Abstract
It is well known that embryos cultured in a group can create a microenvironment through secretion of autocrine and paracrine factors that can support and improve the embryos' development when compared to the embryos cultured individually. In this study, we used a co-culture system for paracrine communication between different kinds of embryos. The results showed that co-culture of porcine parthenogenetic (PA) embryos significantly improved the in vitro development of cloned (nuclear transfer, NT) embryos. To reveal the possible mechanism of communication between the two groups, we isolated exosomes/microvesicles (EXs/MVs) from the PA embryos conditioned medium (PA-CM) through differential centrifugation and identified them through transmission electron microscope and immunoflourescence against exosomal/membrane marker CD9. Furthermore, these EXs/MVs were found to contain mRNA of pluripotency genes (Oct4, Sox2, Klf4, c-Myc, and Nanog), and the PKH67-labeled EXs/MVs could be internalized by the NT embryos. The current study demonstrates that cloned embryos' developmental competence can be improved through co-culturing with PA embryos and revealed, for the first time, that in vitro–produced embryos can secrete EXs/MVs as a possible communication tool within their microenvironment. Moreover, it provides a new paradigm for embryo-to-embryo communication in vitro.
Introduction
Embryo culture environment is a crucial factor that affects embryo development in vitro, gene expression, and postimplantation development in the cultured mammalian embryos (Cheong et al., 2009; Wadhwa et al., 2009; Yamanaka et al., 2009a). The sensitivity of cloned embryos to culture environment was reported as higher than in vitro–fertilized or parthenogenetic (PA) embryos in terms of in vitro development (Yamanaka et al., 2009b). It has been reported that embryos in culture alter the microenvironment through secretion of several trophic autocrine/paracrine growth factors called the “secretome” (Baranao et al., 1997; Bormann et al., 2006; Katz-Jaffe et al., 2006; O'Neill, 1997), and this has been confirmed by embryonic expression of their reciprocal receptors (Dadi et al., 2007; Ishiwata et al., 2000; Richter, 2008).
Porcine cloning and nuclear transfer (NT) have potential application in various aspects of bioscience and biotechnology, such as livestock propagation, endangered species preservation, organ xenotransplantation, and disease model generation. However, the use of this technique is limited owing to its low efficiency (Estrada et al., 2007).
An ever-growing number of studies worldwide has helped to substantiate the essential functions of the cell secreted membrane-derived vesicles, particularly exosomes (EXs) and microvesicles (MVs), and provided new dimensions for the concept of intercellular signaling. These vesicles contain not only proteins but also messenger RNAs (mRNAs) and micro-RNAs (miRNAs) (Lee et al., 2012; Thery, 2011; Valadi et al., 2007), and thereby affect cellular activity via the already made proteins and miRNA or by translation of the transferred mRNAs. Valadi et al. (2007) proposed the name “exosomal shuttle RNA” (esRNA) for those transferred RNAs. These cell membrane-derived vesicles are involved in cell adhesion and signal transfer and provide an important method for cell communication (van der Pol et al., 2012). Evidence of secretion of EXs/MVs has been reported in most cell types, including embryonic stem cells (ESCs) and in vitro–produced embryos (Gardiner et al., 2013; Katsman et al., 2012; Ratajczak et al., 2006).
Several studies have reported embryo co-culture with somatic cells (Desai and Goldfarb, 1998; Funston et al., 1997; Kobayashi et al., 1992), oviduct cells (Bavister, 1988; Lee et al., 2003; Lee et al., 2001; Liu et al., 1998; Mermillod et al., 1993; Van Langendonckt et al., 1996; Xu et al., 2001), and CM (Li et al., 2004a; Li et al., 2004b) to mimic the microenvironment conditions associated with the maternal tract. However, only a single report has studied the co-culture of different kinds of embryos together (Terashita et al., 2011) because of the difficulty in discrimination or separation of different kinds of embryos during the co-culture.
This study was carried out to investigate the effect of co-culture of porcine PA embryos on development of nuclear transfer embryos and whether embryos secrete EXs or MVs as a possible mode of communication between embryos.
Materials and Methods
Chemicals
All chemicals were obtained from Sigma-Aldrich Co. LLC. (St. Louis, MO, USA) unless otherwise stated.
Ovaries and cumulus–oocyte complexes recovery and in vitro maturation
Ovaries were obtained from sows and gilts at a local slaughterhouse and were transported to the laboratory in 0.9% NaCl at 25–30°C. Follicular fluid including cumulus–oocyte complexes (COCs) were aspirated from antral follicles (3–6 mm in diameter) and washed three times with tissue culture medium-199 (TCM-199)-HEPES (Invitrogen, Carlsbad, CA, USA) and selected for in vitro maturation on the basis of morphological features, i.e., a compact multilayered cumulus mass and a dark, evenly granulated cytoplasm. The COCs were cultured in four-well dishes (50 COCs per well; Falcon, Becton Dickinson Ltd, Plymouth, UK) in basic maturation medium, TCM-199 supplemented with 10 ng mL−1 epidermal growth factor (EGF), 0.57 mM cysteine, 0.91 mM sodium pyruvate, 5 μg mL−1 insulin, 1 μg mL−1 follicle-stimulating hormone (FSH) (Antrin, Teikoku, Japan), and 1% (vol/vol) penicillin-streptomycin (Pen-Strep; Invitrogen) at 39°C in a humidified atmosphere of 5% CO2 for 44 h (two stages, with hormonal removal in the second stage). After 44 h, oocytes and expanded cumulus cells were separated by pipetting with 0.1% hyaluronidase in Tyrode's albumin lactate pyruvate (TALP). Denuded oocytes were examined by microscope as free of any attached somatic cells and then were subjected to parthenogenesis or used for nuclear transfer.
Parthenogenetic activation and in vitro culture of matured oocytes
Denuded oocytes were equilibrated sequentially in a gradient concentration (0%, 33%, 66%, and 100%) of mannitol solution (0.25 M) in a four-well dish. The oocytes were then transferred into a two-electrode mannitol chamber connected with a BTX Electrocell Manipulator ECM 2001 (BTX, Inc., San Diego, CA, USA), activated by a single pulse of 1.5 kV/cm for 100 μsec (Okada et al., 2006), and kept for 3 min in the activation medium (Kwon et al., 2011). Electrically activated embryos were equilibrated in reverse order to preactivation to decrease the stress on the oocytes. Activated oocytes were then washed in TALP and transferred into 30-μL microdrops of 4 mM 6-dimethylaminopurine (6-DMAP) covered with mineral oil and cultured for 90 min in an atmosphere of 39°C, 5% CO2, 5% O2, and 90% N2. Presumptive zygotes were washed and randomly distributed to the designed experiment for in vitro culture. Culture medium was serum-free chemically defined porcine zygote medium-5 (PZM-5) (Funakoshi Co., Tokyo, Japan) (Suzuki et al., 2004) covered with mineral oil and cultured in an atmosphere of 39°C, 5% CO2, 5% O2, and 90% N2. On day 2, embryos were evaluated for cleavage to the two-cell stage or beyond. Blastocyst formation was assessed on day 7.
Nuclear transfer
NT was carried out in accordance with our previous protocol (Koo et al., 2009). In brief, in vitro–matured oocytes were enucleated using an aspiration pipette 18 μm in diameter, microinjected with Sinclair male kidney cells, as donor cells, and fused by electrical stimulation. The constructed embryos were then activated, cultured, and checked for development following the same method for PA embryos.
Co-culture of embryos and CM supplementation
PA embryos were co-cultured with cloned (NT) embryos using 24-well plates supported with 0.4-μm Transwell (TW) polyester membrane inserts (Corning Inc., Pittston, PA, USA) to permit embryos communication with a distance of 2 mm between them (Fig. 1) at 39°C in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2. Additionally, we studied the temporal effect of PA embryos CM (PA-CM) supplementation on the development of the same number of cloned embryos. After removing the culture medium, PA-CM of days 2, 4, and 6 was supplemented to the cloned embryos every 2 days using a glass micropipette. The supplementation was either synchronized to the cloned embryos' developmental course (i.e., days 2, 4, and 6 PA-CM were added to the days 2, 4, and 6 cloned embryos, respectively) or preceded by 2 days (i.e., days 2, 4, and 6 PA-CM were added to days 0, 2, and 4 cloned embryos, respectively). Plain PZM-5 was added to the control group in the same manner. The embryo/medium density was one embryo/10 μL in all groups.
FIG. 1.

Embryo co-culture design. Cloned embryos (NT) were kept in 0.4-μm Transwell (TW) Polyester Membrane Inserts with a distance of 2 mm from the PA embryos. The medium covers both groups with a density of one embryo/10 μL of PZM-5 medium.
Isolation of EXs/MVs
PA embryos were cultured in groups (one embryo/1-μL PZM-5 microdrops) covered with mineral oil. PA-CM was collected daily via glass micropipettes until the blastocysts hatched. A total of 750 embryos (six replicates of 125 embryos) were used, and the collected CM were kept in −20°C until use. PA-CM of embryos on days 2, 4, 6, and 7 was subjected to differential centrifugation according to the previous protocols (Lässer et al., 2012; Théry et al., 2006; Witwer et al., 2013). In brief, PA-CM was centrifuged at 300×g for 10 min; the supernatant was centrifuged at 2,000×g for 10 min to remove residual cells and debris and at 10,000×g for 30 min to remove apoptotic bodies and cell debris. An equal amount of phosphate-buffered saline (PBS) was added to the supernatant and centrifuged at 200,000×g for 2 h (OptimaTM TLX ultracentrifuge, Beckman Coulter Inc., Fullerton, CA, USA) in 1-mL thick-wall polycarbonate tubes (Beckman Coulter Inc., item no. 343778) to pellet the EXs/MVs. The pellet, which was very tiny, was finally resuspended in 30 μL of PBS. Any EXs would be contained in these pellets, along with MVs. The resuspended pellets were used for checking the presence of MVs/EXs through the electron microscopy, immunofluorescence, RNA extraction, and EXs/MVs labeling. Porcine follicular fluid (PFF) was used as a control positive (da Silveira et al., 2011); the negative control was the plain PZM-5. The same procedures were applied to punctured PA embryos (PPA-CM) (n=750, six replicates of 125 embryos); a hole was made in the zona pellucida using an 18-μm-diameter needle.
Identification of EXs/MVs through transmission electron microscopy and immunofluorescence
To identify the contents of PA-CM and PPA-CM, the resuspended pellets were examined using transmission electron microscopy (TEM) (Ismail et al., 2012). Briefly, 7.5 μL of the pellet suspension was top loaded on 300-mesh grids and dried. The grids were stained in 2% uranyl acetate and visualized with energy-filtering TEM (Carl Zeiss Microscopy GmbH, Oberkochen, Germany) at 120 kV. The mean diameter of the EXs/ MVs was measured in eight microscopy field images (total 40) using ImageJ 1.47t software (National Institutes of Health, Bethesda, MD, USA). For immunofluorescence (IF), we followed the published protocols (Ng et al., 2013; Théry et al., 2006). In brief, 7.5 μL or 0.5 μg of purified pellet protein (measured by NanoDrop 2000 Spectrophotometer, Thermo Fisher Scientific, Wilmington, DE, USA, according to Desjardins et al., 2009) was incubated with 5 μL of 4-μm aldehyde/sulfate latex beads 4% wt/vol (Life Technologies Corp., Grand Island, NY, USA) in a 30-μL final volume of PBS at room temperature for 15 min. PBS (170 μL) was added, and the mixture was incubated in a test tube rotator for 2.5 h at room temperature. Next 22 μL of 1 M glycine/PBS (i.e., 100 mM final concentration) was added and mixed gently (to block the unbound sites of the latex beads) and then let stand on the bench for 30 min at room temperature. The beads were pelleted by centrifugation at 1,500×g for 3 min at room temperature, washed twice with 1 mL of PBS/0.5% bovine serum albumin (BSA). The EX–bead complex incubated with anti-CD9, immunoglobulin G (IgG) conjugated to fluorescein isothiocyanate isomer 1 (FITC; Abcam, catalog no. ab34162, Dawinbio, Seoul, Korea) for 1 h at room temperature. A negative control antibody reaction was performed using normal mouse IgG. The labeled EX–bead complexes were again pelleted and washed twice as above and finally resuspended in 20 μL of PBS/0.5% BSA. Ten microliters of the final complexes were spread on a microscope slide with a coverslip using Vectashield HardSet Mounting Medium (H-1400, Vector Laboratories Inc., Burlingame, CA, USA), air-dried, cover-slipped, and sealed with nail polish. The slides were examined using a HAL 100 fluorescence microscope (Carl Zeiss Microscopy GmbH). Another 10 μL of complexes were resuspended in 150 μL PBS/0.5% BSA for fluorescence-activated cell sorting (FACS) analysis (analyzed by Cell Lab Quanta SC, Beckman Coulter Inc., Fullerton, CA, USA) to estimate the fluorescence signals of the EX–bead complexes. The control positive was the porcine follicular fluid EXs (da Silveira et al., 2011), and the control negative was the PZM-5 in the above-mentioned procedures.
EXs/MVs labeling and internalization by the embryos
EXs/MVs pellets were subjected to fluorescent labeling using PKH67 dye (Sigma-Aldrich), a green fluorescent dye that labels the lipid membranes, according to the manufacturer's instructions and da Silveira et al. (2011), with some modifications. Briefly, a 5-μL EX suspension in PBS was resuspended in 120 μL of diluent C (provided by the manufacturer), mixed with freshly prepared PKH67 in 125 μL of diluent C to reach a final concentration of 5×10−6 M, and incubated for 5 min. Labeling was stopped by addition of an equal volume of 1% BSA in PBS and incubation for 1 min. The labeled EXs/MVs were washed two times in PZM-5 using ultracentrifugation and resuspended in 25 μL of PZM-5. Embryos were incubated with the labeled EXs/MVs for 22 h (da Silveira et al., 2011), washed twice with PBS, and stained with 25 μg mL−1 bisbenzamide for 10 min at room temperature. For negative controls, the embryos were cultured in plain PZM-5 for 22 h. The embryos were washed twice with PBS, mounted with a coverslip using Vectashield HardSet Mounting Medium (Vector Laboratories Inc.), and dried for 5 min at room temperature. Cellular uptake of EXs/MVs was observed under a confocal microscope (Zeiss, Oberkochen, Germany).
Total blastocyst cell count
Six hatching blastocysts from each experimental group were washed in PBS, and the nuclei were stained with 25 μg mL−1 bisbenzamide for 1 h at 37°C. Stained blastocysts were mounted on a glass slide in a drop of glycerol, gently flattened with a cover glass, and examined for cell counting with a fluorescence microscope using a 346-nm excitation filter. Digital photographs were also taken for total cell counting using ImageJ 1.42q software.
RNA extraction and cDNA synthesis
Total RNA was extracted and eluted from embryos (five blastocysts on day 6 of IVC) and EX pellets (7.5 μL), using the easy-spin™ (DNA-free) Total RNA Extraction Kit (iNtRON Biotechnology, Inc., KyungGi-Do, Korea) according to the manufacturer's instructions. RNA purity was evaluated using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) by estimating the ratios of absorbance at 230 nm, 260 nm, and 280 nm; values of A260/A280 of ≥2.0 and A260/A230 >2.0 are accepted and used for reverse transcription (RT). RT was carried out at 42°C for 60 min. Individual RT reactions of a total of 20 μL per reaction was performed using amfiRivert II cDNA Synthesis Premix (GenDEPOT, Barker, TX, USA) according to the manufacturer's instructions.
Because of the difficulty in obtaining a high number of NT embryos for EXs isolation, we analyzed the exosomal RNA contents of NT CM instead and compared it to the PA-CM. We cultured 60 embryos (one embryo/1-μL PZM-5 microdrops, three replicates of 20 embryos) of both NT and PA embryos. The CM were collected on days 2 and 4 of embryo culture. For CM total RNA extraction, it was expected that the RNA content would be too small; we followed the protocol of Mestdagh et al. (2008) with some modifications. Ten microliters of CM were collected by glass capillary pipette and heated at 95°C for 5 min to lyse and release the EXs contents (Malik et al., 2013). The entire lysate was used for pulsed RT to increase RT efficiency as follows: 40 cycles of 16°C for 2 min, 42°C for 1 min, and 50°C for 1 sec, followed by a final inactivation at 85°C for 5 min.
PCR and real-time PCR
cDNA (1–2 μL) of PA-CM of embryos on days 2, 4, 6, and 7 were subjected to PCR using a Maxime PCR PreMix kit-i-StarTaq (Intron Biotech., Seoul, Republic of Korea). The PCR amplification was carried out for one cycle of denaturation at 95°C for 5 min and a subsequent 40 cycles with denaturation at 95°C, annealing for 30 sec, extension at 72–C for 45 sec, and a final extension at 72°C for 5 min. Ten microliters of PCR products were fractionated on a 1% agarose gel (iNtRON Biotechnology, Inc., Korea) and stained with RedSafe ™ (iNtRON Biotechnology, Inc.). In all assays, cDNA template negative and reactions without RT resulted in negative amplification. The positive control was a cDNA of day-6 PA blastocysts, and the negative control was PZM-5.
For relative mRNA quantification, real-time PCR (qPCR) was done according to Takara Bio Inc. guidelines. A total 20-μL PCR reaction was made by adding 100 ng of cDNA, 1 μM forward primer, 1 μM reverse primer, 10 μL of SYBR Premix Ex Taq with ROX reference (Takara Bio Inc. Shiga, Japan), and 6 μL of nuclease-free water (Ambion Inc., Austin, TX, USA). The reaction was carried out using Applied Biosystems StepOnePlus™ Real-Time PCR Systems (Applied Biosystems, Forest City, CA, USA). The thermal profile for real-time PCR was 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 20 sec. Each transcript was relatively quantified in three replicates by calculation using the 2−ΔΔCt method (Livak and Schmittgen, 2001) for comparison of relative mRNA quantification in embryos after normalizing to the housekeeping gene GAPDH. Each sample was repeated three times. Primer sequences, annealing temperatures, and approximate sizes of the amplified fragments are listed in Table 1. The expressed PCR products were gel purified (QIAquick PCR Purification Kit, QIAGEN, Valencia, CA, USA), and DNA strands were directly sequenced (Macrogen, Seoul, Korea; http://dna.macrogen.com/kor) using the forward primer of the corresponding amplification. The identity of each product (≥95%) was confirmed by sequence homology analysis using the Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information (NCBI) GenBank (http://blast.ncbi.nlm.nih.gov/).
Table 1.
Primer Sequences and Product Size Used for RT–PCR and Real-Time Quantitative PCR
| Gene | Forward (3′→5′) | Reverse (3′→5′) | Size (bp) | Tm °C | Information |
|---|---|---|---|---|---|
| GAPDH | ACACTCACTCTTCTACCTTTG | CAAATTCATTGTCGTACCAG | 90 | 60 | DQ845173.1 (GenBank) |
| Oct4 | GTGAGAGGCAACCTGGAGAG | GAATGGGACCGAGGAGTACA | 297 | 60 | NM_001113060.1 (accession no.) |
| Nanog | TGAGGTTTATGGGCCTGAAG | ATCTGCTGGAGGCTGAGGTA | 297 | 60 | NM_001129971.1 (accession no.) |
| c-Myc | CTGCCAAGAGGGCTAAGTTG | AGCTTTTGCTCCTCTGCTTG | 277 | 60 | X97040.1 (GenBank) |
| Klf4 | CAGCTTCAGCTATCCGATCC | TGCCTTCAACACAAACTTGC | 256 | 60 | DQ000310.1 (GenBank) |
| Sox2 | GCCCTGCAGTACAACTCCAT | GCTGATCATGTCCCGTAGGT | 216 | 60 | EU519824.1 (GenBank) |
Statistical analysis
The ratios (cleavage rates and blastocyst formation rates) were evaluated using Pearson's chi-squared test. The means of relative mRNA quantification were compared using Student's t-test. Statistical significance was considered when the p value was less than 0.05.
Results
PA embryos improve cloned embryos in co-culture
In our initial experiment, co-culture of NT embryos with PA (NT/PA) embryos significantly increased cleavage (CR) and blastocyst formation (BL) rates than those cultured with cloned embryos (NT/NT) (Table 2) with the same embryo density (one embryo/10 μL); CR was 80.64% vs. 65.55% (p=0.02) and BL was 21.5% vs. 8.88% (p=0.017). There was no significant difference in day-6 blastocyst total cell count in both groups, p>0.05 (Table 2). Interestingly, within the NT/PA group, the CR in PA subgroup embryos was statistically higher than NT subgroup embryos; 90.9 % vs. 80.64 (p=0.04), whereas there was no difference between the two subgroups in blastocyst formation rate, 31.31% vs. 21.5% (p=0.12), respectively. On the other hand, PA embryos cultured alone (PAO) showed statistically increased CR% and BL% if compared to the cloned embryos in NT/PA group, 91.39% and 39.78% (p=0.006), respectively. Another experiment was performed using PA embryos that were punctured (PPA) with an 18-μm needle and cultured with cloned embryos; they showed no significant difference (p>0.05) in blastocyst formation when NT embryos were co-cultured with intact zona pellucida PA embryos (Table S1) (Supplementary Data are available at www.liebertpub.com/cell/). Additionally, cloned embryos were cultured with the same number of PA embryos (1XPA) or with double the number of PA embryos (2XPA) with maintaining the same embryo/medium density; they showed no significant difference in BL formation (Table S2).
Table 2.
Comparison Between Cloned and Parthenogenetic Embryo Development Either Cultured Separately or Co-Cultured Together
| NT/PA group | NT/NT group | PAO group | ||||
|---|---|---|---|---|---|---|
| TW-NT | PA | TW-NT | NT | TW-PA | PA | |
| Activated oocytes | 93 | 99 | 90 | 95 | 93 | 93 |
| CR (%) | 75 (80.64)a | 90 (90.9)b,d | 59 (65.55)c | 61 (64.21)c | 85 (91.39)d | 85 (91.39)d |
| BL (%) | 20 (21.5)a | 31 (31.31)a,d | 8 (8.88)b | 7 (7.36)b | 37 (39.78)d | 36 (38.7)d |
| Blastocyst cell count* | 39.3±3.2 | 36.9±5.4 | 38. 1±4.2 | 39.4±4.5 | 39.1±3.3 | 39.5±3.2 |
Values in the same row carrying different superscripts are considered statistically significant at p<0.05.
The mean of cell count of six hatching blastocysts±standard deviation (SD).
NT, nuclear transfer (cloned) embryos; PA, parthenogenetic embryos; PAO, parthenogenetic embryos only; TW, activated oocytes placed in the Transwell Inserts; CR, cleavage rate; BL, blastocyst formation rate.
qPCR analysis of cloned embryos
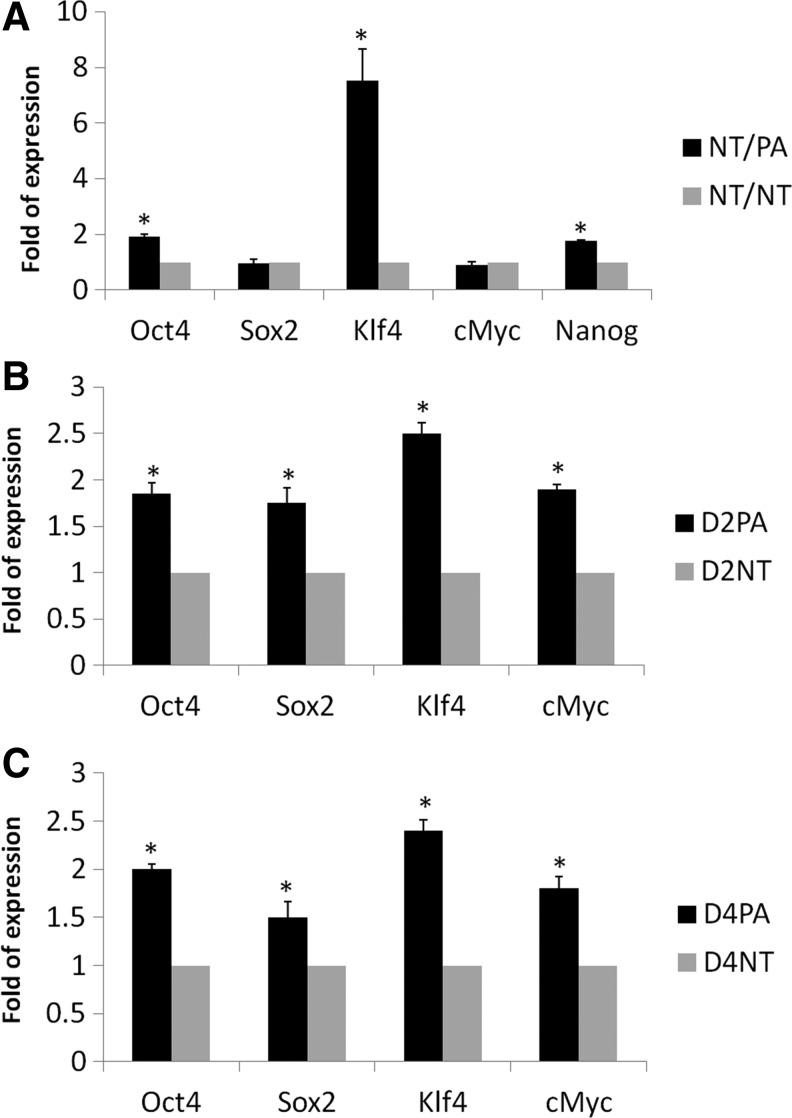
Next, we analyzed the relative mRNA quantification between the NT/PA and NT/NT groups. The mRNA relative expression of each gene in the NT/NT group was arbitrarily set as one-fold. The relative quantitative expression of selected pluripotency genes showed some significant changes between the two groups (Fig. 2A); Oct4 (p=0.003), Klf4 (p=0.01), and Nanog (p=0.001) expression were significantly higher in the NT/PA group, whereas Sox2 (p=0.074) and c-Myc (p=0.11) showed no significant difference when compared with the NT/NT group.
FIG. 2.
Relative quantitation of pluripotency genes in cloned embryos co-cultured with parthenogenetic embryos (NT/PA) (A) compared with the control cloned embryos (NT/NT) and in their conditioned medium (B) by real-time PCR. (A) Relative quantitation of pluripotency mRNAs in cloned blastocysts of NT/PA and NT/NT groups was compared; the expression in NT/NT group was used as an arbitrary unit. (B and C) Relative quantitation of pluripotency mRNAs was compared between the total RNA isolated from NT and PA CM on days 2 and 4, respectively; the expression in NT group was used as an arbitrary unit. (*) The value is significant when p≤0.05.
Presence of EXs/MVs in the PA-CM
On the basis of these results, we cultured the PA embryos alone to investigate the possible secreted factors that might increase the cloned embryo competence and pluripotency mRNA expression during co-culture. CM was subjected to gradient centrifugation to obtain a pellet. The resuspended pellets were subjected to TEM and IF analysis. In negative-staining TEM, the diameter analysis revealed the presence of varied particles or spheres, ranging from 35.4±6.9 nm (in early embryonic stages, Fig. 3A) to 101.66±18.4 nm, along with the small particles in the late stages of embryo development (Fig. 3B). To characterize these particles, and due to their tiny sizes, we used latex beads to adsorb these particles and immunostained them with fluorescent antibodies against CD9. The results showed that these spheres were CD9+ (Fig. 4A) when compared to the control positive sample, which was the porcine follicular fluid (da Silveira et al., 2011) (Fig. 4B), and control negative sample, which was plain PZM-5 (Fig. 4C). Appropriate gating of FACS analysis demonstrated an intensity of 32.78% in PA-CM against 0.02% in the plain PZM (p<0.01). Experiments were repeated with three independent EXs/MVs preparations with similar results.
FIG. 3.
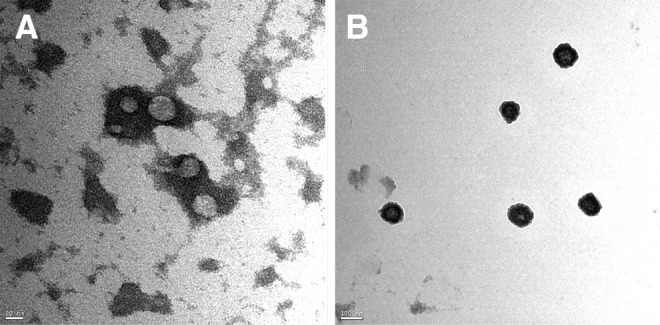
PA embryos–derived EXs/MVs are identified by electron microscope analysis. TEM images show the presence of particles ranged from 30 nm (A) to less than 120 nm (B) in the PA-CM pellet isolated by differential centrifugation after negative staining with uranyl acetate. Bars, 20 nm and 100 nm, respectively.
FIG. 4.
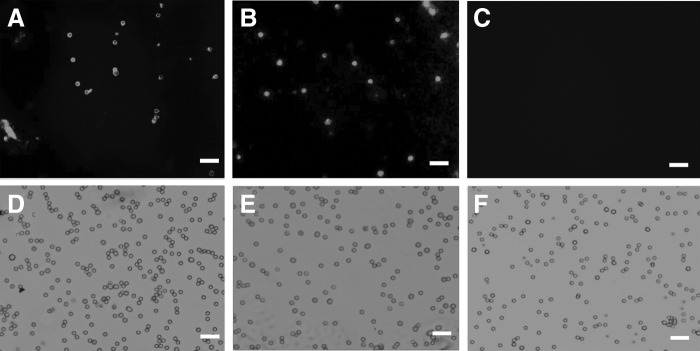
PA embryos–derived EXs/MVs are identified by immunofluourescence. Because the EXs size is too small to be analyzed reliably by direct cell sorting, the pellets containing EXs were bound to beads of a size that is in the detection range of the fluorescent microscopes (4-μm diameter latex beads ). The beads were then bound to fluorescent antibodies against CD9 and demonstrated a strong intensity for the exosomal surface proteins CD9 (A). Controls were the porcine follicular fluid as a positive control (B) (da Silveiraet al., 2011) and the plain PZM-5 as a negative control (C). Images were taken under an ultraviolet (UV) microscope (A–C) and visible light (D–F) showing the 4-μm beads of A–C, respectively. Bars, 20 μm.
Both PA and NT embryos conditioned media contain mRNA of pluripotency genes
Taken into consideration that making an artificial hole in the zona pellucida with an 18-μm diameter needle is sufficient to pass the EXs and MVs, we compared the mRNA contents of pluripotency genes (Oct4, Sox2, Klf4, c-Myc, and Nanog) in the resulting exosomal pellet of intact PA and PPA embryos (Fig. 5). In the early stages (days 2 and 4), there was no significant difference between the expression of all genes except for c-Myc at day-2 embryos which was found in PPA only. In the later stages (days 6 and 7), there was no significant difference between the two groups except in Nanog expression at day 7.
FIG. 5.
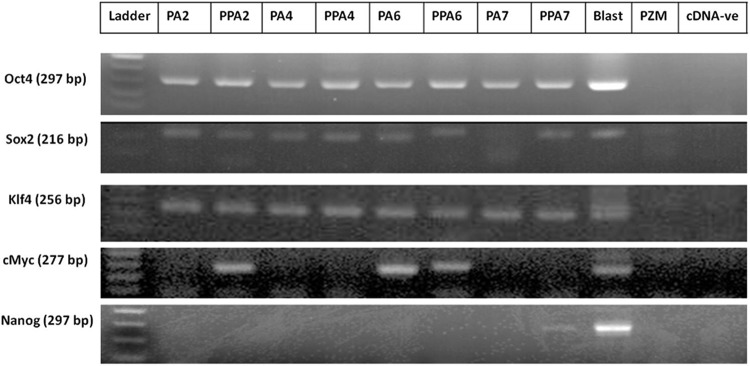
Expression of pluripotency genes in purified EXs/MVs derived from PA embryos. Photomicrograph of gel electrophoresis of PCR products from intact (PA) and punctured (PPA) parthenogenetic embryos on days 2, 4, 6, and 7 of in vitro culture. Day-7 blastocysts (blast) were used as a positive control and plain PZM-5 was used as a negative control (PZM); the other lane was cDNA negative (cDNA-ve) to exclude the primer dimer formation.
Additionally, we compared the relative quantitation of pluripotency mRNAs (Pou5f, Sox2, c-Myc, and Klf4) in CM of small numbers (three replicates of 20 embryos) of PA and NT embryos on days 2 and 4 of embryo culture. The mRNA relative expression of each gene in NT embryos CM was arbitrarily set as one-fold. The relative quantitative expression of selected pluripotency mRNAs in PA embryos was significantly higher than NT embryos either on day 2 or day 4 (Fig. 2B, C).
Labeled EXs/MVs internalization by the developing embryos
To evaluate the possibility of EXs/MVs-mediated transfer of molecules during embryo co-culture, labeled cell membrane EXs/MVs uptake was conducted using the pellets of PA-CM. Purified EXs/MVs were labeled with a green fluorescent PKH67 dye and added to the culture medium of NT embryos. Following a co-incubation period of 22 h, labeled EXs/MVs-treated embryos were washed, fixed, and examined by a confocal microscope. Microscopy results showed effective uptake of PKH67-labeled EXs/MVs by NT embryos (Fig. 6A), whereas NT embryos cultured in plain PZM-5 showed no signals for green fluorescence (Fig. 6B), suggesting that embryo-derived EXs/MVs can be internalized by other embryos sharing the same culture medium.
FIG. 6.
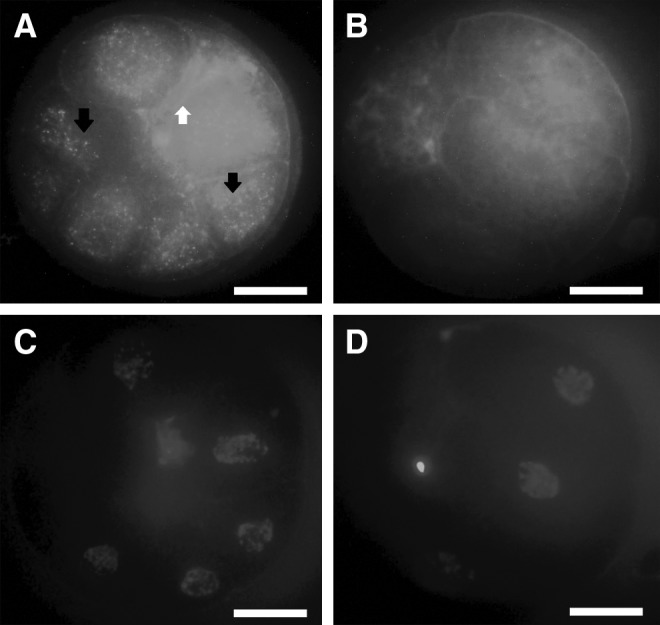
Uptake of PA embryos–derived EXs/MVs by cloned embryos. EXs/MVs purified from PA-CM were labeled with PKH67 dye and added to day-2 cloned embryos for 22 h. PKH67-labeled EXs/MVs emitted fluorescent signals (A). Black arrows indicate internalization of the labeled EXs/MVs by the blastomeres' cytoplasm; the white arrow indicates the blastomere plasma membrane showing no fluorescent signals. Cloned embryos were cultured in plain PZM-5 as a negative control (B). Nuclei are stained blue with 4′,6-diamidino-2-phenylindole (DAPI) (C and D). Scale bar, 20 μm.
PA-CM cannot improve NT embryo developmental competence
PA-CM from days 2, 4, and 6 were supplemented either synchronized to the embryo development course (PA-CM of days 2, 4, and 6 were transferred on days 2, 4, and 6, respectively) or preceded by 2 days (PA-CM of days 2, 4, and 6 were transferred on days 0, 2, and 4, respectively). Supplementation of CM in both groups showed no significant effect on embryo development (Table 3) (p>0.05).
Table 3.
Effect of Temporal Supplementation with Parthenogenetic Embryos Conditioned Medium on Cloned Embryos Development
| Controla | CM 0–2–4b | CM 2–4–6c | |
|---|---|---|---|
| Fused oocytes | 39 | 36 | 39 |
| CR (%) | 30 (76.92) | 28 (77.77) | 29 (74.35) |
| BL (%) | 8 (20.51) | 4 (11.11) | 7 (17.94) |
Control group was supplemented by plain porcine zygote medium-5 (PZM-5) on days 2, 4, and 6.
Conditioned medium of cultured parthogenetic (PA) embryos of days 2, 4, and 6 were transferred on days 0, 2, and 4, respectively.
Conditioned medium of cultured PA embryos of days 2, 4, and 6 were transferred on days 2, 4, and 6, respectively.
CM, conditioned medium; CR, cleavage rate; BL, blastocyst formation rate.
Discussion
The early preimplantation mammalian embryo is known to be relatively autonomous and can regulate its own development independently until hatching in vitro (Schultz and Heyner, 1993). Nevertheless, a number of previous reports proved that in vitro culture of embryos is remarkably successful when the embryos are kept in large groups during the whole culture period (Ferry et al., 1994; Hoelker et al., 2008; Paria and Dey, 1990). Embryos can generate their microenvironment through secretion of complex and enigmatic trophic autocrine/paracrine growth factors, termed the “secretome” (Bormann et al., 2006; Hoelker et al., 2010; Katz-Jaffe et al., 2006; O'Neill, 1998; O'Neill, 2008). Particularly, cloned embryos (NT) are more sensitive to culture (Yamanaka et al., 2009b), and because of the labor required, few embryos are produced and cultured in vitro.
Here, we attempted to improve the porcine cloning efficiency through co-culture with PA embryos. We used a new co-culture system for embryos consisting of Transwell Membrane Inserts (0.4-μm pores) to separate large groups of different kinds of embryos (NT and PA) and permitting dynamic paracrine interaction through the exchange of their microenvironments. In our results, cloned embryos that were co-cultured with PA embryos showed a significant increase in developmental competency in terms of embryo cleavage (CR=80.64%) and blastocyst formation (BL=21.5%). The cloned embryos were cultured separately [65.55% and 8.88%, respectively (p<0.05)] (Table 2). This result is consistent with the result of Terashita et al. (2011), who showed that co-culture of PA but not IVF embryos with cloned ones, using the well-of-the-well (WOW) system, increased blastocyst formation in cloned embryos. However, this system is beneficial only for using few embryos in a small amount of culture medium (Vajta et al., 2008).
Interestingly, in the NT/PA group, NT and PA subgroups showed no significant difference in blastocyst formation rate between them (Table 2); however, there was a significant difference between these subgroups and those of cultured PA embryos (PAO; PA/PA) (Table 2).
Moreover, the RQ (relative quantification) of Oct4, Klf4, and Nanog showed a significant increase in co-cultured cloned embryos than that of control ones (Fig. 2A), which might be the reason for increasing co-cultured cloned embryo competency. Oct4, Sox2, c-Myc, Klf4, and Nanog are all considered the pluripotency genes in most mammals, with differences in temporal and spatial expression in different animals (du Puy et al., 2011). These genes are the key players in reprogramming of the NT–derived embryos, and they were reported to be lower in NT embryos if compared to other in vitro–derived embryos (Niwa et al., 2000; Rodríguez et al., 2012; Zhang et al., 2011; Zhou et al., 2013). The full mechanism of network interaction between these genes is still not elucidated in the pig because of their unique expression in this species (du Puy et al., 2011); however, there were some published reports regarding the mutual interaction of two (Rodda et al., 2005) or more pluripotency genes (du Puy et al., 2011). The increase in RQ of some pluripotency genes in cloned embryos in the current study can be explained simply because of sharing the microenvironment or secretome that was generated by the in vitro–competent PA embryos (CR=90.9% and BL%=31.31%; Table 2).
To investigate the possible reason for the increment in these mRNAs in co-cultured embryos, we cultured the PA embryos separately and collected their CM (PA-CM), which was analyzed through TEM after differential centrifugation. Interestingly, small particles of 30–120 nm diameter (Fig. 3A, B) were observed; they were <40 nm in the two-cell stage until blastocyst formation and <120 nm from the blastocyst stage expansion until hatching. To characterize these particles, the pellet contents were adsorbed to latex beads and were subjected to fluorescent antibodies against CD9. The beads were found to be CD9+, which confirmed that these structures are membrane-derived vesicles, EXs, or MVs (Fig. 4A). The tetraspanin CD9, a membrane-bound protein, is commonly used to identify EXs/MVs (Mathivanan and Simpson, 2009) and is involved in cell packaging of proteins in EXs (Chairoungdua et al., 2010). Moreover, it is expressed in the oocytes and required for fertilization (Barraud-Lange et al., 2012; Li et al., 2004c; Miyado et al., 2000) as well as expressed in the blastocysts (Goissis et al., 2009). We confirmed CD9 expression in different stages of PA embryos (Fig. S1).
Next, we analyzed the mRNA of pluriopotency genes in the contents of the isolated pellets by PCR. We compared the mRNA expression in the pellets derived from intact PA and punctured PA embryos, because the artificial hole that was made by an 18-μm needle is sufficient to pass the EXs/MVs. All of pluriopotency genes (Oct4, Sox2, c-Myc, Klf4) were conspicuously expressed except Nanog, which was found to be expressed in the late stage of blastocysts (Fig. 5); this is consistent with previous studies (du Puy et al., 2011). We compared our mRNA results to the Vesiclepedia (http://microvesicles.org), which is a compendium database of extracellular vesicles and exosomal proteins, RNAs, and lipids (Kalra et al., 2012), to validate our findings and found that all target mRNAs in our study have been identified by other groups in EXs/MVs from different cell types (Kalra et al., 2012).
In our results, some mRNAs were found to exist either along with the embryo developmental course (Oct4, Sox2, and Klf4) or intermittently expressed (c-Myc) in the EXs/MVs. From this comparison, we assume the presence of the same structures that are able to pass either from an artificial hole or from the intact zona pellucida and confirmed by the results of the development of cloned embryos that were co-cultured either with intact (PA) or with punctured (PPA) zona pellucida (Table S2); there was no significant difference between the two groups (p>0.05). Therefore, it can be hypothesized that the mRNA was transferred from PA embryos to cloned embryos through the EXs/MVs.
Moreover, we compared the relative quantification of the candidate mRNAs in PA and NT embryo CM. To release the mRNA cargoes from the EXs/MVs, we heated the CM at 95°C to lyse the EXs/MVs. The whole lysate was used for real-time PCR analysis. The relative quantification of Oct4, Sox2, Klf4, and c-Myc was significantly increased in the CM of PA embryos than in those of cloned ones on days 2 and 4 of embryos culture when normalized to the GAPDH mRNA of both groups (Fig. 2B–C).
Following the confirmation of presence of mRNA in the CM and purified EXs/MVs, we examined the uptake of EXs/MVs by cloned embryos. EXs/MVs were labeled with PKH67 fluorescent dye and were co-cultured with cloned embryos in vitro for 22 h. The fluorescent microscopy revealed the presence of green fluorescent signals in cultured cloned embryos (Fig. 6). Importantly, the fluorescent microscopy results also showed that the labeled EXs/MVs were taken up by the blastomeres; there were no fluorescent signals in plasma membrane of the blastomeres (Fig. 6A), indicating that EXs/MVs have been internalized in to the blastomeres.
During the past decade, membrane-derived EXs/MVs have received much attention regarding intercellular communication because compelling experimental evidence has proved their involvement as bioactive organelles in carrying nucleic acid and protein cargoes between the cells (for review, see Shifrin et al., 2013). But can EXs/MVs pass through the intact zona pellucida? To answer this question, we have to look into the porcine zona pellucida porosity and permeability. The zona pellucida appeared by scanning EM to be a sponge with a complex fibrous network interspersed with numerous pores (Van Soom et al., 2010). The physical dimension of the zona pellucida differed from in vivo– and in vitro–derived zygotes (Funahashi et al., 2001). The zona pellucida was found to be more porous in the latter and made in vitro–produced embryos more prone to pathogen entrapment and passage of nanostructures (Kim et al., 1996). This may be because of the handling of oocytes and embryos during denuding and culture that affects the zona pellucida integrity and porosity. Interestingly, the zona reaction and subsequent changes in the zona surface differs between IVF and PA embryos; the zona pellucida resumed its porous characteristics after parthenogenetic activation of bovine oocytes (Suzuki et al., 2000). Molecule permeability through the zona depends on the molecule's size and physicochemical factors, such as hydrophilic–lipophilic properties; lipid-containing molecules in particular penetrate the zona pellucida with relative ease (Turner and Horobin, 1997). In the pig, some viruses can penetrate the zona and reach inside the embryos (Bane et al., 1990; Mateusen et al., 2006). Moreover, fluorescent inert microspheres with sizes of 20 nm and 26 nm were able to cross the zona; however, 200-nm microspheres cannot (Mateusen et al., 2004). There was a wide range between the particles used in this experiment, around a seven-fold difference. Taken together, according to our findings and the previous reports, we can suggest that the pig zona pellucida can permit the passage of inert particles less than 200 nm in diameter. This is consistent with our findings; EXs/MVs were less than 40 nm in early-stage embryos and less than 120 nm in late-stage embryos (Fig. 3A, B).
In the current results, some embryonic mRNAs were increased significantly (Oct4, Klf4, and Nanog; Fig. 2A), and other genes showed no significant difference with the control group (c-Myc and Sox2; Fig. 2A), suggesting a network interaction/feedback mechanism between the endogenous and exogenous genes to reach cellular balance or homeostasis to adjust the cellular activity after transfer of external mRNAs.
Furthermore, we explored the temporal effect of PA-CM supplementation on the development of the cloned embryos. PA-CM was supplemented either along with the embryo development course or preceded by 2 days showed no significant effect on embryo development (Table 3); however, it showed some tendency (p=0.26) to adversely affect embryo development. This paradox suggests a temporal interaction of the transferred mRNAs (du Puy et al., 2011; Kim et al., 2008; Ng and Surani, 2011) and reflects the dramatic changes in the daily output of the embryos secretome (Katz-Jaffe et al., 2006). This result also provides the importance of embryo co-culture conditions and the continuous supply of embryo-derived factors to improve cloning efficiency in porcine embryos.
We suggest that the increment of the pluripotency genes is caused by continuous transfer of mRNA cargoes from PA to NT embryos via EXs/MVs, but the acute transfer through CM supplementation paradoxically affects embryo development. It was suggested that foreign mRNA's stability in cells is often tightly and coordinately regulated and the transcriptional rates are low; these mRNAs are rapidly turned over with half-lives of 20–40 min (Rajagopalan and Malter, 1996; Wisdom and Lee, 1991). Therefore, continuous supplementation of mRNA, which was achieved by co-culture, is advantageous over the acute transfer of mRNA by the CM, confirming the concept of dynamic microenvironment or the niche among the cultured embryos.
In summary, our result showed the usefulness of co-culturing PA embryos with cloned embryos, as indicated by increasing cleavage rate and blastocyst formation in the cloned embryos. We suggest that in vitro–derived embryos can secrete EXs/MVs in their CM as possible new mediators within the embryonic microenvironment and embryo-to-embryo communication.
Supplementary Material
Acknowledgments
We thank Marek Molas, PhD, for assistance in the statistical analysis. This study was financially supported by grants from IPET (#311011-05-3-SB010), MOTIE (#10033839-2013), Research Institute for Veterinary Science, the BK21 PLUS program, and TS Corporation.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Bane D.P., James J.E., Gradil C.M., and Molitor T.W. (1990). In vitro exposure of preimplantation porcine embryos to porcine parvovirus. Theriogenology 33, 553–561 [DOI] [PubMed] [Google Scholar]
- Baranao R.I., Piazza A., Rumi L.S., and Polak de Fried E. (1997). Determination of IL-1 and IL-6 levels in human embryo culture-conditioned media. Am. J. Reprod. Immunol. 37, 191–194 [DOI] [PubMed] [Google Scholar]
- Barraud-Lange V., Chalas Boissonnas C., Serres C., Auer J., Schmitt A., Lefevre B., Wolf J.P., and Ziyyat A. (2012). Membrane transfer from oocyte to sperm occurs in two CD9-independent ways that do not supply the fertilising ability of CD9-deleted oocytes. Reproduction 144, 53–66 [DOI] [PubMed] [Google Scholar]
- Bavister B.D. (1988). Role of oviductal secretions in embryonic growth in vivo and in vitro. Theriogenology 29, 143–154 [Google Scholar]
- Bormann C.L., Swain J.E., Ni Q., Kennedy R.T., and Smith G.D. (2006). Preimplantation embryo secretome identification. Fertil. Steril. 86, S116–S116 [Google Scholar]
- Chairoungdua A., Smith D.L., Pochard P., Hull M., and Caplan M.J. (2010). Exosome release of beta-catenin: A novel mechanism that antagonizes Wnt signaling. J. Cell Biol. 190, 1079–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong A.W.Y., Lee Y.L., Liu W.M., Yeung W.S.B., and Lee K.F. (2009). Oviductal microsomal epoxide hydrolase (EPHX1) reduces reactive oxygen species (ROS) level and enhances preimplantation mouse embryo development. Biol. Reprod. 81, 126–132 [DOI] [PubMed] [Google Scholar]
- da Silveira J.C., Veeramachaneni D.N.R., Winger Q.A., Carnevale E.M., and Bouma G.J. (2011). Cell-secreted vesicles in equine ovarian follicular fluid contain mirnas and proteins: A possible new form of cell communication within the ovarian follicle. Biol. Reprod. 86, 71–71 [DOI] [PubMed] [Google Scholar]
- Dadi T.D., Li M.W., and Lloyd K.C. (2007). EGF and TGF-alpha supplementation enhances development of cloned mouse embryos. Cloning Stem Cells 9, 315–326 [DOI] [PubMed] [Google Scholar]
- Desai N., and Goldfarb J. (1998). Co-cultured human embryos may be subjected to widely different microenvironments: Pattern of growth factor/cytokine release by Vero cells during the co-culture interval. Hum. Reprod. 13, 1600–1605 [DOI] [PubMed] [Google Scholar]
- Desjardins P., Hansen J. B., Allen M. (2009). Microvolume protein concentration determination using the NanoDrop 2000c spectrophotometer. J. Vis. Exp. (33), e1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Puy L., Lopes S.M.C.D., Haagsman H.P., and Roelen B.A.J. (2011). Analysis of co-expression of OCT4, NANOG and SOX2 in pluripotent cells of the porcine embryo, in vivo and in vitro. Theriogenology 75, 513–526 [DOI] [PubMed] [Google Scholar]
- Estrada J., Sommer J., Collins B., Mir B., Martin A., York A., Petters R.M., and Piedrahita J.A. (2007). Swine generated by somatic cell nuclear transfer have increased incidence of intrauterine growth restriction (IUGR). Cloning Stem Cells 9, 229–236 [DOI] [PubMed] [Google Scholar]
- Ferry L., Mermillod P., Massip A., and Dessy F. (1994). Bovine embryos cultured in serum-poor oviduct-conditioned medium need cooperation to reach the blastocyst stage. Theriogenology 42, 445–453 [DOI] [PubMed] [Google Scholar]
- Funahashi H., Ekwall H., Kikuchi K., and Rodriguez-Martinez H. (2001). Transmission electron microscopy studies of the zona reaction in pig oocytes fertilized in vivo and in vitro. Reproduction 122, 443–452 [DOI] [PubMed] [Google Scholar]
- Funston R.N., Nauta W.J., and Seidel G.E., Jr (1997). Culture of bovine embryos in buffalo rat liver cell-conditioned media or with leukemia inhibitory factor. J. Anim. Sci. 75, 1332–1336 [DOI] [PubMed] [Google Scholar]
- Gardiner C.F., Ferrra J.F., Poli M., Turner K., Child T., and Sargent I.L. (2013). IVF embryos release extracellular vesicles which may act as an indicator of embryo quality. J. Extracell. Vesicles 2, 20826 http://dx.doi.org/10.3402/jev.v2i0.20826
- Goissis M.D., de Barros F.R.O., Marques M.G., Mendes C.M., Milazzotto M.P., Viana C.H.C., Assumpção M.E.O.A., and Visintin J.A. (2009). 276 expression of CD9 and alpha6-integrin in porcine blastocysts. Reprod. Fertil. Dev. 21, 235 [Google Scholar]
- Hoelker M., Rings F., Lund Q., Ghanem N., Phatsara C., Griese J., Schellander K., and Tesfaye D. (2008). Effect of the microenvironment and embryo density on developmental characteristics and gene expression profile of bovine preimplantative embryos cultured in vitro. Reproduction 137, 415–425 [DOI] [PubMed] [Google Scholar]
- Hoelker M., Rings F., Lund Q., Phatsara C., Schellander K., and Tesfaye D. (2010). Effect of embryo density on in vitro developmental characteristics of bovine preimplantative embryos with respect to micro and macroenvironments. Reproduction in Domestic Animals=Zuchthygiene 45, e138–e145 [DOI] [PubMed] [Google Scholar]
- Ishiwata I., Tokieda Y., Kiguchi K., Sato K., and Ishikawa H. (2000). Effects of embryotrophic factors on the embryogenesis and organogenesis of mouse embryos in vitro. Hum. Cell 13, 185–195 [PubMed] [Google Scholar]
- Ismail N., Wang Y., Dakhlallah D., Moldovan L., Agarwal K., Batte K., Shah P., Wisler J., Eubank T.D., Tridandapani S. and others. (2012). Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 121, 984–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra H., Simpson R.J., Ji H., Aikawa E., Altevogt P., Askenase P., Bond V.C., Borras F.E., Breakefield X., Budnik V., et al. (2012). Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 10, e1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsman D., Stackpole E.J., Domin D.R., and Farber D.B. (2012). Embryonic stem cell-derived microvesicles induce gene expression changes in Muller cells of the retina. PloS One 7, e50417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz-Jaffe M.G., Schoolcraft W.B., and Gardner D.K. (2006). Analysis of protein expression (secretome) by human and mouse preimplantation embryos. Fertil. Steril. 86, 678–685 [DOI] [PubMed] [Google Scholar]
- Kim J., Chu J., Shen X., Wang J., and Orkin S.H. (2008). An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N.H., Funahashi H., Abeydeera L.R., Moon S.J., Prather R.S., and Day B.N. (1996). Effects of oviductal fluid on sperm penetration and cortical granule exocytosis during fertilization of pig oocytes in vitro. J. Reprod. Fertil. 107, 79–86 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Takagi Y., Satoh T., Hoshi H., and Oikawa T. (1992). Development of early bovine embryos to the blastocyst stage in serum-free conditioned medium from bovine granulosa cells. In Vitro Cell. Dev. Biol. 28A, 255–259 [DOI] [PubMed] [Google Scholar]
- Koo O.J., Park H.J., Kwon D.K., Kang J.T., Jang G., and Lee B.C. (2009). Effect of recipient breed on delivery rate of cloned miniature pig. Zygote 17, 203–207 [DOI] [PubMed] [Google Scholar]
- Kwon D.K., Koo O.J., Park S.J., Kang J.T., Park H.J., Kim S.J., Moon J.H., Saadeldin I.M., Jang G., and Lee B.C. (2011). Optimizing porcine oocytes electrical activation by adjusting pre- and post-activation mannitol exposure time. Biol. Reprod. 85, 176. [DOI] [PubMed] [Google Scholar]
- Lässer C., Eldh M., and Lötvall J. (2012). Isolation and characterization of RNA-containing exosomes. J. Vis. Exp. e3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., El Andaloussi S., and Wood M.J. (2012). Exosomes and microvesicles: Extracellular vesicles for genetic information transfer and gene therapy. Hum. Mol. Genet. 21, R125–R134 [DOI] [PubMed] [Google Scholar]
- Lee Y.L., Xu J.S., Chan S.T.H., Ho P.C., and Yeung W.S.B. (2001). Vero cells, but not oviductal cells, increase the hatching frequency and total cell count of mouse blastocysts partly by changing energy substrate concentrations in culture medium. J. Assist. Reprod. Gen. 18, 566–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.L., Lee K.F., Xu J.S., Kwok K.L., Luk J.M., Lee W.M., and Yeung W.S. (2003). Embryotrophic factor-3 from human oviductal cells affects the messenger RNA expression of mouse blastocyst. Biol. Reprod. 68, 375–382 [DOI] [PubMed] [Google Scholar]
- Li G.P., Bunch T.D., White K.L., Aston K.I., Meerdo L.N., Pate B.J., and Sessions B.R. (2004a). Development, chromosomal composition, and cell allocation of bovine cloned blastocyst derived from chemically assisted enucleation and cultured in conditioned media. Mol. Reprod. Dev. 68, 189–197 [DOI] [PubMed] [Google Scholar]
- Li G.P., White K.L., Aston K.I., Meerdo L.N., and Bunch T.D. (2004b). Conditioned medium increases the polyploid cell composition of bovine somatic cell nuclear-transferred blastocysts. Reproduction 127, 221–228 [DOI] [PubMed] [Google Scholar]
- Li Y.H., Hou Y., Ma W., Yuan J.X., Zhang D., Sun Q.Y., and Wang W.H. (2004c). Localization of CD9 in pig oocytes and its effects on sperm-egg interaction. Reproduction 127, 151–157 [DOI] [PubMed] [Google Scholar]
- Liu L.P., Chan S.T., Ho P.C., and Yeung W.S. (1998). Partial purification of embryotrophic factors from human oviductal cells. Hum. Reprod. 13, 1613–1619 [DOI] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Malik Z.A., Kott K.S., Poe A.J., Kuo T., Chen L., Ferrara K.W., and Knowlton A.A. (2013). Cardiac myocyte exosomes: Stability, HSP60, and proteomics. Am. J. Physiol. Heart Circ. Physiol. 304, H954–H965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateusen B., Sanchez R.E., Van Soom A., Meerts P., Maes D.G., and Nauwynck H.J. (2004). Susceptibility of pig embryos to porcine circovirus type 2 infection. Theriogenology 61, 91–101 [DOI] [PubMed] [Google Scholar]
- Mateusen B., Soom A.V., Maes D.G.D., Favoreel H., and Nauwynck H.J. (2006). Receptor-determined susceptibility of preimplantation embryos to pseudorabies virus and porcine reproductive and respiratory syndrome virus. Biol. Reprod. 76, 415–423 [DOI] [PubMed] [Google Scholar]
- Mathivanan S., and Simpson R.J. (2009). ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 9, 4997–5000 [DOI] [PubMed] [Google Scholar]
- Mermillod P., Vansteenbrugge A., Wils C., Mourmeaux J.L., Massip A., and Dessy F. (1993). Characterization of the embryotrophic activity of exogenous protein-free oviduct-conditioned medium used in culture of cattle embryos. Biol. Reprod. 49, 582–587 [DOI] [PubMed] [Google Scholar]
- Mestdagh P., Feys T., Bernard N., Guenther S., Chen C., Speleman F., and Vandesompele J. (2008). High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Rese. 36, e143–e143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyado K., Yamada G., Yamada S., Hasuwa H., Nakamura Y., Ryu F., Suzuki K., Kosai K., Inoue K., Ogura A., Okabe M., and Mekada E. (2000). Requirement of CD9 on the egg plasma membrane for fertilization. Science 287, 321–324 [DOI] [PubMed] [Google Scholar]
- Ng H.-H., and Surani M.A. (2011). The transcriptional and signalling networks of pluripotency. Nat. Cell Biol. 13, 490–496 [DOI] [PubMed] [Google Scholar]
- Ng Y.H., Rome S., Jalabert A., Forterre A., Singh H., Hincks C.L., and Salamonsen L.A. (2013). Endometrial exosomes/microvesicles in the uterine microenvironment: A new paradigm for embryo-endometrial cross talk at implantation. PloS One 8, e58502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., and Smith A.G. (2000). Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- O'Neill C. (1997). Evidence for the requirement of autocrine growth factors for development of mouse preimplantation embryos in vitro. Biol. Reprod. 56, 229–237 [DOI] [PubMed] [Google Scholar]
- O'Neill C. (1998). Autocrine mediators are required to act on the embryo by the 2-cell stage to promote normal development and survival of mouse preimplantation embryos in vitro. Biol. Reprod. 58, 1303–1309 [DOI] [PubMed] [Google Scholar]
- O'Neill C. (2008). The potential roles for embryotrophic ligands in preimplantation embryo development. Hum. Reprod. Update 14, 275–288 [DOI] [PubMed] [Google Scholar]
- Okada K., Krylov V., Kren R., and Fulka J., Jr (2006). Development of pig embryos after electro-activation and in vitro fertilization in PZM-3 or PZM supplemented with fetal bovine serum. J. Reprod. Dev. 52, 91–98 [DOI] [PubMed] [Google Scholar]
- Paria B.C., and Dey S.K. (1990). Preimplantation embryo development in vitro: cooperative interactions among embryos and role of growth factors. Proc. Natl. Acad. Sci. USA 87, 4756–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan L.E., and Malter J.S. (1996). Turnover and translation of in vitro synthesized messenger RNAs in transfected, normal cells. J. Biol. Chem. 271, 19871–19876 [DOI] [PubMed] [Google Scholar]
- Ratajczak J., Miekus K., Kucia M., Zhang J., Reca R., Dvorak P., and Ratajczak M.Z. (2006). Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20, 847–856 [DOI] [PubMed] [Google Scholar]
- Richter K.S. (2008). The importance of growth factors for preimplantation embryo development and in-vitro culture. Curr. Opin. Obstet. Gynecol. 20, 292–304 [DOI] [PubMed] [Google Scholar]
- Rodda D.J., Chew J.L., Lim L.H., Loh Y.H., Wang B., Ng H.H., and Robson P. (2005). Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 280, 24731–24737 [DOI] [PubMed] [Google Scholar]
- Rodríguez A., Allegrucci C., and Alberio R. (2012). Modulation of pluripotency in the porcine embryo and iPS cells. PloS One 7, e49079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz G.A., and Heyner S. (1993). Growth factors in preimplantation mammalian embryos. Oxf. Rev. Reprod. Biol. 15, 43–81 [PubMed] [Google Scholar]
- Shifrin D.A., Beckler M.D., Coffey R.J., and Tyska M.J. (2013). Extracellular vesicles: Communication, coercion, and conditioning. Mol. Biol. Cell 24, 1253–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki C., Iwamura S., and Yoshioka K. (2004). Birth of piglets through the non-surgical transfer of blastocysts produced in vitro. J. Reprod. Dev. 50, 487–491 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Ju J.-C., and Yang X. (2000). Surface ultrastructural alterations of bovine oocytes after parthenogenetic activation. Cloning 2, 69–78 [DOI] [PubMed] [Google Scholar]
- Terashita Y.S., Kudo Y., Amano R., and Sato E. (2011). Combination of co-culture with parthenogenetic embryos and aggregation enhances in vitro development and quality of miniature pig somatic cell nuclear transfer embryos Reprodbiotech June28, 2011, published ahead of online issue
- Thery C. (2011). Exosomes: Secreted vesicles and intercellular communications. F1000 Biol. Rep. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Amigorena S., Raposo G., and Clayton A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 3.22.1–3.22.29 [DOI] [PubMed] [Google Scholar]
- Turner K., and Horobin R.W. (1997). Permeability of the mouse zona pellucida: A structure-staining-correlation model using coloured probes. J. Reprod. Fertil. 111, 259–265 [DOI] [PubMed] [Google Scholar]
- Vajta G., Korosi T., Du Y., Nakata K., Ieda S., Kuwayama M., and Nagy Z.P. (2008). The Well-of-the-Well system: An efficient approach to improve embryo development. Reprod. Biomed. Online 17, 73–81 [DOI] [PubMed] [Google Scholar]
- Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., and Lötvall J.O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- van der Pol E., Boing A.N., Harrison P., Sturk A., and Nieuwland R. (2012). Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 64, 676–705 [DOI] [PubMed] [Google Scholar]
- Van Langendonckt A., Vansteenbrugge A., Donnay I., Van Soom A., Berg U., Semple E., Grisart B., Mermillod P., Brem G., Massip A., and Dessy F. (1996). Three year results of in vitro production of bovine embryos in serum-poor bovine oviduct conditioned medium. An overview. Reprod. Nutr. Dev. 36, 493–502 [DOI] [PubMed] [Google Scholar]
- Van Soom A., Wrathall A.E., Herrler A., and Nauwynck H.J. (2010). Is the zona pellucida an efficient barrier to viral infection? Reprod. Fertil. Dev. 22, 21. [DOI] [PubMed] [Google Scholar]
- Wadhwa N., Kunj N., Tiwari S., Saraiya M., and Majumdar S.S. (2009). Optimization of embryo culture conditions for increasing efficiency of cloning in buffalo (bubalus bubalis) and generation of transgenic embryos via cloning. Cloning Stem Cells 11, 387–395 [DOI] [PubMed] [Google Scholar]
- Wisdom R., and Lee W. (1991). The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 5, 232–243 [DOI] [PubMed] [Google Scholar]
- Witwer K.W., Buzás E.I., Bemis L.T., Bora A., Lässer C., Lötvall J., Nolte-‘t Hoen E.N., Piper M.G., Sivaraman S., Skog J., Théry C., Wauben M.H., and Hochberg F. (2013). Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2, 20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.S., Cheung T.M., Chan S.T., Ho P.C., and Yeung W.S. (2001). Temporal effect of human oviductal cell and its derived embryotrophic factors on mouse embryo development. Biol. Reprod. 65, 1481–1488 [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Sugimura S., Wakai T., Kawahara M., and Sato E. (2009a). Acetylation level of histone h3 in early embryonic stages affects subsequent development of miniature pig somatic cell nuclear transfer embryos. J. Reprod. Dev. 55, 638–644 [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Sugimura S., Wakai T., Kawahara M., and Sato E. (2009b). Difference in sensitivity to culture condition between in vitro fertilized and somatic cell nuclear transfer embryos in pigs. J. Reprod. Dev. 55, 299–304 [DOI] [PubMed] [Google Scholar]
- Zhang L., Luo Y.-B., Bou G., Kong Q.-R., Huan Y.-J., Zhu J., Wang J.-Y., Li H., Wang F., Shi Y.-Q., Wei Y.C., and Liu Z.H. (2011). Overexpression nanog activates pluripotent genes in porcine fetal fibroblasts and nuclear transfer embryos. Anat. Rec. 294, 1809–1817 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Huang Y., Xie W., Song Q., Ji Y., Zhang Y., Ouyang H., Lai L., Pang D., and Tang X. (2013). Scriptaid affects histone acetylation and the expression of development-related genes at different stages of porcine somatic cell nuclear transfer embryo during early development. Chinese Sci. Bull. 58, 2044–2052 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.