Abstract
In B cell progenitors, E-proteins E2A and HEB are critical to induce a B-lineage specific program of gene expression and orchestrate the assembly of the immunoglobulin loci. In the thymus E2A and HEB act differently, activating the expression of genes closely associated with the establishment of T cell identity and promoting the rearrangement TCR loci. These findings have raised the question as to how E-proteins exert these different activities. Here we review the distinct regulatory networks that establish B versus T cell identity, and how genomic architecture and location of genes is modulated in these lineage decisions. We conclude by proposing a model wherein stochasticity in the nuclear location of the EBF1 locus in multipotent progenitors determines this lineage choice.
Establishing B and T cell fate
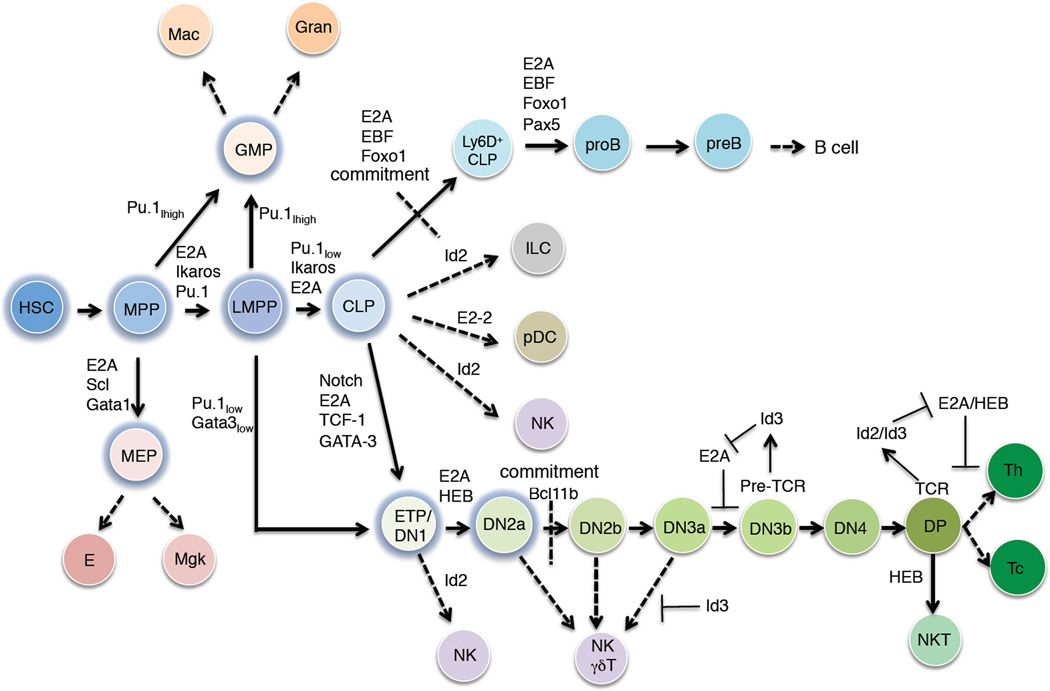
Immune cells are generated from long-term hematopoietic stem cells (LT-HSC) [1]. LT-HSCs are multipotent and self-renew to give rise to multipotent progenitors (MPPs). MPPs give rise to lymphoid primed MPPs (LMPPs) [2]. LMPPs, in turn, give rise to either granulocyte-macrophage progenitor cells (GMPs) or common lymphoid progenitors (CLPs) (Figure 1) [3,4]. GMPs have the ability to differentiate into macrophages or granulocytes whereas CLPs give rise to the plasmacytoid dendritic (pDC), natural killer (NK), innate lymphoid (ILC), B or T cell lineages (Figure 1) [5,6]. Transcriptional regulatory networks underlie these lineage commitment steps.
Figure 1. Transcriptional control of early hematopoiesis.
HSC indicates the hematopoietic stem cell compartment. MPP refers to the multipotent progenitor cell stage. LMPP indicates the lymphoid-primed multipotent progenitor compartment. GMP refers to the granulocyte-macrophage progenitor population. MEP refers to the megakaryocyte-erythrocyte progenitor compartment. E and Mgk refer to erythrocytes and megakaryocytes, respectively. Gran refers to granulocytes. Mac indicates the macrophage compartment. CLP represents the common lymphoid progenitor compartment. ILC refers innate lymphoid cells. pDC refers plasmacytoid dendritic cells. NK indicates natural killer cells. ETP indicates the early thymocyte progenitor population. DN refers to thymocyte progenitors that lack CD4 and CD8 expression. DP refers to immature thymocytes that express both CD4 and CD8. NKT refers to natural killer cells. Th indicates helper T cells. Tc indicates cytotoxic T cells.
The differentiation of myeloid cells is instructed by signaling mediated by the macrophage colony stimulating factor 1 receptor (M-CSFR) or granulocyte-CSFR (G-CSFR) receptors upon exposure of G-CSF, M-CSF and/or GM-CSF respectively [7]. The neutrophil versus macrophage lineage decision is controlled by the relative dosage of the transcriptional regulators PU.1 and CEBPα, with high abundance of PU.1 favoring macrophage development whereas high levels of CEBPα facilitating developmental progression towards the neutrophil lineage [8,9].
CLPs give rise to Ly6D− and Ly6D+ cells [10,11]. Ly6D+ cells differentiate into pre-pro-B cells and ultimately give rise to committed pro-B cells (Figure 1). While DHJH rearrangements are initiated in the CLP compartment, VHDHJH joining begins at the pro-B cell stage. Once a productive Igh chain has been formed, pro-B cells undergo proliferation and differentiation, giving rise to the pre-B cell compartment. CLPs also have the ability to develop into T-lineage cells but this involves an intermediate population named early T cell progenitors (ETPs) (Figure 1) [12]. ETPs have the ability to differentiate into T-lineage as well as dendritic, NK and myeloid cells [4].
The processes the drive lymphocyte development have been examined from multiple directions, all yielding valuable insights. However, there is a great need to integrate the various insights into a comprehensive understanding of how lymphocyte development is achieved at the gene regulation level. Here we review recent findings in two of these areas: the transcriptional networks that impact B and T cell fate decisions, and the changes in nuclear architecture that accompany and enable these transitions and the maintenance of cell fate choices. We conclude by proposing a model that integrates the functions of key developmental regulators with nuclear organization in the B versus T lineage cell choice.
Transcriptional networks orchestrating cell fate decisions
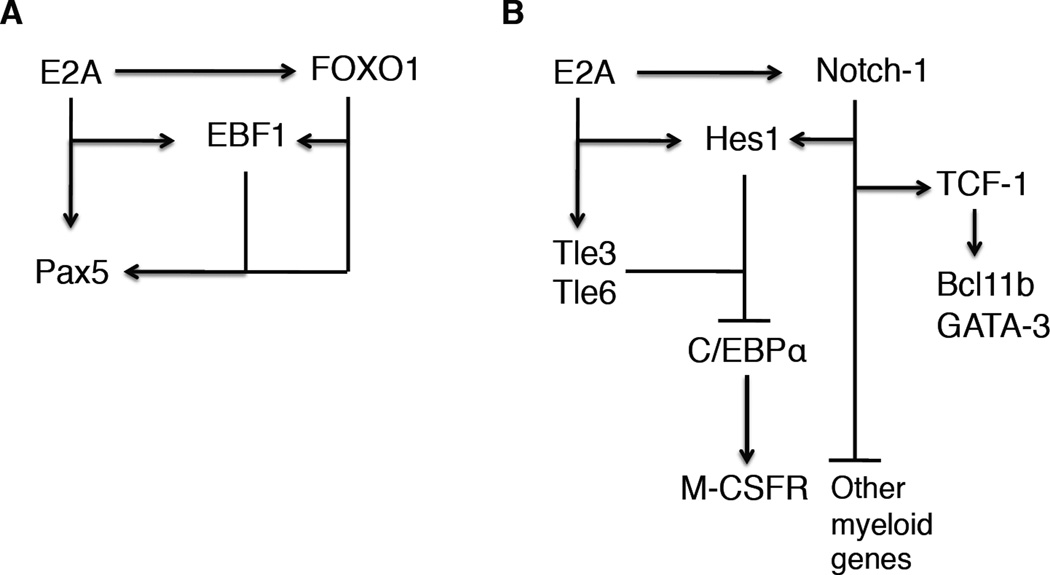
It is now well established that B cell development is specified by the activities of several key regulators. Conspicuous among these are E2A, EBF1, FOXO1 and PAX5 [13–18]. The E2A locus encodes for two isoforms, E12 and E47, which arise through differential splicing of an exon encoding for the helix-loop-helix (HLH) domain. E47 plays a particularly important role in B-lineage specification. B cell development in mice depleted for the expression of E47 is blocked at the CLP cell stage [17]. In CLPs, E47 and HEB act in concert to activate the expression of the Forkhead-containing protein, FOXO1 [19]. E2A and FOXO1, in turn, together induce the expression of EBF1. Once EBF1 and FOXO1 are expressed they establish a positive intergenic feedback circuitry to establish B cell identity [20]. Additionally, EBF1 acts to suppress an innate lymphoid specific program of gene expression by suppressing the induction of expression of the transcriptional regulator Id2 [21]. Finally, E2A, EBF1 and FOXO1 coordinately activate the expression of PAX5 [22]. Once Pax5 is expressed it acts with EBF1 in a positive regulatory feedback loop to establish commitment to the B cell fate [23]. In addition to activating the expression of EBF1 and FOXO1, E47 also activates the expression of components associated with the Notch signaling pathway, such as Notch 1, Notch3, the transcriptional repressors Hes1, Tle3 and Tle6 [24,25,26]. Notch1 is transcribed at high levels in CLPs and pro-B cells whereas Hes1 transcript levels are readily detectable in pro-B cells. However since Notch ligands are not expressed in the bone marrow, the Notch signaling pathway is not activated in B cell progenitors. How then is HES1 involved in orchestrating the developmental progression of lymphoid progenitors? Recent elegant studies have demonstrated that Hes1 antagonizes the expression of C/EBPα to prevent the initiation of a myeloid specific program of gene expression [27]. Similarly, previous studies have shown that the E-proteins act in progenitor cells to suppress a myeloid specific program of gene expression. Specifically, E2A-deficient lymphoid progenitors showed an increased tendency to differentiate into the myeloid cell lineage [24]. Collectively, these studies show that in progenitor cells, the E-proteins induce the expression of HES1, which in turn, coordinately with the Tle proteins antagonize the expression of C/EBPα, linking E2A and HES1 into a regulatory circuitry that acts to suppress the development of myeloid cells (Figure 2). Upon arriving in the thymus, the Notch signaling pathway becomes activated in progenitor cells, being exposed to the Notch ligand, Dll1. Activation of Notch signaling, in turn, leads to the activation of TCF-1, GATA3 and Bcl11b expression (Figure 2). TCF-1 expression is critical to specify the T-lineage cell fate and GATA3 has recently been demonstrated to antagonize the expression of a subset of B-lineage specific genes [28–30]. Furthermore, Notch signaling and E47 act in concert to induce the expression of genes closely associated with a committed T-lineage phenotype [24]. One of the critical targets is the pre-Tα locus, containing binding sites for both E47 and CSL (CBF1, Suppressor of Hairless, Lag-1) [31,32]. That E47 and Notch signaling act collaboratively across cis-regulatory elements to regulate gene expression is not unique to T cell progenitors. In Drosophila melanogaster, development of sensory organs is controlled by daughterless, a homologue of E-proteins, members of the achaete-scute complex and Notch signaling [33,34]. Similarly, HLH proteins acts collaboratively with the Notch signaling cascade to activate Brachyuri expression of the ascidian Ciona intestinalis [35]. Thus, it seems that an ancient regulatory module used to orchestrate Drosophila melanogaster organ development and the induction of a notochord specific transcription signature is also utilized in T cell progenitors to establish T cell identity.
Figure 2. Regulatory networks that orchestrate B and T cell fate.
A Transcription factors that promote early B cell development are indicated. EBF1 functions as a nodal point. Note that E2A acts to induce the expression of FOXO1. E2A and FOXO1 then act collaboratively to activate EBF1 expression. Arrows refer to positive regulation. Bars refer to transcriptional repression. B Transcription factors that promote early T cell development. Notch signaling and E2A, together with Tle3 and Tle6, drive Hes1 transcription, which in turn acts to suppress C/EBPα expression. Notch-mediated signaling leads to the activation of TCF-1, GATA-3 and Bcl11b expression.
A key aspect of T cell fate involves the rearrangement and expression of antigen receptor loci. Upon arriving in the thymus, antigen receptor rearrangement involving the TCRβ, TCRγ and TCRδ loci is activated. Again, prominent participants in this process are the E-proteins [36,37]. How do E-proteins regulate antigen receptor assembly? This has been particularly well studied in the context of the TCRβ locus. E47 proteins occupy multiple sites that span the entire TCRβ locus, involving both the Vβ and DβJβ region clusters [36]. As a working model we propose that the E-proteins function as anchors to sequester the variable regions to the base of a rosette surrounding the DβJβ regions, as proposed for the Igh and Igκ loci [38,39]. A key role for E47 in promoting TCRβ rearrangements was revealed in mice depleted for E47 expression [36]. Specifically, it was shown that the effective concentration of E47 is rate limiting as it relates to the recombination reaction. Furthermore, it was shown that forced E47 expression in developing thymocytes interfered with a feedback mechanism established by the induction of Id3 expression by pre-TCR signaling [40]. These observations imply that the E47/Id3 module regulates TCRβ locus allelic exclusion at two distinct levels: (1) mono-allelic activation and (2) a feedback mechanism involving the activation of Id3 expression. Beyond the pre-TCR checkpoint Id3 levels are essential to maintain the T cell fate [25]. The E2A proteins also function at the TCR checkpoint. Again, both declining levels of E2A and increasing levels of Id3 modulate transition through the TCR checkpoint to promote positive selection [40–42]. Thus, a common theme has emerged in which E-proteins promote the assembly of antigen receptor loci, which upon expression act in a negative feedback loop to antagonize continued antigen receptor locus rearrangement and to promote developmental progression.
In addition to orchestrating T-lineage specification, E-protein activity is critical to orchestrate lineage choice during thymocyte development. Id3 limits the expansion of innate NK-γδ T cells, while HEB plays an essential role in the development of NKT cells [43–45]. Furthermore, the relative dosages of E-proteins versus Id-proteins enforce the TCR checkpoint and promote positive selection [41,46]. In sum, E-proteins not only act as critical determinants of T-lineage specification but also function as gatekeepers at critical checkpoints throughout thymocyte development. Interestingly, these transcriptional regulators are also involved in defining nuclear architecture via enhancer-promoter interactions. In the following section we discuss nuclear organization in the context of transcriptional activation and repression, and explore how nuclear positioning of key regulators impacts lymphocyte cell fate decisions.
The dynamic lymphoid genome
The chromatin fiber consists of nucleosomes that are organized as beads-on-a-string. Nucleosomes consist of DNA that surrounds an octamer of histones, packaged as a 30 nm polymer chain. Early microscopic studies showed that the chromosomes in Drosophila are organized into topological domains closely associated with active transcription [47,48]. However, it remained largely unknown as to how chromosomes are organized in 3D-space. Initial studies predominantly involved comparisons of experimental and simulated measurements assuming distinct topologies as starting configurations. Prominent among such topologies were the Random Walk/Giant Loop, the Multi-Loop-Subcompartment and the Random-Loop Model. The Random Walk/Giant Loop model (RW/GL) involves random walk behaviour adopted by large genomic segments that are connected by distinct anchors [49]. The Multi-Loop-Subcompartment (MLS) predicts a chromatin architecture in which the genome is folded into ~1 Mbp chromatin domains consisting of bundles of loops [50]. The Random-Loop (RL) model assumes a configuration of both small and large loops that vary in size [51]. Recent studies have compared experimentally derived spatial distances to those observed during simulations using the RW/GL, MLS and RL models as starting topologies. Specifically, measurements across the immunoglobulin heavy chain (Igh) locus were most consistent with the chromatin fiber folded into bundles of loops separated by linkers, consistent with that originally proposed for the Igh locus [52–54]. Recently, global formaldehyde-cross linking studies using chromosome capture approaches reached similar conclusions, showing that the genomes of vertebrate organisms are organized as transcriptionally active domains, consisting of globules, that are interspersed with transcriptionally inactive regions that comprise, at least in part, the heterochromatic compartment [39,55–58].
The transcriptional active globules consist of clusters of loops organized by tethers. Most prominent among these tethers is the transcriptional regulator CTCF, which forms homodimers and in this way loops associated DNA, and can in addition anchor associated chromatin regions to the nuclear lamina [56,59]. In addition to CTCF the globules are structured by enhancer-promoter interactions that involve lineage-specific transcriptional regulators, such as E2A, EBF1, FOXO1, HES1 etc. Although it is now evident that these factors contribute to the folding patterns of lymphoid genomes it remains to be clarified as to how they promote the organization of the genome. Recent advances in super resolution microscopy should make it possible to gain insight into the structures of chromatin globules, how they are assembled and reorganized during active transcription.
Genes frequently switch nuclear location upon in developing lymphocytes. For example, a large spectrum of genes switches nuclear compartments in differentiating embryonic stem cells as well as progenitor B cells [39,60]. Conspicuous among those genes that switch nuclear environments in early B cell progenitors are the Ebf1, Foxo1, Igκ and Igl loci. Changing the nuclear neighborhood of genes correlates well with changes in patterns of gene expression, with transitions to a transcriptionally repressive compartment - marked by heterochromatin and packed nucleosomes - correlating with transcriptional silencing of a locus, and vice versa. However, there are exceptions. During the pre-pro-B to the pro-B cell transition, a cluster of genes repositions from the transcriptionally repressive compartment to the transcriptionally permissive compartment, yet remain transcriptionally inert [39]. They remain inert by recruitment to polycomb bodies, nuclear foci defined by the presence of the Polycomb group (PcG) proteins, a family of transcriptional repressors with chromatin modification and nucleosome remodeling activities, and that have also been associated with regulation of nuclear architecture. PcG proteins mediate the methylation of histone 3 at lysine 27 (H3K27me3), a silencing epigenetic mark, at their target loci. Thus, at least two distinct mechanisms mediate transcriptional silencing during the developmental progression of lymphoid progenitors: nuclear positioning and association with polycomb bodies. We note that previous observations have indicated a role for polycomb in repression of EBF1 transcritption in hematopoietic progenitors [61]. These data indicate that EBF1 expression is silenced in hematopoietic progenitors at multiple levels. Why is transcriptional repression exerted by distinct mechanisms? We propose that the recruitment of genes to the heterochromatic compartment permits efficient repression across a large genomic region. Such a mechanism may prevent the aberrant activation of genes encoding for key regulators since the entire regions would be in a transcriptionally inert environment. On the other hand, genes silenced by being localized across polycomb bodies might readily associate with active transcription factories upon developmental progression since they are already localized in the transcriptionally permissive compartment.
Finally, it seems likely that controlling the nuclear location would be a strategy utilized in other developmental pathways. Recently, we found that the Bcl11b gene, encoding for a transcription factor controlling T cell fate, is also positioned at the heterochromatic compartment of multipotent progenitors [39]. Thus, we are now faced with the question as to how the EBF1 and Bcl11b loci are sequestered at the nuclear lamina and how their release from the heterochromatin is regulated during the progression of developing hematopoietic progenitors. Factors that control the nuclear location of genes encoding for key developmental regulators might be the key to understanding how multipotency is enforced and how B and T lineage development is initiated. We suggest that the positioning of key developmental regulators at the nuclear lamina in progenitor cells is a general principle to prevent the premature activation of lineage-specific programs of gene expression and developmental progression.
Stochasticity and Establishment of the B versus T Cell Fate
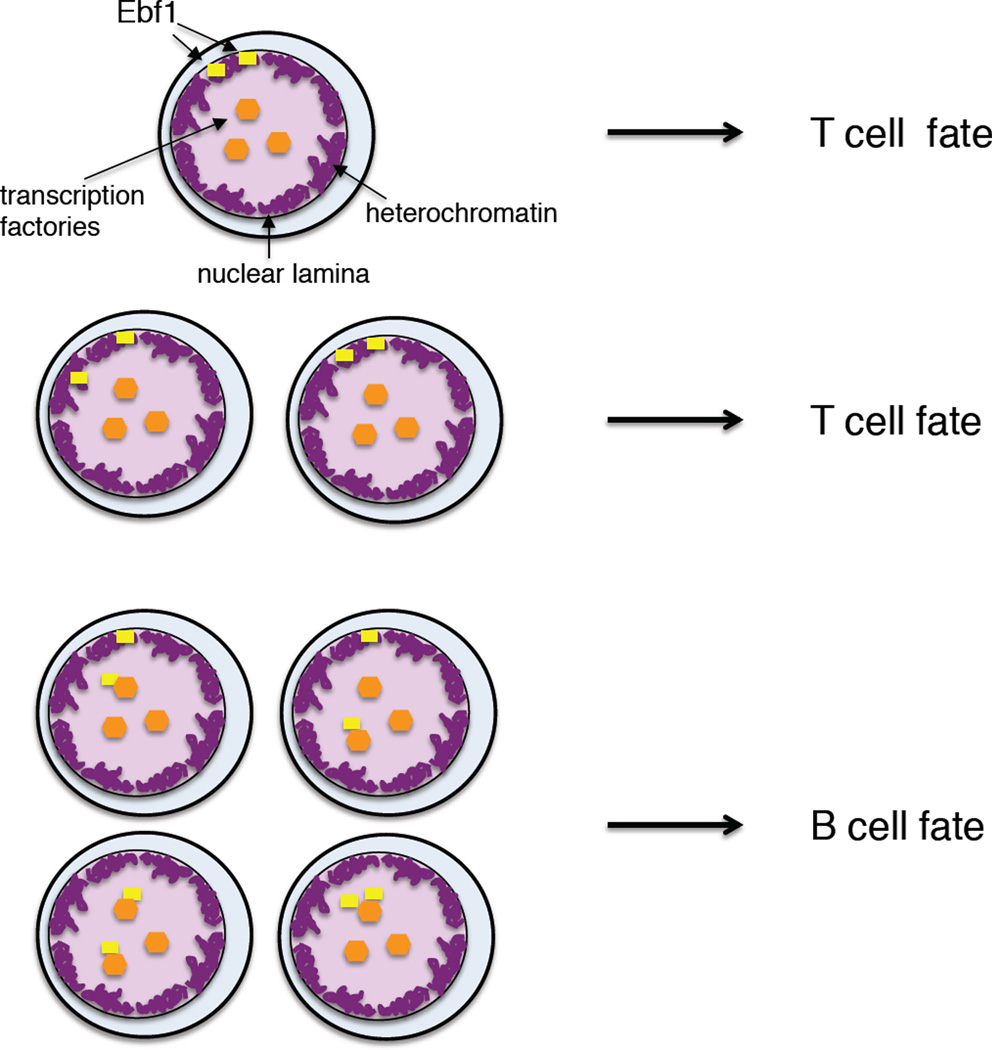
While it is now established that the E-proteins in the multipotent progenitors specify the B and T cell fate it remains unclear as to how they instruct the B versus T cell fate choice. E2A and HEB, in a subset of cells activate the expression of FOXO1 and EBF1 to orchestrate B cell fate. How then is the T cell fate established? The critical step may involve a failure of progenitor cells to induce FOXO1 and EBF1 transcription. Cells that do not succeed in activating FOXO1 and EBF1 expression may become specified to the T cell fate. In such a model, T cell development is viewed as a default pathway. The question then becomes how progenitor cells decide whether or not to activate FOXO1 and EBF1 expression. E2A and HEB levels vary slightly within the HSC, LMPP and CLP populations [62]. However, it seems unlikely that small differences in E2A and HEB levels affect the B versus T cell fate choice. Rather, we propose an alternative mechanism, as recently described for olfactory receptor cell choice. Olfactory receptor loci are located at the heterochromatic compartment and are transcriptionally silent [63,64]. In developing olfactory neurons, a singular olfactory receptor allele is released from the heterochromatic compartment to associate with existing transcription factories. The release of olfactory receptor alleles from the lamina is stochastic. As aforementioned, in the majority of multipotent progenitor cells the EBF1 locus is located at the heterochromatic compartment whereas in committed pro-B cells the EBF1 locus is associated with the euchromatic compartment [39]. However, in a small but significant fraction of multipotent progenitors, one or two EBF1 alleles have switched nuclear location [39]. Hence, the association of the EBF1 locus with the lamina in multipotent progenitors varies within the population. Multipotent progenitors, in which one or two EBF1 alleles have been released from the lamina, now become specified to the B cell lineage (Figure 3). In contrast, we propose that multipotent progenitors that fail to release EBF1 alleles from the lamina ultimately give rise to T, NK or ILC progenitors (Figure 3).
Figure 3. Stochasticity in the nuclear location of the EBF1 locus may determine B versus T cell identity.
Multipotent progenitors are shown in which one or two alleles of the EBF1 locus are sequestered at the nuclear lamina. Release of one or two EBF1 alleles from the lamina promotes developmental progression towards the B cell fate. In contrast, multipotent progenitors that do not succeed to release EBF1 alleles from the lamina differentiate into T/NK/ILC cell progenitors. EBF1 alleles are represented by yellow dots. Heterochromatin associated with the nuclear lamina is shown in purple. Transcription factories are indicated by orange dots.
Concluding Remarks
There is now ample evidence that E2A and HEB proteins orchestrate both B and T cell identity. These findings have raised the question as to how E-proteins act in multipotent progenitors to specify the B versus T fate choice. Here we propose that in the adult bone marrow stochastic mechanisms involving the nuclear repositioning of genes encoding for key developmental regulators determine whether within a population of progenitor cells E-proteins activate a B-lineage versus T-lineage specific programs of gene expression. We suggest that the nuclear location of the EBF1 locus is critical. However, as aforementioned, although detailed single cells analysis is required, genomic regions associated with the FOXO1 locus are also associated with the heterochromatic compartment in multipotent progenitors. It is conceivable that stochasticity in the nuclear positioning of both the EBF1 and FOXO1 loci determines the B versus T cell fate. The critical question now is to determine how the EBF1 and FOXO1 loci are sequestered at the nuclear lamina and how recruitment and release from the lamina is regulated. It will be important to determine the underlying mechanism of stochasticity. Does the abundance of factors that control sequestration change during developmental progression and is it rate limiting in the multipotent progenitor cell stage? It seems unlikely that the mechanisms that relate to nuclear positioning and the control of cell fate are limited to the B versus T cell identity. Rather, we suggest that the findings obtained from studying the B versus T cell fate choice will serve as a paradigm for the specification and commitment of other developmental pathways across a wide spectrum of organisms. Finally, we note that additional complexity may be provided by the differential recruitment of the EBF1 and FOXO1 alleles to polycomb bodies. Future studies should reveal how the individual contributions of nuclear location and recruitment of polycomb bodies of the EBF1 and FOXO1 loci relate to the enforcement of multipotency and the establishment of B versus T cell identity.
Highlights.
Transcriptional regulatory networks establish lymphocyte lineage decisions
E-protein transcription factors are critical for establishing both B and T cell identities
E-proteins also define nuclear architecture via enhancer-promoter interactions
Stochastic nuclear positioning of EBF1 may determine B versus T lineage choice
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weissman IL. Stem cells: Units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 2.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Kondo M, et al. Identification of clonogenic common lymphoid lrogenitors in mouse bone marrow. Nat. Immunol. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 4.Wada H, et al. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 5.Pronk CJH, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Lai AY, Kondo M. Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J. Exp. Med. 2006;203:1867–1873. doi: 10.1084/jem.20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecchini MG, et al. Role of colony stimulating factor-1 in the establishment and regulation of tissue macrophages during postnatal development of the mouse. Development. 1994;120:1357–1372. doi: 10.1242/dev.120.6.1357. [DOI] [PubMed] [Google Scholar]
- 8.DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- 9.Laslo P, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 10.Inlay MA, et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Devel. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mansson R, et al. Single-cell analysis of the common lymphoid progenitor compartment reveals functional and molecular heterogeneity. Blood. 2010;115:2601–2609. doi: 10.1182/blood-2009-08-236398. [DOI] [PubMed] [Google Scholar]
- 12.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 13.Bain G, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 14.Zhuang Y, et al. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 15.Lin HH, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor Ebf. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 16.Nutt SL, et al. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. [PubMed] [Google Scholar]
- 17.Beck K, et al. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J. Exp. Med. 2009;17:2271–2284. doi: 10.1084/jem.20090756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murre C. Developmental trajectories in early hematopoiesis. Genes Dev. 2009;23:2366–2370. doi: 10.1101/gad.1861709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welinder E, et al. The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc. Nat.l Acad. Sci. U.S.A. 2011;108:17402–17407. doi: 10.1073/pnas.1111766108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mansson R, et al. Positive intergenic feedback circuitry, involving EBF1 and FOXO1, orchestrates B-cell fate. Proc. Natl. Acad. Sci. U.S.A. 2012;108:17402–17407. doi: 10.1073/pnas.1211427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nechanitzky R, et al. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat. Immunol. 2013;14:868–876. doi: 10.1038/ni.2641. [DOI] [PubMed] [Google Scholar]
- 22.Mercer EM, et al. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity. 2011;35:413–425. doi: 10.1016/j.immuni.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medina KL, et al. Assembling a gene regulatory network for specification of the B cell fate. Dev. Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Ikawa T, et al. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J. Exp. Med. 2006;203:1329–1342. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyazaki M, et al. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naïve fate of T cells. Nat. Immunol. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatakeyama J, et al. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- 27.De Obaldia ME, et al. T cell development requires constraint of the myeloid regulator C/EBPα by the Notch target and transcriptional repressor Hes1. Nat. Immunol. 2013;12:1277–1284. doi: 10.1038/ni.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ting CN, et al. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–478. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 29.Weber BN, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.García-Ojeda ME, et al. GATA-3 promotes T cell specification by repressing B cell potential in pro-T cells in mice. Blood. 2013;121:1749–1759. doi: 10.1182/blood-2012-06-440065. [DOI] [PubMed] [Google Scholar]
- 31.Reizis B, Leder P. The upstream enhancer is necessary and sufficient for the expression of the pre-T cell receptor α gene is immature T lymphocyte. J. Exp. Med. 2001;194:979–990. doi: 10.1084/jem.194.7.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reizis B, Leder P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 2002;16:295–300. doi: 10.1101/gad.960702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kageyama R, et al. Roles of bHLH genes in neural stem cell differentiation. Exp. Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 34.Nellesen DT, et al. Discrete enhancer elements mediate selective responsiveness of Enhancer of split complex genes to common transcriptional activators. Dev. Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- 35.Corbo JC, et al. Suppressor of hairless activates Brachyuri expression in the Ciona embryo. Dev. Biol. 1998;203:358–368. doi: 10.1006/dbio.1998.9067. [DOI] [PubMed] [Google Scholar]
- 36.Agata Y, et al. Regulation of T cell receptor β gene rearrangements and allelic exclusion by the helix-loop-helix protein, E47. Immunity. 2007;27:871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 37.Yashiro-Ohtani Y, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lucas JL, et al. Transcription and recombination factories: Common features? Curr. Opin. Cell Biol. 2011;2:318–324. doi: 10.1016/j.ceb.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin YC, et al. Global changes in nuclear positioning of genes and intra- and inter-domain genomic interactions that orchestrate B cell fate. Nat. Immunology. 2012;12:1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engel I, et al. Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 2001;194:733–745. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones ME, et al. E protein transcription factors are required for the development of CD4(+) lineage T cells. Immunity. 2012;36:348–361. doi: 10.1016/j.immuni.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lauritsen JP, et al. Marked induction of the helix-loop-helix protein Id3 promotes the γδ T cell fate and renders their functional maturation Notch independent. Immunity. 2009;31:565–575. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ueda-Hayakawa I, et al. Id3 restricts the developmental potential of gamma delta lineage during thymopoiesis. J. Immunol. 2007;182:5306–5316. doi: 10.4049/jimmunol.0804249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verykokakis M, et al. Inhibitor of DNA binding 3 limits development of murine slam-associated adpator protein-dependent "innate" gd T cells. PLoS One. 2010;15:e9303. doi: 10.1371/journal.pone.0009303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D'Cruz LM, et al. An essential role for the transcription factor HEB in thymocyte survival, Tcrα rearrangement and the development of natural killer T cells. Nat. Immunol. 2010;3:240–249. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones ME, Zhuang Y. Acquisition of a functional TCR during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;6:860–868. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sedat J, Manuelidis L. A direct approach to the structure of eukaryotic chromosomes. Cold Spring Harbor Sym Quant Biol. 1978;42:331–350. doi: 10.1101/sqb.1978.042.01.035. [DOI] [PubMed] [Google Scholar]
- 48.Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- 49.Sachs RK, et al. A random-walk/giant-loop model for interphase chromosomes. Proc. Nat. Acad. Sci. U.S.A. 1995;92:2710–1714. doi: 10.1073/pnas.92.7.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Münkel C, et al. Compartmentalization of interphase chromosomes observed in simulation and experiment. J. Mol. Biol. 1999;285:1053–1065. doi: 10.1006/jmbi.1998.2361. [DOI] [PubMed] [Google Scholar]
- 51.Bohn M, et al. Random loop model for long polymers. Physical review. E. 2007;76:051805. doi: 10.1103/PhysRevE.76.051805. [DOI] [PubMed] [Google Scholar]
- 52.Jhunjhunwala S, et al. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo C, et al. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy chain locus. Cell. 2011;147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo C, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477:424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fullwood MJ, et al. An estrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 58.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li G, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kind J, van Steensel B. Genome-nuclear lamina interactions and gene regulation. Curr Opin Cell Biol. 2010;22:320–325. doi: 10.1016/j.ceb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 61.Oguro, et al. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein, bmi. Cell Stem Cell. 2010;6:279–286. doi: 10.1016/j.stem.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 62.Semerad CL, et al. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc. Natl. Acad. Sci. U.S. A. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clowney EJ, et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Magklara A, Lomvardas S. Stochastic gene expression in mammals: lessons from olfaction. Trends Cell Biol. 2013;23:449–456. doi: 10.1016/j.tcb.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]