Summary
Association and disassociation of gene loci with respect to specific nuclear compartments accompany changes in gene expression, yet little is known concerning the mechanisms by which this occurs or its functional consequences. Previously we showed that tethering acidic activators to a peripheral chromosome site led to movement of the chromosome site away from the nuclear periphery, but the physiological relevance of this movement was unclear [1]. Nuclear speckles, or interchromatin granule clusters, are enriched in factors involved in RNA processing [2], and the association of a subset of active genes at their periphery suggests speckles may play a role in gene expression [3, 4]. Here we show an actin dependent association of Hsp70 transgenes with nuclear speckles after heat shock. We visualized Hsp70 transgenes moving curvilinearly towards nuclear speckles over ~0.5–6 μm distances at velocities of 1–2 μm min−1. Chromatin stretching in the direction of movement demonstrates a force generating mechanism. Transcription in nearly all cases increased noticeably only after initial contact with a nuclear speckle. Moreover, blocking new Hsp70 transgene/speckle association by actin depolymerization prevented significant heat-shock induced transcriptional activation in transgenes not associated with speckles, although robust transcriptional activation was observed for Hsp70 transgenes associated with nuclear speckles. Our results demonstrate the existence of a still to be revealed machinery for moving chromatin in a direct path over long distances towards nuclear speckles in response to transcriptional activation; moreover this speckle association enhances the heat-shock activation of these Hsp70 transgenes.
Results and Discussion
Hsp70 transgenes associate with nuclear speckles [5, 6], recapitulating the heat shock dependent speckle association of the endogenous locus [7]. Consistent with previous demonstrations of speckle association for only ~50% of 25 active genes surveyed [3, 4], no speckle association of MT (metallothionein) or DHFR transgenes was observed after their transcriptional activation [5, 6]. Moreover, neither MT nor DHFR transgenes associated with nuclear speckles after heat shock (data not shown). Promoter swapping experiments showed that Hsp70 speckle association was conferred specifically by the Hsp70 upstream regulatory region [6]. Hsp70 transgene speckle association was specific to heat shock, as transcriptional activation by cadmium did not result in speckle association [6].
Our previous work indicated that speckle association occurred mostly through apparent de novo formation of a nuclear speckle or association with a nearby, pre-existing speckle [5]. Hints of long-range movements of the transgenes to nuclear speckles were limited by technical factors. In particular, long-range chromosome movements are profoundly sensitive to phototoxicity, yet our previous live-cell imaging [5] used light levels shown to inhibit long-range movements [1]. In most cells the BAC transgene array localized near a pre-existing nuclear speckle even before heat shock activation, but we were limited to live cell microscopy of 1–2 cells per experiment, reducing the number of observations for statistical analysis. Our speckle marker showed a poor speckle to nuclear background ratio, making it difficult to detect small speckles. Finally, we had no direct, live-cell readout of transcriptional activity, complicating efforts to relate changes in transgene nuclear position to changes in transcriptional activity.
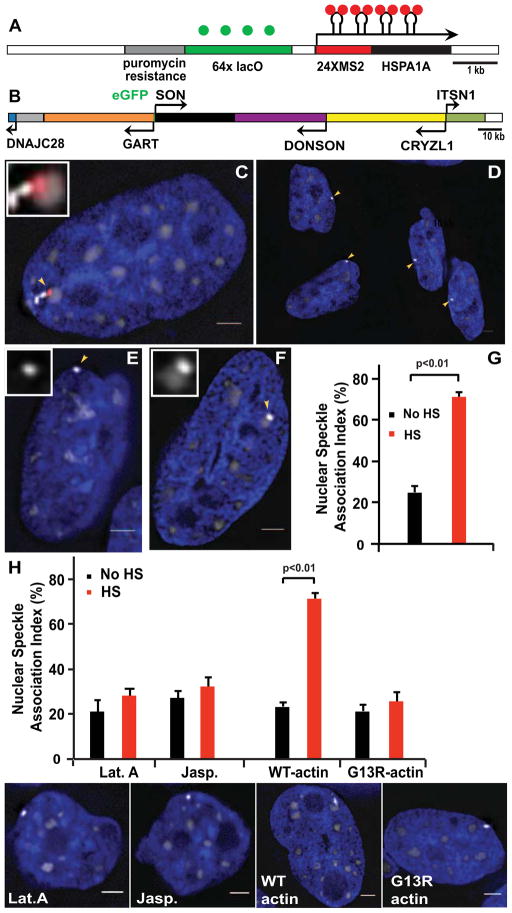
Here, an Applied Precision OMX microscope provided reduced phototoxicity and increased data acquisition speeds. A plasmid containing a 64-mer lac operator repeat, selectable marker, and the HSPA1A gene [6] was modified to insert 24 MS2 repeats into the HSPA1A 5′ UTR and stably transfected into CHO DG44 cells expressing EGFP-LacI (Fig. 1A,C). Expressing EGFP-SON from a BAC transgene, recombineered to insert the EGFP in frame into the SON NH2 terminus, provided uniform expression and visualization of nuclear speckles (Fig. 1B–C). Uniform EGFP-SON expression allowed us to simultaneously localize the higher intensity EGFP tagged Hsp70 transgenes relative to the lower intensity EGFP labeled nuclear speckles in stably transfected cell clones (Fig. 1C–F). A second transfection yielded cells stably expressing MS2 binding protein- mCherry for labeling of nascent transcripts (Fig. 1C).
Figure 1. Simultaneous visualization of Hsp70 transgenes, nascent transcripts, and nuclear speckles.
(A–B) Hsp70 plasmid pSP14-14-5′UTR-MS2 (A) and the modified SON BAC (B) transgenes. (C–F) DAPI-stained nuclei (blue) versus Hsp70 transgenes (GFP-LacI, bright grey-scale), speckles (GFP-SON, less bright grey-scale) and Hsp70 transcripts (MS2-binding protein-mCherry, red): After heat shock (HS), nascent transcripts (arrowhead) accumulate near speckle (C). Peripheral transgenes (arrowheads) prior to HS (D). Hsp70 transgenes (arrowheads) not associated (E) or associated (F) with speckles. (G) Speckle association index before or after 30 min HS. (H) Disrupting nuclear actin polymerization abolishes HS-induced speckle association. SEM error bars, scale bar = 2μm.
Nuclear speckle location is biased towards the nuclear interior [8]. Using cell clone C16_C4_1 in which the Hsp70 plasmid transgene array is preferentially positioned at the nuclear periphery (Fig. 1D), we increased the fraction of cells in which the array is not close to any speckle prior to heat shock. For this cell clone, containing ~700 plasmid copies as estimated by qPCR, the 25% to 71% increased array association with nuclear speckles (Fig. 1E–G) and Hsp70 transcript accumulation within nuclear speckles after heat shock (data not shown) were similar to that previously observed for plasmid Hsp70 transgenes [6]. In previous work where active interphase chromosomal motion was suspected, an either direct or indirect dependence on actin was implicated through dominant negative and/or inhibitor studies [1, 9, 10]. Similarly, speckle association of the Hsp70 transgene array after heat shock was blocked by expression of a nonpolymerizable NLS-RFP-actin G13R mutant, incapable of actin polymerization, but not by expression of the corresponding wild type actin construct (Fig. 1H). Because these actin fusion proteins are concentrated in the nucleus, a dependence specifically on nuclear actin is suggested. Hsp70 transgene association with nuclear speckles was also blocked by treatment with latrunculin A, which depolymerizes F-actin, and by jasplakinolide, which blocks F-actin depolymerization (Fig 1H).
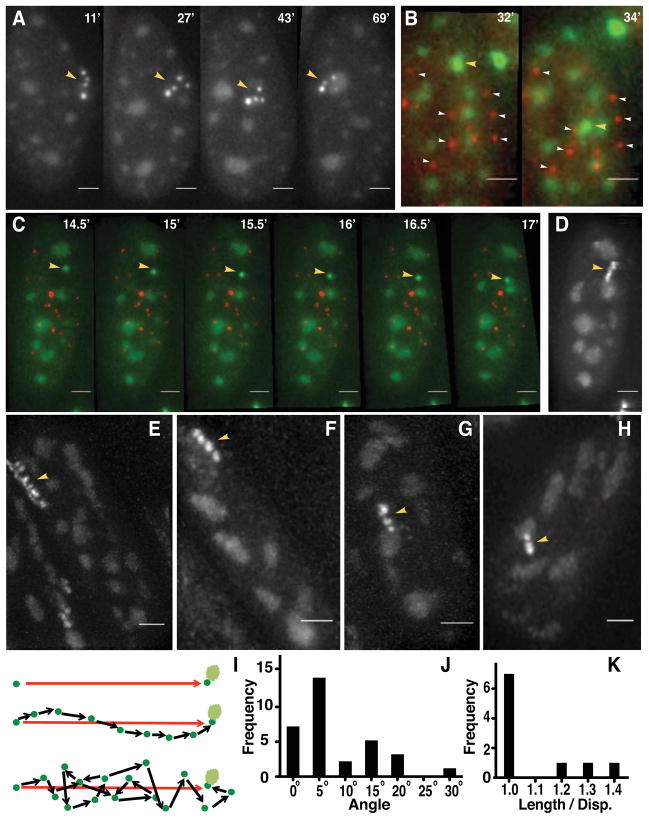
Directed chromosome motion is difficult to distinguish from the rapid, “constrained diffusion” motion [11] observed at most chromosome loci in a wide range of eukaryotic cells, including mammalian [12]. We focused on movements at least two-fold longer-range than the ~ 0.2 μm “radius of constraint” derived from mean square displacement (MSD) versus time measurements at both 37 and 44°C (Fig. S1A). We collected 3D images every 2 min for 60 min for ~900 cells (~400 movies) identifying 90 cells in which the starting distance between the transgene array and the nearest nuclear speckle was at least 0.4 μm. Inspection of 2D projections versus time revealed long-range movements exceeding 0.4 μm in ~50% of these 90 cells. Nearly all of these movements terminated at a nuclear speckle (Fig. 2A), although in several cases the transgene array moved to one speckle and then to another. (Speckle-transgene association in ~20% of these 90 cells occurred through de novo formation or enlargement of a speckle, as previously described [5], or, in ~10% of these cells, through speckle protrusion or motion. Otherwise, either no speckle association was observed or association occurred through a complex combination of events.)
Figure 2. Live-cell imaging reveals long-range, directed motion of Hsp70 transgenes towards nuclear speckles.
(A–C) Hsp70 transgene (yellow arrowheads) movements during heat shock. (B–C) Transgene movement relative to centromeres (mCherry-CENPA, white arrowheads) rules out nuclear rotation. (D–H) Transgene trajectories projected over time. (I) Directed (middle) versus random motion (bottom). (J) Histogram of angular deviations between trajectory direction at each time point and vector (I, top) drawn between initial and final transgene positions. (K) Histogram of total trajectory lengths (sum of vector lengths; I, bottom) divided by net displacement distances (I, top). Time is minutes (‘) after heat shock. Scale bar= 2 μm. See also Figure S1 and Movies S1, S2, and S3.
Long-range movements, typically over 2–4 min, ranged from 0.5 to 6 μm (median 1.7 μm) (Movie S1). We created a derivative cell line stably expressing CENPA-mCherry together with EGFP-SON and EGFP-LacI. The CENPA-mCherry labeled centromeres served as 3D fiducial marks, demonstrating that transgene movements were not due to nuclear rotation (Fig. 2B–C, Movie S2). Similar long-range movements were not observed at 37°or at 44°C in cells carrying lac operator/repressor tagged, MT BAC transgene arrays. In cells showing long-range movement, ~20% of Δd values, measuring the change in position during a single, 2 min time interval, exceeded the maximum Δd value observed in cells at 37°C or, after heat shock, in CHO DG44 cells containing a control metallothionein BAC transgene array (cell clone MT1_1_4, reference [5]) (Fig. S1B–D). No examples of long-range movements to nuclear speckles or long-range movements per se after heat shock were observed in movies of ~200 cells carrying the control, MT BAC transgene array (data not shown) or in ~350 cells at 37°C carrying the Hsp70 plasmid transgene array.
Linear trajectories of transgenes directed towards nuclear speckles were revealed using shorter 30 sec time intervals (Fig. 2C–H, Movie S3); the mean velocity measured in these trajectories during motion was 1.14 μm min−1. Linearity was indicated by both the absolute angular offset between consecutive steps (14.2° mean, 15.7° sd, Fig. 2J) and by the ratio between the sum of all vector step lengths and the net vector length (1.094 mean, 0.147 sd, Fig. 2K) for 10 trajectory examples. Directionality towards nuclear speckles was supported by two measures. First, we measured the magnitude of the angular deviation of each step direction from the vector defined by the line connecting the starting transgene position and its end position adjacent to the nuclear speckle (7.9°mean, 29.1° max, 7.3° sd). Trajectories varied from 2–10 steps (4.2 mean). Assuming all directions are equally likely, the probability in a random walk for all 4 steps deviating less than or equal to the ~30° observed maximum from the speckle direction is ~0.0008 ((30/180)4) and becomes vanishingly smaller with angular deviations closer to the mean. Second, we measured the sum of all angles represented by possible trajectories that would intercept a speckle based on the geometry observed at the first time point in the trajectory (139° mean, 33.1° sd). The probability of all 10 long-range trajectories terminating at a speckle over the 360° possible directions for each would be ~0.00007 ((139/360)10). These numbers strongly support directed movement towards speckles rather than a “speckle capture” model in which trajectories end when they hit a speckle.
Transient chromatin stretching in the trajectory direction demonstrates a force-generating mechanism underlying these long-range movements and further supports their directionality. Approximately 40% of long-range movements (18/46) were accompanied by chromatin stretching during and sometimes immediately preceding movement, while relaxation of stretching was typically observed after the movement stopped (Fig. 3A–D, Movies S4). The angle between the estimated directions of stretching versus movement averaged 3.5 ° (7.6° sd) for 14 examples (Fig. 3E). No examples of stretching were observed in 350 cells at 37°C or 200 cells carrying metallothionine gene arrays at 44°C. In 3/90 cells, dramatic chromatin stretching occurred without actual movement to a speckle (Movie S5).
Figure 3. Transgene chromatin stretching coincident with and parallel to long-range movements.
(A–D) Hsp70 transgenes (arrowheads) show stretching during (A,B) or immediately proceeding (C) long-range movements; stretching relaxes after long-range movement stops (A,D). Insets (A–D) show 2-fold enlarged regions surrounding the transgene array. (C) Increasing distance between two spots (arrowheads) demonstrated with display (bottom) of 8 consecutive optical sections (OS), separated by 200 nm focus steps. Time shown is minutes (‘) after heat shock. Scale bar= 2 μm. (E) Histogram (bottom) showing near-zero angle (top) between direction of transgene stretching and movement direction. See also Movies S4, S5.
Stretching, rather than re-orientation of a fiber segment from parallel to perpendicular to the optical axis, was inferred from inspection of optical sections and seeing a round spot change to an elliptical or linear shape. Limited resolution along the optical axis complicates unambiguous discrimination between stretching versus reorientation for structures ≤ ~0.5 μm in length. However, stretching rather than reorientation is clearly established by multiple examples in which distinct deformations occur perpendicular to the optical axis (Fig. 3B–C).
All of the preceding experiments were done using a high copy number plasmid transgene array, raising questions of whether similar long-range motion would be observed with a low copy number Hsp70 BAC transgene array, which more closely approximates the behavior of the endogenous locus [5, 6]. We deliberately used a Hsp70 BAC in which two of the three Hsp70 genes contained at this locus were deleted [6] (Fig. S2A). We also used a CHO cell clone containing only ~1–3 copies (as measured by qPCR) of this Hsp70 BAC; therefore the total number of Hsp70 genes approximates that of the endogenous locus. Unfortunately, in this clone nearly all cells showed the BAC transgene array prior to heat shock already too close to a nuclear speckle to distinguish subsequent motions to these nearby speckles as either directed or resulting from “constrained diffusion”. Significantly, however, we were able to capture several examples of similar long-range trajectories towards nuclear speckles using this low-copy Hsp70 BAC transgene (Fig. S2B–D).
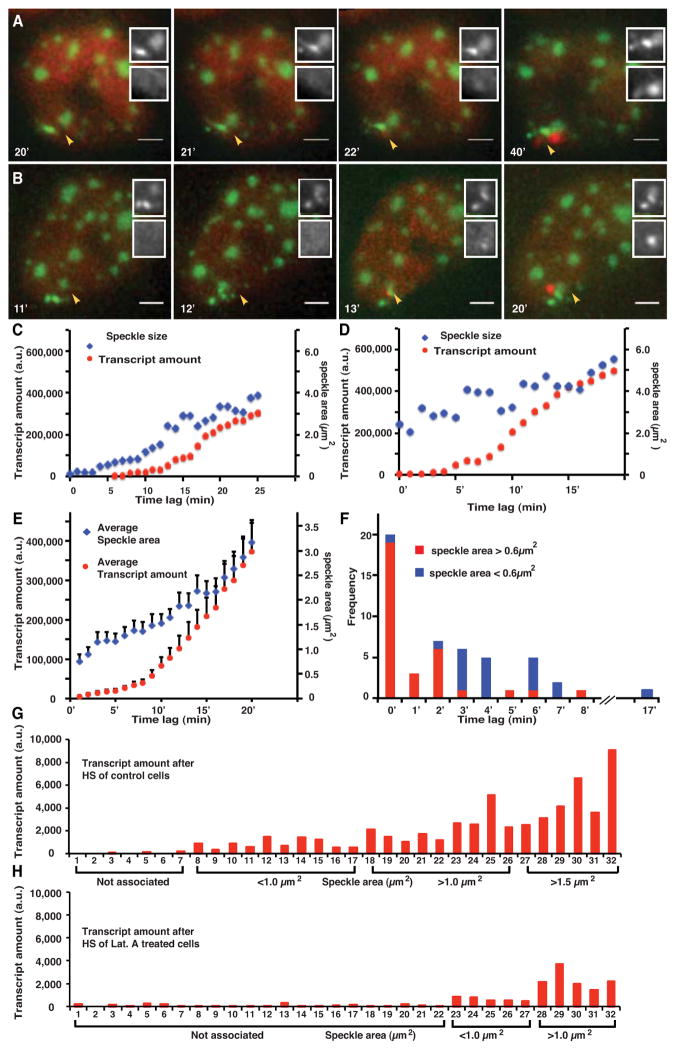
In fixed cells we observed a strong positive correlation between the nascent RNA (either MS2 or RNA FISH) signal at the plasmid transgene array and speckle size (data not shown). In several cases in which the Hsp70 plasmid transgene array was split into spatially distinct spots, only the spot in contact with a nuclear speckle showed a measurable nascent RNA signal (Fig. 1C). Stable expression of the MS2-binding protein in these cells perturbs nascent transcript processing. Specifically, without expression of the MS2-binding protein, RNA FISH demonstrates transcript accumulation within the speckle (data not shown) as previously observed [5, 6]. With expression of the MS2-binding protein, Hsp70 transcripts accumulate adjacent to rather than inside nuclear speckles (Fig. 1C) and appear to progressively accumulate over time as if release from the transgene array is inhibited (Fig. 4A–B). The timing of transcriptional induction, however, parallels what we observe by RNA FISH in cells not expressing the MS2 binding protein. MS2- mCherry signals therefore likely reflect the integrated transcription amount over time, but as a consequence provide a sensitive assay for transcriptional output.
Figure 4. Hsp70 transgene transcription is facilitated by nuclear speckle association.
(A–B) Transcript signal (mCherry-MS2-binding protein, red) increases above background levels shortly after first contact of transgene array (bright green) with nuclear speckle (lighter green). Top inset- green channel; Bottom inset- red channel. Time is min after heat shock. Scale bar= 2 μm. (C–D) Two examples of speckle area (blue) plotted versus integrated MS2-binding protein signal (red, “transcript amount”) in arbitrary units (a.u.) as function of time after initial transgene contact with a nuclear speckle. (E) Speckle size versus transcript signal as function of time after speckle first contact averaged over all observations. (F) Histograms for the delay between first contact with speckle and appearance of transcript signal plotted for transgene arrays that contact small speckles (< 0.6 μm2, blue) versus large speckles (>0.6 μm2, red). Speckle size is measured at time of first contact. (G–H) Integrated transcript signals from each of 32 different cells (x-axis) 30 min after heat shock ordered by increasing speckle size. Low transcript levels are observed for transgene arrays not associated with nuclear speckles. A much larger fraction of transgenes arrays were not associated with nuclear speckles after latrunculin A treatment (H) as compared to control cells (G). Speckle associated transgene arrays show comparable transcript levels between control (G) and latrunculin A treated cells (H) if transgene arrays are grouped according to the size of their associated nuclear speckles. See also Movie S6.
A priori, this correlation between speckle size and nascent RNA signal at the associated Hsp70 plasmid transgene array is consistent with either facilitated Hsp70 transgene transcription through association with a nuclear speckle and/or with Hsp70 transgene transcription causing an increase in the size of associated nuclear speckles.
We addressed the causal relationship between transcription and speckle association by using live cell microscopy to determine the temporal relationship between Hsp70 plasmid transgene transcription and speckle association. In these experiments we counted all examples in which the transgene array initially shows no speckle association. Live cell microscopy revealed that in 31/32 movements of Hsp70 transgene arrays to nuclear speckles, the MS2 binding protein- mCherry signal first increased above the nucleoplasmic background only after initial contact of the transgene array with a nuclear speckle (Fig. 4A–B, Movie S6). As previously noted, occasionally the transgene array makes transient contact, comes off, and then reattaches to the same (Fig. 4A, Movie S6) or even a different speckle. Therefore we cannot exclude the possibility that in the one example in which transcription begins prior to nuclear speckle association that a transient speckle association occurred between adjacent time points.
Plotting speckle size versus integrated MS2-binding protein signal as a function of time after first contact showed a range of patterns. However, a general trend was suggested in which when transgenes contacted a small speckle, a jump in speckle size preceded the onset of significant transgene transcript accumulation (Fig. 4C); in contrast, when transgene arrays contacted larger speckles there was a more immediate rise in transcript accumulation (Fig. 4D). Plotting speckle size and transcript levels averaged over all cells for each time revealed the average trend in which initially speckle size increases more rapidly than transcript followed by a coordinated increase in both at later times (Fig. 4E). Overall, transgene arrays that made initial contact with a larger speckle (> 0.6 μm2) showed shorter time lags (typically 0–2 min) between first contact and transcript accumulation above background than the time lags (typically 3–7 min) observed for transgene arrays that made initial contact with a smaller speckle (<0.6 μm2) (Fig. 4F).
Our results suggest a dose response relationship in which the ability of nuclear speckles to initially boost Hsp70 transgene transcription increases with speckle size. At higher levels of transcription, transcript accumulation levels increase in parallel with speckle size, suggesting again some type of functional connection between nuclear speckle size and transcription levels of surrounding chromatin.
We next asked whether preventing the increased frequency of Hsp70 transgene association with nuclear speckles after heat shock would effect transgene expression. We used latrunculin A treatment which blocks the normal increase in Hsp70 transgene association from ~30% to ~70% after heat shock (Fig. 1H). Whereas the average level of transgene transcription is greatly decreased, this correlates with the reduced percentage of transgenes associated after heat shock with nuclear speckles (Fig. 4G–H). After latrunculin A treatment, the low level of transcription for the ~70% of transgene arrays not associated with nuclear speckles (Fig. 4H) is similar to the transcription levels observed in the small fraction of transgene arrays not associated with nuclear speckles in control cells (Fig. 4G). Robust transcription at similar levels to control cells, when comparing association with similar size speckles, is still observed for the ~30% of transgene arrays in contact with nuclear speckles (Fig. 4G–H). Importantly, latrunculin A treatment does not appear to inhibit transcription per se. These results further support a model in which Hsp70 transgene association with nuclear speckles facilitates their transcriptional activation.
In summary, we have demonstrated long-range, directed movements of Hsp70 transgenes towards nuclear speckles occurring through an active, force-generating mechanism, and an increased transcription following nuclear speckle contact.
Previously, long-range motion of a peripheral chromosome site away from the nuclear periphery was observed after artificial tethering of an acidic transcriptional activator [1], but the physiological relevance to natural gene activation was questionable. Here, we have demonstrated long-range movements with nearly 4-fold higher velocity (1.14 versus 0.31 μm/min) through activation of a transgene from a naturally occurring promoter sequence.
Long-range movement is not synonymous with directed movement. Varied mechanisms could be imagined as giving rise to long-range, but randomly directed motion. Coherent movements of chromosome regions over micron-scale distances has been suggested as due to elastic coupling, through chromatin, within the nucleus [13]. A “search and capture” type model in which random motion was terminated by transgene binding to nuclear speckles could give the appearance of directed motion. A long-range, directed movement of a U2 snRNA mini-gene array towards coiled bodies has been reported [9], yet this study was based largely on two examples of long-range movement, unaccompanied by any statistical analysis, making it difficult to conclude directed versus random motion. Here, statistical analysis of multiple trajectories supported a vectorial directionality to long-range movements. Moreover, statistical analysis taking into account the measured solid angle of all trajectories that would result in contact versus no contact with nuclear speckles strongly supported a directed movement towards nuclear speckles rather than a search and capture model.
The strongest indication of a directed chromosome movement towards nuclear speckles is the accompanying chromatin stretching tangential to the direction of movement that appears shortly before or during the actual period of chromosome motion. This stretching strongly argues for a force pulling on the chromatin in the direction of motion. In contrast, an alternative model in which a chromatin fiber stretched between another anchor point and the nuclear speckle might recoil elastically towards the nuclear speckle after release of the chromatin from the anchor point instead would predict maximum stretching prior to motion, with a progressive relaxation of tension and stretching during movement.
Finally, our results may bridge two previous models for nuclear speckle function. Alternatively, nuclear speckles have been proposed to act as hubs for a subset of transcriptionally active genes [4] versus as storage sites for RNA processing components [14]. During heat shock we see evidence for both roles. As with other conditions resulting in a general transcriptional inhibition, heat shock is accompanied by a reduced number and rounding of nuclear speckles. Yet, our results demonstrate a functional enhancement of Hsp70 transgene transcription resulting from proximity with nuclear speckles through an as yet unknown mechanism. We note that the SRSF2/SC35 SR-splicing factor, concentrated in nuclear speckles, has been shown to play a role in transcriptional pause release and transcriptional activation [15]. It is tempting to speculate that the initial rise in speckle size we see frequently preceding the first detection of nascent transcripts above background, as well as the coordinated increase in speckle size and transcription we observe at later times, reflects a flux of speckle components back and forth between the transcription site and the speckle, with local storage and accumulation of factors at the speckle. Cycles of modification of speckle components in the speckle followed by reversal of these modifications at the site of transcription might explain the functional coupling between speckles and transcription at neighboring genes. These cycles could include, for instance, phosphorylation of SR proteins in the speckle and dephosphorylation at the transcription site as previously suggested [16].
Several special features of our cell system likely facilitated our experiments. By using a plasmid transgene array that prior to heat shock targets preferentially to the nuclear periphery, we were able to maximize the average distance of the array to nearby speckles. This was crucial for distinguishing directed movements from the rapid “constrained diffusion” type movements [11] observed at all interphase chromosome sites. Second, previously we were puzzled by the delayed and asynchronous transcriptional induction of plasmid Hsp70 transgene arrays as compared to BAC Hsp70 transgene arrays. Now it appears this delay may be related to the time required for these plasmid transgene arrays to become associated with nuclear speckles. We suspect the heterochromatin nature of the plasmid transgene arrays accentuates the positive impact of speckle proximity for transcriptional activation, thus highlighting the capacity of nuclear speckles to enhance transcription. Future experiments will examine the relationship between transcriptional induction and speckle proximity for BAC and endogenous Hsp70 genes.
We anticipate that our results and experimental system will facilitate future experiments aimed at elucidating the molecular mechanism underlying this movement and the transcriptional enhancement resulting from speckle association. A significant fraction of endogenous genes associate with nuclear speckles when transcriptionally active; we speculate that similar active movements may play a role in the transcriptional regulation of those genes that change their nuclear localization upon transcriptional activation. We also anticipate that the machinery enabling such long-range, active movements may be involved in multiple aspects of genome organization and nuclear architecture.
Experimental Procedures
Plasmids and BACs were stably transfected into CHO DG44 cells expressing EGFP-LacI (clone 7) cells [5]. The pSP14-14-5′UTR-MS2 plasmid was stably transfected using 7.5μg/ml puromycin (Sigma) for selection to generate the cell clone pSP14_5′UTR_MS2_HSP70_42 (C42). C42 was transfected with the EGFP labeled SON-BAC and selected using 200 μg/ml Zeocin to generate a cell clone C42-165J2-C16_C4_1. The above generated cell clone was transfected with either CENPA-mCherry or Ub-MS2bp-mCherry and selected using 400 μg/ml G418 selection, to generate either C42-165J2-CENPA-mCh-C36 or C42-165J2-UbMS2mCh-B respectively. The constructs mRFP-actin-NLS and mRFP-G13R actin-NLS[1] were transiently transfected into cells and speckle association analyzed after 48 hrs. Cells were treated with either Latrunculin A (Calbiochem) for 5min or Jasplakinolide (Calbiochem) for 30min at a final concentration of 1μM, followed by heat shock for 30 min. An Applied Precision OMX microscope with EMCCD cameras was used for live cell microscopy and a Personal Deltavision for fixed cell microscopy. Data was deconvolved using Applied Precision SoftWorx software.
Supplementary Material
Highlights.
Hsp70 transgenes move unidirectionally to nuclear speckles
Movement is accompanied by chromatin stretching parallel to the motion
High transcription levels appear nearly always only after speckle contact
New speckle association and transcription boost requires actin polymerization
Acknowledgments
We thank Dr. Paula Bubulya for providing us with a cDNA EGFP-SON expression vector plasmid that led to our choice of SON as a speckle marker. We thank Dr. Robert Singer for the pSL-MS2-24 plasmid, Dr. Roger Tsien for the pRSETB-mCherry plasmid, and Dr. Yaron Shav-Tal for the MS2bp-mCherry plasmid. Light microscopy was performed in the MCB Light Microscopy Facility funded with support from the Carver Foundation. This work was supported by grant GM58460 from the National Institute of General Medical Sciences awarded to ASB.
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 2.Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. Proteomic analysis of interchromatin granule clusters. Mol Biol Cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol. 2003;162:981–990. doi: 10.1083/jcb.200303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall LL, Smith KP, Byron M, Lawrence JB. Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:664–675. doi: 10.1002/ar.a.20336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y, Kireev I, Plutz MJ, Ashourian N, Belmont AS. Large-scale chromatin structure of inducible genes- transcription on a linear template. J Cell Biol. 2009;185:87–100. doi: 10.1083/jcb.200809196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Y, Plutz M, Belmont AS. Hsp70 gene association with nuclear speckles is Hsp70 promoter specific. J Cell Biol. 2010;191:711–719. doi: 10.1083/jcb.201004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jolly C, Vourc’h C, Robert-Nicoud M, Morimoto RI. Intron-independent association of splicing factors with active genes. J Cell Biol. 1999;145:1133–1143. doi: 10.1083/jcb.145.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter KC, Bowman D, Carrington W, Fogarty K, McNeil JA, Fay FS, Lawrence JB. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- 9.Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Q, Kwon YS, Nunez E, Cardamone MD, Hutt KR, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- 12.Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol. 2002;12:439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- 13.Zidovska A, Weitz DA, Mitchison TJ. Micron-scale coherence in interphase chromatin dynamics. Proc Natl Acad Sci U S A. 2013;110:15555–15560. doi: 10.1073/pnas.1220313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 15.Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misteli T, Caceres JF, Clement JQ, Krainer AR, Wilkinson MF, Spector DL. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. The Journal of cell biology. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.