Abstract
During interphase, Rab11-GTPase-containing endosomes recycle endocytic cargo. However, little is known about Rab11 and endosomes in mitosis. Here we show that Rab11 localizes to the mitotic spindle and regulates dynein-dependent endosome localization at poles. We found that mitotic recycling endosomes bind γ-TuRC components and associate with tubulin in vitro. Rab11-depletion or dominant-negative Rab11 expression disrupts astral microtubules, delays mitosis, and redistributes spindle pole proteins. Reciprocally, constitutively-active Rab11 increases astral microtubules, restores γ-tubulin spindle pole localization and generates robust spindles. This suggests a fundamental role for Rab11 activity in spindle pole maturation during mitosis. Rab11 depletion causes misorientation of the mitotic spindle and the plane of cell division. These findings suggest a molecular mechanism for the organization of astral microtubules and the mitotic spindle through Rab11-dependent control of spindle pole assembly and function. We propose that Rab11 and its associated endosomes co-contribute to these processes through retrograde transport to poles by dynein.
Keywords: Rab11, centrosome, spindle orientation, endosomes, γ-tubulin, Ran GTPase, Dynein, Astral Microtubules, Spindle Assembly Checkpoint, microtubules, mitosis, FIP3, spindle maturation, spindle assembly
INTRODUCTION
We recently showed that the recycling endosome (RE) GTPase, Rab11, binds to mother centriole appendages in interphase cells. RE vesicles interact with and organize around these appendages. Endosome dissociation from centrosomes disrupts endosome recycling (Hehnly et al., 2012). We hypothesized that REs may, in turn, play a role in centrosome function. In fact, Rab11 was identified in a screen for mitotic regulators of microtubule (MT) dynamics in Caenorhabditis elegans (Zhang et al., 2008) and was shown to require dynein for this function (Ai et al., 2009). Rab11 is also an important regulator of endosome asymmetric distribution in sensory organ precursor (SOP) cells during cell division (Emery G, Cell 2005). Rab11-vesicles have recently been implicated in asymmetric spindle positioning in mouse oocytes (Holubcová et al., 2013), but how Rab11-vesicles can directly contribute to this process was not thoroughly investigated. However, the authors do propose that these vesicles function as cytoskeletal modulators (Holubcová et al., 2013). Here we explore possible functions for Rab11 in the formation of spindle poles, assembly of MT nucleating, anchoring and regulatory proteins at poles, organization of astral MTs, and orientation of mitotic spindles and the plane of cell division.
RESULTS
Rab11-associated endosomes associate with mitotic spindle poles and the mitotic spindle
Little is known of Rab11 and endosome function during spindle formation in mitosis. We found that GFP-Rab11-decorated-endosomes and the Rab11 effector, FIP3, clustered at spindle poles and along the mitotic spindle (Figure 1A, B, S1A; Movies S1, S2; noted in (Hobdy-Henderson et al., 2003)). Rab11 also localized to isolated spindle poles demonstrating that it is a bona fide centrosome protein (Figure S1D). In cells that were cold treated (Figure 1C), which destabilizes dynamic microtubules while preserving kinetochore MTs (Meunier and Vernos, 2011), Rab11 localized predominantly to kinetochore MTs (Figure 1C). This suggested that Rab11-associated endosomes were bound to MTs and that they might possibly be transported along spindle fibers to and from the pole. We established that these Rab11 endosomes were in fact recycling endosomes by staining for several additional RE proteins (Figure 1A–C, 2A–B). However, the early endosome Rab5 effector, EEA1, and Golgi complex proteins did not localize to these organelles (Figure 2C, S2B, Movie S2; Golgi positioning reviewed in (Yadav and Linstedt, 2011)).
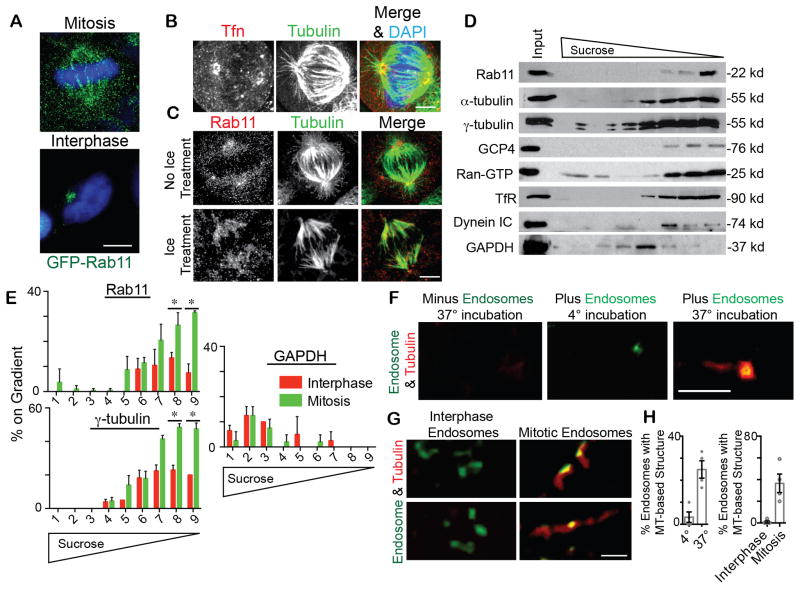
Figure 1. Recycling (transferrin-loaded) endosomes (REs) localize along the spindle, contain γ-tubulin and associate with a microtubule (MT)-based structure.
(A)Cells expressing GFP-Rab11 were fixed during mitosis or interphase to visualize Rab11. DAPI, blue. Bar, 5 μm.
(B, C) Alexa Fluor 594-Tfn filled endosomes (B; red) and Rab11 (C; red) localize to the mitotic spindle and spindle poles. MTs, green. DAPI, blue. Bar, 5 μm for B,C. In cold-treated cells that maintain stable kinetochore-attached-MTs, Rab11 localized to MTs (C).
(D) Immunoblots of endosomes were isolated by floatation in a sucrose gradient and labeled for transferrin receptor (TfR) and Rab11. Endosomes co-fractionated with the centrosome proteins α– and γ– tubulin, GCP4, Ran-GTP, and dynein intermediate chain (IC), but not with the cytosolic component, GAPDH.
(E) Mitotic and interphase endosomes isolated by floatation were analyzed for associated Rab11, γ-tubulin, and GAPDH. Mitotic endosomes contained significantly more Rab11 and γ-tubulin than interphase endosomes (n=3 experiments; (*) depicts p-value of <0.01; representative immunoblots, Figure S1E).
(F) 10 μM tubulin was incubated with or without isolated mitotic endosomes (green, decorated with FIP3-GFP), spun onto coverslips, and tested for MT-association. Endosomes showed MT-association at 37°C, but not 4°C. Tubulin alone did not form a MT-based structure without endosomes present. Bar, 3 μm.
(G) Isolated mitotic versus interphase endosomes (GFP-FIP3, Green) were spun onto coverslips and tested for MT association/assembly (Tubulin, Red). Two examples are shown for each. Bar, 3 μm.
(H) The percent of endosomes forming MT-based structures was quantified. Left, 25% of mitotic endosomes formed MT-structures at 37°C, compared to ~4% at 4°C (n=3 experiments, p<0.01). Right, 25% of mitotic endosomes formed MT-based structures compared to 0% in control (n=4 experiments, p<0.01).
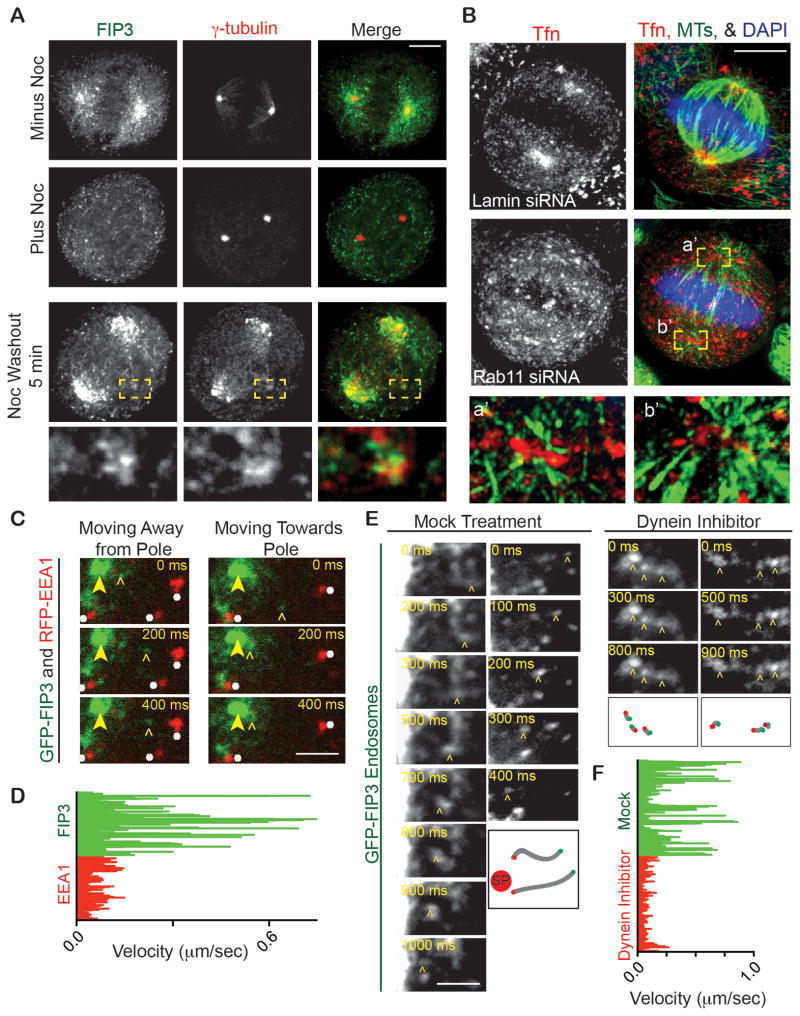
Figure 2. REs exhibit Rab11-dependent spindle pole localization and dynein-mediated transport.
(A) Maximum projections of mitotic cells after nocodazole- or mock-treatment, and 5 minutes after nocodazole washout. Cells were fixed, stained for γ-tubulin (red) and FIP3 (green). Mock-treated cells show FIP3 having spindle and pole localization, whereas nocodazole-mediated MT disruption dispersed FIP3 from poles. 5 minutes after nocodazole washout FIP3 refocuses at the spindle pole and at acentrosomal foci (inset, labeled with γ-tubulin, red).
(B) Control (lamin siRNA) and Rab11-depleted cells incubated with Alexa Fluor 594-conjugated Tfn (red) to label endosomes. Most endosomes focus around poles (MTs; green) in control cells, but are dispersed in Rab11-depleted cells. DAPI, blue. MT-organization around poles was also disrupted after Rab11 depletion (insets a′, b′, disrupted endosomes with MTs, 5x original image). Bar, 5 μm.
(C) Selected images from time-lapse movies of GFP-FIP3 labeled REs (marked by open arrowhead) moving towards and away from spindle poles (spindle pole marked by closed arrowhead) and dispersed RFP-EEA1 early endosomes remaining fairly immobile (marked by white asterisk). Refer to Movie S2. Bar, 1 μm.
(D) GFP-FIP3 and RFP-EEA1 decorated endosome track velocities were characterized. RFP-EEA1 tracks moved at a decreased velocity compared to GFP-FIP3 tracks (n>30 tracks in n=3 cells for each condition).
(E) Selected still images from time-lapse movies of GFP-FIP3 labeled REs moving towards spindle poles in prometaphase cells. Left, is a mock control (DMSO only) prometaphase cell showing minus-end directed motion of FIP3-decorated RE toward pole (marked by open arrowhead; Movie S3). Right, is a dynein-inhibited (Firestone et al., 2012) prometaphase cell with no minus-end mediated transport occurring (endosomes marked by open arrowhead). Schematic representation of shown puncta is depicted below time course. Green point represents when the vesicle starts moving, red point is when it stopped, and grey line is the path it took. Red circle labeled “SP” depicts where spindle pole is. Bar, 1 μm.
(F) GFP-FIP3 decorated recycling endosome tracks were characterized in either a control or a dynein inhibitor treated prometaphase cell. The dynein inhibitor significantly decreased GFP-FIP3 endosome velocity (n>40 measured tracks for each treatment, 3 cells measured per treatment).
The selective mitotic MT-localization of Rab11 and its associated endosomes (Figure 1A–C, S1A) was confirmed by biochemical assessment of microtubule pellets (Figure S1B). Mitotic MTs bound significantly more Rab11 than interphase MTs due most likely to degradation of the Rab11 GTPase activating protein (GAP), Evi5, which converts Rab11 to its GDP-binding state (Laflamme et al., 2012; Dabbeekeh et al., 2007; Eldridge et al., 2006; Figure S1B). Consistent with the MT association and GTP status of Rab11 was the observation that forced expression of constitutively-active Rab11(Q70L) increased MT-association over dominant-negative Rab11(S25N), (Figure S1C). This suggested that the enhanced MT-association of mitotic Rab11 could result from reduced Evi5 levels ((Eldridge et al., 2006); Figure 1A, S1B) subsequently causing an increase in active Rab11.
REs contain MT-nucleating components and dynein
To better understand the differences between endosomes in mitotic versus interphase cells, we examined the molecular composition of isolated endosomes. Endosomes detected by the presence of GFP-FIP3, a Rab11 effector that localizes to spindle poles, and Rab11 itself (Figure S1A), were isolated via flotation upward through a sucrose step gradient ((Ori-McKenney, 2012); Figure 1D). Biochemical analysis of the mitotic endosomes revealed proteins involved in MT-organization (e.g. dynein) as well as MT-nucleation, spindle pole organization and regulators of these processes (e.g. γ-tubulin, α-tubulin, GCP4, Ran-GTP; Figure 1D). The specificity of spindle pole proteins localization to endosomes was confirmed by showing that GAPDH did not cofractionate with endosomes (Figure 1D, S1E). The presence of α-tubulin in these isolated endosome fractions (Figure 1D) is consistent with Rab11 pelleting in MT pull-down experiments (Figure S1B).
We found that isolated mitotic endosomes contained significantly more Rab11 and γ-tubulin than interphase endosomes (Figure 1E, S1E). Notably, mitotic endosomes are enriched at mitotic spindle poles/centrosomes compared to interphase endosomes (Tfn-filled; Movie S1). This suggested that mitotic membranes may recruit more MT-nucleating components, which is consistent with their potential role in centrosome maturation. To directly address this, we tested MT-nucleation from isolated mitotic and interphase endosomes, as done for Golgi vesicle MT-nucleation assays in previous studies (Ori-McKenney et al., 2012). Similar to Golgi MT nucleation, about 25% of mitotic endosomes assembled linear MT elements, which were not present at 4°C (Figure 1F, H) or when nocodazole was added (Figure S1F). Endosomes isolated from interphase cells showed little to no detectable MT nucleation (Figure 1G–H).
Depletion of Rab11 or FIP3 disrupts endosome organization at spindle poles
To further test the mechanism of endosome localization to spindle poles, we depleted Rab11 or FIP3 using previously characterized siRNAs ((Hehnly et al., 2012), Figure S2C). In both cases, the association of REs with spindle pole (Figure 2B, S2B–C) was decreased ~3-fold compared with controls. Depolymerization of MTs similarly decreased spindle pole localization of REs (Figure 2A–B, S2A) demonstrating a requirement for MTs in organization or MT-dependent transport of endosomes.
REs require dynein-based transport for spindle pole localization
To more directly distinguish between a) binding of REs to spindle poles via MTs at this site or b) transport of REs to spindle poles via MTs, RE dynamics (TfR in Figure S2A; GFP-FIP3 in Figure 2C–F and 2A; Tfn-filled in Movie S1) were examined in living cells or in fixed cells over a time-course. After spindle MTs were disrupted by nocodazole, spindle re-assembly was imaged over a time course after drug washout. Spindle re-assembly in control cells was initiated by re-nucleation of MTs from spindle poles and reorganization of endosomes around the poles. We found that endosomes reorganized around acentrosomal peripheral microtubule clusters containing γ-tubulin (Figure 2A, modeled in Figure 3B) that formed in the cytoplasm and moved vectorially to spindle poles (Figure 2A, S2A). These observations suggested that endosomes were being transported on spindle MTs as part of these MT clusters (Figure 2A, S2A, modeled in Figure 3B), to become incorporated into spindle poles. In our previous work we showed that these MT clusters carried centrosome proteins to spindle poles (Delaval et al., 2011). Consistent with this notion is the observation that the MT clusters in this study appear to be transporting endosomes that we show contain centrosome proteins identified in our biochemical assays (Figure 1D, colocalization between FIP3 and γ-tubulin in Figure 2A).
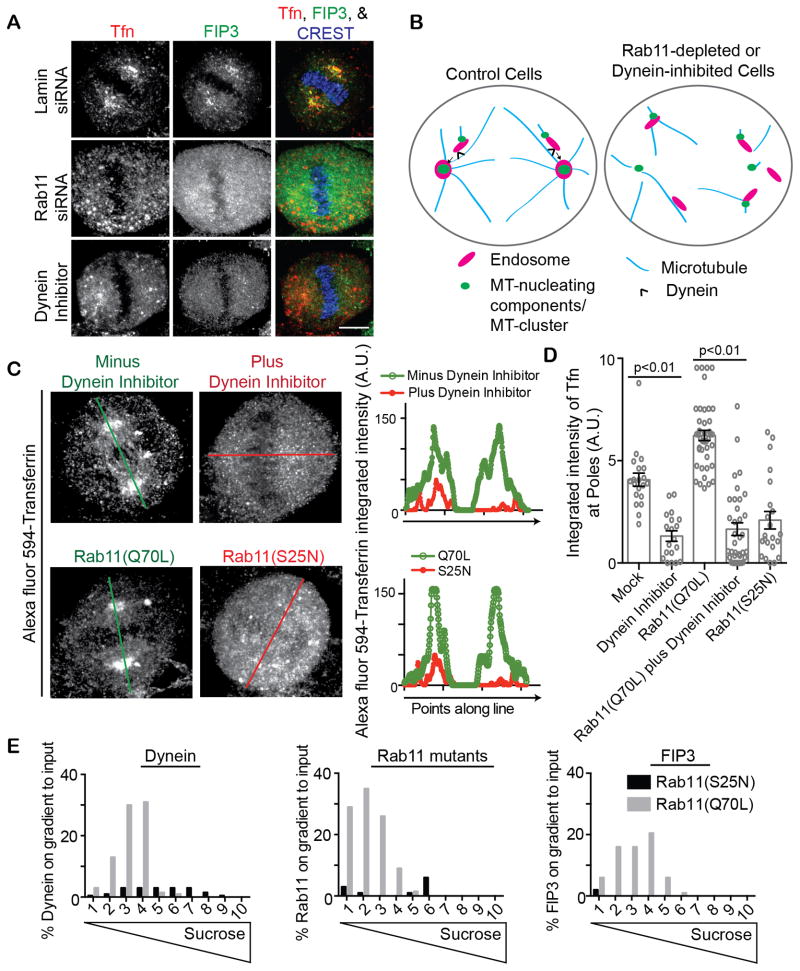
Figure 3. Rab11 and dynein coordinately organize REs at mitotic spindle poles.
(A) Cells treated with a dynein inhibitor (Firestone et al., 2012), or depleted of lamin (control) or Rab11 were incubated with Alexa Fluor 594-conjugated Tfn (red) to label endosomes. Mitotic cell maximum projections are shown and cells were stained for the Rab11-effector FIP3 (green) specifically labeling REs. REs were dispersed with dynein inhibition and Rab11-depletion. Bar, 5 μm.
(B) Model depicting REs together with MT-clusters contributing to spindle organization and function by acting as a carrier to transport spindle pole components to spindle poles by dynein-mediated transport. RE transport to the spindle pole is disrupted by either dynein inhibition or Rab11-depletion.
(C) Cells treated with dynein inhibitor or expressed Rab11(S25N) had significantly less Alexa Fluor 594-conjugated Tfn (red) labeled endosomes organized at mitotic spindle poles compared to either a mock control (DMSO treated) or cells expressing Rab11(Q70L). Bar, 5 μm. A line scan through the mitotic spindle poles (oriented by γ-tubulin staining, not shown) is drawn and the integrated intensity is plotted for Tfn.
(D) Shown is quantification of Tfn intensity at spindle poles (n=25 poles per treatment, p-values marked on graph. Representative of n=3 experiments). A significant increase in spindle pole localized Tfn was observed in cells expressing Rab11(Q70L) when compared to Rab11(S25N). However, Rab11(Q70L) did not cause an increase in Tfn in the presence of a dynein inhibitor.
(E) Endosomes isolated by floatation from cells expressing Rab11(S25N) or Rab11(Q70L) were compared for amounts of Rab11, Dynein, and FIP3 associated. Mitotic endosomes containing Rab11(Q70L) recruited significantly more dynein and the Rab11-effector FIP3 than membranes isolated from cells expressing Rab11(S25N), (representative of n=3 experiments, example western blots shown in Figure S3B).
We contend that endosomes are part of the MT-cluster-based transport pathway, as REs recruit the same/similar centrosome proteins and move to spindle poles. Work on MT-clusters suggests that dynein may be the driving force for this process (Delaval et al., 2011; Tulu et al., 2003). To directly test the role of dynein in endosome movements, we examined the dynamics of GFP-FIP3 labeled REs in mitotic cells. We observed movement toward and away from the mitotic spindle poles (Figure 2C; Movie S2–3). Endosomes moving toward the spindle pole had an average velocity of 0.8 μm/s (Figure 2E; Movie S3). REs were more motile than RFP-EEA1 labeled early endosomes, which rarely organized at poles or moved in a pole-ward direction (Figure 2C–D; Movie S2, S3). To test the role of dynein in these movements, we acutely inactivated the motor with a specific membrane permeable dynein inhibitor (Firestone et al., 2012). This essentially abolished vectorial retrograde motion of the GFP-FIP3 REs (Figure 2E–F).
Rab11 regulates endosome organization at mitotic spindle poles through dynein
To further analyze the role of Rab11 and dynein in recycling endosome organization we either depleted Rab11, expressed a dominant-negative Rab11 mutant, Rab11(S25N), expressed a constitutive-active mutant, Rab11(Q70L), and/or treated cells with a dynein inhibitor. We discovered that when Rab11 activity was inhibited by either depletion or Rab11(S25N) expression, the spindle pole localization of REs was disrupted (Tfn-loaded, FIP3 labeled, Figure 2B, S2B, 3A–D, S3A, and modeled in Figure 3B). Similar results were obtained with the dynein inhibitor (Figure 3A–C). In contrast, expression of constitutive-active Rab11(Q70L) induced tighter organization of endosomes at spindle poles (Figure 3C, S3A) and in some cases fewer endosomes were observed along the mitotic spindle (compare minus dynein inhibition to Rab11(Q70L) expression in images and line scan of Figure 3C).
Based on these findings, we hypothesized that Rab11 recruited dynein to mitotic endosome membranes. To test this, we assessed whether Rab11(Q70L) could rescue endosome organization at mitotic spindle poles when dynein was inhibited. First, we showed that Rab11(Q70L) expression increased spindle pole associated endosomes (Figure 3C, quantification in 3D). We next determined that endosomes in cells expressing Rab11(Q70L) and treated with a dynein inhibitor (Firestone et al., 2012), was decreased to the same level as treatment alone (Figure 3C–D), suggesting that active-Rab11 recruits dynein to mitotic endosomes. To address this more directly, we examined mitotic endosomes isolated from cells expressing either Rab11(Q70L) or Rab11(S25N), (Figure 3E, S3B). Endosomes from cells expressing constitutive-active Rab11(Q70L) recruited Rab11(Q70L) and dynein but those from cells expressing dominant-negative Rab11(S25N) recruited neither Rab11(S25N) nor dynein. As a positive control, Rab11(Q70L) recruited its effector, FIP3, to mitotic membranes where Rab11(S25N) did not (Figure 3E, S3B). These results suggest that active-Rab11 assists in dynein recruitment to endosomes during mitosis (Figure 3E, Model Figure 3B).
Rab11 is involved in mitotic progression
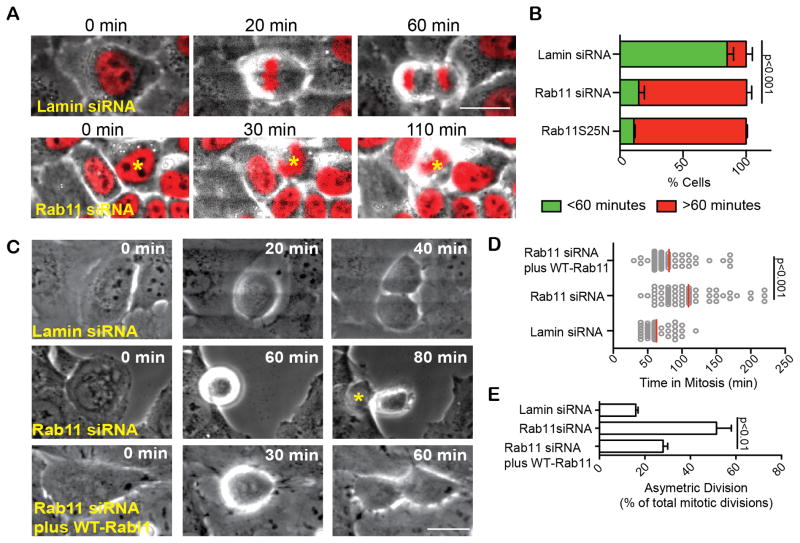
During the course of these studies, we noticed that the prevalence of mitotic cells was increased when Rab11-activity was disrupted. To formally address the origin of this phenotype, we performed time lapse imaging to determine the time spent in mitosis in cells depleted of Rab11 or expressing dominant-negative Rab11 (Rab11(S25N); HeLa cells in Figure 4A–B, U2OS cells in Figure 4C–E, S4A–B). Cells were engineered to express histone 2B-RFP (H2B; Figure 4A; Movie S4) to identify chromosome organization and mitotic stage. 80% of control cells (Figure 4B) or 60% of Rab11-depleted cells rescued by expression of a siRNA resistant Rab11 (Figure 4D, S2C) progressed into anaphase within 60 minutes. In contrast, only ~20% of Rab11-depleted or Rab11(S25N)-expressing cells entered anaphase within this time frame (average 100–130 minutes; Figure 4B–D, S4B). This mitotic delay was confirmed by imaging live cells stably expressing cyclin B-GFP to distinguish prometaphase from metaphase cells. As expected, cyclin B levels in control cells were highest in metaphase and decreased sharply at anaphase onset (Lindqvist et al., 2009; Chen et al., 2012). In contrast, cyclin B levels in Rab11-depleted cells (Figure S4C) remained elevated for prolonged time periods, demonstrating a metaphase delay.
Figure 4. Rab11 plays a role in mitotic progression.
(A) Time lapse imaging show Rab11-depleted HeLa cells stably expressing histone 2B RFP (H2B, red), entering mitosis as in control (lamin siRNA). However, Rab11-depleted cells become delayed in prometa/metaphase. Bar, 20 μm. Refer to Movie S4.
(B) Quantification of cells that spent > or < 60 minutes in mitosis following lamin (control) or Rab11-depletion, or after expression of dominant-negative (S25N) Rab11. (~5-fold decrease in <60 min, n>3 experiments; p<0.001 between lamin siRNA and Rab11 siRNA).
(C–D) Rab11-depleted cells spent more time in mitosis (C, D, 2-fold, n>75 cells, representative of n=3 experiments, p-value depicted on graph) compared with lamin (control). The increased time spent in mitosis and asymmetric divisions can be rescued by an RNAi-resistant Rab11 construct. Bar, 20 μm.
(E) Asymmetric division, determined by cell flattening is increased in Rab11-depleted cells. (p<0.001, n=3 experiments, asterisk indicating daughter cell that flattened first, C).
Rab11 plays a role in spindle orientation
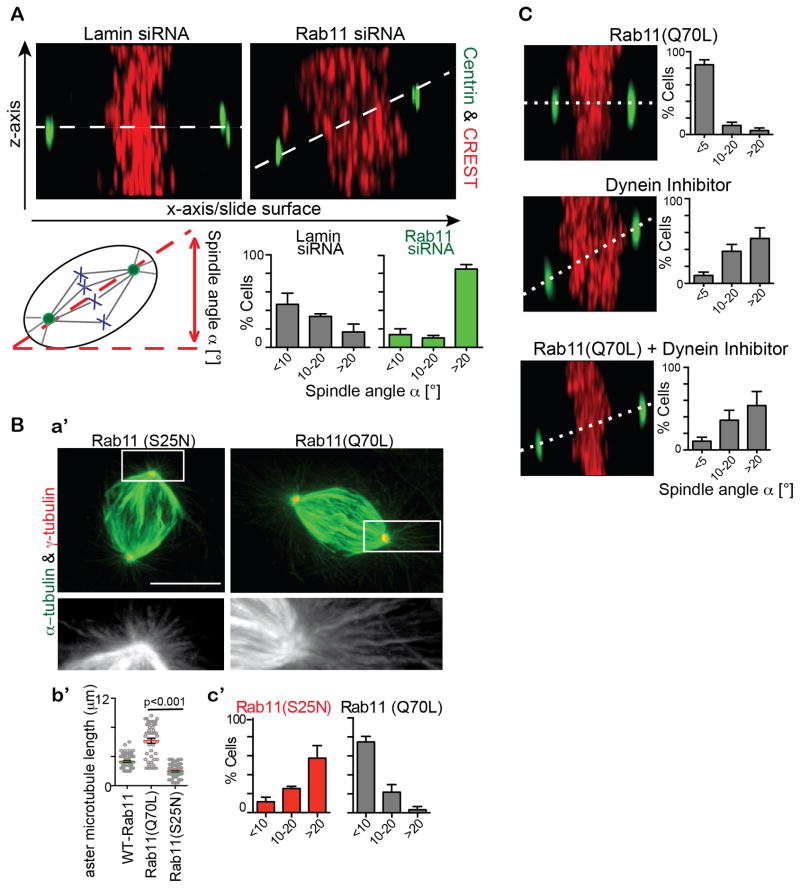
The mitotic delay following Rab11 disruption suggested a defect in mitotic spindle organization and indicated that the GTPase may have additional mitotic functions. Closer inspection of mitotic spindles in Rab11-depleted cells revealed several defects in spindle organization and function. The most prevalent phenotype was misorientation of the mitotic spindle and the plane of cell division. More specifically, the spindle angle relative to the cell-substrate adhesion plane in ~60% of Rab11-depleted cells was greater than 20°, whereas most control spindles were parallel to the substratum (~80%, Figure 5A). In addition, we found that the cell division plane was oriented more vertically in Rab11-depleted cells compared to control cells where the division plane was horizontal to the substratum (Figure 4C). The misoriented division plane caused a delay in flattening of the daughter cell farthest away from the substrate (Figure 4C, quantification 4E). To determine the cellular and molecular basis of spindle and cell division misorientation, we examined spindles in more detail.
Figure 5. Rab11 is involved in orientation of the mitotic spindle.
(A) Top, Rab11-depletion showed increased asymmetric divisions by measuring spindle angle (dotted line) relative to substrate (maximum projection of the z-axis, centrin (green), and kinetochores (CREST, red)). Bottom, quantification showing a significant increase (>20°) of spindle angle in cells depleted of Rab11 (~2-fold, n=3 experiments, p<0.001 comparing spindle angles >20° across treatments. n>20 cells counted/experiment.)
(B) Longer astral MTs in constitutively active Rab11(Q70L) expressing cells compared to dominant negative Rab11(S25N) expressing cells revealed by staining for α-tubulin (green; a′). Shown is quantification of astral MT length (n>50 asters per treatment, p<0.001; b′). Quantification showing a significant increase (>20°) of spindle angle in cells expressing Rab11(S25N) compared to Rab11(Q70L). (n=3 experiments. n>20 cells counted/experiment; c′)
(C) Z-axis maximum projections of a cell expressing Rab11(Q70L) and/or incubated with a dynein inhibitor. (n=3 experiments. n>20 cells counted/experiment)
Rab11 plays a role in astral MT-organization
Astral MT arrays in cells depleted of Rab11 (data not shown) or expressing dominant-negative Rab11(S25N) were disrupted when compared to wild-type or constitutively-active Rab11(Q70L) expressing cells (Figure 5B). This was consistent with the increased spindle angle in either Rab11-depleted cells (Figure 5A) or Rab11(S25N) expressing cells (Figure 5B). Constitutively-active Rab11(Q70L) enhanced astral MT-networks, substantiating the importance of the GTPase state of Rab11 in the formation of the astral MT-network, which is the subpopulation of spindle MTs involved in contacting dynein at the cell cortex and mediating spindle reorientation through dynein motor activity (Kotak and Gönczy, 2013). Consistent with this is our finding that dynein-inhibition causes an increased spindle angle similar to Rab11-depleted cells (Figure 5C). Constitutively-active Rab11(Q70L) could not rescue the spindle asymmetry phenotypes observed under dynein inhibition (Figure 5C) compatible with dynein and Rab11 coordinately regulating symmetric division.
Cell and molecular mechanism of spindle defects following Rab11 depletion
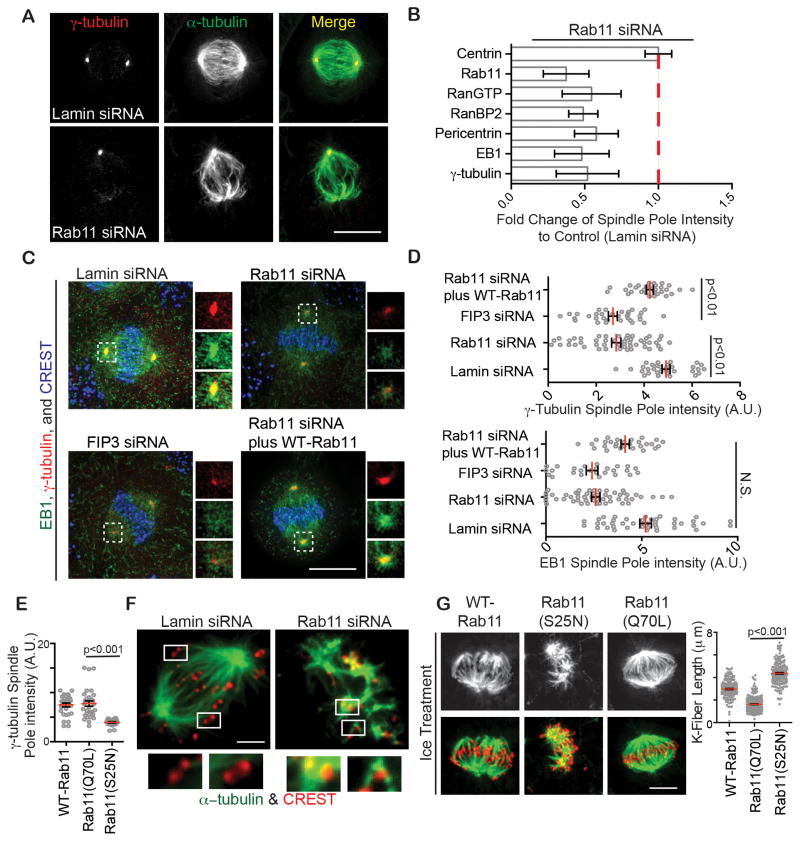
The localization of Rab11 to spindle poles and spindle MTs (Figure 1 and S1) and its role in mitotic progression and spindle assembly (Figure 4, 5) suggested a role for the GTPase in spindle pole assembly and function. Rab11 depletion (Figure 6A–D, S2C) or dominant negative Rab11(S25N) expression (Figure 6E) decreased spindle pole levels of major PCM proteins (γ-tubulin and pericentrin), MT-anchoring and plus-end binding proteins (EB1) and MT-nucleation regulators (Ran-GTP and its binding protein RanBP2), (~2-fold, Figure 6A–E, S5A). It is important to reiterate that most of these proteins are, at least in part, bound to endosomes (Figure 1). Equally important, was the observation that spindle pole levels of the centriolar protein centrin (Figure 5A) and the MT-motor, Kif2a (Figure S5B) were not affected by Rab11-depletion. These results demonstrate a role for Rab11 in the organization of a selective subset of spindle pole proteins, primarily those involved in centrosome maturation, the process of centrosome protein recruitment initiated during progression into mitosis and completed by metaphase (Mahen and Venkitaraman, 2012). This rapid recruitment of centrosome proteins drives a dramatic increase in MT-nucleating activity (Bornens, 2012).
Figure 6. Rab11 contributes to the organization of the mitotic spindle.
(A) Control or Rab11 depleted metaphase cells were stained for γ-tubulin (red) and MTs (green). Bar, 10 μm.
(B) Loss of spindle pole proteins as indicated, in Rab11-depleted cells was quantified (n=3 experiments). Fold change was calculated over control (lamin siRNA). A significant loss of γ-tubulin, EB1, RanBP2, RanGTP, and pericentrin was observed in Rab11-depleted cells (n=3, p<0.001).
(C) Cells were depleted of Lamin-, FIP3-, Rab11-, or Rab11 complimented with siRNA resistant Rab11. Metaphase cells were stained for γ-tubulin (red), EB1 (green), and CREST (blue). Bar, 10 μm.
(D) Spindle pole associated γ-tubulin and EB1 were decreased in Rab11- and FIP3-depleted cells, but not in control cells (lamin siRNA) or cells treated with Rab11-siRNA rescued with an siRNA resistant Rab11 vector (n>25 poles per treatment, p-values depicted on graphs, N.S., Not Significant).
(E) A significant increase in γ-tubulin at spindle poles was observed in cells expressing Rab11(Q70L) when compared to Rab11(S25N), (n=25 poles per treatment, p<0.001).
(F) Control prometaphase cells 5 min after nocodazole washout had robust MT-growth (green) from poles, whereas Rab11-depleted cells had a significant number of MT-clusters emanating from kinetochores (CREST, red) and little from poles (data not shown). Insets show kinetochores with MT-clusters in Rab11-depleted cells compared to none in control. Bar, 3 μm.
(G) Kinetochore fibers in cells expressing WT-Rab11, Rab11(S25N), and Rab11(Q70L) after cold-treatment, fixation and staining with anti-α-tubulin (MTs, green) and kinetochores (CREST, red). Bar, 3 μm. Right, quantification of kinetochore fiber disruption following Rab11(S25N) expression. (n>100 K-fibers per treatment, p-value depicted on graph.)
To further examine the role of Rab11 in centrosome maturation, we performed a functional test monitoring both assembly of centrosome proteins onto spindle poles and spindle-pole-mediated MT-nucleating activity over time. Spindles were disassembled with nocodazole, then examined at different times after nocodazole washout for protein recruitment and MT-nucleation ((Delaval et al., 2011); Figure S5C). At 1 minute of regrowth, spindle poles in control cells showed an increased ability to nucleate MTs and recruit the spindle pole proteins γ-tubulin, pericentrin, and EB1 (Figure S5C). These activities were impaired in Rab11-depleted cells, (Figure S5C). At 5 minutes of regrowth, control cells nucleated MTs robustly from the poles. In contrast, Rab11-depleted cells showed little nucleation at poles. Instead, MT-clusters assembled at kinetochores (Figure 6F). This finding suggests that spindle poles in Rab11-depleted cells are compromised in their ability to nucleate MTs thus allowing the kinetochore-mediated MT-formation pathway to be activated or dominant. In control cells, when spindle poles are intact, no kinetochore MTs are generated. This result also demonstrates that kinetochores are intact and functional in Rab11-depleted cells and are not likely candidates for chromosome misalignment. Eventually after 40 minutes, spindles were assembled with non-congressed chromosomes whereas control spindles had tight metaphase plates (Figure S5D). It is important to note that the kinetochore-based process of spindle assembly in Rab11-depleted cells is similar to that in acentrosomal cells (Theurkauf and Hawley, 1992), where spindle MTs grow from sites around kinetochores then self-organize into a spindle (Heald et al., 1996; Theurkauf and Hawley, 1992). Based on these findings and our live cell video microscopy (Figure 2), we conclude that Rab11 and its associated endosomes assist in spindle pole function by organizing MT-nucleating components at this site (Figure 6).
To more closely examine kinetochore fiber integrity we employed cold-treatment to specifically eliminate dynamic MTs from mitotic cells (Meunier and Vernos, 2011; Firestone et al., 2012). Kinetochore fibers in control cells were well organized and robust (Figure 6G, S5E). Following Rab11-depletion or dominant-negative Rab11(S25N) expression, kinetochore fibers were reduced to collections of small-disorganized linear segments (Figure 6G, S5E). These results support the idea that Rab11-depleted spindle poles are compromised in MT-nucleation and –anchoring and likely release MTs, as observed (Figure 6G, S5E). Finally expression of constitutively-active Rab11(Q70L) generated longer kinetochore-based MTs than cells expressing WT-Rab11 or Rab11(S25N), (Figure 6G). These results demonstrate that the GTP-state of Rab11 is important for spindle pole-mediated MT-nucleation and anchoring.
Rab11 is involved in chromosome orientation
In addition to spindle misorientation, Rab11 depleted cells showed a significant increase in misaligned chromosomes (~60%, Figure S6A). Chromosome congression and alignment requires both attachment of chromosomes to MTs and tension on the chromosomes. It is important to note that tension on chromosomes requires robust MT attachments, not only at kinetochores but also at spindle poles. MT-attachment to poles is an integral but often unappreciated part of the spindle assembly process. We propose that disruption of spindle poles and spindle pole generated MTs in Rab11-depleted cells (Figure 6), disrupts chromosome attachments to spindle pole MTs (Figure 7). Consistent with this are results showing that kinetochores are not compromised by Rab11-depletion as they can induce MT-formation (Figure 6F) and activate the spindle assembly checkpoint (SAC, Figure 7). These compromised MT-attachments at spindle poles could prevent the generation of tension at the chromosomes and thus, activate the SAC. In fact, our data show that the SAC is activated, as Mad1, ZW10, and BubR1 remained at kinetochores in Rab11-depleted cells. There was no change in the kinetochore kinesin, MCAK (Figure 7A, 7B), suggesting that the kinetochores were structurally intact. We further found a significant increase in dynein localization to kinetochores in Rab11-depleted cells (Figure S6B).
Figure 7. Rab11 is involved in chromosome congression/alignment.
(A) The spindle assembly checkpoint proteins BubR1, ZW10, and MAD1 (green) are maintained on kinetochores (CREST, blue) in Rab11-depleted cells compared to control; no difference in MCAK was observed.
(B) Labeled checkpoint proteins were quantified on individual kinetochores and normalized to CREST. A significant increase in MAD1, BUBR1, and ZW10 was observed (n>200 kinetochores per treatment, p-values marked on graph, N.S. is non-significant).
(C) MT attachments and tension were examined by measuring the distance between two CREST (red) puncta for n>300 kinetochores, n>25 cells. Rab11 depleted cells displayed a 2-fold decrease in the distance between kinetochores (~0.5 μm). Refer to p-values on graph.
(D) Non-congressed chromosomes in Rab11-depleted cells revealed by staining for Aurora A (blue), Aurora B (green, see insets from boxes), and kinetochores (CREST, red). Representative of n=10 cells. (A,D) Bar, 5 μm.
We propose a model for chromosome alignment that is controlled by Rab11. First, kinetochores in Rab11-depleted cells are functional in that they are able to activate SAC and generate and bind to MTs. It is unlikely that they are the cause of SAC activation. In contrast, spindle pole MT-nucleation and anchoring are disrupted, which can prevent or compromise MT-spindle pole attachments, (perhaps through the MT-nucleating protein γ-tubulin). This, in turn, could activate the SAC through the inability to maintain tension at spindle poles. In fact, diminished tension was revealed as a dramatic reduction in inter-kinetochore distances (Figure 7C) and the presence of Aurora B at kinetochores flanked by two CREST-labeled puncta (Figure 7D), where it senses tension (Howell et al., 2004; Famulski and Chan, 2007; Lampson and Cheeseman, 2011).
DISCUSSION
This work has led us to unexpected functions of the Rab11 GTPase and its associated endosomes. First, our results provide evidence for Rab11 function in the construction of spindle poles. We propose that this occurs through dynein-mediated transport of endosomes that, in turn, carries MT nucleating/anchoring and regulatory proteins to the poles. In essence, Rab11-endosomes act as carriers to recruit and organize MT-nucleating material at spindle poles through dynein (Figure 1, 2, 3). We believe that this is particularly significant because previous studies suggested that endosomes, like Golgi complexes and endoplasmic reticulum disperse throughout the cytoplasm during mitosis, and that membrane trafficking is thought to be halted until cytokinesis (Foley and Kapoor, 2013; Yadav and Linstedt, 2011; Chen et al., 2012; Takatsu et al., 2013). Second, when Rab11 is disrupted, the molecular and functional integrity of spindle poles is compromised. This diminishes astral MT integrity and inhibits MT attachment to cortical sites essential for orienting spindles. This work reveals a function for Rab11 in spindle pole assembly and spindle orientation. Third, compromised MTs at spindle poles in Rab11-disrupted cells can explain the misaligned chromosomes, mitotic delay and activation of the SAC. Our results support a pathway for activation of the SAC through poor MT-attachments to spindle poles. This pathway is conventionally thought to sense defects in kinetochores-MT attachments, but we contend that poor MT-attachments can occur at kinetochores and/or spindle poles (Zimmerman and Doxsey, 2000; Delaval et al., 2011; Firestone et al., 2012). Compromised attachments at either site will prevent MT-tension, which is required to satisfy the SAC and permit anaphase onset. The most compelling data for this additional aspect of an old pathway is that kinetochore integrity and function is normal in Rab11 cells (MT nucleation, SAC activation, presence of kinetochore proteins-MCAK, CREST, p150 (data not shown)) and is sufficient to drive spindle assembly (at least the early stages) presumably through activation/dominance of the acentrosomal kinetochore/chromatin-based pathway. Within the same Rab11-depleted cells lies the centrosome, which usually acts dominantly in vertebrate cells to drive spindle assembly. However, the deleterious effects of Rab11-depletion compromise the centrosome presumably rendering it inadequate for spindle assembly.
EXPERIMENTAL PROCEDURES
Reagents and siRNAs are discussed in Supplemental Experimental Procedures.
Immunofluorescence
A human osteosarcoma cell line, U20S, HeLa-GFP-FIP3 (kind gift from Dr. Rytis Prekeris, University of Colorado), HeLa-GFP-Dynein Heavy Chain (DHC, kind gift from Dr. Cheeseman, MIT), HeLa-GFP-cyclin B/H2B-RFP (kind gift from Dr. Gerlich), or parental HeLa cells were cultured in D-minimal essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin-streptomycin. HeLa (GFP-FIP3) was kept under selection conditions with Hydromycin B (Sigma). U2OS or HeLa cells were grown to subconfluence on glass coverslips and prepared for immunofluorescence, fixed using either saponin extraction with formaldehyde or methanol (Hehnly et al., 2009). Images were taken on a Perkin Elmer Ultraview spinning disk confocal microscope: Zeiss Axiovert 200, 100 × Plan-APOCROMAT NA1.4 Oil, DIC lens and Hamamatsu ORCA-ER camera. The entire fixed cell volume was imaged and displayed as a two-dimensional projection (Meta-Morph, Molecular Devices) to ensure that all stained material was visible in two-dimensional images.
Live Cell Imaging
Cells were cultured on 35-mm dishes containing a central 14-mm #1.5 glass coverslip (MatTek). Confocal Microscopy was performed on a Solamere Technology Group (Salt Lake City, UT) CSU10B Spinning Disk Confocal System attached to a Nikon TE2000-E2 motorized inverted fluorescence microscope. For GFP and RFP imaging frames were acquired every 100 ms using a Rolera MGi EMCCD 14-bit camera (Qimaging, Surrey, BC, Canada). MetaMorph (Molecular Devices) software was used for equipment control, image acquisition and image analysis.
Tracking Vesicle movement
GFP-FIP3 particles were semi-automatically monitored for 50 to 100 frames taken every 100 ms in a single z-axis using MetaMorph. After configuring MetaMorph to determine intensity centroids for each particle, up to 20 GFP-FIP3 particles paths and velocities were measured per cell. 50 μm of ciliobrevin was used for dynein inhibition (Firestone et al., 2012).
Transferrin Endocytosis
U2OS or Hela cells were treated with 10 μg/ml Alexa-Fluor 594- transferrin (Tfn) as described (Hehnly et al., 2012, 2006). The cells were placed at 37°C. The amount of internalized Tfn was detected using fluorescent microscopy.
Image Analysis
Z-stacks are shown as 2D maximum projections (MetaMorph) or processed for 3D rendering (Imaris). Spindle pole fluorescence was quantified as described for the kinetochore, along with kinetochore fluorescence (Howell et al., 2004). Concentric circle regions defined by Metamorph software (Molecular Devices) were used to define the inner (centrosome or kinetochore) and outer area (local background area was calculated as the difference between the outer and inner circle areas). These circular areas contain the calculated integrated intensities. Cells were counted for 3 independent experiments as described. Graphs were created and statistical analysis was completed using GraphPad Prism software. Bars represent SE, p<0.05 was considered as statistically significant.
Membrane Isolation (Float-up)
HeLa GFP-FIP3 cells were washed with a 8% sucrose, 10 mM Tris-HCl, 100 μM GTP, 100 μM ATP, 10 μM BME, and 1 μM PMSF (pH 7.4) buffer, homogenized with a dounce homogenizer, and spun at 5,000 × g for 3 min to generate postnuclear supernatant. The postnuclear supernatant was spun at 50,000 × g to generate a membrane pellet, the membrane pellet was then resuspended to a final concentration of 50% sucrose and layered beneath a sucrose step gradient from 40% to 15%. The gradient was placed in the TLA 100.3 rotor and spun at 45,000 × g for 2 h at 4 °C (Hehnly et al., 2010). Fractions were collected and run on an SDS-PAGE gel and the fraction containing endosomes saved for in vitro MT regrow experiments.
In Vitro Microtubule Nucleation from membranes
The microtubule nucleation solution assays were performed as described with a few modifications (Ori-McKenney et al., 2012; Macurek et al., 2008). GFP-labeled FIP3 endosomes were partially purified from mitotic cells by membrane flotation through a sucrose step gradient, then incubated with 10 μM tubulin/tubulin-Cy3 (Cytoskeleton; at a 10:1 ratio) and 1 mM GTP at 37° for 20 min. The solution was incubated with 4% PFA and spun onto a coverslip. GFP-FIP3 labeled endosomes were counted for Cy3 tubes.
Supplementary Material
Highlights.
Endosomes are not rendered inactive during mitosis as previously envisioned.
Endosomes are MT-nucleating, -anchoring, and spindle pole material carriers.
Rab11 regulates mitotic progression and spindle symmetry.
Rab11-activity regulates astral microtubule organization.
Acknowledgments
This work was supported by NIH grants: GM051994 (S.D.) and K99GM107355 (H.H.). We are grateful to Alison Bright, Chun-Ting Chen (S.D. laboratory), Felix Rivera-Molina, and Derek Toomre (Yale) for their revisions to this manuscript. We thank Mark Stamnes (University of Iowa) for his advice and assistance in the MT-membrane in vitro reassembly experiments. We thank Dr. Paul Furcinitti of the Digital Light Microscopy Core Facility at the UMass Medical School for assistance with live-cell spinning disk confocal microscopy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES CITED
- Ai E, Poole DS, Skop AR. RACK-1 Directs Dynactin-dependent RAB-11 Endosomal Recycling during Mitosis in Caenorhabditis elegans. 2009;20:1629–1638. doi: 10.1091/mbc.E08-09-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M. The Centrosome in Cells and Organisms. Science (80-) 2012;335:422–426. doi: 10.1126/science.1209037. [DOI] [PubMed] [Google Scholar]
- Chen C-T, Hehnly H, Doxsey SJ. Orchestrating vesicle transport, ESCRTs and kinase surveillance during abscission. Nat Rev Mol Cell Biol. 2012 doi: 10.1038/nrm3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbeekeh JTS, Faitar SL, Dufresne CP, Cowell JK. The EVI5 TBC domain provides the GTPase-activating protein motif for RAB11. Oncogene. 2007;26:2804–2808. doi: 10.1038/sj.onc.1210081. [DOI] [PubMed] [Google Scholar]
- Delaval B, Bright A, Lawson ND, Doxsey S. The cilia protein IFT88 is required for spindle orientation in mitosis. Nat Cell Biol. 2011;13:461–468. doi: 10.1038/ncb2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge AG, Loktev AV, Hansen DV, Verschuren EW, Reimann JDR, Jackson PK. The evi5 oncogene regulates cyclin accumulation by stabilizing the anaphase-promoting complex inhibitor emi1. Cell. 2006;124:367–380. doi: 10.1016/j.cell.2005.10.038. [DOI] [PubMed] [Google Scholar]
- Famulski JK, Chan GK. Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores. Curr Biol. 2007;17:2143–2149. doi: 10.1016/j.cub.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Firestone AJ, Weinger JS, Maldonado M, Barlan K, Langston LD, O’Donnell M, Gelfand VI, Kapoor TM, Chen JK. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature. 2012;484:125–129. doi: 10.1038/nature10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Hehnly H, Chen CT, Powers CM, Liu HL, Doxsey S. The centrosome regulates the rab11- dependent recycling endosome pathway at appendages of the mother centriole. Curr Biol. 2012;22:1944–1950. doi: 10.1016/j.cub.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Longhini KM, Chen JL, Stamnes M. Retrograde Shiga toxin trafficking is regulated by ARHGAP21 and Cdc42. Mol Biol Cell. 2009;20:4303–4312. doi: 10.1091/mbc.E09-02-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Sheff D, Stamnes M. Shiga toxin facilitates its retrograde transport by modifying microtubule dynamics. Mol Biol Cell. 2006;17:4379–4389. doi: 10.1091/mbc.E06-04-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehnly H, Xu W, Chen JL, Stamnes M. Cdc42 regulates microtubule-dependent Golgi positioning. Traffic. 2010;11:1067–1078. doi: 10.1111/j.1600-0854.2010.01082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobdy-Henderson KC, Hales CM, Lapierre La, Cheney RE, Goldenring JR. Dynamics of the apical plasma membrane recycling system during cell division. Traffic. 2003;4:681–693. doi: 10.1034/j.1600-0854.2003.00124.x. [DOI] [PubMed] [Google Scholar]
- Holubcová Z, Howard G, Schuh M. Vesicles modulate an actin network for asymmetric spindle positioning. Nat Cell Biol. 2013;15:937–947. doi: 10.1038/ncb2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- Kotak S, Gönczy P. Mechanisms of spindle positioning: cortical force generators in the limelight. Curr Opin Cell Biol. 2013;25:741–748. doi: 10.1016/j.ceb.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Laflamme C, Assaker G, Ramel D, Dorn JF, She D, Maddox PS, Emery G. Evi5 promotes collective cell migration through its Rab-GAP activity. J Cell Biol. 2012;198:57–67. doi: 10.1083/jcb.201112114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011;21:133–140. doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A, Rodríguez-Bravo V, Medema RH. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J Cell Biol. 2009;185:193–202. doi: 10.1083/jcb.200812045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macurek L, Dráberová E, Richterová V, Sulimenko V, Sulimenko T, Dráberová L, Marková V, Dráber P. Regulation of microtubule nucleation from membranes by complexes of membrane-bound gamma-tubulin with Fyn kinase and phosphoinositide 3-kinase. Biochem J. 2008;416:421–430. doi: 10.1042/BJ20080909. [DOI] [PubMed] [Google Scholar]
- Mahen R, Venkitaraman AR. Pattern formation in centrosome assembly. Curr Opin Cell Biol. 2012;24:14–23. doi: 10.1016/j.ceb.2011.12.012. [DOI] [PubMed] [Google Scholar]
- Meunier S, Vernos I. K-fibre minus ends are stabilized by a RanGTP-dependent mechanism essential for functional spindle assembly. Nat Cell Biol. 2011;13:1406–1414. doi: 10.1038/ncb2372. [DOI] [PubMed] [Google Scholar]
- Ori-McKenney KM, Jan LY, Jan YN. Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron. 2012;76:921–930. doi: 10.1016/j.neuron.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu H, Katoh Y, Ueda T, Waguri S, Murayama T, Takahashi S, Shin HW, Nakayama K. Mitosis-coupled, microtubule-dependent clustering of endosomal vesicles around centrosomes. Cell Struct Funct. 2013;38:31–41. doi: 10.1247/csf.12028. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE, Hawley RS. Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J Cell Biol. 1992;116:1167–1180. doi: 10.1083/jcb.116.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulu US, Rusan NM, Wadsworth P. Peripheral, non-centrosome-associated microtubules contribute to spindle formation in centrosome-containing cells. Curr Biol. 2003;13:1894–1899. doi: 10.1016/j.cub.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Yadav S, Linstedt AD. Golgi positioning. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Squirrell JM, White JG. RAB-11 permissively regulates spindle alignment by modulating metaphase microtubule dynamics in Caenorhabditis elegans early embryos. Mol Biol Cell. 2008;19:2553–2565. doi: 10.1091/mbc.E07-09-0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman W, Doxsey SJ. Construction of centrosomes and spindle poles by molecular motor-driven assembly of protein particles. Traffic. 2000;1:927–934. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.