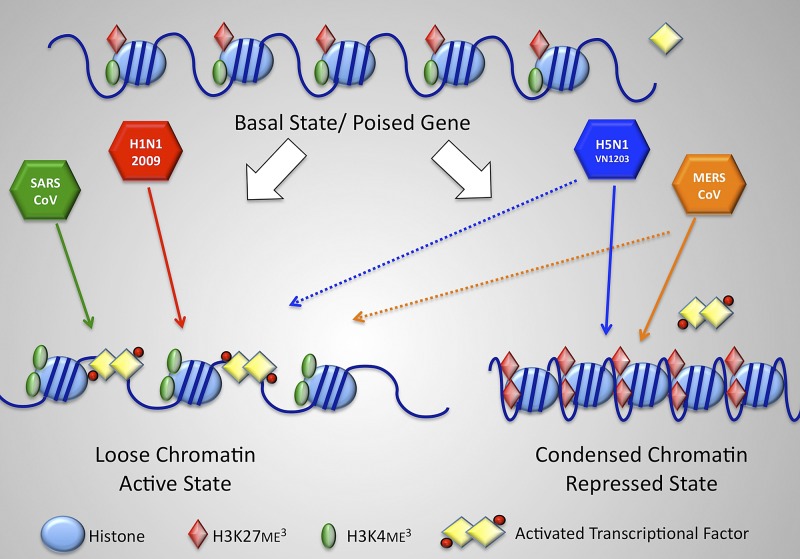
In addition to identifying differential ISG induction profiles, this study also introduces a novel avenue for viral antagonism of host gene expression via altered histone modification (Fig. 6). A large number of host enzymes catalyze methylation and demethylation of histones, a process that is essential for regulating transcriptional programs (26, 34). In particular, innate immune responses, including ISG responses, are highly regulated by epigenetic control mechanisms, including alteration of methylation and acylation patterns in chromatin (35, 36). By catalyzing the placement or removal of histone methyl marks at H3K4 (transcriptionally active) and H3K27 (transcriptionally repressive), histone lysine methyltransferases (MLL2, MLL3, and EZH2) and demethylases (RBP2, UTX, and JMJD3) modify the chromatin state to maintain and fine-tune transcriptional control (26). During H1N1-09 or SARS-CoV infection, the host cell recognizes the viruses, produces type I and type III IFN, and induces histone modulation complexes to remove the repressive histone marks (H3K27me3) and add activating modifications (H3K4me3); this process shifts targeted chromosomal locations to a more open, activated state and permits the binding of activated transcription factors like STAT1 and IRF7, allowing robust ISG expression. In contrast, during MERS-CoV and H5N1-VN1203 infection, some histones undergo the activating modifications resulting in robust expression of a subset of ISGs; however, for another subset of genes, chromatin remodeling complexes incorporate the repressive histone modification (H3K27me3) while also removing the active marker (H3K4me3), resulting in a condensed chromatin structure. This closed structure likely excludes transcription factors and reduces RNA expression of target ISGs. While differences in transcriptional factor phosphorylation state or binding sites may also contribute, the altered histone modification approach suppresses expression of target genes. While histone modification has been shown to play a role in a variety of host processes, including cell development (37), host metabolism (38), and ISG induction (36), these findings represent additional evidence that pathogens can exploit the same repression processes to modulate and control the host IFN response (39, 40); similar modulation by pathogens likely impacts other critical host processes, including inflammation, antigen presentation, and aging (26, 34). The presence of this antagonism in disparate, highly virulent respiratory pathogens not only supports earlier studies involving the H3N2 NS1 histone mimic (24) but also suggests that the approach may be conserved across multiple viral families and contribute to enhanced disease virulence. The large number of cellular proteins present within histone modification complexes potentially provides a rich diversity of targets for pathogen-coded control and manipulation (26). Importantly, antagonizing histone regulation may also prove to be an important factor in the emergence of both H5N1-VN1203 and MERS-CoV; broad evolutionary conservation of both histone modifications and machinery suggests that host targets may be conserved across diverse species, potentially easing cross-species infection by modulation of the host response (41). However, the effect may also be reducing transmission efficiency by removing portions of the adaption requirements for host shifting. Future studies explore avian- and bat-derived cells for conservation of virus-mediated histone modification.
FIG 6 .
Model of histone modification changes during highly virulent respiratory virus infection. Viral infection induces changes in the basal state of host chromatin. For H1N1-09 and SARS-CoV, infection results in H3K4me3 incorporation (green ovals), reduced H3K27 (red diamonds), and an open confirmation. In contrast, for H5N1-VN1203 and MERS-CoV, infection augments H3K27me3 incorporation and reduces H3K4me3 for a subset of genes, resulting in a closed conformation that excludes activated transcription factors and represses ISG expression.
For influenza viruses, ISG expression differences between H5N1-VN1203 and H1N1-09 highlighted possible virulence determinants. Numerous reports have shown that specific influenza proteins impede the type I IFN response by targeting host signaling molecules and transcription factors (9, 10); together, the studies imply that disruption of these components results in a loss or dampening of the entire pathway’s downstream components. However, robust IFN production following H5N1-VN1203 infection confounds this interpretation (21–23); instead, these data argue that during H5N1-VN1203 infection, host recognition and IFN signaling remain largely intact and a hierarchy in ISG activation exists. In contrast, the less virulent H1N1-09 virus lacks similar ISG antagonism both at the RNA and protein levels. Several HPAI viral protein functions may have contributed to ISG downregulation prior to mRNA production, including cap snatching, cleavage and polyadenylation specificity factor (CPSF) interference, and/or microRNA targeting (42–44). However, each possibility would lead to nonspecific degradation of mRNA and thus could not explain the differential ISG expression observed in these studies. In contrast, significant changes in histone activation and repression marks, coupled with interaction with histone components (21–23) and the recent discovery of a histone mimic in H3N2 infection (17, 24), provided a rational avenue for the observed contrast in ISG antagonism.
Mechanistically, C-terminal truncation of NS1 ablated global ISG antagonism for H5N1-VN1203. While mutations in PB1-F2 and PB2 resulted in significant ISG augmentation, the majority of ISGs maintain low or downregulated expression. In contrast, NS1 truncation halted ISG downregulation by H5N1-VN1203 and enhanced expression of a significant subset of ISGs at both the RNA and protein levels. This ISG stimulation occurred despite the presence of both the RNA-binding domain and part of the effector domain for NS1 (45). Infection with the NS1 truncation mutant also resulted in reduced association with the repressive H3K27me3 and increased association with the active H3K4me3, identifying a role for NS1 in altered histone modification. Together, the data indicate a role for NS1 beyond just IFN pathway inhibition and identifies histone modification as a new target for ISG antagonism in H5N1-VN1203 infection.
While NS1 histone mimicry has been identified in an H3N2 virus, the responsible binding motif is not contained in either H5N1-VN1203 or 1918 H1N1 (24). This suggests that the C-terminal portion of NS1 may mediate selective ISG subset downregulations through different mechanisms depending on strain. For H5N1-VN1203, NS1 may target a similar functional pathway as H3N2 by mimicking different histones, targeting histone-modifying enzymes, or disrupting a histone adaptor protein complex. For H1N1-09, it suggests that the absent 11-amino-acid C-terminal truncation of NS1 may contribute to its relative lack of ISG antagonism; this absent tail has multiple functional domains in addition to possible histone modification elements (17, 46, 47). Together, these possibilities illustrate the complexity of NS1 and the caveats of analyzing its contribution to histone modification; however, further study with the plethora of H5N1-VN1203 and H1N1-09 viral mutants provides a golden opportunity to further characterize NS1’s role in ISG manipulation.
The data also lent the possibility that NS1 from other pathogenic influenza strains may mediate differential antagonism of the ISG subset in addition to interfering with host recognition. Meta-analysis of NS1 from 1918 H1N1 and a seasonal influenza virus supports this possibility; while derived from Calu3 cells, the expanded ISG list demonstrated that control was altered between virulent and avirulent strains based on NS1, and the data indicated that the directions of expression changes were not uniformly in favor of the more virulent strain. Variant responses in viruses encoding different NS1 genes imply the existence of categories of NS1 genes that may target different aspects of the host immune response. This fact, coupled with highly species-specific virulence determinants (48, 49), suggests that NS1 genes may circulate and contribute to species-specific severe disease outcomes, perhaps by coordinating unique patterns of histone modification that regulate the ISG in addition to standard IFN pathway antagonism. Importantly, exploring the global mechanism of ISG manipulation by NS1 genes within the context of infection, in addition to transfection-based approaches, will provide significant insights into disease severity across different hosts, tissues, and species.
The data also highlight the ability of both SARS-CoV and MERS-CoV to inhibit recognition, delay IFN induction, and sequester ISG production until after peak viral titers had been achieved. This ability effectively neutralizes the innate immune response and permits both CoVs to establish a foothold within the lung prior to recognition. However, MERS-CoV and SARS-CoV also utilize additional divergent approaches to limit the host IFN response. While SARS-CoV IFN antagonism is well established (11), MERS-CoV appears to employ different, less effective antagonists, resulting in increased sensitivity to type I IFN pretreatment (15, 16). However, altered histone modification represents a novel CoV antagonism approach. While newly emergent, several MERS-CoV proteins have been identified as IFN antagonists (50, 51); notably, ORF4b, a MERS accessory protein with no homologue in SARS-CoV, maintains a purported nuclear localization signal, exhibits nuclear localization based on transfection, and may play a role in IFN antagonism (51). Further studies are under way utilizing the MERS-CoV infectious clone (52) and attempt to identify viral components required to mediate altered histone modification.
Overall, this study combines a systems biology approach that integrates complex RNA and proteomics datasets across different virus infections with detailed molecular and cell biology validation approaches to identify novel patterns and mechanisms of virus-host interaction and innate immune regulation. While ISGs have been previously known to be an important factor during viral infection, global analysis has been limited by the lack of uniform platforms, infection conditions, and data collection methods. Establishing conditions that afford comparisons across systems virology datasets alleviated these problems and permitted analysis beyond a single treatment or infection condition. The resulting study revealed similarities and stark contrasts in terms of ISG induction between SARS-CoV, MERS-CoV, an HPAI virus, and a 2009 pandemic influenza virus strain, which likely contribute to differences in disease outcomes and may help our understanding of these infections. Importantly, discovery-based studies informed molecular approaches that identified a novel avenue of immune antagonism utilized by two diverse viral families, providing a new area for exploration and study. This study highlights the power and utility of combining systems biology and standard molecular-based approaches in emerging infectious disease research.
MATERIALS AND METHODS
Cells and viruses.
Calu3 cells were utilized as previously described (5–7). Viral titrations and propagation of SARS-CoV, MERS-CoV, and influenza virus were performed in VeroE6 and MDCK cells using standard methods. Wild-type (WT) icSARS-CoV was derived from the Baric laboratory’s infectious clone (ic) constructs (53). MERS-CoV (EMC 2012 strain) was kindly provided by Bart L. Haagmans, Erasmus Medical Center, Rotterdam, Netherlands, via Material Transfer Agreement. Influenza A/California/04/2009 (H1N1-09), influenza A/Vietnam/1203/2004 (H5N1-VN1203), and H5N1-VN1203 mutants (NS1trunc124, PB1-F2del, and PB2-K627E) were derived from a plasmid-based reverse-genetic system and subsequently amplified in MDCK cells (54). Detailed information about construction of mutant viruses is included in the supplemental material (see Text S1).
Infections.
Experiments using HPAI H5N1, SARS-CoV, or MERS-CoV were performed in containment laboratories at the University of Wisconsin-Madison or University of North Carolina-Chapel Hill as previously described (5, 53). Influenza and CoV infections were carried out at an MOI of 1 (H5N1-VN1203), MOI of 3 (H1N1-09), and MOI of 5 (SARS- and MERS-CoV) as described (5–7).
RNA isolation, microarray processing, and identification of differential expression.
RNA isolation and microarray processing from Calu3 cells were carried out as previously described (5). Differential expression (DE) was determined by comparing virus-infected replicates to time-matched mock replicates. Criteria for DE in determining the consensus ISG list were an absolute log2 FC of >1.5 and a false discovery rate (FDR)-adjusted P value of <0.05 for a given time point.
Proteomics reagents, sample preparation, database construction, and data processing.
Proteomics preparation was carried out as previously described (7). Detailed experimental approaches have been included in the supplemental material (see Text S1).
ChIP-PCR.
ChIP analysis was performed using the EpiTect ChIP OneDay kit (Qiagen). Briefly, infected Calu3 cells were cross-linked, harvested, and frozen at −80°C. Cells were then lysed and chromatin sheared via sonication to generate chromatin fragments between 250 and 1,000 bp. Sonicated samples were then immunoprecipitated with anti-STAT1 (clone C-24; Santa Cruz Biotechnology), anti-H3K4me3 (Qiagen), anti-H3K27me3 (Qiagen), or anti-mouse IgG (Qiagen) as a control. To determine the histone modification distribution, quantitative real-time PCR was performed targeting the 5′UTR (−750 bp—TSS [region 750 base pairs upstream of the transcriptional start site]) of select genes; target and primer information is included in the supplemental material. ChIP results were reported as fold difference (or differential occupancy), allowing comparison across multiple samples. For this, each sample was normalized to the input and then the fold enrichment was calculated using the ΔΔCT method. The fold difference for each gene was determined by dividing the appropriate time-matched mock by the experimental group. Finally, the fold difference values were converted to log2 and plotted. Data presented are the means ± standard errors for triplicate samples.
Meta-analysis of A549 cells infected with H1N1 1918 viruses.
RNA expression data from studies with H1N1 1918 virus infections (33) were reanalyzed, and ISG expression was examined. Two-channel analysis was performed using R statistical programming language and bioconductor package LIMMA. To lower variance, two channel arrays were background corrected with a zero value and normalized with the Loess method. A contrast matrix file was used to direct which conditions and files should be used to perform differential expression analysis. LIMMA produced a gene list which was carried further for downstream functional analysis.
Accession numbers.
Raw microarray data have been deposited in the National Center for Biotechnology Information’s (NCBI) Gene Expression Omnibus database and are accessible through GEO series GSE33267, GSE37571, GSE28166, and GSE45042 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi). Raw proteomics data corresponding to peptide identifications used to populate the AMT tag database are available at the PRoteomics IDEntification (PRIDE) database (http://www.ebi.ac.uk/pride/) under the project name “A Systems Biology Approach to Emerging Respiratory Viral Diseases” in the PRIDE Public Projects folder and correspond to PRIDE accession numbers 19877 to 19890. The raw quantitative proteomics data can be accessed at the Pacific Northwest National Laboratory’s (PNNL) Biological Mass Spectrometry (MS) Data and Software Distribution Center (http://omics.pnl.gov/) in the “Systems Virology Contract Data” folder within the “Browse Available Data” folder.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download
Proteomic analysis validated ISG antagonism following CoV and influenza virus infection. (A) Diagram illustrating significantly upregulated proteins for all time points and number of identified ISGs within H5N1-VN1203 (blue), H1N1-09 (red), SARS-CoV (green), and MERS-CoV (orange). (B) Protein abundance log2 FC for STAT1 and PKR following each infection condition relative to time-matched mock. (C and D) RNA and protein expression plots following H1N1-09 and SARS-CoV infection. RNA expression, on the left axis, represents log2 FC relative to mock following H1N1-09 (red solid line; C) and SARS-CoV (D; green solid line). Linear, mean protein abundance for mock (black dashed line) compared to H1N1-09 (C; red dashed)- or SARS-CoV (D; green dashed)-infected samples. Download
Transcriptional factor analysis indicated partial induction of downstream genes in HPAI virus infection. (A) Ingenuity pathway analysis (IPA) network diagram of H5N1-VN1203 (right) consensus ISG expression connected to STAT1 at 12 h postinfection. Increases (red) or decreases (green) in expression level are indicated by color intensity and are relative to genes included within each individual plot. (B to D) IPA downstream transcription factor activation of STAT1 (B), IRF7 (C), and NF-κB1 (D). Z scores representing activation (value of >2.0) or repression (value of <−2.0) of transcriptional factor based on downstream gene expression plotted over time for H5N1-VN1203 (blue), H1N1-09 (red), and type I IFN (black). Download
STAT1 binding corresponds with differential ISG expression. (A) RNA expression of consensus ISGs linked to STAT1 following MERS-CoV infection. Values represent log2 FC relative to time-matched mocks. (B) Chromatin immunoprecipitation with antibodies against phospho-STAT1 (Santa Cruz) followed by qPCR of the 5′UTR of upregulated genes CXCL10 and IFIT1 or downregulated genes CFHR1 and APOL6 within the context of H5N1-VN1203 infection. Values represent fold increase binding compared to mock on a linear scale. Download
Increased activating histone modifications at late times after SARS-CoV infection. Chromatin immunoprecipitation with antibodies against H3K4me3 (black) or H3K27me3 (white) followed by qPCR of the 5′UTR of the identified genes 36 h postinfection with SARS-CoV. Values represent fold increase binding compared to mock on a log2 scale. Download
H5N1-VN1203 mediates control over the global ISG response primarily through the activity of NS1. (A) Viral replication in Calu3 cells following an MOI of 1 with wild-type H5N1-VN1203 (blue), PB1-F2del (black), PB2-K627E (gray), or NS1trunc124 (red) mutants. (C) RNA expression (left axis) and linear, mean protein abundance (right axis) for H5N1 NS1trunc124-infected samples compared to time-matched mocks. Download
Consensus Calu3 type I IFN-stimulated genes. Included genes have expression >1.5 log2 fold change over mock at more than one time point after type I IFN treatment. Unannotated genes are not included in graphical depictions. Other genes grouped by log2 fold change in H5N1-VN1203 as follows: upregulated (>0.75), minimal stimulation (0.75 to −0.75), and downregulated (<−0.75). Italicized genes are downregulated in MERS-CoV.
ACKNOWLEDGMENTS
Research was supported by grants from NIAID of the NIH (U19AI100625 to R.S.B., U19AI106772 to Y.K., and F32AI102561 to V.D.M.) and under contract no. HHSN272200800060C (to M.G.K.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
The proteomics was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE Office of Biological and Environmental Research and located at PNNL and used capabilities developed under efforts supported by the National Institute of General Medical Sciences (8 P41 GM103493-10). PNNL is operated by Battelle Memorial Institute for the DOE under contract number DE-AC05-76RLO1830.
Footnotes
Citation Menachery VD, Eisfeld AJ, Schäfer A, Josset L, Sims AC, Proll S, Fan S, Li C, Neumann G, Tilton SC, Chang J, Gralinski LE, Long C, Green R, Williams CM, Weiss J, Matzke MM, Webb-Robertson B-J, Schepmoes AA, Shukla AK, Metz TO, Smith RD, Waters KM, Katze MG, Kawaoka Y, Baric RS. 2014. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio 5(3):e01174-14. doi:10.1128/mBio.01174-14.
REFERENCES
- 1. Perlman S. 1998. Pathogenesis of coronavirus-induced infections. Review of pathological and immunological aspects. Adv. Exp. Med. Biol. 440:503–513 [PubMed] [Google Scholar]
- 2. Gillim-Ross L, Subbarao K. 2006. Emerging respiratory viruses: challenges and vaccine strategies. Clin. Microbiol. Rev. 19:614–636. 10.1128/CMR.00005-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3(6):e00473-12. 10.1128/mBio.00473-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ng WF, To KF, Lam WW, Ng TK, Lee KC. 2006. The comparative pathology of severe acute respiratory syndrome and avian influenza A subtype H5N1—a review. Hum. Pathol. 37:381–390. 10.1016/j.humpath.2006.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li C, Bankhead A, Eisfeld AJ, Hatta Y, Jeng S, Chang JH, Aicher LD, Proll S, Ellis AL, Law GL, Waters KM, Neumann G, Katze MG, McWeeney S, Kawaoka Y. 2011. Host regulatory network response to infection with highly pathogenic H5N1 avian influenza virus. J. Virol. 85:10955–10967. 10.1128/JVI.05792-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Josset L, Menachery VD, Gralinski LE, Agnihothram S, Sova P, Carter VS, Yount BL, Graham RL, Baric RS, Katze MG. 2013. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio 4(3):e00165-00113. 10.1128/mBio.00165-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sims AC, Tilton SC, Menachery VD, Gralinski LE, Schäfer A, Matzke MM, Webb-Robertson BJ, Chang J, Luna ML, Long CE, Shukla AK, Bankhead AR, Burkett SE, Zornetzer G, Tseng CT, Metz TO, Pickles R, McWeeney S, Smith RD, Katze MG, Waters KM, Baric RS. 2013. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J. Virol. 87:3885–3902. 10.1128/JVI.02520-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Menachery VD, Baric RS. 2013. Bugs in the system. Immunol. Rev. 255:256–274. 10.1111/imr.12092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Katze MG, He Y, Gale M., Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675–687. 10.1038/nri888 [DOI] [PubMed] [Google Scholar]
- 10. García-Sastre A. 2011. Induction and evasion of type I interferon responses by influenza viruses. Virus Res. 162:12–18. 10.1016/j.virusres.2011.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Totura AL, Baric RS. 2012. SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2:264–275. 10.1016/j.coviro.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rockx B, Baas T, Zornetzer GA, Haagmans B, Sheahan T, Frieman M, Dyer MD, Teal TH, Proll S, van den Brand J, Baric R, Katze MG. 2009. Early upregulation of acute respiratory distress syndrome-associated cytokines promotes lethal disease in an aged-mouse model of severe acute respiratory syndrome coronavirus infection. J. Virol. 83:7062–7074. 10.1128/JVI.00127-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mi Z, Ma Y, Tong Y. 2012. Avian influenza virus H5N1 induces rapid interferon-beta production but shows more potent inhibition to retinoic acid-inducible gene I expression than H1N1 in vitro. Virol. J. 9:145. 10.1186/1743-422X-9-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zielecki F, Semmler I, Kalthoff D, Voss D, Mauel S, Gruber AD, Beer M, Wolff T. 2010. Virulence determinants of avian H5N1 influenza A virus in mammalian and avian hosts: role of the C-terminal ESEV motif in the viral NS1 protein. J. Virol. 84:10708–10718. 10.1128/JVI.00610-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan RW, Chan MC, Agnihothram S, Chan LL, Kuok DI, Fong JH, Guan Y, Poon LL, Baric RS, Nicholls JM, Peiris JS. 2013. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J. Virol. 87:6604–6614. 10.1128/JVI.00009-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zielecki F, Weber M, Eickmann M, Spiegelberg L, Zaki AM, Matrosovich M, Becker S, Weber F. 2013. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J. Virol. 87:5300–5304. 10.1128/JVI.03496-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hale BG, Randall RE, Ortín J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376. 10.1099/vir.0.2008/004606-0 [DOI] [PubMed] [Google Scholar]
- 18. Decker T, Kovarik P, Meinke A. 1997. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res. 17:121–134. 10.1089/jir.1997.17.121 [DOI] [PubMed] [Google Scholar]
- 19. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129:823–837. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- 20. Bonham AJ, Wenta N, Osslund LM, Prussin AJ, Vinkemeier U, Reich NO. 2013. STAT1:DNA sequence-dependent binding modulation by phosphorylation, protein:protein interactions and small-molecule inhibition. Nucleic Acids Res. 41:754–763. 10.1093/nar/gks1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alfonso R, Lutz T, Rodriguez A, Chavez JP, Rodriguez P, Gutierrez S, Nieto A. 2011. CHD6 chromatin remodeler is a negative modulator of influenza virus replication that relocates to inactive chromatin upon infection. Cell. Microbiol. 13:1894–1906. 10.1111/j.1462-5822.2011.01679.x [DOI] [PubMed] [Google Scholar]
- 22. Garcia-Robles I, Akarsu H, Müller CW, Ruigrok RW, Baudin F. 2005. Interaction of influenza virus proteins with nucleosomes. Virology 332:329–336. 10.1016/j.virol.2004.09.036 [DOI] [PubMed] [Google Scholar]
- 23. Zhirnov OP, Klenk HD. 1997. Histones as a target for influenza virus matrix protein M1. Virology 235:302–310. 10.1006/viro.1997.8700 [DOI] [PubMed] [Google Scholar]
- 24. Marazzi I, Ho JS, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, Lee K, García-Sastre A, Roeder RG, Tarakhovsky A. 2012. Suppression of the antiviral response by an influenza histone mimic. Nature 483:428–433. 10.1038/nature10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 26. Cloos PA, Christensen J, Agger K, Helin K. 2008. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 22:1115–1140. 10.1101/gad.1652908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hargreaves DC, Horng T, Medzhitov R. 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138:129–145. 10.1016/j.cell.2009.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu H, Zhu S, Zhou B, Xue H, Han JD. 2008. Inferring causal relationships among different histone modifications and gene expression. Genome Res. 18:1314–1324. 10.1101/gr.073080.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schat KA, Bingham J, Butler JM, Chen LM, Lowther S, Crowley TM, Moore RJ, Donis RO, Lowenthal JW. 2012. Role of position 627 of PB2 and the multibasic cleavage site of the hemagglutinin in the virulence of H5N1 avian influenza virus in chickens and ducks. PLoS One 7:e30960. 10.1371/journal.pone.0030960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ozawa M, Basnet S, Burley LM, Neumann G, Hatta M, Kawaoka Y. 2011. Impact of amino acid mutations in PB2, PB1-F2, and NS1 on the replication and pathogenicity of pandemic (H1N1) 2009 influenza viruses. J. Virol. 85:4596–4601. 10.1128/JVI.00029-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varga ZT, Grant A, Manicassamy B, Palese P. 2012. Influenza virus protein PB1-F2 inhibits the induction of type I interferon by binding to MAVS and decreasing mitochondrial membrane potential. J. Virol. 86:8359–8366. 10.1128/JVI.01122-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267. 10.1016/j.cell.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Billharz R, Zeng H, Proll SC, Korth MJ, Lederer S, Albrecht R, Goodman AG, Rosenzweig E, Tumpey TM, García-Sastre A, Katze MG. 2009. The NS1 protein of the 1918 pandemic influenza virus blocks host interferon and lipid metabolism pathways. J. Virol. 83:10557–10570. 10.1128/JVI.00330-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anonymous. 2010. Epigenetic control of the immune system: histone demethylation as a target for drug discovery. Drug Discov. Today Technol. 7:e1–e94. 10.2174/157016310791162721 [DOI] [PubMed] [Google Scholar]
- 35. Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, Carson WF, Cavassani KA, Li X, Lukacs NW, Hogaboam CM, Dou Y, Kunkel SL. 2009. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114:3244–3254. 10.1182/blood-2009-04-217620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fang TC, Schaefer U, Mecklenbrauker I, Stienen A, Dewell S, Chen MS, Rioja I, Parravicini V, Prinjha RK, Chandwani R, MacDonald MR, Lee K, Rice CM, Tarakhovsky A. 2012. Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J. Exp. Med. 209:661–669. 10.1084/jem.20112343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cedar H, Bergman Y. 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 10:295–304. 10.1038/nrg2540 [DOI] [PubMed] [Google Scholar]
- 38. Kaelin WG, Jr, McKnight SL. 2013. Influence of metabolism on epigenetics and disease. Cell 153:56–69. 10.1016/j.cell.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fonseca GJ, Thillainadesan G, Yousef AF, Ablack JN, Mossman KL, Torchia J, Mymryk JS. 2012. Adenovirus evasion of interferon-mediated innate immunity by direct antagonism of a cellular histone posttranslational modification. Cell Host Microbe 11:597–606. 10.1016/j.chom.2012.05.005 [DOI] [PubMed] [Google Scholar]
- 40. Gómez-Díaz E, Jordà M, Peinado MA, Rivero A. 2012. Epigenetics of host-pathogen interactions: the road ahead and the road behind. PLoS Pathog. 8:e1003007. 10.1371/journal.ppat.1003007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Woo YH, Li WH. 2012. Evolutionary conservation of histone modifications in mammals. Mol. Biol. Evol. 29:1757–1767. 10.1093/molbev/mss022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim MJ, Latham AG, Krug RM. 2002. Human influenza viruses activate an interferon-independent transcription of cellular antiviral genes: outcome with influenza A virus is unique. Proc. Natl. Acad. Sci. U. S. A. 99:10096–10101. 10.1073/pnas.152327499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen Z, Krug RM. 2000. Selective nuclear export of viral mRNAs in influenza-virus-infected cells. Trends Microbiol. 8:376–383. 10.1016/S0966-842X(00)01794-7 [DOI] [PubMed] [Google Scholar]
- 44. Li Y, Chan EY, Li J, Ni C, Peng X, Rosenzweig E, Tumpey TM, Katze MG. 2010. MicroRNA expression and virulence in pandemic influenza virus-infected mice. J. Virol. 84:3023–3032. 10.1128/JVI.02203-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Noah DL, Twu KY, Krug RM. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307:386–395. 10.1016/S0042-6822(02)00127-7 [DOI] [PubMed] [Google Scholar]
- 46. Chen Z, Li Y, Krug RM. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273–2283. 10.1093/emboj/18.8.2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. U. S. A. 104:1853–1858. 10.1073/pnas.0610977104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Korth MJ, Tchitchek N, Benecke AG, Katze MG. 2013. Systems approaches to influenza-virus host interactions and the pathogenesis of highly virulent and pandemic viruses. Semin. Immunol. 25:228–239. 10.1016/j.smim.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Uraki R, Kiso M, Shinya K, Goto H, Takano R, Iwatsuki-Horimoto K, Takahashi K, Daniels RS, Hungnes O, Watanabe T, Kawaoka Y. 2013. Virulence determinants of pandemic A(H1N1)2009 influenza virus in a mouse model. J. Virol. 87:2226–2233. 10.1128/JVI.01565-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, Suliman T, Barchet W, Weber F, Drosten C, Müller MA. 2013. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J. Virol. 87:12489–12495. 10.1128/JVI.01845-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Matthews KL, Coleman CM, van der Meer Y, Snijder EJ, Frieman MB. 2014. The ORF4b-encoded accessory proteins of MERS-coronavirus and two related bat coronaviruses localize to the nucleus and inhibit innate immune signaling. J. Gen. Virol. 95:874–882. 10.1099/vir.0.062059-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, Menachery VD, Graham RL, Swanstrom J, Bove PF, Kim JD, Grego S, Randell SH, Baric RS. 2013. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 110:16157–16162. 10.1073/pnas.1311542110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yount B, Curtis KM, Fritz EA, Hensley LE, Jahrling PB, Prentice E, Denison MR, Geisbert TW, Baric RS. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. U. S. A. 100:12995–13000. 10.1073/pnas.1735582100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345–9350. 10.1073/pnas.96.16.9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download
Proteomic analysis validated ISG antagonism following CoV and influenza virus infection. (A) Diagram illustrating significantly upregulated proteins for all time points and number of identified ISGs within H5N1-VN1203 (blue), H1N1-09 (red), SARS-CoV (green), and MERS-CoV (orange). (B) Protein abundance log2 FC for STAT1 and PKR following each infection condition relative to time-matched mock. (C and D) RNA and protein expression plots following H1N1-09 and SARS-CoV infection. RNA expression, on the left axis, represents log2 FC relative to mock following H1N1-09 (red solid line; C) and SARS-CoV (D; green solid line). Linear, mean protein abundance for mock (black dashed line) compared to H1N1-09 (C; red dashed)- or SARS-CoV (D; green dashed)-infected samples. Download
Transcriptional factor analysis indicated partial induction of downstream genes in HPAI virus infection. (A) Ingenuity pathway analysis (IPA) network diagram of H5N1-VN1203 (right) consensus ISG expression connected to STAT1 at 12 h postinfection. Increases (red) or decreases (green) in expression level are indicated by color intensity and are relative to genes included within each individual plot. (B to D) IPA downstream transcription factor activation of STAT1 (B), IRF7 (C), and NF-κB1 (D). Z scores representing activation (value of >2.0) or repression (value of <−2.0) of transcriptional factor based on downstream gene expression plotted over time for H5N1-VN1203 (blue), H1N1-09 (red), and type I IFN (black). Download
STAT1 binding corresponds with differential ISG expression. (A) RNA expression of consensus ISGs linked to STAT1 following MERS-CoV infection. Values represent log2 FC relative to time-matched mocks. (B) Chromatin immunoprecipitation with antibodies against phospho-STAT1 (Santa Cruz) followed by qPCR of the 5′UTR of upregulated genes CXCL10 and IFIT1 or downregulated genes CFHR1 and APOL6 within the context of H5N1-VN1203 infection. Values represent fold increase binding compared to mock on a linear scale. Download
Increased activating histone modifications at late times after SARS-CoV infection. Chromatin immunoprecipitation with antibodies against H3K4me3 (black) or H3K27me3 (white) followed by qPCR of the 5′UTR of the identified genes 36 h postinfection with SARS-CoV. Values represent fold increase binding compared to mock on a log2 scale. Download
H5N1-VN1203 mediates control over the global ISG response primarily through the activity of NS1. (A) Viral replication in Calu3 cells following an MOI of 1 with wild-type H5N1-VN1203 (blue), PB1-F2del (black), PB2-K627E (gray), or NS1trunc124 (red) mutants. (C) RNA expression (left axis) and linear, mean protein abundance (right axis) for H5N1 NS1trunc124-infected samples compared to time-matched mocks. Download
Consensus Calu3 type I IFN-stimulated genes. Included genes have expression >1.5 log2 fold change over mock at more than one time point after type I IFN treatment. Unannotated genes are not included in graphical depictions. Other genes grouped by log2 fold change in H5N1-VN1203 as follows: upregulated (>0.75), minimal stimulation (0.75 to −0.75), and downregulated (<−0.75). Italicized genes are downregulated in MERS-CoV.