ABSTRACT
During their nuclear replication stage, influenza viruses hijack the host splicing machinery to process some of their RNA segments, the M and NS segments. In this review, we provide an overview of the current knowledge gathered on this interplay between influenza viruses and the cellular spliceosome, with a particular focus on influenza A viruses (IAV). These viruses have developed accurate regulation mechanisms to reassign the host spliceosome to alter host cellular expression and enable an optimal expression of specific spliced viral products throughout infection. Moreover, IAV segments undergoing splicing display high levels of similarity with human consensus splice sites and their viral transcripts show noteworthy secondary structures. Sequence alignments and consensus analyses, along with recently published studies, suggest both conservation and evolution of viral splice site sequences and structure for improved adaptation to the host. Altogether, these results emphasize the ability of IAV to be well adapted to the host’s splicing machinery, and further investigations may contribute to a better understanding of splicing regulation with regard to viral replication, host range, and pathogenesis.
INTRODUCTION
Influenza viruses A, B, and C (IAV, IBV, and ICV, respectively) belong to the family Orthomyxoviridae. Among these three types of viruses that infect humans, only IAV and IBV are pathogenic, with IAV being the most predominant worldwide. IAVs are classified into subtypes depending on their surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA) (e.g., H1N1, H3N2, etc.) (1, 2). Influenza viruses constitute a serious public health problem, causing severe illness and death within high-risk populations during seasonal epidemics and, more rarely but recurrently, IAV-related pandemics.
Influenza viruses contain a segmented genome of single-stranded negative-sense RNA. Each viral RNA (vRNA) segment is included in viral ribonucleoprotein complexes (vRNPs) that are encapsidated by viral nucleoproteins (NP) in association with a complex of three viral proteins (PB1, PB2, and PA for IAV/IBV and PB1, PB2, and P3 for ICV), forming the RNA-dependent RNA polymerase (RdRp) (Fig. 1A) (3, 4). For all influenza viruses, after virus entry into the host cell, incoming vRNPs are transported toward the nucleus, where they are transcribed into viral mRNAs and replicated into new vRNAs by the viral polymerase with the support of the host RNA polymerase II and transcriptosome machinery (Fig. 1B) (5–7). Such a nuclear localization is uncommon for RNA viruses and highlights that influenza virus can directly interfere with the host nuclear machineries and their components. Evidence for this is illustrated during infection by a major remodeling of host nuclear ultrastructures (3, 8) and by multiple functional interactions between IAV and host nuclear factors, an example being the 5′-cap snatching from host pre-mRNAs to viral ones (reviewed in reference 9). Interestingly, influenza viruses have developed various strategies to optimize the coding potential of their segmented genome. Indeed, IAV and IBV possess eight vRNA segments, which have been shown to encode up to 17 and 11 proteins, respectively. ICV has seven vRNA segments that encode nine viral proteins (10, 11). In the case of IAV, segment 2 encodes the polymerase basic (PB) proteins PB1, PB1-F2, and PB1-N40 by using alternative translation initiation sites (12, 13); segment 3 encodes the polymerase acidic (PA) proteins PA and PA-X by a ribosomal frameshift, as well as two additional N-terminally truncated forms (PA-N155 and PA-N182) by using alternative translation initiation sites (14, 15); and finally, segment 7 encodes the matrix (M) protein M1 and ion channel proteins M2 and M42 and segment 8 encodes the nonstructural (NS) protein NS1, nuclear export protein NS2/NEP, and NS3 by alternative mRNA splicing. Most knowledge about cellular and viral genome splicing comes from studies performed with adenoviral and retroviral models, which revealed the rich proteomic variety offered by a single viral genomic molecule (16, 17). In contrast, more limited data are available for other viral models, such as influenza virus.
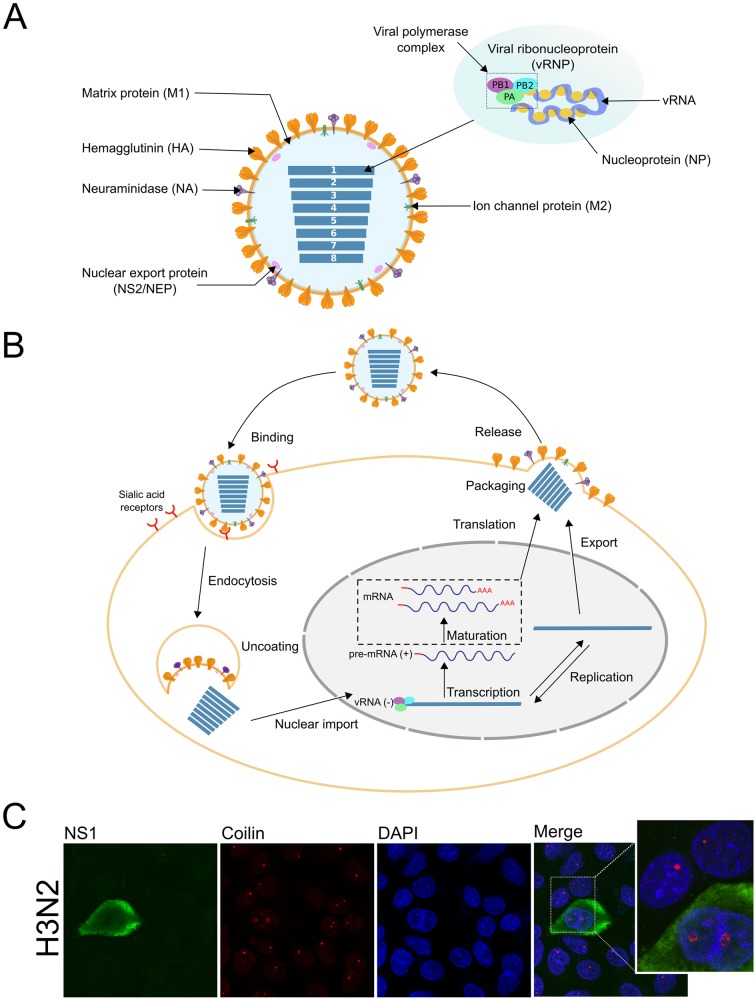
FIG 1 .
(A) Influenza A virus (IAV) particle. The IAV genome is composed of eight ribonucleoprotein complexes (vRNPs). Each one consists of single-stranded negative-sense viral RNA (vRNA) encapsidated by viral nucleoprotein (NP) and a viral polymerase complex (PA, PB1, and PB2) positioned at the extremity of the vRNA segment (depicted at top right). Three viral proteins are embedded within the viral membrane, hemagglutinin (HA), neuraminidase (NA), and ion channel protein (M2). Matrix protein 1 (M1) underlies the viral envelope and holds the vRNPs inside the virion. NS2/NEP is involved in vRNP packaging into viral particles and remains inside, and it is associated with M1. (B) Influenza viral cycle. The viral particle binds to sialic acid receptors and enters the cell via receptor-mediated endocytosis. Acidification of the endocytic vesicles leads to virus uncoating mediated by the M2 ion channel. vRNPs are then released into the cytoplasm and transported into the nucleus. There, the viral RNA-dependent RNA polymerase complex snatches the host mRNA caps (red line) to initiate the negative vRNA [vRNA(-)] transcription. Transcribed vRNAs then need to undergo an mRNA maturation phase, including the pre-mRNA splicing (depicted in dotted-line box), before export to the cytoplasm to be translated. vRNAs are also replicated in the nucleus to generate new vRNPs in association with neosynthesized viral proteins. Progeny vRNPs are transported toward the cytoplasmic membrane with viral components to be packaged into new infectious particles which are formed by cellular envelope budding. Panels A and B are based on data from reference 3. (C) Coilin is used as an example to show IAV-induced modifications of host splicing factors in the nucleus. A549 cells were infected with influenza virus A/Moscow/10/99 (H3N2) at a multiplicity of infection of 0.1 for 24 h, fixed, and immunostained to study the localization of IAV NS1 (green) and coilin (red). DAPI was used to stain cell nuclei (blue). Coilin expression is strongly affected within infected cells compared with its expression in noninfected cells (well-defined spots).
In this review, we provide an overview of the current knowledge gathered on the interplay between influenza viruses and the cellular spliceosome, with a special focus on IAVs, which are better documented in the literature. While the splicing events of the two smallest IAV RNA segments were well described soon after the description of its genome, much remains unknown, and current studies working toward unraveling the fine regulation mechanisms of viral splicing are necessary to a better understanding of viral replication, host range, and pathogenesis mechanisms.
CLASSICAL SIGNALS, EFFECTORS, AND REGULATORS OF PRE-mRNA SPLICING IN HUMANS
Splicing is an essential step for eukaryotic gene expression. To generate mature mRNAs for translation, introns/noncoding sequences have to be removed to allow exons, the genuine coding sequences, to be joined together accurately. Moreover, pre-mRNAs can undergo alternative splicing that provides a large assortment of mRNAs from the same nascent sequence and which contributes toward widening the proteomic diversity (18). The splicing process is performed by the coordinated actions of a large ribonucleoprotein (RNP) complex known as the spliceosome, which is composed of five small nuclear RNAs (snRNAs) (U1, U2, U4, U5, and U6) in association with numerous regulating factors. The accurate activity of the spliceosome is supported by mRNA cis-acting classical signals that are specifically recognized by the ribonucleoprotein complex; these are the donor splice site (5′ ss), the branch point site, the polypyrimidine tract, and the acceptor splice site (3′ ss). Additional RNA motifs are also recognized by processing and splicing factors, including serine–arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) (Fig. 2) (19). SR proteins are characterized as splicing enhancers and act to stabilize the spliceosome assembly and modulate the selection of alternative 5′ ss in a concentration-dependent manner. The best-characterized SR proteins, SF2/ASF, SC35, SRp40, and SRp55, are represented in Fig. 2 (20, 21). hnRNPs constitute a family of pre-mRNA-binding proteins that are associated with different functions, such as preventing the folding of pre-mRNA and exporting mRNA out of the nucleus (22).
FIG 2 .
Classical signals, effectors, and regulators of pre-mRNA splicing in human cells. Four intronic motifs are required for splicing of pre-mRNA: the donor site GU at the intronic 5′ end (5′ ss), the branch point A, the polypyrimidine tract (Y)n upstream from the acceptor site AG dinucleotide at the intronic 3′ end, and the acceptor splice site (3′ ss). The sequences surrounding both the donor and acceptor dinucleotides are highly conserved, as illustrated by the consensus sequence representations (20). For each sequence motif, the size of a nucleotide is proportional to its frequency at the given position obtained from sequence alignments. Colored nucleotides show the exon-intron boundaries. The spliceosome is composed of small nuclear RNAs and polypeptides in small nuclear ribonucleoproteins (snRNPs), including the subunit U1, U2, U4, U5, and U6 snRNPs. In the first stages, U1 snRNP binds to the acceptor site and U2 snRNP to the donor site, and then, with the help of splicing factors such as U2AF, the other subunits are recruited. The splicing process is also directed by the action of numerous regulators, which are serine–arginine-rich proteins (SR proteins), such as SRp40, SRp55, and SF2/ASF, and heterogeneous nuclear ribonucleoproteins (hnRNPs). Splicing regulators recognize cis elements in the pre-mRNA sequence. SR proteins bind to enhancing signals, whereas hnRNPs recognize silencing ones (100). ESS, exonic splicing silencer; ISS, intronic splicing silencer; ISE, intronic splicing enhancer; ESE, exonic splicing enhancer.
Even though the major consensus motifs for classical splicing signals in the human genome are now well established (Fig. 2), a complete picture of the integrated regulatory code that drives the splicing process remains to be deciphered (18, 20, 23–25). Indeed, splicing is considered one of the most complex processes occurring in the cell. Furthermore, this machinery is closely coupled with the transcriptosome, in which RNA polymerase II recruits splicing factors while nuclear export receptors are recognizing spliced mRNAs (reviewed in references 23 and 24). In this context, the ability of influenza viruses to hijack these processes demonstrates their impressive adaptation to their host.
ALTERATION OF HOST RNA MATURATION PROCESS BY INFLUENZA VIRUS
IAV interactions with the spliceosome are well illustrated by the delocalization of components within the nucleus, such as SC35 splicing factor and p80/coilin from speckle domains and Cajal bodies, respectively (Fig. 1C) (26). Such a remodeling makes it possible for IAV to alter cellular splicing and, thus, impact the biogenesis and maturation of host mRNAs. Indeed, numerous studies, including ours, have described a marked impact of influenza virus infection on cellular gene expression (3, 27, 28). In addition to RdRp and cap-snatching activity, the other most influential viral factor contributing toward such an impact is suggested to be the NS1 protein, as it interacts with numerous cellular components through its RNA-binding and effector domains (29). Interestingly, NS1 can interact with several spliceosome subunits, including U2 and U6 snRNA in the classical complex but also U6atac, a U6 snRNA counterpart, in the case of minor donor-acceptor site combinations (Fig. 2) (30, 31). Thus, NS1 protein would appear to block cellular gene expression by inhibiting complete spliceosome recruitment and, therefore, the transition to an active splicing complex. However, as viral splice sites have been shown to match human consensus sequences, it was hypothesized that there is a mechanism that preferentially drives the splicing machinery toward viral mRNAs that need to be spliced, for which it would be worth considering the role that NS1 protein may have. Indeed, NS1 protein can efficiently bind the cellular 30-kDa subunit of CPSF (CPSF30), which is an essential component of the cellular 3′-end processing machinery (32). Through this interaction, NS1 protein globally inhibits the polyadenylation of cellular mRNA but not that of viral mRNA, which occurs independently of the cellular 3′-end processing machinery (33). In addition, NS1 also interacts with components of the mRNA export machinery, such as NXF1/Tap, p15, and Rae1, and with factors related to nuclear pores, like nucleoporin and Nup98, and has been shown to induce a blockage of host mRNA export (34).
Viral alteration of host RNA splicing and maturation steps is suggested to be a crucial point that balances cellular and viral expression throughout influenza virus infection, since it has been demonstrated that silencing of these cellular factors has a direct impact on viral production (35).
SPLICING: A NECESSARY STEP FOR INFLUENZA REPLICATION
Among the classical stages of pre-mRNA transcription and maturation in eukaryotic cells, the influenza viruses have developed strategies to replace the host RNA polymerase II with a viral counterpart, as well as the 5′ capping and 3′ polyadenylation, but not the splicing process. Indeed, influenza virus mRNA splicing remains dependent on the host machinery.
Up to three overlapping coding sequences in the NS segment.
Splicing of IAV mRNA was first considered in the late 1970s after the discovery of a new 11-kDa viral polypeptide, named NS2 (also NEP), within influenza virus-infected cells (36). Using reassortant viruses and RNA hybridization techniques, Lamb and Choppin demonstrated that this ninth influenza virus protein was encoded by the NS segment, already known to encode NS1 (37). Further analyses, including S1 nuclease mapping of the NS segment, have highlighted its potential to produce two overlapping mRNAs from the same nascent sequence (38). Subsequent cloning and sequencing strategies then provided a more accurate understanding about these two distinct mRNAs (39). NS1 is encoded in the NS segment by a colinear transcript of 864 nucleotides (nt) in which the open reading frame (ORF) runs from codon position 15–17 to 738–740 in the A/Udorn/72 reference strain (Fig. 3A). The second mRNA, encoding the NS2 protein, is an interrupted transcript of 394 nt (Fig. 3A). The first section of the NS2 transcript (up to position 56) is identical to the NS1 transcript, and the second section corresponds with the end of the NS segment from position 526. At this position, the ORF shifts to a different alignment, which leads to the translation of NS2/NEP.
FIG 3 .
Splicing of influenza NS and M segments. (A) There are up to three overlapping coding sequences in the NS segment. A full-length NS transcript is usually 864 nucleotides (nt) long and is terminated by a short polyadenylation motif at the 3′ extremity. NS1 protein is encoded by a colinear transcript from the initiation codon at position 27–29 to the termination codon at position 738–740. NS2/NEP protein is encoded by an interrupted transcript that is identical to the first section of the NS1 transcript (up to position 56), with a second section that is similar to the end of the NS segment from position 526 to 864. The black boxes at the 5′ ends represent leader nonviral sequences of 10 or 11 nt. The viral transcript encoding NS3 contains a novel 5′ ss (GG/GUA) and the same 3′ ss as NS2 mRNA. This novel 5′ ss was only observed in a limited number of IAV strain sequences. The noncoding and coding sequences are represented as thin lines and colored boxes, respectively. The positions and sequences of both splice sites are detailed. “A(n)” at the 3′ extremity is the polyadenylation motif. Consensus motifs corresponding to the 5′ ss and 3′ ss were generated using weblogo (http://weblogo.berkeley.edu/) (101) from a sequence alignment of 541 complete NS sequences (human and avian origin, 2011 to 2012) obtained from the Influenza Research Database (http://www.fludb.org/) (102). (B) One M segment template has four transcripts. The full-length M transcript is 1,004 nt. The M1 protein is encoded by the colinear transcript from the initiation codon at position 26–28 to the termination codon at position 782–784. The M2 protein originates from the interrupted mRNA M2, which has a sequence in common with that of the mRNA M1 until position 51 and then, at the 3′ end, consists of the full-length transcript from position 740. mRNA M2 shares the same initiation codon with mRNA M1 but switches to another open reading frame (ORF) in the last part of the mRNA. mRNA3 is interrupted from position 11 to 740, and mRNA M4 from position 145 to 740. Only mRNA M4 codes for a third protein (M42), using another initiation codon at position 114–116. Black boxes at the 5′ ends represent nonviral leader sequences of 10 or 11 nt. The noncoding and coding sequences are represented as thin lines and colored boxes, respectively. The positions and sequences of all efficient splice sites are detailed. “A(n)” at the 3′ extremity is the polyadenylation motif. Using the same methodology as described in the legend to panel A, consensus motifs for the different 5′ ss and the 3′ ss were obtained from sequence alignment of 669 complete M sequences (human and avian origin, 2011 to 2012) obtained from the Influenza Research Database. (A and B) Stars above the mRNA NS3 and mRNA M4 consensus sequences indicate the positions of mutations described in the literature. Nucleotide positions are based on A/Udorn/72 (H3N2) sequences.
Further analysis of the interrupting sequence (positions 56 to 526) revealed similarities between the intervening sequences at both the 5′ and 3′ extremities and major cellular splicing consensus motifs. This provided support to the hypothesis for nuclear splicing of the NS transcript through recognition of joining sequences by the host spliceosome complex (39). This hypothesis was validated by Lamb and Lai through the construction of a simian virus 40 (SV40) recombinant vector with a chimeric NS/β-globin segment (40, 41). Using uninfected HeLa nuclear extracts in vitro, the NS segment was efficiently processed, proving that its joining sites were indeed splicing motifs that were recognized as classical cellular consensus motifs. However, a larger amount of spliced products was found upon the replacement of the NS 3′ ss by its β-globin counterpart, indicating that the NS 3′ ss was not optimal in the absence of infection (42). Based on the alignment of more than 500 complete NS sequences chosen from recent human and avian strains (2011 to 2012), we obtained the consensus motifs for both the 5′ donor and 3′ acceptor splice sites. The donor and acceptor dinucleotides (GU and AG motifs) and nucleotides in close proximity to these motifs were remarkably conserved (Fig. 3A) and very similar to the human consensus splice sites (Fig. 2). More recently, Selman and colleagues (43) have demonstrated the existence of a third NS transcript in the context of adaptation of a human virus (A/Hong Kong/1/1968 H3N2) within a mouse host. This viral transcript, encoding NS3, contains a novel 5′ ss (GG/GUA) and the same 3′ ss as NS2 mRNA. Nevertheless, the authors performed a bioanalysis of the Influenza Research Database and have only found this additional NS 5′ splice site in a limited number of IAV strain sequences (43).
One M segment template and four transcripts.
As with the NS segment, mapping strategies have been performed on the IAV M segment to confirm the description of three transcripts (mRNA M1, mRNA M2, and mRNA3) for only two expressed proteins, M1 and M2 (44, 45). The M1 protein is encoded by the M segment colinear transcript mRNA M1, in which the ORF runs from codon position 26–28 to 783–785 (Fig. 3B). The second protein (M2) is encoded by an interrupted M2 mRNA transcript that is similar to mRNA M1 until position 51. At this point, the sequence is interrupted up to position 740, resulting in an ORF shift during translation such as that described for the NS2 transcript (Fig. 3B). Based on the unraveling of the splicing process occurring for the NS segment, the extremities of the interrupting sequence of mRNA M2, AC/GUA in the 5′ ss and CAG/G in the 3′ ss, were described as matching well with human consensus splice site motifs (45). In addition to these two coding mRNAs, a third transcript, mRNA3, has been described for segment M (45). mRNA3 consists of the first 11 nucleotides and the last part of the colinear sequence, from position 740 (Fig. 3B). The mRNA3 5′ joining sequence (CAG/GUAGAU) is located upstream from the mRNA M2 counterpart and has been described to be a much better match to the human consensus 5′ ss than the M2 5′ ss (40). Although a potential initiation codon is localized at position 755–757, no corresponding protein product has ever been detected for mRNA3. The construction of chimeric M/β-globin sequences in a recombinant SV40 vector has confirmed that all of these viral sites undergo nuclear splicing using the host machinery in the absence of infection, similar to the results for the NS study (40). Additionally, a fourth mRNA product (mRNA M4) was later discovered in A/WSN/33 (H1N1) and in a limited number of other IAV strains (46). This mRNA M4 corresponds to the first 145 nt of the colinear transcript followed by the last part of mRNA M2 from position 740 to the poly(A) site and encodes M42 (Fig. 3B). Although a small peptide of 54 amino acids was expected from ORF1 initiation, it has never been detected in infected cells, but more recently, another coding sequence was predicted from an additional initiation codon at position 114–116 (Fig. 3B). Wise and colleagues have suggested that M42 could play the role of a functional alternative to the ion channel in M2-deficient strains (47).
Based on the alignment of more than 600 recently completed M sequences (2011 to 2012), we obtained the consensus motifs for all previously described 5′ and 3′ splice sites (Fig. 3B). Our analysis highlighted a marked conservation of the 3′ ss motifs among the influenza virus sequences studied. This motif is, however, less closely matched to the human consensus 3′ ss than its NS counterpart, with a notable G>A polymorphism localized just after the acceptor site dinucleotide AG at the end of the 3′ ss (Fig. 2 and 3B). The three distinct 5′ ss present different levels of conservation, with the motif corresponding to mRNA M4 being the least conserved. Interestingly, the comparison of the 5′ ss motifs for mRNA M2 and mRNA3 with the human consensus sequences confirmed the observation made by Lamb and Lai indicating that the mRNA3 5′ ss motif matches the human consensus 5′ ss more closely than the M2 5′ ss (40).
Expression of the spliced NS and M products is highly regulated.
The amounts of cellular mRNAs and proteins from each spliced viral product evolve differently during IAV infection. Indeed, NS1 protein is detected early after the initial infection, and then M1 appears a little later, followed by the expression of NS2/NEP and M2 (48). More precisely, while it is assumed that the ratio of spliced to unspliced mRNA of the NS segment is maintained in cells at around 10% throughout the infection cycle, the quantity of NS1 protein increases continuously with time until the later stages of infection (49). Alternatively, it has been shown that higher levels of M1 mRNA are detected during the early stages of infection compared with the levels of M2 mRNA, the ratio of unspliced to spliced decreasing as M2 mRNA is more highly expressed later during infection (50). Wise and colleagues showed that strain-specific single-nucleotide changes in the 5′ ss result in marked alterations of splice site usage (47). It is worth noting that the splicing efficiency seems to be strongly dependent on the virus strain considered. Indeed, Robb and Fodor reported a completely different pattern of the unspliced-to-spliced ratio with another influenza A virus strain, A/WSN/33 (51). In this case, the major mRNA detected is the spliced M2 transcript, followed by mRNA3 and mRNA M4, whereas M1 mRNA represents only 8% of the mRNA products from the M segment. Throughout infection, these viral proteins strongly affect the outcome of viral replication: NS2/NEP is required for nucleocytoplasmic transport of vRNPs, and M2 is an important factor in viral pathogenicity, as demonstrated by splice site mutations resulting in deficient viruses (52–54). Thus, the occurrence of an adapted and specific regulation of viral splicing in infected cells appears essential for viral replication.
UNDERSTANDING THE MECHANISM AND REGULATION OF NS AND M SPLICING
Using actinomycin D-induced inhibition of RNA polymerase II, it has been determined that each viral mRNA has an equivalent half-life, and therefore, the possibility of regulation via a difference in their stability and fate in the cytoplasm can be ruled out (50). Several hypotheses remain and seem to differ according to the nature of the viral transcripts and the stage of infection; they include (i) direct regulation by cis- and/or trans-acting elements targeting the splicing process, (ii) indirect regulation via differential export of viral mRNA, and (iii) a combination of both.
Clues for direct regulation of viral splicing.
The first studies investigating the regulation of viral splicing focused on the stability of the splicing complexes required for accurate viral processing. During splicing progression, the spliceosome dynamically recruits factors that form different complexes at each step of the process. The first one sediments at around 55S on a sucrose gradient and is considered a marker for efficient splicing. In vitro experiments in the absence of infection reported that the NS1 transcript can form more stable 55S complexes than cellular mRNAs and could therefore alter the dynamic progress of the machinery, resulting in low or absent production of NEP mRNAs (55). In fact, using a chimeric β-globin–NS transcript, a large intronic sequence has been identified as being responsible for such a blockage (56). Although this is not an inhibition dependent on the nature of the nucleotide sequence, it has been hypothesized that such a cis element would be necessary for regulating an mRNA conformation that resists spatial rearrangements and thereby interferes with splicing progression (56). In addition to their previous observations about the NS2 3′ ss being less efficient than its β-globin counterpart, Nemeroff and Krug hypothesized that a secondary structure, including 3′ ss, inhibits the splicing mechanism. However, this blockage is partially lifted during infection, as NS2 mRNA is produced in a 1:10 ratio, confirming that a virus-specific factor and/or infection context are involved, as previously suggested (57), and this factor is most likely a product from NS segment. Indeed, splicing can occur even if only an NS-expressing plasmid is transfected into cells, demonstrating that an NS element is sufficient to allow viral splicing but not to its full extent (55).
However, experiments using a vRNP reconstitution assay to mimic infection gave contradictory results. NS splicing was shown to be down-regulated by the NS1 protein via its RNA-binding domain (58). In contrast, another study using more sensitive detection methods demonstrated that the presence or absence of NS1 protein does not affect the levels of spliced and unspliced NS transcripts (49). Considering all of these findings, we can assume that these contradictory results may be due to the different experimental strategies used (transient expression versus infection) and the viral origin of genomic and proteomic materials. Nevertheless, further investigations will need to consider NS1 protein the obvious viral candidate involved in viral splicing regulation.
On the other hand, splicing regulation is assumed to be more complex for M mRNAs than for NS mRNAs. First, it is important to note that the mRNA3 5′ ss is located prior to M2 mRNA on the colinear sequence (Fig. 3B). In this way, it could be considered to be the main 5′ ss that best matches to the human consensus donor motifs, as illustrated by our sequence analysis (Fig. 3B). However, only the expression of M2 protein is relevant for viral replication, whereas the function of mRNA3 remains unknown. Thus, it has been postulated that regulation in infected cells occurs to promote the use of M2 5′ ss in preference to the distal 5′ ss. The viral polymerase complex has been proposed by Shih and colleagues to fulfill this role, and this idea has been supported by more recent studies (59, 60). In fact, RdRp could interact with a specific sequence at the 5′ end of the M transcript, blocking mRNA3 5′ ss recognition and directing the spliceosome onto the alternative M2 5′ ss (Fig. 4A). However, as the viral polymerase can interact with all the viral RNAs to perform transcription and replication of the influenza virus genome, this interaction being shown to be relatively unsteady, this particular role for the polymerase complex has recently been challenged. Alternatively, Robb and Fodor have proposed that NS1 also has the ability to bind to the same region with increased stability and thus spatially obstruct the mRNA3 5′ ss (Fig. 4A) (51), suggesting its main contribution in the regulation of M1 mRNA splicing.
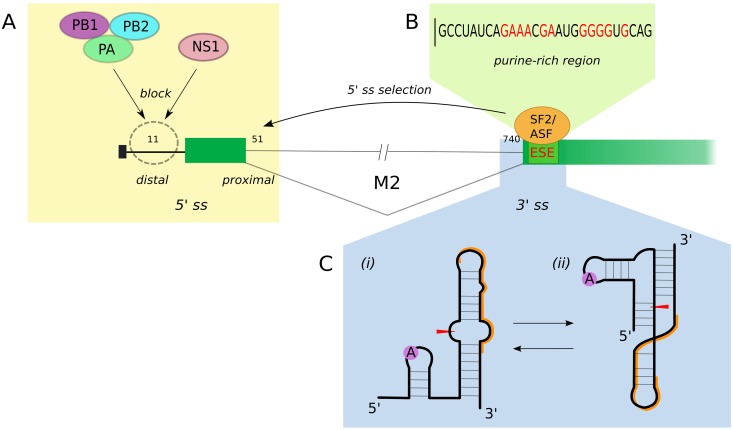
FIG 4 .
Model of the regulation of segment M splicing. (A) Blocking of distal 5′ ss. The switch from the distal mRNA3 5′ ss (11 nt) to the proximal M2 alternative 5′ ss (51 nt) by obstruction of the spliceosome binding by a viral product (viral polymerase complex or NS1 protein) is depicted. (B) Selection of the alternative 5′ ss is obtained by stabilization of the SF2/ASF interaction in the 3′ ss region. SF2/ASF recognizes an exonic splicing enhancer (ESE) composed of a purine-rich sequence (shown in red). (C) Stable secondary structures formed around the 3′ ss region. (i) Hairpin conformation opens access to the branch point site (A, highlighted in purple) and 3′ ss (red arrowhead) for the spliceosome assembly and to the ESE motif (orange line) for the SF2/ASF factor. (ii) Pseudoknot conformation tends to prevent 3′-ss processing by the spliceosome.
While each interrupted M transcript has a different 5′ ss, they all use the same 3′ ss, thus highlighting this site as an important region to stabilize RNA splicing. Indeed, a purine-rich sequence located in this region corresponds to an exonic splicing enhancer (ESE) motif recognized by SF2/ASF, a well-known splicing factor belonging to the SR protein family (Fig. 4B) (61). In uninfected cells, SF2/ASF functions in a concentration-dependent manner to influence the selection of alternative competitive 5′ ss (62, 63). To support this evidence, increasing levels of SF2/ASF in vivo are correlated with increased M2 mRNA during infection compared to the levels of other M transcripts (61). SF2/ASF thus seems to be an important factor for M splicing stabilization and could also play a role in establishing the rate of unspliced/spliced M products.
Structural basis for regulation.
The importance of secondary structures in molecular mechanisms is now well described, most particularly for processes involving RNA, such as alternative splicing (64). Therefore, it is important to consider that structural conformation is an important element for the regulation of viral splicing. Interestingly, based on sequence analyses and thermodynamic stability studies, it has been predicted that there are regions within IAV mRNAs that are unusually stable and conserved, particularly around M and NS segment 3′ ss regions (65–67). From phylogenetic analysis coupled to biochemical and biophysical studies, a single-nucleotide substitution near the 3′ splice site of the NS segment of H5N1 strains was shown to be critical for the equilibrium between (i) a hairpin and (ii) a pseudoknot conformation, with potential consequences for splicing regulation (68, 69). In the case of M segment transcripts, recent studies indicate that the region around the 3′ ss can also switch between these two distinct secondary structures (Fig. 4C) (70, 71). The folding of this region into one or the other conformation changes the accessibility to multiple important sites for splicing, including the site of interaction with SF2/ASF, the 3′ ss, and the branch point site previously discussed (Fig. 4C) (71). Considering the 3-dimensional structure, the hairpin conformation would reveal recognition sites for accurate spliceosome assembly, whereas the pseudoknot configuration would inversely inhibit 3′ ss processing (71). Altogether, these recent developments illustrate the most-probably underestimated role of RNA secondary structures in influenza virus biology and, notably, in viral splicing regulation.
Implication of nucleocytoplasmic transport.
To address the question of unbalanced viral transcript splicing, it is also important to consider indirect regulation via the export of viral mRNAs, as the nucleocytoplasmic export machinery is closely interlinked with pre-mRNA splicing (72). There are two distinct cases in term of IAV transcript export, (i) viral intronless transcripts (six of the eight viral RNA segments produce only colinear mRNA) and (ii) spliced viral mRNA, such as the NS2/NEP transcript. Thus, it would seem evident that the viral mRNAs that are spliced by the host machinery are favored and more rapidly transported into the cytoplasm than the intronless transcripts, on the basis of host transcript export. In any case, viral proteins derived from spliced transcripts are expressed in lower quantities than proteins from unspliced mRNAs. Several publications have highlighted interconnections with the host NFX1/Tap export system (reviewed in reference 73). Indeed, it has been shown that the coadaptor NFX1/Tap, which facilitates the recruitment of mRNA to the nuclear pore complex (NPC), coimmunoprecipitates with the IAV mRNAs, and Read and Digard have shown that IAV mRNAs present differential dependence on the NXF1/Tap pathway for their nuclear export (74). Moreover, silencing of NFX1/Tap expression induces inefficient IAV replication, which provides conclusive evidence of the importance of this export system for IAV (74, 75). Different studies suggest that this phenomenon is possible thanks to the action of the multifunctional NS1 protein on the regulation of export. Indeed, NS1 protein can interact with NFX1/Tap, p15, and Rae1, among other export coadaptors, components of the nuclear pore complex, such as nucleoporin Nup98, and several viral mRNAs, by its RNA-binding domain (34, 76). Moreover, by recognizing an NS2-specific sequence, NS1 protein also retains NS2 mRNA within the nucleus (77). Thus, the less efficient transport of NS2 spliced mRNA combined with a blockage of NS splicing by the cis element may be a consistent explanation for the small amount of and delay in NS2 protein expression during infection (78, 79). While the whole mechanism is not yet fully known, these results illustrate how IAV is able to differentially regulate nucleocytoplasmic export of viral mRNAs and, especially, the spliced one, which could permit host and viral protein expression regulation in time.
Numerous host-splicing factors are necessary for influenza A virus infection.
As previously described, host factors, notably those interacting with NS1, would be involved in the global down-regulation of host cellular expression, while at the same time, they could allow the completion and regulation of viral splicing during infection. The characterization of host factors that interact with NS1 has highlighted a cellular protein named NS1-binding protein (NS1-BP) (80). In the absence of infection, this protein colocalizes with the SC35 factor in speckle domains, which are involved in spliceosome assembly. Upon infection, as NS1 contributes to a remodeling of the nuclear compartment, and notably, the speckle domains (26), the NS1-BP protein is dispersed inside the whole nucleus. If the physiological function of NS1-BP is unknown, it seems to achieve an important aim throughout influenza virus infection. Indeed, a recent study showed that nuclear proteins NS1-BP and hnRNP K have a privileged function in the regulation of M2 splicing (60). After infection of an NS1-BP-depleted cell line by an A/WSN/33 strain, in which the spliced M2 mRNA is the most highly represented, the M2-to-M1 mRNA ratio is strongly impaired, whereas it remains at steady state for the other M-derived transcripts. This effect is directly related to the presence of hnRNP K, which acts as an adaptor protein between M transcript and NS1-BP, which is unable to directly bind RNA (60). In this example, the presence of these two host factors is essential for the expression of viral M2 protein and, thus, for the viral replication.
In addition, over the past 5 years, several genome-wide screening studies have identified important host cell factors in the influenza virus replication cycle (35, 75, 81–83). In a short meta-analysis of hundreds of host factors identified based on these different strategies, we have highlighted up to 49 key host factors involved in RNA maturation and splicing processes (Table 1). Among them, several splicing factors appear to be essential for effective IAV infection, such as pre-mRNA processing factor 8 (PRPF8), splicing factor 3A (SF3A1), the 70-kDa small nuclear ribonucleoprotein (SNRP70), and polypyrimidine tract-binding protein 1 (PTBP1) (35, 81, 82). Interestingly, despite differences between the cellular and viral models used for these high-throughput studies (e.g., Homo sapiens versus Drosophila models), more than 18% (9/49) of splicing-associated factors were commonly identified in at least two studies (Table 1). The importance of these host factors was highlighted in particular in the genome-wide RNA interference (RNAi) screen published by Karlas and colleagues, in which gene ontology (GO) term enrichment analyses revealed that the U2-dependent spliceosome and the spliceosome were among the most enriched terms (35). This subclass of host factors may constitute a very interesting list of potential therapeutic targets for influenza A virus infection, as illustrated by the pioneering work from retroviral models (84).
TABLE 1 .
List of cellular splicing factors identified using high-throughput approaches that are important for viral replication
| High-throughput approach(es) used | Cellular factor identified | Complete name | Influenza virus(es)—cellular model(s) | Reference |
|---|---|---|---|---|
| Genome-wide RNAi screening | AQR | Aquarius homolog (mouse) | A/Puerto Rico/8/34 (H1N1)—U2OS cells, modified A/WSN/33 (H1N1)—D-Mel2 Drosophila cells | 75, 81 |
| BCAS2 | Breast carcinoma amplified sequence 2 | Modified A/WSN/33 (H1N1)—D-Mel2 Drosophila cells | 75 | |
| C19orf29 | Cactin | A/Puerto Rico/8/34 (H1N1)—U2OS cells | 81 | |
| CLK1 | CDC-like kinase 1 | A/Puerto Rico/8/34 (H1N1)—U2OS cells, A/WSN/33 (H1N1)—A549 cells | 35, 81 | |
| CLK3 | CDC-like kinase 3, dual-specificity protein kinase CLK3 | A/Puerto Rico/8/34 (H1N1)—U2OS cells | 81 | |
| CRNKL1 | Crooked neck pre-mRNA splicing factor-like 1 (Drosophila) | |||
| CWC22 | Pre-mRNA-splicing factor CWC22 homolog | |||
| DHX8 | ATP-dependent RNA helicase DHX8 | |||
| EFTUD2 | Elongation factor Tu GTP binding domain containing 2 | |||
| FUS | Fused in sarcoma (HNRNPP2) | |||
| HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | Modified A/WSN/33 (H1N1)—D-Mel2 Drosophila cells | 75 | |
| HNRNPU | Heterogeneous nuclear ribonucleoprotein U | A/Puerto Rico/8/34 (H1N1)—U2OS cells | 81 | |
| LSM2 | LSM2 homolog, U6 small nuclear RNA associated | |||
| NHP2L1 | NHP2 nonhistone chromosome protein 2-like 1 | A/Puerto Rico/8/34 (H1N1)—U2OS cells, A/WSN/33 (H1N1)—A549 cells | 81, 82 | |
| POLR2H | Polymerase (RNA) II (DNA directed) polypeptide H | A/WSN/33 (H1N1)—A549 cells | 35 | |
| POLR2L | Polymerase (RNA) II (DNA directed) polypeptide L, 7.6 kDa | |||
| PPAN | Peter pan homolog (Drosophila) | A/Puerto Rico/8/34 (H1N1)—U2OS cells | 81 | |
| PPWD1 | Peptidylprolyl isomerase domain and WD repeat-containing 1 | Modified A/WSN/33 (H1N1)—D-Mel2 Drosophila cells | 75 | |
| PRPF31 | PRP31 pre-mRNA processing factor 31 homolog | A/Puerto Rico/8/34 (H1N1)—U2OS cells | 81 | |
| PRPF8 | PRP8 pre-mRNA processing factor 8 homolog | A/Puerto Rico/8/34 (H1N1)—U2OS cells, A/WSN/33 (H1N1)—A549 cells | 35, 81 | |
| RBM5 | RNA binding motif protein 5 | A/WSN/33 (H1N1)—A549 cells | 82 | |
| RNPS1 | RNA binding protein S1, serine-rich domain | Modified A/WSN/33 (H1N1)—D-Mel2 Drosophila cells | 75 | |
| SART1 | Squamous cell carcinoma antigen recognized by T cells | A/Puerto Rico/8/34 (H1N1)—U2OS cells | 81 | |
| SF3A1 | Splicing factor 3a, subunit 1, 120 kDa | A/WSN/33 (H1N1)—A549 cells, A/WSN/33 (H1N1)—A549 cells | 35, 82 | |
| SF3B1 | Splicing factor 3b, subunit 1, 155 kDa | A/Puerto Rico/8/34 (H1N1)—U2OS cells, A/WSN/33 (H1N1)—A549 cells | 35, 81 | |
| SF3B14 | Splicing factor 3b, 14-kDa subunit | A/WSN/33 (H1N1)—A549 cells | 35 | |
| SF3B2 | Splicing factor 3b, subunit 2, 145 kDa | A/Puerto Rico/8/34 (H1N1)—U2OS cells | 81 | |
| SF3B3 | Splicing factor 3b, subunit 3, 130 kDa | |||
| SLU7 | Pre-mRNA-splicing factor SLU7 | |||
| SNRP70 | Small nuclear ribonucleoprotein 70-kDa polypeptide (RNP antigen) | A/WSN/33 (H1N1)—A549 cells, A/WSN/33 (H1N1)—A549 cells | 35, 82 | |
| SNRPA1 | Small nuclear ribonucleoprotein polypeptide A′ | A/WSN/33 (H1N1)—A549 cells | 82 | |
| SNRPB | Small nuclear ribonucleoprotein polypeptides B and B1 | A/Puerto Rico/8/34 (H1N1)—U2OS cells | 81 | |
| SNRPC | Small nuclear ribonucleoprotein polypeptide C | Modified A/WSN/33 (H1N1)—D-Mel2 Drosophila cells | 75 | |
| SNRPD1 | Small nuclear ribonucleoprotein D1 polypeptide, 16 kDa | |||
| SNRPD2 | Small nuclear ribonucleoprotein D2 polypeptide, 16.5 kDa | A/Puerto Rico/8/34 (H1N1)—U2OS cells | 81 | |
| SNRPD3 | Small nuclear ribonucleoprotein D3 polypeptide, 18 kDa | |||
| SNRPF | Small nuclear ribonucleoprotein polypeptide F | A/WSN/33 (H1N1)—A549 cells | 35 | |
| SNRPG | Small nuclear ribonucleoprotein polypeptide G | Modified A/WSN/33 (H1N1)—D-Mel2 Drosophila cells | 75 | |
| THOC4 | THO complex 4 | |||
| TXNL4A | Thioredoxin-like 4A | A/WSN/33 (H1N1)—A549 cells | 35 | |
| YTHDC1 | Putative splicing factor YT521 | A/Puerto Rico/8/34 (H1N1)—U2OS cells | 81 | |
| Genome-wide RNAi screening, random homozygous gene perturbation | PTBP1 | Polypyrimidine tract-binding protein 1 | A/Puerto Rico/8/34 (H1N1)—U2OS cells, A/Udorn/72 (H3N2)—MDCK cells | 81, 83 |
| Random homozygous gene perturbation | DDX17 | DEAD (Asp–Glu–Ala–Asp) box polypeptide 17 | A/Udorn/72 (H3N2)—MDCK cells | 83 |
| LSM4 | LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) | |||
| PAPOLA | Poly(A) polymerase alpha | |||
| SF3A3 | Splicing factor 3a, subunit 3, 60 kDa | |||
| SNW1 | SNW domain-containing 1 | |||
| Yeast two-hybrid system, siRNA screening | UAP56 | DEAD (Asp–Glu–Ala–Asp) box polypeptide 39B | A/Puerto Rico/8/34 (H1N1) HeLa Cells, A/Bratislava/79/H7N7 (H7N7)—A549 and 293T cells | 103, 104 |
WHAT ABOUT INFLUENZA B AND C VIRUSES?
Whereas most of our knowledge about the cellular and molecular biology of influenza viruses relies on studies dedicated to IAV, we know comparatively little about IBV and ICV. With both of their genomes composed of eight vRNA segments, IAV and IBV share numerous features. However, these viruses have weak percentages of homology between analogous proteins, reflecting divergent evolution, particularly concerning their coding strategy (11, 85). Actually, only the eighth segment of IBV, encoding its NS1 (B/NS1) and NS2/NEP (B/NS2) proteins, undergoes splicing in a way similar to its IAV counterpart (86, 87). Indeed, B/NS1 is encoded by a colinear transcript in an ORF running from position 43–45 to the termination codon at position 886–888 in IBV reference strain B/Lee/40, and B/NS2 is encoded by an interrupted transcript of nearly 360 nt (87). In contrast, the IBV segment 7 encodes two proteins, matrix protein M1 and ion channel protein BM2, via a translational stop-start mechanism in place of the IAV alternative splicing strategy (88).
The ICV genome is composed of only seven vRNA segments and has been described to go through a splicing mechanism in the case of segments 6 and 7. Whereas ICVs use a splicing strategy similar to that of IAV to express both NS1 and NS2 protein from its seventh segment, the processing of segment 6, which encodes the ICV M1 (C/M1) and C/M2, differs more (89, 90). Indeed, the C/M1 transcript needs to be spliced to introduce a termination codon (nucleotides 753, 754, and 983 for reference strain C/Yamagata/1/88), whereas C/M2 protein is produced after the cleavage of a P42 peptide encoded by an unspliced segment 6 transcript. In the case of ICV, it has been shown that the spliced M transcript is expressed predominantly compared to the unspliced one, which represents only 13% of the amount of spliced M1 mRNA (91–94).
As we have reported above, several studies presume a central role of the IAV NS1 protein (A/NS1) in the regulation of the viral and host splicing process. First, it is interesting to note that, despite the low percentage of identity between A/NS1 and B/NS1, many similar functions and interactions, such as type 1 interferon antagonism or inhibition of PKR kinase, have been described (11). Surprisingly, in contrast to A/NS1, little is known about the possible role of B/NS1 in splicing regulation. However, at early stages of infection, B/NS1 was found to colocalize with the splicing factor SC35 in nuclear speckles, suggesting a possible role in splicing (95).
Interestingly, influenza C virus NS1 protein (C/NS1) differs strongly from A/NS1, particularly in the C-terminal half of the protein. It seems that, in contrast to A/NS1, C/NS1 could upregulate the splicing of viral mRNAs in ICV-infected cells, which explains the higher rate of spliced mRNAs than of unspliced ones in this context. In addition, C/NS1 was also shown to facilitate the splicing of IAV M transcripts in vitro (94). For now, there is no certainty about the mechanism used by C/NS1 to enhance viral splicing. Further studies are necessary for a better understanding of the nuclear function of C/NS1 and its possible role in spliceosome recruitment.
IS SPLICING A KEY DETERMINANT FOR VIRAL REPLICATION, HOST RANGE, AND PATHOGENESIS?
Recent studies have hypothesized that the splicing efficiency of the M and NS segments of IAV could be a determinant in their ability to replicate or to adapt to a new host, denoting an important concern for pathogenicity (79, 96). Chua and colleagues have recently demonstrated that suboptimal splicing of the NS segment plays a role as a “molecular timer” that coordinates the timing of infection (79). Moreover, results obtained by Backström Winquist and colleagues have shown that the splicing efficiency of M and NS mRNAs varies between different influenza viruses (96). For example, the NS segment from A/Brevig Mission/1919/1 (H1N1) has been shown to be inefficiently spliced compared to those of other influenza viruses, possibly due to the differential binding properties of cellular splicing factors (e.g., SR proteins), resulting in the production of higher levels of functional NS1. The authors hypothesized that this could have contributed to the pathogenicity of the 1918 pandemic Spanish influenza virus (96). Another argument in favor of the importance of splicing strategy for influenza virus fitness was brought forward by Selman and colleagues with the description of NS3 protein expression in response to mouse adaptation (43). Their study indicates that one of the features of the adaptation of the influenza virus genome to a new host is the creation of new splice sites and that, in consequence, the virus can produce a novel protein, possibly conferring an advantage for viral replication. As it is easy to conceive the potential for the modulation of splicing strategy to optimize the host range and pathogenicity of influenza viruses, it is actually too early to estimate the importance of this mechanism among others.
CONCLUSION AND PERSPECTIVES
During their replication cycles, influenza viruses hijack the host splicing machinery to process their smallest segments. Influenza viruses have developed accurate regulation mechanisms to utilize the host spliceosome to enable the expression of specific spliced influenza virus products throughout infection. Indeed, in the case of IAV, viral spliced segments contain splice sites that display high levels of similarity with human consensus splice sites, as well as interaction capacities evolved to best fit the host machinery. Our bioinformatic analysis, along with recently published studies, suggest both a conservation and a possible evolution of viral splice site sequences and structure for improved adaptation to the host (67, 70). Even though the mechanism of regulation of viral splicing is still not fully understood, the currently available data highlight some interesting potential applications. Such applications are particularly notable for the NS segment, encoding the multifunctional NS1 protein, which has been shown to affect a diversity of cellular mechanisms, including the cellular interferon type I response (29). In fact, the major role of the NS1 protein in cellular processes has been demonstrated by the modification of the NS segment to produce a truncated or deleted NS1 protein (97, 98). A recent study reported that the NS segment can be modified at the level of the 5′ ss and 3′ ss regions to induce efficient secretion of exogenous interleukin-2 cytokine in addition to NS products, in a vaccine based on an NS1-deficient viral vector (99). It highlights one of the interesting applications offered by exploiting IAV hijacking of the cellular nuclear machinery. Future studies are required to further explore and evaluate the biological significance of splicing of viral segments, notably in terms of viral replication, host range, and pathogenicity.
ACKNOWLEDGMENTS
We thank Juan Ortin (Centro Nacional de Biotecnologia [CSIC], Madrid, Spain) for his help and his greatly appreciated comments. We also thank the two anonymous reviewers for their constructive remarks and suggestions.
Footnotes
Citation Dubois J, Terrier O, Rosa-Calatrava M. 2014. Influenza viruses and mRNA splicing: doing more with less. mBio 5(3):e00070-14 doi:10.1128/mBio.00070-14.
REFERENCES
- 1. Bouvier NM, Palese P. 2008. The biology of influenza viruses. Vaccine 26:D49–D53. 10.1016/j.vaccine.2008.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lamb RA, Krug R. 2001. Orthomyxoviridae: the viruses and their replication, p 1487–1579 In Knipe DM, Howley PM. (ed), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3. Josset L, Frobert E, Rosa-Calatrava M. 2008. Influenza A replication and host nuclear compartments: many changes and many questions. J. Clin. Virol. 43:381–390. 10.1016/j.jcv.2008.08.017 [DOI] [PubMed] [Google Scholar]
- 4. Neumann G, Kawaoka Y. April 2011. Influenza viruses: molecular virology. In eLS John Wiley & Sons Ltd, Chichester, United Kingdom: http://www.els.net.gate1.inist.fr. 10.1002/9780470015902.a0001031.pub3 [DOI] [Google Scholar]
- 5. Resa-Infante P, Jorba N, Coloma R, Ortin J. 2011. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 8:207–215. 10.4161/rna.8.2.14513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Engelhardt OG, Fodor E. 2006. Functional association between viral and cellular transcription during influenza virus infection. Rev. Med. Virol. 16:329–345. 10.1002/rmv.512 [DOI] [PubMed] [Google Scholar]
- 7. Hutchinson EC, Fodor E. 2013. Transport of the influenza virus genome from nucleus to nucleus. Viruses 5:2424–2446. 10.3390/v5102424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Terrier O, Moules V, Carron C, Cartet G, Frobert E, Yver M, Traversier A, Wolff T, Riteau B, Naffakh N, Lina B, Diaz JJ, Rosa-Calatrava M. 2012. The influenza fingerprints: NS1 and M1 proteins contribute to specific host cell ultrastructure signatures upon infection by different influenza A viruses. Virology 432:204–218. 10.1016/j.virol.2012.05.019 [DOI] [PubMed] [Google Scholar]
- 9. Amorim MJ, Digard P. 2006. Influenza A virus and the cell nucleus. Vaccine 24:6651–6655. 10.1016/j.vaccine.2006.05.066 [DOI] [PubMed] [Google Scholar]
- 10. Muraki Y, Hongo S. 2010. The molecular virology and reverse genetics of influenza C virus. Jpn. J. Infect. Dis. 63:157–165 [PubMed] [Google Scholar]
- 11. Jackson D, Elderfield RA, Barclay WS. 2011. Molecular studies of influenza B virus in the reverse genetics era. J. Gen. Virol. 92:1–17. 10.1099/vir.0.026187-0 [DOI] [PubMed] [Google Scholar]
- 12. Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O’Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306–1312. 10.1038/nm1201-1306 [DOI] [PubMed] [Google Scholar]
- 13. Wise HM, Foeglein A, Sun J, Dalton RM, Patel S, Howard W, Anderson EC, Barclay WS, Digard P. 2009. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol. 83:8021–8031. 10.1128/JVI.00826-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza a virus segment 3 modulates the host response. Science 337:199–204. 10.1126/science.1222213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muramoto Y, Noda T, Kawakami E, Akkina R, Kawaoka Y. 2013. Identification of novel influenza A virus proteins translated from PA mRNA. J. Virol. 87:2455–2462. 10.1128/JVI.02656-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cavallari I, Rende F, D’Agostino DM, Ciminale V. 2011. Converging strategies in expression of human complex retroviruses. Viruses 3:1395–1414. 10.3390/v3081395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoeben RC, Uil TG. 2013. Adenovirus DNA replication. Cold Spring Harb. Perspect. Biol. 5:a013003. 10.1101/cshperspect.a013003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136:701–718. 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- 19. Sperling J, Azubel M, Sperling R. 2008. Structure and function of the pre-mRNA splicing machine. Structure 16:1605–1615. 10.1016/j.str.2008.08.011 [DOI] [PubMed] [Google Scholar]
- 20. Cartegni L, Chew SL, Krainer AR. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3:285–298. 10.1038/nrg775 [DOI] [PubMed] [Google Scholar]
- 21. Wu JY, Maniatis T. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061–1070. 10.1016/0092-8674(93)90316-I [DOI] [PubMed] [Google Scholar]
- 22. Han SP, Tang YH, Smith R. 2010. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 430:379–392. 10.1042/BJ20100396 [DOI] [PubMed] [Google Scholar]
- 23. Mount SM. 1982. A catalogue of splice junction sequences. Nucleic Acids Res. 10:459–472. 10.1093/nar/10.2.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mount SM. 1983. RNA processing. Sequences that signal where to splice. Nature 304:309–310. 10.1038/304309a0 [DOI] [PubMed] [Google Scholar]
- 25. Wang Z, Burge CB. 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14:802–813. 10.1261/rna.876308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fortes P, Lamond AI, Ortín J. 1995. Influenza virus NS1 protein alters the subnuclear localization of cellular splicing components. J. Gen. Virol. 76(Pt 4):1001–1007. 10.1099/0022-1317-76-4-1001 [DOI] [PubMed] [Google Scholar]
- 27. Schmolke M, Viemann D, Roth J, Ludwig S. 2009. Essential impact of NF-kappaB signaling on the H5N1 influenza A virus-induced transcriptome. J. Immunol. 183:5180–5189. 10.4049/jimmunol.0804198 [DOI] [PubMed] [Google Scholar]
- 28. Terrier O, Josset L, Textoris J, Marcel V, Cartet G, Ferraris O, N’guyen C, Lina B, Diaz JJ, Bourdon JC, Rosa-Calatrava M. 2011. Cellular transcriptional profiling in human lung epithelial cells infected by different subtypes of influenza A viruses reveals an overall down-regulation of the host p53 pathway. Virol. J. 8:285. 10.1186/1743-422X-8-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hale BG, Randall RE, Ortín J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376. 10.1099/vir.0.2008/004606-0 [DOI] [PubMed] [Google Scholar]
- 30. Qiu Y, Nemeroff M, Krug RM. 1995. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA 1:304–316 [PMC free article] [PubMed] [Google Scholar]
- 31. Wang W, Krug RM. 1998. U6atac snRNA, the highly divergent counterpart of U6 snRNA, is the specific target that mediates inhibition of AT-AC splicing by the influenza virus NS1 protein. RNA 4:55–64 [PMC free article] [PubMed] [Google Scholar]
- 32. Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30-kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991–1000. 10.1016/S1097-2765(00)80099-4 [DOI] [PubMed] [Google Scholar]
- 33. Noah DL, Twu KY, Krug RM. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307:386–395. 10.1016/S0042-6822(02)00127-7 [DOI] [PubMed] [Google Scholar]
- 34. Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. U. S. A. 104:1853–1858. 10.1073/pnas.0610977104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Mäurer AP, Müller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822. 10.1038/nature08760 [DOI] [PubMed] [Google Scholar]
- 36. Lamb RA, Etkind PR, Choppin PW. 1978. Evidence for a ninth influenza viral polypeptide. Virology 91:60–78. 10.1016/0042-6822(78)90355-0 [DOI] [PubMed] [Google Scholar]
- 37. Lamb RA, Choppin PW. 1979. Segment 8 of the influenza virus genome is unique in coding for two polypeptides. Proc. Natl. Acad. Sci. U. S. A. 76:4908–4912. 10.1073/pnas.76.10.4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lamb RA, Choppin PW, Chanock RM, Lai CJ. 1980. Mapping of the two overlapping genes for polypeptides NS1 and NS2 on RNA segment 8 of influenza virus genome. Proc. Natl. Acad. Sci. U. S. A. 77:1857–1861. 10.1073/pnas.77.4.1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lamb RA, Lai CJ. 1980. Sequence of interrupted and uninterrupted mRNAs and cloned DNA coding for the two overlapping nonstructural proteins of influenza virus. Cell 21:475–485. 10.1016/0092-8674(80)90484-5 [DOI] [PubMed] [Google Scholar]
- 40. Lamb RA, Lai CJ. 1982. Spliced and unspliced messenger RNAs synthesized from cloned influenza virus M DNA in an SV40 vector: expression of the influenza virus membrane protein (M1). Virology 123:237–256. 10.1016/0042-6822(82)90258-6 [DOI] [PubMed] [Google Scholar]
- 41. Lamb RA, Lai CJ. 1984. Expression of unspliced NS1 mRNA, spliced NS2 mRNA, and a spliced chimera mRNA from cloned influenza virus NS DNA in an SV40 vector. Virology 135:139–147. 10.1016/0042-6822(84)90124-7 [DOI] [PubMed] [Google Scholar]
- 42. Plotch SJ, Krug RM. 1986. In vitro splicing of influenza viral NS1 mRNA and NS1-beta-globin chimeras: possible mechanisms for the control of viral mRNA splicing. Proc. Natl. Acad. Sci. U. S. A. 83:5444–5448. 10.1073/pnas.83.15.5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Selman M, Dankar SK, Forbes NE, Jia J-J, Brown EG. 2012. Adaptive mutation in influenza A virus non-structural gene is linked to host switching and induces a novel protein by alternative splicing. Emerg. Microbes Infect. 1:e42. 10.1038/emi.2012.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Inglis SC, Brown CM. 1981. Spliced and unspliced RNAs encoded by virion RNA segment 7 of influenza virus. Nucleic Acids Res. 9:2727–2740. 10.1093/nar/9.12.2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lamb RA, Lai CJ, Choppin PW. 1981. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc. Natl. Acad. Sci. U. S. A. 78:4170–4174. 10.1073/pnas.78.7.4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shih SR, Suen PC, Chen YS, Chang SC. 1998. A novel spliced transcript of influenza A/WSN/33 virus. Virus Genes 17:179–183. 10.1023/A:1008024909222 [DOI] [PubMed] [Google Scholar]
- 47. Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, Gog JR, Taubenberger JK, Digard P. 2012. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 8:e1002998. 10.1371/journal.ppat.1002998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Inglis SC, Brown CM. 1984. Differences in the control of virus mRNA splicing during permissive or abortive infection with influenza A (fowl plague) virus. J. Gen. Virol. 65(Pt 1):153–164. 10.1099/0022-1317-65-1-153 [DOI] [PubMed] [Google Scholar]
- 49. Robb NC, Jackson D, Vreede FT, Fodor E. 2010. Splicing of influenza A virus NS1 mRNA is independent of the viral NS1 protein. J. Gen. Virol. 91:2331–2340. 10.1099/vir.0.022004-0 [DOI] [PubMed] [Google Scholar]
- 50. Valcárcel J, Portela A, Ortín J. 1991. Regulated M1 mRNA splicing in influenza virus-infected cells. J. Gen. Virol. 72(Pt 6):1301–1308. 10.1099/0022-1317-72-6-1301 [DOI] [PubMed] [Google Scholar]
- 51. Robb NC, Fodor E. 2012. The accumulation of influenza A virus segment 7 spliced mRNAs is regulated by the NS1 protein. J. Gen. Virol. 93:113–118. 10.1099/vir.0.035485-0 [DOI] [PubMed] [Google Scholar]
- 52. Cheung TK, Guan Y, Ng SS, Chen H, Wong CH, Peiris JS, Poon LL. 2005. Generation of recombinant influenza A virus without M2 ion-channel protein by introduction of a point mutation at the 5′ end of the viral intron. J. Gen. Virol. 86:1447–1454. 10.1099/vir.0.80727-0 [DOI] [PubMed] [Google Scholar]
- 53. Chiang C, Chen GW, Shih SR. 2008. Mutations at alternative 5′ splice sites of M1 mRNA negatively affect influenza A virus viability and growth rate. J. Virol. 82:10873–10886. 10.1128/JVI.00506-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P. 2008. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J. Virol. 82:11869–11879. 10.1128/JVI.01634-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Agris CH, Nemeroff ME, Krug RM. 1989. A block in mammalian splicing occurring after formation of large complexes containing U1, U2, U4, U5, and U6 small nuclear ribonucleoproteins. Mol. Cell. Biol. 9:259–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nemeroff ME, Utans U, Krämer A, Krug RM. 1992. Identification of cis-acting intron and exon regions in influenza virus NS1 mRNA that inhibit splicing and cause the formation of aberrantly sedimenting presplicing complexes. Mol. Cell. Biol. 12:962–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith DB, Inglis SC. 1985. Regulated production of an influenza virus spliced mRNA mediated by virus-specific products. EMBO J. 4:2313–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Garaigorta U, Ortín J. 2007. Mutation analysis of a recombinant NS replicon shows that influenza virus NS1 protein blocks the splicing and nucleo-cytoplasmic transport of its own viral mRNA. Nucleic Acids Res. 35:4573–4582. 10.1093/nar/gkm230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shih SR, Nemeroff ME, Krug RM. 1995. The choice of alternative 5′ splice sites in influenza virus M1 mRNA is regulated by the viral polymerase complex. Proc. Natl. Acad. Sci. U. S. A. 92:6324–6328. 10.1073/pnas.92.14.6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tsai PL, Chiou NT, Kuss S, García-Sastre A, Lynch KW, Fontoura BM. 2013. Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza A virus RNA splicing. PLoS Pathog. 9:e1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shih SR, Krug RM. 1996. Novel exploitation of a nuclear function by influenza virus: the cellular SF2/ASF splicing factor controls the amount of the essential viral M2 ion channel protein in infected cells. EMBO J. 15:5415–5427 [PMC free article] [PubMed] [Google Scholar]
- 62. Cáceres JF, Krainer AR. 1993. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 12:4715–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Krainer AR, Conway GC, Kozak D. 1990. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell 62:35–42. 10.1016/0092-8674(90)90237-9 [DOI] [PubMed] [Google Scholar]
- 64. Warf MB, Berglund JA. 2010. The role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 35:169–178. 10.1016/j.tibs.2009.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gog JR, Afonso EDS, Dalton RM, Leclercq I, Tiley L, Elton D, von Kirchbach JC, Naffakh N, Escriou N, Digard P. 2007. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 35:1897–1907. 10.1093/nar/gkm087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Moss WN, Priore SF, Turner DH. 2011. Identification of potential conserved RNA secondary structure throughout influenza A coding regions. RNA 17:991–1011. 10.1261/rna.2619511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Priore SF, Moss WN, Turner DH. 2012. Influenza A virus coding regions exhibit host-specific global ordered RNA structure. PLoS One 7:e35989. 10.1371/journal.pone.0035989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gultyaev AP, Heus HA, Olsthoorn RC. 2007. An RNA conformational shift in recent H5N1 influenza A viruses. Bioinformatics 23:272–276. 10.1093/bioinformatics/btl559 [DOI] [PubMed] [Google Scholar]
- 69. Gultyaev AP, Olsthoorn RC. 2010. A family of non-classical pseudoknots in influenza A and B viruses. RNA Biol. 7:125–129. 10.4161/rna.7.2.11287 [DOI] [PubMed] [Google Scholar]
- 70. Moss WN, Dela-Moss LI, Kierzek E, Kierzek R, Priore SF, Turner DH. 2012. The 3′ splice site of influenza A segment 7 mRNA can exist in two conformations: a pseudoknot and a hairpin. PLoS One 7:e38323. 10.1371/journal.pone.0038323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Moss WN, Dela-Moss LI, Priore SF, Turner DH. 2012. The influenza A segment 7 mRNA 3′ splice site pseudoknot/hairpin family. RNA Biol. 9:1305–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Reed R, Hurt E. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108:523–531. 10.1016/S0092-8674(02)00627-X [DOI] [PubMed] [Google Scholar]
- 73. York A, Fodor E. 2013. Biogenesis, assembly and export of viral messenger ribonucleoproteins in the influenza A virus infected cell. RNA Biol. 10:1274–1282. 10.4161/rna.25356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Read EK, Digard P. 2010. Individual influenza A virus mRNAs show differential dependence on cellular NXF1/TAP for their nuclear export. J. Gen. Virol. 91:1290–1301. 10.1099/vir.0.018564-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893. 10.1038/nature07151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang W, Cui ZQ, Han H, Zhang ZP, Wei HP, Zhou YF, Chen Z, Zhang XE. 2008. Imaging and characterizing influenza A virus mRNA transport in living cells. Nucleic Acids Res. 36:4913–4928. 10.1093/nar/gkn475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Alonso-Caplen FV, Krug RM. 1991. Regulation of the extent of splicing of influenza virus NS1 mRNA: role of the rates of splicing and of the nucleocytoplasmic transport of NS1 mRNA. Mol. Cell. Biol. 11:1092–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fortes P, Beloso A, Ortín J. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 13:704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chua MA, Schmid S, Perez JT, Langlois RA, Tenoever BR. 2013. Influenza A virus utilizes suboptimal splicing to coordinate the timing of infection. Cell Rep. 3:23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wolff T, O’Neill RE, Palese P. 1998. NS1-binding protein (NS1-BP): a novel human protein that interacts with the influenza A virus nonstructural NS1 protein is relocalized in the nuclei of infected cells. J. Virol. 72:7170–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. 10.1016/j.cell.2009.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. König R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, García-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. 2010. Human host factors required for influenza virus replication. Nature 463:813–817. 10.1038/nature08699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sui B, Bamba D, Weng K, Ung H, Chang S, Van Dyke J, Goldblatt M, Duan R, Kinch MS, Li WB. 2009. The use of random homozygous gene perturbation to identify novel host-oriented targets for influenza. Virology 387:473–481. 10.1016/j.virol.2009.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tazi J, Bakkour N, Marchand V, Ayadi L, Aboufirassi A, Branlant C. 2010. Alternative splicing: regulation of HIV-1 multiplication as a target for therapeutic action. FEBS J. 277:867–876. 10.1111/j.1742-4658.2009.07522.x [DOI] [PubMed] [Google Scholar]
- 85. Almond JW, Haymerle HA, Felsenreich VD, Reeve P. 1979. The structural and infected cell polypeptides of influenza B virus. J. Gen. Virol. 45:611–621. 10.1099/0022-1317-45-3-611 [DOI] [PubMed] [Google Scholar]
- 86. Briedis DJ, Lamb RA, Choppin PW. 1981. Influenza B virus RNA segment 8 codes for two nonstructural proteins. Virology 112:417–425. 10.1016/0042-6822(81)90289-0 [DOI] [PubMed] [Google Scholar]
- 87. Briedis DJ, Lamb RA. 1982. Influenza B virus genome: sequences and structural organization of RNA segment 8 and the mRNAs coding for the NS1 and NS2 proteins. J. Virol. 42:186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Horvath CM, Williams MA, Lamb RA. 1990. Eukaryotic coupled translation of tandem cistrons: identification of the influenza B virus BM2 polypeptide. EMBO J. 9:2639–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nakada S, Graves PN, Palese P. 1986. The influenza C virus NS gene: evidence for a spliced mRNA and a second NS gene product (NS2 protein). Virus Res. 4:263–273. 10.1016/0168-1702(86)90005-5 [DOI] [PubMed] [Google Scholar]
- 90. Alamgir AS, Matsuzaki Y, Hongo S, Tsuchiya E, Sugawara K, Muraki Y, Nakamura K. 2000. Phylogenetic analysis of influenza C virus nonstructural (NS) protein genes and identification of the NS2 protein. J. Gen. Virol. 81:1933–1940 [DOI] [PubMed] [Google Scholar]
- 91. Yamashita M, Krystal M, Palese P. 1988. Evidence that the matrix protein of influenza C virus is coded for by a spliced mRNA. J. Virol. 62:3348–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pekosz A, Lamb RA. 1998. Influenza C virus CM2 integral membrane glycoprotein is produced from a polypeptide precursor by cleavage of an internal signal sequence. Proc. Natl. Acad. Sci. U. S. A. 95:13233–13238. 10.1073/pnas.95.22.13233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hongo S, Sugawara K, Nishimura H, Muraki Y, Kitame F, Nakamura K. 1994. Identification of a second protein encoded by influenza C virus RNA segment 6. J. Gen. Virol. 75(Pt 12):3503–3510. 10.1099/0022-1317-75-12-3503 [DOI] [PubMed] [Google Scholar]
- 94. Muraki Y, Furukawa T, Kohno Y, Matsuzaki Y, Takashita E, Sugawara K, Hongo S. 2010. Influenza C virus NS1 protein upregulates the splicing of viral mRNAs. J. Virol. 84:1957–1966. 10.1128/JVI.01627-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Schneider J, Wolff T. 2009. Nuclear functions of the influenza A and B viruses NS1 proteins: do they play a role in viral mRNA export? Vaccine 27:6312–6316. 10.1016/j.vaccine.2009.01.015 [DOI] [PubMed] [Google Scholar]
- 96. Backström Winquist E, Abdurahman S, Tranell A, Lindström S, Tingsborg S, Schwartz S. 2012. Inefficient splicing of segment 7 and 8 mRNAs is an inherent property of influenza virus A/Brevig Mission/1918/1 (H1N1) that causes elevated expression of NS1 protein. Virology 422:46–58. 10.1016/j.virol.2011.10.004 [DOI] [PubMed] [Google Scholar]
- 97. Fernandez-Sesma A, Marukian S, Ebersole BJ, Kaminski D, Park MS, Yuen T, Sealfon SC, García-Sastre A, Moran TM. 2006. Influenza virus evades innate and adaptive immunity via the NS1 protein. J. Virol. 80:6295–6304. 10.1128/JVI.02381-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Haye K, Burmakina S, Moran T, García-Sastre A, Fernandez-Sesma A. 2009. The NS1 protein of a human influenza virus Inhibits type I interferon production and the induction of antiviral responses in primary human dendritic and respiratory epithelial cells. J. Virol. 83:6849–6862. 10.1128/JVI.02323-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wolschek M, Samm E, Seper H, Sturlan S, Kuznetsova I, Schwager C, Khassidov A, Kittel C, Muster T, Egorov A, Bergmann M. 2011. Establishment of a chimeric, replication-deficient influenza A virus vector by modulation of splicing efficiency. J. Virol. 85:2469–2473. 10.1128/JVI.01650-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Krainer A. 1997. Eukaryotic mRNA processing. Frontiers in molecular biology, vol 17. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 101. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Squires RB, Noronha J, Hunt V, García-Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN, Ramsey A, Zhou L, Zaremba S, Kumar S, Deitrich J, Klem E, Scheuermann RH. 2012. Influenza Research Database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respir. Viruses 6:404–416. 10.1111/j.1750-2659.2011.00331.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Momose F, Basler CF, O’Neill RE, Iwamatsu A, Palese P, Nagata K. 2001. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 75:1899–1908. 10.1128/JVI.75.4.1899-1908.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wisskirchen C, Ludersdorfer TH, Müller DA, Moritz E, Pavlovic J. 2011. The cellular RNA helicase UAP56 is required for prevention of double-stranded RNA formation during influenza A virus infection. J. Virol. 85:8646–8655. 10.1128/JVI.02559-10 [DOI] [PMC free article] [PubMed] [Google Scholar]