Abstract
Here we review the rapidly growing toolbox of transgenic mice and rats that exhibit functional expression of engineered opsins for neuronal activation and silencing with light. Collectively, these transgenic animals are enabling neuroscientists to access and manipulate the many diverse cell types in the mammalian nervous system in order to probe synaptic and circuitry connectivity, function, and dysfunction. The availability of transgenic lines affords important advantages such as stable and heritable transgene expression patterns across experimental cohorts. As such, the use of transgenic lines precludes the need for other costly and labor-intensive procedures to achieve functional transgene expression in each individual experimental animal. This represents an important consideration when large cohorts of experimental animals are desirable as in many common behavioral assays. We describe the diverse strategies that have been implemented for developing transgenic mouse and rat lines and highlight recent advances that have led to dramatic improvements in achieving functional transgene expression of engineered opsins. Furthermore, we discuss considerations and caveats associated with implementing recently developed transgenic lines for optogenetics-based experimentation. Lastly, we propose strategies that can be implemented to develop and refine the next generation of genetically modified animals for behaviorally-focused optogenetics-based applications.
Keywords: optogenetics, channelrhodopsin, halorhodopsin, archaerhodopsin, transgenic mice, engineered opsin
1. Introduction
The past decade has yielded remarkable breakthroughs in methodologies that impart functional control of genetically targeted cell types using light, particularly with respect to applications involving the nervous system. As a result, this field of research called ‘optogenetics’ has rapidly evolved from a specialized technique into a mainstay of modern neuroscience. This progression is not surprising given the fact that the ability to manipulate neuronal activity with light greatly improves spatial and temporal precision as compared to classical methods such as physical lesion or chemical inactivation for analyzing brain function. In addition, optogenetic approaches enable unprecedented rapid reversibility, a design that is apt to produce especially compelling experimental evidence. Such refinements are well-suited to behavioral neuroscientists seeking to probe the causal relationship between defined brain circuits and complex behaviors. Indeed, an abundance of recent investigations have harnessed the power of optogenetics to glean invaluable insights into the neural underpinnings of innate behaviors such as sociability [1], aggression [2], anxiety [3], sleep and arousal [4, 5], habits [6], feeding and hunger [7–9], and conditioned learning and memory [10–15], as well as pathological behaviors relevant to reward-seeking and addiction [16–22], depression [23, 24], and motor dysfunction [25, 26]. These pioneering studies employing optogenetics methodologies demonstrate the feasibility and remarkable utility of using light to deconstruct complex behaviors circuit-by-circuit and cell type-by-cell type.
Optogenetics-based investigations in vivo require effective strategies for functional transgenic expression of the proteins that impart optogenetic control, which we refer to collectively as ‘engineered opsins’. Several strategies have been employed with success, including in utero electroporation, viral gene delivery, and manipulation of the mouse genome (e.g. developing transgenic lines). Notably, progress in the development of transgenic lines has lagged behind other expression strategies such as viral vector based approaches, which is to be expected given the relatively prolonged time frame required to design, establish and validate a novel transgenic line. Nonetheless, several new transgenic lines have recently been established for optogenetics-based research (Table 1). Here we described the diverse strategies for making transgenic mouse and rat lines that are suitable for optogenetic studies and highlight the recent innovations that have led to dramatic improvements in achieving functional transgenic expression of engineered opsins. Additional reviews of alternative opsin expression strategies can be found elsewhere [27–29].
Table 1.
Transgenic lines for optogenetic manipulation of mammalian nervous system function
| Line name | Species | Strategy | Opsin variant | Reporter | Cellular promoter | Reported opsin expression/induction | Line availability | Citation |
|---|---|---|---|---|---|---|---|---|
| Thy1.2-ChR2-EYFP line 9 | mouse | Plasmid transgenic | ChR2 | EYFP fusion | Thy1.2 | Layer V cortical pyramidal neurons,
hippocampal CA1 and CA3 pyramidal neurons, various nuclei of the thalamus, midbrain and brainstem, retinal ganglion cells |
JAX stock 007615 | [42.43] |
| Thy1.2-ChR2-EYFP line 18 | mouse | Plasmid transgenic | ChR2 | EYFP fusion | Thy1.2 | Layer V cortical pyramidal neurons,
hippocampal CA1 and CA3 pyramidal neurons, various nuclei of the thalamus, midbrain and brainstem, cerebellar mossy fibers, retinal ganglion cells |
JAX stock 007612 | [42.43] |
| Thy1.2-ChR2(H134R)-EYFP line 20 | mouse | Plasmid transgenic | ChR2(H134R) | EYFP fusion | Thy1.2 | Cortex, CA1, thalamus, superior colliculus, inferior colliculus, brainstem, amygdala and cerebellum | JAX stock 012350 | unpublished |
| WTChR2V4 | Wistar rat | Plasmid transgenic | ChR2 | Venus fusion | Thy1.2 | Retinal ganglion cell layer and inner
plexiform layer of the retina, mechanoreceptive dorsal root ganglion
neurons, hippocampal neurons. |
contact investigator | [57,58,59] |
| Thy1.2-VChR1-EYFP line 1 | mouse | Plasmid transgenic | VChR1 | EYFP fusion | Thy1.2 | Layer V cortical pyramidal neurons,
hippocampus, thalamus, superior colliculus, inferior
colliculus, brainstem, amygdala, and cerebellum. (Note: sparse labeling) |
JAX stock 012341 | unpublished |
| Thy1.2-VChR1-EYFP line 8 | mouse | Plasmid transgenic | VChR1 | EYFP fusion | Thy1.2 | Cortical layer II/III and V pyramidal neurons,
hippocampus, thalamus, superior colliculus, inferior
colliculus, brainstem, amygdala, and cerebellum. (Note: widespread labeling) |
JAX stock 012348 | unpublished |
| Omp-ChR2-EYFP (ORC-M) | mouse | Plasmid transgenic | ChR2 | EYFP fusion | Omp | Mature olfactory sensory neurons of the olfactory epithelium, vomeronasal organ and accessory olfactory bulb | contact investigator | [60] |
| Thy1.2-NpHR-EYFP line 6 | mouse | Plasmid transgenic | NpHR | EYFP fusion | Thy1.2 | Layer V cortical pyramidal neurons,
hippocampal CA1 and CA3 pyramidal neurons, dentate granule
cells, superior and inferior colliculus, thalamus and brain stem. (Note: sparse labeling and intracellular aggregation) |
no longer available | [88] |
| Thy1.2-eNpHR-EYFP line 2 | mouse | Plasmid transgenic | eNpHR2.0 | EYFP fusion | Thy1.2 | Layer V cortical pyramidal neurons,
hippocampal CA1 and CA3 pyramidal neurons, dentate granule
cells, subiculum, thalamus, superior and inferior colliculus, brainstem, amygdala, and cerebellum |
JAX stock 012332 | unpublished |
| Thy1.2-eNpHR-EYFP line 4 | mouse | Plasmid transgenic | eNpHR2.0 | EYFP fusion | Thy1.2 | Layer II/III and V cortical pyramidal neurons,
hippocampus (interneuron), subiculum, thalamus, superior and inferior colliculus, brainstem, amygdala, globus pallidus (interneuron), and cerebellum |
JAX stock 012334 | unpublished |
| Vglut2-ChR2(H134)-EYFP | mouse | BAC transgenic | ChR2(H134R) | EYFP fusion | Vglut2 | Glutamatergic neurons in the hindbrain and brainstem | JAX stock 017978 | [61] |
| VGAT-ChR2(H134R)-EYFP line 8 | mouse | BAC transgenic | ChR2(H134R) | EYFP fusion | VGAT | GABAergic neurons throughout the brain
(cortex, hippocampus, olfactory bulb, thalamic reticular
nucleus, superior and inferior colliculus, the molecular layer of the cerebellum, brain stem) |
JAX stock 014548 | [62] |
| TPH2-ChR2(H134R)-EYFP line 5 | mouse | BAC transgenic | ChR2(H134R) | EYFP fusion | TPH2 | Serotonergic neurons in the dorsal raphe nucleus, median raphe nucleus, and interpeduncular nucleus | JAX stock 014555 | [62] |
| ChAT-ChR2(H134R)-EYFP line 5 | mouse | BAC transgenic | ChR2(H134R) | EYFP fusion | ChAT | Cholinergic neurons in striatum (weak expression) | JAX stocl 014545 | [62] |
| ChAT-ChR2(H134R)-EYFP line 6 | mouse | BAC transgenic | ChR2(H134R) | EYFP fusion | ChAT | Cholinergic neurons in striatum, basal forebrain, facial nucleus, trochlear nucleus, medial habenula, cortex | JAX stock 014546 | [62,67] |
| Pvalb-ChR2(H134R)-EYFP line 15 | mouse | BAC transgenic | ChR2(H134R) | EYFP fusion | Pvalb | Thalamic reticular nucleus neurons, cerebellum Purkinje cells, basket and stellate molecular layer interneurons | JAX stock 012355 | [62] |
| Mrgprd-ChR2(H134R)-Venus | mouse | Knock-in | ChR2(H134R) | Venus fusion | Mrgprd locus | Non-peptidergic spinal cord sensory neuron subset | contact investigator | [70] |
| Omp-ChR2(H134R)-EYFP | mouse | Knock-in | ChR2(H134R) | EYFP fusion | Omp locus | Mature olfactory sensory neurons of the olfactory epithelium | JAX stock 014173 | [72] |
| R26-ChR2(H134R)-EGFP | mouse | Cre-inducible knock-in | ChR2(H134R) | EGFP fusion | CAG from Rosa26 locus | Cortical interneurons (Gad2-CreERT2) | contact investigator | [73] |
| Ai27 | mouse | Cre-inducible knock-in | ChR2(H134R) | tdTomato fusion | CAG from Rosa26 locus | Cortical and hippocampal pyramidal neurons (Emx1-Cre), cholinergic neurons (ChAT-Cre) | JAX stock 012567 | [75] |
| Ai32 | mouse | Cre-inducible knock-in | ChR2(H134R) | EYFP fusion | CAG from Rosa26 locus | Cortical and hippocampal pyramidal neurons
(Emx1-Cre), cholinergic neurons (ChAT-Cre), hippocampal CA1 basket cells, thalamic reticular nucleus neurons (Pvalb-IRES-Cre) |
JAX stock 012569 | [75] |
| Ai35 | mouse | Cre-inducible knock-in | Arch | EGFP fusion | CAG from Rosa26 locus | Cortical and hippocampal pyramidal neurons (Emx1-Cre), cortical pyramidal neurons (CamKIIa-CreERT2) | JAX stock 012735 | [75] |
| Ai39 | mouse | Cre-inducible knock-in | eNpHR3.0 | EYFP fusion | CAG from Rosa26 locus | Cortical pyramidal neurons (Emx1-Cre), cortical pyramidal neurons (CamKIIa-CreERT2) | JAX stock 014539 | [75] |
| R26-2XChETA | mouse | Cre-inducible knock-in | ChR2(E123T/H134R) | tdTomato (cytosolic) | CAG from Rosa26 locus | Cortical fast-spiking interneurons
(Pvalb-IRES-Cre, PPP1R2-Cre), direct pathway-projecting MSNs, dorsal
and ventral striatum (D1R-Cre), indirect pathway-projecting MSNs, dorsal and ventral striatum (A2AR-Cre), cerebellum Purkinje cells, basket cells, stellate cells (Pvalb-IRES-Cre) |
JAX stock 017455 | [76] |
| BTR6 (bitetO-rhodopsin line 6) | mouse | tTA-inducible plasmid transgenic | ChR2 NpHR | mCherry fusion EGFP fusion | tetO | Dorsal striatum medium spiny neurons (ChR2,
functional) Dorsal striatum medium spiny neurons (NpHR, non-functional) |
contact investigator | [81] |
| TetO-ChR2(C128S)-EYFP | mouse | tTA-inducible knock-in | ChR2(C128S) | EYFP fusion | tetO from beta-actin locus | Cerebellum Purkinje cells (Gad67-tTA),
habenula neurons (Htr5B-tTA), inferior olive neurons
(Htr5B-tTA), microglia (Iba1-tTA), cortical astrocytes and Bergmann glia (Mlc1-tTA), oligodendrocytes (PLP-tTA), hippocampal CA1 pramidal neurons (CaMKII-tTA), striatal MSNs (PDE10A2-tTA) |
contact investigator | [82, 84, 85] |
2. Overview of strategies for developing transgenic lines for optogenetics research
Numerous genetic modification strategies have been implemented to develop transgenic mouse lines for optogenetics research. These strategies can be easily parsed into distinct categories: (1) plasmid transgenic approaches, (2) bacterial artificial chromosome (BAC) transgenic approaches, and (3) knock-in approaches. Examples of these transgene expression strategies are illustrated as they pertain to developing transgenic lines for optogenetics research (Figure 1).
Figure 1. Diverse strategies for developing transgenic lines with functional expression of engineered opsins in the mammalian nervous system.
(A–C) Plasmid transgenic strategy. (A) An engineered opsin gene fused to a reporter gene (XFP) is inserted into a plasmid just downstream of the defined promoter region and upstream of a polyadenylation (pA) transcription termination sequence. (B) The Thy1.2 vector directs neuron-specific transgene expression. The opsin-XFP transgene is inserted downstream of the 4.2 Kb Thy1.2 promoter region. Deletion of exon III and flanking introns prevents expression in non-neuronal tissues. (C) Sagittal brain sections showing the broad transgene expression pattern for Th1.2-ChR2-EYFP line 18 (top) and Thy1.2-eNpHR2.0-EYFP line 2 (bottom). (D–F) BAC transgenic strategy. (D) Selection of a suitable 200 Kb clone from a BAC library which contains ~100 Kb of upstream sequence and ~50 Kb of downstream sequence flanking the ‘Target Gene.’ Note the presence of loxP and mlox511 sites in the BAC vector sequence, as well as hypothetical extra genes (Gene X, Gene Y). (E) BAC recombineering steps in the E. coli host cell, including homologous recombination using two short homology arms (Box A and Box B) to target the transgene cassette into the BAC DNA at a defined site, and Flp –mediated excision for removal of the FRT flanked selection cassette (NEO). (F) Sagittal brain sections from BAC transgenic mouse lines showing restricted cell-type specific transgene expression in cholinergic (left), serotonergic (middle), and Pvalb-expressing (right) neuronal subsets. (G) Knock-in strategy. Gene targeting involves homologous recombination in mouse ES cells. The transgene cassette is targeted to a specific endogenous chromosomal locus using a short homology arm (SHA) and long homology arm (LHA). The diphtheria toxin-A (DTA) gene is for negative selection against random integration events and the loxP flanked NEO cassette is for positive selection of correctly targeted clones. The NEO cassette is excised by expression of Cre.
Pronuclear injection of plasmid DNA represent the most straightforward and commonly used approach for creating a transgenic line [30]. Production of plasmid transgenic lines is limited by the availability of relatively short, well-defined promoters to direct transgene expression in specific cell populations. It is often the case that plasmid transgenic lines exhibit broad transgene expression in multiple brain regions or variable expression patterns across multiple founder lines derived from a single pronuclear injection [31, 32]. These features can often be exploited to establish several lines with unique utility, but ultimately it remains a challenge to restrict transgene expression to small subsets of cells in only one or a few brain regions with this approach. In contrast, BAC transgenesis is a powerful approach for directing transgene expression to highly restricted neuronal subsets. BACs are large (generally 100–300 Kb) genomic DNA fragments that can be propagated and manipulated in bacteria. BACs often span entire genes as well as significant portions of flanking sequences that are required to direct the faithful expression pattern of a particular gene in vivo. The Gene Expression Nervous System ATlas (GENSAT) project at Rockefeller University has developed hundreds of GFP reporter and Cre driver mouse strains using BAC transgenesis, and the success of this large scale project has provided an extensive knowledge base for selecting specific mouse BAC clones to target transgene expression to specific domains of the nervous system [33–35]. Moreover, BAC clone libraries spanning the mouse, rat, and human genome are currently available, along with wellestablished BAC recombineering protocols [36–38]. Lastly, the knock-in approach utilizes traditional gene targeting to introduce a transgene expression cassette into the mouse genome by homologous recombination in mouse embryonic stem cells [39]. The knock-in transgene expression strategy ensures that a single copy of the transgene is expressed from a defined genomic locus and is particularly valuable for comparing the properties of diverse transgenes that have been targeted into the genome using identical methodology.
3. Transgenic lines for neuronal activation with Channelrhodopsin
Following the seminal reports in 2005 demonstrating optical control of neuronal firing with Channelrhodopsin-2 (ChR2) in cultured neurons [40, 41], our laboratory and several others immediately began working to develop ChR2 transgenic mouse lines. From these collective initial efforts it was clear that there were significant challenges to overcome to achieve functional ChR2 expression in genetically modified mice. To impart adequate control of neuronal firing with light required very high ChR2 expression level in vivo with minimal toxicity, proper targeting to the neuronal membrane, and sustained expression throughout the lifespan of the animal. In particular, achieving adequate high-level expression has posed the largest obstacle to success.
3.1 Plasmid Transgenic mice and rats
In 2007 we reported the first transgenic mouse lines with functional expression of ChR2 [42, 43]. This was achieved by inserting the ChR2-EYFP transgene into a plasmid containing the mouse Thy1.2 expression cassette and successful integration of the transgenic cassette into the mouse genome. The Thy1.2 promoter drives neuron-specific transgene expression postnatally, peaking around 1–2 months of age, and is sustained in the adult. Thy1.2 is among the strongest defined neuronal promoters and has been previously used to create many other mouse lines with high level transgene expression in the nervous system [31, 44]. Notably, transgene expression from the Thy1.2 promoter often results in distinct lines exhibiting either broad neuronal expression or more restricted and random expression pattern in subsets of neurons in the central and/or peripheral nervous system.
Seven unique Thy1.2-ChR2-EYFP founder lines were initially described that showed regionally restricted expression of ChR2-YFP in the mouse nervous system. Two of these lines, Thy1.2-ChR2-EYFP lines 9 and 18, were deemed to be most broadly useful. Both lines exhibit ChR2 expression in layer V cortical pyramidal neurons, CA1 and CA3 pyramidal neurons of the hippocampus, and various nuclei of the thalamus, midbrain and brainstem. The lines differ slightly in that line 9 has more extensive expression of ChR2 in retinal ganglion cells and line 18 has expression in cerebellar mossy fibers. A third line, Thy1.2-ChR2(H134R)-EYFP line 20, was developed for expression of the ChR2(H134R) variant that is reported to have enhanced photocurrent amplitudes relative to wild-type ChR2. Additional motifs were added to the ChR2 coding region to enhance sub-cellular trafficking, though it seems these alterations did not lead to notable improvements. Thy1.2-ChR2(H134R)-EYFP line 20 exhibits a similar but notably more diffuse pattern of expression compared to the earlier lines (indicating relatively lower ChR2(H134R)-EYFP expression in a broader subset of neurons rather than higher expression in a restricted subset of neurons), and there is no evidence to suggest significant improvement in the functionality of this line. As such, Thy1.2-ChR2-EYFP line 9 and line 18 remain the preferred lines and are much more widely used in the neuroscience field.
Several investigators have demonstrated successful optogenetic manipulation of neuronal function in brain slices derived from both Thy1.2-ChR2-EYFP line 9 and line 18 for diverse applications including technical innovations in photostimulation methods [45] [46], cortical circuit mapping [43], analysis of cortical information processing [47], and demonstration of integrated optical stimulation and voltage-sensitive dye imaging [48]. In addition, ChR2 expression in a subset of retinal ganglion cells was sufficient to restore light responsiveness in retina isolated from a mouse model of retinal degeneration crossed to Thy1-ChR2-EYFP transgenic mice [49], but was not sufficient to restore visual function in vivo.
The Thy1.2-ChR2-EYFP transgenic lines have also been implemented in many in vivo studies. Using anesthetized Thy1.2-ChR2-EYFP line 18 mice it was shown that superficial illumination of the exposed cortex or olfactory bulb was sufficient to induce robust neuronal firing in cortical layer V pyramidal neurons and mitral cells, respectively [42]. This finding demonstrated that optic fiber implantation is not required for optogenetic manipulation in living mice. Anesthetized Thy1.2-ChR2- EYFP line 18 mice were also used to map brain activity in sensory cortex using combined optical stimulation and functional magnetic resonance imaging (opto-fMRI), thereby demonstrating a powerful new non-invasive technique for in vivo analysis of brain function [50]. A complementary approach was also implemented to extensively map activity patterns and connectivity of sensory and motor cortex using combined optical stimulation and voltage-sensitive dye imaging in vivo [51, 52]. Similarly, combined optogenetic simulation and Ca2+ dye-based imaging in vivo was used to explore mechanisms of visual map development during the critical period using Thy1.2-ChR2-EYFP line 18 mice [53].
Optogenetic manipulation of animal behavior has also been achieved using awake as opposed to anesthetized Thy1.2-ChR2-EYFP line 18 transgenic mice [25, 54]. In a landmark study examining the therapeutic potential of optogenetics for treating brain disorders, high frequency (but not low frequency) photostimulation of cortical layer V pyramidal neurons in the motor cortex or the axonal projection to the subthalamic nucleus (STN) was sufficient to fully ameliorate motor dysfunction in transgenic mice with chemical lesion of the nigrostriatal pathway [25]. These findings in hemiparkinsonian rodents provide crucial insights into the therapeutic mechanism of deep brain stimulation in debilitating human disorders such as Parkinson’s disease. The Thy1.2-ChR2-EYFP transgenic mice were also instrumental in developing a potentially therapeutic strategy for motor control using optical neural control of the peripheral nervous system. Photostimulation applied to the isolated sciatic nerve by an optical cuff was effective at achieving orderly recruitment of motor units in the physiological sequence [55], a feat that was not possible using electrical stimulation methods. These findings suggest new avenues of therapeutic intervention for individuals with severe motor deficits such as paralysis.
VChR1 is a channelrhodopsin isolated from Volvox carteri that has a red-shifted activation spectrum relative to ChR2 [56]. VChR1 can be strongly activated with yellow light but is only moderately activated with blue light, thereby providing a potentially useful tool for dual-wavelength, separable optical control of distinct neuronal populations using the combination of ChR2 and VChR1. Three Thy1.2-VChR1-EYFP transgenic mouse lines (line 1, line 4, and line 8) were established. Line 1 has very sparse VChR1-EYFP expression in cortical layer 5 pyramidal neurons, hippocampus, thalamus, superior colliculus, inferior colliculus, brainstem, amygdala, and cerebellum. Lines 4 and 8 are highly similar and have very widespread expression in the brain regions mentioned for line 1. The initial characterization of Thy1.2-VChR1-EYFP line 8 transgenic mice revealed that the fidelity of lightevoked firing of cortical and hippocampal pyramidal neurons expressing VChR1 declined sharply at frequencies >10 Hz due to the very slow kinetics of channel closure upon light offset (Ting and Feng, unpublished data). In addition, the activation spectrum of VChR1 is notably broader than that of ChR2, suggesting that despite the prior demonstration of feasibility [56], VChR1 is not ideal for dualwavelength optical stimulation experiments.
The first ChR2 transgenic rat lines were also developed using the Thy1.2 plasmid to drive expression of the ChR2-Venus transgene. Four unique lines of transgenic Wistar rats were described based on the target selection criteria of transgene expression in the retina. Of these lines, only line 4 (W-TChR2V4) was selected for detailed functional analysis as this line exhibited expression in the retinal ganglion cell layer and inner plexiform layer, a pattern that was most suitable for the particular investigation at hand [57]. ChR2 expression in retinal ganglion cells was sufficient to impart light responsiveness and to restore a very limited form of vision in rats with photoreceptor degeneration. In a separate study, the same W-TChR2V4 rats were found to have functional expression of ChR2 in mechanoreceptive dorsal root ganglion neurons and their peripheral sensory nerve endings [58]. The expression of ChR2 in this unique neuronal population was sufficient to impart the ability to sense touch in response to blue light applied to the bare plantar skin of forepaws or hindpaws. Thus, the W-TChR2V4 transgenic rat line may prove useful for investigating the various aspects of touch perception. Given the use of the Thy1.2 promoter to drive transgene expression, it is likely that some of these existing W-TChR2V lines also have functional ChR2-Venus in various additional neuronal populations of the nervous system. It was recently reported that W-TChR2V4 has functional expression throughout the hippocampal formation and was useful for opto-fMRI mapping of neuronal connectivity following optical stimulation of dentate gyrus granule cells [59]. Further detailed characterization is necessary to clarify the full potential of these transgenic lines for optogenetic investigations in rat.
One other ChR2 plasmid transgenic mouse line was developed using the 12 Kb fragment of the rat olfactory marker protein (Omp) gene promoter sequence to drive expression of the ChR2- EYFP gene fusion [60]. Three distinct lines were reported (line 20, 32, and 33) with transgene expression detected in the vomeronasal organ and accessory olfactory bulb. Line 20 (named ORCM) had additional expression in the olfactory epithelium and proved to have functional ChR2-EYFP expression in all olfactory sensory neurons. Photostimulation of olfactory neuron axon terminals within individual glomeruli generated strong synaptic excitation and allowed for precise mapping of mitral and tufted cells that share common inputs—so-called sister cells. This approach facilitated detailed analysis of odor responses measured simultaneously from sister pairs in vivo to reveal distinctions in correlations of phase but not rate of sensory-evoked firing, indicating that sister cells likely convey non-redundant information. In addition to olfactory circuit mapping, these transgenic mice may also be useful for behavioral studies exploring neural mechanisms of odor perception and discrimination.
3.2 BAC Tg mice
BAC transgenesis is a reliable method for restricting transgene expression to only one or a few genetically-specified neuronal subtypes. The large DNA fragments (~100–300 Kb) contained in BAC clones are far more likely to contain all of the necessary elements for proper specification of gene expression as compared to plasmids containing relatively short promoter fragments (generally <10 Kb). However, few endogenous gene promoters are predicted to drive the high level of transgene expression that is required to achieve functionality with ChR2 or other engineered opsins. Despite this concern, several useful lines have been established.
The first successful ChR2 BAC transgenic line was developed using a BAC clone that spanned the genomic sequence of the vesicular glutamate transporter-2 (Vglut2) gene and 95 Kb of upstream and 56 Kb of downstream sequence [61]. The ChR2(H134R)-EYFP sequence was inserted in place of the Vglut2 gene in order to prevent overexpression of Vglut2. Vglut2-ChR2(H134R)-EYFP BAC transgenic mice exhibited extensive ChR2(H134R)-EYFP expression in glutamatergic neurons of the spinal cord and hindbrain regions consistent with the known expression pattern of Vglut2. Vglut2 is also expressed in glutamatergic neurons located outside of the hindbrain and spinal cord but it remains to be determined if Vglut2-ChR2(H134R)-EYFP transgenic mice exhibit functional ChR2 expression in any of these other regions of the nervous system.
The Vglut2-ChR2(H134R)-EYFP transgenic mice were used to demonstrate that photostimulation of glutamatergic neurons in the lumbar region of the spinal cord is sufficient to drive rhythmic locomotor output. In addition, photostimulation of glutamatergic neurons in the caudal hindbrain regions could activate the spinal locomotor network by a direct descending synaptic command. In contrast, photostimulation of the hindbrain region containing the pre-Bötzinger complex and para-facial nucleus did not alter locomotor activity, but was sufficient to entrain respiratory rhythms. Thus, Vglut2-ChR2(H134R)-EYFP transgenic mice can play an important role in the optogenetic dissection of diverse motor-related animal behaviors.
Our laboratory developed a collection of cell-type specific BAC transgenic mouse lines with functional expression of ChR2(H134R)-EYFP in several major classes of neurons [62]. VGAT-ChR2(H134R)-EYFP BAC transgenic mice have strong expression of ChR2(H134R)-EYFP specified by the vesicular γ-aminobutyric acid (GABA) transporter (VGAT) gene regulatory elements to direct transgene expression in GABAergic neurons. ChR2(H134R)-EYFP expression was detected in the olfactory bulb, thalamic reticular nucleus (TRN), superior and inferior colliculus, the molecular layer of the cerebellum, and brainstem. We also detected ChR2(H134R)-EYFP expression in cortical and hippocampal interneurons (all sub-regions) and demonstrated robust neuronal firing induced by light in these regions in brain slices from hemizygous transgenic mice. Of particular note, cortical fast-spiking interneurons were able to sustain light-evoked firing rates up to 80 Hz with high fidelity, and photostimulation of interneurons in cortical microcircuits elicited powerful synaptic inhibition of pyramidal neurons. We also demonstrated successful axon terminal field photostimulation of GABA release within the medial habenula [62], a region that is reported to be devoid of local GABAergic interneurons [63]. In a separate study, VGAT-ChR2(H134R)-EYFP transgenic mice were used to demonstrate that optical activation of the TRN switches the firing mode of thalamocortical relay neurons from tonic firing to burst firing and generates cortical spindle oscillations in freely-behaving animals [64]. Further work with this line may help to clarify the importance of cortical spindle generation to certain behavioral state-transitions and in sleep-dependent memory consolidation.
The enzyme tryptophan hydroxylase 2 (TPH2) is the rate-limiting enzyme in the synthesis of serotonin in the central nervous system. TPH2-ChR2(H134R)-EYFP BAC transgenic mice have expression of ChR2(H134R)-EYFP specified by the TPH2 gene regulatory elements to direct transgene expression in serotonergic neurons [62]. ChR2(H134R)-EYFP expression was detected in the dorsal raphe nucleus (DRN), median raphe nucleus (MnR) and interpeduncular nucleus (IPN) brainstem regions. We demonstrated light-evoked spiking in serotonergic neurons of the dorsal raphe nucleus (DRN) using brain slices from hemizygous transgenic mice. ChR2 functionality in other serotonergic populations was not assessed. A previous study using viral expression of ChR2 in brainstem serotonergic neurons was able to show dual component light-evoked serotonin/glutamate release from axon projections of serotonergic neurons that terminated onto GABAergic interneurons of the hippocampus [65]. In contrast, we were unable to provide evidence for axon terminal photostimulation in projection targets of serotonergic neurons such as the hippocampus and medial prefrontal cortex in the TPH2-ChR2(H134R)-EYFP BAC transgenic line. Based on these findings it seems that direct illumination of the raphe nucleus may be the only means to achieve successful optogenetic control of serotonergic transmission in this line, probably due to the moderate level of transgene expression.
To enable optogenetic control over parvalbumin (Pvalb)-expressing neurons in the nervous system, the ChR2(H134R)-EYFP transgene was expressed under the control of the Pvalb gene regulatory elements [62]. Pvalb-ChR2(H134R)-EYFP BAC transgenic mice have strong transgene expression in the TRN, brainstem, inferior colliculus, and the molecular layer of the cerebellum. Light-evoked neuronal spiking was confirmed in the TRN and cerebellum (Purkinje cells and molecular layer basket and stellate cells). Despite the known expression of parvalbumin in cortical, striatal and hippocampal interneuron subsets, we observed no functional ChR2(H134R)-EYFP expression in these areas, likely reflecting the relatively weak activity of the parvalbumin promoter in these regions. Pvalb-ChR2(H134R)-EYFP BAC transgenic mice have been used to dissect cerebellar circuitry [66]. Photostimulation of cerebellar molecular layer interneurons in brain slices from Pvalb-ChR2(H134R)- EYFP BAC transgenic mice elicited robust inhibitory synaptic input onto Purkinje cells, whereas, no inhibitory responses were detected from simultaneously recorded Golgi cells.
Choline acetyltransferase (ChAT) is the enzyme responsible for acetylcholine synthesis. ChAT-ChR2(H134R)-EYFP BAC transgenic mice have expression of ChR2(H134R)-EYFP specified by the ChAT gene regulatory elements to direct transgene expression in cholinergic neurons of the nervous system [62]. Two distinct lines were established, one with high level transgene expression (line 6) and one with lower transgene expression (line 5). In ChAT-ChR2(H134R)-EYFP line 5 functional transgene expression was only demonstrated in striatal cholinergic interneurons. The relatively weak ChR2 photocurrents were sufficient to drive sustained moderate potentiation of spiking activity over tens of seconds without inducing depolarization block, a feature that may be desirable for behavioral testing paradigms involving prolonged or repeated bouts of photostimulation. Notably, EYFP reporter fluorescence is not readily visible without signal enhancement in this line. In contrast, ChAT-ChR2(H134R)-EYFP line 6 has ChR2(H134R)-EYFP expression in the striatum, basal forebrain, facial nucleus, trochlear nucleus, medial habenula, cortex, and various other brainstem motor nuclei, consistent with the known distribution of cholinergic neurons. Robust light-evoked neuronal spiking was confirmed for striatal cholinergic interneurons, medial septal neurons, and neurons in the ventral two-thirds of the medial habenula using brain slices from line 6 hemizygous transgenic mice.
The cholinergic neurons located in the ventral two-thirds of the medial habenula send a massive axonal projection to the interpeduncular nucleus via the fasciculus retroflexus. This pathway shows remarkably high transgene expression in the ChAT-ChR2(H134R)-EYFP line 6 BAC transgenic mice, which enabled a clear demonstration of successful photostimulation of axons by applying a restricted light stimulus to the fasciculus retroflexus [62] or directly to the axon terminal field in the interpeduncular nucleus in acute brain slices [67]. This approach yielded direct evidence for corelease of glutamate and acetylcholine at the habenulo-peduncular synapse [67]. Optogenetic stimulation of this pathway in vivo may help to unravel the physiological significance of the dual transmitter phenotype in the context of discrete animal behaviors.
In order to demonstrate the utility of the ChAT-ChR2(H134R)-EYFP line 6 BAC transgenic mice for precise optogenetic control of spiking in vivo, we implanted a tetrode/optic fiber assembly into the striatum of hemizygous line 6 mice. We reported that optogenetic drive of striatal cholinergic activity at 30 Hz exerted a depressive effect on putative medium spiny neuron activity in awake, behaving animals [62]. In a separate study, optogenetic activation of cholinergic projection neurons in the horizontal limb of the diagonal band of Broca inhibited spontaneous activity of all major cell types in the main olfactory bulb in anesthetized ChAT-ChR2(H134R)-EYFP line 6 mice [68]. This finding is consistent with the hypothesis that cholinergic activity enhances olfactory discrimination in vivo.
3.3 Knock-in mice
The Mas-related G-protein-coupled receptor D (Mrgprd) is a molecular marker for a subset of non-peptidergic spinal cord sensory neurons that transmit polymodal pain information [69]. Mrgprd-ChR2(H134R)-Venus knock-in mice were developed to investigate spinal circuitry connectivity of Mrgprd-expressing neurons [70]. Light-evoked firing was recorded from dissociated dorsal root ganglion (DRG) neurons of heterozygous and homozygous Mrgprd-ChR2(H134R)-Venus knock-in mice. In spinal cord slices, photostimulation of DRG axon terminals was sufficient to evoke excitatory synaptic transmission onto substantia gelatinosa (SG) neurons. Successful optogenetic control enabled ChR2-assisted circuit mapping to directly demonstrate that Mrgprd-expressing neurons form connections onto virtually all known classes of SG neurons, although only half of the SG neurons receive direct afferent input from Mrgprd-expressing neurons. These findings suggest that SG modules exist and may represent a mechanism for modality-selective sensory information processing [70]. Importantly, because ChR2(H134R)-Venus transgene insertion at the Mrgprd locus replaced the endogenous Mrgprd gene, heterozygous knock-in mice have reduced, while homozygous knock-in mice have ablated Mrgprd expression. Although Mrgprd null mice were reported to have no overt behavioral or anatomical abnormalities [69], Mrgprd deletion did alter DRG neuron excitability [71]. This finding may place some constraints on the use of Mrgprd-ChR2(H134R)-Venus knock-in mice for optogenetics-based behavioral studies aimed at understanding mechanistic aspects of pain information processing.
A second Omp-ChR2-EYFP transgenic mouse line was reported in which the ChR2(H134R)-EYFP transgene replaced the endogenous Omp gene [72]. This knock-in line has transgene expression in all mature olfactory sensory neurons and their axon terminals in glomeruli of the olfactory bulb, which is identical to the pattern reported for the Omp-ChR2-EYFP plasmid transgenic mice [60]. The main difference between lines is that the knock-in strategy disrupts the endogenous Omp gene, whereas the plasmid transgenic strategy leaves the endogenous gene unaltered. Thus, mice that are heterozygous for the Omp-ChR2(H134R)-EYFP knock-in allele will have a 50% reduction in the expression of Omp, which may present a confound for the analysis of olfactory sensory functions. Despite this caveat, the knock-in mice were used to obtain evidence that mice can be trained to behaviorally report the perception of sniff phase of olfactory input (delivered as direct patterned photostimulation of the olfactory sensory neurons), which to some extent complements the findings of Dhawale et al. [60].
3.4 Cre/loxP intersectional expression strategy
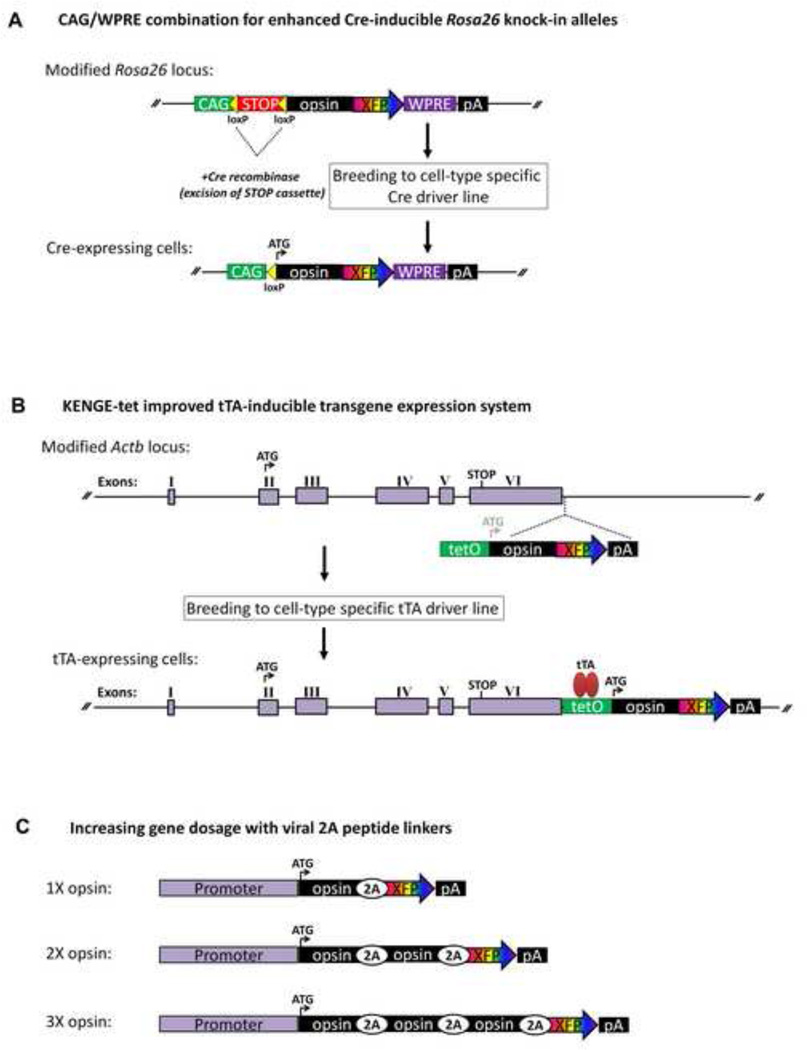
A particularly exciting area of progress in transgenic lines for optogenetics research has been the rapid progress in developing mice with Cre-inducible expression of engineered opsins. This approach typically involves a ‘Cre reporter’ line specifying the transgene of interest with an upstream floxed transcriptional STOP cassette (loxP-STOP-loxP) to prevent transgene expression, and a second ‘Cre driver’ line with Cre expression driven by a cellular promoter to dictate the cell-type specificity. Crossing the two lines leads to excision of the loxP-STOP-loxP cassette to allow transgene expression in cells that have at any point in time expressed Cre. This approach has immense potential because it can leverage the ever-expanding repository of well-characterized Cre driver mice to hypothetically achieve functional opsin expression in any neuronal or even nonneuronal cell type. It is also possible to use virus injection to achieve restricted Cre expression in Cre reporter lines, thereby adding another layer of versatility with this technology.
The first Cre-inducible ChR2 knock-in line was developed by targeting a CAG promoter-loxPSTOP-loxP-ChR2(H134R)-EGFP cassette into the Rosa26 locus of the mouse genome [73]. When crossed to a cre driver mouse line the transcriptional STOP cassette is excised (leaving one residual loxP site) to activate ChR2(H134R)-EGFP expression only in Cre-expressing cells. Thus, this intersectional expression strategy can enable transgene expression in diverse genetically-defined cell types. Unfortunately, only weak functional transgene expression was demonstrated with this initial Cre-inducible knock-in line. To achieve functional ChR2(H134R)-EGFP expression it was necessary to use mice homozygous for the Cre-inducible ChR2 allele and heterozygous for a transgene directing expression of tamoxifen-inducible Cre in cortical interneurons. This enabled ChR2-assisted circuit mapping of inhibitory connections onto cortical excitatory neurons in brain slices. However, even two copies of the Cre-inducible allele were not sufficient for light-evoked neuronal firing in mice expressing Cre in cortical pyramidal neurons. These findings indicate that additional strategies are necessary to achieve robust ChR2 expression from the Rosa26 locus in order to gain access to broad classes of neurons in the nervous system.
A simple but highly effective improvement was recently described for creating Rosa26 locus knock-in mouse lines with enhanced Cre-dependent expression of reporter genes by combining the strong ubiquitous CAG promoter and woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) sequence in the targeting vector design [74]. This approach proved to be the much-needed solution for creating improved Cre-inducible ChR2 knock-in mouse lines. Two new lines were described with robust cre-inducible expression of ChR2(H134R)-tdTomato (line Ai27) or ChR2(H134R)-EYFP (line Ai32) [75]. The functionality of ChR2 was demonstrated by photostimulation of cortical pyramidal neurons in brain slices from Emx1-Cre;Ai27 (E-Ai27) and Emx1-Cre;Ai32 (E-Ai32) mice. Detailed comparative analysis revealed stronger light-evoked responses with the E-Ai32 line compared to the E-Ai27 line. No differences were found in the expression level or membrane localization, suggesting that perhaps the bulkier tdTomato fluorophore interferes with ChR2 function with expressed as a fusion protein. Despite this issue the Ai27 line may be desirable for some applications since the tdTomato fluorophore can be excited with minimal photostimulation of ChR2.
The potential for axon stimulation with light was assessed in E-Ai27 and E-Ai32 mice. Responses originating from axonal versus somato-dendritic photostimulation in local circuits were distinguished by analyzing the charging phase preceding an action potential threshold. Antidromic action potentials that originate in axons do not have a typical charging phase. This approach was used to demonstrated that ChR2(H134R)-expressing cortical neurons in brain slices from both the EAi27 and E-Ai32 transgenic lines can be efficiently stimulated in axons using light [75]. Lastly, the Ai27 and Ai32 lines were evaluated for optogenetic control of neuronal firing in vivo. High reliability of light-induced spiking in putative hippocampal CA1 basket cells and putative TRN neurons was confirmed in awake, behaving Pvalb-IRES-Cre;Ai32 mice. In addition, light-evoked spiking was also observed for putative cortical pyramidal neurons in awake, head-fixed E-Ai27 and E-Ai32 mice. The magnitude of photoactivation was significantly higher in E-Ai32 versus E-Ai27 mice, consistent with the results obtained in brain slices. Although optogenetic control of neuronal firing was only empirically demonstrated in a limited range of neuronal populations, the available evidence suggests that these lines will be useful for optogenetic control of many other diverse cell types.
We recently developed a Rosa26 locus knock-in mouse line for Cre-inducible expression of the ChETA variant of ChR2 to enable ultrafast optical control of neuronal firing in genetically defined cell types [76]. In light of the significantly reduced photocurrents conferred by the ChETA mutation relative to wild type ChR2 [77], we implemented several measures to achieve functional transgene expression. We employed the strong and ubiquitous CAG promoter combined with WPRE to enhance transgene expression (as mentioned above). We also selected the E123T/H134R double mutant (ChETATR) to try to maximize photocurrent amplitude. Lastly, we inserted two copies of ChETATR instead of a single copy. At present, this line is the only transgenic animal for optogenetics research that incorporates a physical uncoupling of the fluorophore from the opsin gene, a feature that improves visualization of transgene expressing neurons in living brain tissue and greatly simplifies targeted patch clamp electrophysiology experiments. The choice of the tdTomato fluorophore allows for fluorophore excitation that is largely independent of ChR2 stimulation while searching for transgene-expressing cells in living tissue while avoiding constraints imposed by the gene fusion design.
Previous work has demonstrated ultrafast light-evoked firing in ChETA-expressing cortical fast-spiking interneurons up to 200 Hz [77, 78]. In addition, the fast kinetic properties of ChETA variants lead to improvements in several features of optical control of neuronal function, including (1) elimination of plateau potentials during trains of stimulation due to faster neuronal membrane repolarization between light-evoked spikes, (2) increased fidelity of light-evoked spiking (elimination of extra spikes), and (3) greater consistency of successful spiking across time for extended bouts of photostimulation [78]. Each of these attributes was validated with the cre-inducible ChETATR knock-in mice following induction of ChETA expression in cortical fast-spiking interneurons. Functional ChETATR expression was also obtained for additional cell types and was adequate for light-evoked spiking in medium spiny neurons (MSNs) of the striatum (both direct and indirect pathway projecting MSNs) [76]. We are currently investigating if the line is suitable for in vivo optogenetic applications. Notably, although this line has some advantages over existing ChR2 lines, the photocurrent amplitudes attainable are considerably smaller as compared to the Ai32 line. Thus Ai32 remains the Cre-inducible ChR2 line of choice for highly reliable optical control of neuronal firing. Furthermore, preliminary evidence suggests that sustained high level expression of ChETATR is toxic for some cell types (Ting and Feng, unpublished data).
3.5 TetO/tTA intersectional transgene expression strategy
The tetracycline-controlled gene induction system is a transcriptional amplification system with proven utility for enabling inducible and high-level transgene expression in the nervous system of transgenic mice [79, 80]. Two complementary forms of control exist, one for tetracycline-mediated induction of gene expression (tet-on), and one for tetracycline-mediated suppression of gene expression (tet-off). For tet-off applications in transgenic animals, the system requires one transgenic line with a cellular promoter driving expression of the tetracycline transactivator (tTA) and a second transgenic line with a tTA-responsive promoter linked to the gene of interest. The use of cell-type specific promoters to drive tTA expression imparts specificity of transgene induction in genetically-defined cell types. If desired, tetracycline or the analog doxycycline can be used to repress transgene expression.
A tet-off strategy was used to create a plasmid transgenic mouse line for tTA-inducible expression of ChR2-mCherry and NpHR-EGFP from a bidirectional tetO promoter [81]. The line was designated BTR6 for ‘bidirectional tetO-rhodopsin line 6,’ and BTR6;CaMKII-tTA mice exhibited ChR2-mCherry and NpHR-EGFP expression restricted to a subset of dorsal striatum MSNs. The NpHR-EGFP subcellular localization revealed extensive aggregation indicative of poor trafficking and tolerability in mammalian cells, and this correlated with virtually no hyperpolarization with yellow light. In contrast, ChR2-mCherry showed the expected membrane localization, and blue light produced strong depolarization and firing of MSNs. Thus, functionally the BTR6 line is regarded solely as a tTA-inducible ChR2-mCherry transgenic mouse line.
ChR2-assisted circuit mapping was performed in brain slices of BTR6;CaMKII-tTA mice to map the functional connectome of striatal MSNs. Within the striatum, MSNs formed synaptic connections with other MSNs and cholinergic interneurons, but not with fast-spiking interneurons. Axon terminal photostimulation in the two major projection targets of striatal MSNs revealed connections with one of two identified classes of neurons in the globus pallidus, and with GABAergic (but not dopaminergic) neurons in the substania nigra. These findings demonstrate the utility of the BTR6 line for circuit mapping; however, the NpHR-EGFP intracellular aggregates may be detrimental to cellular function in a manner that could preclude meaningful behavioral analysis with these mice.
Recently, an improved tetracycline-controlled gene induction system was described (KENGE-tet) where the tetO-ChR2(C128S)-EYFP transgene was inserted immediately downstream of the beta-actin gene polyadenylation signal to create tTA-inducible ChR2(C128S)-EYFP knock-in mice [82]. This strategy was shown to be superior to plasmid and BAC transgenic approaches for tTA-mediated expression from similar but not identical tetO-ChR2 cassettes [81, 82]. The differences are presumably due to relatively more permissive chromatin structure at the site of transgene insertion for the knock-in line relative to the plasmid and BAC transgenic lines. Several new cell-type specific tTA driver lines were developed to expand the range of cell types that could be accessed using the tetO/tTA system. The tetO-ChR2(C128S)-EYFP knock-in mice were crossed with the various tTA driver lines and functional ChR2(C128S)-EYFP expression was demonstrated in Purkinje cells (Gad67-tTA driver), habenula neurons (Htr5B-tTA driver), and inferior olive neurons (Htr5B-tTA driver) in brain slices. Photocurrents were activated by brief blue light and terminated with yellow light, a defining feature of the ChR2(C128S) variant [83]. ChR2-mediated photocurrents were also evoked in non-neuronal cell types, including microglia (Iba1-tTA driver), Bergmann glia (Mlc1-tTA driver), and oligodendrocytes (PLP-tTA driver), indicating a much broader utility of this line.
In vivo optogenetic manipulation of hippocampal CA1 pyramidal neuron firing was demonstrated using head-fixed, anesthetized tetO-ChR2(C128S)-EYFP;CaMKII-tTA mice. A rundown of light evoked firing was observed with repeated photostimulation trials, but light-evoked firing was still observed in later trials. In freely behaving mice, prolonged unilateral photoactivation of the CA1 region was sufficient to induce pronounced locomotion following a delay period of about one minute. This delay argues against a direct motor response to photostimulation, but may instead reflect a motor response linked to some form of evoked cognitive response. The in vivo photoactivation corresponded with biochemical detection of c-fos induction throughout the hippocampal subregions and in both hemispheres. Induction of c-fos expression was also observed in cortical astrocytes when light was applied to the overlying region through the skull of tetO-ChR2(C128S)-EYFP; Mlc1-tTA mice. This finding empirically demonstrates the heightened light sensitivity of the ChR2(C128S) variant and the feasibility of non-invasive light delivery to avoid damage to brain tissue.
Bigenic tetO-ChR2(C128S)-EYFP;PDE10A2-tTA mice were utilized to investigate optogenetic induction of immediate early genes in the striatum region of freely moving animals [84]. The PDE10A2-tTA driver enabled functional ChR2(C128S)-EYFP expression in all virtually striatal MSNs, and brief blue light stimulation in vivo was found to induce rapid upregulation of Npas4 but not c-fos. This approach may represent an effective strategy for investigating differential induction of various immediate early genes in the brain. Most recently, bigenic tetO-ChR2(C128S)-EYFP;Mlc1-tTA mice were further utilized to evaluate the impact of optogenetic stimulation of Bergmann glia on synaptic function in cerebellar circuits [85]. Photostimulation was sufficient to trigger glutamate release from glial cells, resulting in perturbed neuronal activity and synaptic plasticity in cerebellar circuits. This glial optogenetic stimulation paradigm was also sufficient to alter cerebellar-modulated motor behavior. Although the physiological significance of these findings is not clear at present, tetO-ChR2(C128S)-EYFP;Mlc1-tTA mice seem to be an excellent new model to help further our mechanistic understanding of neuron-glia interactions in the brain.
Collectively, these recent results are very promising and strongly suggest that this improved tTA-inducible knock-in line should be further evaluated for the potential to achieve functional opsin expression in diverse neuronal subtypes using additional cell-type specific tTA driver lines. In addition, particular attention should be paid to the functionality of the ChR2(C128S) variant, a first generation step-function opsin [83]. Earlier studies have described issues relating to permanent inactivation produced by exposure to common laboratory white light sources and reduced efficacy of photostimulation with repeated trials [86], which may pose some important limitations for behavioral testing with these mice.
4. Transgenic lines for neuronal silencing with Halorhodopsin and Archaerhodopsin
Of the engineered opsin variants described for optogenetic neural silencing [27, 87], only halorhodopsin from Natronomonas pharaonisin (NpHR) and archaerhodopsin-3 from Halorubrum sodomense (Arch) have been successfully used in developing transgenic mouse lines for optogenetics research. NpHR is a light-activated chloride pump, whereas, Arch is a light-activated proton pump. The distinct modes of action are each effective at hyperpolarizing neuronal membranes and provide alternative means for implementing optogenetic silencing in different experimental contexts.
4.1 Plasmid Tg lines
In 2008 we reported five distinct Thy1.2-NpHR-EYFP transgenic mouse lines designed for optogenetic silencing [88]. These mice are viable but developed extensive intracellular aggregates of NpHR-EYFP in the endoplasmic reticulum (ER). Although it was possible to demonstrate lightinduced hyperpolarization in cortical and hippocampal neurons expressing the transgene, the degree of hyperpolarization was very modest, consistent with poor trafficking of the NpHR to the membrane. Thus, these mice were not suitable for further in vivo experimentation. In light of these findings, we and others designed rational modifications to improve cellular trafficking to the membrane. Addition of an N-terminal signal peptide sequence alone did not fully eliminate intracellular aggregation; however, addition of an ER export motif either alone or in combination with the N-terminal signal peptide sequence (eNpHR2.0) was sufficient to eliminate intracellular aggregates [88, 89]. eNpHR2.0 allows for high-level expression in mammalian neurons without toxicity and confers stronger light-induced neuronal silencing.
Two distinct Thy1.2-eNpHR2.0-EYFP transgenic mouse lines have been developed (line 2 and line 4) that exhibit improved functionality and no overt toxicity. Line 2 and Line 4 have broad transgene expression in cortical layer V pyramidal neurons, hippocampus, thalamus, superior and inferior colliculus, brainstem, amygdala, and cerebellum. Line 4 has additional modest transgene expression in cortical layer II/III pyramidal neurons. Interestingly, Line 4 is an atypical Thy1.2 transgenic mouse line in that eNpHR2.0-EYFP is more highly expressed in interneuron populations relative to principle neurons in some brain regions (e.g. hippocampus and globus pallidus). For example, in the hippocampus of Line 4 mice there is virtually no functional transgene expression in CA1 or CA3 pyramidal neurons or in dentate granule cells. In contrast, interneuron populations have robust eNpHR2.0-EYFP expression in all hippocampal subregions—including strong labeling in the hilus. Alternatively, Line 2 exhibits robust functional eNpHR2.0-EYFP expression primarily in subiculum and CA1 pyramidal neurons and subsets of CA3 pyramidal neurons and dentate granule cells. Average recorded light-induced hyperpolarization is relatively weaker across all brain regions in line 4 as compared to line 2, with the strongest functionality detected in principle neurons of the basolateral amygdala and subiculum in both lines. Thus, Thy1.2-eNpHR2.0-EYFP line 2 is the more broadly useful line for optogenetic silencing studies and exhibits a more conventional Thy1.2 expression pattern; however, Line 4 may be uniquely suited for specialized studies requiring optogenetic silencing of hippocampal interneuron populations. Importantly, modest but functional transgene expression is detected in subsets of cortical and hippocampal interneurons for Line 2 (Ting and Feng, unpublished data), and although under-reported in the literature, this is fairly common for transgene expression from the Thy1.2 promoter.
A plasmid transgenic mouse line with expression of NpHR-EGFP under the control of the human prepro-orexin promoter fragment was developed to enable optogenetic control over orexinexpressing neurons of the lateral hypothalamic area [4]. Three independent lines were reported with varying extent of transgene expression in orexin neurons (line 5, line 7, and line 8) with no overt intracellular NpHR aggregation detected, and line 5 was selected for further use because it had transgene expression in nearly all orexin neurons. The ability of NpHR photoactivation with orange light to inhibit orexin neuron spontaneous firing was validated in brain slices from orexin-NpHR-EGFP transgenic mice, with maximal activation causing hyperpolarization leading to greater than 90% inhibition in spontaneous firing rate. The photoinhibition was effective over extended photostimulation bouts lasting minutes, although inhibition was not complete throughout the duration of light presentation. These results suggested it may be possible to sustain NpHR-mediated silencing of orexin neurons over behaviorally-relevant time scales in vivo. Indeed, acute one-minute bouts of optogenetic inhibition delivered to hypothalamic orexin neurons in vivo was found to induce slow-wave sleep brain activity patterns in freely-behaving orexin-NpHR-EGFP transgenic mice. Optogenetic silencing of orexin neurons with simultaneous extracellular recording from putative dorsal raphe serotonergic neurons, a major projection target of hypothalamic orexin neurons, showed that induction of SWS activity was associated with decreased firing rate of serotonergic neurons. The ability for optogenetic silencing to induce SWS activity patterns was restricted to the day time period, perhaps because the homeostatic drive for sleep is higher in rodents during this period. These findings demonstrate the usefulness of orexin-NpHR-EGFP transgenic mice for optogenetic induction of behavioral state-transitions, in this case the transition from wakefulness to sleep.
4.2 Cre/loxP intersectional expression strategy
The Madisen et al, 2012 study [75] describing Ai27 and Ai32 Cre-inducible ChR2 mouse lines also introduced two new Rosa26 Cre-inducible knock-in lines for neuronal silencing that were created using an identical targeting strategy. The Ai35 line has Cre-inducible expression of ss-Arch-EGFPER2, a modified version of the Arch-EGFP fusion protein with enhanced membrane trafficking [87]. The Ai39 line has Cre-inducible expression of eNpHR3.0-EYFP, a functionally improved third generation NpHR that was created by adding an additional trafficking signal sequence derived from Kir2.1 to eNpHR2.0-EYFP [90]. A detailed comparative analysis was performed using brain slices from Emx1-Cre; Ai35 (E-Ai35) and Emx1-cre;Ai39 (E-Ai39) mice [75]. Photostimulation of cortical pyramidal neurons demonstrated that the E-Ai35 line exhibits two to three times higher hyperpolarizing photocurrents than the E-Ai39 line in this neuronal population using yellow light; however, both lines were highly effective for silencing of neuronal firing induced by current injection. In addition, both lines produced nearly identical hyperpolarizing photocurrent amplitudes and equally effective neuronal silencing with photostimulation using either a white LED or red laser light. One notable difference in performance is that the E-Ai39 line showed diminished hyperpolarization of cortical pyramidal neurons with repeated photostimulation trials, whereas, no such diminished hyperpolarization was observed for the E-Ai35 line.
E-Ai35 mice were further analyzed to demonstrate the potential for optogenetic manipulation of neuronal networks. Persistent population bursting was induced in hippocampal slices and activity was monitored with an extracellular electrode in the CA1 pyramidal cell layer. Photostimulation of the CA3 region with a white LED caused a dramatic reduction of burst frequency in CA1, indicating effective optogenetic silencing of the propagation of synaptic activity in this well-defined network. Furthermore, effective silencing of cortical pyramidal neuron activity with green light was demonstrated in vivo using the extracellular recording technique in awake, head-fixed mice for both the Ai35 and Ai39 lines crossed to CamKIIa-CreERT2 tamoxifen-inducible driver mice (C-Ai35 and C-Ai39). The neuronal silencing was more effective using C-Ai35 mice compared to C-Ai39 mice, similar to the findings in the same neuronal population using yellow light in brain slices from E-Ai35 and E-Ai39 mice. Although optogenetic silencing was only empirically demonstrated in cortical and hippocampal pyramidal neurons, the available evidence suggests that these lines will be useful for optogenetic silencing of many other diverse cell types.
A second Rosa26 knock-in mouse line has been established for either Cre or Flp-inducible expression of eNpHR2.0-EYFP [91]. The original founder line with adjacent loxP-STOP-loxP and FRT-STOP-FRT cassettes upstream of the eNpHR2.0-EYFP transgene was bred to germline Cre or germline Flp deleter strains to produce two distinct derivative strains to enable either Cre or Flpinducible transgene expression. Functional silencing with yellow light was demonstrated in cortical pyramidal neurons or cortical interneurons in acute brain slices prepared from mice resulting from a cross to either the Math2-Cre or Dlx5/6-Flpo driver line, respectively. Although these findings demonstrate adequate optical silencing in brain slices, the available evidence suggests that the hyperpolarizing photocurrents attainable with these new lines are approximately 50% less than those measured from E-Ai39 mice (Cre-inducible eNpHR3.0-EYFP line) under similar conditions. This suggests that some aspect of the targeting design results in suboptimal expression from the Rosa26 locus. Consistent with this interpretation, the authors reported insufficient functionality for a second Rosa26 knock-in line for Cre or Flp-inducible ChR2(C128A)-mCherry expression that was developed using an identical targeting design [91], but the contribution of the C128A mutation must also be taken into account. Although there are few well-characterized Flp driver mouse lines available at present, the Flp-inducible eNpHR2.0-EYFP line may prove to be a valuable tool in the near future since in principle it could be implemented in conjunction with Cre/loxP technology for simultaneous labeling of non-overlapping, genetically-defined cellular populations with Cre and Flp.
5. Utilization of transgenic lines for optogenetics-based behavioral studies: considerations and caveats
5.1 Are all neuronal subtypes accessible for optogenetic manipulation using existing transgenic lines?
In principle, virtually any cell population in the nervous system may be accessed for optogenetic manipulation using Cre-inducible ChR2, NpHR, and Arch transgenic mice [75], assuming a suitable Cre driver line is available. However, at the present time there is no direct evidence to substantial that all cell types will exhibit robust functional opsin expression following Cre-mediated recombination in these new mouse lines. Furthermore, the optimal level of opsin expression required to achieve dynamic functional manipulation of neuron activity is likely to vary extensively across diverse neuron types, and thus, it may not be possible to achieve optimal ChR2 expression in all neuron types with a single ‘universal’ inducible ChR2 knock-in line. Similar caveats apply for the recently described tTA-inducible ChR2(C128S) mouse line [82]. Only a handful of driver lines and select neuronal populations have been evaluated thus far, and as such, much work remains over the coming years to delineate how broadly useful each of these lines may truly be.
5.2 Potential confounds relating to random transgene integration
Pronuclear injection of plasmid or BAC DNA results in the integration of linear DNA into the host genome at a random location. With this in mind, the use of homozygous transgenic animals is never recommended in any experimental context since it is possible that the transgene integration disrupts or alters the expression of an endogenous gene (or genes), and thus the homozygous transgenic animal is in the worst case a null for this unknown gene. It has been estimated that insertional mutations are present in 5–10% of transgenic lines [92]. It is also possible that the pattern of transgene expression may be altered in homozygous versus hemizygous transgenic animals, or that transgene expression may become toxic with increased expression level in a homozygous animal. However, where feasible, it is acceptable to maintain plasmid and BAC transgenic lines in the homozygous state for the purposes of breeding.
5.3 A hidden confound: overexpression of ‘extra’ genes in BAC transgenic mice
Perhaps the most serious yet underappreciated confound is that of ‘extra’ genes that may be present and overexpressed in BAC transgenic lines. BACs contain large segments of genomic DNA, are readily propagated in E. coli, and are easily manipulated using standard recombination-based genetic engineering methods. These features make BACs an excellent choice for inserting transgenes under the control of cell-type specific promoter elements spanning tens of kilobases or more. Unfortunately, the presence of large genomic segments also poses a significant concern. It can be challenging or sometimes impossible to identify BAC clones that contain sufficient length of regulatory elements for directing faithful patterns of transgene expression but that don’t contain ‘extra’ genes. Failure to remove these extra genes is likely to result in elevated expression of genes unrelated to the target transgene. The extra genes may be expressed in the same populations of cells as the target transgene and/or in various other cellular populations throughout the animal. In the most worrisome cases the extra genes are part of a gene cluster based on function. For example, a BAC clone encompassing the ChAT gene that was modified to create BAC transgenic mouse and rat lines harbors multiple extra genes including the vesicular acetylcholine transporter (VAChT) gene that is important for transport of acetylcholine into vesicles for synaptic release [34, 62, 93]. The VAChT gene is nested within the first intron of the ChAT gene and is not easily deleted without disrupting the promoter region for the ChAT gene. This potential confound should be carefully considered when selecting a BAC transgenic line to employ in a particular study, as this has important implications for the selection of appropriate control groups and in determining the limitations of the study.
5.4 Choosing between transgenic lines and virus-based strategies for opsin expression
The availability of transgenic lines affords important advantages such as stable and heritable transgene expression patterns across experimental cohorts. As such, the use of transgenic lines precludes the need for costly and labor-intensive procedures to achieve functional transgene expression in each individual experimental animal—an obligatory step when utilizing the viral gene delivery approach. Furthermore, the viral gene delivery method can prove technically challenging depending on the location and size of the targeted injection site, which leads to several important limitations. Every experimental animal requires surgical stereotaxic delivery of virus encoding the opsin transgene. Inevitably, not every injected mouse is suitable for experimentation owing to experimenter error. Even the mice deemed suitable may have variable spread of the virus at the injection site, gradients of transgene expression in infected brain regions, or potential tissue damage. This represents an important consideration when large cohorts of experimental animals are desirable as in many common behavioral assays. Conversely, if a particular experimental or behavioral paradigm requires a mammalian species other than mouse, then viral gene delivery would be the method of choice due to the insufficient availability of transgenic lines for other species at present.
The ability to achieve robust axon stimulation with light has emerged as an especially desirable feature for ChR2 expression systems. In general, axon terminal field photostimulation is most readily achieved with viral vectors to deliver ChR2 to discrete brain regions and cell types because viral gene delivery methods can achieve exceptionally high transgene expression levels. In addition, the viral gene delivery method is inherently well-suited for restricting transgene expression to restricted volumes of tissue and for targeting discrete cellular populations or nuclei. However, several transgenic lines have achieved sufficient opsin expression to enable robust axon stimulation with light in at least some of the brain regions examined, and several transgenic lines either exhibit or allow for highly restricted transgene expression patterns. To improve the utility of future transgenic lines for axon stimulation experiments in behaving animals it will be ideal to implement variants exhibiting more robust photocurrents such as the recently described ChR2(T159C) mutant [94]. Although axon terminal field stimulation experiments are now widely implemented in the optogenetics field, it is important to note that the physiological relevance of ChR2-mediated axon terminal field stimulation with light is not well established at this time, particularly with respect to behavioral testing. Backpropagating action potentials, unintended axon collateral stimulation, and enhanced synaptic release probability are some of the potential confounds that may accompany axon terminal photostimulation.
The exceptionally high expression levels obtained with viral gene delivery of ChR2 and other opsin variants can be undesirable in some experimental contexts. Care must be taken to select appropriate stimulation paradigms as depolarization block of neuronal firing is readily observed when exceedingly strong depolarization is evoked for more than a few milliseconds. For experiments involving viral gene delivery of ChR2 and optogenetic brain stimulation in behaving animals without feedback from simultaneous electrophysiological recordings, an inappropriate light stimulus (e.g. 1-s constant blue light) may elicit pseudo-silencing in a subpopulation of illuminated cells due to depolarization block of firing (see also [95]). This potential confound has rarely been observed with transgenic lines developed to date. Viral vectors for opsin expression utilizing strong cellular promoters also have the potential for causing toxicity or morphological perturbations with prolonged expression times in vivo, although to a lesser extent as compared with the in utero electroporation method [96]. Although this can generally be overcome by opting for shorter expression times, reducing viral titers, or employing alternative cellular promoter elements, again these confounds appear to be less pronounced with transgenic lines, even with sustained expression of the opsin genes throughout the lifespan of the animal. It is clear that both viral gene delivery methods and transgenic lines each have several benefits and pitfalls, and these approaches should be viewed as complementary rather than competing. Indeed, some recent studies have implemented a combination of these methods to strengthen the central findings of the work [25, 53].
5.5 Silencing modality can perturb normal cellular and circuit function
The development and application of optogenetic silencing strategies has notably lagged behind that of optogenetic stimulation with ChR2 and related variants. One potential reason is that the first implemented optogenetic silencing tool, the light-activated chloride pump NpHR (also called Halorhodopsin), was not very effective due to poor trafficking to the neuronal membrane and propensity for intracellular aggregation. Subsequently, rational molecular engineering has produced highly functional, enhanced versions of NpHR (eNpHR2.0 and eNpHR3.0) [90]. More recently, other silencing opsins have been introduced, including the light-activated proton pumps Arch, ArchT, and Mac [87, 97]. Direct comparative analysis of eNpHR3.0, eArch3.0, eArchT3.0, and eMac3.0 revealed that all are well tolerated in neurons, localize efficiently to the surface membrane, and confer robust hyperpolarizing photocurrents [77], though eMac3.0 was the least effective silencing tool.
Which silencing tool is the most favorable for in vivo applications? Recent work has revealed that NpHR variants may be a poor choice for in vivo applications, including use in the development of transgenic lines. Sustained photostimulation of NpHR-expressing hippocampal pyramidal neurons dramatically perturbs the reversal potential for chloride ions and transiently converts GABA action from hyperpolarizing to depolarizing [98]. This phenomenon has serious implications for analysis of animal behavior using sustained NpHR-based silencing with light. Thus, photoactivation of NpHR should not be regarded as pure silencing mediated by membrane hyperpolarization. In contrast, a transient depolarizing action of GABA can produce enhanced neuronal firing rather than true neuronal silencing during NpHR photostimulation. No effect on the chloride reversal potential was detected with sustained photostimulation of Arch-expressing neurons, as expected since Arch is a proton pump instead of a chloride pump. These findings suggest that Arch and ArchT may be the best suited for optogenetic silencing in vivo, although it is not clear at this time whether extended photostimulation of a light-activated proton pump may have other presently unrecognized adverse impacts on neuronal function. It is suggested that the presence of multiple mechanisms for pH homeostasis may allow neurons to better tolerate proton pumping as opposed to chloride pumping across the cell membrane [98].
6. Future Prospects: next generation optogenetic tools
6.1 SSFO and relevance to behavioral testing with extended time frames and non-invasive light delivery
The parameter of light sensitivity is also of prime importance for in vivo applications with optogenetics. In this regard, an important barrier has already been broken with the introduction of the stabilized step-function opsin (SSFO) variant of ChR2 [1]. This slow kinetic mutant can sustain depolarizing photocurrents for tens of minutes following a single flash of blue light and is inactivated by green/yellow light. This feature enables sustained activation in the absence of sustained light pulses, which may be important for avoiding brain alterations from extended light delivery (e.g. heat related artifacts or damage). It is also well suited for manipulating neuronal activity in behavioral testing paradigms that last tens of minutes or longer for each trial without the need for continuous light delivery during testing. In contrast, conventional ChR2 stimulation with pulsed light throughout a behavioral trial lasting tens of minutes may be cumbersome. Moreover, the SSFO variant is an order of magnitude more sensitive to light activation, which enables functional activation of SSFO-expressing cortical neurons in vivo with light delivery through the surface of the intact skull [1]. This demonstrates that invasive procedures for light delivery are not an obligatory component of optogenetic experimentation (see also [82]).
While many deep brain structures still remain well beyond the reach of light delivered through the skull even when utilizing highly light-responsive engineered opsins, it would seem that SSFO-expressing cells in virtually all cortical layers, and perhaps superficial regions of olfactory bulb and cerebellum may be accessible in rodents using this light delivery method. Further work will be needed to define the maximal volume of brain tissue that can be functionally manipulated with light dispersed across the skull. In principle it may be possible to modulate large areas of widely distributed cortical neuron populations such as cortical fast-spiking interneurons, and this may have implications for optogenetics-based behavioral analysis in animal models of schizophrenia (and perhaps many other brain disorders). It also seems that more work will be required to fully understand the implications and limitations of enhanced light sensitivity for the SSFO variants, especially given that there is currently insufficient evidence to address whether the light sensitivity for channel closure with green/yellow light exhibits a proportional enhancement relative to activation with blue light. Our preliminary work using viral expression of SSFO variants suggests that despite the enhanced sensitivity of activation with blue light, a relatively higher irradiance with yellow light is required to ensure complete and rapid channel closure.
6.2 Further development of engineered opsins for activation and silencing
The recent resolution of the crystal structure of ChR2 [99] now paves the way for rational modification of gene function using mutagenesis. Already a wide array of ChR2 variants have been described which collectively span an impressive range of biophysical properties [100]. The development of fast and slow kinetic mutants of ChR2 enables control of neuronal firing on a timescale spanning several orders of magnitude, from the sub-millisecond (ChETA variants) to tens of minutes with a single light pulse (SSFO variants). Complementary tools for optogenetic neuronal silencing would be of exceptional value in order to gain bidirectional control of neuronal activity on matched time scales and with similar effectiveness. The hyperpolarizing chimeric light-gated channel Hy-Lighter seems a promising candidate non-opsin based transgene for achieving extended bouts of silencing with a single brief light pulse [101]; however, the requirement for co-application of the photoswitched tethered ligand presents a formidable limitation at present.
Secondly, much effort has been extended towards developing engineered opsins with variations in spectral properties. Developing engineered opsins with distinct peak activation wavelengths and narrow activation spectra is a major goal for achieving two-wavelength independent control of neuronal activity using multiple opsins in combination. Proof-of-principle has already been established for activation and silencing of a single population using the combination of ChR2 and NpHR [102], as well as for independent activation of distinct neuronal populations using the combination of ChR2 and VChR1 [56] or ChR2 and the chimera C1V1 [1]. Further refinements will expand the capabilities and robustness of dual-wavelength optogenetic control, as well as potentially enhance practicality of all-optical approaches such as combined optigenetic neuronal control and imaging of neuronal activity or intracellular calcium dynamics. In the context of behavioral studies, it will be especially valuable to develop engineered opsins with peak activation spectra shifted more toward the infrared (IR) as opposed to opsins which are merely capable of some degree of responding to near-IR light. This will enable deeper penetration of light through neural tissue in order to better access deep brain structures, nuclei embedded in heavy myelination, and distributed neuronal populations. One such example is Halo57, a variant of NpHR that exhibits stronger hyperpolarizing photocurrent in response to near-IR light [103]. Given the potential for neuronal modulation with near-IR light, it will be important to carefully evaluate such potential confounds when stimulating opsins with near-IR light.
The most promising new variants of engineered opsins should be rapidly implemented in developing new transgenic lines with cell-type specific expression, or with expression that can be induced by Cre or tTA. As we have discussed, several unique molecular-genetic engineering strategies have been devised in recent years to attempt to overcome the limitation of insufficient opsin expression in transgenic lines (Figure 2), and continued efforts along these lines will ensure rapid progress and continued success in developing highly useful transgenic lines for optogenetics research and beyond. One important feature of these designs is that they are ‘transgene-independent’ and can be generalizable to any transgene of interest requiring high-level expression to achieve full functionality. Additional rigorous testing is warranted to sufficiently validate each of these promising new strategies.
Figure 2. Transgene-independent strategies for enhanced opsin expression in transgenic animals.
(A) Combination of the CAG promoter and WPRE sequence enhances transgene expression from Cre-inducible Rosa26 knock-in alleles. The presence of a floxed STOP cassette prevents opsin-XFP expression in the absence of Cre. Following breeding of the Cre reporter and Cre driver lines, the floxed STOP is excised in Cre-expressing cells to allow for opsin-XFP expression. Please see ref. 74 and ref. 75 for full details. (B) the KENGE-tet improved tTA-inducible transgene expression system utilizes the gene targeting method to insert a tetO-opsin-XFP-pA cassette immediately downstream of the beta-actin (Actb) pA sequence. Following breeding of the tet responsive line with the a cell-type specific tTA driver line, opsin-XFP expression is activated in tTA-expressing cells (ref. 82). (C) Controlling gene dosage with viral 2A peptide linkers. One (1X), two (2X), or three (3X) copies of the opsin gene are linked to an upstream cellular promoter region and downstream reporter gene and polyadenylation (pA) sequence. The 2A peptide linkers ensure separate stoichiometric expression of the opsin and XFP protein. This strategy was modified from ref. 76.
6.3 Development of transgenic rats for optogenetics research
To date there is only a single well-characterized transgenic rat line with functional expression of ChR2 [57]. Despite the immense value of the expanding array of transgenic mouse lines to enable optogenetic probing of brain function, there are notable limitations of using mice in behavioral assays. In particular, it is widely asserted that rats are a superior model for behavioral research involving training paradigms for goal-directed tasks (i.e. operant conditioning). Thus, it is anticipated that efforts directed at developing transgenic rat lines with expression of engineered opsins will greatly facilitate progress in behavioral neuroscience and would represent a welcome addition to existing viral gene delivery methods for achieving functional opsin expression in rats. The recent introduction of the first transgenic rat lines with functional expression of Cre recombinase in the nervous system was a major advance toward enabling precise targeting of opsin expression to genetically-defined neuronal populations in the rat [93]. The ChAT-Cre and tyrosine hydroxylase-Cre (TH-Cre) driver lines were developed by pronuclear injection of BAC DNA that was originally created by the GENSAT project to derive ChAT-Cre and TH-Cre transgenic mouse lines. Stereotaxic injection of Cre-inducible ChR2- EYFP virus into the ventral tegmental area (VTA) of TH-Cre driver rats resulted in functional ChR2 expression in VTA dopamine neurons, thereby imparting the ability to manipulate dopamine neuron activity and behavior with light [93]. This study further demonstrated that optical stimulation of VTA dopamine neurons was sufficient to support robust intracranial self-stimulation.
The successful development of BAC transgenic Cre driver rats establishes the feasibility of using BAC DNA derived from a mouse BAC library to instruct faithful transgene expression in defined cell types in rats. Although BAC libraries of rat genomic DNA are available as source material to create BAC transgenic rats, progress in this endeavor may be rapidly accelerated by re-purposing modified BAC clones that have been successfully utilized to create existing ChR2 BAC transgenic mice. It is also conceivable that this strategy could be implemented to create transgenic lines for ChR2 expression in other mammalian species such as non-human primates. Importantly, the same caveats that apply to BAC transgenic mice also apply to BAC transgenic lines for other species. For example, both the TH-Cre and ChAT-Cre rat lines are predicted to overexpresses ‘extra’ genes based on the selected BAC clones. It will be important to utilize strategies to eliminate unwanted genes from modified BAC DNA in order to develop cleaner transgenic lines with fewer potential confounds. Although Cre-dependent opsin expression has many advantages not adequately discussed herein, the availability of transgenic rat lines with direct opsin expression under cell-type specific promoters would circumvent the need for repeated viral injections in each experimental animal and should be a particularly welcome resource. This avenue represents one of the most exciting future prospects for realizing the full potential of optogenetics in behavioral neuroscience.
Highlights.
Diverse transgenic mouse and rat lines were developed for optogenetic neural control
These lines can help establish a causal role of brain circuits to specific behaviors
Transgenic lines have clear benefits as well as caveats for behavioral studies
New avenues are proposed for developing future lines for optogenetics-based research
Acknowledgements
Portions of the unpublished work included in this manuscript were supported by the Poitras Center for Affective Disorders Research and an American Recovery and Reinvestment Act grant from the US National Institute of Mental Health (RC1-MH088434) to G.F., a National Alliance for Research on Schizophrenia and Depression: The Brain and Behavior Research Foundation Young Investigator award and US National Institutes of Health Ruth L. Kirschstein National Research Service award (F32MH084460) to J.T.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Related Resources
The Optogenetics Wiki http://www.openoptogenetics.org
Optogenetics Resource Center (Deisseroth Lab at Stanford University) http://www.stanford.edu/group/dlab/optogenetics/
Optogenetics – The Jackson Laboratory http://research.jax.org/grs/optogenetics.html
References
- 1.Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O'Shea DJ, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin D, Boyle MP, Dollar P, Lee H, Lein ES, Perona P, et al. Functional identification of an aggression locus in the mouse hypothalamus. Nature. 2011;470:221–226. doi: 10.1038/nature09736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tye KM, Prakash R, Kim SY, Fenno LE, Grosenick L, Zarabi H, et al. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471:358–362. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsunematsu T, Kilduff TS, Boyden ES, Takahashi S, Tominaga M, Yamanaka A. Acute optogenetic silencing of orexin/hypocretin neurons induces slow-wave sleep in mice. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:10529–10539. doi: 10.1523/JNEUROSCI.0784-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter ME, Yizhar O, Chikahisa S, Nguyen H, Adamantidis A, Nishino S, et al. Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nature neuroscience. 2010;13:1526–1533. doi: 10.1038/nn.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith KS, Virkud A, Deisseroth K, Graybiel AM. Reversible online control of habitual behavior by optogenetic perturbation of medial prefrontal cortex. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18932–18937. doi: 10.1073/pnas.1216264109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atasoy D, Betley JN, Su HH, Sternson SM. Deconstruction of a neural circuit for hunger. Nature. 2012;488:172–177. doi: 10.1038/nature11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domingos AI, Vaynshteyn J, Voss HU, Ren X, Gradinaru V, Zang F, et al. Leptin regulates the reward value of nutrient. Nature neuroscience. 2011;14:1562–1568. doi: 10.1038/nn.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aponte Y, Atasoy D, Sternson SM. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nature neuroscience. 2011;14:351–355. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letzkus JJ, Wolff SB, Meyer EM, Tovote P, Courtin J, Herry C, et al. A disinhibitory microcircuit for associative fear learning in the auditory cortex. Nature. 2011;480:331–335. doi: 10.1038/nature10674. [DOI] [PubMed] [Google Scholar]
- 11.Alonso M, Lepousez G, Sebastien W, Bardy C, Gabellec MM, Torquet N, et al. Activation of adult-born neurons facilitates learning and memory. Nature neuroscience. 2012;15:897–904. doi: 10.1038/nn.3108. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber D, Petreanu L, Ghitani N, Ranade S, Hromadka T, Mainen Z, et al. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature. 2008;451:61–64. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haubensak W, Kunwar PS, Cai H, Ciocchi S, Wall NR, Ponnusamy R, et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–276. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goshen I, Brodsky M, Prakash R, Wallace J, Gradinaru V, Ramakrishnan C, et al. Dynamics of retrieval strategies for remote memories. Cell. 2011;147:678–689. doi: 10.1016/j.cell.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 16.Witten IB, Lin SC, Brodsky M, Prakash R, Diester I, Anikeeva P, et al. Cholinergic interneurons control local circuit activity and cocaine conditioning. Science. 2010;330:1677–1681. doi: 10.1126/science.1193771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobo MK, Covington HE, 3rd, Chaudhury D, Friedman AK, Sun H, Damez-Werno D, et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330:385–390. doi: 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuber GD, Sparta DR, Stamatakis AM, van Leeuwen WA, Hardjoprajitno JE, Cho S, et al. Excitatory transmission from the amygdala to nucleus accumbens facilitates reward seeking. Nature. 2011;475:377–380. doi: 10.1038/nature10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamantidis AR, Tsai HC, Boutrel B, Zhang F, Stuber GD, Budygin EA, et al. Optogenetic interrogation of dopaminergic modulation of the multiple phases of reward-seeking behavior. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:10829–10835. doi: 10.1523/JNEUROSCI.2246-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Zessen R, Phillips JL, Budygin EA, Stuber GD. Activation of VTA GABA neurons disrupts reward consumption. Neuron. 2012;73:1184–1194. doi: 10.1016/j.neuron.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai HC, Zhang F, Adamantidis A, Stuber GD, Bonci A, de Lecea L, et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim KM, Baratta MV, Yang A, Lee D, Boyden ES, Fiorillo CD. Optogenetic mimicry of the transient activation of dopamine neurons by natural reward is sufficient for operant reinforcement. PloS one. 2012;7:e33612. doi: 10.1371/journal.pone.0033612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhury D, Walsh JJ, Friedman AK, Juarez B, Ku SM, Koo JW, et al. Rapid regulation of depression-related behaviours by control of midbrain dopamine neurons. Nature. 2013;493:532–536. doi: 10.1038/nature11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tye KM, Mirzabekov JJ, Warden MR, Ferenczi EA, Tsai HC, Finkelstein J, et al. Dopamine neurons modulate neural encoding and expression of depression-related behaviour. Nature. 2013;493:537–541. doi: 10.1038/nature11740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324:354–359. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kravitz AV, Freeze BS, Parker PR, Kay K, Thwin MT, Deisseroth K, et al. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Zhang F, Gradinaru V, Adamantidis AR, Durand R, Airan RD, de Lecea L, et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nature protocols. 2010;5:439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gradinaru V, Thompson KR, Zhang F, Mogri M, Kay K, Schneider MB, et al. Targeting and readout strategies for fast optical neural control in vitro and in vivo. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:14231–14238. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng G, Lu J, Gross J. Generation of transgenic mice. Methods in molecular medicine. 2004;99:255–267. doi: 10.1385/1-59259-770-X:255. [DOI] [PubMed] [Google Scholar]
- 31.Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 32.Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, et al. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- 33.Gong S, Kus L, Heintz N. Rapid bacterial artificial chromosome modification for large-scale mouse transgenesis. Nature protocols. 2010;5:1678–1696. doi: 10.1038/nprot.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 36.Gong S, Yang XW, Li C, Heintz N. Highly efficient modification of bacterial artificial chromosomes (BACs) using novel shuttle vectors containing the R6Kgamma origin of replication. Genome research. 2002;12:1992–1998. doi: 10.1101/gr.476202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic acids research. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nature biotechnology. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 39.Griep AE, John MC, Ikeda S, Ikeda A. Gene targeting in the mouse. Methods Mol Biol. 2011;770:293–312. doi: 10.1007/978-1-61779-210-6_11. [DOI] [PubMed] [Google Scholar]
- 40.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nature neuroscience. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 41.Li X, Gutierrez DV, Hanson MG, Han J, Mark MD, Chiel H, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arenkiel BR, Peca J, Davison IG, Feliciano C, Deisseroth K, Augustine GJ, et al. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–218. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Peca J, Matsuzaki M, Matsuzaki K, Noguchi J, Qiu L, et al. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8143–8148. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caroni P. Overexpression of growth-associated proteins in the neurons of adult transgenic mice. Journal of neuroscience methods. 1997;71:3–9. doi: 10.1016/s0165-0270(96)00121-5. [DOI] [PubMed] [Google Scholar]
- 45.Andrasfalvy BK, Zemelman BV, Tang J, Vaziri A. Two-photon single-cell optogenetic control of neuronal activity by sculpted light. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11981–11986. doi: 10.1073/pnas.1006620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katz Y, Yizhar O, Staiger J, Lampl I. Optopatcher-An electrode holder for simultaneous intracellular patch-clamp recording and optical manipulation. Journal of neuroscience methods. 2013 doi: 10.1016/j.jneumeth.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 47.Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Airan RD, Hu ES, Vijaykumar R, Roy M, Meltzer LA, Deisseroth K. Integration of light-controlled neuronal firing and fast circuit imaging. Current opinion in neurobiology. 2007;17:587–592. doi: 10.1016/j.conb.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thyagarajan S, van Wyk M, Lehmann K, Lowel S, Feng G, Wassle H. Visual function in mice with photoreceptor degeneration and transgenic expression of channelrhodopsin 2 in ganglion cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:8745–8758. doi: 10.1523/JNEUROSCI.4417-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desai M, Kahn I, Knoblich U, Bernstein J, Atallah H, Yang A, et al. Mapping brain networks in awake mice using combined optical neural control and fMRI. Journal of neurophysiology. 2011;105:1393–1405. doi: 10.1152/jn.00828.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim DH, Mohajerani MH, Ledue J, Boyd J, Chen S, Murphy TH. In vivo Large-Scale Cortical Mapping Using Channelrhodopsin-2 Stimulation in Transgenic Mice Reveals Asymmetric and Reciprocal Relationships between Cortical Areas. Frontiers in neural circuits. 2012;6:11. doi: 10.3389/fncir.2012.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ayling OG, Harrison TC, Boyd JD, Goroshkov A, Murphy TH. Automated light-based mapping of motor cortex by photoactivation of channelrhodopsin-2 transgenic mice. Nature methods. 2009;6:219–224. doi: 10.1038/nmeth.1303. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Ackman JB, Xu HP, Crair MC. Visual map development depends on the temporal pattern of binocular activity in mice. Nature neuroscience. 2012;15:298–307. doi: 10.1038/nn.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wentz CT, Bernstein JG, Monahan P, Guerra A, Rodriguez A, Boyden ES. A wirelessly powered and controlled device for optical neural control of freely-behaving animals. Journal of neural engineering. 2011;8:046021. doi: 10.1088/1741-2560/8/4/046021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Llewellyn ME, Thompson KR, Deisseroth K, Delp SL. Orderly recruitment of motor units under optical control in vivo. Nature medicine. 2010;16:1161–1165. doi: 10.1038/nm.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang F, Prigge M, Beyriere F, Tsunoda SP, Mattis J, Yizhar O, et al. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nature neuroscience. 2008;11:631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomita H, Sugano E, Fukazawa Y, Isago H, Sugiyama Y, Hiroi T, et al. Visual properties of transgenic rats harboring the channelrhodopsin-2 gene regulated by the thy-1.2 promoter. PloS one. 2009;4:e7679. doi: 10.1371/journal.pone.0007679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ji ZG, Ito S, Honjoh T, Ohta H, Ishizuka T, Fukazawa Y, et al. Light-evoked somatosensory perception of transgenic rats that express channelrhodopsin-2 in dorsal root ganglion cells. PloS one. 2012;7:e32699. doi: 10.1371/journal.pone.0032699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abe Y, Sekino M, Terazono Y, Ohsaki H, Fukazawa Y, Sakai S, et al. Opto-fMRI analysis for exploring the neuronal connectivity of the hippocampal formation in rats. Neuroscience research. 2012;74:248–255. doi: 10.1016/j.neures.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 60.Dhawale AK, Hagiwara A, Bhalla US, Murthy VN, Albeanu DF. Non-redundant odor coding by sister mitral cells revealed by light addressable glomeruli in the mouse. Nature neuroscience. 2010;13:1404–1412. doi: 10.1038/nn.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hagglund M, Borgius L, Dougherty KJ, Kiehn O. Activation of groups of excitatory neurons in the mammalian spinal cord or hindbrain evokes locomotion. Nature neuroscience. 2010;13:246–252. doi: 10.1038/nn.2482. [DOI] [PubMed] [Google Scholar]
- 62.Zhao S, Ting JT, Atallah HE, Qiu L, Tan J, Gloss B, et al. Cell type-specific channelrhodopsin-2 transgenic mice for optogenetic dissection of neural circuitry function. Nature methods. 2011;8:745–752. doi: 10.1038/nmeth.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim U, Chung LY. Dual GABAergic synaptic response of fast excitation and slow inhibition in the medial habenula of rat epithalamus. Journal of neurophysiology. 2007;98:1323–1332. doi: 10.1152/jn.00575.2007. [DOI] [PubMed] [Google Scholar]
- 64.Halassa MM, Siegle JH, Ritt JT, Ting JT, Feng G, Moore CI. Selective optical drive of thalamic reticular nucleus generates thalamic bursts and cortical spindles. Nature neuroscience. 2011;14:1118–1120. doi: 10.1038/nn.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Varga V, Losonczy A, Zemelman BV, Borhegyi Z, Nyiri G, Domonkos A, et al. Fast synaptic subcortical control of hippocampal circuits. Science. 2009;326:449–453. doi: 10.1126/science.1178307. [DOI] [PubMed] [Google Scholar]
- 66.Hull C, Regehr WG. Identification of an inhibitory circuit that regulates cerebellar Golgi cell activity. Neuron. 2012;73:149–158. doi: 10.1016/j.neuron.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ren J, Qin C, Hu F, Tan J, Qiu L, Zhao S, et al. Habenula "cholinergic" neurons co-release glutamate and acetylcholine and activate postsynaptic neurons via distinct transmission modes. Neuron. 2011;69:445–452. doi: 10.1016/j.neuron.2010.12.038. [DOI] [PubMed] [Google Scholar]
- 68.Ma M, Luo M. Optogenetic activation of Basal forebrain cholinergic neurons modulates neuronal excitability and sensory responses in the main olfactory bulb. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:10105–10116. doi: 10.1523/JNEUROSCI.0058-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zylka MJ, Rice FL, Anderson DJ. Topographically distinct epidermal nociceptive circuits revealed by axonal tracers targeted to Mrgprd. Neuron. 2005;45:17–25. doi: 10.1016/j.neuron.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 70.Wang H, Zylka MJ. Mrgprd-expressing polymodal nociceptive neurons innervate most known classes of substantia gelatinosa neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:13202–13209. doi: 10.1523/JNEUROSCI.3248-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rau KK, McIlwrath SL, Wang H, Lawson JJ, Jankowski MP, Zylka MJ, et al. Mrgprd enhances excitability in specific populations of cutaneous murine polymodal nociceptors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:8612–8619. doi: 10.1523/JNEUROSCI.1057-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smear M, Shusterman R, O'Connor R, Bozza T, Rinberg D. Perception of sniff phase in mouse olfaction. Nature. 2011;479:397–400. doi: 10.1038/nature10521. [DOI] [PubMed] [Google Scholar]
- 73.Katzel D, Zemelman BV, Buetfering C, Wolfel M, Miesenbock G. The columnar and laminar organization of inhibitory connections to neocortical excitatory cells. Nature neuroscience. 2011;14:100–107. doi: 10.1038/nn.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Madisen L, Mao T, Koch H, Zhuo JM, Berenyi A, Fujisawa S, et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nature neuroscience. 2012;15:793–802. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ting JT, Peca J, Daigle TL, Zhao S, Caron MG, Deisseroth K, et al. Ultrafast optogenetic control of diverse neuronal populations with with cre-inducible ChETA knock-in mice. Society for Neuroscience Abstracts. 2012;208.11 [Google Scholar]
- 77.Mattis J, Tye KM, Ferenczi EA, Ramakrishnan C, O'Shea DJ, Prakash R, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nature methods. 2012;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, Hegemann P. Ultrafast optogenetic control. Nature neuroscience. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 79.Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- 80.Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lubbert H, et al. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:10933–10938. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chuhma N, Tanaka KF, Hen R, Rayport S. Functional connectome of the striatal medium spiny neuron. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:1183–1192. doi: 10.1523/JNEUROSCI.3833-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanaka KF, Matsui K, Sasaki T, Sano H, Sugio S, Fan K, et al. Expanding the repertoire of optogenetically targeted cells with an enhanced gene expression system. Cell reports. 2012;2:397–406. doi: 10.1016/j.celrep.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 83.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nature neuroscience. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 84.Bepari AK, Sano H, Tamamaki N, Nambu A, Tanaka KF, Takebayashi H. Identification of optogenetically activated striatal medium spiny neurons by npas4 expression. PloS one. 2012;7:e52783. doi: 10.1371/journal.pone.0052783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sasaki T, Beppu K, Tanaka KF, Fukazawa Y, Shigemoto R, Matsui K. Application of an optogenetic byway for perturbing neuronal activity via glial photostimulation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20720–20725. doi: 10.1073/pnas.1213458109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schoenenberger P, Gerosa D, Oertner TG. Temporal control of immediate early gene induction by light. PloS one. 2009;4:e8185. doi: 10.1371/journal.pone.0008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao S, Cunha C, Zhang F, Liu Q, Gloss B, Deisseroth K, et al. Improved expression of halorhodopsin for light-induced silencing of neuronal activity. Brain cell biology. 2008;36:141–154. doi: 10.1007/s11068-008-9034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain cell biology. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Imayoshi I, Tabuchi S, Hirano K, Sakamoto M, Kitano S, Miyachi H, et al. Light-induced silencing of neural activity in Rosa26 knock-in mice conditionally expressing the microbial halorhodopsin eNpHR2.0. Neuroscience research. 2012 doi: 10.1016/j.neures.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 92.Palmiter RD, Brinster RL. Germ-line transformation of mice. Annual review of genetics. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Witten IB, Steinberg EE, Lee SY, Davidson TJ, Zalocusky KA, Brodsky M, et al. Recombinase-driver rat lines: tools, techniques, and optogenetic application to dopamine-mediated reinforcement. Neuron. 2011;72:721–733. doi: 10.1016/j.neuron.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Berndt A, Schoenenberger P, Mattis J, Tye KM, Deisseroth K, Hegemann P, et al. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7595–7600. doi: 10.1073/pnas.1017210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kravitz AV, Kreitzer AC. Optogenetic manipulation of neural circuitry in vivo. Current opinion in neurobiology. 2011;21:433–439. doi: 10.1016/j.conb.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miyashita T, Shao YR, Chung J, Pourzia O, Feldman DE. Long-term channelrhodopsin-2 (ChR2) expression can induce abnormal axonal morphology and targeting in cerebral cortex. Frontiers in neural circuits. 2013;7:8. doi: 10.3389/fncir.2013.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Han X, Chow BY, Zhou H, Klapoetke NC, Chuong A, Rajimehr R, et al. A high-light sensitivity optical neural silencer: development and application to optogenetic control of non-human primate cortex. Frontiers in systems neuroscience. 2011;5:18. doi: 10.3389/fnsys.2011.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Raimondo JV, Kay L, Ellender TJ, Akerman CJ. Optogenetic silencing strategies differ in their effects on inhibitory synaptic transmission. Nature neuroscience. 2012;15:1102–1104. doi: 10.1038/nn.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kato HE, Zhang F, Yizhar O, Ramakrishnan C, Nishizawa T, Hirata K, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–374. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annual review of neuroscience. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Janovjak H, Szobota S, Wyart C, Trauner D, Isacoff EY. A light-gated, potassium-selective glutamate receptor for the optical inhibition of neuronal firing. Nature neuroscience. 2010;13:1027–1032. doi: 10.1038/nn.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 103.Klapoetke NC. Novel classes of optogenetic reagent derived from screening genomic and ecological diversity Society for Neuroscience Abstracts. 2010 106.1/MMM8. [Google Scholar]