Abstract
Curcumin, a natural bioactive polyphenol, has been widely investigated as a conventional medicine for centuries. Over the past two decades, major pre-clinical and clinical trials have demonstrated its safe therapeutic profile but clinical translation has been hampered due to rapid degradation, poor water solubility, bioavailability and pharmaco-kinetics. To overcome such translational issues, many laboratories have focused on developing curcumin nanoformulations for cancer therapeutics. In this review, we discuss the evolution of curcumin nanomedicine in cancer therapeutics, the possible interactions between the surface of curcumin nanoparticles and plasma proteins, the role of nanoparticle-protein complex architecture parameters, and the rational design of clinically useful curcumin nanoformulations. Considering all the biologically relevant phenomena, curcumin nanoformulations can be developed as a new neutraceutical or pharmaceutical agent.
Keywords: Polyphenol, drug delivery, nanomedicine, cancer therapeutics, bioavailability, protein corona
1. CURCUMIN IN CANCER THERAPY
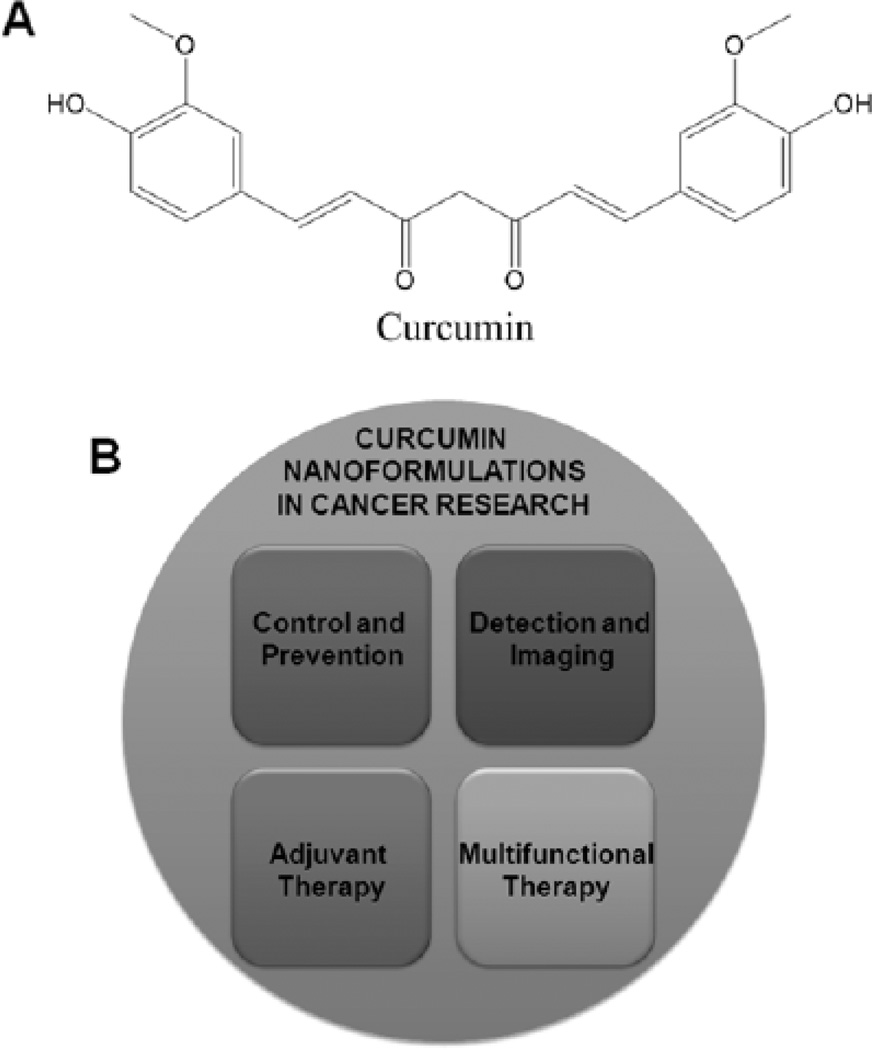
Cancer remains the second leading cause of death worldwide. The American Cancer Society estimated there were approximately 1,638,910 new diagnoses and 577,190 deaths in 2012 due to cancer in the United States [1]. Present treatment modalities such as surgery, chemotherapy, hormone therapy, adjuvant therapy and radiation have been successful to reduce the morbidity or mortality of cancer. Chemotherapy is a common means of treatment for all stages of cancers. Eventhough the use of chemotherapy typically results in an improved survival rate, there are numerous side effects associated with it. Since ancient days many natural products have been used to improve human health outcomes. In addition, due to low toxicity and low hypersensitive reactions, use of these natural products increases the patient's compliance and strength. Therefore, there are a growing number of scientists and clinicians interested in exploring the use of traditional or herbal medicine for preventive and therapeutic purposes to control cancer growth. Traditional medicines derived from natural sources come under preventive treatment options according to complementary and alternative medicine by the National Cancer Institute (NCI) of the National Institutes of Health (NIH, USA). In fact, three-fourths of drugs approved by the Food and Drug Administration (FDA, USA) for cancer treatment are either natural compounds or their analogues [2] which exhibit favorable therapeutic potential. Significant data among the herbal medicinal molecules demonstrates that curcumin exhibits antitumor effects against various cancer cell lines and animal models. Curcumin (CUR) is principally a bioactive polyphenol (chemical structure Fig. 1A), derived from the rhizome of Curcuma longa Linn that has proven to have a pleiotropic property. This molecule can target and modulate a number of carcinogenic intracellular signaling pathways that regulate cell growth, inflammation and apoptosis in cancer therapeutics [3–5]. Additionally, curcumin enhances the potency of chemotherapeutic agents and radiation therapies in cancer treatments [6–7]. More importantly, curcumin is nontoxic to healthy cells and therefore does not hinder health care practitioners from using it. Many clinical trials for cancer treatments have investigated the roles of curcumin both alone and in combination with standard therapeutic agents [8, 9]. Ongoing clinical and animal studies have demonstrated the therapeutic significance of curcumin. Curcumin could also emerge as a potential therapeutic and chemopreventive agent in cancer therapeutics, by increasing its solubility, absorption, bioavailability, pharmaco-kinetics and specificity to improve plasma and tissue levels. Recent investigations focusing on developing new approaches based on nanotechnology such as selfassemblies, polymer complexes, curcumin loaded nanoparticles (NPs) or layers of micelles, aim to increase the systemic availability for efficient therapeutics at the tumor site to (1) control and prevent the cancer from progression, (2) detect and image simultaneously, (3) provide adjuvant therapy in addition to conventional therapies, and (4) have multi-functional therapeutic purposes [10] (Fig. 1B).
Fig. (1).
Multi-functional role of curcumin nanoformulations in cancer therapeutics.
2. CURCUMIN-HUMAN SERUM ALBUMIN IN DRUG DELIVERY
Orally administered curcumin was found in the liver, kidney and blood in rats due to albumin, an effective carrier [11]. Subsequent investigation further confirmed that curcumin interacts with human serum albumin (HSA) at two distinct binding sites [12]. A Leung and Kee study [13] delineates that among plasma proteins, curcumin binds to HSA and fibrinogen with a binding constant of 1.22 ± 0.35 × 105 M−1 and 5.99 ± 1.75 × 104 M−1, respectively. Such binding phenomena reduce hydrolytic degradation of curcumin by 95% compared to curcumin in PBS. Similarly, a significant reduction of curcumin degradation was also observed in the presence of a 6–8% serum protein in a buffer solution [14]. Curcumin has shown to bind the sub-domain IIA site of HSA [15]. Curcumin-HSA interactions in synchronous fluorescence spectral analysis are 2 fold higher for tryptophan than tyrosine residue in HSA, and proves that curcumin-HSA interaction mainly takes place at tryptophan residue in HSA [16]. The curcumin-HSA binding interaction occurs at 2.8 nm förster critical distance, 3.1 nm binding distance to the tryptophan residue of protein with 2.209 × 10−14 cm3 m−1 overlap integral and 0.35 energy transfer efficiency [17]. Upon curcumin binding, it alters the content of the secondary structure of HSA, i.e., α-helix structure increased while the random coil percent decreased. All investigations and literature proposed that HSA can act as an efficient transporter for the curcumin molecule [18]. The physiological role of HSA is widely known and it can bind to numerous lipophilic molecules that may lead to an increase in its pharmacology profile [19]. In a review article, Xiao et al. [20] concluded that the effect of the polyphenol-HSA interaction on biological activity is not equivocal and depends entirely on the structure-affinity relationship. Although attempts have been made to improve the solubility, stability, and degradation of curcumin in vitro, delivering it in vivo in the presence of organic solvent/surfactant (needed to dissolve curcumin) which are may be toxic to humans. Additionally, achieving optimal therapeutic concentration of curcumin at the tumor site is highly difficult using the HSA-binding or HSA-complex transportation method because no specific target mechanism is involved. Alternatively, nanoparticle (NP) carriers have been chosen for effective delivery of curcumin [10]. However, a number of studies showed that relatively small fractions of NPs reached the bloodstream due to hazardous agglomeration or cluster formations [21]. In fact, many curcumin and anti-cancer therapeutic nanoformulations have a bioavailability less than 5% injected dose volume (IDV) at tumor sites. Therefore, bioavailability of curcumin NPs is strongly dependent on the extension of adsorption of proteins in blood. The Centre for BioNano Interaction at University College Dublin, Belfield (Dublin, Ireland) has initiated studies which examine the interaction of NPs with various plasma proteins [22]. Based on these observations, a detailed interaction of curcumin NPs with plasma proteins and implications for their use in cancer therapeutics will be discussed in forthcoming sections.
3. NANOPARTICLES-PLASMA PROTEINS BINDING: GENERAL VIEW
3.1. Nanoparticles-plasma Proteins Interaction in Drug Delivery Applications
The use of NPs has tremendously increased for drug delivery applications. However, it is a highly challenging task to design a NP system that exhibits superior in vivo behavior. Many of the conventional nanoformulations may be compromised by the reticuloendothelial system (RES) prior to reaching the tumor tissue or the blood brain barrier. For improving such biological phenomenon, several methods have been implemented to control particle size and increase the stealthiness of nanoformulations. Curcumin NPs are considered to be a novel alternative therapeutic option in cancer therapeutics [20, 23, 24]. Studies have confirmed that NPs are robustly bound with plasma proteins, cells or tissues and are never found in the form of parent particles [21]. After adsorption of plasma proteins, NPs may either lead to beneficial or harmful biological actions. Therefore, in order to develop effective translational therapeutic outcomes for curcumin NPs, it is essential to learn its complex role in the biological plasma system.
In general, NPs contain a larger active surface area than their origin bulk material. Active surfaces of NPs are first exposed and begin to interact with human plasma proteins when they are in blood circulation [25, 26]. During this process, a selective exchange and adsorption of proteins occurs via supramolecular interactions that lead to the formation of corona on the surface of NPs [26]. This dynamic process may give rise to hard/permanent corona or soft corona depending on the nature of the NPs and their composition [27–29]. These interactions may determine the biological entity of the NPs. The biomolecular interface of the protein corona on NPs are constructed either with “short” or “long” lived structures which can be determined by the state of exchange of proteins. For superior drug delivery applications, NPs must be generated to allow the formation of hard corona which will likely improve the interface between NPs, formulations and biological milieu.
3.2. Evaluation of Nanoparticles-plasma Proteins Interaction
Dell’Orco et al. [30] proposed a model after considering the time evolution of the NP-protein corona formation in body fluids (HSA, high density lipoprotein and fibrinogen) with 70 nm polymer NPs. This model provides binding data fitted by a simple binding model of inter-protein interaction phenomena and the dynamics of the NP -protein interactions of a reversible biochemical equation (Equation 1). The overall biochemical system differential equation and the time evolution of individual proteins and NP complexation was assessed by numerical kinetic models carried out by SBTOOLBOX2 (Matlab, http://www.sbtoolbox2.org) (Equation 2). This modeling technique can further extend to evaluate the specific molecular interactions of nanoparticle-cell/tissues. NP, PR, and t represent concentration of nanoparticles, proteins and time of measurement, respectively. The association and dissociation of NPPR complexes were represented as concentrations, individual values of the rate constants κon, κoff, and the stoichiometry in a single layer.
| (1) |
| (2) |
The charge/density of NPs and plasma protein coronas may also have a major biological impact for in vivo applications [31]. Fibrinogen showed higher association with NPs at a higher membrane charge density while apolipoproteins and C4b-binding proteins bound NPs showed less membrane charge density. Charge density can be calculated from the relative amount of charge on the protein corona by multiplying the total charge on the protein κ (qκ), probability threshold (Φ) by protein abundance, molecular weight of normalized scaffold for protein κ (MWNSC κ):
| (3) |
Recently, quantitative proteomics analysis has gained much attention as a means to evaluate the adsorbed plasma proteins on NPs [32–34]. Zhang et al. [32] have studied different surface modifications and particle sizes of NP interactions with plasma proteins. All NPs showed significant binding with 42 plasma proteins. Only one out of three clusters from these 42 proteins exhibited a different protein binding pattern with different surface modifications. Two clusters of proteins presented opposite patterns of relative abundance changes. This method also revealed that different sizes of NPs are exclusively able to alter protein abundance. A revolutionary proteomics and nanotechnology concept was introduced to identify new therapeutic targets in cancer via proteins bound to NPs [35].
As expected, when NPs make contact with biological fluids, the surfaces are coated with proteins which change their surface charge and properties. However, high NP protein binding is not always disadvantageous. In some cases, these interactions may lead to the delivery of NPs to a specific organ or tissue. For instance, a paclitaxel nanoformulation [(albumin-coated paclitaxel nanoformulation (nab-PTX) (Abraxane®)] showed specific interactions with secreted acidic and rich extratumoral proteins help with targeted delivery of paclitaxel [36, 37]. Further, such interactions efficiently modulate the transportation of paclitaxel across endothelial barriers through gp60 receptor and caveolin-1 mediated pathways [38].
Dell'Orco et al. proposed that theoretical systems-level analysis of the protein corona of plasma protein formations on the engineered and functionalized surface of NPs which have specific biological targets is often neglected [39]. Their suggestion for effective targeted delivery of drug(s), in addition to the biophysical quantities of early/hard/dynamic corona, is that systems-level mathematical approach generated stoichiometry and the kinetics of the relative interactions must be evaluated with a particular tissue(s). Global sensitivity analyses of selected dynamic conditions of the network models will improve the targeted drug delivery success rate through nanomedicine. We believe such global sensitive models can also be implemented for effective delivery of curcumin to the tumor site for an enhanced therapeutic potential.
3.3. Techniques Used to Investigate Nanoparticles-plasma Proteins Interaction
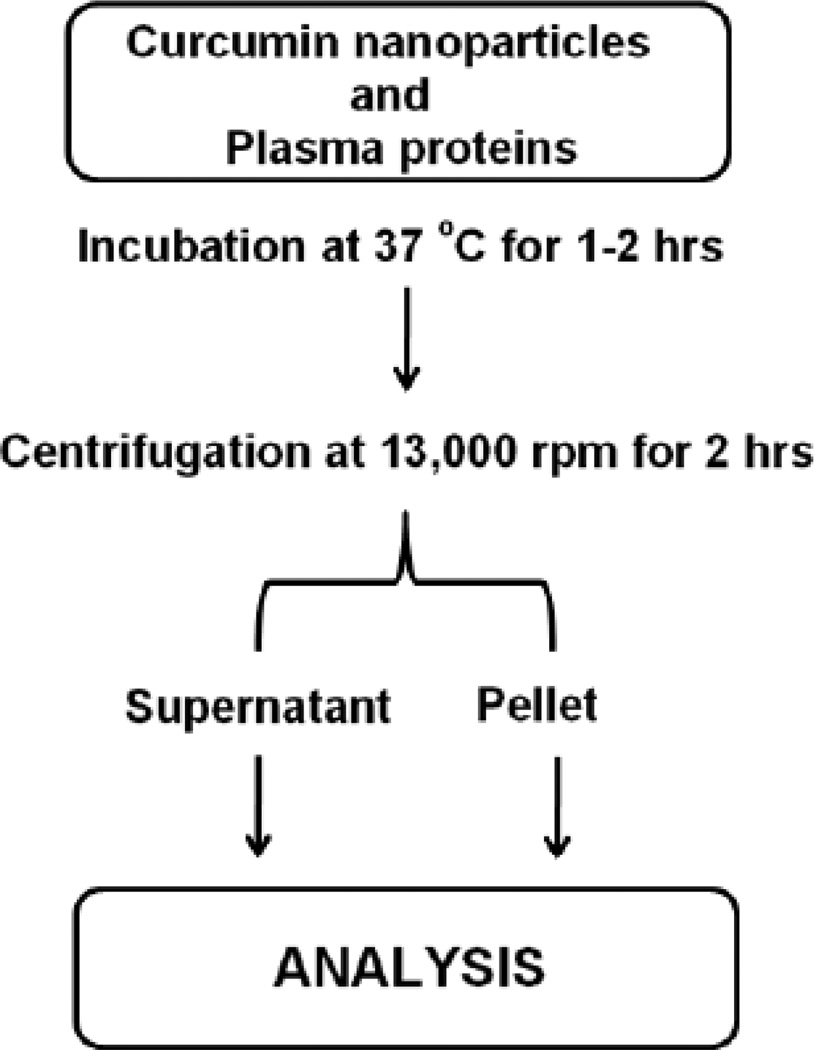
The molecular structure and affinity relation of curcumin with serum proteins, observed as non-covalent binding, can be investigated by the binding constants obtained from spectral, dynamic light scattering, microscopic analysis and other analytic methods [22, 40–43]. In the case of curcumin nanoparticles, bound plasma proteins were separated by normal or gradient centrifugation, and then pellets containing curcumin nanoparticle-plasma protein corona (Fig. 2) are subjected to various analytical methods for accurate evaluation. Gebauer and co-workers demonstrated that the protein corona of HSA around citrate-functionalized silver NPs stabilized the particles without inducing agglomeration and protein structure changes [44]. CD spectral analysis can provide signals of proteins arising from electronic transitions in secondary structural elements such as α-helix, β-helix, turn and random coils. The quantitative analysis of these data provides insight into the alterations which occurred in the original protein structure by the interaction. Atomic force microscopy is a relatively new technique that can provide additional information on currently used methods. This technique delineates the amount of proteins present on NPs and the force of adhesion and surface free energy on the particles before and after modification with proteins [45]. Hydrophobicity in NP composition is also highly favored for extensive binding of plasma proteins [46]. Lindman et al., [46] demonstrated that a fully covered HSA layer was achieved by hydrophobic NPs but at a lower curvature. These results were confirmed experimentally by isothermal titration calorimetry, yielding stoichiometry, affinity, and enthalpy changes from the NP and HSA binding. Most of the NP and plasma protein interaction studies were evaluated by fluorescence quenching or the decay of fluorescence of proteins. These measurements can be achieved by steady-state absorption, synchronous fluorescence, three-dimensional (3D) fluorescence and circular dichroism (CD) spectroscopic techniques [43]. In addition, one and two-dimensional polyacrylamide gel electrophoresis (1D and 2D-PAGE), liquid chromatography mass spectrometry (LC-MS) and liquid chromatography mass spectrometry/mass spectrometry (LCMS/MS) can also help to quantify NPs and plasma proteins interaction. Table 1 illustrates various analytical techniques used to evaluate nanoparticles and plasma proteins interactions.
Fig. (2).
Schematic representation of sample preparation from nanoparticles-plasma proteins corona for estimation of bound plasma proteins. Incubation time, centrifugation time, and number of washing for obtaining nanoparticle-plasma protein corona varies depend on the specific protocol.
Table 1.
Various Analytical Techniques Used to Evaluate Nanoparticles and Plasma Proteins Interactions
| Analytic Method | Type of Evaluation and Comments |
|---|---|
| 1D-polyacrylamide gel electrophoresis (PAGE) and 2D PAGE | Identification of proteins in corona on nanoparticles. The order of protein adsorbed onto the surface of nanoparticles can be determined. With the help of densitometry, the half-max protein adsorption and hill slope for protein. Highly laborious and not helpful/useful for quick kinetic adsorption evaluations. |
| BSA Quantification | Half-max protein adsorption. Hill slope for protein adsorption. |
| UV-Vis extinction spectroscopy | Half-max protein adsorption. Hill slope for protein adsorption. Apparent thickness, orientation and refractive index of the adsorbed proteins. Simultaneous protein adsorption measurements. |
| Gel permeation chromatography | Determination of molecular weight of plasma proteins bound to nanoparticles. Comparative evaluation of bound proteins on nanoparticles. Requires larger volumes of samples for analysis. |
| LC-MS/MS | Efficient separation of bound plasma proteins. Identification of protein/peptide sequence. Accurate molecular weight distribution. Requires larger volumes of samples for analysis. |
| Capillary electrophoresis | Analysis of protein–nanoparticle interaction with high degree of resolution. Efficient separation of plasma proteins and very small sample volumes are required for analysis. Determination of the fate of nanoparticles in vivo including delivery to tumors due to their interactions with specific plasma proteins. Stable nanoparticle-protein complexes and transient complexes were resolved by capillary zone electrophoresis and affinity capillary electrophoresis. This technique is also used in combination with other analytical methods to evaluate each type and percentage of various bound proteins on nanoparticles. |
| Atomic force microscope | Measure of change of indention before and after protein corona in situ modification. Determined the work of adhesion. Evaluation of surface free energy during protein corona formation. Force of adhesion involved in protein corona formation. |
| Fluorescence correlation spectroscopy | Used to quantitatively monitor protein binding or adsorption onto fluorescent nanoparticles. Correlation analysis of the fluorescence emission time traces yields a characteristic time scale of diffusion from the hydrodynamic radius. Size of the nanoparticles after protein deposition on their surfaces can be monitored. Nanomolar concentrations of the nanoparticles in microlitre-sized volumes required. |
| Particle Size and Distribution (TEM and DLS) | Plasma protein corona structure examination. Particle’s dispersibility before and after protein corona formation on nanoparticles. Particle’s zeta potential before and after protein corona formation on nanoparticles. |
4. CURCUMIN NANOPARTICLES-PLASMA PROTEIN CORONA IN CANCER THERAPEUTICS
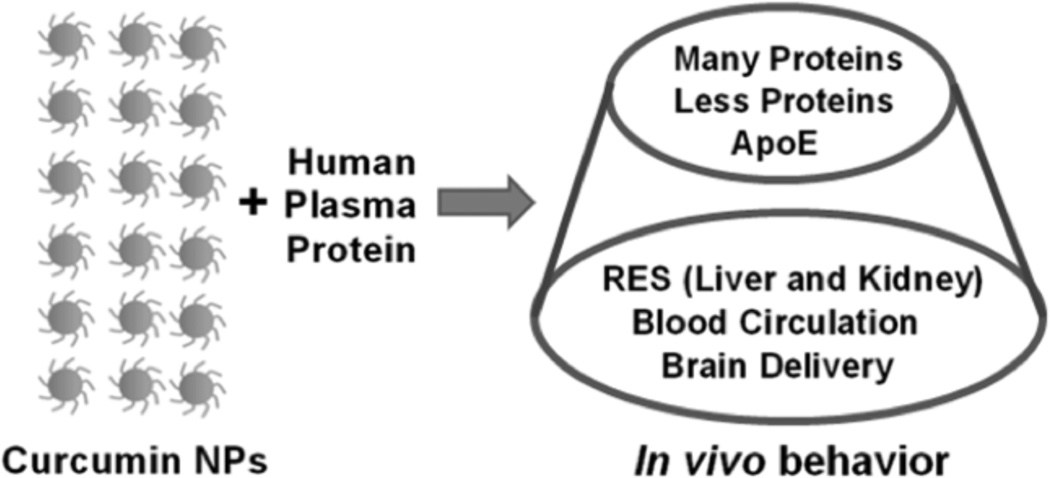
Deeper penetration of curcumin NPs into cancer cells can be achieved through proper protein corona formation on the surface of NPs. Based on the plasma protein interaction with curcumin NPs, their in vivo biologically relevance can be determined (Fig. 3). Table 2 illustrates how curcumin NPs can significantly show increased bioavailability in vivo [10, 49–63] due to their favorable plasma protein corona formation i.e., less number of proteins are required to increase the NPs in blood circulation [47, 48]. The NP-protein corona is highly associated with cellular uptake in cancer cells [35]. Modulation of protein corona around NPs is a promising approach and an effective therapeutic target. Serum proteins have ensured a role in the feeding process which permits cellular entry through contacting bare NP surfaces. Such types of internalization include the uptake of NPs or drug loaded NPs in living cells followed by (1) passive diffusion, (2) endocytosis, and/or (3) phagocytosis. Phagocytosis occurs when NP-plasma protein interactions make particles larger in size, typically several hundred nanometers to microns, due to aggregation. Endocytosis of NPs is possible through an internalization mechanism in which NPs are coated slightly with human serum proteins.
Fig. (3).
In vivo fate of curcumin nanoparticles after interacting with human serum or human plasma proteins.
Table 2.
Improved Bioavailability of Curcumin with Curcumin Nanoformulations
| Formulation | Study Outcome | |
|---|---|---|
| Dose and Type of Administration | Improved Bioavailability | |
| Curcumin loaded poly(lactide-co-glycolide) (PLGA) NPs [49] | 250 mg/kg (curcumin), 250 mg/kg (curcumin) + 10 mg/kg(piperine), and 100 mg/kg (PLGA NPs) body weight of rat Oral | The in vivo oral bioavailability was increased up to 9-fold compared to curcumin+piperine composition. |
| PLGA and a stabilizer polyethylene glycol (PEG)-5000 [50] | 2.5 mg/kg body weight of mice Intravenous | Curcumin NPs or curcumin were maintained at 430–100 ng/ml and 275–25 ng/ml in serum for 1–24 hrs. All time points tested curcumin NPs showed at least 2 fold more bioavailability. |
| Nanocrystal solid dispersion (CSD-Cur), amorphous solid dispersion (ASD-Cur), and nanoemulsion (NE-Cur) [51] | 100 mg/kg body weight of rat for curcumin and 20 mg/kg body weight of rat for CSD-CUR, ASD-CUR, and NE-CUR Oral | Oral bioavailability was increased from 0.9 (cur-cumin) to 14.3, 10.7, and 7.9% for CSD-CUR, ASD-CUR, and NE-CUR formulations, respectively. |
| Curcumin nanosuspension with D-α -tocopheryl polyethylene glycol 1000 succinate (TPGS) [52] | 15 mg/kg body weight of rabbit Intravenous | Approximately 3.8-fold greater bioavailability (145.42 ± 9.29 µg/mL min) was achieved with a mean residence time of 194.57 ± 32.18 (11.2-fold longer vs. curcumin). |
| Glycerol monooleate-poly(vinyl alco-hol)-pluronic (F127) polymer NPs [53] | 30 mg/kg body weight of mice Intravenous | At 1–24 hrs time points, curcumin NPs exhibited 17.5-4 µg/ml curcumin in serum while native cur-cumin was barely detected (0.2 µg/ml serum). |
| Nanocur™ [54] | 25 mg/kg body weight of mice Intraperitoneal | > 8 µg/ml plasma levels of Nanocur™ was present between 1 to 8 hrs whereas free curcumin in corn oil was barely detectable. |
| Solid lipid NPs C-SLNs: very high (VH); high (H); medium (M); and small (S) and free curcumin (C-S) [55] | 50, 50, 25, 12.5, and 1 mg/kg body weight of rat for C-S, VH, H, M, and S, respectively Oral | Approximately, 0.292, 14.29, 8.00, 7.87, and 1.00 µg/ ml of maximum of curcumin was present in serum by C-S, VH, H, M and S administrations, respectively. |
| Organogel-based nanoemulsion [56] | 240 mg/kg body weight of mice Oral | 8.5–9.8 fold increase in bioavailability. |
| PLGA-CUR LMw and HMw nanofor-mulations [57] | 50 mg/kg body weight of rat for LMw and HMw nanocurcumin formulations and 1000 mg/kg body weight of rat for free curcumin Oral | No significant difference in bioavailability between LMw and HMw curcumin nanoformulations but 1.67 and 40-fold higher bioavailability compared to free curcumin |
| Solid lipid NPs [58] | 400 mg/kg body weight of mice Intraperitoneal | 20 micro molar curcumin was noticed at 30 min. |
| Poly(ε–caprolactone)-b-poly(ethylene glycol)-b-poly(ε–caprolactone) triblock copolymer NPs [59] | 15 mg/kg body weight of rat Intravenous | Increased mean residence time from 0.169 to 40.148 hours and the area under the concentration– time curve increased 4.178-fold. |
| PLGA-CUR [60] | 7.5 mg/kg body weight of rat Intravenous | PLGA-CUR was found to be 6.139 mg/L h. |
| PLGA and PLGA-poly(ethylene glycol) (PEG) [61] | 50 mg/kg body weight of rat Oral | PLGA and PLGA-PEG NPs increased the curcu-min bioavailability by 15.6- and 55.4-fold, respectively. |
| Silica-coated flexible liposomes as a nanohybrid [62] | 50 mg/kg body weight of rat Oral | The bioavailability of CUR-SLs and CUR-FLs was 7.76- and 2.35-fold higher, respectively. |
| Lipid based oral formulation [63] | 250 mg/kg rat Oral | Improved Cmax and AUC0–∞ by 11.6 and 35.8 fold, respectively, over control. |
Lundqvist et al. [28] suggested a possible NP trafficking scenario in which the role of protein corona formation is highly significant. The adsorbed-protein complexes on NPs are dynamically changed from one biological environment into another by replacing proteins from the new biological fluid. In the end, the NP and adsorbed protein may control signals that can be explained from the protein corona. In contrast, Jiang et al. [64] revealed that in the presence of 100 µM transferrin or HSA coating on 1 nM NPs, the amount of NPs on the cell surface, as well as in the intracellular binding, was substantially reduced. This behavior was further verified through confocal analysis of a group of 15 cells from two to three independent experiments. Interestingly, lower intracellular fluorescence was observed from the NPs than the membraneassociated particles. The media used for in vitro experiments has a major role in internalization of curcumin NPs in cancer cells. Gold NPs suspended in DMEM media display a red shift and increased band intensity over time [65]. Such significant changes were not noticed in RPMI, which indicated DMEM endorsed abundant protein corona on the NPs. This in turn influenced the uptake and safe internalization in U937 and HeLa cells [65].
Recent findings have reported on the roles of particle aggregation and plasma protein coating on cell uptake by the iron content estimation [66]. According to this study, at low plasma concentrations NPs form clusters by proteins like fibrinogen, whereas at high plasma concentrations NPs stabilized by apolipoprotein coating. However, magnetic fractionation was useful to separate clusters and stabilize NPs which have a specific protein fingerprint. Overall, these results indicate that administration of a single population of NPs (protein-NP complexes) may be helpful to increase the availability in bloodstream. A drug delivery system with an entrapped cargo with transferrin corona spontaneously bound to a transferrin receptor (TfR) and displayed a superior cellular uptake and delivered payload to the cytosol [67]. Mulik et al. [68] demonstrated a significant increase in the uptake of curcumin NPs covered with transferrin protein corona in MCF-7 breast cancer cells. This condition greatly promoted curcumin activity by inducing increased loss of mitochondrial membrane potential and generation of excessive reactive oxygen species leading to superior apoptotic characteristics. Another formulation, namely superparamagnetic iron oxide NPs with transferrin composition, showed efficient targeting of curcumin against K562 myeloid leukemia cancer cells [69]. Further, curcumin loaded fibrinogen nanocarriers showed a preferential internalization and stimulated the apoptosis pathway in MCF-7 cancer cells compared to L929 fibroblast cells [70]. Similarly, a number of curcumin NPs decorated with plasma proteins have been documented as possessing improved therapeutic potential in cancer cells [10].
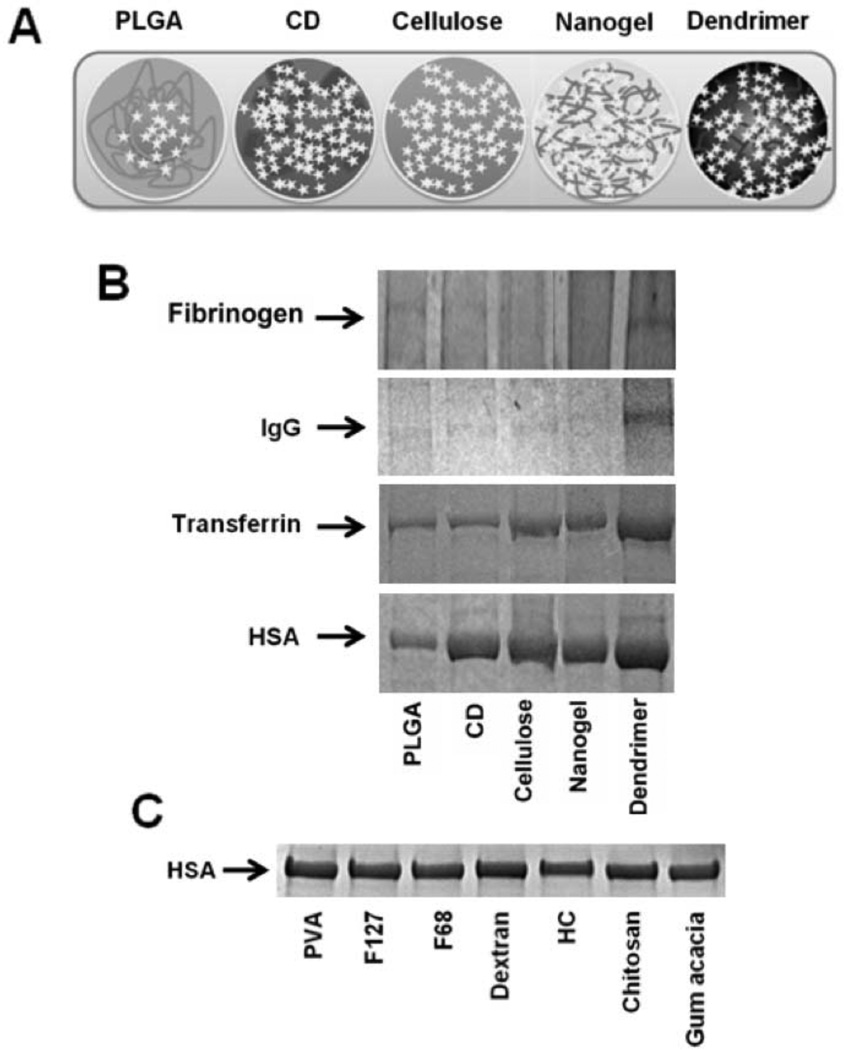
One of our studies has delineated the efficient internalization of PLGA-CUR NPs in cancer cells [71]. The serum corona drives curcumin NPs in a time and cell type dependant internalization. This implies that internalized NPs may release curcumin in its native active form for therapeutic purpose. Additionally, the NPs designed by PLGA with a polyvinyl alcohol stabilizer have the capability to escape from late endosomes and deliver a larger amount of drug to the cytosol [72]. Among five curcumin nanoformulations (Fig. 4A) tested for HSA protein binding, the PLGA based nanoformulation has the lowest protein binding which suggests it is a good carrier for delivering curcumin to target site(s) over an extended period (Fig. 4B) [73]. Higher HSA protein binding was noticed with dendrimer curcumin formulations which can lead to aggregation of the formulations over time and probably elimination from the body. However, the adsorption of different plasma proteins onto curcumin NPs varies, and in all types of formulations fibrinogen and immunoglobulin G adsorption was found to be very low. Overall, the degree of protein adsorption to curcumin NPs is as followed: HSA > transferrin > immunoglobulin G > fibrinogen. Furthermore, our un-published data on HSA binding on PLGA NPs stabilized by PVA in combination with other polymers, F127, F68, dextran, hydroxyl cellulose, chitosan, and gum acacia, did not show significant differences (Fig. 4C).
Fig. (4).
Influence of plasma proteins interaction with curcumin nanoformulations. (A) Structurally varied curcumin nanoformulations generated by different methods. Curcumin nanoformulations (20 µM) were incubated in 100 µg human plasma proteins and after 2 hours of incubation, adsorbed proteins were separated by ultra centrifugation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis was run at 150 V for 60 minutes and resultant stained with Coomassie® G-250 stain. (B) Fibrinogen, immunoglobulin G (IgG), transferrin, and serum albumin bound to curcumin nanoformulations. (C) HSA associated with PLGA nanoformulations which are prepared in the presence of various stabilizers. CD: β-cyclodextrin; CUR: curcumin; PLGA: poly(lactide-co-glycolide); Nanogel: poly(N-isopropyl acrylamide); Dendrimer: polyaminoamide (4 generation) based curcumin NPs. F127 and F68 are Pluronic polymers and HC indicates hydroxy cellulose. Figure 3A–B has been reprinted with permission from Dove Medical Press (Copyright Clearance Center, Order License No: 3059490805921) See Ref. 73.
Jedlovszky-Hajdú et al. [74] investigated the influence of composition and structure of magnetic NP interactions with human plasma. The citric acid (CA), poly(acrylic acid) (PAA), and oleic acid double layer (OAOA) coated magnetic NPs have shown increased NP size in soft (10% serum) and hard corona (55% serum) determined by Dynamic Light Scattering (DLS), NP Tracking Analysis (NTA), and Differential Centrifugal Sedimentation (DCS) methods [74]. More importantly, OAOA and CA have clearly shown a variation in protein coronas associated with lipid carrier proteins, complement factors and immunoglobulin after exposure with 10% and 55% plasma [74]. From this it can be considered that oleic acid layered magnetic NPs have enriched lipoproteins and albumin while complement and immunoglobulin proteins likely escape the immune system and RES. With this in mind, we have generated oleic acid/cyclodextrin layers with Pluronic polymer stabilizers for delivery of curcumin/doxorubicin [75, 76].
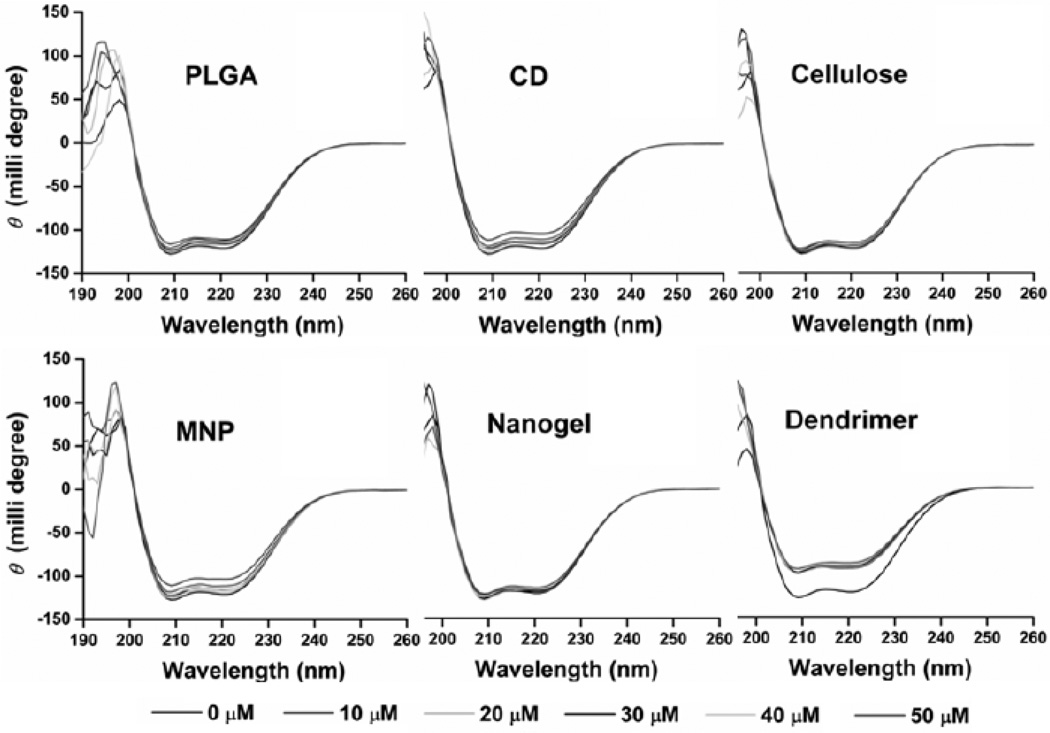
It was demonstrated that the structure of proteins was significantly altered upon incubation with polystyrene NPs [77]. No charge and negatively charged NPs encouraged formation of the helical structure in apoAI or HDL (negatively charged) while positively charged NPs decreased the amount of helical structure. Similarly, the secondary structure is affected by NPs carrying an opposite surface charge relative to the protein. This indicates that surface charge of NPs and proteins are critical parameters for predicting protein adsorption and corona formation. In our comparative plasma protein binding, the dendrimer curcumin nanoformulation significantly alters secondary structures while other formulations did not have considerable influence (Fig. 5). This severe binding potential is achieved because of the positive nature of dendrimer curcumin formulations.
Fig. (5).
Curcumin NPs and serum albumin complex formation in PBS solution. Serum albumin (12.5 µM) was incubated with curcumin nanoformulations (0–50 µM) and the circular dichroism spectra were obtained at room temperature. CD: β-cyclodextrin; CUR: curcumin; PLGA: poly(lactide-co-glycolide); MNP: magnetic NPs; Nanogel: poly(N-isopropyl acrylamide); Dendrimer: polyaminoamide (4 generation) based curcumin NPs. Note: Large variation in circular dichroism spectra of dendrimer curcumin formulation indicates significant structural changes occurred in serum albumin.
5. FACTORS INFLUENCE CURCUMIN NANOPARTICLEPLASMA PROTEIN CORONA
Design and engineering of the NP–protein interface itself is a broad field in drug delivery. In our opinion, protein corona formation on curcumin NPs is dependent on the physicochemical properties of NPs (Fig. 6). A recent review [78] presents that the protein corona formation on nanoparticles depends on (1) the characteristics of the nanomaterial, such as nanoparticle size, shape, and composition, (2) the physiological environment, such as blood, interstitial fluid, cell cytoplasm, etc., and (3) the physiological response, including kind of transport, signaling mechanism, pharmacokinetic, accumulation process, and toxicity. This review also discussed in detail approximately 125 plasma proteins that have higher association with various nanoparticles.
Fig. (6).
Schematic representation of the influence of various structural, chemical and biological parameters on protein plasma corona formation on curcumin NPs.
The bound plasma protein corona on curcumin NPs is strictly a highly complex phenomenon, as such, the corona contained bound protein cannot be presumed based on profusion in the plasma and particle size. Commonly, PEGylated NPs protect from over plasma binding and avoid clearance by the RES mechanism. Kodiyan et al. [79] successfully designed NPs coated with cysteine-functionalized alginate based polymers which exist up to 4 weeks in circulation of peripheral blood in mice. Such plasma protein repellent stabilizers can be employed as an alternative to overcome the existence of limitations of PEG coating or PEGylation in therapeutic applications. A recent report of NP–histodine complexes demonstrated that the orientation, geometry and stoichiometry and its specificity can be regulated by the size and binding sites on the NP’s surface [80].
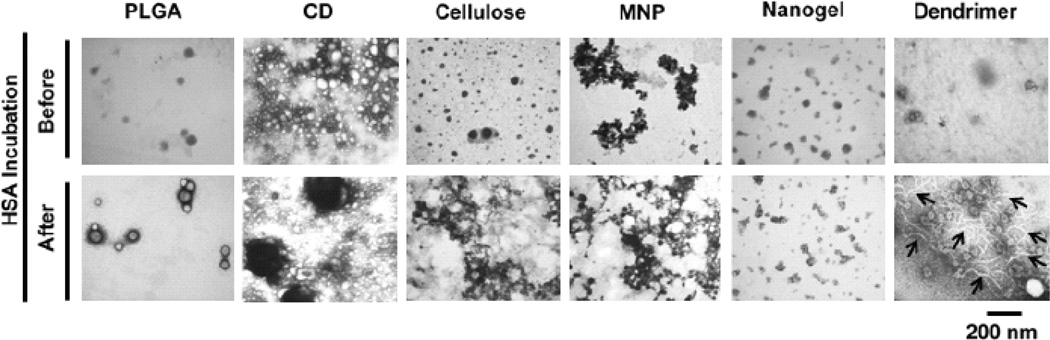
In some instances, plasma proteins adsorbed on the surfaces of NPs enhance the interaction with partially unfolded proteins that are directed toward cluster formation [81, 82]. These radical transformations favor extensive development of fibrils that promote pathological conditions ranging from neurodegenerative disorders to systemic amyloidoses [83]. Our studies have demonstrated that curcumin nanoformulations made with PLGA NPs, cyclodextrin assembly, cellulose NPs, magnetic NPs, and nanogel do not show any signs of fibril formation in a transmission electron microscopy evaluation [73] (Fig. 7). However, the dendrimer curcumin formulation exhibited linear filament formation but not fibrils. In addition, chemical surface modification of NPs with small molecules that can target proteins of the associated corona in biological milieu can stimulate the protein misfolding phenomenon [84]. This misfolding phenomenon allows access for cell specific internalization.
Fig. (7).
Human serum interaction leads to change in the morphology of curcumin nanoformulations. Curcumin nanoformulations (1 mg/mL) were incubated with and without human serum albumin (100 µg) for 2 hrs and after centrifugation and washing with water, curcumin NPs were viewed under a transmission electron microscopy. CD: β-cyclodextrin; CUR: curcumin; PLGA, poly(lactide-co-glycolide); MNP: magnetic NP; Nanogel: poly(N-isopropyl acrylamide); Dendrimer: polyaminoamide (4 generation) based curcumin NPs. Filament formation was observed only in dendrimer curcumin formulation (bar arrows represent filaments). Reprinted with permission from Dove Medical Press (Copyright Clearance Center, Order License No: 3059490805921) See Ref. 73.
PLGA NPs have exhibited the adsorption of three major plasma proteins: fibrinogen > albumin > globulin (γ-globulin) [85]. Thiol modified PLGA NPs (57.16 ± 6 8.5 µg –SH groups/100 mg of particles) resulted in maximum adsorption reduction with globulin (up to 20% reduction) while only 10% reduced adsorption was noticed with albumin and fibrinogen. PLGA-CUR NPs with smaller sizes had a significantly higher uptake (2–6 fold) in A2780CP and MDA-MD-231 cancer cells which is responsible for the superior anticancer potential [71]. A recent study suggested that proteins can adopt different orientations depending on the size of the NPs [86]. Different orientations of adsorbed proteins affect the level of biological activity of the formulation. Small particle size formulations were found not to be precipitated or bound to protein A, which illustrates the principle that the size of NPs can strongly influence the binding activity of adsorbed proteins. An extensive Pathway and Network Analysis Software of Genespring GX and the Ingenuity Study revealed that not only does the particle size of formulations determine its protein binding capacity but also its various functional components involved in the biological system such as immune response, lipid and cholesterol metabolism, activation, coagulation and acute response [34].
We have shown in our recent publication that surface chemistry plays a significant role in the formation of protein corona on NPs [41]. These structural variations lead to different uptake profiles in MDA-MB-231, HPAF-II and SKBR-3 cancer cells. Wiogo et al. [87]. validated that surface chemistry assisted protein corona formation. Branched polyethylene glycol coated magnetic NPs binds ~ 95% serum protein while polymethacrylic acid coated magnetic NPs binds ~ 10% serum protein. Linear polyethylene glycol coating also helps to reduce serum binding to 40% compared to un-coated NPs. The higher serum bound protein corona on the magnetic NPs can also cause a reduction to their MRI characteristics. Additionally, these inherent properties are also dependent on the type of corona formed on NPs. Casals et al. [42] examined the hard and soft corona density in terms of thickness by the dynamic light scattering method. They concluded that soft corona has more thickness than hard corona because soft corona is achieved by prolonged incubation times. They predicted that particles protein corona densities were correlated with particle size.
Hemolysis is a measure of an acute toxicity assay that indicates the extent of damage to red blood cells via release of hemoglobin. This assay is widely used to evaluate the hemo-compatibility of the nanoformulation. To assess the curcumin nanoformulations for human use, the formulations must be tested for hemolytic compatibility. Curcumin nanoformulations demonstrated almost no hemolysis existed at the concentrations tested compared to the positive controls (up to 1 mg/mL) mL) [70, 76, 88]. The noted hemolytic ratio of curcumin formulations was < 3–4% (below 5%) which is considered a critical safe ratio for nanomaterials or biomaterials according to ASTM F756 and ISO/TR 7406. This suggests that the risk of hemolysis by curcumin nanoformulations is completely absent. Blood-NP interactions by TEM and SEM studies revealed that NPs can interact with platelets, red blood cells (RBCs), and white blood cells (WBCs) [89]. A study showed that gold NPs not only attached to these cell surfaces but were engulfed in small channels in the cell membranes regardless of their surface properties [89]. However, RBC membrane damage may lead to increased spleen uptake due to the damaged RBCs recycling process. Therefore, design of curcumin NPs that are highly associated with RBCs without damage would be highly suitable for cancer therapeutics. PLGA NPs are widely employed for curcumin delivery. PLGA NPs have shown a lower aggregation tendency in blood cell fractions and are almost equivalent to the control RBCs in terms of membrane integrity [85].
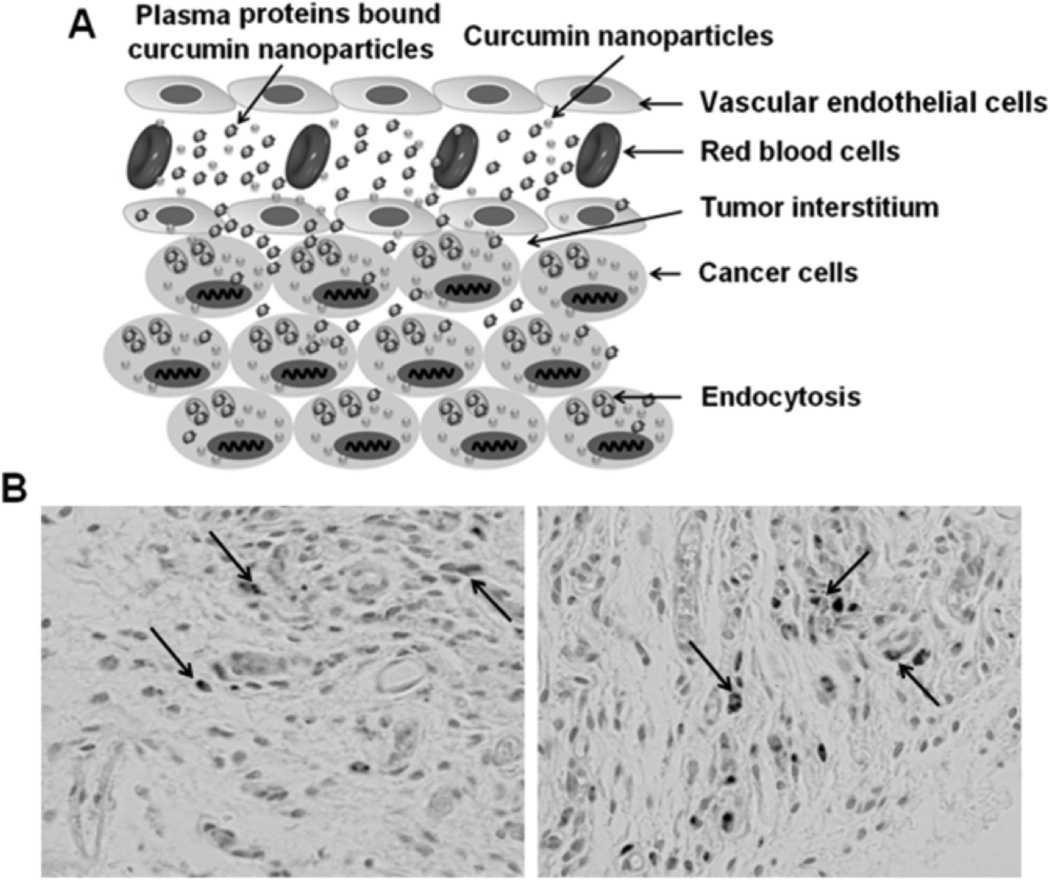
Targeted motif mediated delivery of curcumin NPs is reportedly limited at this time and their plasma protein binding is unknown. However, a combination of inert fresh anti-fouling surfactants and bioactive moieties (via ligand, aptamer, penetrating peptide or protein, and antibody mediation) of particular delivery mechanisms will increase cancer therapeutic potential [89]. Our preliminary data on curcumin loaded magnetic particles showed improved tumor uptake that could be due to optimum plasma protein corona formation in the HPAF-II xenograft mouse model (Fig. 8). The targeting can be further improved by antibody or aptamer conjugation to NPs for treatment of pancreatic cancer.
Fig. (8).
Curcumin loaded magnetic nanoparticle (MNP-CUR) formulation targets tumor cells in HPAF-II xenograft mouse model. (A) Schematic representation of possible targeting behavior of curcumin NPs to tumor cells. (B) MNP-CUR internalization in tumor cells in vivo. MNP-CUR (20 µg per mice) was administered via intraperitoneal injection and internalization in tumor cells was detected with Prussian blue staining. Black arrows indicate MNP-CUR NPs internalization in tumor cells.
6. FUTURE CONSIDERATIONS FOR CANCER THERAPEUTICS
Understanding interactions of curcumin NPs and plasma proteins in the pharmaceutical fields of research is important. Structural, functionality, configuration and morphological alterations occur during the formation process of plasma protein coronas which involves a number of biological phenomena that determines the in vivo fate of curcumin NPs. Some of these interactions can protect the integrity of curcumin NPs and their ability to reach the intended target organs. However, to determine such phenomena by any one specific method is difficult because NPs are made with different chemical substances and surfactants. The secondary interactions may be dominant in some cases. Studying complex curcumin NP and plasma protein biological interactions should account for: (i) physicochemical properties of curcumin NPs, (ii) thermodynamic feasibilities (enthalpy and entropy driven process where ΔH < 0, ΔS > 0 and Gibbs free energy ΔG < 0), and (iii) electrostatic interactions [43]. Further, it is important to develop unique rational methods to evaluate NP and protein bio-interfaces. These studies may become landmark modules to evaluate curcumin NP-plasma protein interactions which can be extrapolated for their in vivo utility and for cancer therapeutics. Investigation of complement system recognition protein (C1q, ficolins, mannose binding lactin) interactions with nanocurcumin particles will determine its opsonise characteristics. This information will enhance the specificity towards tumors, provided the NP avoids interactions with complement system recognition proteins.
There are 200 types of cells with distinguished membranes responsible for various cellular uptake. Plasma protein corona on NPs recognized by the cell may depend on the extent of internalization by the “sees-cell observer” mechanism [90]. In this way, uptake of encapsulated drugs in NPs is robustly connected to their transport through the cellular membrane. However, no known specific incubation time of NPs and plasma proteins have been reported for predicting such internalization in cancer cells.
Both legislative policies and the pharmaceutical industry need to encourage academic and translational research that is focused on the NP mediated delivery of curcumin. These policies can expedite novel findings related to the use of curcumin nanoformulations for cancer therapeutics. Further, these policies can also help to increase the utilization of various polyphenols, such as resveratrol, quercetin, epigallocatechin-3-gallate, and genistein. The McNeil group [91] proposed an overview of the number of in vitro assays in addition to protein binding NPs that needed to be tested for NP compatibility with the immune system, including hemolysis, platelet aggregation, plasma coagulation, complement activation, phagocytosis, leukocyte proliferation, nitric oxide production, and chemotaxis CFU-GM test. The next important consideration is to develop a curcumin nanoformulation utlizing existing and approved technology, such as Abraxane or Doxil, which follows good laboratory and manufacturing (cGLP and GMP) practices. This process will help to obtain the FDA’s approval for curcumin NPs as a New Drug Application (NDA), provided there is enough clinical and preclinical significance for the treatment of cancer(s).
CONCLUSION
Interaction of curcumin nanoformulations with serum plasma proteins can provide a new platform for the interpretation of NP-interfaces from the biological perspective and to estimate the formulation’s fate in vivo. The method, polymer, and surfactants used to prepare curcumin nanoformulations, as well as the particle size, aggregation phenomenon and surface charge of particles can influence the binding of plasma proteins. It is necessary to develop a standard analytical procedure and optimization for evaluating nanoformulation and plasma protein binding. The discussed curcumin nanoformulations and plasma protein binding and interaction studies are foundational to establish a rational basis for developing improved curcumin nanoformulations for cancer therapeutics.
ACKNOWLEDGEMENTS
The authors thank Cathy Christopherson and Jenna Klepatz (Sanford Research/University of South Dakota) for editorial assistance. This work was partially supported by grants from Governor’s Cancer 2010, the National Institutes of Health Research Project Grant Program (RO1) (CA142736), and the Centers of Biomedical Research Excellence (P20 GM103548–02 and a pilot grant through P20 GM103548-02).
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J. Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period. J. Nat. Prod. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 3.Bar-Sela G, Epelbaum R, Schaffer M. Curcumin as an anticancer agent: review of the gap between basic and clinical applications. Curr. Med. Chem. 2010;17:190–197. doi: 10.2174/092986710790149738. [DOI] [PubMed] [Google Scholar]
- 4.Marathe SA, Dasgupta I, Gnanadhas DP, Chakravortty D. Multifaceted roles of curcumin: two sides of a coin! Expert Opin. Biol. Ther. 2011;11:1485–1499. doi: 10.1517/14712598.2011.623124. [DOI] [PubMed] [Google Scholar]
- 5.Basnet P, Skalko-Basnet N. Curcumin: an anti-inflammatory molecule from a curry spice on the path to cancer treatment. Molecules. 2011;16:4567–4598. doi: 10.3390/molecules16064567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta SC, Prasad S, Kim JH, Patchva S, Webb LJ, Priyadarsini IK, Aggarwal BB. Multitargeting by curcumin as revealed by molecular interaction studies. Nat. Prod. Rep. 2011;28:1937–1955. doi: 10.1039/c1np00051a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garg AK, Buchholz TA, Aggarwal BB. Chemosensitization and radiosensitization of tumors by plant polyphenols. Antioxid. Redox. Signal. 2005;7:1630–1647. doi: 10.1089/ars.2005.7.1630. [DOI] [PubMed] [Google Scholar]
- 8.Shehzad A, Wahid F, Lee YS. Curcumin in cancer chemoprevention: molecular targets, pharmacokinetics, bioavailability, and clinical trials. Arch. Pharm. 2010;343:489–499. doi: 10.1002/ardp.200900319. [DOI] [PubMed] [Google Scholar]
- 9.Fan X, Zhang C, Liu DB, Yan J, Liang HP. The Clinical Applications of Curcumin: Current State and the Future. Curr. Pharm. Des. 2013;19:2011–2031. [PubMed] [Google Scholar]
- 10.Yallapu MM, Jaggi M, Chauhan SC. Curcumin nanoformulations: a future nanomedicine for cancer. Drug Discov. Today. 2012;17:71–80. doi: 10.1016/j.drudis.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravindranath V, Chandrasekhara N. Metabolism of curcumin-- studies with [3H]curcumin. Toxicology. 1981;22:337–344. doi: 10.1016/0300-483x(81)90027-5. [DOI] [PubMed] [Google Scholar]
- 12.Pulla Reddy AC, Sudharshan E, Appu Rao AG, Lokesh BR. Interaction of curcumin with human serum albumin-a spectroscopic study. Lipids. 1999;34:1025–1029. doi: 10.1007/s11745-999-0453-x. [DOI] [PubMed] [Google Scholar]
- 13.Leung MH, Kee TW. Effective stabilization of curcumin by association to plasma proteins: human serum albumin and fibrinogen. Langmuir. 2009;25:5773–5777. doi: 10.1021/la804215v. [DOI] [PubMed] [Google Scholar]
- 14.Wang YJ, Pan MH, Cheng AL, Lin LI, Ho YS, Hsieh CY, Lin JK. Stability of curcumin in buffer solutions and characterization of its degradation products. J. Pharm. Biomed. Anal. 1997;15:1867–1876. doi: 10.1016/s0731-7085(96)02024-9. [DOI] [PubMed] [Google Scholar]
- 15.Barik A, Mishra B, Kunwar A, Priyadarsini KI. Interaction of curcumin with human serum albumin: Thermodynamic properties, Fluorescence energy transfer and Denaturation effects. Chem. Phys. Lett. 2007;436:239–243. [Google Scholar]
- 16.Patra D, Barakat C, Tafech RM. Study on effect of lipophilic curcumin on sub-domain IIA site of human serum albumin during unfolded and refolded states: a synchronous fluorescence spectroscopic study. Colloids Surf. B: Biointerfaces. 2012;94:354–361. doi: 10.1016/j.colsurfb.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 17.Mohammadi F, Bordbar AK, Divsalar A, Mohammadi K, Saboury AA. Analysis of binding interaction of curcumin and diacetylcurcumin with human and bovine serum albumin using fluorescence and circular dichroism spectroscopy. Protein. J. 2009;28:189–196. doi: 10.1007/s10930-009-9184-1. [DOI] [PubMed] [Google Scholar]
- 18.Kratz F. Albumin as a drug carrier: design of prodrugs, drug conjugates and nanoparticles. J. Control. Release. 2008;132:171–183. doi: 10.1016/j.jconrel.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 19.Elsadek B, Kratz F. Impact of albumin on drug delivery--new applications on the horizon. J. Control. Release. 2012;157:4–28. doi: 10.1016/j.jconrel.2011.09.069. [DOI] [PubMed] [Google Scholar]
- 20.Xiao JB, Kai GY. A review of dietary polyphenol-plasma protein interactions: Characterization, influence on the bioactivity, and structure-affinity relationship. Crit. Rev. Food Sci. Nutr. 2012;52:85–101. doi: 10.1080/10408398.2010.499017. [DOI] [PubMed] [Google Scholar]
- 21.De Jong WH, Borm PJ. Drug delivery and nanoparticles: applications and hazards. Int.J. Nanomedicine. 2008;3:133–149. doi: 10.2147/ijn.s596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch I, Cedervall T, Lundqvist M, Cabaleiro-Lago C, Linse S, Dawson KA. The nanoparticle-protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century. Adv. Colloid. Interface. Sci. 2007;134–135:167–174. doi: 10.1016/j.cis.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Chen S, Lv L, Song L, Guo S. Recent Progresses in Studying Curcumin and Its Nano-preparations for Cancer Therapy. Curr. Pharm. Des. 2013;19:1974–1993. [PubMed] [Google Scholar]
- 24.Ji JL, Huang XF, Zhu HL. Curcumin and its formulations: potential anti-cancer agents. Anticancer Agents Med. Chem. 2012;12:210–218. doi: 10.2174/187152012800228733. [DOI] [PubMed] [Google Scholar]
- 25.Albanese A, Tang PS, Chan WC. The effect of nanoparticle size, shape, and surface chemistry on biological systems. Annu. Rev. Biomed. Eng. 2012;14:1–16. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 26.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Understanding the nanoparticle- protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. USA. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treuel L, Nienhaus GU. Toward a molecular understanding of nanoparticle–protein interactions. Biophys. Rev. 2012;4:137–147. doi: 10.1007/s12551-012-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundqvist M, Stigler J, Cedervall T, Berggard T, Flanagan MB, Lynch I, Elia G, Dawson K. The evolution of the protein corona around nanoparticles: a test study. ACS Nano. 2011;5:7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- 29.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. U S A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dell'Orco D, Lundqvist M, Oslakovic C, Cedervall T, Linse S. Modeling the time evolution of the nanoparticle-protein corona in a body fluid. PLoS One. 2010;5:e10949. doi: 10.1371/journal.pone.0010949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capriotti AL, Caracciolo G, Cavaliere C, Foglia P, Pozzi D, Samperi R, Lagana A. Do plasma proteins distinguish between liposomes of varying charge density? J. Proteomics. 2012;75:1924–1932. doi: 10.1016/j.jprot.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Burnum KE, Luna ML, Petritis BO, Kim JS, Qian WJ, Moore RJ, Heredia-Langner A, Webb-Robertson BJ, Thrall BD, Camp DG, 2nd, Smith RD, Pounds JG, Liu T. Quantitative proteomics analysis of adsorbed plasma proteins classifies nanoparticles with different surface properties and size. Proteomics. 2011;11:4569–4577. doi: 10.1002/pmic.201100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simberg D, Park JH, Karmali PP, Zhang WM, Merkulov S, McCrae K, Bhatia SN, Sailor M, Ruoslahti E. Differential proteomics analysis of the surface heterogeneity of dextran iron oxide nanoparticles and the implications for their in vivo clearance. Biomaterials. 2009;30:3926–3933. doi: 10.1016/j.biomaterials.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tenzer S, Docter D, Rosfa S, Wlodarski A, Kuharev J, Rekik A, Knauer SK, Bantz C, Nawroth T, Bier C, Sirirattanapan J, Mann W, Treuel L, Zellner R, Maskos M, Schild H, Stauber RH. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS Nano. 2011;5:7155–7167. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- 35.Arvizo RR, Giri K, Moyano D, Miranda OR, Madden B, McCormick DJ, Bhattacharya R, Rotello VM, Kocher JP, Mukherjee P. Identifying new therapeutic targets via modulation of protein corona formation by engineered nanoparticles. PLoS One. 2012;7:e33650. doi: 10.1371/journal.pone.0033650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desai N, Trieu V, Damascelli B, Soon-Shiong P. SPARC Expression Correlates with Tumor Response to Albumin-Bound Paclitaxel in Head and Neck Cancer Patients. Transl. Oncol. 2009;2:59–64. doi: 10.1593/tlo.09109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desai NP, Trieu V, Hwang LY, Wu R, Soon-Shiong P, Gradishar WJ. Improved effectiveness of nanoparticle albuminbound (nab) paclitaxel versus polysorbate-based docetaxel in multiple xenografts as a function of HER2 and SPARC status. Anticancer Drugs. 2008;19:899–909. doi: 10.1097/CAD.0b013e32830f9046. [DOI] [PubMed] [Google Scholar]
- 38.Desai N. Drug Delivery Report. Oxford OX4 4GA, UK: PharmaVentures Ltd; 2007. nab-Paclitaxel (Abraxane®): an albumin-bound cytotoxic exploiting natural delivery mechanisms into tumors; pp. 37–41. [Google Scholar]
- 39.Dell'orco D, Lundqvist M, Cedervall T, Linse S. Delivery success rate of engineered nanoparticles in the presence of the protein corona: a systems-level screening. Nanomedicine. 2012;8:1271–1281. doi: 10.1016/j.nano.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Xie X, Tao Q, Zou Y, Zhang F, Guo M, Wang Y, Wang H, Zhou Q, Yu S. PLGA nanoparticles improve the oral bioavailability of curcumin in rats: characterizations and mechanisms. J. Agric. Food Chem. 2011;59:9280–9289. doi: 10.1021/jf202135j. [DOI] [PubMed] [Google Scholar]
- 41.Aleksenko SS, Shmykov AY, Oszwaldowski S, Timerbaev AR. Interactions of tumour-targeting nanoparticles with proteins: potential of using capillary electrophoresis as a direct probe. Metallomics. 2012;4:1141–1148. doi: 10.1039/c2mt20141k. [DOI] [PubMed] [Google Scholar]
- 42.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Time evolution of the nanoparticle protein corona. ACS Nano. 2010;4:3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- 43.Paul BK, Bhattacharjee K, Bose S, Guchhait N. A spectroscopic investigation on the interaction of a magnetic ferrofluid with a model plasma protein: effect on the conformation and activity of the protein. Phys. Chem. Chem. Phys. 2012;14:15482–15493. doi: 10.1039/c2cp42415k. [DOI] [PubMed] [Google Scholar]
- 44.Gebauer JS, Malissek M, Simon S, Knauer SK, Maskos M, Stauber RH, Peukert W, Treuel L. Impact of the nanoparticle-protein corona on colloidal stability and protein structure. Langmuir. 2012;28:9673–9679. doi: 10.1021/la301104a. [DOI] [PubMed] [Google Scholar]
- 45.Schaefer J, Schulze C, Marxer EE, Schaefer UF, Wohlleben W, Bakowsky U, Lehr CM. Atomic force microscopy and analytical ultracentrifugation for probing nanomaterial protein interactions. ACS Nano. 2012;6:4603–4614. doi: 10.1021/nn202657q. [DOI] [PubMed] [Google Scholar]
- 46.Lindman S, Lynch I, Thulin E, Nilsson H, Dawson KA, Linse S. Systematic investigation of the thermodynamics of HSA adsorption to N-iso-propylacrylamide/N-tert-butylacrylamide copolymer nanoparticles. Effects of particle size and hydrophobicity. Nano Lett. 2007;7:914–920. doi: 10.1021/nl062743+. [DOI] [PubMed] [Google Scholar]
- 47.Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv. Drug. Deliv. Rev. 2009;61:428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahon E, Salvati A, Baldelli Bombelli F, Lynch I, Dawson KA. Designing the nanoparticle-biomolecule interface for"targeting and therapeutic delivery". J. Control. Release. 2012;161:164–174. doi: 10.1016/j.jconrel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 49.Shaikh J, Ankola DD, Beniwal V, Singh D, Kumar MN. Nanoparticle encapsulation improves oral bioavailability of curcumin by at least 9-fold when compared to curcumin administered with piperine as absorption enhancer. Eur. J. Pharm. Sci. 2009;37:223–230. doi: 10.1016/j.ejps.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 50.Anand P, Nair HB, Sung B, Kunnumakkara AB, Yadav VR, Tekmal RR, Aggarwal BB. Design of curcumin-loaded PLGA nanoparticles formulation with enhanced cellular uptake, and increased bioactivity in vitro and superior bioavailability in vivo. Biochem. Pharmacol. 2010;79:330–338. doi: 10.1016/j.bcp.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Onoue S, Takahashi H, Kawabata Y, Seto Y, Hatanaka J, Timmermann B, Yamada S. Formulation design and photochemical studies on nanocrystal solid dispersion of curcumin with improved oral bioavailability. J. Pharm. Sci. 2010;99:1871–1881. doi: 10.1002/jps.21964. [DOI] [PubMed] [Google Scholar]
- 52.Gao Y, Li Z, Sun M, Li H, Guo C, Cui J, Li A, Cao F, Xi Y, Lou H, Zhai G. Preparation, characterization, pharmacokinetics, and tissue distribution of curcumin nanosuspension with TPGS as stabilizer. Drug. Dev. Ind. Pharm. 2010;36:1225–1234. doi: 10.3109/03639041003695139. [DOI] [PubMed] [Google Scholar]
- 53.Mohanty C, Sahoo SK. The in vitro stability and in vitro pharmacokinetics of curcumin prepared as an aqueous nanoparticulate formulation. Biomaterials. 2010;31:6597–6611. doi: 10.1016/j.biomaterials.2010.04.062. [DOI] [PubMed] [Google Scholar]
- 54.Bisht S, Mizuma M, Feldmann G, Ottenhof NA, Hong SM, Pramanik D, Chenna V, Karikari C, Sharma R, Goggins MG, Rudek MA, Ravi R, Maitra A. Systemic administration of polymeric nanoparticle-encapsulated curcumin (NanoCurc) blocks tumor growth and metastases in preclinical models of pancreatic cancer. Mol. Cancer. Ther. 2010;9:2255–2264. doi: 10.1158/1535-7163.MCT-10-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kakkar V, Singh S, Singla D, Kaur IP. Exploring solid lipid nanoparticles to enhance the oral bioavailability of curcumin. Mol. Nutr. Food. Res. 2011;55:495–503. doi: 10.1002/mnfr.201000310. [DOI] [PubMed] [Google Scholar]
- 56.Yu H, Huang Q. Improving the oral bioavailability of curcumin using novel organogel-based nanoemulsions. J. Agric. Food. Chem. 2012;60:5373–5379. doi: 10.1021/jf300609p. [DOI] [PubMed] [Google Scholar]
- 57.Tsai YM, Chang-Liao WL, Chien CF, Lin LC, Tsai TH. Effects of polymer molecular weight on relative oral bioavailability of curcumin. Int. J. Nanomedicine. 2012;7:2957–2966. doi: 10.2147/IJN.S32630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W, Zhu R, Xie Q, Li A, Xiao Y, Li K, Liu H, Cui D, Chen Y, Wang S. Enhanced bioavailability and efficiency of curcumin for the treatment of asthma by its formulation in solid lipid nanoparticles. Int. J. Nanomedicine. 2012;7:3667–3677. doi: 10.2147/IJN.S30428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feng R, Song Z, Zhai G. Preparation and in vivo pharmacokinetics of curcumin-loaded PCL-PEG-PCL triblock copolymeric nanoparticles. Int. J. Nanomedicine. 2012;7:4089–4098. doi: 10.2147/IJN.S33607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ranjan AP, Mukerjee A, Helson L, Vishwanatha JK. Scale up, optimization and stability analysis of Curcumin C3 complex-loaded nanoparticles for cancer therapy. J. Nanobiotechnology. 2012;10:38. doi: 10.1186/1477-3155-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khalil NM, Nascimento TC, Casa DM, Dalmolin LF, Mattos AC, Hoss I, Romano MA, Mainardes RM. Pharmacokinetics of curcumin-loaded PLGA and PLGA-PEG blend nanoparticles after oral administration in rats. Colloids Surf. B: Biointerfaces. 2013;101:353–360. doi: 10.1016/j.colsurfb.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 62.Li C, Zhang Y, Su T, Feng L, Long Y, Chen Z. Silica-coated flexible liposomes as a nanohybrid delivery system for enhanced oral bioavailability of curcumin. Int. J. Nanomedicine. 2012;7:5995–6002. doi: 10.2147/IJN.S38043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pawar YB, Purohit H, Valicherla GR, Munjal B, Lale SV, Patel SB, Bansal AK. Novel lipid based oral formulation of curcumin: development and optimization by design of experiments approach. Int. J. Pharm. 2012;436:617–623. doi: 10.1016/j.ijpharm.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 64.Jiang X, Weise S, Hafner M, Rocker C, Zhang F, Parak WJ, Nienhaus GU. Quantitative analysis of the protein corona on FePt nanoparticles formed by transferrin binding. J. R. Soc. Interface. 2010;7(1):S5–S13. doi: 10.1098/rsif.2009.0272.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maiorano G, Sabella S, Sorce B, Brunetti V, Malvindi MA, Cingolani R, Pompa PP. Effects of cell culture media on the dynamic formation of protein-nanoparticle complexes and influence on the cellular response. ACS Nano. 2010;4:7481–7491. doi: 10.1021/nn101557e. [DOI] [PubMed] [Google Scholar]
- 66.Lartigue L, Wilhelm C, Servais J, Factor C, Dencausse A, Bacri JC, Luciani N, Gazeau F. Nanomagnetic sensing of blood plasma protein interactions with iron oxide nanoparticles: impact on macrophage uptake. ACS Nano. 2012;6:2665–2678. doi: 10.1021/nn300060u. [DOI] [PubMed] [Google Scholar]
- 67.Dittrich C, Burckhardt CJ, Danuser G. Delivery of membrane impermeable cargo into CHO cells by peptide nanoparticles targeted by a protein corona. Biomaterials. 2012;33:2746–2753. doi: 10.1016/j.biomaterials.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mulik RS, Monkkonen J, Juvonen RO, Mahadik KR, Paradkar AR. Transferrin mediated solid lipid nanoparticles containing curcumin: enhanced in vitro anticancer activity by induction of apoptosis. Int. J. Pharm. 2010;398:190–203. doi: 10.1016/j.ijpharm.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 69.Dilnawaz F, Singh A, Sahoo SK. Transferrin-conjugated curcumin-loaded superparamagnetic iron oxide nanoparticles induce augmented cellular uptake and apoptosis in K562 cells. Acta Biomater. 2012;8:704–719. doi: 10.1016/j.actbio.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 70.Rejinold NS, Muthunarayanan M, Chennazhi KP, Nair SV, Jayakumar R. Curcumin loaded fibrinogen nanoparticles for cancer drug delivery. J. Biomed. Nanotechnol. 2011;7:521–534. doi: 10.1166/jbn.2011.1320. [DOI] [PubMed] [Google Scholar]
- 71.Yallapu MM, Gupta BK, Jaggi M, Chauhan SC. Fabrication of curcumin encapsulated PLGA nanoparticles for improved therapeutic effects in metastatic cancer cells. J. Colloid. Interface. Sci. 2010;351:19–29. doi: 10.1016/j.jcis.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 72.Panyam J, Zhou WZ, Prabha S, Sahoo SK, Labhasetwar V. Rapid endo-lysosomal escape of poly(DL-lactide-co-glycolide) nanoparticles: implications for drug and gene delivery. FASEB J. 2002;16:1217–1226. doi: 10.1096/fj.02-0088com. [DOI] [PubMed] [Google Scholar]
- 73.Yallapu MM, Ebeling MC, Chauhan N, Jaggi M, Chauhan SC. Interaction of curcumin nanoformulations with human plasma proteins and erythrocytes. Int. J. Nanomedicine. 2011;6:2779–2790. doi: 10.2147/IJN.S25534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jedlovszky -Hajdu A, Bombelli FB, Monopoli MP, Tombacz E, Dawson KA. Surface coatings shape the protein corona of SPIONs with relevance to their application in vivo. Langmuir. 2012;28:14983–14991. doi: 10.1021/la302446h. [DOI] [PubMed] [Google Scholar]
- 75.Yallapu MM, Othman SF, Curtis ET, Bauer NA, Chauhan N, Kumar D, Jaggi M, Chauhan SC. Curcumin-loaded magnetic nanoparticles for breast cancer therapeutics and imaging applications. Int. J. Nanomedicine. 2012;7:1761–1779. doi: 10.2147/IJN.S29290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yallapu MM, Othman SF, Curtis ET, Gupta BK, Jaggi M, Chauhan SC. Multi-functional magnetic nanoparticles for magnetic resonance imaging and cancer therapy. Biomaterials. 2011;32:1890–1905. doi: 10.1016/j.biomaterials.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cukalevski R, Lundqvist M, Oslakovic C, Dahlback B, Linse S, Cedervall T. Structural changes in apolipoproteins bound to nanoparticles. Langmuir. 2011;27:14360–14369. doi: 10.1021/la203290a. [DOI] [PubMed] [Google Scholar]
- 78.Walkey CD, Chan WC. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 2012;41(7):2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 79.Kodiyan A, Silva EA, Kim J, Aizenberg M, Mooney DJ. Surface modification with alginate-derived polymers for stable, protein-repellent, long-circulating gold nanoparticles. ACS Nano. 2012;6:4796–4805. doi: 10.1021/nn205073n. [DOI] [PubMed] [Google Scholar]
- 80.Hu M, Qian L, Brinas RP, Lymar ES, Hainfeld JF. Assembly of nanoparticle-protein binding complexes: from monomers to ordered arrays. Angew. Chem. Int. Ed. Engl. 2007;46:5111–5114. doi: 10.1002/anie.200701180. [DOI] [PubMed] [Google Scholar]
- 81.Colvin VL, Kulinowski KM. Nanoparticles as catalysts for protein fibrillation. Proc. Natl. Acad. Sci. U S A. 2007;104:8679–8680. doi: 10.1073/pnas.0703194104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hirano A, Yoshikawa H, Matsushita S, Yamada Y, Shiraki K. Adsorption and disruption of lipid bilayers by nanoscale protein aggregates. Langmuir. 2012;28:3887–3895. doi: 10.1021/la204717c. [DOI] [PubMed] [Google Scholar]
- 83.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 84.Prapainop K, Witter DP, Wentworth P., Jr A chemical approach for cell-specific targeting of nanomaterials: small-moleculeinitiated misfolding of nanoparticle corona proteins. J. Am. Chem. Soc. 2012;134:4100–4103. doi: 10.1021/ja300537u. [DOI] [PubMed] [Google Scholar]
- 85.Thasneem YM, Sajeesh S, Sharma CP. Effect of thiol functionalization on the hemo-compatibility of PLGA nanoparticles. J. Biomed. Mater. Res A. 2011;99:607–617. doi: 10.1002/jbm.a.33220. [DOI] [PubMed] [Google Scholar]
- 86.Kaur K, Forrest JA. Influence of particle size on the binding activity of proteins adsorbed onto gold nanoparticles. Langmuir. 2012;28:2736–2744. doi: 10.1021/la203528u. [DOI] [PubMed] [Google Scholar]
- 87.Wiogo HT, Lim M, Bulmus V, Gutiérrez L, Woodward RC, Amal R. Insight into Serum Protein Interactions with Functionalized Magnetic Nanoparticles in Biological Media. Langmuir. 2012;28:4346–4356. doi: 10.1021/la204740t. [DOI] [PubMed] [Google Scholar]
- 88.Mangalathillam S, Rejinold NS, Nair A, Lakshmanan VK, Nair SV, Jayakumar R. Curcumin loaded chitin nanogels for skin cancer treatment via the transdermal route. Nanoscale. 2012;4:239–250. doi: 10.1039/c1nr11271f. [DOI] [PubMed] [Google Scholar]
- 89.Shah NB, Vercellotti GM, White JG, Fegan A, Wagner CR, Bischof JC. Blood-Nanoparticle Interactions and in vivo Biodistribution: Impact of Surface PEG and Ligand Properties. Mol. Pharm. 2012;9:2146–2155. doi: 10.1021/mp200626j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sohaebuddin SK, Thevenot PT, Baker D, Eaton JW, Tang L. Nanomaterial cytotoxicity is composition, size, and cell type dependent. Part. Fibre. Toxicol. 2010;7:22. doi: 10.1186/1743-8977-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dobrovolskaia MA, Aggarwal P, Hall JB, McNeil SE. Preclinical studies to understand nanoparticle interaction with the immune system and its potential effects on nanoparticle biodistribution. Mol. Pharm. 2008;5:487–495. doi: 10.1021/mp800032f. [DOI] [PMC free article] [PubMed] [Google Scholar]