SUMMARY
mTORC1 promotes cell growth in response to nutrients and growth factors. Insulin activates mTORC1 through the PI3K-Akt pathway, which inhibits the TSC1-TSC2-TBC1D7 complex (the TSC complex) to turn on Rheb, an essential activator of mTORC1. However, the mechanistic basis of how this pathway integrates with nutrient-sensing pathways is unknown. We demonstrate that insulin stimulates acute dissociation of the TSC complex from the lysosomal surface, where subpopulations of Rheb and mTORC1 reside. The TSC complex associates with the lysosome in a Rheb-dependent manner, and its dissociation in response to insulin requires Akt-mediated TSC2 phosphorylation. Loss of the PTEN tumor suppressor results in constitutive activation of mTORC1 through the Akt-dependent dissociation of the TSC complex from the lysosome. These findings provide a unifying mechanism by which independent pathways affecting the spatial recruitment of mTORC1 and the TSC complex to Rheb at the lysosomal surface serve to integrate diverse growth signals.
INTRODUCTION
Cells within multicellular organisms simultaneously sense both cell autonomous and systemic growth signals in the form of nutrients and endocrine factors. The ability to properly integrate these signals is key to coordinating the growth of individual cells with the needs of both the local cellular niche and the whole organism. As such, the pathways sensing and relaying the status of cellular growth conditions are frequently dysregulated in common human diseases with underlying genetic and environmental influences, including cancer and diabetes.
The mechanistic target of rapamycin (mTOR) complex 1 (mTORC1) is a highly conserved regulator of cell growth and is one of the most highly integrated signaling nodes present in all cells (Dibble and Manning, 2013; Laplante and Sabatini, 2012). mTORC1 is comprised of the protein kinase mTOR, in complex with two other essential core components, Raptor and mLST8. Upon activation, mTORC1 shifts the metabolic program of the cell from catabolic metabolism to growth-promoting anabolic metabolism, stimulating the synthesis of proteins, lipids, and nucleotides (Howell et al., 2013). As the cellular processes downstream of mTORC1 are costly with respect to the carbon, nitrogen, oxygen, and ATP required, it is not surprising that cells have evolved exquisite mechanisms by which the intracellular availability of nutrients and energy influence the activation state of mTORC1 (Dibble and Manning, 2013). In addition, mTORC1 is regulated by a large variety of secreted factors, including growth factors, cytokines, and hormones, such as insulin and insulin-like growth factor 1 (IGF1), which relay systemic metabolic signals and stimulate signaling cascades upstream of mTORC1. In this manner, mTORC1 responds to diverse local and systemic growth cues to control anabolic metabolism and the growth of cells, tissues, and organisms.
The progress made in understanding how mTORC1 senses these diverse signals stems from the discovery of two classes of Ras-related small G proteins lying directly upstream of mTORC1, the Rag and Rheb GTPases. Rag proteins function as a heterodimer of a RagA or B subunit complexed with a RagC or D subunit and are required for mTORC1 to sense amino acids (Kim et al., 2008; Sancak et al., 2008). The Rag heterodimer is held at the lysosomal surface by a complex of proteins referred to as the Ragulator (Sancak et al., 2010). Importantly, amino acids influence the GTP/GDP-loading state of the RagA/B subunit through the combined action of a GTPase-activating protein (GAP) complex called GATOR1 (Bar-Peled et al., 2013) and a guanine-nucleotide exchange factor (GEF) activity inherent to the Ragulator (Bar-Peled et al., 2012). In the presence of amino acids, the RagA/B subunit is converted to its GTP-bound form, and the Ragulator-Rag complex recruits mTORC1 to the lysosomal surface through direct interactions between the Rag heterodimer and Raptor (Bar-Peled et al., 2013; Bar-Peled et al., 2012; Kim et al., 2008; Sancak et al., 2010; Sancak et al., 2008; Zoncu et al., 2011). This dynamic regulation of mTORC1 localization by amino acid availability, while essential, is not sufficient for the activation of mTORC1, which also requires the presence of Rheb (Sancak et al., 2010). Rheb has been described to localize on multiple endomembrane compartments, including the lysosome, and this is believed to require the C-terminal farnesylation of Rheb (Buerger et al., 2006; Clark et al., 1997; Saito et al., 2005; Sancak et al., 2010; Takahashi et al., 2005). The GTP/GDP-loading state of Rheb is controlled by the presence of secreted growth factors, rather than amino acids, and GTP-bound Rheb is a potent and essential direct activator of mTORC1 (Dibble and Manning, 2013).
Rheb is controlled by a complex of three core proteins, referred to as the TSC complex, comprised of the tuberous sclerosis complex (TSC) tumor suppressors, TSC1 and TSC2, and Tre2-Bub2-Cdc16-1 domain family member 7 (TBC1D7) (Dibble and Manning, 2013). Within the TSC complex, TSC2 acts as a GAP for Rheb and TSC1 scaffolds TSC2 and TBC1D7 together and stabilizes these proteins. Through its Rheb-GAP activity, the TSC complex is an essential inhibitor of mTORC1 signaling, and loss of any of the three components of the complex results in growth factor-independent activation of mTORC1. Importantly, insulin and growth factors activate mTORC1 primarily through the stimulation of a pathway involving class I phosphatidylinositol-3-kinase (PI3K) and its downstream effector Akt, a protein kinase that directly phosphorylates multiple serine and threonine residues on TSC2 within the TSC complex (Inoki et al., 2002; Manning et al., 2002; Potter et al., 2002). The Akt-mediated phosphorylation of all five mapped phosphorylation sites on TSC2 appears to be required for maximal activation of mTORC1 by insulin (Inoki et al., 2002; Zhang et al., 2009). Therefore, insulin signaling negatively regulates the TSC complex to promote the Rheb-dependent activation of mTORC1 (Figure 1A).
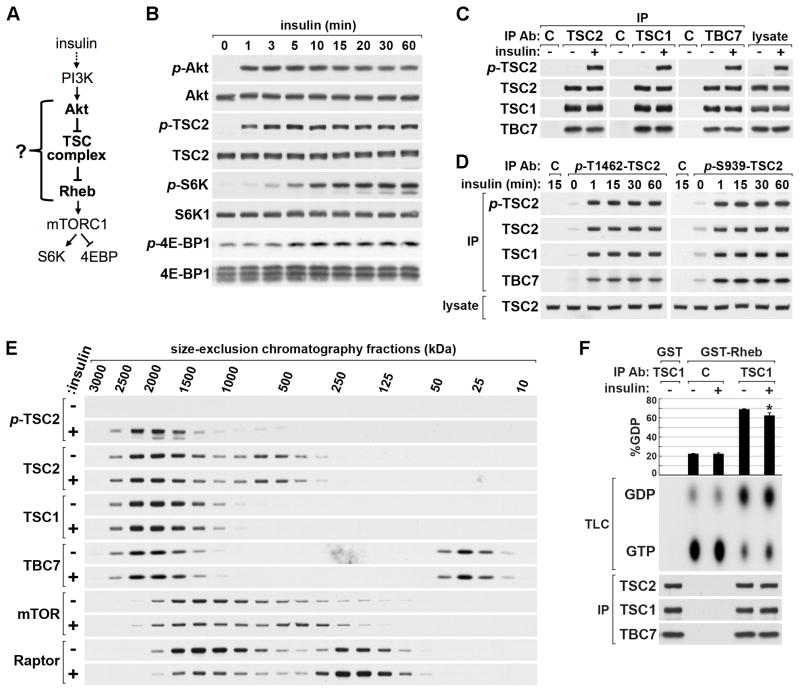
Figure 1. Insulin signaling acutely stimulates mTORC1 without effects on TSC complex stability or a block in Rheb-GAP activity.
(A) Schematic of the Akt-TSC complex-Rheb circuit through which insulin activates mTORC1.
(B) HeLa cells were serum starved then stimulated with a time course of insulin.
(C,D) HeLa cells were serum starved then stimulated with insulin (15 min) prior to lysis and immunoprecipitation with antibodies to TSC2, TSC1, and TBC1D7 (C) or phospho-TSC2-T1462 and phospho-TSC2-S939 (D).
(E) Lysates from cells treated as in (C) were fractionated using size-exclusion chromatography. Estimated molecular weights (kDa) for fractions were calculated from a standard curve (Figure S1E).
(F) Endogenous TSC complexes immunoprecipitated with a TSC1 antibody, or IgG control, from lysates of cells treated as in (C) were subjected to Rheb-GAP assays using recombinant GST or GST-Rheb preloaded with GTP[α-32P]. Rheb-bound GTP and GDP were separated by thin-layer chromatography (TLC). The percentage of conversion to GDP (GDP/GTP + GDP) are graphed as the mean of three independent experiments ±SEM. *p<0.02 compared to TSC1 immunoprecipate from unstimulated cells.
Despite the establishment of the TSC complex and Rheb as the major signaling components regulating the activation state of mTORC1 by growth factors (Figure 1A), the mechanistic basis for this regulation remains poorly defined. It is widely believed that TSC2 phosphorylation by Akt turns off its Rheb-GAP activity, but this has never been shown. Some studies and models have suggested that insulin signaling inhibits the TSC complex by causing disassembly of the complex (Cai et al., 2006; Inoki et al., 2002; Potter et al., 2002), while this has not been observed in other studies (Dong and Pan, 2004; Manning et al., 2002). Additional studies have suggested that the Akt-mediated phosphorylation of TSC2 causes its degradation (Dan et al., 2002; Plas and Thompson, 2003). Finally, how the TSC complex-Rheb circuit is spatially integrated with the recruitment of mTORC1 to the lysosome by the Ragulator-Rag complex in response to amino acids is unknown. Here, through the localization of endogenous proteins, we demonstrate that the TSC complex localizes to the lysosome in the absence of growth factors and acutely dissociates from this location in response to insulin. The lysosomal localization of the TSC complex requires farnesylated Rheb, and the insulin-stimulated release of the TSC complex from the lysosomal surface requires the Akt-mediated phosphorylation of TSC2. Our data demonstrate that this spatial regulation of the TSC complex is independent of the spatial control of mTORC1 and provides a unifying mechanism by which mTORC1 integrates diverse growth signals at the lysosome.
RESULTS
Insulin signaling does not influence TSC complex stability or block its GAP activity
To facilitate an investigation into the molecular nature of signaling through the Akt-TSC complex-Rheb circuit, we first established the dose-dependent and temporal nature of signaling between Akt and mTORC1 in HeLa cells. We found that maximal pathway activation occurs at 1-μM insulin (Figure S1A) and that Akt is rapidly activated (<1 min) with maximal downstream phosphorylation of TSC2 occurring almost simultaneously (Figure 1B). Subsequent activation of mTORC1, as monitored by phosphorylation of its classic substrates S6K1 and 4E-BP1, occurs several minutes later, reaching near maximum at 15 min. Therefore, the mechanism through which Akt acutely inhibits the TSC complex to activate Rheb and mTORC1 must occur within this time frame.
Contradictory to previously proposed models (Cai et al., 2006; Dan et al., 2002; Inoki et al., 2002; Plas and Thompson, 2003; Potter et al., 2002), no effects of acute insulin stimulation on total endogenous TSC2 levels or complex stability were detected, even out to an hour of stimulation (Figure 1B–E, S1B–E). To assess effects on complex integrity, we immunopurified TSC complexes from unstimulated or insulin-stimulated (15 min) cells using antibodies to each component of the complex (Figure 1C) or from cells stimulated with a time course of insulin using a second TSC1 antibody (Figure S1B). Regardless of the antibody used or time point, there was no change in the association of complex components upon insulin stimulation. Additional immunoprecipitations with antibodies that only recognize TSC2 phosphorylated by Akt on either S939 or T1462, confirmed that TSC1 and TBC1D7 readily associate with phosphorylated TSC2 and remain stably bound even after an hour of insulin stimulation (Figure 1D). We also failed to detect effects of insulin signaling and TSC2 phosphorylation on TSC complex stability in MEFs (Figure S1C,D). As a complementary approach to assess effects on the TSC complex, including any higher-order quaternary structure, we used size-exclusion chromatography (Figure 1E). We previously showed, using density-gradient fractionation, that loss of any of the three TSC complex components results in a shift of the remaining subunits to fractions of lower molecular mass (Dibble et al., 2012). Likewise, any effects of insulin signaling on TSC complex integrity should be evident in the shifting of complex components between fractions. In serum-starved cells, TSC2, TSC1, and TBC1D7 co-fractionated with a peak mass of ~2 MDa (as calculated from a standard curve, Figure S1E), representing a complex that is considerably larger than the predicted mass of a single heterotrimeric unit (~365 kDa). Consistent with our previous observations (Dibble et al., 2012), all TSC1 species appear to be part of the TSC complex, whereas smaller pools of both TSC2 and TBC1D7 exist that are not part of the complex, with the TBC1D7 pool fractionating near its predicted mass of ~34 kDa and the TSC2 pool running at an apparent mass of ~600 kDa. Importantly, in lysates from insulin-stimulated cells, TSC2 phosphorylation was largely restricted to TSC2 within the TSC complex, and the fractionation patterns of TSC complex components were identical to those from unstimulated cells, indicating that higher-order complex integrity was unaffected. As a control, we examined the fractionation pattern of the mTORC1 components mTOR and Raptor, which are known to loosen their intermolecular contacts upon activation (Kim et al., 2002). We found that mTOR and Raptor cofractionated at ~1 MDa, consistent with mTORC1 forming a higher-order dimer (Wang et al., 2006; Yip et al., 2010), and a subset of both mTOR and Raptor shifted to lower molecular mass fractions upon insulin stimulation (Figure 1E). Finally, we found that endogenous TSC complexes display a small, but reproducible, decrease in Rheb-GAP activity when isolated from insulin-stimulated cells (Figure 1F). However, the GAP activity of the TSC complex is largely intact upon insulin stimulation. Therefore, using multiple independent approaches, we establish that regulation of the TSC complex by insulin does not involve even small effects on complex stability or the turning off of its GAP activity, suggesting alternative inhibitory mechanisms underlying the robust and rapid downstream activation of mTORC1.
Insulin activates mTORC1 by inducing dissociation of the TSC complex from the lysosome
To determine whether insulin signaling might alter the spatial distribution of the TSC complex, we localized endogenous TSC2, with an antibody validated previously for immunofluorescence (Dibble et al., 2012). The primary localization of TSC2 under basal, serum-starved conditions was found to be at the lysosomal surface, colocalizing with the lysosomal marker LAMP2 (Figure 2A). The remainder of TSC2 localized diffusely to punctate structures throughout the cytosol, exhibiting some small overlap with Golgi (Figure S2A) and no discernable colocalization with markers of either the mitochondria (Figure S2B) or peroxisomes (Figure S2C,D). Strikingly, the extensive TSC2-LAMP2 colocalization observed under serum starvation conditions was acutely and significantly reduced in response to insulin (Figure 2A graphs, Figure S2E immunoblots, and Figure 2B high-resolution confocal imaging).
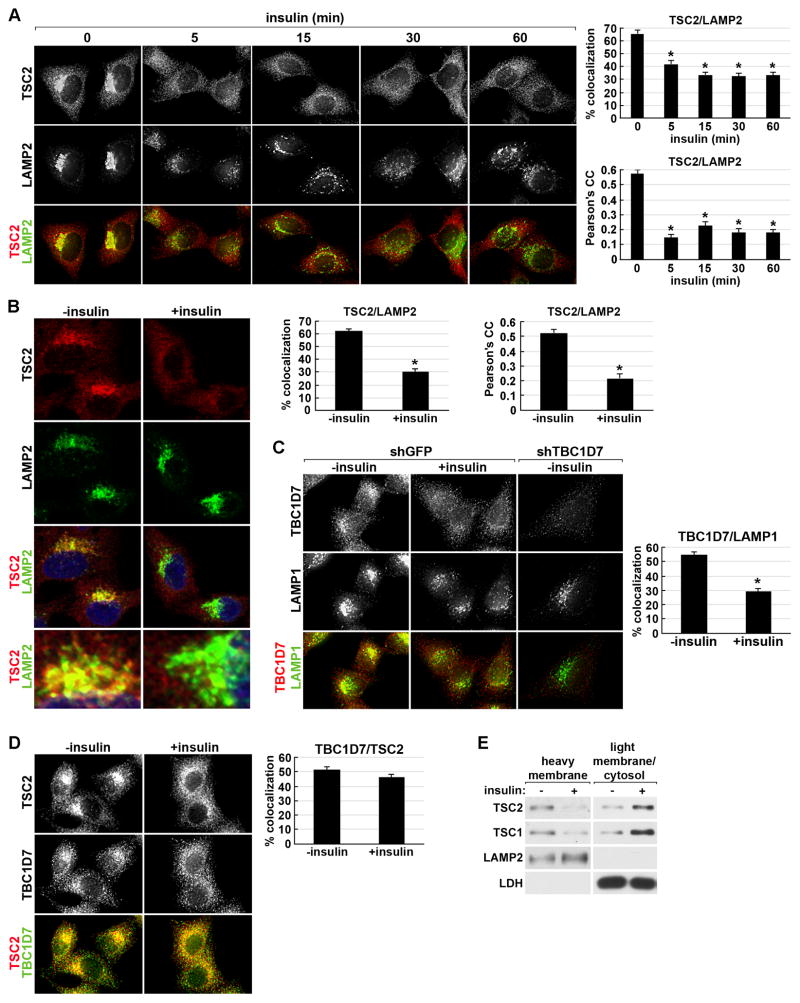
Figure 2. Insulin acutely disrupts the lysosomal localization of the TSC complex.
(A) HeLa cells were serum starved then stimulated with a time course of insulin prior to immunofluorescent labeling of endogenous TSC2 (red) and LAMP2 (green). Representative cells are shown where yellow or orange pixels indicate colocalization in the merged images. Percent colocalization and Pearson’s Correlation Coefficient (PCC) are graphed as a mean±SEM (right). *p< 1X10−6 (% colocalization) and *p< 1X10−12 (PCC), compared to unstimulated (0 min).
(B) Cells were treated and labeled as in (A), but with a single 15-min insulin stimulation, and imaged by confocal microscopy. An enlarged view of the LAMP2-containing compartment from the bottom cells in the two images is shown below. Percent colocalization and PCC are presented as in (A). *p< 1X10−13 (% colocalization) and *p< 1X10−10 (PCC) compared to unstimulated (0 min).
(C) HeLa cells stably expressing a control shRNA (shGFP) or one targeting TBC1D7 (shTBC1D7) were treated as in (B) and labeled for TBC1D7 (red) and LAMP1 (green). Representative cells are shown and percent colocalization is presented as in (A). *p< 1X10−10.
(D) Cells treated as in (B) were labeled for TSC2 (red) and TBC1D7 (green), with percent colocalization presented as in (A).
(E) Cells treated as in (B) and lysates were separated into heavy membrane and light membrane/cytosolic fractions.
Consistent with the dose response of the Akt-TSC complex-mTORC1 pathway to insulin (Figure S1A), increasing doses of insulin stimulated more dissociation of TSC2 from the LAMP2 compartment (Figure S2F). Furthermore, the standard error of the mean for TSC2-LAMP2 colocalization decreased with increasing doses of insulin, indicating the stochastic nature of this response. Hence, the dose-dependency of insulin signaling detected on immunoblots results from both stronger responses within individual cells and more cells simultaneously responding with increasing insulin doses. A similar reduction in TSC2-LAMP1 colocalization was observed upon insulin stimulation in MEFs (Figure S2G). We also noted that insulin stimulation led to a more dispersed cellular distribution of lysosomes in both HeLa cells and MEFs, an effect described previously (Korolchuk et al., 2011). To determine whether the loss of TSC2 from its lysosomal localization is secondary to effects on lysosome clustering, we used nocodazole to physically disperse the lysosomes by disrupting the microtubule cytoskeleton (Figure S2H,I). Unlike insulin stimulation, TSC2-LAMP2 colocalization was unaffected by nocodazole-induced dispersion of the lysosomes. To determine whether it is the entire TSC complex that is being spatially regulated in this manner, we validated (via shRNA knockdown) a TBC1D7 antibody for immunofluorescence and found that under serum starvation conditions a large subpopulation of endogenous TBC1D7 also colocalizes with LAMP1 (Figure 2C). Like TSC2, insulin stimulation acutely reduced the lysosomal localization of TBC1D7. However, insulin did not affect the colocalization of TBC1D7 with TSC2 (Figure 2D), consistent with the fact that their association, which is mediated through their interactions with TSC1 (Dibble et al., 2012), is unaffected by insulin (Figures 1C–E and S1B–E). Finally, in lysates of cells stimulated with insulin for 15 min, both TSC1 and TSC2 levels were reduced in the heavy membrane fraction containing lysosomes, with a concomitant increase in the fraction containing cytosolic and light membrane compartments (Figure 2E), further indicating that it is the intact TSC complex that acutely dissociates from the lysosomal surface in response to insulin.
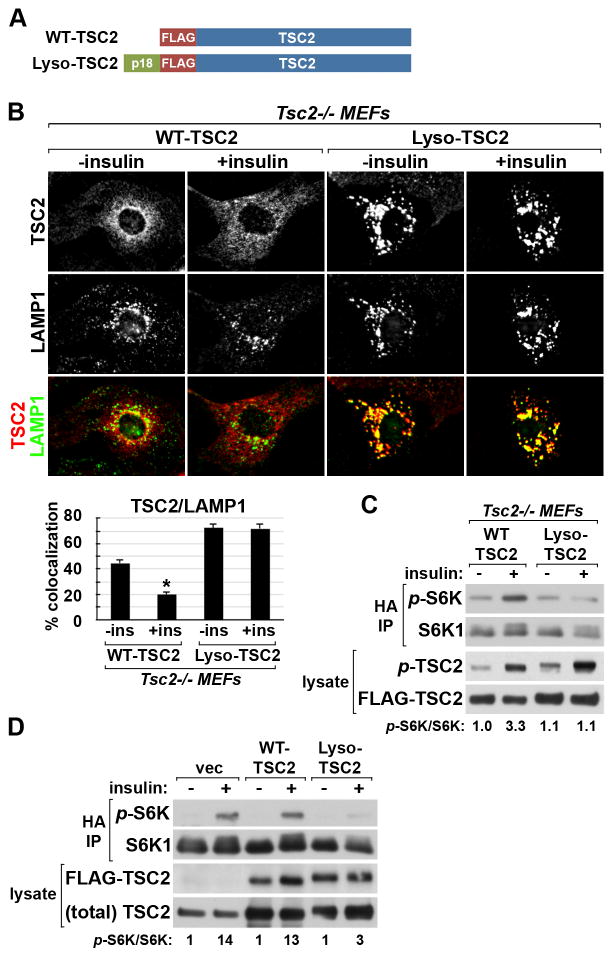
Taken together with studies demonstrating the importance of lysosomal localization for mTORC1 activation, the above findings suggest that the insulin-stimulated dissociation of the TSC complex from the lysosome could represent a mechanism by which insulin signaling activates mTORC1. To further test this idea, we tethered TSC2 to the lysosomal surface through a fusion with the amino-terminus of p18/LAMTOR1 (Lyso-TSC2; Figure 3A), which localizes the Ragulator complex to the lysosome through myristoylation and palmitoylation moieties (Nada et al., 2009; Sancak et al., 2010). Lyso-TSC2 was indistinguishable from wild-type TSC2 in its ability to assemble into the TSC complex (Figure S3A). The constitutive lysosomal localization of this fusion protein was confirmed inTsc2−/− MEFs, where insulin induced the release of exogenously expressed wild-type TSC2 but not Lyso-TSC2 from the lysosome (Figure 3B). Importantly, unlike in wild-type TSC2-expressing cells, insulin failed to stimulate the phosphorylation of T389 on co-expressed S6K1 in the Lyso-TSC2-expressing cells, despite similar phosphorylation of wild-type and Lyso-TSC2 (Figure 3C). Exogenous expression of Lyso-TSC2, but not wild-type TSC2, also attenuated mTORC1 signaling to both endogenous and co-expressed S6K1 in 293E cells grown in full serum (Figure S3B-D) and suppressed the insulin-stimulated phosphorylation of S6K1 (Figure 3D). Therefore, the ability of growth factors to induce dissociation of the TSC complex from the lysosomal surface is required for the proper stimulation of mTORC1 signaling.
Figure 3. Forced targeting of TSC2 to the lysosome suppresses insulin-stimulated mTORC1 signaling.
(A) Schematic of FLAG-tagged WT-TSC2 and Lyso-TSC2, a fusion of TSC2 with the lysosome- targeting signal of p18/LAMTOR1.
(B) Tsc2−/− MEFs were infected with lentiviruses encoding WT-TSC2 or Lyso-TSC2 and were serum starved then insulin stimulated (15 min). Representative TSC2-expressing cells are shown colabeled for TSC2 (red) and LAMP1 (green). Percent colocalization is graphed as the mean±SEM (below). *p< 1X10−12 compared to unstimulated WT-TSC2.
(C) WT-TSC2 or Lyso-TSC2 were cotransfected with HA-S6K1 in Tsc2−/− MEFs, followed by serum starvation and insulin stimulation (100 nM, 15 min). Phosphorylation of the HA-S6K1 reporter was detected in anti-HA immunoprecipitates, and the relative ratios of phospho-S6K1 to total S6K1 are shown normalized to unstimulated WT-TSC2 cells.
(D) 293E cells were cotransfected with empty vector (vec), WT-TSC2, or Lyso-TSC2 and HA-S6K1 and were treated and analyzed as in (C), with relative phospho-S6K1 levels normalized to the unstimulated vector cells.
Insulin and amino acids independently regulate localization of the TSC complex and mTORC1 to the lysosome and Rheb
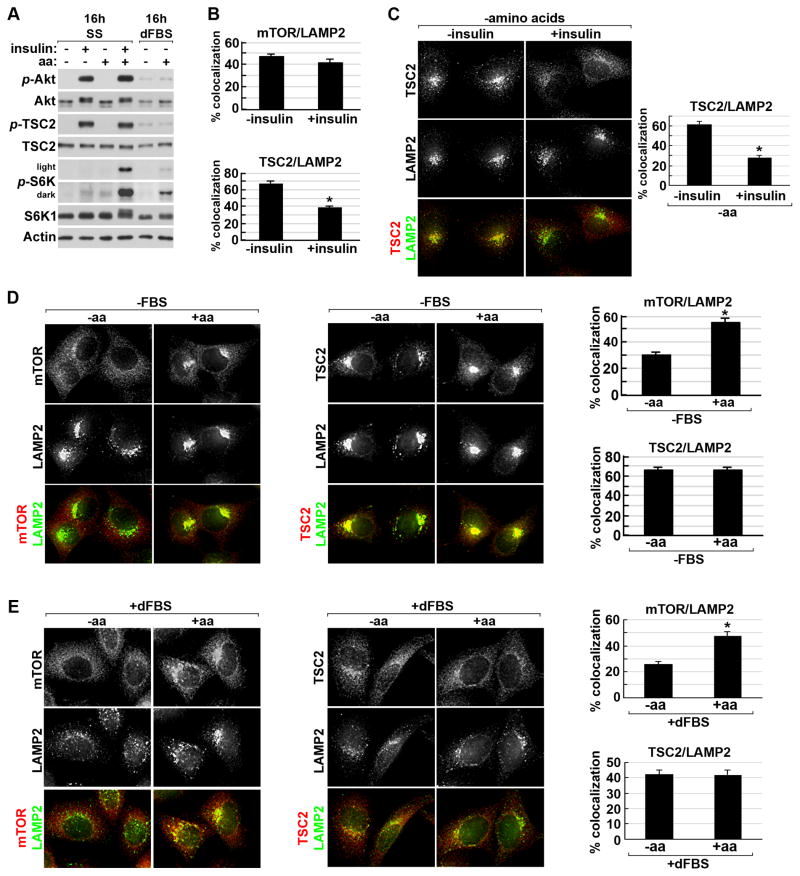
Neither amino acids nor insulin alone are sufficient to robustly stimulate mTORC1 (Hara et al., 1998) (Figure 4A). To understand the relationship between these signals and the spatial regulation of both mTORC1 and the TSC complex to the lysosomal surface, we compared the parallel effects of acute insulin or amino acid stimulation. As described previously (Sancak et al., 2010; Zoncu et al., 2011), the population of mTOR at the lysosome represents mTORC1, as mTOR-LAMP2 colocalization is lost upon knockdown of Raptor (data not shown). Unlike TSC2, the colocalization of mTOR with LAMP2 was unaffected by insulin stimulation (Figure 4B and S4A). The insulin-stimulated release of TSC2 from the LAMP2 compartment was similar in the presence (Figure 4B and S4A) or absence (Figure 4C) of amino acids. As anticipated, mTOR translocated to the lysosome within 10 minutes of refeeding cells with amino acids in either the absence (Figure 4D) or presence (Figure 4E) of serum. In contrast, the lysosomal localization of TSC2 was unaffected by amino acid starvation and acute refeeding under these conditions (Figure 4D,E). Therefore, acute signals from amino acids and insulin independently and reciprocally regulate the association of mTORC1 and the TSC complex with the lysosome.
Figure 4. Reciprocal effects of amino acids and insulin on localization of mTORC1 and the TSC complex to the lysosome.
(A) HeLa cells were grown in the absence (ss) or presence of dialyzed serum (dFBS) and then starved of all amino acids (50 min) prior to stimulation with insulin (15 min), amino acids (10 min), or insulin (15 min) plus amino acids (final 10 min). Light and dark exposures are shown for phospho-S6K1.
(B) HeLa cells were serum starved and stimulated with insulin (15 min) prior to immunofluorescent co-labeling of TSC2 or mTOR with LAMP2 (images shown in Figure S4A). Percent colocalization is graphed as the mean±SEM. *p< 1X10−11.
(C) Cells starved of serum (16 h) and amino acids (50 min) were stimulated with insulin (15 min) prior to co-labeling of TSC2 (red) and LAMP2 (green) and quantification of colocalization as in (B) (graph at right). *p< 1X10−11.
(D) Cells starved of serum (16 h) and amino acids (50 min) were stimulated with amino acids (10 min) prior to labeling and quantification of colocalization as in (B) *p< 1X10−8.
(E) HeLa cells grown in 10% dFBS (16 hr) and starved of amino acids (50 min) were stimulated with amino acids (10 min) prior to labeling and quantification of colocalization as in (B). *p< 1X10−8 See supporting data in Figure S4.
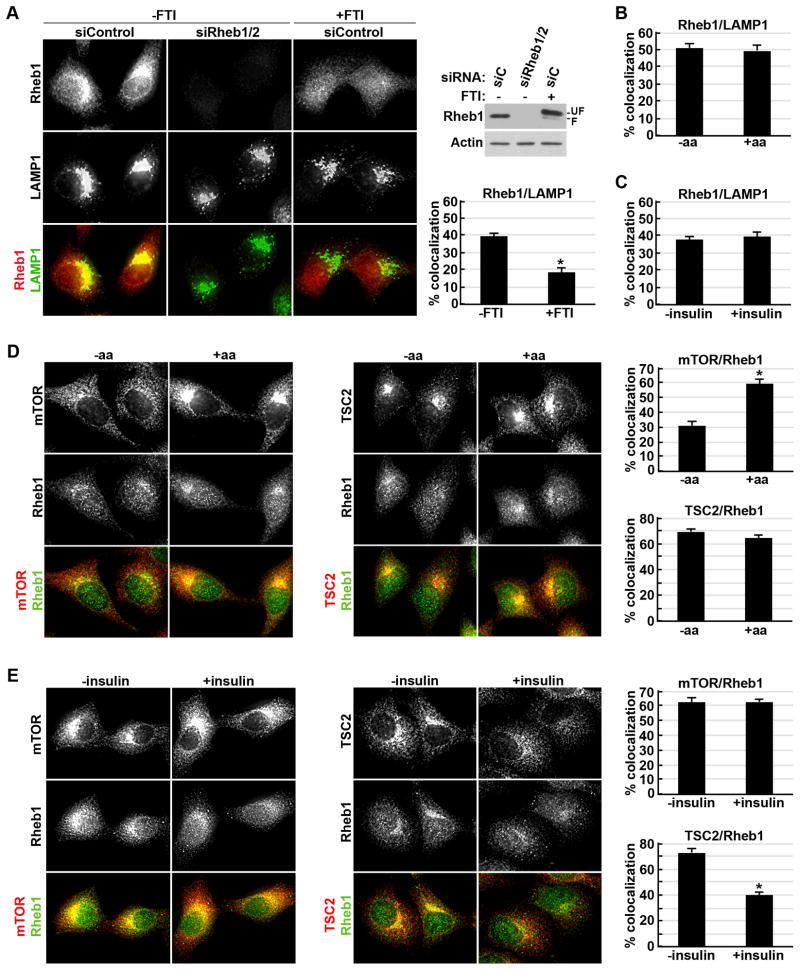
As the TSC complex regulates mTORC1 through Rheb, we sought to characterize the subcellular localization of endogenous Rheb. A Rheb antibody for which the immunostaining pattern was abolished upon siRNA-mediated knockdown of Rheb1 and Rheb2/RhebL1 was identified (Figure 5A). Under serum starvation conditions, endogenous Rheb was detected as punctate labeling throughout the cell with concentrated labeling in a perinuclear region that colocalized with LAMP1, indicating that like mTOR and TSC2, a subset of endogenous Rheb localizes to the lysosome. The lysosomal localization of Rheb requires membrane anchoring through its C-terminal farnesyl group, as a farnesyl-transferase inhibitor (FTI) disrupted the Rheb-LAMP1 colocalization (Figure 5A). The effects of FTI treatment on Rheb farnesylation were evident by a decrease in Rheb mobility in SDS-PAGE, indicating that nearly all cellular Rheb was farnesylated and following FTI treatment ~90% of Rheb became unfarnesylated (Figure 5A, blot).
Figure 5. A subpopulation of Rheb localizes to the lysosome where its colocalization with the TSC complex and mTORC1 is respectively regulated by insulin and amino acids.
(A) HeLa cells with siRNA-mediated knockdown of Rheb1 and 2, or control siRNAs, were serum starved (16 hr) with or without FTI-277 (10 μM) prior to co-labeling of Rheb1 (red) and LAMP1 (green). Representative cells are shown, along with an immunoblot of parallel lysates (right) showing effects on Rheb levels and modification (farnesylated (F) and un-farnesylated (UF)) and the percent colocalization graphed as a mean±SEM. *p< 1X10−7.
(B) HeLa cells starved of serum (16 hr) and amino acids (50 min) were stimulated with amino acids (10 min) prior to co-labeling of Rheb1 and LAMP1 (images in Figure S5A). The percent colocalization is graphed as in (A).
(C) Cells were serum starved then stimulated with insulin (15 min) prior to co-labeling as in (B) (images in Figure S5B). The percent colocalization is graphed as in (A).
(D) Cells were treated as in (B) prior to co-labeling of mTOR or TSC2 (red) and Rheb1 (green). Representative cells are shown, and the percent colocalization is graphed as in (A). *p< 1X10−9.
(E) Cells were treated as in (C) prior to the labeling and analysis described in (D). *p< 1X10−9. See supporting data in Figure S5.
To determine whether the lysosomal localization of Rheb, like that of mTORC1 and the TSC complex, is dynamically regulated, we examined the effects of amino acid and insulin stimulation. Neither amino acid starvation and refeeding (Figures 5B and S5A) nor growth factor withdrawal followed by acute insulin stimulation (Figures 5C and S5B in HeLa and Figure S5C in MEFs) influenced the colocalization of Rheb and LAMP1. Therefore, the lysosomal localization of Rheb is stable and not influenced by cellular growth conditions that alter the activation state of mTORC1. Consistent with the independent spatial regulation of mTORC1 and the TSC complex at the lysosome, described above, acute amino acid stimulation greatly increased mTOR-Rheb colocalization but left TSC2-Rheb colocalization unaffected (Figure 5D), while acute insulin stimulation had no effect on mTOR-Rheb colocalization but greatly diminished TSC2-Rheb colocalization (Figure 5E and S5D for confocal image). These results indicate that converging signals from amino acids and insulin differentially influence the binding and release of mTORC1, the downstream effector of Rheb, and the TSC complex, the upstream regulator of Rheb, from the lysosomal surface where a subpopulation of Rheb resides.
Rheb is required for localization of the TSC complex, but not mTORC1, to the lysosome
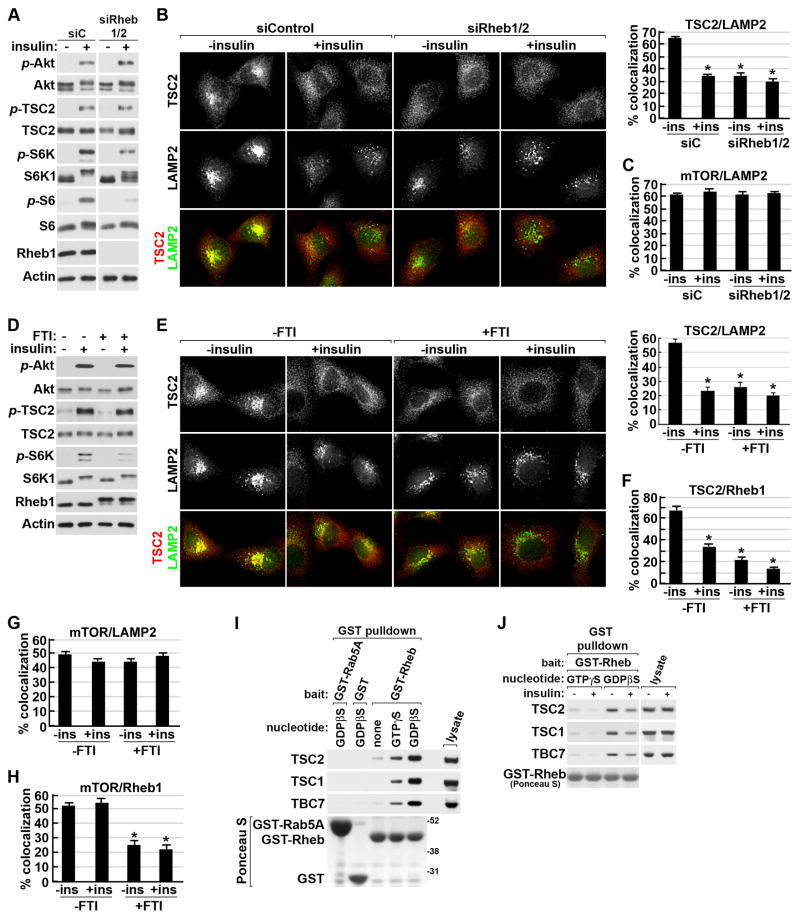
As Rheb is the only protein at the lysosome known to interact with TSC2, we determined whether it was involved in the lysosomal localization of the TSC complex. Upon siRNA-mediated knockdown of Rheb, the insulin-stimulated activation of Akt and its phosphorylation of TSC2 were unaffected, but the expected decrease in mTORC1 signaling was observed (Figure 6A). Interestingly, the perinuclear clustering of TSC2 and its pronounced colocalization with LAMP2, typical of serum starved cells, was disrupted upon Rheb knockdown, resulting in TSC2 localization that resembled the insulin-stimulated state (Figure 6B). This effect was specific to the TSC complex, as mTOR-LAMP2 colocalization was unaffected by Rheb depletion (Figure 6C and S6A). Dissociation of Rheb from the lysosomal membrane by FTI treatment also decreased TSC2-LAMP2 colocalization without effects on Akt activation or TSC2 phosphorylation (Figure 6D and 6E). Consistent with the predominant interactions between the TSC complex and Rheb occurring on the lysosomal surface, FTI treatment also greatly reduced TSC2-Rheb colocalization (Figure 6F and S6B). Association of mTORC1 with the lysosome was not affected by FTI treatment (Figure 6G and S6C), although loss of Rheb from the lysosome was reflected in a decrease in mTOR-Rheb colocalization (Figure 6H and S6D). Therefore, the ability of the TSC complex to associate with the lysosome under growth factor withdrawal conditions is dependent upon Rheb and its proper membrane anchoring, while the lysosomal localization of mTORC1 is independent of Rheb.
Figure 6. Localization of the TSC complex, but not mTORC1, to the lysosome is dependent on farnesylated Rheb.
(A) HeLa cells with siRNA-mediated knockdown of Rheb1 and 2, or control siRNAs (siC), were serum starved then stimulated with insulin (100 nM, 15 min). Note: the siC and siRheb1/2 samples are from the same blot and exposure.
(B) Cells treated as in (A) were co-labeled for TSC2 (red) and LAMP2 (green). Representative cells are shown and percent colocalization is graphed as a mean±SEM (right). *p< 1X10−12 for comparison with unstimulated siC cells.
(C) Cells treated as in (A) were co-labeled for mTOR and LAMP2 (images in Figure S6A) and quantified as in (B).
(D) HeLa cells serum starved (16 hr) with or without FTI-277 (FTI; 10 μM) were left unstimulated or were stimulated with insulin (15 min).
(E) Cells were treated as in (D) prior to labeling and quantification of colocalization as in (B) (graph to right). *p< 1X10−10 for comparison with untreated, serum-starved cells.
(F) Cells treated as in (D) were co-labeled for TSC2 and Rheb1 (images in Figure S6B). Colocalization was quantified as in (B). *p< 1X10−9 for comparison with untreated cells.
(G) Cells treated as in (D) were co-labeled for mTOR and LAMP2 (images in Figure S6C). Colocalization was quantified as in (B).
(H) Cells treated as in (D) were co-labeled for mTOR and Rheb1 (images in Figure S6D). Colocalization was quantified as in (B). *p< 1X10−8 for comparison with untreated cells.
(I) Endogenous TSC complex components were pulled down from lysates of serum starved HeLa cells with recombinant purified GST-Rheb preloaded with no nucleotide, GTPγS, or GDPβS compared to control pull downs with GST or GST-Rab5A preloaded with GDPβS. GST-fused bait proteins were detected with Ponceau S stain.
(J) GST-Rheb pulldowns were performed as in (I), but cells were stimulated with insulin (15 min). See supporting data in Figure S6.
To further characterize the molecular nature of the interaction between Rheb and the TSC complex, we determined how the nucleotide-binding state of Rheb influenced this interaction. Interestingly, we found that the endogenous TSC complex binds most strongly to the GDP-loaded form of Rheb (Figure 6I), a property unusual amongst Ras-family GAPs (see discussion). This binding was dependent on TSC2 (Figure S6E) and sensitive to insulin stimulation, which weakened the ability of endogenous TSC complexes to bind both GDP- and GTP-bound Rheb (Figure 6J). These data suggest a regulated interaction with Rheb contributes to the spatial control of the TSC complex.
The PI3K-Akt pathway induces dissociation of the TSC complex from the lysosome through phosphorylation of TSC2
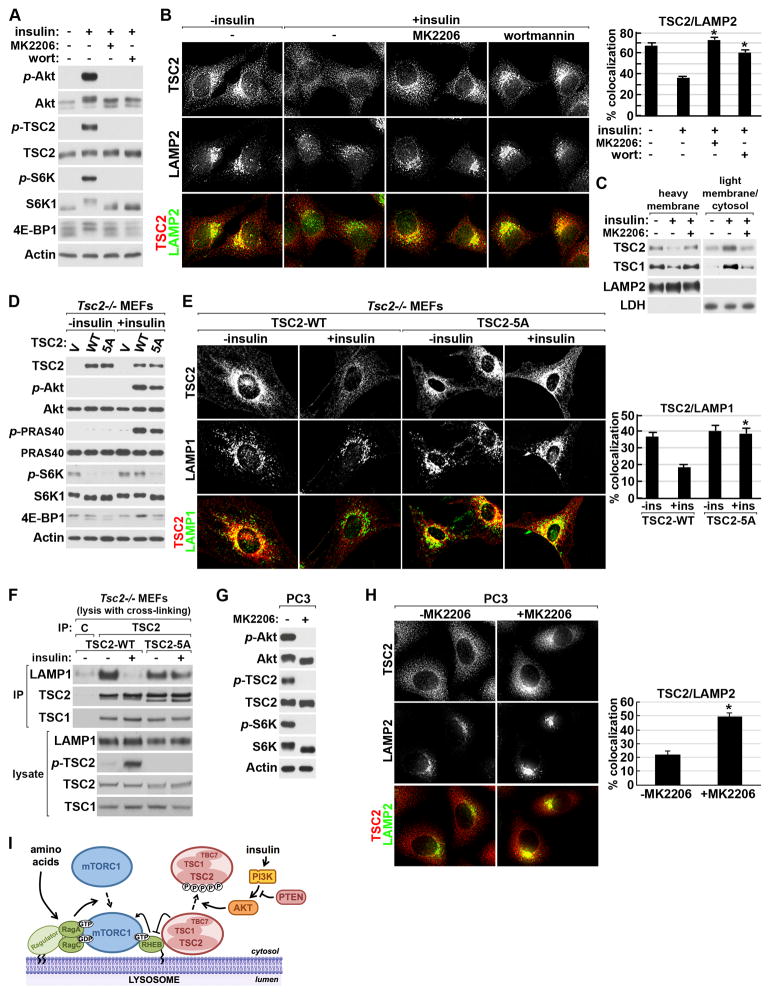
In response to insulin, the direct phosphorylation of TSC2 by Akt occurs in the same acute fashion as dissociation of the TSC complex from the lysosome (see Figures 1B, 2A, S2E and S2G), suggesting involvement of this modification in the spatial regulation of the complex. Brief pretreatment of cells with the PI3K inhibitor wortmannin or the Akt-specific inhibitor MK2206 completely blocked insulin-stimulated Akt activation, TSC2 phosphorylation, and mTORC1 signaling in both HeLa cells and MEFs (Figure 7A and S7A). Importantly, this loss of insulin-stimulated mTORC1 signaling coincided with an inability of insulin to induce the dissociation of TSC2 from the lysosome in these cells (Figure 7B and S7B). Likewise, the insulin-induced movement of TSC1 and TSC2 from heavy membrane fractions to those containing cytosol and light membranes was blocked by Akt inhibition (Figure 7C). The weakened association of the TSC complex with recombinant Rheb-GDP upon insulin stimulation was also attenuated by inhibition of Akt (Figure S7C).
Figure 7. Akt-mediated phosphorylation of TSC2 results in dissociation of the TSC complex from the lysosome in response to insulin or PTEN loss.
(A) Serum starved HeLa cells were pretreated (30 min) with vehicle (DMSO), wortmannin (100 nM), or MK2206 (2 μM) and then stimulated with insulin (15 min).
(B) Cells treated as in (A) were co-labeled for TSC2 (red) and LAMP2 (green). Representative cells are shown and the percent colocalization is graphed as a mean±SEM. *p< 1X10−8 for comparison with insulin stimulation in vehicle-treated cells.
(C) Cells were treated as in (A) and lysates were separated into heavy membrane and light membrane/cytosolic fractions.
(D) Tsc2−/− MEFs expressing empty vector (V), wild-type TSC2 (WT), or the Akt-phosphorylation-site mutant of TSC2 (5A) were serum starved and stimulated with insulin (15 min).
(E) Tsc2−/− MEFs were treated as in (D) prior to co-labeling for reconstituted TSC2 (red) and endogenous LAMP1 (green). Colocalization was quantified as in (B). *p< 1X10−8 for comparison with insulin-stimulated, TSC2-WT-expressing cells.
(F) Tsc2−/− MEFs reconstituted with wild-type TSC2 (WT) or mutant TSC2 (5A) were treated as in (D) prior to lysis in hypotonic buffer with chemical crosslinking followed by immunoprecipitation with protein A/G agarose alone (C) or with TSC2 antibody.
(G) Serum starved (16 hr) PC3 cells were treated (30 min) with vehicle (DMSO) or MK2206 (2 μM).
(H) PC3 cells were treated as in (G) prior to co-labeling for TSC2 (red) and LAMP2 (green). Colocalization was quantified as in (B). *p< 1X10−10.
(I) Model of the spatial regulation of mTORC1 and the TSC complex at the lysosome. See text for details. See supporting data in Figure S7.
Previous studies in wild-type (Inoki et al., 2002) or Tsc2 null (Zhang et al., 2009) cells have indicated that phosphorylation of all five Akt-targeted residues on TSC2 is required for full activation of mTORC1 signaling in response to insulin. To test the role of these phosphorylation events in the spatial regulation of the TSC complex, we reconstituted Tsc2 null MEFs with wild-type TSC2 (TSC2-WT) or TSC2 lacking these 5 phospho-acceptor sites (TSC2-5A). In TSC2-WT-expressing cells, the growth factor-independent activation of mTORC1 signaling was blocked and its sensitivity to insulin was restored (Figure 7D). In TSC2-5A-expressing cells, the constitutive activation of mTORC1 was similarly blocked, but mTORC1 signaling became unresponsive to insulin, despite proper activation of Akt and signaling to other Akt targets, such as PRAS40. Consistent with the proper suppression of mTORC1 signaling in the absence of growth factors, a similar percentage of TSC2-WT and TSC2-5A were found to associate with the lysosome under serum starvation conditions (Figure 7E). As with endogenous TSC2 in wild-type MEFs (Figure S2G), the lysosomal localization of TSC2-WT was disrupted by insulin stimulation.
However, TSC2-5A association with the lysosome remained unchanged upon insulin stimulation (Figure 7E). To further examine effects of the Akt sites on TSC2 for dissociation of the TSC complex from the lysosomal surface, we immunopurified TSC2-associated complexes from reconstituted Tsc2−/− cells that were lysed in the presence of a cross-linking agent. Consistent with our immunofluorescence findings, insulin stimulated release of TSC2-WT, but not TSC2-5A, from the compartment containing LAMP1 (Figure 7F). It is worth noting that we do not detect association of the TSC complex with LAMP1 or 2 under standard lysis conditions, suggesting lack of a direct interaction with these lysosomal membrane proteins. These data demonstrate that the insulin-induced dissociation of the TSC complex from the lysosome and subsequent Rheb-dependent activation of mTORC1 requires the phosphorylation of TSC2 by Akt, thereby providing a mechanism of TSC complex inhibition by the PI3K-Akt pathway.
The PI3K-Akt pathway is frequently activated in human cancers, and this commonly occurs through loss of the PTEN tumor suppressor. Consistent with constitutive PI3K signaling, PTEN-deficient prostate cancer cells (PC3, Figure 7G) and MEFs (Figure S7D) displayed growth factor-independent activation of Akt, phosphorylation of TSC2, and mTORC1 activation, signaling events that were completely blocked by Akt inhibition. Importantly, these cells displayed dissociation of TSC2 from the lysosome even under serum starvation conditions, and Akt inhibition led to rapid translocation of TSC2 to the lysosome, demonstrating the acutely reversible nature of this regulation (Figure 7H and S7E). Therefore, in addition to physiological stimuli, common pathological mutations leading to growth factor-independent activation of the PI3K-Akt pathway in cancer induce mTORC1 signaling through dissociation of the TSC complex from the lysosomal surface.
DISCUSSION
Signals from amino acids and growth factors are both necessary, but not sufficient, for full activation of mTORC1 (Dibble and Manning, 2013; Hara et al., 1998; Laplante and Sabatini, 2012). Our findings suggest a unifying model for integrated co-regulation of mTORC1 by these inputs (Figure 7I). It has been established that the amino acid signal through the Ragulator and Rag proteins serves to localize mTORC1 to the lysosome without directly stimulating its activation (Kim et al., 2008; Sancak et al., 2010; Sancak et al., 2008). Localization of mTORC1 to the lysosome brings it into proximity with its essential activator Rheb (Sancak et al., 2010), which in its GTP-bound form can directly stimulate mTORC1 kinase activity (Sancak et al., 2007). Previous studies have localized exogenously expressed Rheb fusion proteins to endomembrane compartments, including the ER, golgi, and lysosomes (Buerger et al., 2006; Clark et al., 1997; Saito et al., 2005; Sancak et al., 2010; Takahashi et al., 2005). We find that a subpopulation of endogenous Rheb does indeed localize, through its farnesyl lipid modification, to the same LAMP1/2-containing compartment as mTORC1, and this localization is unaffected by amino acids or insulin. The only currently established direct regulator of Rheb is the TSC complex, which we find localizes to the lysosome in the absence of growth factors, at least in part, through its association with Rheb. In contrast to previous studies using the isolated GAP domain of TSC2 (Castro et al., 2003; Long et al., 2005), we found that endogenous TSC2 within the intact TSC complex has a strong binding preference for Rheb-GDP, a property that is atypical for GAP proteins, which normally bind preferentially to the GTP-bound or transition states of their small G protein targets (Bos et al., 2007; Chen et al., 2011; Daumke et al., 2004). Our data are consistent with a model where under growth factor withdrawal conditions, the TSC complex stimulates the intrinsic GTPase activity of Rheb on the lysosomal surface, and through the conversion to Rheb-GDP creates a strong binding partner that holds the complex at this location. Whether the TSC complex also acts as a guanine-nucleotide dissociation inhibitor (or GDI) in this state, thereby preventing nucleotide exchange on Rheb, will be interesting to explore in future studies. Such a model would also help explain the rapid accumulation of the TSC complex at the lysosome and inhibition of mTORC1 signaling that we observe upon inhibition of the PI3K-Akt pathway. We show that insulin acutely stimulates dissociation of the TSC complex from Rheb at the lysosomal surface and that this requires the direct phosphorylation of TSC2 by Akt. This induced release of the TSC complex from lysosomal Rheb allows Rheb-GTP loading and is required for the proper activation of mTORC1 by insulin.
In contrast to previously suggested models (Cai et al., 2006; Dan et al., 2002; Inoki et al., 2002; Plas and Thompson, 2003; Potter et al., 2002), we demonstrate through a variety of biochemical and immunofluorescence assays with endogenous TSC complexes that acute insulin stimulation has no effect on TSC complex integrity within the time frame in which mTORC1 is fully activated. It remains possible that there are longer-term effects of growth factors on complex assembly or stability. However, such effects have not been detected in time courses out to 12 hours of stimulation (Manning et al., 2002) and cannot underlie the rapid Akt-dependent activation of mTORC1 by insulin and other growth factors. In our biochemical characterization of the TSC complex, we found the endogenous complex to be an estimated 2 MDa in size, much larger than the predicted mass of a single TSC1-TSC2-TBC1D7 unit (365 kDa), but in agreement with evidence of multiple TSC1 and TSC2 subunits within the quaternary structure (Hoogeveen-Westerveld et al., 2012b). This size is consistent with the heterotrimeric unit, which we fail to detect in cell lysates, perhaps forming a higher order pentameric structure with five of each component. Coincidently, pentameric arrangements were observed in recent crystal structures of the N-terminal domain of yeast TSC1 (Sun et al., 2013).
Another misconception about TSC complex regulation was that growth factor signaling acts primarily through inhibitory effects on the GAP activity of TSC2. GAP assays on endogenous TSC complexes did indeed reveal a minor, but significant and reproducible, reduction in GAP activity of complexes from insulin-stimulated cells. However, following insulin stimulation, the GAP activity of the TSC complex is largely intact, indicating that phosphorylation of TSC2 on Akt-targeted sites does not constitute an on/off switch for its GAP activity. As we found that insulin and Akt signaling weaken the ability of the TSC complex to associate with Rheb even in a cell-free system, the slight decrease in GAP activity measured in these assays might reflect this decreased binding efficiency. Akt signaling exerts a stronger effect on release of the TSC complex from Rheb and the lysosomes in intact cells, suggesting that more complex physical interactions are involved in this spatial regulation. Given our data that the TSC complex exists as a large oligomeric structure, it seems likely that the higher order complex interacts with multiple Rheb proteins on the lysosomal surface simultaneously and, perhaps, cooperatively. While we find that Rheb is required for the lysosomal localization of the TSC complex, it is possible that interactions with other lysosomal surface proteins or lipids also contribute to this localization and regulated release. Currently, the organizational structure of the TSC complex is completely unknown, but it is worth noting that in the primary structure of TSC2, the five Akt phosphorylation sites lie outside of its GAP domain. Structural information on how the domains of the three proteins are spatially positioned within the oligomeric complex is required to understand the molecular contributions of the individual Akt phosphorylation sites, and other regulatory sites, to disrupting the binding interactions with Rheb and possibly other constituents of the lysosomal membrane.
A vast network of oncogenes and tumor suppressors lie upstream of mTORC1, and its activation is common in genetic tumor syndromes and sporadic cancers (Menon and Manning, 2009). While loss of function mutations in GATOR1, upstream of the Rag GTPases (Bar-Peled et al., 2013), can contribute to tumorigenesis, the majority of genetic events leading to aberrant activation of mTORC1 in human tumors either directly or indirectly affect the TSC complex. TSC1 and TSC2 are themselves tumor suppressors that, while more rarely mutated in sporadic cancers, are disrupted in the cancer-like syndromes tuberous sclerosis complex (TSC) and lymphangioleiomyomatosis (LAM), which are characterized by widespread tumors exhibiting elevated mTORC1 signaling (Crino et al., 2006). Additionally, loss of TBC1D7 gives rise to a megalencephaly syndrome accompanied by increased mTORC1 signaling (Capo-Chichi et al., 2013). In TSC and LAM patients, over a hundred distinct missense mutations or in frame deletions have been identified in TSC2 alone (Hoogeveen-Westerveld et al., 2012a; Hoogeveen-Westerveld et al., 2013; Nellist et al., 2005). It seems likely that the pathogenic mutations that do not affect TSC complex stability or GAP activity might influence proper localization of the complex to the lysosomal surface where it regulates Rheb. In the majority of sporadic cancers with elevated mTORC1 signaling, frequently activated oncogenic signaling pathways, such as the PI3K-Akt pathway, activate mTORC1, at least in part, by promoting constitutive dissociation of the TSC complex from the lysosome. Reciprocally, pharmacological compounds targeting these upstream pathways would drive the TSC complex to Rheb at the lysosome and inhibit mTORC1 activation.
The recognition of this spatial regulation as a major mechanism of control over the TSC complex and Rheb provides a valuable tool for characterizing the mechanistic basis for newly identified upstream regulators, mutations, or compounds found to influence mTORC1 signaling. In this context, investigators can take advantage of the independent mechanisms affecting the translocation of mTORC1 and the TSC complex to Rheb at the lysosome. Altered lysosomal localization of mTOR would indicate effects on the Ragulator-Rag branch, while altered localization of TSC2 would indicate effects upstream of the TSC complex or on Rheb itself. For instance, decreases in cellular ATP levels inhibit mTORC1 signaling through a variety of independent pathways, including those involving AMPK (Dibble and Manning, 2013). We found that the widely-used AMPK-activating compound, A769662 (Cool et al., 2006), blocks the insulin-stimulated dissociation of the TSC complex from the lysosome and inhibits mTORC1 activation, without effects on mTOR localization (data not shown). However, using cells with knockout or knockdown of AMPKα1 and α2, we were surprised to find that the effects of this compound on mTORC1 signaling are independent of AMPK. Interestingly, a recent study suggested that eccentric muscle contractions activate mTORC1 and that this correlates with a decrease in the colocalization of TSC2 and LAMP2 in mouse tissue sections (Jacobs et al., 2013). Finally, our findings suggest that spatial regulation of GAP proteins may be a general control mechanism for other classes of small G proteins, where the GAP proteins are acutely regulated by phosphorylation without known effects on GAP activity, including several direct targets of Akt (Chen et al., 2011; Miinea et al., 2005).
EXPERIMENTAL PROCEDURES
Cell Culture
Cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) with 10% fetal bovine serum (FBS). For amino acid-stimulation experiments, subconfluent cells were starved for 50 min in DMEM lacking all amino acids, with or without 10% dialyzed FBS (dFBS), followed by replacement with either fresh amino acid-free DMEM or standard DMEM for 10 min, with or without dFBS. For insulin stimulation, subconfluent cells were serum starved in DMEM (16 hr) and stimulated with 1-μM insulin for 15 min, unless otherwise noted. Further details on cell lines and reagents are provided in the Extended Experimental Procedures.
Immunofluorescence
Cells grown on glass coverslips were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton x-100, and blocked with Odyssey blocking buffer, all in PBS. Coverslips were incubated with primary antibody overnight at 4°C, incubated with labeled secondary antibodies for 1 h at room temperature and mounted. Images were acquired through 63X or 40X oil immersion objectives with either a Zeiss Axiotome fluorescence microscope with Apotome feature engaged or a Zeiss LSM 510META confocal laser microscope. Representative cells are shown in all figures at the same exposure and magnification. Quantitative analyses were done using Axiovision (for pseudo-confocal images) or Volocity (for confocal images) software, with calculation of thresholded Pearson’s correlation coefficients and colocalization percentages. The latter is derived from the number of pixels in the red channel that overlap with pixels from the green channel divided by the total number of pixels detected in the red channel above the threshold value (x 100). Identical settings were used to capture images across five separate fields (25 to 50 cells) per condition, with the data presented being representative of at least two independent experiments. Antibody details and methods for image acquisition and analyses are provided in the Extended Experimental Procedures.
DNA Constructs, RNAi, and Biochemical Analyses
Details regarding all constructs and RNAi reagents and their introduction into cells, antibodies used for immunoblotting and immunoprecipitation, and detailed biochemical methods are provided in the Extended Experimental Procedures.
Statistics
Data are expressed as mean ± SEM. Significance of differences in GAP assays and colocalization experiments were determined using an unpaired two-tailed Student’s t test assuming equal variance.
Supplementary Material
Highlights.
Insulin triggers acute dissociation of the TSC complex from Rheb at the lysosome
Release of the TSC complex from the lysosome is required to activate mTORC1
Dissociation of TSC complex from the lysosome requires Akt phosphorylation of TSC2
The TSC complex is constitutively dissociated from the lysosome in PTEN-null cells
Acknowledgments
We thank J. Howell for critical comments on the manuscript, S.G. Kim and J. Blenis for antibody advice, L. Bar-Peled and D.M. Sabatini for the p18-N-terminal construct, D.J. Kwiatkowski for the Tsc2−/− MEFs, and P.P. Pandolfi for the Pten−/− MEFs. This work was supported by NIH grants T32-HL007893 (C.C.D.), R01-CA122617 (B.D.M.), and P01-CA120964 (B.D.M. and L.C.C.), and The Ellison Medical Foundation (B.D.M.).
Footnotes
Supplemental information includes Extended Experimental Procedures and seven supplemental figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem Biophys Res Commun. 2006;344:869–880. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- Cai SL, Tee AR, Short JD, Bergeron JM, Kim J, Shen J, Guo R, Johnson CL, Kiguchi K, Walker CL. Activity of TSC2 is inhibited by AKT-mediated phosphorylation and membrane partitioning. J Cell Biol. 2006;173:279–289. doi: 10.1083/jcb.200507119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-Chichi JM, Tcherkezian J, Hamdan FF, Decarie JC, Dobrzeniecka S, Patry L, Nadon MA, Mucha BE, Major P, Shevell M, et al. Disruption of TBC1D7, a subunit of the TSC1-TSC2 protein complex, in intellectual disability and megalencephaly. J Med Genet. 2013 doi: 10.1136/jmedgenet-2013-101680. [DOI] [PubMed] [Google Scholar]
- Castro AF, Rebhun JF, Clark GJ, Quilliam LA. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J Biol Chem. 2003;278:32493–32496. doi: 10.1074/jbc.C300226200. [DOI] [PubMed] [Google Scholar]
- Chen XW, Leto D, Xiong T, Yu G, Cheng A, Decker S, Saltiel AR. A Ral GAP complex links PI 3-kinase/Akt signaling to RalA activation in insulin action. Mol Biol Cell. 2011;22:141–152. doi: 10.1091/mbc.E10-08-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GJ, Kinch MS, Rogers-Graham K, Sebti SM, Hamilton AD, Der CJ. The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J Biol Chem. 1997;272:10608–10615. doi: 10.1074/jbc.272.16.10608. [DOI] [PubMed] [Google Scholar]
- Cool B, Zinker B, Chiou W, Kifle L, Cao N, Perham M, Dickinson R, Adler A, Gagne G, Iyengar R, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell metabolism. 2006;3:403–416. doi: 10.1016/j.cmet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355:1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- Dan HC, Sun M, Yang L, Feldman RI, Sui XM, Ou CC, Nellist M, Yeung RS, Halley DJ, Nicosia SV, et al. Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J Biol Chem. 2002;277:35364–35370. doi: 10.1074/jbc.M205838200. [DOI] [PubMed] [Google Scholar]
- Daumke O, Weyand M, Chakrabarti PP, Vetter IR, Wittinghofer A. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature. 2004;429:197–201. doi: 10.1038/nature02505. [DOI] [PubMed] [Google Scholar]
- Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Pan D. Tsc2 is not a critical target of Akt during normal Drosophila development. Genes Dev. 2004;18:2479–2484. doi: 10.1101/gad.1240504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Hoogeveen-Westerveld M, Ekong R, Povey S, Karbassi I, Batish SD, den Dunnen JT, van Eeghen A, Thiele E, Mayer K, Dies K, et al. Functional assessment of TSC1 missense variants identified in individuals with tuberous sclerosis complex. Hum Mutat. 2012a;33:476–479. doi: 10.1002/humu.22007. [DOI] [PubMed] [Google Scholar]
- Hoogeveen-Westerveld M, Ekong R, Povey S, Mayer K, Lannoy N, Elmslie F, Bebin M, Dies K, Thompson C, Sparagana SP, et al. Functional assessment of TSC2 variants identified in individuals with tuberous sclerosis complex. Hum Mutat. 2013;34:167–175. doi: 10.1002/humu.22202. [DOI] [PubMed] [Google Scholar]
- Hoogeveen-Westerveld M, van Unen L, van den Ouweland A, Halley D, Hoogeveen A, Nellist M. The TSC1-TSC2 complex consists of multiple TSC1 and TSC2 subunits. BMC Biochem. 2012b;13:18. doi: 10.1186/1471-2091-13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell JJ, Ricoult SJ, Ben-Sahra I, Manning BD. A growing role for mTOR in promoting anabolic metabolism. Biochem Soc Trans. 2013;41:906–912. doi: 10.1042/BST20130041. [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Jacobs BL, You JS, Frey JW, Goodman CA, Gundermann DM, Hornberger TA. Eccentric Contractions Increase TSC2 Phosphorylation and Alter the Targeting of TSC2 and mTOR to the Lysosome. J Physiol. 2013 doi: 10.1113/jphysiol.2013.256339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Saiki S, Lichtenberg M, Siddiqi FH, Roberts EA, Imarisio S, Jahreiss L, Sarkar S, Futter M, Menzies FM, et al. Lysosomal positioning coordinates cellular nutrient responses. Nat Cell Biol. 2011;13:453–460. doi: 10.1038/ncb2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR Signaling in Growth Control and Disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2009;27:S43–S51. doi: 10.1038/onc.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miinea CP, Sano H, Kane S, Sano E, Fukuda M, Peranen J, Lane WS, Lienhard GE. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem J. 2005;391:87–93. doi: 10.1042/BJ20050887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada S, Hondo A, Kasai A, Koike M, Saito K, Uchiyama Y, Okada M. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 2009;28:477–489. doi: 10.1038/emboj.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellist M, Sancak O, Goedbloed MA, Rohe C, van Netten D, Mayer K, Tucker-Williams A, van den Ouweland AM, Halley DJ. Distinct effects of single amino-acid changes to tuberin on the function of the tuberin-hamartin complex. Eur J Hum Genet. 2005;13:59–68. doi: 10.1038/sj.ejhg.5201276. [DOI] [PubMed] [Google Scholar]
- Plas DR, Thompson CB. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J Biol Chem. 2003;278:12361–12366. doi: 10.1074/jbc.M213069200. [DOI] [PubMed] [Google Scholar]
- Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- Saito K, Araki Y, Kontani K, Nishina H, Katada T. Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. J Biochem. 2005;137:423–430. doi: 10.1093/jb/mvi046. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Sun W, Zhu YJ, Wang Z, Zhong Q, Gao F, Lou J, Gong W, Xu W. Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations. Nat Commun. 2013;4:2135. doi: 10.1038/ncomms3135. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Nakagawa M, Young SG, Yamanaka S. Differential membrane localization of ERas and Rheb, two Ras-related proteins involved in the phosphatidylinositol 3-kinase/mTOR pathway. J Biol Chem. 2005;280:32768–32774. doi: 10.1074/jbc.M506280200. [DOI] [PubMed] [Google Scholar]
- Wang L, Rhodes CJ, Lawrence JC., Jr Activation of mammalian target of rapamycin (mTOR) by insulin is associated with stimulation of 4EBP1 binding to dimeric mTOR complex 1. J Biol Chem. 2006;281:24293–24303. doi: 10.1074/jbc.M603566200. [DOI] [PubMed] [Google Scholar]
- Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell. 2010;38:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HH, Huang J, Duvel K, Boback B, Wu S, Squillace RM, Wu CL, Manning BD. Insulin stimulates adipogenesis through the Akt-TSC2-mTORC1 pathway. PLoS One. 2009;4:e6189. doi: 10.1371/journal.pone.0006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.