Abstract
Protein kinase D1 (PKD1) is a serine-threonine kinase that regulates various functions within the cell, including cell proliferation, apoptosis, adhesion, and cell motility. In normal cells, this protein plays key roles in multiple signaling pathways by relaying information from the extracellular environment and/or upstream kinases and converting them into a regulated intracellular response. The aberrant expression of PKD1 is associated with enhanced cancer phenotypes, such as deregulated cell proliferation, survival, motility, and epithelial mesenchymal transition. In this review, we summarize the structural and functional aspects of PKD1 and highlight the pathobiological roles of this kinase in cancer.
Introduction
Protein kinase D1 (PKD1) is a ubiquitous serine-threonine protein kinase that is involved in a multitude of functions in both normal and diseased states (1–3). This evolutionarily well-conserved protein, with homologs in mice (Mus musculus), rats (Rattus norvegicus), flies (Drosophila melanogaster), worms (Caenorhabditis elegans), and yeast (Saccharomyces cerevisiae), was initially recognized as a member of the protein kinase C (PKC) family and named PKCμ (2, 3). However, distinct differences in the protein structure, variation in substrate(s) and inhibitor specificity, and low homology of the kinase domain to other members of the PKC family resulted in its reclassification. PKD1 is now classified as a member of the protein kinase D (PKD) family, a distinct branch under the calcium/calmodulin-dependent protein kinase (CaMK) branch of the cellular kinome (4). The PKD family possesses substrate specificity and characteristics that combine the structural features of the PKC and CaMK protein kinase families, uniquely positioning it to perform key functions in multiple signaling pathways. PKD serves to integrate signaling information from multiple upstream stimuli that activate PKC or generate diacylglycerol (DAG). In addition to possessing substrate specificity similar to that of CaMK, PKD family members also contain a pleckstrin homology (PH) domain, differentiating them from other members of the PKC family. This allows PKDs to interact with and modulate the functions of proteins involved in a multitude of signaling pathways, and to disseminate information from the PKC signaling pathway as well. Further, PKD family members exhibit diverse subcellular localizations (such as cytosol, membrane, nucleus, Golgi, and mitochondria), which allow them to influence different signaling pathways (5–7).
The PKD family contains 3 members that are homologous in structure and function, namely, PKD1, PKD2, and PKD3 (5–9). PKD1, the most-studied member of this family, is also referred to by other names, such as PRKD1 [to differentiate it from polycystic kidney disease (PKD)] and PKCμ.
The PKD1 gene, located on human chromosome 14q11, is broadly expressed in many organs, including the thyroid, brain, heart, and lungs, with the highest expression in the prostate and testis germ cells (1, 3, 10). PKD1 has been shown to play important roles in a variety of cellular functions that regulate intracellular signal transduction pathways, cell survival, proliferation, motility, invasion, angiogenesis, and apoptosis (1, 5–9). PKD1 also plays a critical role in the formation and consolidation of memory in the neurons (11), in cardiac cell functioning and maintenance of cardiovascular health (12), and in the regulation of the immune system (13, 14). Thus, the deregulation of PKD1 has been connected with the development of cancers, cardiovascular hypertrophy, and other diseases. In this review, we will focus on PKD1 and its role in cancer development and cancer cell motility.
Structural Characteristics of PKD1
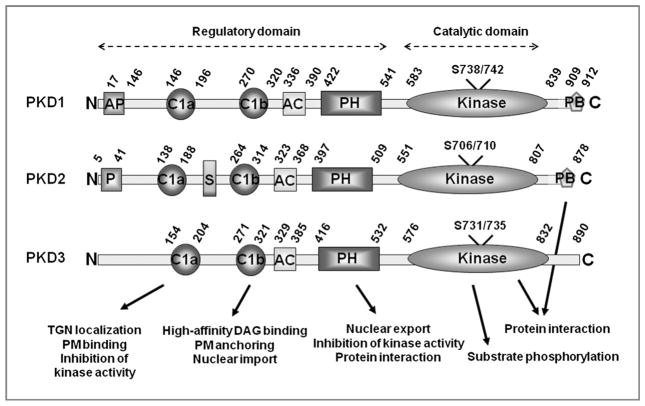
All 3 members of the PKD family share distinct structural homology. The human PKD1 is the largest member, with 912 amino acids and a molecular mass of 115 kDa. The other 2 members are PKD2 with 878 amino acids (molecular mass 105 kDa) and PKD3 (previously called PKCν/PKCnu) with 890 amino acids (molecular mass 110 kDa) (5). The PKD family members possess a common modular structure consisting of an N-terminal region with regulatory domains and a C-terminal region with a kinase domain [Fig. 1; reviewed by Rozengurt and colleagues (5)]. The proteins are maintained in an inactive state through autoinhibition of the kinase domain by its regulatory domains (5). The regulatory region of PKD1 contains an alanine-proline–rich region (AP domain; amino acids 17–146) at the N-terminus, followed by a cysteine-rich domain (CRD; amino acids 146–320) that consists of 2 tandem cysteine-rich Zn-finger like domains (C1a and C1b) separated by a long spacer, an acidic amino-acid–rich region (AC domain; amino acids 336–390), and a PH domain (amino acids 422–541) (5, 7, 15). The catalytic kinase domain present at the C-terminus extends from amino acids 585–839 (Fig. 1). The type 1 PDZ (PSD-95/Discs large/ZO-1) binding motif is present at the most C-terminal end of PKD1 (16). Both the CRD region and the PH domain play important roles in intramolecular inhibition of the kinase activity. Deletion of either of these domains or mutation of critical residues within the PH domain results in constitutively active PKD1 (5, 7). The CRD domain also binds and responds to lipid second messenger diacyl glycerol (DAG) or DAG analog phorbol esters, and determines the membrane localization of this protein. Whereas C1a participates in Golgi DAG binding and localization, C1b participates in high-affinity DAG binding, as well as membrane and nuclear localization of the protein (5). The PH domain in PKD1 has been shown to interact with other proteins and play a role in subcellular localization of this protein (5). The PH domain also participates in the nuclear export of PKD1. Unlike PH domains in other proteins that bind lipids, no lipid-binding ability has yet been shown for this domain of PKD1 (6). The type 1 PDZ binding domains of PKD1 have been shown to be essential for interaction with proteins (16, 17) and for proper surface localization of Kidins220, a PKD1 substrate molecule (16). The 3 amino acid consensus sequences, containing Ser or Thr at the −2 position and a hydrophobic amino acid at the zero position (S/T.X.Φ; Φ-hydrophobic amino acid), are present at the most C-terminal end of proteins and interact with PDZ domains containing proteins (16). Human cells contain >200 proteins with at least a PDZ domain that appears to mainly aid in protein-protein interaction and the assembly of huge protein complexes. Thus, it is likely that the PDZ binding domain of PKD1 plays an essential role in the proper surface localization of proteins involved in cell signaling pathways, cell-cell adhesion, and cell polarization. The PDZ domain may also determine the subcellular localization of PKD1 within the cells by interacting with specific protein scaffolds.
Figure 1.
Molecular structure of PKD. PKD family members possess an N-terminal regulatory region and a C-terminal catalytic domain. The schematic representation shows the modular structure of PKD family members and the major functions carried out by the domains. PKD1 is the largest member, with 912 amino acids; PKD2 contains 878 amino acid residues, and PKD3 contains 890 amino acids. All 3 PKDs show a high degree of homology within the various domains; however, only PKD1 and PKD2 contain an alanine-proline or proline-rich region at the N-terminus and a PDZ binding domain at the C-terminus. The numbers represent the amino acid positioning of each domain in human PKD1, PKD2, and PKD3. The serine residues marked in the kinase domain represent the activation loop residues phosphorylated by nPKC members. The highly homologous mouse PKD1 contains 918 amino acids, whereas mouse PKD2 is made of 875 amino acids, and mouse PKD3 contains 889 amino acids. AP, alanine proline–rich domain; C1a and C1b, cysteine-rich domain 1a and 1b; AC, acidic domain; PH, pleckstrin homology domain; kinase, kinase domain; PB, PDZ binding domain; P, proline-rich region; S, serine-rich region; TGN, trans-Golgi network; PM, plasma membrane; DAG, Diacyl glycerol.
Although the regulatory regions of PKD1 have some features similar to those of members of the PKC family, the catalytic domain of PKD1 possesses features that distantly resemble CaMK, such as structural homology, and substrate and inhibitor specificity. The substrate consensus sequence for PKD1 is L.X.R.(Q/K/E/M).(M/L/K/E/Q/A).S*.X.X.X. X, and unlike members of the PKC family, PKD1 exhibits a high requirement for leucine at the −5 position for substrate phosphorylation (7, 18–20). PKD2 and PKD3 also have very similar modular arrangements with minor structural differences. Whereas PKD3 lacks an N-terminal AP-rich region and a C-terminal PDZ binding domain, the N-terminus of PKD2 possesses a proline-rich region instead of an AP-rich region. In addition, PKD2 also contains a serine-rich region in between the C1a and C1b regions of the CRD domain (ref. 5; Fig. 1).
All 3 members of the PKD family exhibit extensive homology within these structural domains. However, the 3 PKD proteins carry out independent and diverse functions due to different subcellular localization and expression patterns in different tissues (5). PKD1 is localized predominantly in the cytoplasm and is found in minor quantities in various organelles, such as the Golgi apparatus and mitochondria (6). After stimulation of the cell, PKD1 is rapidly recruited to the cell membrane and activated. This is followed by a quick translocation of PKD1 back into the cytoplasm and its redistribution to other organelles. In contrast, PKD2 is predominantly cytoplasmic and accumulates in the nucleus following cell membrane recruitment and activation (21). PKD3, on the other hand, is both nuclear and cytoplasmic in nature, even under resting conditions (22). All 3 isoforms show nuclear cytoplasmic shuttling following activation, indicating that PKD serves vital functions as a messenger between these subcellular structures. The subtle differences in the structural organization of the isoforms, combined with their different sub-cellular localization and expression patterns, allow these proteins to participate in multiple signaling pathways of various cells.
Activation and Regulation of PKD1
PKD1 resides primarily as an inactive kinase in the cytoplasm under resting conditions. A small fraction also exists in the Golgi apparatus (6). In certain special cell types, PKD1 has been found to have other subcellular distributions. For example, in neurons, PKD1 has been shown to reside in transport vesicles (23), whereas in B cells it has been shown to be localized at the mitochondria (6). PKD1 is activated by a number of different agents, including pharmacological agents (e.g., Bryostatin1), phorbol esters, and/or physiological stimuli, such as tumor necrosis factor (TNF), neuropeptides, angiotensin II, and platelet derived growth factors (PDGF) (5, 6). Investigators have identified multiple mechanisms that regulate the activity of PKD1. The best-characterized PKD1 activation mechanism is the phospholipase C/protein kinase C (PLC/PKC) signaling pathway (5). The activation of PLC through the engagement of surface receptors by various physiological stimuli, such as neuropeptides and G-protein coupled receptor (GPCR) agonists, results in the generation of DAG, which mediates the recruitment and activation of PKCs at the membrane. DAG also participates in the recruitment of PKD1 to the plasma membrane through its interaction with the C1b domain. This is followed by phosphorylation of PKD1 in the activation loop at residues Ser-744 and Ser-748 in mouse (equivalent to human Ser-738 and Ser-742) by the members of PKC family (1, 5, 7). Activation loop phosphorylation releases PH-mediated inhibition of PKD1 activity and stabilizes the protein in its active form (5). Thus, biochemical studies suggest sequential steps in the activation of PKD1 by PKC as follows: (1) engagement of surface receptors in response to various stimuli, (2) generation of DAG, (3) PKC recruitment and activation at the membrane, and (4) activation of PKD1 by PKC. Activation by DAG/phorbol ester is completely suppressed in the presence of PKC inhibitors or through the mutation of active site residues in PKD1 (PKD1Ser744/748Ala) (6, 7). Conversely, replacement of both serine residues with glutamic acid (Ser744/748Glu) markedly increased activity without phorbol ester stimulation (6, 7). These results indicate that activation loop phosphorylation is a key mechanism in PKD1 activation. The novel PKCs PKCδ, PKCε, PKCη, and PKCθ have been shown to be involved in PKD1 activation (6, 7). Following phosphorylation, PKD1 rapidly transduces the DAG-PKC signals from the cell surface by quickly dissociating from the plasma membrane, relocating to the cytosol, and eventually moving to the nucleus, which is facilitated by the C1b domain of PKD1. Activation loop phosphorylation of PKD1 has also been shown to occur on the mitochondrial surface in response to oxidative stress (6, 7). During oxidative stress conditions, mitochondrial DAG (mDAG) generated through the action of phospholipase D1 recruits PKD1 to the mitochondrial surface through its interaction with the CRD domain (24). The recruited PKD1 is tyrosine phosphorylated by Src-Abl kinases, eventually leading to PKD1 activation by PKCδ (25).
Mechanisms other than PKC-dependent phosphorylation also appear to be involved in PKD1 activation. For example, although Ser-255 of PKD1 is also phosphorylated in response to phorbol esters, the Ser255Glu PKD1 mutant is not constitutively active. Of interest, this mutant is capable of phorbol ester–mediated activation even in the presence of the PKC inhibitor, indicating the existence of PKD1 activation mechanisms that do not involve PKC (1). Caspase-3 (Casp3) mediates activation of PKD1 by cleaving the protein between the acidic domain and the PH domain, a process that is especially active during apoptosis induced by genotoxic drugs (1). Studies have also revealed long-term activation of PKD1 by a GPCR agonist in the presence of the PKC inhibitor. The slow phase activation of PKD1 in the presence of the PKC inhibitor clearly indicates a PKC-independent activation mechanism (26, 27). PKD1 activation by slow autophosphorylation of Ser-748 also represents another PKC-independent mode of activation (28).
Several regulatory mechanisms modulate PKD1 activity, including autoinhibition, phosphorylation, proteolytic degradation, subcellular localization, and various cell-context–dependent PKD1 activation mechanisms (1, 5–7). The autoinhibition of PKD1 kinase activity that is mediated by the CRD and PH domains can be abrogated by interactions of these domains with other proteins, such as PKC and Gβγ, or by phosphorylation within these domains, resulting in structural changes within PKD1 (5–7). Interactions with other proteins can also lead to change in the subcellular localization of the protein. The activity of phosphorylated PKD1 can be attenuated by the binding of 14-3-3 proteins to the C1a regulatory domain, resulting in altered subcellular localization of the protein (1). The differential expression of various PKC isoforms that activate PKD1 (i.e., PKCδ, PKCε, PKCη, and PKCθ) determines the cell context–dependent activation mechanisms that operate in a particular cell type (7). Activation of PKD1 by phosphorylation at Ser-738/742 (equivalent to mouse PKD1 Ser-744/748) is often followed by autophosphorylation at other sites, including Ser-910 (equivalent to mouse PKD1 Ser-916), which is a characteristic step that follows PKD1 activation (1, 5, 8). Recent evidence indicates that phosphorylation of Ser-916 may be an intermolecular reaction (28). Although phosphorylation at Ser-916 may play a role in structural modification of the protein and has been used as a tool to monitor active PKD1, the role of this autophosphorylation is only partly recognized. For example, phosphorylation of Ser-916 residue in the PDZ binding motif of PKD is essential for the interaction and appropriate localization of Kidins220 on the membrane (16). In addition, the phosphorylation of Ser-916 has been suggested to have a potential role in terminating/stunting PKD1-mediated substrate phosphorylation, because the Ser916Ala mutant shows increased resistance to proteolysis and dephosphorylation (28, 29).
Cancer-Related Signaling Pathways Modulated by PKD1
PKD1 modulates a diverse array of signaling pathways and therefore regulates multiple biological functions that are critical for the normal functioning of the cell. The biological functions regulated by PKD1 include (1) DNA synthesis, chromatin remodeling, and cell proliferation; (2) cell-cell adhesion and cell polarization, migration, and invasion; (3) apoptosis; (4) detoxification of oxidative stress signals; (5) Golgi organization and transport of vesicles from the trans-Golgi network (TGN); (6) angiogenesis; (7) immune cell response; and (8) insulin secretion and survival of pancreatic β cells (1, 9). As a downstream component of the PLC/PKC pathway, PKD1 integrates signaling from multiple stimulating factors and activates diverse downstream substrates/pathways. Thus, its deregulation affects a multitude of signaling pathways, resulting in diseases such as cancer, cardiovascular hypertrophy, and diabetes. In this section, we will briefly discuss the signaling pathways that are modulated by the aberrant expression of PKD1 in cancer (Fig. 2).
Figure 2.
Schematic representation of signaling pathways modulated by PKD1 in cancer. The deregulated expression of PKD1 results in the development of cancer. The schematic representation shows the pathways that activate PKD1, and the various downstream pathways and functions modulated by this kinase (maroon ovals). The activation of various membrane receptors, such as GPCR and other growth factor receptors (GR), leads to the activation of PKD1 by the PLC-PKC pathway through the formation of DAG. DAG modulates PKD1 function by binding and quickly recruiting it to the cell membrane for activation by PKCs. PKD1 can also be activated by the Golgi Gβγ-PKCε in the Golgi apparatus, by proteolytic cleavage by caspase 3, by oxidative stress resulting in PKD1 activation on mitochondrial surface through the action of SRC-Abl kinase and PKCδ, by UVB ray-activated SRC kinase, and by a PKC-independent self-activation mechanism. Activated PKD1 is rapidly translocated from the membrane to the cytoplasm and eventually to the nucleus, where it regulates downstream pathways. Activated PKD1 also regulates the process of vesicle trafficking from the Golgi to the membrane, which eventually controls cell surface proteins that are involved in cell adhesion, cell polarity, and motility. Depending on the cell type, PKD1 functions as either a tumor suppressor or an oncogene within the cell. A, tumor suppressor functions of PKD1. PKD1 has been shown to inhibit cancer in the prostate, breast, and GI tract. PKD1 inhibits tumorigenesis by enhancing cell adhesion and inhibiting the function of proteins involved in cell migration, cell invasion, cell proliferation, and EMT. PKD1 phosphorylates E-cadherin and β-catenin, thereby enhancing cell-cell adhesion. PKD1 helps to maintain cellular polarity by phosphorylating Par-1 polarity–associated kinase and thus enhancing its cytoplasmic sequestration by 14-3-3 protein. Activated PKD1 also helps to establish cell polarity by positively regulating the TGN carriers to the basolateral membrane. Activated PKD1 can also inhibit the transcriptional activity of β-catenin and AR, resulting in reduced cell proliferation. It also inhibits EMT by regulating the activity of snail transcription factor. PKD1 negatively regulates cell invasion by influencing the levels of MMPs through the modulation of HDACs. It also negatively regulates actin remodeling and thus cell motility through the phosphorylation of SSH-1L and cortactin. B, prooncogenic role of PKD1. The upregulation of PKD1 has been linked to the development of pancreatic and skin cancer, and it may also be involved in prostate cancer (although this notion is controversial). Some of the pathways that are modulated by PKD1 and may result in a prooncogenic role are as follows: Activated PKD1 enhances cell survival and proliferation by enhancing DNA synthesis and upregulating the function of Erk1/2 protein in the MAPK pathway, leading to the accumulation of c-Fos. Activated PKD1 decreases apoptosis by suppressing the function of the JNK pathway, resulting in decreased c-Jun levels. Under oxidative stress conditions, PKD1 enhances cell survival through activation of the NFkB pathway. PKD1 expression contributes to oncogenesis by enhancing angiogenesis by regulating the activity of HDAC5 and HDAC7.
Modulation of Mitogen-Activated Protein Kinase Pathway by PKD1
Aberration in the mitogen-activated protein kinase (MAPK) signaling pathway is associated with the development and progression of many types of cancer (30). The activation of various surface receptors, such as GPCR, by their ligands results in activation of the MAPK pathway. Different lines of evidence also support a role for GPCR in the development and progression of many types of cancers (5, 30). GPCR agonists activate PKD1 in many different cell types. Activation of Swiss 3T3 cells overexpressing PKD1 by GPCR agonist results in prolonged activation of the MEK/ERK/RSK pathway compared with cells over-expressing kinase-dead mutant PKD1. This role for PKD1 in activating the MAPK pathway may occur through the phosphorylation of RIN1 (a competitive Ras effector protein), resulting in sustained stimulation of the Ras/Raf/MEK/ERK/RSK pathway (reviewed in refs. 1 and 6). The resulting accumulation of c-Fos leads to increased DNA synthesis, cell cycle progression, and cell proliferation (8). This pathway is prominently seen in pancreatic cancer cells (8).
Long-term activation of the c-Jun N-terminal kinase (JNK) pathway by the epidermal growth factor (EGF) results in phosphorylation of the Ser-63 residue of c-Jun and induction of apoptosis (31). Although PKD1 was found to directly interact and phosphorylate c-Jun at its N-terminus, it has also been suggested to decrease apoptosis via modulation of JNK functions and suppression of c-Jun phosphorylation (5, 6). The outcome of PKD1-mediated downregulation of the JNK signaling pathway is dependent on the cell type and the stimulus (6, 26). For example, this pathway is activated upon exposure to hydrogen peroxide, but not ceramide or TNF-α–induced cell death (1). Also, although attenuation of JNK signaling was observed following overexpression of PKD1 in HEK cells, this was not observed in A549 non–small cell lung carcinoma cells (32). In addition, whereas PKD1-mediated inhibition of this pathway appears to play a prominent role in pancreatic cancer cell line survival (33), PKD1 was found to enhance apoptosis in the renal tubular epithelial cells by activating JNK (34). Thus, more studies are required to elucidate the contrasting outcomes of PKD1-mediated activation of the JNK pathway in different cell types.
Activation of NFκB by PKD1
NFκB transcription factors are heterodimeric proteins that play critical roles in regulating stress-induced inflammatory and immune responses. Mutation in the NFκB gene has been linked to enhanced cell survival and proliferation in various cancers (25). PKD1 acts as a sensor for mitochondrial oxidative stress and regulates the cellular response by activating the NFκB pathway (25). PKD1 activates NFκB by the phosphorylation and activation of inhibitory κ kinase (IKK) (25). This results in the degradation of inhibitory protein IκB and the release of NFκB from the inhibitory complex, followed by accumulation in the nucleus and induction of downstream target genes (25), which in turn causes cell survival, cell proliferation, and inflammation (8, 35). PKD1 may also be involved in IKK-independent mechanisms that activate the NFκB pathway within cells (25). PKD1-mediated NFκB activation has been shown in different cell lines, including immune cells, intestinal epithelial cells, and lung cells (8). However, only PKD1 activation following tyrosine phosphorylation is capable of activating the NFκB pathway, which may point to specific requirements, including specific structural changes in PKD1, for NFκB activation (25). More work is needed to understand and establish the underlying structural requirements and functional mechanism involved in PKD1-mediated NFκB activation.
Modulation of androgen receptor signaling by PKD1
The androgen receptor (AR) is a ligand-dependent transcription factor that is present in many types of cells (36). The binding of AR to its ligand (androgen hormones) leads to the translocation of the AR-ligand complex into the nucleus, where it binds to androgen response element (ARE) regions in the DNA to trigger transcription of various downstream genes involved in cell survival and proliferation (36). In prostate cancer, somatic mutation in AR results in progression of tumor from an androgen-sensitive (AS) stage to an androgen-insensitive (AI) stage that is refractory to androgen-depletion treatment (36). Recently, it was shown that PKD1 exists in a transcription complex along with AR and a promoter sequence for prostate specific antigen (PSA) in prostate cancer cells (37). PKD1 negatively regulates the function of AR in prostate cancer cells, because the overexpression of wild-type PKD1 or kinase-dead PKD1 attenuates ligand-dependent AR function. Alternatively, PKD1 knockdown enhances ligand-dependent AR activity (37). Studies have also revealed that PKD1 interacts and phosphorylates the Ser-82 residue of heat-shock protein 27 (Hsp27, a molecule that is necessary for nuclear translocation of AR) and represses AR functions in prostate cancer cells (38). The AR function is also modulated by interaction with other proteins, such as β-catenin, which augments AR functions (39). Because PKD1 interacts with and downregulates both nuclear β-catenin and AR transcription activity, deregulated expression of PKD1 may play a critical role in the initiation and progression of prostate cancer (1, 40–42). A better understanding of how PKD1 modulates AR signaling at the molecular level will facilitate the development of new strategies for the treatment of prostate cancer.
Regulation of histone deacetylases by PKD1
The DNA-binding proteins, histones, control protein expression by regulating the access of transcription factors to the DNA sequence. Orchestrated acetylation of lysine residues by histone acetyl transferase (HAT) and its deacetylation by histone deacetylase (HDAC) enzymes determine the epigenetic regulation of genes (43). Deacetylation of lysine residues of histones by HDACs results in a tighter chromatin structure and transcriptional repression of genes. Aberrant histone deacetylation has been found to correlate with pathological gene repression and neoplastic transformation (43). PKD1 modulates the phosphorylation and transportation of class II HDACs (HDAC5 and HDAC7) from the nucleus to the cytoplasm, which in turn mitigates the transcriptional repression of silenced genes (1, 5). PKD1 that is activated in endothelial cells by vesicular endothelial growth factor (VEGF) phosphorylates HDAC5 at Ser259/498 and induces cell proliferation and angiogenesis (44). Studies have also revealed that the phosphorylation of HDAC7 at Ser-178, Ser-344, and Ser-479 by PKD1 causes transcription of angiogenic genes (45). PKD1-mediated modulation of HDACs has also been shown to play an essential role in B-cell response and muscle formation by regulating the transcription activity of myocyte enhancer factor-2 (9). These studies indicate a vital role for PKD1 in HDAC modulation. However, a further understanding of this pathway would help delineate the role of PKD1 in epigenetic regulation.
PKD1 modulates cell polarity and cell adhesion
Recent studies have suggested an important role for PKD1 in maintaining cell polarity and a critical role in enhancing cell-cell adhesion and decreasing motility. Cellular polarization is critical for differentiation, proliferation, and tissue homeostasis, which are some of the vital characteristics that are lost in cancer cells. Partitioning-defective (Par) proteins are highly conserved serine-threonine kinases that play a critical role in maintaining cell polarity. PKD1 was recently shown to phosphorylate Par1B and thereby regulate its presence on the membrane (46). Thus, modulating the levels or function of PKD1 may be important for maintaining cell polarity.
The adhesion complex formed by the E-cadherin-β-catenin complex plays a vital role in maintaining cell-cell contact (1). Also, the aberrant expression and distribution of these proteins have been associated with cancer (1, 40). In addition to its role in cell adhesion, β-catenin functions as a cotranscription factor with T-cell factor (TCF) and plays an important role in the Wnt signaling pathway. Aberrant subcellular localization of β-catenin in the nucleus leads to enhanced transcription of genes such as c-myc and cyclin D1, resulting in oncogenic transformation of the cells. In prostate cancer cells, PKD1 modulates E-cadherin and β-catenin function (1, 42, 47). Studies from our laboratory have shown that PKD1 interacts, phosphorylates, and modulates the functions of E-cadherin, resulting in increased cell-cell adhesion and decreased cellular motility, suggesting a pivotal role of PKD1 in prostate cancer progression and metastasis (47). Our studies have also shown that PKD1 directly interacts, phosphorylates, and alters the functions of β-catenin (42). The activation of PKD1 by Bryostatin1 decreased nuclear β-catenin and also enriched membrane localization of β-catenin, resulting in increased cellular aggregation and decreased motility (42). Thus, activation of PKD1 inhibits the oncogenic signals produced by β-catenin’s cotranscription factor activity. Of interest, both β-catenin and E-cadherin, like PKD1, have been shown to be aberrantly expressed in prostate cancer (40, 48). In addition to prostate cancer cells, PKD1 also seems to play an important role in cell adhesion in other cancer cell types. For example, in the MDA-MB-435 breast cancer cell line, PKD1 enhanced cell adhesion following stimulation with cis-polyunsaturated fatty acids (49). Moreover, PKD1 regulated cell adhesion by directly interacting with focal adhesion molecule α-v-β-3 and promoted its localization at focal adhesion sites (49). These studies suggest that PKD1 may be a potential target for therapeutic intervention to prevent cancer cell metastasis.
PKD1 in actin remodeling and cell migration
Dynamic remodeling of actin cytoskeleton is crucial for cell motility and migration, and it plays a central part not only in pathological conditions, such as cancer metastasis, but also in a variety of normal biological processes, such as morphogenesis and wound healing. Accumulating evidence suggests a prominent role for PKD1 as a negative regulator of cell motility/migration through F-actin reorganization (42, 50–53). The dynamic and complex process of actin remodeling involves the synergistic action of a number of proteins that are active in the polymerization of actin and depolymerization/severing of actin polymer to generate actin monomer for further polymerization (53). In brief, actin monomer nucleation attracts a number of actin-binding proteins that allow the elongation and formation of F-actin filaments. However, the process is limited by the availability of G-actin monomers that are generated through the severing of F-actin by an actin depolymerizing factor (ADF/cofilin) (53). The activity of cofilin is negatively regulated by LIMKinase and positively regulated by Slingshot phosphatases (SSH). This process not only releases actin monomers that are recycled for polymerization at the leading edge but also produces a new barbed end that promotes further growth and branching of F-actin fiber by binding of the Arp2/3 complex. The process of nucleation and branching is enhanced by cortactin and WASP proteins, and the newly generated actin filaments at the branch points form the membrane protrusion that enables cell motility (53).
PKD1 has been shown to directly interact with F-actin, and activated PKD1 colocalizes at the leading edge following activation of a signal transduction cascade (50). In various cancer cell line models, including breast, prostate, and pancreatic cancer, inhibition of PKD1 function by the overexpression of kinase-inactive PKD1 or siRNA-mediated inhibition shows enhanced cell migration, whereas over-expression of PKD1 shows reduced migration, suggesting a prominent role of PKD1 in cell migration (42, 50–52). Dissection of the mechanism revealed that activated PKD1 not only colocalizes at the leading edge but also binds to and phosphorylates a number of actin interacting proteins, such as cortactin, Arp2/3, and Slingshot-like 1 (SSH-1L) phosphatase, which subsequently inhibit actin polymerization (50, 52, 54). In PKD1-mediated modulation of many actin-binding proteins, the modus operandi seems to involve a phosphorylation-induced structural change in the proteins that exposes the 14-3-3 protein-binding site within these proteins. This allows quick binding, sequestration, and translocation of these proteins from the leading edge into the cytoplasm by the protein 14-3-3 (54–56). This mechanism of action was very clearly shown in the PKD1-mediated phosphorylation and regulation of cortactin at Ser-298 (56). Following its phosphorylation, cortactin was unavailable for participation in lamellipodia extension due to its translocation into the cytoplasm from the leading edge. In addition, the overexpression of phosphorylation-deficient cortactin-S298A protein in pancreatic cancer cells resulted in enhanced lamellipodia extension and directed cell migration due to faster Arp-cortactin–mediated synergistic actin poly-merization, underscoring a negative role for PKD1 in cell migration (56). Contrary to this, De Kimpe and colleagues (57) showed that PKD1 phosphorylation of cortactin at Ser-298 and Ser-348 does not result in subcellular changes in cortactin localization or affect lysophosphatidic acid (LPA)-induced cell migration. The process of actin severing is carried out by activated cofilin and requires the function of SSH-1L phosphatase. The phosphorylation of SSH1L by PKD1 structurally modulates the protein, thereby generating a 14-3-3 binding site and subsequent sequestration into cytoplasm, resulting in SSH-1L and, thus, cofilin inactivation (55). Therefore, activators of PKD1 indirectly reduce the levels of active cofilin. Although further analysis is required to establish the role of PKD1 in cell motility, overwhelming evidence points toward the involvement of PKD1 in the modulation of proteins involved in actin remodeling. As a result, this protein could play a very important role in cell migration and cancer metastasis.
PKD1 in Cancer
The role of PKD1 in cancer is not surprising given its involvement in many cellular functions, such as cell proliferation, apoptosis, cell adhesion, invasion, and vesicle trafficking (1). Similar to the intricate roles played by many kinases, PKD1 has a complex relationship with respect to cancer development. PKD1 has been shown to be down-regulated in prostate cancer (41, 58), breast cancer (52), gastric cancer (59), and colon cancer (60). However, the overexpression of PKD1 has been shown to play a role in the development of pancreatic cancer (61) and skin cancers (refs. 62 and 63; Table 1). Therefore, the consequence of upregulation or downregulation of PKD1 in cancer development is dependent on the tissue type. Because PKD1 functions as a critical kinase that integrates extracellular signals into intracellular processes by modulating a multitude of signaling pathways, the regulation of PKD1 levels and/or activity through pharmacological or genetic intervention might aid in cancer treatment (Fig. 3). The expression pattern of PKD1 in different cancers and its role in cancer development are discussed in this section.
Table 1.
PKD1 expression and its roles in various cancers
| No. | Cancer type | PKD1 status | Modulated signaling pathway | Reference |
|---|---|---|---|---|
| 1 | Breast cancer | Downregulated | MMPs | Eisler et al (52) |
| Actin remodeling and cell motility | Peterburs et al (54) | |||
| EMT | Du et al (65) | |||
| 2 | Prostate cancer | Contentious | E-cadherin/β-catenin | Jaggi et al (40, 42) |
| Downregulated | AR signaling | Mak et al (37) | ||
| HSP27 | Hassan et al (38) | |||
| EMT | Du et al (65) | |||
| ERK/MAPK/MMPs | Biswas et al (66) | |||
| Upregulated | Chen et al (70) | |||
| 3 | Gastrointestinal cancer | Downregulated | β-catenin | Kim et al (59) |
| Cox-2 | Jepperson et al (60) | |||
| Rodriguez Perez et al (72) | ||||
| 4 | Pancreatic cancer | Upregulated | Ras/Raf/MEK/ERK/RSK | Guha et al (61, 73) |
| HSP27 | Yuan et al (78) | |||
| EGF/JNK | Kisfavli et al (33) | |||
| Apoptosis | Trauzold et al (32) | |||
| 5 | Basal cell cancer | Upregulated | Ristich et al (63) | |
| ERK/MAPK | Jadali et al (75) | |||
| Apoptosis | Arun et al (76) |
Figure 3.
Proposed modes of action of PKD1 modulators in cancer therapeutics. PKD1 is aberrantly regulated in many cancers. It is downregulated in prostate, breast, and gastroenteric cancers, and upregulated in pancreatic and skin cancers. Targeted suppression of PKD1 in pancreatic and skin cancer with the use of siRNA or specific small-molecule inhibitors might aid cancer treatment by modulating NFκB, Erk, JNK, and HDAC signaling. In contrast, tumor-specific delivery of the PKD1 gene or PKD1 activators may be useful for prostate, breast, and gastroenteric cancer treatment. This would suppress AR and β-catenin signaling, invasion, motility, and EMT, and enhance cell-cell adhesion via increased E-cadherin and β-catenin localization at the membrane. Pathways that are activated following treatment are shown in green boxes, and those that are inhibited are shown in red boxes.
PKD1 in prostate cancer
Prostate cancer is one of the leading causes of cancer-related deaths in men in the United States. Although early detection of prostate cancer has greatly decreased the mortality rate, this cancer is often fatal at later stages because it can progress from an androgen-dependent (AD) to androgen-independent (AI) phenotype, for which there is no effective treatment. The highest expression of PKD1 is observed in prostate tissues, suggesting a crucial role of this protein in normal prostate functions (64). Studies from our laboratory and others suggest important roles for PKD1 in prostate cancer development and metastasis. Immunohistochemical (IHC) analyses revealed severe downregulation of PKD1 in prostate cancer cells and an incremental decrease in PKD1 expression following progression from AD to AI prostate cancer (41). Additionally, an AI C4-2 cell line showed significantly lower PKD1 expression compared with AD LNCaP prostate cancer cells. These data suggest a potential role of PKD1 in the progression of prostate cancer from an AD to an AI state (41). Furthermore, our studies show that PKD1 interacts, phosphorylates, and positively regulates the functions of E-cadherin and β-catenin (cadherin-catenin complex), enhancing cell-cell adhesion and decreasing cellular motility, strongly suggesting a major role for PKD1 in prostate cancer progression and metastasis (42, 47). Activated PKD1 also decreases nuclear β-catenin levels, resulting in the attenuation of oncogenic signaling by β-catenin’s cotranscription factor activity. Additionally, while overexpression of PKD1 and E-cadherin suppresses the cancer cell phenotype, simultaneous coexpression of PKD1 and E-cadherin in prostate cancer cells results in a cumulative decrease in cancer phenotypes. These data strongly support a pivotal role of PKD1 in prostate cancer (51). Recent studies in xenograft mouse models with PKD1 overexpressing prostate cancer cells revealed significantly reduced tumor growth compared with control, supporting a tumor suppressor role of PKD1 (65).
PKD1 also interacts and modulates the function of ARs in prostate cancer cells (37, 38, 51). This is especially significant given that ARs are ligand-dependent transcription factors that play a critical role in prostate biology and carcinogenesis (36). Studies with overexpression or knockdown cell line models of PKD1 showed that PKD1 negatively modulates the function of AR (37) through the modulation of Hsp27-mediated AR functions (38). Additionally, PKD1 was found to be involved in the invasion of prostate cancer cells via modulation of matrix metalloproteinase (MMP)-2 and MMP-9 (66) or metallothionin 2A, a protein that is involved in cell proliferation and chemoresistance of cancer cells (67, 68). A recent study revealed that PKD1 plays a vital role in epithelial-mesenchymal transition (EMT) by modulating the function of transcription factor snail, a central regulator of EMT (65). PKD1 phosphorylates the Ser-11 residue of snail, resulting in nuclear export of this protein and thus alterations in the expression of proteins involved in EMT (65).
Despite all the accumulating evidence supporting a tumor suppressor function for PKD1 in prostate cancer growth, invasion, and metastasis, other studies suggest upregulation of PKD1 and PKD3 in prostate cancer (69, 70). IHC analysis of patient tissue samples revealed upregulation of PKD1 and PKD3 compared with control samples (70). Inhibition of PKD kinase activity using chemical inhibitors with higher specificity for PKD family members revealed inhibition of cell proliferation, motility, and invasion (65, 66). These data indicate an oncogenic role for PKD family members in prostate cancer, although experimental evidence generated in studies using inhibitors are fraught with errors due to cross-reactivity with other kinases (66, 69, 70). PKD3 overexpression was shown to enhance the tumorigenicity of prostate cancer cells by enhancing the ERK1/2 and AKT signaling pathways; however, the molecular basis for the oncogenic role of PKD1 has not yet been probed in prostate cancer cells (65). In addition, an independent study suggested a prooncogenic role for PKD1 in prostate cancer (48). Experiments with prostate cell lines overexpressing Wnt5a suggest the involvement of PKD1 in establishing enhanced cell migration and invasion (48). Inhibition of PKD1 suppressed Wnt5a-dependent cell migration and invasion (48). Hence, further detailed investigations, including analyses of proteomic and metabolomic changes and IHC analyses in a large cohort of human prostate cancer samples, are necessary to unequivocally establish the pro- or anticancer role of this protein in prostate cancer.
PKD1 in breast cancer
Breast cancer affects both males and females, and it is one of the leading causes of cancer-related deaths in females. Only 10 to 15% of breast cancers are due to inherited genetic mutations (i.e., mutations in BRCA1 and BRCA2). The majority of incidences occur due to somatic mutations that result in uncontrolled cell proliferation and metastasis. Recent accumulating evidence suggests a potential role of PKD1 in breast cancer progression (52). Tissue microarray analysis showed that, whereas PKD1 was highly expressed in ductal epithelial cells of normal breast tissues, its expression was significantly reduced in 95% of the 40 invasive breast cancer tissue samples. Of importance, no changes in PKD2 and PKD3 levels were observed, indicating an important role for PKD1 function in breast cancer. In breast cancer cell lines, PKD1 expression was detected only in noninvasive or minimally invasive breast cancer cell lines, such as MCF-7, whereas no or low expression was detected in highly invasive cell lines, such as MDA-MB-231 (9, 52). This suppression was attributed to DNA methylation, which supports the involvement of epigenetic mechanisms in breast cancer progression (52). In 2D and 3D cell culture models, PKD1 expression was shown to drastically alter the invasive and proliferative ability of breast cancer cells (52). At the molecular level, PKD1 has been shown to interact with cortactin and paxillin at invadopodia (71), and to inhibit cellular motility and invasion in breast cancer cells via phosphorylation and inactivation of the SSH-1L protein (50, 52, 54). Additionally, PKD1 was found to suppress the expression of many proinvasion MMPs, such as MMP-2, MMP-7, MMP-9, MMP-10, MMP-11, MMP-13, and MMP-14 (52). Of interest, PKD1 was recently implicated in EMT via inhibition of snail functions and modulation of mesenchymal-epithelial markers, such as vimentin and E-cadherin (65). Thus, the reexpression or activation of PKD1 might serve as a potential therapeutic strategy for breast cancer treatment.
PKD1 in gastrointestinal tract cancers
Research from our laboratory and others has suggested a negative role for PKD1 in the development of gastrointestinal (GI) tract cancers (59, 60). In gastric carcinoma, PKD1 was found to be downregulated in >70% of cell lines and in ~60% of patient tissue samples (59). Target-specific silencing of PKD1 using siRNA increased the invasion and motility of gastric carcinoma cells, confirming a negative role for PKD1 in gastric cancer. PKD1 was found to be epigenetically downregulated in gastric cancer cells expressing low levels of PKD1 (59). In colon cancer tissues, IHC analysis revealed downregulation of PKD1 in ~70% of higher Dukes stages (stages II, III, and IV), moderately, and poorly differentiated colon cancer samples compared with nonneoplastic colon samples (60). The overexpression of PKD1 in SW480 colon cancer cells inhibits nuclear β-cate-nin accumulation and reduces β-catenin/TCF transcriptional activity, suggesting a suppressive role of PKD1 in colon cancer (60). On the other hand, a recent study revealed an important role for PKD1 in potentiating Cox-2 production in colonic myofibroblasts following TNF-α and lysophosphatidic acid treatment, suggesting a role for PKD1 in modifying the tumor microenvironment to promote tumor growth (72). Thus, thorough investigations are warranted to confirm the role of PKD1 in GI tract malignancies.
PKD1 in pancreatic cancer
Pancreatic cancer is one of the most aggressive cancers, and it is highly resistant to cancer chemotherapy. The overexpression of PKD1 has been shown to play a role in pancreatic cancer progression (8). Using the PANC-1 human pancreatic ductal adenocarcinoma cell line as a model system, Guha and colleagues (61, 73) showed the involvement of a functional PKD1 signaling pathway in mitogenic signals initiated by neurotensin (NT). In vitro and in vivo assays following NT treatment revealed rapid PKC-dependent activation of PKD1, which in turn led to rapid activation of MAPK/ERK kinase 1 (MEK1/2), activation and nuclear translocation of extracellular signal regulated kinase 1 (ERK-1) and 2 (ERK-2), and eventually increased DNA synthesis (61, 73). PKD1, ERK-1, and ERK-2 activation was inhibited by the specific PKC inhibitors bisindolylmaleimide 1 (GF109203X, also called Gö6850) and bisindolylmaleimide IX (Ro31-8220), suggesting the involvement of a PKC/PKD signaling process in human ductal pancreatic carcinoma cells (61, 73). IHC analyses of a small cohort of tissue samples revealed the overexpression of PKD1 in pancreatic cancer compared with normal pancreatic tissues (32). The overexpression of PKD1 has been suggested to confer enhanced proliferation and higher antiapoptotic activity to the pancreatic cancer cells. The overexpression of PKD1 in the Colo357 pancreatic cancer cell line, which shows low PKD1 expression, not only decreased the sensitivity of the cells to CD95-mediated apoptosis but also enhanced cell growth and telomerase activity, suggesting a correlation between PKD1 expression and resistance to apoptosis (32). The antiapoptotic proteins c-FLIPL and survivin were upregulated in PKD1 overexpressing cells, and their enhanced presence could be a possible reason for the prosurvival role of PKD1 in pancreatic cells. Inhibition of the PKC/PKD pathway in these PKD1-overexpressing cells with the use of the broad-spectrum PKC/PKD inhibitor Gö6983 restored the sensitivity of cells to apoptosis, suggesting a procarcinogenic role for this protein in pancreatic malignancy (8, 32). A recent in vitro and in vivo animal study involving the use of a new PKD1-specific, small-molecule inhibitor (CRT0066101) showed inhibition of pancreatic cancer growth, suggesting the development of PKD1 inhibitors as a novel therapeutic target for treatment of pancreatic cancer (74).
PKD1 in skin cancer and other cancers
Basal cell carcinoma (BCC) is the most common type of skin cancer in the world. Aberrant distribution and upregulation of PKD1 has been shown to be associated with BCC and neoplastic mouse keratinocytes, suggesting a putative role of PKD1 in skin cancer (63). More specifically, PKD1 was found to be associated with repression of keratinocyte differentiation and upregulation of cell proliferation, possibly through the involvement of the ERK/MAPK pathway (75). In vitro analysis of primary mouse keratinocytes exposed to UVB radiation (a key risk factor for development of BCC) revealed activation of PKD1 by the Src tyrosine kinase family without the involvement of the canonical PKC activation pathway (76). Overexpression of wild-type PKD, but not mutant PKD, attenuated UVB-induced apoptosis, suggesting a role of PKD1 in BCC (76).
The role of PKD1 in the development of other cancers is not well defined; however, some experimental evidence implicates PKD1 in the development of certain types of cancers. For example, the rapid activation of PKD1 by PKCs was detected in small cell lung carcinoma (SCLC) cell lines in response to treatment with phorbol esters or diffusible DAG. This suggests a possible role of PKD1 in the conversion of external carcinogenic stimuli into an intracellular response in SCLC (77). PKDs may also play a role in renal cell carcinoma and metastasis (34). Using a combination of PKC and PKD inhibitors, Brenner and colleagues (34) showed that PKDs play a role in the initial steps of tumor progression by regulating the adhesion of renal carcinoma cells to endothelial cells. Studies of human malignant lymphoma cells revealed no detectable levels of PKD1 in these tissues, whereas PKD2 expression was documented with no change in expression level in either normal or malignant tissues. However, further studies are necessary to confirm the role of PKD1 in skin and other cancers and to unveil the mechanisms behind its deregulated expression.
Conclusions
PKD1 is emerging as an important molecule in cancer development. Because many types of cancer show dysregulation of PKD1 expression, it is being explored as a biomarker for cancer diagnosis and prognosis. Although the outcome of PKD1 deregulation seems to be dependent on the type of tissues that present aberrant levels of PKD1, pharmacological interference to regulate PKD1 levels may prove to be an important additional tool to treat cancer. Thus, regulation of PKD1 levels and/or activity may aid cancer treatment because of its role as a critical kinase that integrates extracellular signals into intracellular processes. The active role of PKD1 in cell motility, actin cytoskeleton remodeling, and EMT suggests its potential involvement in cancer metastasis. A further understanding of the molecular basis of PKD1 deregulation and the contrasting outcome of this deregulation in various tissues will help investigators strategically design and formulate novel targeted therapies for cancer treatment.
Acknowledgments
The authors thank Cathy Christopherson and Jenna Hultgren for editorial assistance; Dr. Keith Miskimins for valuable advice; and Dr. Diane Maher, Joshua Hughes, and Sarah Radel for their suggestions.
Grant Support
Sanford Research/USD, Department of Defense (PC073887 to S.C. Chauhan, PC073643 to M. Jaggi); South Dakota Governor’s Cancer 2010 (M. Jaggi).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
V. Sundram drafted the manuscript. S.C. Chauhan and M. Jaggi participated in revising the manuscript.
References
- 1.Jaggi M, Du C, Zhang W, Balaji KC. Protein kinase D1: a protein of emerging translational interest. Front Biosci. 2007;12:3757–67. doi: 10.2741/2349. [DOI] [PubMed] [Google Scholar]
- 2.Johannes FJ, Prestle J, Eis S, Oberhagemann P, Pfizenmaier K. PKCu is a novel, atypical member of the protein kinase C family. J Biol Chem. 1994;269:6140–8. [PubMed] [Google Scholar]
- 3.Valverde AM, Sinnett-Smith J, Van Lint J, Rozengurt E. Molecular cloning and characterization of protein kinase D: a target for diacylglycerol and phorbol esters with a distinctive catalytic domain. Proc Natl Acad Sci USA. 1994;91:8572–6. doi: 10.1073/pnas.91.18.8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 5.Rozengurt E, Rey O, Waldron RT. Protein kinase D signaling. J Biol Chem. 2005;280:13205–8. doi: 10.1074/jbc.R500002200. [DOI] [PubMed] [Google Scholar]
- 6.Van Lint J, Rykx A, Maeda Y, Vantus T, Sturany S, Malhotra V, et al. Protein kinase D: an intracellular traffic regulator on the move. Trends Cell Biol. 2002;12:193–200. doi: 10.1016/s0962-8924(02)02262-6. [DOI] [PubMed] [Google Scholar]
- 7.Rykx A, De Kimpe L, Mikhalap S, Vantus T, Seufferlein T, Vandenheede JR, et al. Protein kinase D: a family affair. FEBS Lett. 2003;546:81–6. doi: 10.1016/s0014-5793(03)00487-3. [DOI] [PubMed] [Google Scholar]
- 8.Guha S, Tanasanvimon S, Sinnett-Smith J, Rozengurt E. Role of protein kinase D signaling in pancreatic cancer. Biochem Pharmacol. 2010;80:1946–54. doi: 10.1016/j.bcp.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.LaValle CR, George KM, Sharlow ER, Lazo JS, Wipf P, Wang QJ. Protein kinase D as a potential new target for cancer therapy. Biochim Biophys Acta. 2010;1806:183–92. doi: 10.1016/j.bbcan.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu Y, Ren M, Feng H, Chen L, Altun ZF, Rubin CS. Neuronal and intestinal protein kinase d isoforms mediate Na+ (salt taste)-induced learning. Sci Signal. 2009;2:ra42. doi: 10.1126/scisignal.2000224. [DOI] [PubMed] [Google Scholar]
- 12.Avkiran M, Rowland AJ, Cuello F, Haworth RS. Protein kinase d in the cardiovascular system: emerging roles in health and disease. Circ Res. 2008;102:157–63. doi: 10.1161/CIRCRESAHA.107.168211. [DOI] [PubMed] [Google Scholar]
- 13.Murphy TR, Legere HJ, 3rd, Katz HR. Activation of protein kinase D1 in mast cells in response to innate, adaptive, and growth factor signals. J Immunol. 2007;179:7876–82. doi: 10.4049/jimmunol.179.11.7876. [DOI] [PubMed] [Google Scholar]
- 14.Hao Q, Wang L, Tang H. Vascular endothelial growth factor induces protein kinase D-dependent production of proinflammatory cytokines in endothelial cells. Am J Physiol Cell Physiol. 2009;296:C821–7. doi: 10.1152/ajpcell.00504.2008. [DOI] [PubMed] [Google Scholar]
- 15.UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39(Database issue):D214–9. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Ruiloba L, Cabrera-Poch N, Rodríguez-Martínez M, López-Menéndez C, Jean-Mairet RM, Higuero AM, et al. Protein kinase D intracellular localization and activity control kinase D-interacting substrate of 220-kDa traffic through a postsynaptic density-95/discs large/zonula occludens-1-binding motif. J Biol Chem. 2006;281:18888–900. doi: 10.1074/jbc.M603044200. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel MT, Garcia EL, Kajimoto T, Hall RA, Newton AC. The protein scaffold NHERF-1 controls the amplitude and duration of localized protein kinase D activity. J Biol Chem. 2009;284:24653–61. doi: 10.1074/jbc.M109.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, et al. A rapid method for determining protein kinase phosphorylation specificity. Nat Methods. 2004;1:27–9. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 19.Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J Biol Chem. 1997;272:952–60. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- 20.Döppler H, Storz P, Li J, Comb MJ, Toker A. A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J Biol Chem. 2005;280:15013–9. doi: 10.1074/jbc.C400575200. [DOI] [PubMed] [Google Scholar]
- 21.Auer A, von Blume J, Sturany S, von Wichert G, Van Lint J, Vandenheede J, et al. Role of the regulatory domain of protein kinase D2 in phorbol ester binding, catalytic activity, and nucleocytoplasmic shuttling. Mol Biol Cell. 2005;16:4375–85. doi: 10.1091/mbc.E05-03-0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rey O, Yuan J, Young SH, Rozengurt E. Protein kinase C nu/protein kinase D3 nuclear localization, catalytic activation, and intracellular redistribution in response to G protein-coupled receptor agonists. J Biol Chem. 2003;278:23773–85. doi: 10.1074/jbc.M300226200. [DOI] [PubMed] [Google Scholar]
- 23.Bisbal M, Conde C, Donoso M, Bollati F, Sesma J, Quiroga S, et al. Protein kinase d regulates trafficking of dendritic membrane proteins in developing neurons. J Neurosci. 2008;28:9297–308. doi: 10.1523/JNEUROSCI.1879-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowell CF, Döppler H, Yan IK, Hausser A, Umezawa Y, Storz P. Mitochondrial diacylglycerol initiates protein-kinase D1-mediated ROS signaling. J Cell Sci. 2009;122:919–28. doi: 10.1242/jcs.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Storz P. Mitochondrial ROS—radical detoxification, mediated by protein kinase D. Trends Cell Biol. 2007;17:13–8. doi: 10.1016/j.tcb.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Hurd C, Rozengurt E. Uncoupling of protein kinase D from suppression of EGF-dependent c-Jun phosphorylation in cancer cells. Biochem Biophys Res Commun. 2003;302:800–4. doi: 10.1016/s0006-291x(03)00268-7. [DOI] [PubMed] [Google Scholar]
- 27.Jacamo R, Sinnett-Smith J, Rey O, Waldron RT, Rozengurt E. Sequential protein kinase C (PKC)-dependent and PKC-independent protein kinase D catalytic activation via Gq-coupled receptors: differential regulation of activation loop Ser(744) and Ser(748) phosphorylation. J Biol Chem. 2008;283:12877–87. doi: 10.1074/jbc.M800442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rybin VO, Guo J, Steinberg SF. Protein kinase D1 autophosphorylation via distinct mechanisms at Ser744/Ser748 and Ser916. J Biol Chem. 2009;284:2332–43. doi: 10.1074/jbc.M806381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vertommen D, Rider M, Ni Y, Waelkens E, Merlevede W, Vandenheede JR, et al. Regulation of protein kinase D by multisite phosphorylation. Identification of phosphorylation sites by mass spectrometry and characterization by site-directed mutagenesis. J Biol Chem. 2000;275:19567–76. doi: 10.1074/jbc.M001357200. [DOI] [PubMed] [Google Scholar]
- 30.Rozengurt E. Mitogenic signaling pathways induced by G protein-coupled receptors. J Cell Physiol. 2007;213:589–602. doi: 10.1002/jcp.21246. [DOI] [PubMed] [Google Scholar]
- 31.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–49. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 32.Trauzold A, Schmiedel S, Sipos B, Wermann H, Westphal S, Röder C, et al. PKCmu prevents CD95-mediated apoptosis and enhances proliferation in pancreatic tumour cells. Oncogene. 2003;22:8939–47. doi: 10.1038/sj.onc.1207001. [DOI] [PubMed] [Google Scholar]
- 33.Kisfalvi K, Hurd C, Guha S, Rozengurt E. Induced overexpression of protein kinase D1 stimulates mitogenic signaling in human pancreatic carcinoma PANC-1 cells. J Cell Physiol. 2010;223:309–16. doi: 10.1002/jcp.22036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brenner W, Beitz S, Schneider E, Benzing F, Unger RE, Roos FC, et al. Adhesion of renal carcinoma cells to endothelial cells depends on PKCmu. BMC Cancer. 2010;10:183. doi: 10.1186/1471-2407-10-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storz P, Toker A. Protein kinase D mediates a stress-induced NF-kappaB activation and survival pathway. EMBO J. 2003;22:109–20. doi: 10.1093/emboj/cdg009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isaacs JT, Isaacs WB. Androgen receptor outwits prostate cancer drugs. Nat Med. 2004;10:26–7. doi: 10.1038/nm0104-26. [DOI] [PubMed] [Google Scholar]
- 37.Mak P, Jaggi M, Syed V, Chauhan SC, Hassan S, Biswas H, et al. Protein kinase D1 (PKD1) influences androgen receptor (AR) function in prostate cancer cells. Biochem Biophys Res Commun. 2008;373:618–23. doi: 10.1016/j.bbrc.2008.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassan S, Biswas MH, Zhang C, Du C, Balaji KC. Heat shock protein 27 mediates repression of androgen receptor function by protein kinase D1 in prostate cancer cells. Oncogene. 2009;28:4386–96. doi: 10.1038/onc.2009.291. [DOI] [PubMed] [Google Scholar]
- 39.Wang G, Wang J, Sadar MD. Crosstalk between the androgen receptor and beta-catenin in castrate-resistant prostate cancer. Cancer Res. 2008;68:9918–27. doi: 10.1158/0008-5472.CAN-08-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaggi M, Johansson SL, Baker JJ, Smith LM, Galich A, Balaji KC. Aberrant expression of E-cadherin and beta-catenin in human prostate cancer. Urol Oncol. 2005;23:402–6. doi: 10.1016/j.urolonc.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 41.Jaggi M, Rao PS, Smith DJ, Hemstreet GP, Balaji KC. Protein kinase C mu is down-regulated in androgen-independent prostate cancer. Biochem Biophys Res Commun. 2003;307:254–60. doi: 10.1016/s0006-291x(03)01161-6. [DOI] [PubMed] [Google Scholar]
- 42.Jaggi M, Chauhan SC, Du C, Balaji KC. Bryostatin 1 modulates beta-catenin subcellular localization and transcription activity through protein kinase D1 activation. Mol Cancer Ther. 2008;7:2703–12. doi: 10.1158/1535-7163.MCT-08-0119. [DOI] [PubMed] [Google Scholar]
- 43.Müller S, Krämer OH. Inhibitors of HDACs—effective drugs against cancer? Curr Cancer Drug Targets. 2010;10:210–28. doi: 10.2174/156800910791054149. [DOI] [PubMed] [Google Scholar]
- 44.Ha CH, Wang W, Jhun BS, Wong C, Hausser A, Pfizenmaier K, et al. Protein kinase D-dependent phosphorylation and nuclear export of histone deacetylase 5 mediates vascular endothelial growth factor-induced gene expression and angiogenesis. J Biol Chem. 2008;283:14590–9. doi: 10.1074/jbc.M800264200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ha CH, Jhun BS, Kao HY, Jin ZG. VEGF stimulates HDAC7 phosphorylation and cytoplasmic accumulation modulating matrix metalloproteinase expression and angiogenesis. Arterioscler Thromb Vasc Biol. 2008;28:1782–8. doi: 10.1161/ATVBAHA.108.172528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watkins JL, Lewandowski KT, Meek SE, Storz P, Toker A, Piwnica-Worms H. Phosphorylation of the Par-1 polarity kinase by protein kinase D regulates 14-3-3 binding and membrane association. Proc Natl Acad Sci USA. 2008;105:18378–83. doi: 10.1073/pnas.0809661105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaggi M, Rao PS, Smith DJ, Wheelock MJ, Johnson KR, Hemstreet GP, et al. E-cadherin phosphorylation by protein kinase D1/protein kinase Cmu is associated with altered cellular aggregation and motility in prostate cancer. Cancer Res. 2005;65:483–92. [PubMed] [Google Scholar]
- 48.Yamamoto H, Oue N, Sato A, Hasegawa Y, Yamamoto H, Matsubara A, et al. Wnt5a signaling is involved in the aggressiveness of prostate cancer and expression of metalloproteinase. Oncogene. 2010;29:2036–46. doi: 10.1038/onc.2009.496. [DOI] [PubMed] [Google Scholar]
- 49.Woods AJ, White DP, Caswell PT, Norman JC. PKD1/PKCmu promotes alphavbeta3 integrin recycling and delivery to nascent focal adhesions. EMBO J. 2004;23:2531–43. doi: 10.1038/sj.emboj.7600267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eiseler T, Schmid MA, Topbas F, Pfizenmaier K, Hausser A. PKD is recruited to sites of actin remodelling at the leading edge and negatively regulates cell migration. FEBS Lett. 2007;581:4279–87. doi: 10.1016/j.febslet.2007.07.079. [DOI] [PubMed] [Google Scholar]
- 51.Syed V, Mak P, Du C, Balaji KC. Beta-catenin mediates alteration in cell proliferation, motility and invasion of prostate cancer cells by differential expression of E-cadherin and protein kinase D1. J Cell Biochem. 2008;104:82–95. doi: 10.1002/jcb.21603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eiseler T, Döppler H, Yan IK, Goodison S, Storz P. Protein kinase D1 regulates matrix metalloproteinase expression and inhibits breast cancer cell invasion. Breast Cancer Res. 2009;11:R13. doi: 10.1186/bcr2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Storz P. Protein kinase D1: a novel regulator of actin-driven directed cell migration. Cell Cycle. 2009;8:1975–6. [PubMed] [Google Scholar]
- 54.Peterburs P, Heering J, Link G, Pfizenmaier K, Olayioye MA, Hausser A. Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res. 2009;69:5634–8. doi: 10.1158/0008-5472.CAN-09-0718. [DOI] [PubMed] [Google Scholar]
- 55.Eiseler T, Döppler H, Yan IK, Kitatani K, Mizuno K, Storz P. Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat Cell Biol. 2009;11:545–56. doi: 10.1038/ncb1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eiseler T, Hausser A, De Kimpe L, Van Lint J, Pfizenmaier K. Protein kinase D controls actin polymerization and cell motility through phosphorylation of cortactin. J Biol Chem. 2010;285:18672–83. doi: 10.1074/jbc.M109.093880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Kimpe L, Janssens K, Derua R, Armacki M, Goicoechea S, Otey C, et al. Characterization of cortactin as an in vivo protein kinase D substrate: interdependence of sites and potentiation by Src. Cell Signal. 2009;21:253–63. doi: 10.1016/j.cellsig.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 58.Du C, Jaggi M, Zhang C, Balaji KC. Protein kinase D1-mediated phosphorylation and subcellular localization of beta-catenin. Cancer Res. 2009;69:1117–24. doi: 10.1158/0008-5472.CAN-07-6270. [DOI] [PubMed] [Google Scholar]
- 59.Kim M, Jang HR, Kim JH, Noh SM, Song KS, Cho JS, et al. Epigenetic inactivation of protein kinase D1 in gastric cancer and its role in gastric cancer cell migration and invasion. Carcinogenesis. 2008;29:629–37. doi: 10.1093/carcin/bgm291. [DOI] [PubMed] [Google Scholar]
- 60.Jepperson T, Hughes J, Hausser A, Balaji K, Chauhan S, Jaggi M. Protein kinase D1 reduces cellular proliferation and invasiveness via suppression of beta-catenin/T-cell factor activity in SW480 colon cancer cells [abstract 4256]. Proceedings of the 100th Annual Meeting of the American Association for Cancer Research; 2009 Apr 18–22; Denver, CO. Philadelphia (PA): AACR; 2009. [Google Scholar]
- 61.Guha S, Rey O, Rozengurt E. Neurotensin induces protein kinase C-dependent protein kinase D activation and DNA synthesis in human pancreatic carcinoma cell line PANC-1. Cancer Res. 2002;62:1632–40. [PubMed] [Google Scholar]
- 62.Bollag WB, Dodd ME, Shapiro BA. Protein kinase D and keratinocyte proliferation. Drug News Perspect. 2004;17:117–26. doi: 10.1358/dnp.2004.17.2.829045. [DOI] [PubMed] [Google Scholar]
- 63.Ristich VL, Bowman PH, Dodd ME, Bollag WB. Protein kinase D distribution in normal human epidermis, basal cell carcinoma and psoriasis. Br J Dermatol. 2006;154:586–93. doi: 10.1111/j.1365-2133.2005.07073.x. [DOI] [PubMed] [Google Scholar]
- 64.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du C, Zhang C, Hassan S, Biswas MH, Balaji KC. Protein kinase D1 suppresses epithelial-to-mesenchymal transition through phosphorylation of snail. Cancer Res. 2010;70:7810–9. doi: 10.1158/0008-5472.CAN-09-4481. [DOI] [PubMed] [Google Scholar]
- 66.Biswas MH, Du C, Zhang C, Straubhaar J, Languino LR, Balaji KC. Protein kinase D1 inhibits cell proliferation through matrix metalloproteinase-2 and matrix metalloproteinase-9 secretion in prostate cancer. Cancer Res. 2010;70:2095–104. doi: 10.1158/0008-5472.CAN-09-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rao PS, Jaggi M, Smith DJ, Hemstreet GP, Balaji KC. Metallothionein 2A interacts with the kinase domain of PKCmu in prostate cancer. Biochem Biophys Res Commun. 2003;310:1032–8. doi: 10.1016/j.bbrc.2003.09.118. [DOI] [PubMed] [Google Scholar]
- 68.Smith DJ, Jaggi M, Zhang W, Galich A, Du C, Sterrett SP, et al. Metallothioneins and resistance to cisplatin and radiation in prostate cancer. Urology. 2006;67:1341–7. doi: 10.1016/j.urology.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 69.Lavalle CR, Bravo-Altamirano K, Giridhar KV, Chen J, Sharlow E, Lazo JS, et al. Novel protein kinase D inhibitors cause potent arrest in prostate cancer cell growth and motility. BMC Chem Biol. 2010;10:5. doi: 10.1186/1472-6769-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen J, Deng F, Singh SV, Wang QJ. Protein kinase D3 (PKD3) contributes to prostate cancer cell growth and survival through a PKCepsilon/PKD3 pathway downstream of Akt and ERK 1/2. Cancer Res. 2008;68:3844–53. doi: 10.1158/0008-5472.CAN-07-5156. [DOI] [PubMed] [Google Scholar]
- 71.Bowden ET, Barth M, Thomas D, Glazer RI, Mueller SC. An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene. 1999;18:4440–9. doi: 10.1038/sj.onc.1202827. [DOI] [PubMed] [Google Scholar]
- 72.Rodriguez Perez CN, Nie W, Sinnett-Smith J, Rozengurt E, Yoo J. TNF-alpha potentiates lysophosphatidic acid-induced COX-2 expression via PKD in human colonic myofibroblasts. Am J Physiol Gastrointest Liver Physiol. 2011;300:G637–46. doi: 10.1152/ajpgi.00381.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guha S, Lunn JA, Santiskulvong C, Rozengurt E. Neurotensin stimulates protein kinase C-dependent mitogenic signaling in human pancreatic carcinoma cell line PANC-1. Cancer Res. 2003;63:2379–87. [PubMed] [Google Scholar]
- 74.Harikumar KB, Kunnumakkara AB, Ochi N, Tong Z, Deorukhkar A, Sung B, et al. A novel small-molecule inhibitor of protein kinase D blocks pancreatic cancer growth in vitro and in vivo. Mol Cancer Ther. 2010;9:1136–46. doi: 10.1158/1535-7163.MCT-09-1145. Erratum in Mol Cancer Ther 2010, 9, 2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jadali A, Ghazizadeh S. Protein kinase D is implicated in the reversible commitment to differentiation in primary cultures of mouse keratinocytes. J Biol Chem. 2010;285:23387–97. doi: 10.1074/jbc.M110.105619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arun SN, Kaddour-Djebbar I, Shapiro BA, Bollag WB. Ultraviolet B irradiation and activation of protein kinase D in primary mouse epidermal keratinocytes. Oncogene. 2010;30:1586–96. doi: 10.1038/onc.2010.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paolucci L, Rozengurt E. Protein kinase D in small cell lung cancer cells: rapid activation through protein kinase C. Cancer Res. 1999;59:572–7. [PubMed] [Google Scholar]
- 78.Yuan J, Rozengurt E. PKD, PKD2, and p38 MAPK mediate Hsp27 serine-82 phosphorylation induced by neurotensin in pancreatic cancer PANC-1 cells. J Cell Biochem. 2008;103:648–62. doi: 10.1002/jcb.21439. [DOI] [PubMed] [Google Scholar]