Abstract
Purpose
Allogeneic glioma cell lines that are partially matched to the patient at class I human leukocyte antigen (HLA) loci and that display tumor-associated antigens (TAA) or antigenic precursors [tumor antigen precursor proteins (TAPP)] could be used for generating whole tumor cell vaccines or, alternatively, for extraction of TAA peptides to make autologous dendritic cell vaccines.
Experimental Design
Twenty human glioma cell lines were characterized by molecular phenotyping and by flow cytometry for HLA class I antigen expression. Twelve of the 20 cell lines, as well as analyses of freshly resected glioma tissues, were further characterized for protein and/or mRNA expression of 16 tumor antigen precursor proteins or TAA.
Results
These 20 human glioma cell lines potentially cover 77%, 85%, and 78% of the U.S. Caucasian population at HLA-A, HLA-B, and HLA-C alleles, respectively. All cells exhibited multiple TAA expressions. Most glioma cells expressed antigen isolated from immunoselected melanoma-2 (Aim-2), B-cyclin, EphA2, GP100, β1,6-N-acetylglucosaminyltransferase V (GnT-V), IL13Rα2, Her2/neu, hTert, Mage, Mart-1, Sart-1, and survivin. Real-time PCR technology showed that glioblastoma specimens expressed most of the TAA as well. Tumor-infiltrating lymphocytes and CD8+ CTL killedT2 cells when loaded with specific HLA-A2+ restricted TAA, or gliomas that were both HLA-A2+ and also positive for specific TAA (Mart-1, GP100, Her2/neu, and tyrosinase) but not those cells negative for HLA-A2 and/or lacking the specific epitope.
Conclusions
These data provide proof-in-principle for the use of allogeneic, partially HLA patient – matched glioma cells for vaccine generation or for peptide pulsing with allogeneic glioma cell extracts of autologous patient dendritic cells to induce endogenous CTL in brain tumor patients.
Glioblastoma multiforme patients usually undergo debulking surgery followed by radiation and aggressive rounds of chemotherapy. The recently revised standard of care radiotherapy and chemotherapy for newly diagnosed glioblastoma multiforme patients has only improved the survival from 12.1 to 14.6 months following diagnosis (1). Despite the majority of the tumors being removed or killed, no lasting and effective antitumor immunity is generated and the patients die from the tumor cells escaping or resisting those therapies. These failures lead many to believe that an aggressive combination of standard therapies, along with other biologically based therapies, is needed to successfully treat this cancer.
Some glioma patients were successfully treated by immunotherapy (2-7). Adjuvant immunotherapy modalities may be particularly useful for treating gliomas located next to critical brain control regions (brain stem or the thalamus) where surgery or radiation cannot be used, provided inflammation can be controlled. The advantage of generating an immune response toward the cancer cells is that the immunized T cells can now seek and destroy the remaining tumor cells that resisted the therapies or were localized to sites that were inaccessible to the traditional treatments.
Human glial tumors have been shown to express a wide variety of tumor-associated antigens (TAA; refs. 8-10). Some glioma-specific antigens have been found, such as processed fragments derived from the interleukin-13 receptor α2 (IL13Rα2, ref. 11). Many TAA found within gliomas are also expressed on other cancers (10); i.e., melanomas: Mage-1 (12-16), Aim-2 (17, 18), tyrosinase (13), tyrosinase-related protein 1 (Trp-1; also known as Trp-75; ref. 13), Trp-2 (13, 14, 19), Gage (14), human melanoma-associated antigen p97/GP100 (melano-transferrin; refs. 12, 15, 16); breast and ovarian cancers: Her2/neu (15, 16); multiple cancer types: B-cyclin (20), EphA2/Eck (21, 22), telomerase reverse transcriptase (hTert; ref. 23), Sart-1 (24), survivin (25, 26), and GnT-V (27). All of these antigens have elicited human immune responses, primarily in the form of CTL responses. Many of these antigenic peptides are restricted to the HLA-A2 allele: B-cyclin, EphA2, Gage-1, GP100, Her2/neu, hTert, IL13Rα2, Mage-1, Trp-1, tyrosinase, and survivin (10). Aim-2 is restricted to the HLA-A1 locus (17), Sart-1 is restricted to HLA-A24 and A26, whereas survivin and hTert are also HLA-A24 restricted (10). Effective immunotherapy will require that the patients’ neoplasm displays the tumor antigens properly associated with their restricted HLA alleles. This will allow T-cell clonal expansion after their recognition. One therapeutic scenario would involve obtaining the antigenic profile of the patient glioma. Another possibility would involve HLA typing of the individual as that might be predictive of the TAA on the patient’s glioma. With that knowledge, an allogeneic vaccine could be customized so that the relevant antigens are contained within the vaccine to stimulate the patient’s immune system.
Several possible vaccination approaches can be envisioned for treating glioblastoma multiforme patients. In one method, the patients’ own tumor cells are isolated, inactivated, and used either as a whole tumor vaccine or by genetically engineering them with immunostimulatory molecules. For slow-growing cancers, this approach may be feasible, but, for glioblastoma multiforme patients, it has largely failed because the patient relapses before the autologous glioma cells can be established in tissue culture. A second method uses whole tumor homogenates made from surgically resected tumor. The homogenates are incubated with the patient’s dendritic cells. These peptide-pulsed dendritic cells act as antigen-presenting cells and are then infused back into the patient to engender host T-cell responses. Liau et al. (28) and Liu et al. (16) used this approach in phase I trials with 12 and 14 patients, respectively. In both trials, there was an increased T-cell infiltration into the remaining tumor, an expansion of CD8+ antigen–specific T-cell clones, and an increase in IFN-γ expression within the in vitro activated T cells. No evidence of induced autoimmunity after these immunizations was observed. As a result of these antiglioma immune responses, an improvement in survival time occurred. A third method uses well-established glioma cell lines either as allogeneic genetically modified tumor vaccines or as a source of minor tumor antigens for vaccination purposes. Within a month of tumor debulking surgery and chemotherapy and radiation therapy, Mahaley et al. (2) used irradiated unmodified U-251 glioma cells as a monthly vaccine to achieve longer survival (>950 days) in six of nine patients. When they used D-54 glioma cells as the vaccine in another set of patients, no increased survival resulted compared with their historical controls. This work suggests that U-251 cells were intrinsically more immunogenic than D-54 glioma cells.
In this report, we HLA phenotyped 20 human glioma cell lines. We further evaluated a dozen from this panel to assess for their expression of 16 known TAA/tumor antigen precursor proteins (TAPP), most of which we showed were also associated with primary glioma specimens. HLA-A2+/TAA–restricted CTLs and tumor-infiltrating lymphocytes (TIL) were able to lyse a number of HLA-A2+ gliomas cells expressing EphA2, GP100, Her2/neu, Mart-1, and tyrosinase, which was consistent with their antigenic profiles. These data should allow neuro-oncologists and immunologists to customize generic allogeneic whole tumor cell vaccine(s) for initial vaccination purposes, or allow for autologous dendritic cell vaccines to be generated with TAA peptides derived from allogeneic glioma cells, for glioblastoma multiforme patients.
Materials and Methods
Glioblastoma tumor material and glioma cell culture
Freshly resected surgical specimens collected at the University of Colorado Health Sciences Center (Denver, CO) between 1986 and 1998 were snap frozen and kept at −70°C. By histopathologic criteria, diagnoses consistent with glioblastoma multiforme were made on all specimens chosen for TAA analysis.
U-251 and SF767 were obtained from Dr. Dennis Deen (University of California, San Francisco, CA). Human glioma cells, U-87, U-118, A172, T98G, SNB19, and LNZ308, were obtained from either Dr. Thomas Chen (University of Southern California, Los Angeles, CA) or William Welch (University of Pittsburgh, PA). Dr. Darell Bigner (Duke University, Durham, NC) supplied us with the D-645, D-54, and U-373 glioma cells. LN18 and LN229 were purchased from the American Tissue Type Collection (Manassas, VA). DBTRG05-MG, 04-11-MG, 12-11-MG, and 13-06-MG were established in the Carol Kruse laboratory. NR203MG, NR206MG, and NR213MG were established in the Habib Fakhrai laboratory. The glioma cells were grown in DMEM (Sigma Chemical Co., St. Louis, MO) supplemented with 5% to 10% fetal bovine serum (Gemini, Calabassas, CA; Life Technologies, San Diego, CA; and Hyclone, Logan, UT).
Antibodies for neuroglial typing and HLA expression
Expressions of neurofilament 160 (for neural cells), glial fibrillary acidic protein (for astrocytes/astroglial), vimentin (for astrocytes/astroglial cells, vascular endothelium and mesenchyme), galactocerebroside (for oligodendrocytes/oligodendroglial), and fibronectin (extracellular matrix marker also associated with for glial cells and fibroblasts) were determined by immunofluorescence microscopy. Staining with monoclonal antibodies (Neural Cell Typing Kit, Boehringer Mannheim Co., Indianapolis, IN) was as described (29). Isotype-matched control antibodies were used to distinguish background staining.
The glioma cells were stained for either the specific HLA-A2 allele or for the HLA-ABC framework using mouse monoclonal antibodies purchased from PharMingen BD Biosciences (San Diego, CA).
Molecular HLA typing
The glioma cells were either submitted to the University of California, Irvine HLA Histocompatibility Laboratory (Orange, CA) for low-resolution PCR HLA class I typing or to ClinImmune Laboratories (Aurora, CO) where HLA class I and II typing was determined by molecular analyses using PCR sequence-specific primers and sequence-based typing. The U-251 glioma was previously HLA typed (30). The HLA phenotype of the LN18 glioma cells was provided by Dr. A.C. Diserens (Lucerne, Switzerland).
Intracellular flow cytometry
Exponentially growing glioma cells (10) were prepared with the reagents and protocols of Santa Cruz Biotechnology (Santa Cruz, CA). The fixed cells were washed twice in ice-cold PBS. The cells were permeabilized for 15 min on ice. The cells were washed twice. The resuspended cells were then divided into 106 cell aliquots and then incubated with the primary antibody for 1 h. The anti-IL13Rα2 monoclonal antibody was obtained from Cell Sciences (Canton, MA). Anti–Eck/EphA2, anti–B cyclin, anti-GP100, anti–Mage-1, anti–Mart-1, anti–Trp-1/Trp-75, and anti–Her2/neu monoclonal antibodies were from Lab Vision (Fremont, CA). Anti-survivin and anti–GnT-V polyclonal antibodies were purchased from Santa Cruz Biotechnology. The source of the monoclonal antibody directed against the hTert was either Novus Biologicals (Littleton, CO) or Santa Cruz Biotechnology. The cells were washed twice and the secondary antibody conjugated with FITC (Vector Labs, Burlingame, CA) was incubated on ice for another hour. After washing the cells twice, the cells were analyzed with a Coulter XL flow cytometer. The staining profiles were scored and normalized scores of 0 to 4, where 0 reflected no staining above background and 4 was the most intense.
Immunofluorescence microscopy
U-251 cells were allowed to grow on sterile collagen-coated coverslips overnight. The coverslips were dipped in a 2% formalin solution and then acetone treated for 2 min. The antibodies (as described above) were diluted 1:100 in PBS. The tissue was incubated 2 h with the primary antibody in a humidified chamber at 4°C. The slides were washed thrice in PBS and the 1:100 diluted secondary FITC-labeled anti–primary antibody (Vector Labs, Carpinteria, CA) was incubated at room temperature for 30 min. The slides were washed thrice in PBS. The staining of the cells was visualized with a Nikon Eclipse E600 microscope equipped with epifluorescence. Photomicrographs were taken with an RT-KE digital camera (Diagnostics Instruments, Inc., Sterling Heights, MI).
Real-time PCR analysis
Total RNA was isolated from the cells using Trizol reagent (Sigma Chemical). Any possible DNA contamination in the sample was eliminated by incubation with RNase-free DNase I digestion (Boehringer Mannheim). cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA) containing 1 Ag of total RNA per sample. Real-time PCR reactions were done on an iCycler iQ detection system (Bio-Rad Laboratories) in conjunction with the SYBR Green kit (Stratagene, San Diego, CA). The thermal profile was 95°C for 15 min, followed by 40 cycles of 95°C for 15 s and 58°C for 30 s, finally holding at 4°C. The following primers were synthesized by Operon Biotech (Germantown, MD): Aim-2, forward 5′-GTGTATGCCCCACCAACC-3′, reverse 5′-CCTGGTGACCTAGTCATTGG-3′; Gage-1, forward 5′-CCACGGAAACCTTGAGTG-3′, reverse 5′-CAGCCTGCAACATTTCAGC-3′; Sart-1, forward 5′-GACAAGTACAGCCGGAGG-3′, reverse 5′-CCGTCTCATCCACGTATTCG-3′; Trp-2, forward 5′-CCTGTCTCTCCAGAAGTTTG-3′, reverse 5′-CAGAGTCCCATCTGCTTTATC-3′.
Samples were run in triplicate, and a reaction without cDNA was used to establish baseline fluorescence levels with 18S RNA. The fluorescent signal was plotted versus cycle number, and the threshold cycle (CT) was determined at the cycle number where an increase above background fluorescence could be reliably detected. Each PCR run also included nontemplate controls containing all reagents except cDNA. After cycling, a melting curve was produced by slow denaturation of the PCR end products to validate the specificity of amplification. The relative quantification of expression of the gene was determined by 2−ΔCT as described by Pfaffl (31). Very strong mRNA expression occurred when the ΔΔCT was between 10 and 14. Positive signals occurred when the ΔCT score was <14 to 24, whereas negative signals were defined when the ΔCT score was >25.
Induction and evaluation of CTL or TIL against glioma antigen peptides
Glioma patient–derived peripheral blood mononuclear cells were stimulated in vitro with peptide-pulsed autologous dendritic cells as previously described (11, 21). Briefly, to generate dendritic cells, the plastic adherent cells from peripheral blood mononuclear cells were cultured in AIM-V medium (Invitrogen/Life Technologies) supplemented with 1,000 units/mL rhGM-CSF and 500 units/mL recombinant human (rh) interleukin (IL)-4 (Cell Sciences) at 37°C in a humidified, 5% CO2 incubator. Six days later, the immature dendritic cells were stimulated with recombinant human tumor necrosis factor-α, IL-6, and IL-1β (10 ng/mL each). Mature dendritic cells were harvested on day 8, resuspended in AIM-V medium at 106 cells/mL with peptide (10 μg/mL), and incubated for 2 h at 37°C. Populations of CD8+ T cells, autologous to the dendritic cell donors, were enriched from peripheral blood mononuclear cells using magnetic microbeads (Miltenyi Biotech, Auburn, CA). CD8+ T cells (2 × 106 per well) were cocultured with peptide-pulsed dendritic cells (2 × 105 per well) in 2 mL of AIM-V medium supplemented with 5% human AB serum (Invitrogen, San Diego, CA), 10 units/mL rhIL-2 (R&D Systems, Minneapolis, MN), and 10 units/mL rhIL-7 (Cell Sciences) in each well of 24-well tissue culture plates. On day 15, to increase CTL frequency, the lymphocytes were restimulated with autologous dendritic cells pulsed with peptide in AIM-V medium supplemented with 5% human AB serum (Life Technologies), rhIL-2, and rhIL-7 (10 units/mL each). The CD8+ cultured cells were analyzed for CTL activity 5 days after second stimulation in 4-h 51Cr-release assays. The peptides used for stimulation of peripheral blood mononuclear cells were as follows: Mart-1 (27-35: AAGIGILTV; ref. 32), GP100 (209-217[2M]: IMDQVPFSV; refs. 33, 34), EphA2 (883-891: TLADFDPRV; ref. 21), influenza M1 (58-66: GILGFVFTL), and Her2/neu (369-377: KIFGSLAFL; ref. 10). Positive cytotoxic responses were defined as follows: the specific lysis by the responder cells against antigen-positive target cells was at least 15% and 2-fold higher than lytic levels displayed by corresponding control conditions in at least two of the effector-to-target ratios.
Tumor-infiltrating lymphocyte lines, TIL 771 and TIL 1374, known to be HLA-A2 restricted to GP100 (154-162: KTWGQYWQV) and tyrosinase (369-377: YMDGTMSQV), respectively, were used as effector cells to test the lysis of target cells with and without those specific TAA (35). The TIL were maintained in RPMI 1640 supplemented with 10% human AB serum (Omega Scientific, Tarzana, CA), 50 units/mL penicillin and 50 mg/mL streptomycin, and 6,000 IU IL-2/mL (Chiron, Emeryville, CA). In addition, the T2 clone (ATCC CRL1992), a hybrid B-T lymphoblastic cell line often used as a model system for class I antigen presentation purposes, was loaded with or without the relevant glioma TAA peptide as described (34) and used as target cells with the effector CTL generated as described above.
Results
Neuroglial cell marker profiling of the human glioma cell lines
Twenty established glioma cell lines were profiled for various markers representative of central nervous system tumors by immunofluorescence staining. A summary of this work is shown in Table 1 (column 2). The majority of the glioma cell lines (16 of 20) tested strongly for vimentin whereas only of a quarter of the cultured gliomas (5 of 20) expressed glial fibrillary acidic protein. Variable expression of the neural cell marker, neurofilament 160 (neurofilament 160), was detected within 11 of the 20 gliomas. Fibronectin was detected in 7 of the 20 glioma cell lines. Expression of the oligodendrocyte marker, galactocerebroside, was nil in all. The staining patterns of these gliomas are consistent with the histopathologic diagnoses of glioma/astrocytoma that possess no oligodendroglial component.
Table 1.
Human glioma cell line neuroglial profile and HLA Class I and II phenotypesa
| Glioma | Neuroglial b | MOLECULAR PHENOTYPE | ||||
|---|---|---|---|---|---|---|
| Class I | Class II | |||||
| HLA-A | HLA-B | HLA-C | HL-DR | HLA-DQ | ||
| 04-11-MG | V, F | 1 | 8,57,w4,w6 | w6,w7 | 13,17,52 | 2,6 |
| 12-11-MG | N,GF | 24,32 | 27,64,w4,w6 | w2,w8 | 4,7 | 2,3 |
| 13-06-MG | N, GF, V | 1,2 | 44,57,w4 | NT | NT | NT |
| A172 | N, V | 1,3 | 8,42 | w7 | NT | NT |
| D-54MG | N, V | 1,3 | 7,8 | w7 | NT | NT |
| D-645MG | V | 2,23 | 35,49,w4,w6 | w4,w7 | 1,4,53 | 5,8 |
| DBTRG05-MG | V, F | 2,68 | 35,38,w4,w6 | w12,w15 | 4,14,52,53 | 5,8 |
| NR203 | V, F | 2 | 7,60,w6 | w7,w10 | 4,15,51,53 | 6,7 |
| NR206 | GF, V | 2 | 62,w6 | w10 | 4,53 | 8 |
| NR213 | GF, V | 2 | 44,55,w4,w6 | w5,w9 | 4,15,51,53 | 6,7 |
| LN-18 | NT | 2,9 | 5,w35 | NT | w3 | NT |
| LN-229 | NT | 3 | 35 | w4, w12? | NT | NT |
| LNZ308 | N, V, F | 24 | 35,51 | w4,14 | NT | NT |
| SF767 | N | 1,32 | 8,44 | w5,w7 | NT | NT |
| SNB19 | N, V | 2 | 18 | w5 | NT | NT |
| T98G | N, V, F | 2 | 35,39,w6 | w4,w7 | 8,12,52 | 4,7 |
| U-87MG | N, V, F | 2 | 44 | w5 | NT | NT |
| U-118MG | N, V, F | 24,29 | 39,44 | w7,16 | NT | NT |
| U-138MG | N, V, F | w24 | 27,25 | NT | NT | NT |
| U-251MG | N, V | 2 | 18,w6 | w4 | NT | NT |
| U-373MG | GF, V | 2 | 18 | w5 | NT | NT |
The gliomas were typed by a PCR-based molecular approach. NT: not tested; w4,w12?: indicates that we cannot rule out w12 in the presence of w4.
The gliomas were profiled for neuroglial markers: NF 160 (N); GFAP (GF); Galactocerebroside (GC); vimentin (V); and fibronectin (F).
- 04-11-MG: A*01:01; B*08:01/19N,*57:01; Cw*06:02, *07:01/06/18; DPB1*01:01, *03:01 DRB1*03:01, *13:02; DRB3*01:01, *03:01; DQB1*02:01, *06:04
- D-645MG: A*02:05/14, *23:01/02; B*35:01/,42, *49:01; Cw*04:01/09N, *07:01/06/18; DRB1*01:01,*04:05, DRB4*01:03; DQB1*03:02,*05:01
- DBTRG05-MG: A*02:01/04/09, *68:01/11N/23; B*35:01/42, *38:01; Cw*12:03/04, *15:02/07; DRB1*04:02, *14:01/39; DRB3, DRB4; DQB1*03:02, *05:03
- NR203: A*02:01, B*07:02/05/06, *40:01/33; Cw*03:04, *07:02; DPB1*04:01, DRB1*04:08, *15:01; DRB4*01:03, DRB5*01:01; DQB1*03:01, *06:02
- NR206: A*02:01; B*15:01; Cw*03:04; DPB1*13:01; DRB1*04:05; DRB4*01:05; DQB1*03:02
- NR213: A*02:01; B*44:02/10N/27, *55:01; Cw*03:03, *05:01/03; DPB1*04:01, *11:01; DRB1*04:01, *15:01; DRB4*01:03, DRB5*01:01; DQB1*03:01, *06:02
- T98G: A*02:01; B*35:03, *39:06; Cw*04:01/09N, *07:02; DPB1*03:01,*04:01; DRB1*08:01, *12:01; DRB3*02:02; DQB1*03:01, *04:02
HLA phenotype of and expression by the human glioma cell lines
The glioma cell lines were molecularly phenotyped for HLA antigen expression for class I HLA-A, HLA-B, and HLA-C determinants (Table 1). Eight of the 20 were also typed for class II HLA-DR, HLA-DP, and HLA-DQ determinants. Twelve of the 20 gliomas were HLA-A2+ and, additionally, 8 of the 12 were homozygous at that allele. Five of the 20 were HLA-A1+, 3 were HLA-A3+, and 3 were HLA-A24+. Six glioma cell lines displayed other less common HLA-alleles. By χ2 analysis, these cells reflect the general frequencies of the HLA-A, HLA-B, and HLA-C alleles present in the U.S. Caucasian population.
We verified that the panel of glioma cells expressed the HLA class I proteins by flow cytometry. All of the cells within the glioma cell lines were positive for HLA-ABC. Twelve of the glial tumor cell lines that were molecularly phenotyped as HLA-A2 positive also proved positive for the HLA-A2 protein using a monoclonal antibody that specifically recognized this allele (data not shown). A similar pattern of HLA expression also occurred when we used the monoclonal antibody that detects the HLA-ABC framework. All the glioma cells that were HLA-A2 negative by molecular phenotyping failed to stain with the anti–HLA-A2 antibody. However, all of the HLA-A2 negative cells stained strongly with the anti-HLA-ABC antibody. The generally abundant HLA expression on all the glioma cell lines examined should allow for T-cell recognition to occur.
Immunofluorescence microscopy and intracellular flow cytometry show that common TAPP are present within the glioma cell lines
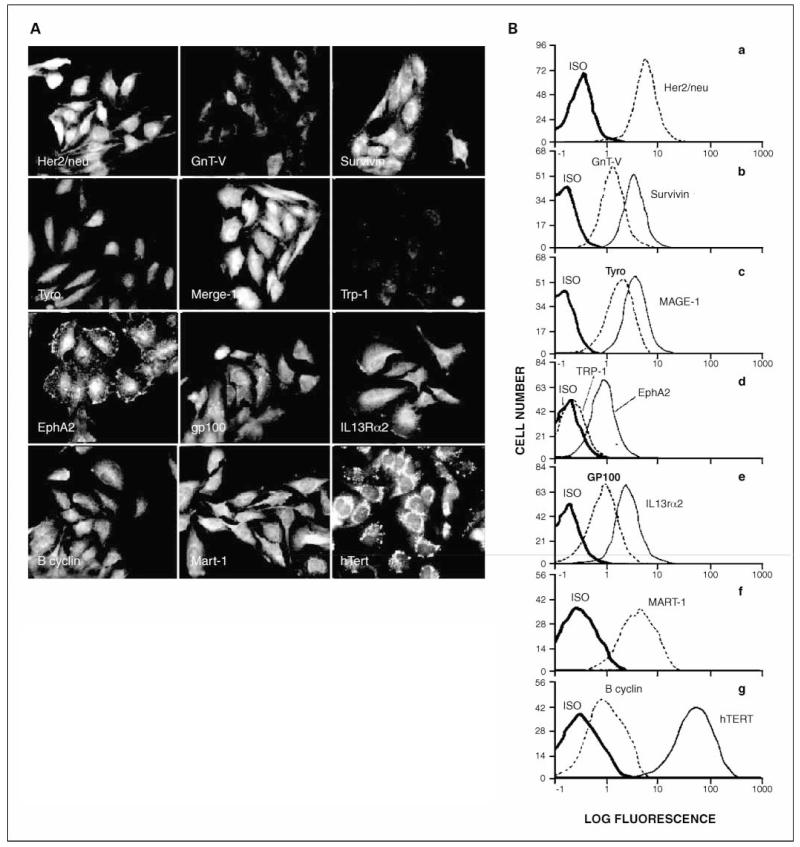
A subset of the glioma panel (n = 17) was examined for expression of 12 TAPP: B1-cyclin, EphA2, GnT-V, GP100, Her2/neu, hTert, IL13Rα2, Mage-1, Mart-1, survivin, Trp-1, and tyrosinase. We confirmed, as visualized by immunofluorescence microscopy, that TAPP were present within U-251 glioma cells as appropriately localized membrane-associated and/or intracellular proteins (Fig. 1A), as specified by the provider/manufacturer of the antibodies. The microscopy findings were in agreement with the staining intensities determined by intracellular flow cytometry. Figure 1B shows the fluorescent profiles of the TAPP within U-251 cells. All the TAPP tested were positive, with the exception of the Trp-1 antigen (Fig. 1D). The strongest TAPP expression within U-251 cells was hTert (Fig. 1G). The cells exhibited moderate antigen expression for Her2/neu, survivin, Mage-1, IL13Rα2 and GP100 (Fig. 1A, B, C, and E, respectively). GnT-V (Fig. 1B), Tyrosinase (Fig. 1C) and EphA2 (Fig. 1D) intensities were less and B-cyclin (Fig. 1G) had low expression.
Fig. 1.
TAPP associated with U-251cells by immunofluorescence microscopy and by intracellular flow cytometry. A, immunofluorescence of U-251cells shows that the TAPP are present within their predicted intracellular locations. U-251gliomas were allowed to attach to coverslips overnight. The cells were fixed, permeabilized, and then stained with the specific antibodies. Top, left to right, Her2/neu, GnT-V, and survivin. Top middle, tyrosinase Mage-1andTrp-1 (which is as bright as the negative control; not shown). Bottom middle, EphA2, GP100, and IL13Rα2. Bottom, B-cyclin, Mart-1, and hTert. B, intracellular flow cytometry of TAPP within U-251glioma cells. U-251glioma cells (106) were fixed and permeabilized. The permeabilized cells were incubated with the primary antibodies for 1h, washed, and subsequently incubated with a FITC-labeled secondary antibody directed against the primary antibody. Flow cytometric analysis of 104 glioma cells is shown. The isotypic control (dark line) and the anti-TAPP staining (either a fine line or dashed line) are shown. a, Her2/neu; b, GnT-V and survivin; c, tyrosinase and Mage-1; d, Trp-1 and EphA2; e, GP100 and IL13Rα2; f, Mart-1; g, B-cyclin and hTert.
Table 2A semiquantitatively summarizes the expression of the TAPP within 12 of the glioma cell lines. A score was assigned between 0 and 4, which was based on the mean peak channel intensity obtained by flow cytometry. As a negative control, human peripheral blood lymphocytes were shown to be negative or weakly positive (+1 for survivin, GnT-V, and Her2/neu) for these proteins (data not shown). B-cyclin, GnT-V, Her2/neu, hTert, IL13Rα2, Mage-1, and survivin were detected within every glioma line in our panel, and EphA2 was in all cell lines except T98G. GP100 was found within 9 of 12, whereas tyrosinase was seen in 8 of 12 glioma cell lines. Trp-1 expression was seen in six cell lines of the panel. Only one cell line, SNB19, possessed all of the TAPP relevant to glioma immunotherapy. Quantitative scoring of the 12 TAPP (Σ scorea in the bottom row) is provided to give an idea of which cell lines had the strongest overall expression profiles. Those with scores of ≥28 included A172, LNZ308, SNB19, U-251, and U-373.
Table 2.
Summary of TAPP expression profiles by glioma cell lines
| (A) Intracellular flow cytometric analysis of 12 TAPP expressed within the glioma cells* | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antigen | Glioma |
|||||||||||
| A172 | D-54 | LN18 | LN229 | LNZ308 | SF767 | SNB19 | T98G | U-118 | U-251 | U-373 | U-87 | |
| B-cyclin | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 1 | 1 | 1 | 2 | 1 |
| EphA2 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 0 | 1 | 2 | 3 | 1 |
| Her2/neu | 2 | 3 | 3 | 3 | 3 | 2 | 3 | 2 | 2 | 3 | 3 | 3 |
| IL13Rα2 | 3 | 4 | 2 | 2 | 3 | 2 | 3 | 2 | 3 | 3 | 3 | 3 |
| GnT-V | 2 | 1 | 2 | 2 | 1 | 2 | 2 | 3 | 1 | 3 | 2 | 2 |
| GP100 | 3 | 2 | 0 | 0 | 3 | 2 | 3 | 3 | 0 | 3 | 4 | 3 |
| Mage-1 | 4 | 3 | 2 | 3 | 3 | 3 | 3 | 2 | 2 | 3 | 2 | 3 |
| Mart-1 | 3 | 0 | 3 | 3 | 3 | 2 | 3 | 3 | 0 | 3 | 3 | 3 |
| Survivin | 3 | 2 | 2 | 2 | 3 | 2 | 2 | 3 | 1 | 3 | 2 | 3 |
| hTert | 4 | 4 | 3 | 3 | 4 | 4 | 2 | 4 | 4 | 4 | 3 | 4 |
| Trp-1 | 0 | 2 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 |
| Tyrosinase | 2 | 0 | 2 | 3 | 1 | 2 | 2 | 3 | 0 | 2 | 0 | 0 |
| Σ score | 28 | 23 | 23 | 26 | 28 | 24 | 28 | 27 | 15 | 30 | 28 | 26 |
| (B) Real-time PCR expression of Aim-2, Gage-1, Sart-1, and Trp-2 mRNA† | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antigen | Glioma |
|||||||||||
| A172 | D-54 | LN18 | LN229 | LNZ308 | SF767 | SNB19 | T98G | U-118 | U-251 | U-373 | U-87 | |
| Aim-2 | 2 | 3 | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 3 | 3 | 2 |
| Gage-1 | nd | nd | nd | nd | nd | nd | nd | nd | 2 | nd | nd | nd |
| Sart-1 | 2 | 2 | 3 | 2 | nd | 1 | 2 | 2 | 2 | 1 | 2 | nd |
| Trp-2 | nd | nd | nd | 2 | nd | 2 | nd | 2 | 2 | nd | 1 | nd |
| Σ score | 4 | 5 | 6 | 7 | 3 | 5 | 5 | 7 | 9 | 4 | 6 | 2 |
| Total‡ | 32 | 28 | 29 | 33 | 31 | 29 | 33 | 34 | 24 | 34 | 34 | 28 |
The cells were fixed, permeabilized, and stained with the antibodies directed toward the various tumor antigens. Cells were scored for the intensity of immunofluorescence staining obtained by flow cytometry. A score of 0 was given when the staining was not above that of the negative control cells. A score of 1+ to 4+ reflects the mean peak channel intensity of the cells. In the last row, we assign a cumulative score for each of the 12 cell lines (A score to summarize the overall strength of the 12 TAPP analyzed).
RNA was isolated from gliomas in exponential growth. The RNA was analyzed by real-time PCR after the RNA was converted into cDNA. The ΔCT value was calculated by taking the experimental values of the tumor antigen and subtracting the 18S value. nd indicates mRNA was not detected; the CT score was >35 cycles. A ΔCT value of 10 to 14 was a high degree of expression and assigned a 4; a value of 14 to 18 was assigned a 3; a value of 18 to 22 was assigned a 2; and a value of 22 to 26 was assigned a 1. In the next to last row, we assign a score for each of the 12 cell lines (A score) to summarize the overall strengths of the 4 TAPP analyzed.
The sum total of the Σ score from A + Σ score from B = total Σ score‡. The Σ score provides overall evaluation of the strength of expression of the 16 TAPP associated with each of the glioma cell lines.
Real-time reverse transcription-PCR analysis confirmed the presence of TAPP mRNA in glioma cell lines
We genetically confirmed that the TAPP mRNA was present for each glioma cell line using quantitative real-time reverse transcription-PCR technique. This eliminated the possibility that other cross-reactive proteins were being detected by the antibody staining. RNA was extracted from the exponentially growing glioma cells and analyzed by real-time PCR techniques using the SYBR green method. The real-time PCR data (data not shown) confirmed the intracellular flow cytometry data. Cells that failed to make TAPP mRNA also failed to make the protein.
Real-time reverse transcription-PCR suggests that four other TAA can be made by the glioma cell lines
The real-time reverse transcription-PCR technique also allowed us to determine whether the glioma cell lines make mRNA for four other glioma TAA. Human CTL likely can recognize the TAA, but commercially available antibodies to test for the proteins do not exist; they include Aim-2, Gage-1, Sart-1, and Trp-2 (Table 2B). All of the 12 glioma cell lines analyzed had mRNA expression for Aim-2 (ΔCT values ranged between 14.6 and 20.3) with U-251 having the most mRNA (ΔCT = 14.6). Sart-1 mRNA was weakly seen within all gliomas except for two, LNZ308 and U-87. Five of the 12 glioma lines made mRNA for Trp-2. Only U-118 cells uniquely made Gage-1 transcripts (ΔCT = 20.4). Again, we provide quantitative scoring by converting the ΔCT values of the 4 TAPP; they are provided in the second to last row (Σ scoreb).
The scores for the four TAPP were combined with the scores for the 12 TAPP to provide an overall assessment of TAPP expression (i.e., Σ scoreb + Σ scorea = Σ scorec). These scores are given in Table 2B (bottom row). The expression scores when totaled showed that most of the glioma cell lines are good TAA expressors, but the better choices for inclusion into cell-based immunotherapy regimens would include those with a Σ scorec of ≥31; these include A172 (HLA-A1), LN229 (HLA-A3), LNZ308 (HLA-A24), SNB19, T98G, U-251, and U-373 (HLA-A2). Based on the distribution of HLA-A alleles present in individuals comprising the U.S. Caucasian population, we can successfully match 77% of them with one or more of the glioma cell lines to use for an allogeneic vaccine or as a source for peptide pulsing of their dendritic cell, should the need arise for immunotherapy of their diagnosed brain tumor. With the same panel of glioma cells, we can match 61% of the population for alleles at HLA-B and 91% of the population for alleles at HLA-C.
TAPP mRNA expression within surgical glioblastoma multi-forme specimens
To be confident that the TAPP expressed by cell lines maintained in tissue culture are equivalent to that expressed in clinically derived material, we examined for TAPP mRNA using real-time reverse transcription-PCR in freshly resected surgical glioblastoma multiforme specimens. The extracted RNA from the surgical specimens was considered sufficient for protein expression in 9 of 11 specimens (Table 3, column 4). The TAPP included Aim-2, EphA2, Her2/neu, IL13Rα2, GnT-V, GP100, Mage, Sart-1, survivin, Trp-1, Trp-2, and tyrosinase. The frequencies of expression correlated fairly well with the data we collected with the glioma cell lines (Table 3, column 2). Also shown in Table 3, within parentheses of columns 2 and 4, respectively, are frequencies of these TAPP protein and molecularly detected TAPP. Finally, in the right hand column are data obtained from a microarray analysis using Affymetrix U96Av2 chips obtained from RNA derived from 14 “classic” glioblastoma multiforme patients (36). Interestingly, these latter values differed significantly from our data and those of other studies.
Table 3.
Frequency comparisons of TAPP protein and mRNA in glioma cell lines and in surgically resected glioblastoma multiforme tissues as found in the studies presented here and elsewhere
| TAPP gene |
Glioma cell lines/protein detection |
GBM specimens/ protein detection |
GBM specimens/ qPCR detection |
Glioma cell lines/qPCR detection |
GBM specimens/ molecular detection |
GBM specimens/ microarray detection |
|---|---|---|---|---|---|---|
| Positive/ total analyzed (% frequency) |
Positive/ total analyzed (% frequency) reference source |
Positive/ total analyzed (% frequency) |
Positive/ total analyzed (% frequency) |
Positive/ total analyzed (% frequency) reference source |
Positive/ total analyzed (% frequency) |
|
| B-cyclin | 17/17 (100%) | 10/10 (100%) | 6/14 (43%) | |||
| EphA2 | 16/17 (94%) | 13/14 (93%), ref. 22 9/9 (100%), ref. 21 |
10/11 (91%) | 10/14 (71%) | ||
| Her2/neu | 17/17 (100%) | (76%) ref. 16 | 11/11 (100%) | N = 43 (81%), ref. 16 | 2/14 (14%) | |
| IL13Rα2 | 17/17 (100%) | 11/11 (100%), ref. 44 | 10/11 (91%) | 11/11 (100%), ref. 44 | 4/14 (29%) | |
| GnT-V | 17/17 (100%) | 11/11 (100%) | 6/13 (46%), ref. 27 | 0/14 (0%) | ||
| GP100 | 10/17 (59%) | (45%) ref 16 | 9/11 (82%) | 8/21 (38%), ref. 12; 43 (46%), ref 16 |
0/14 (0%) | |
| Mage-1 | 17/17 (100%) | 12/14 (86%), ref. 45; (38%) ref. 16 |
11/11 (100%) | 8/21 (38%), ref. 12 43 = n (40%), ref. 16 |
3/14 (21%) | |
| Mart-1 | 13/17 (76%) | 4/11 (36%) | 0/14 (0%) | |||
| Survivin | 17/17 (100%) | 45/56 (80%), ref. 25 (83%) ref. 26 |
10/11 (91%) | 1/14 (7%) | ||
| hTert | 17/17 (100%) | 10/10 (100%) | 30/43 (70%), ref. 23 | 0/14 (0%) | ||
| Trp-1 | 6/17 (35%) | 10/11 (91%) | 11/21 (52%), ref. 12 | 0/14 (0%) | ||
| Tyrosinase | 11/17 (65%) | 10/11 (91%) | 8/21 (38%), ref. 12 | 14/14 (100%) | ||
| Aim-2 | 10/11 (91%) | 17/17 (100%) | N = 43 (93%), ref. 18 | 0/14 (0%) | ||
| Gage-1 | 0/11 (0%) | 2/17 (12%) | 13/20 (65%), ref. 13 | 14/14 (100%) | ||
| Sart-1 | 5/10 (50%), ref. 24 | 10/11 (91%) | 17/17 (100%) | 14/14 (100%) | ||
| Trp-2 | 9/11 (82%) | 7/17 (100%) | 13/21 (62%), ref. 12 | 0/14 (0%) |
NOTE: Microarray based on http://www.broad.mit.edu/mpr/publications/projects/Cancer_Susceptibility/glioma_nutt_combo.gct.
Abbreviations: GBM, glioblastoma multiforme; qPCR, quantitative PCR.
CTL can recognize the functional HLA-A2–restricted immunotherapeutic peptide on the appropriate tumor target cells
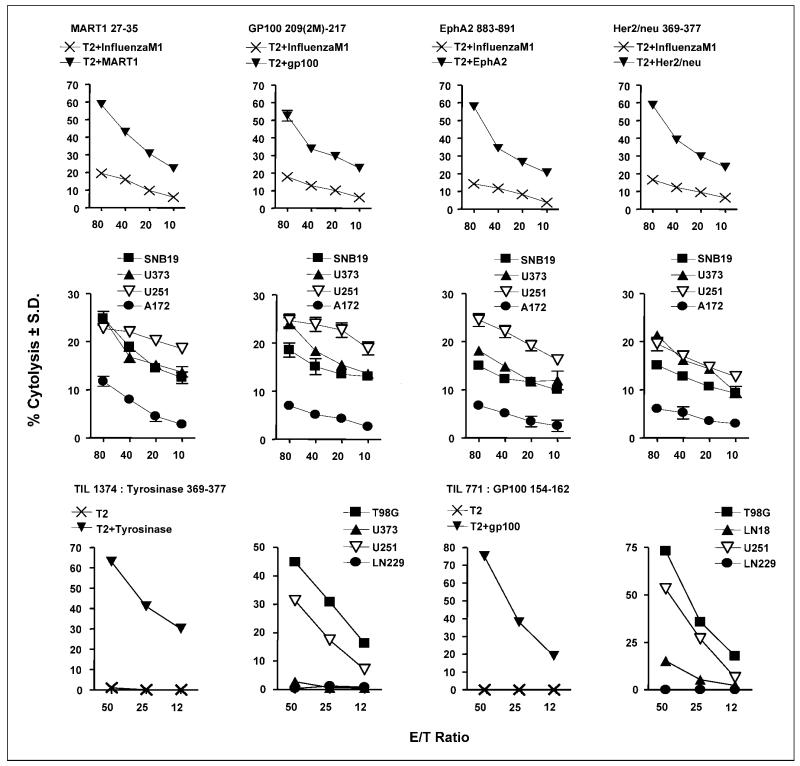
CTL effector cells were generated from CD8+ T cells derived from glioma patients by stimulation with autologous dendritic cells loaded with individual TAA peptides. We showed that glioma antigens expressed by glioma cells and by peptide-loaded T2 target cells are lysed by specifically sensitized CTL effector cells and TIL cells in cell-mediated cytotoxicity experiments (Fig. 2). In Fig. 2 (top row), T2 target cells loaded with Mart-1 (27-35), GP100 (209-217), EphA2 (883-891), or Her2/neu (369-377) were killed in a dose-dependent manner, whereas T2 cells loaded with an irrelevant influenza M1 antigen were not. The lytic values for lysis of targets displaying relevant antigen were statistically distinct (P < 0.05) at all effector-to-target ratios compared with those obtained with targets displaying irrelevant antigen. In Fig. 2 (middle row), we similarly showed that these same CTLs lysed the allogeneic HLA-A2+ glioma cells expressing Mart-1, GP100, EphA2, or Her2/neu HLA-A2–restricted antigens. The glioma cell lines (SNB19, U-373, U-251, and A172; see Table 2) tested in the cytotoxicity assays were positive for these antigens. In contrast, A172 glioma cells that were HLA-A1+, HLA-A3+ were not lysed (≤10% specific lysis) although they expressed the TAPP. The lysis of A172 was statistically different from each of the other glioma cell lines tested (P < 0.05).
Fig. 2.
Cytotoxicity assays done with HLA-A2–restricted CTL or TIL effectors and peptide-loaded T2 cells or glioma target cells. CTL (top and middle rows) were directed against Mart-1, GP100, EphA2, and Her/neu peptides and incubated with peptide-loaded T2 target cells (top row) or glioma target cells (middle row). TIL (bottom row) were incubated with either peptide-loaded T2 cells or the glioma target cells. The effector TIL 1374 cells are specific for tyrosinase (366-377; left) and TIL 771 cells are specific for GP100 (154-162; right) and incubated with the target glioma cells. The phenotypes of the glioma cells used are, for SNB19: HLA-A2+, Mart-1+, GP100+, EphA2+, Her2/neu+; U-251: HLA-A2+, Mart-1+, GP100+, EphA2+, Her2/neu+, tyro+; U-373: HLA-A2+, Mart-1+, GP100+, EphA2+, Her2/neu+, tyro−; A172:HLA-A2−, Mart-1+, GP100+, EphA2+;T98G: HLA-A2+, tyro+; LN229: HLA-A2−, tyro+.
We also did cytotoxicity experiments with TIL effector cells derived from melanoma patients (Fig. 2, bottom row). TIL 1374 cells are specific to tyrosinase (369-377) whereas TIL 771 cells are specific for an epitope of GP100 peptide (154-162) different from that tested earlier. Again, T2 peptide-unloaded or tyrosinase- or GP100-loaded cells were used as targets (bottom row, first and third columns), or glioma cell lines with known HLA and tyrosinase or GP100 TAA expressions were used as targets (bottom row, second and fourth columns). In Fig. 2 (bottom row, first column), T2 tyrosinase peptide–loaded cells were lysed by TIL 1374 effectors, whereas the unloaded T2 cells were not. Similar findings were obtained with TIL 771 effectors and T2 GP100 peptide-loaded and unloaded cells (bottom row, third column). In Fig. 2 (bottom row, second column), TIL 1374 cells mediated the lysis of HLA-A2+ T98G and U-251 glioma cells that expressed tyrosinase. In contrast, the HLA-A2+ but tyrosinase-negative cell line U-373 was not recognized by TIL 1374 cells. TIL 1374 cells did not lyse the tyrosinase-positive but HLA-A2–negative LN229 glioma cells. The GP100-restricted TIL 771 effectors killed HLA-A2+ U-251 and T98G glioma cells, both of which are also positive for GP100 (Fig. 2; bottom row, fourth column). These data cumulatively indicate that U-251 cells display two GP100 epitopes (154-162 and 209-217). TIL 771 cells failed to lyse the HLA-A2+ but GP100-negative LN18 glioma cells. In addition, LN229 glioma cells that are HLA-A2 and GP100 negative were not lysed. Similar findings with the two peptide-restricted TIL indicate that the critical recognition of the HLA molecule with the peptide is necessary for lysis to occur.
In summary, the CTL and TIL data indicate that functional epitopes of six of the TAPP that were found to be associated with glioma cells have the potential to be lysed by effector CTLs specific to those antigens.
Discussion
We confirmed that the 20 glioma cell lines we studied possessed one or more characteristic neuroglial markers expected on glioma cells of neuroectodermal origin (Table 1) and notably lacked the marker for oligodendrogliomas (37, 38). The glioma cell lines were phenotyped for their HLA haplotype specificities by PCR techniques (Table 1). Some of the more well-known cell lines were profiled decades ago by less sensitive methods (39), and some were never completely described for their exact HLA profiles (i.e., HLA typed at one allele). This report updates and compiles the HLA phenotypic data into a single source that can quickly be referenced. The 20 glioma cell lines in our panel provide a repertoire of 40 different class I HLA alleles (i.e., 9 different HLA-A, 19 HLA-B, and 12 HLA-C alleles). This extensive representation would allow for flexibility in matching HLA on glioma cell lines to patient HLA for making individualized patient vaccines. These glioma cell lines share some histocompatibility with 77%, 85%, and 78% of the U.S. Caucasian population at HLA-A, HLA-B, and HLA-C loci, respectively.
We showed by flow cytometry that the majority of the glioma cell lines expressed adequate levels of HLA that can stimulate CTL effector function. Strong HLA-A, HLA-B, and HLA-C expression is important because whole-cell vaccines with low expression likely would not stimulate T cells as well. The glioma cell lines we tested should elicit T-cell responses. Additionally, we characterized 12 of the human glioma cell lines for the presence of 16 possible glioma tumor antigens (TAPP and TAA; Tables 2 and 3). The particular TAPP that we studied were selected based on their ability to elicit human immune responses (10, 11, 14-19). As well, the Aim-2, Gage-1, Sart-1, and Trp-2 TAA were previously reported to be made by gliomas (12, 13, 17-19, 24). Importantly, individual glioma cell lines in our panel express multiple TAPP and/or TAA (Fig. 2; Tables 2 and 3) that are also expressed within primary glioblastoma multiforme specimens. Therefore, their substitution for autologous tumor in generating cell-based immunotherapeutics is validated. The presence of the whole protein does not guarantee that these antigens are properly processed into antigenic peptides that functional CTLs can respond to, however. When TAPP are processed, or when TAA peptides are present, they are loaded onto the HLA motif for presentation to T cells. Previously, one coauthor’s laboratory showed that CTLs can kill U-251, T98G, or SNB19 gliomas in a HLA-A2-restricted manner for either IL13Rα2 (11) or EphA2 (21) antigens. We expanded these studies and showed that functional glioma antigens are recognized by CTL or TIL effector cells by doing cell-mediated cytolysis experiments. Importantly, we showed that HLA-A2+ gliomas or T2 targets were killed by CTL or TIL that were restricted to Mart-1, Her2/neu, GP100, tyrosinase, or EphA2 (Fig. 2). When glioma cells either lack the HLA-A2 molecule or lack the peptide, no cell killing occurs. Our study of cell-mediated cytotoxicity with Mart-1–specific CTLs is the first to show that these CTLs can kill human gliomas expressing this antigen. U-251 cells were also killed via two different epitopes (154-162 and 209-217) derived from the GP100 TAPP. Thus, multiple targets are available for CTL recognition of glioma cells. These functional cytotoxicity data provide proof in principle of the allogeneic tumor vaccine approach.
The human glioma cell lines we characterized could be used for creating a universal allogeneic vaccine or, alternatively, for individualizing an allogeneic glioma vaccine for a particular patient. Each glioma cell line expressed multiple TAPP or TAA. Because not one cell line possessed all of the TAPP or TAA (Tables 2 and 3) relevant for glioma immunotherapy, it would be necessary to combine several of the cell lines to create a universal allogeneic vaccine to comprise all or a majority of the “glioma-specific” antigens tested here. This approach would be similar to that previously used by Hsueh and Morton (40). Their polyvalent melanoma vaccine (Canvaxin) was composed of three melanoma cell lines capable of exposing multiple “melanoma” antigens to a patient’s immune system.
Our work provides an explanation for some successful results reported 23 years ago by Mahaley et al. (2). Use of the inactivated, unmodified U-251 glioma cells as a vaccine to treat glioblastoma multiforme patients resulted in documented responders. After standard debulking surgeries and chemotherapy, patients were vaccinated with monthly s.c. injections of killed tissue culture–derived glioma cell lines. When U-251 cells were used as the vaccine, the survival of six of nine patients was more than 950 days. In contrast, when another cohort of 11 glioblastoma multiforme patients was vaccinated with D-54 glioma cells, there was little difference in survival from that of their historical controls. There are several plausible explanations for these results. U-251 cells may be more immunogenic than D-54 cells. It is possible that the responding patients had HLA and TAA matching those in U-251 cells. U-251 cells are homozygous at HLA-A2, which may provide enhanced recognition, and roughly half of the general U.S. population is positive for HLA-A2. D-54 cells display HLA-A1 and A3. Because both alleles roughly represent a quarter of the population, theoretically up to 50% of the population could respond. Because U-251 and D-54 cells possessed equivalent relative antigen densities of HLA-A, HLA-B, and HLA-C, the expression of the HLA antigens is not likely the limiting factor for response with the D-54 vaccine. The treated patients were not HLA typed, so it is impossible to know what the precise match was between the HLA of the patients and the vaccine that they received. Another possibility is that U-251 cells possessed more TAA, inducing a more efficient immune response. From our comparative analysis of 12 TAPP, U-251 had a slightly higher expression of the different TAPP (Σ score = 34) compared with D-54 (Σ score = 28). One might presume that the U-251 vaccine resulted in more peptides being processed into different epitopes and loaded onto different HLA molecules (10), thus stimulating stronger T-cell responses. Another possibility is that the number of tumor antigens is a limiting factor. Currently, there are more known tumor antigens restricted to HLA-A2 than to other alleles like HLA-A1 or HLA-3. Thus, genetically some people might be more capable of responding to target antigens with an allogeneic vaccine. A prospective study is warranted to determine if a relationship between HLA and survival exists. Finally, another possible explanation for the response was that U-251 cells produced a stimulation that drove a process called “epitope spreading” (41). Here immune responses toward weaker, cryptic epitopes are produced because a much stronger immune response occurred to the immunodominant epitope. Epitope spreading has been observed in the rodent glioma model (42, 43). Understanding the mechanism(s) by which the vaccination with U-251 cells increased glioblastoma multiforme survival would be informative for the successful treatment of this cancer with new allogeneic vaccines.
In summary, our current report characterizes a panel of well-known established glioma cell lines. We have identified their HLA phenotypes and profiled their expression of a plethora of glioma-associated antigens known to stimulate human immune responses. One major ramification of our work is that resected primary gliomas could individually be screened for multiple antigens. A customized allogeneic vaccine, or autologous dendritic cell vaccine prepared with allogeneic glioma cell peptides, could be produced by first HLA phenotyping the patient and matching the appropriate tumor cell lines that possess as many immunotherapeutic antigens with the patient. Alternatively, based on HLA type of the patient, one could predict which TAA might be commonly associated with the patient glioma. This knowledge would allow neuro-oncologists and immunologists the ability to match glioma cell lines with their patients and begin immunotherapy as soon as possible, while tumor burden and serum transforming growth factor-β levels are low, to improve patient survival.
Acknowledgments
We thank Alina Miller and Hao Trinh of the University of California, Irvine HLA typing lab for their help on phenotyping the glioma cells; Dr. J. Wunderlich (National Cancer Institute, Bethesda, MD) for kindly providing tumor-infiltrating lymphocyte lines TIL 771 and TIL 1374; Dr. Peter Cresswell (Yale University, New Haven, CT) for the kind supply of T2 cells; Dr. Catherine Nutt (Harvard University) for her useful discussions and for helping us clarify several key issues with her microarray analysis; and Mehrtash Hashemzadeh for assisting with the various statistical analyses that were presented within this article.
Grant support: Veterans Affairs Medical Center (M.R. Jadus and H.T. Wepsic), the Avon Breast Cancer Foundation via the University of California at Irvine Cancer Research Program (M.R. Jadus), NIH/National Institute of Neurological Disorders and Stroke grant P01NS40923 (H. Okada), Clinical Scientist Development Award from Doris Duke Charitable Foundation (H. Okada), 21st Century Scientist Award from James S. McDonnell Foundation (H. Okada), the Copeland Fund of the Pittsburgh Foundation (J. Eguchi and H. Okada), a grant from the Brain Tumor Society (B. Minev), NIH/National Cancer Institute grant R44-CA105964 (H. Fakhrai), NIH grant F31 CA94834 (G.G. Gomez), NIH/National Institute of Neurological Disorders and Stroke grants NS046463 and NS056300 (C.A. Kruse), the R. Herbert and Alma S. Manweiler Memorial Research Fund (C.A. Kruse), and the La Jolla Foundation for Molecular Medicine Research (C.A. Kruse).
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Mahaley MS, Jr., Bigner DD, Dudka LF, et al. Immunobiology of primary intracranial tumors. Part 7. Active immunization of patients with anaplastic human glioma cells: a pilot study. J Neurosurg. 1983;59:201–7. doi: 10.3171/jns.1983.59.2.0201. [DOI] [PubMed] [Google Scholar]
- 3.Plautz GE, Miller DW, Barnet GH, et al. T Cell adoptive immunotherapy of newly diagnosed gliomas. Clin Cancer Res. 2000;6:2209–18. [PubMed] [Google Scholar]
- 4.Hayes RL, Arbit E, Odaimi M, et al. Adoptive cellular immunotherapy for the treatment of malignant gliomas. Crit Rev Oncol Hematol. 2001;39:31–42. doi: 10.1016/s1040-8428(01)00122-6. [DOI] [PubMed] [Google Scholar]
- 5.Quattrocchi KB, Miller CH, Cush S, et al. Pilot study of local autologous tumor infiltrating lymphocytes for the treatment of recurrent malignant gliomas. J Neurooncol. 1999;45:141–57. doi: 10.1023/a:1006293606710. [DOI] [PubMed] [Google Scholar]
- 6.Kruse CA, Rubinstein D. Cytotoxic T lymphocytes reactive to patient major histocompatibility proteins for therapy of recurrent primary brain tumors. In: Liau LM, Becker DP, Cloughsey TF, Bigner D, editors. Brain tumor immunotherapy. Humana Press; New York: 2001. pp. 149–70. [Google Scholar]
- 7.Parney IF, Hao C, Petruk KC. Glioma immunology and immunotherapy. Neurosurgery. 2000;46:778–91. doi: 10.1097/00006123-200004000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Sahin U, Koslowski M, Tureci O, et al. Expression of cancer testis genes in human brain tumors. Clin Cancer Res. 2000;6:3916–22. [PubMed] [Google Scholar]
- 9.Ueda R, Iizuka Y, Yoshida K, et al. Identification of a human glioma antigen, SOX6, recognized by patients’ sera. Oncogene. 2004;23:1420–7. doi: 10.1038/sj.onc.1207252. [DOI] [PubMed] [Google Scholar]
- 10.Renkvist N, Castelli C, Robbins PF, et al. A listing of human tumor antigens recognized by T cells. Cancer Immunol Immunother. 2001;50:3–15. doi: 10.1007/s002620000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okano F, Storkus WJ, Chambers WH, et al. Identification of a novel HLA-A* 0201 restricted cytotoxic T lymphocyte epitope in human glioma-associated antigen, interleukin-13 receptor α2 chain. Clin Cancer Res. 2002;8:2851–5. [PubMed] [Google Scholar]
- 12.Chi DD, Merchant RE, Rand R, et al. Molecular detection of tumor-associated antigens shared by human cutaneous melanomas and gliomas. Am J Pathol. 1997;150:2143–52. [PMC free article] [PubMed] [Google Scholar]
- 13.Scarcella DL, Chow CW, Gonzales MF, et al. Expression of MAGE and GAGE in high grade brain tumors: a potential target for specific immunotherapy and diagnostic markers. Clin Cancer Res. 1999;5:335–41. [PubMed] [Google Scholar]
- 14.Yu JS, Wheeler CJ, Zeltzer PM, et al. Vaccination of malignant glioma patients with peptide-pulsed dendritic cells elicits systemic cytotoxicity and intracranial T cell infiltration. Cancer Res. 2001;61:842–7. [PubMed] [Google Scholar]
- 15.Yu JS, Liu G, Ying H, et al. Vaccination with tumor lysate-pulsed dendritic cells elicits antigen-specific, cytotoxic T cells in patients with malignant glioma. Cancer Res. 2004;64:4973–9. doi: 10.1158/0008-5472.CAN-03-3505. [DOI] [PubMed] [Google Scholar]
- 16.Liu G, Ying H, Zeng G, et al. Her-2, gp100 and MAGE-1 are expressed in human glioblastoma and recognized by cytotoxic T cells. Cancer Res. 2004;64:4980–6. doi: 10.1158/0008-5472.CAN-03-3504. [DOI] [PubMed] [Google Scholar]
- 17.Harada M, Li YF, El-Gamil M, et al. Melanoma-reactive CD8+ T cells recognize a novel tumor antigen expressed in a wide variety of tumor types. J Immunother. 2001;24:323–33. doi: 10.1097/00002371-200107000-00008. [DOI] [PubMed] [Google Scholar]
- 18.Liu G, Yu JS, Zeng G, et al. AIM-2: a novel tumor antigen is expressed and presented by human glioma cells. J Immunother. 2004;27:220–6. doi: 10.1097/00002371-200405000-00006. [DOI] [PubMed] [Google Scholar]
- 19.Liu G, Khong HT, Wheeler CJ, et al. Molecular and functional analysis of tryosinase-related protein (TRP-2) as a cytotoxic T lymphocyte target in patients with malignant glioma. J Immunother. 2003;26:301–12. doi: 10.1097/00002371-200307000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Kakino S, Sasaki K, Kurose A, Ito H. Intracellular localization of cyclin B1 during cell cycle in glioma cells. Cytometry. 1996;24:49–54. doi: 10.1002/(SICI)1097-0320(19960501)24:1<49::AID-CYTO6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 21.Hatano M, Eguchi J, Tatsumi T, et al. EphA2 as a glioma-associated antigen: a novel target for glioma vaccines. Neoplasia. 2005;7:717–22. doi: 10.1593/neo.05277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wykosky J, Gibo DM, Stanton C, Debinski W. EphA2 as a novel molecular marker and target in glioblastoma multiforme. Mol Cancer Res. 2005;3:541–51. doi: 10.1158/1541-7786.MCR-05-0056. [DOI] [PubMed] [Google Scholar]
- 23.Tchirkov A, Rolhion C, Kemeny JL, et al. Clinical implications of quantitative real-time RT-PCR analysis of hTERT gene expression in human gliomas. Br J Cancer. 2003;88:516–20. doi: 10.1038/sj.bjc.6600754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imaizumi T, Kuramoto T, Matsunaga K, et al. Expression of the tumor-rejection antigen SART-1 in brain tumors. Int J Cancer. 1999;83:760–4. doi: 10.1002/(sici)1097-0215(19991210)83:6<760::aid-ijc11>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 25.Chakravarti A, Noll E, Black PM, et al. Quantitatively determined survivin expression levels are of prognostic value in human gliomas. J Clin Oncol. 2002;20:1063–8. doi: 10.1200/JCO.2002.20.4.1063. [DOI] [PubMed] [Google Scholar]
- 26.Xie D, Zeng YX, Wang HJ, et al. Expression of cytoplasmic and nuclear survivin in primary and secondary human glioblastoma. Br J Cancer. 2006;94:108–14. doi: 10.1038/sj.bjc.6602904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto H, Swoger J, Greene S, et al. β1,6-N-Acetylglucosamine-bearing N-glycans in human gliomas: implications for a role in regulating invasivity. Cancer Res. 2000;60:134–42. [PubMed] [Google Scholar]
- 28.Liau LM, Prins RM, Kiertscher SM, et al. Dendritic cell vaccination in glioblastoma patients induces systemic and intracranial T-cell responses modulated by the local central nervous system tumor microenvironment. Clin Cancer Res. 2005;11:5515–25. doi: 10.1158/1078-0432.CCR-05-0464. [DOI] [PubMed] [Google Scholar]
- 29.Gjerset RA, Fakhrai H, Shawler DL, et al. Characterization of a new human glioblastoma cell line that expresses mutant p53 and lacks activation of the PDGF pathway. In Vitro Cell Dev Biol. 1995;31:207–14. doi: 10.1007/BF02639435. [DOI] [PubMed] [Google Scholar]
- 30.Jadus MR, Chen Y, Boldaji MT, et al. Human U251MG glioma cells expressing the membrane form of macrophage colony-stimulating factor (mM-CSF) are killed by human monocytes in vitro and are rejected within immunodeficient mice via paraptosis that is associated with increased expression of three different heat shock proteins. Cancer Gene Ther. 2003;10:411–20. doi: 10.1038/sj.cgt.7700583. [DOI] [PubMed] [Google Scholar]
- 31.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawakami Y, Eliyahu S, Sakaguchi K, et al. Identification of the immunodominant peptides of the MART-1human melanoma antigen recognized by the majority of HLA-A2-restricted tumor infiltrating lymphocytes. J Exp Med. 1994;180:347–52. doi: 10.1084/jem.180.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawakami Y, Eliyahu S, Jennings C, et al. Recognition of multiple epitopes in the human melanoma antigen gp100 by tumor-infiltrating T lymphocytes associated with in vivo tumor regression. J Immunol. 1995;154:3961–8. [PubMed] [Google Scholar]
- 34.Parkhurst MR, Salgaller ML, Southwood S, et al. Improved induction of melanoma-reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A* 0201-binding residues. J Immunol. 1996;157:2539–48. [PubMed] [Google Scholar]
- 35.Minev BR, Chavez FL, Dudoue BM, et al. Synthetic insertion signal sequences enhance MHC class I presentation of a peptide from the melanoma antigen MART-1. Eur J Immunol. 2000;30:2115–24. doi: 10.1002/1521-4141(2000)30:8<2115::AID-IMMU2115>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 36.Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlated better with survival than histological classification. Cancer Res. 2003;63:1602–7. [PubMed] [Google Scholar]
- 37.McKeever PE, Davenport RD, Shakui P. Patterns of antigenic expression of human glioma cells. Crit Rev Neurobiol. 1991;6:119–47. [PubMed] [Google Scholar]
- 38.Yung WK, Luna M, Borit A. Vimentin and glial fibrillary acidic protein in human brain tumors. J Neurooncol. 1985;3:35–8. doi: 10.1007/BF00165169. [DOI] [PubMed] [Google Scholar]
- 39.Bigner SH, Bullard DE, Pegram CN, et al. Relationship of in vitro morphologic and growth characteristics of established human glioma-derived cell lines to their tumorigenicity in athymic nude mice. J Neuropathol Exp Neurol. 1981;40:390–409. doi: 10.1097/00005072-198107000-00004. [DOI] [PubMed] [Google Scholar]
- 40.Hsueh EC, Morton DL. Antigen-based immunotherapy of melanoma: Canvaxin therapeutic polyvalent cancer vaccine. Semin Cancer Biol. 2003;13:401–7. doi: 10.1016/j.semcancer.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 41.El-Shami K, Tirosh B, Bar-Haim E, et al. MHC class I-restricted epitope spreading in the context of tumor rejection following vaccination with a single immunodominant CTL epitope. Eur J Immunol. 1999;29:3295–301. doi: 10.1002/(SICI)1521-4141(199910)29:10<3295::AID-IMMU3295>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 42.Liau LM, Jensen ER, Kremen TJ, et al. Tumor immunity within the central nervous system stimulated by recombinant Listeria monocytogenes vaccination. Cancer Res. 2002;62:2287–93. [PubMed] [Google Scholar]
- 43.Sanchez R, Williams CC, Daza JL, et al. T9 glioma cells expressing membrane macrophage colony stimulating factor produce CD4+ associated protective immunity against T9 intracranial gliomas and systemic immunity against different syngeneic glioma cells. Cell Immunol. 2002;216:1–11. doi: 10.1016/s0008-8749(02)00011-4. [DOI] [PubMed] [Google Scholar]
- 44.Kawakami M, Kawakami K, Takahashi S, et al. Analysis of interleukin-13 receptor α2 expression in human pediatric brain tumors. Cancer. 2004;101:1036–42. doi: 10.1002/cncr.20470. [DOI] [PubMed] [Google Scholar]
- 45.Kuramoto T. Detection of MAGE-1 tumor antigen in brain tumor. Kurume Med J. 1997;44:43–5. doi: 10.2739/kurumemedj.44.43. [DOI] [PubMed] [Google Scholar]