Abstract
microRNAs (miRNAs) are regulators of differentiation and development of inner ear cells. Mutations in miRNAs lead to deafness in humans and mice. Among inner ear pathologies, inflammation may lead to structural and neuronal defects and eventually to hearing loss and vestibular dysfunction. While the genetic factors of these pathways have not been defined, autoimmunity participates in these processes. We report that inflammatory stimuli in the inner ear induce activation of the innate immune system via miR-224 and pentraxin 3 (Ptx3). miR-224 is a transcriptional target of nuclear factor κB, a key mediator of innate immunity. Ptx3 is a regulator of the immune response. It is released in response to inflammation and regulated by nuclear factor κB. We show that miR-224 and Ptx3 are expressed in the inner ear and we demonstrate that miR-224 targets Ptx3. As a model of the innate immune response, we injected lipopolysaccharide into the scala tympani of mouse inner ears. This resulted in changes in the levels of miR-224 and Ptx3, in addition to activation of the complement system, as measured by immune cell infiltration and activated C3. This suggests that while miR-224 regulates Ptx3 under normal conditions, upon inflammation, both are recruited to offer a front line of defense in acting as responders to inflammation in the inner ear. miR-224 diminishes the innate immune response by down-regulating Ptx3 expression, while Ptx3 stimulates the innate immune response. An understanding of the molecular components of the inflammatory pathway may help develop therapeutics for reducing inflammation associated with inner ear injury.
INTRODUCTION
The roles of miRNAs in the auditory and vestibular systems have been of particular interest since hundreds of miRNAs are expressed in the inner ear and mutations in the seed regions of specific miRNAs lead to hearing and balance dysfunction (1–3). miRNAs are ∼23 nucleotide long non-coding RNA molecules that regulate gene expression by binding to the 3′UTR of its mRNA target. This post-transcriptional regulation results in translational repression or, and most commonly, mRNA destabilization (4).
The mammalian ear contains outer, middle and inner ears, each with a specialized role for collecting and converting sounds to enable hearing and for sensing movement and linear acceleration. Within the inner ear, the cochlea and vestibular end organs are responsible for hearing and balance, respectively. Among ear pathologies, inflammation, a response of the innate immune system, may lead to structural, neuronal and vestibular defects (5). In the middle ear, chronic otitis media can, if left untreated, lead to inflammation of the inner ear and sensorineural hearing loss (6). In the inner ear, vestibular neuritis and labyrinthitis can result from infections and lead to both hearing loss and vertigo. For example, hearing loss that occurs in patients with bacterial meningitis results in loss of hair cells or damage to the auditory nerve (7). Several forms of immune-mediated hearing loss are associated with inflammation of the inner ear (5). While the genetic factors that play a role in these forms of hearing loss are not yet elucidated, miRNAs may be involved in regulating gene expression and influence the outcome of inflammation in the ear.
To identify regulatory networks mediating inner ear inflammation, we searched for miRNAs previously identified in a microarray screen derived from cochlear and vestibular sensory epithelium (8) that might be involved in inflammation. We focused on potential miRNA–target interactions. We identified miR-224 as a regulator of Pentraxin 3 (Ptx3) by demonstrating that Ptx3 specifically binds miR-224 and overexpression of miR-224 decreases Ptx3 levels. miR-224 has been implicated previously in cancer, including pancreatic ductal adenocarcinomas (9), colorectal cancer (10) and prostate cancer (11), and linked to tumor invasion and metastasis (12). In particular, it was found to be the most up-regulated miRNA in hepatocellular carcinoma (HCC) (13). The miR-224 promoter has a binding site for the nuclear factor κB (NF-κB), a key transcription factor in the immune response, which activates miR-224 transcription in HCC cells (14). Moreover, miR-224 up-regulation is associated with activation of the lipopolysaccharide (LPS) and NF-κB-dependent LTα and TNFα inflammatory pathways. The pentraxin superfamily of conserved proteins are components of the humoral arm of innate immunity, consisting of both, a short arm that includes pentraxin C reactive protein and serum amyloid P component, together with a long arm that includes Ptx3 (15). The Ptx3 promoter contains an NF-κB binding site and NF-κB is involved in Ptx3 regulation (16). Ptx3 expression is rapidly induced upon response to pro-inflammatory cytokines in various cell types, including epithelial cells and fibroblasts (15). Our study reveals that in the inner ear, Ptx3 is a target of miR-224 and both are mediators of inflammation. As the first miRNA regulatory pathway elucidated for inflammation in the inner ear, this pair is implicated in the inflammatory response of the inner ear as a defense against disease.
RESULTS
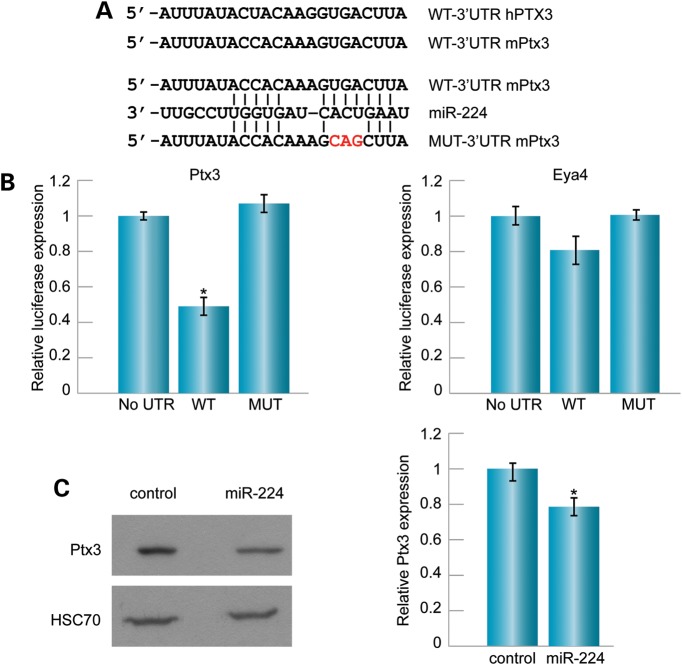
microRNAs previously found to be differentially expressed in auditory and vestibular epithelia (8) were submitted to the publicly available bioinformatic target prediction programs TargetScan, miRanda and Diana, in order to identify miRNA targets in inner ear disease pathways. The computational analysis provided a list of 60 putative targets of miR-224 that overlapped between the three prediction programs (Supplementary Material, Table S1). Immune response-related targets were further defined by the PANTHER Classification System (Supplementary Material, Table S2), and included Ptx3, known to play a key role in the innate immune system and inflammatory response in other systems (15). Both the mouse and human Ptx3 3′UTR possess a putative binding site bearing a high sequence complementarity to the miR-224 seed region, exhibiting high conservation among mammals (Fig. 1A).
Figure 1.
Ptx3 is a target of miR-224. (A) Alignment of miR-224 and mouse Ptx3 binding site in the Ptx3 mRNA 3′UTR. The mutant 3′UTR of Ptx3 for the luciferase activity assay is indicated. (B) HEK-293T cells were co-transfected with the luciferase-expressing vector containing either the wild-type (WT) or mutant (MUT) 3′UTR of Ptx3 or Eya4 and miR-224. For Ptx3, the relative luciferase activity was lower in the WT when compared with the mutant and an empty vector containing no 3′UTR (no UTR), which served as a control. For Eya4, the relative luciferase activity was similar in the WT and the mutant, indicating it is not a target of miR-224. P < 0.05. (C) miR-224 was overexpressed in NIH3T3 cells by transfection with mirVEC-224. Left: a representative western blot demonstrates that Ptx3 was reduced upon miR-224 overexpression relative to transfected cells transfected with an empty vector (control), as measured using the Image J software. HSC70 was used as a loading control. Right: relative Ptx3 expression was measured using the Image J software and following averaging of three independent experiments.
To determine whether miR-224 directly regulates Ptx3 mRNA, the 3′UTR of Ptx3 was cloned into the pGL3 luciferase reporter vector, downstream to the luciferase reporter gene. This construct, together with a miRVEC-224 expression vector and a Renilla vector, was transfected into mammalian HEK293T cells. As seen in Figure 1, overexpression of miR-224 inhibited luciferase expression from the reporter plasmid (Fig. 1B). Mutations introduced into the putative miR-224 binding site of the 3′UTR of Ptx3 abolished the repression, indicating specificity of the binding sites. Another candidate target identified by TargetScan, miRanda and Diana, Eya4, which is associated with human deafness (17), was also tested. The 3′UTR was cloned into the pGL3 luciferase reporter vector, but in this case, the overexpression of miR-224 did not show significant differences in luciferase activity between the wild-type and the mutant vectors (Fig. 1B). To test whether miR-224 affects the level of endogenous Ptx3 protein, NIH3T3 cells were transfected with the miRVEC-224 expression vector. Overexpression of miR-224 led to a 21.6% reduction in Ptx3 levels compared with cells transfected with an empty vector (Fig. 1C).
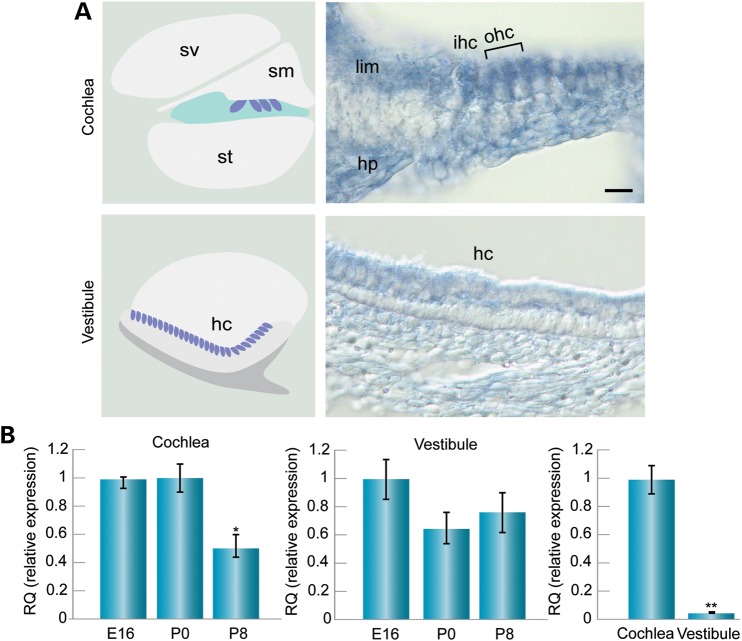
To determine which developmental stage and cell type of the inner ear are influenced by miR-224 and Ptx3 mRNA regulation, we determined the expression levels of both factors using quantitative real-time (qRT) PCR, in situ hybridization and immunohistochemistry. In situ hybridization of miR-224 on whole mount preparations of mouse inner ears using mirCURY™ LNA probes revealed expression in mouse sensory epithelia of the cochlea and vestibule, with higher expression in the cochlea (Fig. 2A). In the cochlea, miR-224 was localized specifically to the hair cells and surrounding supporting cells. Expression was also seen in the spiral limbus, a thickened mass of periosteal connective tissue of the osseous spiral lamina and in the region of the habenula perforate, where the nerve fibers reach or leave the organ of Corti. In the saccule, signal was detected specifically in the hair and supporting cell layer of the sensory epithelium, while the underlying mesenchymal cells were devoid of signal. To further analyze expression during different developmental stages, miR-224 levels were assessed from fetal to neonatal stages of sensory epithelia. In the cochlea, miR-224 was expressed at embryonic day (E)16, post-natal day (P)0 and P8, with a reduction at P8 relative to E16 and P0 (Fig. 2B). In the vestibular organs, expression did not change significantly over time (Fig. 2B). However, comparing the expression in the cochlea versus the vestibule at P0, revealed a 33-fold higher miR-224 expression in the cochlea (P < 0.005) (Fig. 2B).
Figure 2.
miR-224 expression in the mouse inner ear sensory epithelia. (A) Schematic diagram of the cochlea and vestibule sensory epithelia (left) and representative miR-224 in situ hybridization (right) of P0 mouse inner ear. In the cochlea, miR-224 is expressed in the hair and supporting cells, as well as in the spiral limbus and the region of the habenula perforate. In the vestibule, expression is detected in the hair cells of the saccule. Scale bar: 20 mm. (B) qRT–PCR analysis of miR-224 in the mouse cochlea sensory epithelia (left panel). miR-224 exhibited higher expression at younger ages, at E16 and P0, and lower expression at P8 in cochlea sensory epithelia. *P < 0.05, whereas the levels were similar at these ages in the vestibule (middle panel). At P0, expression was 33-fold higher in the cochlear than the vestibular sensory epithelia **P < 0.005 (right panel). sv, scala vestibuli; sm, scala media; st, scala tympani; ihc, inner hair cells; ohc, outer hair cells; hc, hair cells; lim, spiral limbus; hp, habenula perforate.
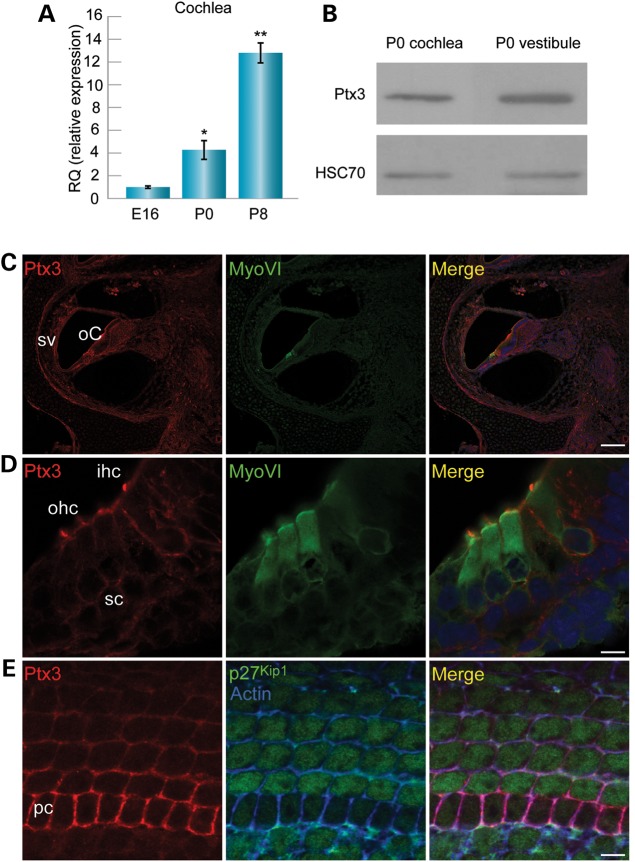
Temporal expression of Ptx3 mRNA was evaluated by qRT–PCR in the cochlear sensory epithelium. Ptx3 expression increased with age in the cochlea; at P0, expression was 4-fold higher than at E16 and at P8 expression was 13-fold higher than at E16 (Fig. 3A). To define the protein expression pattern of Ptx3 in the inner ear, an antibody against Ptx3, previously used in corpus luteum (18) and parietal peritoneum (19), was applied on cochlear and vestibular tissues. Expression of Ptx3 was detected in protein extracted from cochlea and vestibule separated from dissected inner ears (Fig. 3B). In sections of P0 mouse inner ears, Ptx3 was found in the organ of Corti and stria vascularis (Fig. 3C). The antibody stained the boundaries between hair and supporting cells, as well as hair cell stereocilia (Fig. 3D). In whole mount preparations, a view of the supporting cell layer, indicated by P27kip1 staining in the supporting cells, shows that Ptx3 appears strongest in the lateral membrane of the supporting cells, and in particular, in the pillar cells (Fig. 3E).
Figure 3.
Ptx3 expression in the mouse inner ear. (A) qRT–PCR showing the relative Ptx3 transcript expression in mouse cochlear sensory epithelia. Ptx3 exhibited increasing expression with age. At P0, expression was four-fold higher than at E16; at P8 expression was 13-fold higher than at E16. n = 5; *P < 0.05. **P < 0.005. (B) Ptx3 protein expression as measured by western blot analysis. A representative blot comparing Ptx3 expression in cochlear and vestibular tissues. HSC70 used as a loading control. (C) Immunohistochemistry of Ptx3 in P0 inner ear sections. Ptx3 (red) was localized to the sensory epithelium in the organ of Corti and to the stria vascularis. Hair cells stained by myosin VI (green) and nuclei stained by Draq5 (blue). Scale bar: 100 µm. (D) Ptx3 (red) was found in the cellular outline of hair and supporting cells. Hairs cells stained by myosin VI (green) and nuclei stained by Draq5 (blue). Scale bar: 7.5 µm. (E) Ptx3 localization is most prominent surrounding the supporting pillar cells. Scale bar: 7.5 µm.Ptx3 stained with an anti-Ptx3 antibody (red), supporting cells stained by p27kip1 (green), and actin stained by phalloidin (blue). sv, stria vascularis; oC, organ of Corti; ihc, inner hair cells; ohc, outer hair cells; hc, hair cells; sc, supporting cells; pc, pillar cells.
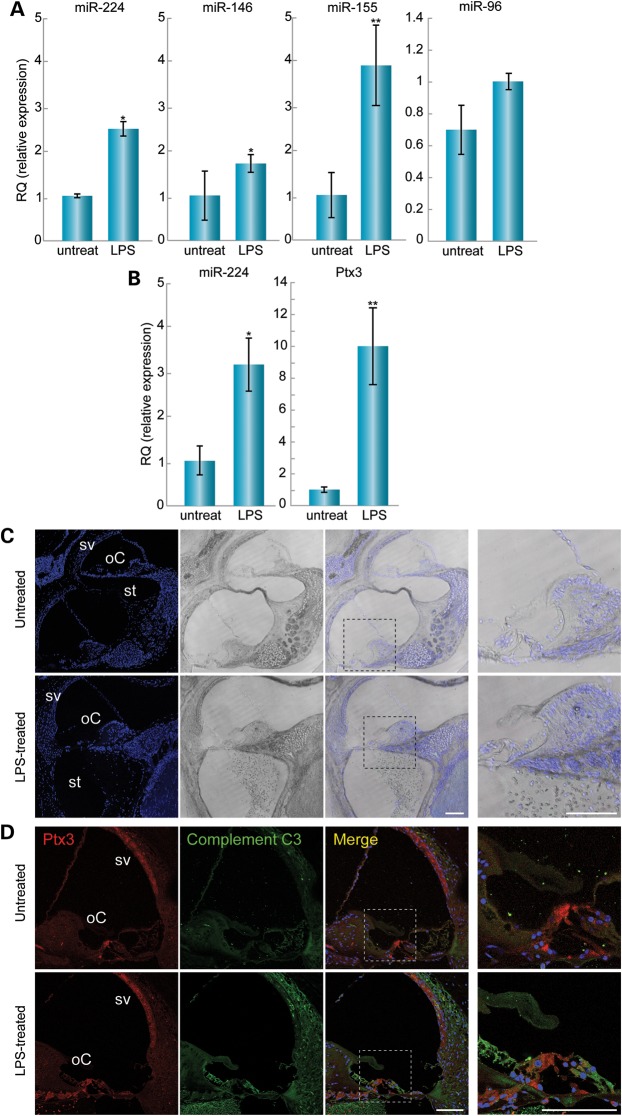
The innate immune response is known to regulate miRNA levels (20). Since Ptx3 is involved in the inflammatory response and in the recruitment of the complement system (15), we aimed to induce inflammation in the inner ear in order to examine the involvement of miR-224 and Ptx3 in this process. LPS was injected directly into the scala tympani of adult mice in one ear, with the contralateral ear serving as a control. The control was untreated, since a mock injection and drilling into the bulla would lead to inflammation in the control ear as well. Inner ears were harvested 24 or 72 h following injection, followed by RNA extraction. We assessed the expression by qRT–PCR of miR-224, as well as miR-146 (21) and miR-155 (20), two miRNAs known to be up-regulated as a consequence of exposure to inflammatory mediators (Fig. 4A). Twenty-four hours after LPS treatment, miR-224 levels were 2.5-fold (P < 0.05) higher in the LPS-treated ears, when compared with the non-treated ears. miR-146 and miR-155 were increased by 1.7-fold (P < 0.05) and by 4-fold (P < 0.005), respectively. Seventy-two hours after LPS injection, miR-224 expression levels increased by 3-fold (P < 0.05) (Fig. 4B). To confirm that there was no bias in the up-regulation of the miRNAs examined, miR-96, a hair-cell-specific miRNA, showed no significant difference between the LPS-treated and the non-treated ears (Fig. 4A). Levels of Ptx3 mRNA, evaluated by qRT–PCR, increased by 10-fold in the LPS treated versus the control ears (P < 0.005) (Fig. 4B).
Figure 4.
LPS-induced inflammation in the inner ear leads to changes in RNA expression and activation of the immune system. (A) At 24 h after LPS injection, miR-224 expression was increased by 2.5-fold in the LPS-treated (n = 4) when compared with non-treated (n = 4) ears. *P < 0.05. miR-146 expression was increased by 1.7-fold in the LPS-treated (n = 6) when compared with non-treated (n = 6) ears. *P < 0.05. miR-155 expression was increased by four-fold in the LPS treated (n = 6) when compared with non-treated (n = 6) ears. (**) P < 0.005. There was no significant change in miR-96 expression between the LPS treated (n = 3) and non-treated (n = 3) ears. (B) At 72 h after LPS injection, miR-224 expression was increased by three-fold in the LPS-treated (n = 5) when compared with non-treated (n = 5) ears. (*) P < 0.05. Ptx3 expression was increased by 10-fold in the LPS treated (n = 3) when compared with non-treated (n = 3) ears. (**) P < 0.005. (C) Draq5-stained nuclei in the LPS-treated and non-treated inner ear sections. No morphological changes were observed. Cells were present in the scala tympani in the LPS-treated ears, indicating immune cell infiltration. The inset represents the organ of Corti enlarged for optimal visualization (far right images). Scale bar: 100 µm. (D) At 24 h after LPS injection, Ptx3 staining was observed in the sensory epithelia, mostly in pillar cells in the organ of Corti and in the stria vascularis. Stronger staining was seen in the LPS treated when compared with the non-treated inner ears. Staining with a complement C3 antibody was stronger in the LPS treated when compared with the non-treated inner ears, indicating activation of the immune system. The inset represents the organ of Corti enlarged for optimal visualization (far right images). oC, organ of Corti; sv, stria vascularis. Scale bar: 7.5 µm.
To further evaluate the changes in Ptx3 protein during inflammation, LPS-treated and non-treated ears were embedded in paraffin blocks and sectioned. No morphological changes were observed by immunohistochemistry in these sections. Infiltration of immune cells into the scala tympani was observed in the LPS-treated ears (Fig. 4C). Ptx3 staining was observed in the sensory epithelia, mostly in the pillar cells, and in the stria vascularis (Fig. 4D). Stronger staining was seen in the LPS-treated ears, specifically in the sensory epithelia, stria vascularis and the spiral ganglia, thus ruling out the possibility that the increased Ptx3 levels are due to the infiltration of Ptx3 positive immune cells upon LPS stimulation. Although there is 10-fold increase in Ptx3 mRNA (Fig. 4B), there does not appear to be as great of an increase in Ptx3 protein, consistent with the increase in miR-224 expression and its role in repressing Ptx3 protein expression. As expected, C3 deposition, indicative of activation of the complement system, was stronger in the LPS-treated ears (Fig. 4D), thereby indicating that the dosage of LPS used in the study was sufficient to induce robust inflammation.
DISCUSSION
We suggest that miR-224, along with NF-κB transcriptional regulation, is part of the inflammation pathway in the inner ear and that miR-224 and Ptx3 form a feedback loop to protect the inner ear against the damage induced by inflammation. Our findings demonstrate that miR-224 targets Ptx3 and provides partial inhibition of its expression. Under normal conditions, miR-224 may help control the levels of Ptx3, along with NF-κB transcriptional regulation. miRNAs are considered to function in coordination with transcriptional regulation to further define development and cellular identity, providing robustness (22).
Our data also demonstrated that LPS-induced inflammation in the inner ear led to an increase in both miR-224 and Ptx3. This is in line with the fact that upon exposure to inflammation, cells release Ptx3 upon stimulation with pro-inflammatory cytokines (IL-1, TNF) or with microbial components (such as LPS) (15). Furthermore, inflammatory stimuli have an influence on gene expression via the NF-κB family of transcription factors, key players in the immune response (23). One of the multiple connections between the innate immune response and the tight regulation that is required is via the NF-κB binding site in the promoter of Ptx3.
Our study proposes new players in the inner ear inflammatory response. Ptx3, a key regulator of the immune response in other systems, is presented here as a functional protein in the inner ear immune response as well. Its role in the inner ear may be similar to other systems where it acts in the recruitment of the complement system and initiates the immune cascade (24). We show here that Ptx3 is expressed in both cell types of the sensory epithelia, the hair cells and supporting cells. Hair cells are highly specialized sensory cells, while supporting cells are non-sensory cells that reside between hair cells with a wide range of functions, including a role in regeneration (25). Our data provide evidence that miR-224 plays a role in the regulation of Ptx3 expression by directly targeting its 3′UTR and thus, mir-224 is a participant in the cell's inflammatory response.
Both miR-224 and Ptx3 have a co-localized expression pattern in the organ of Corti, as demonstrated by qRT–PCR analysis, but while miR-224 decreases with age, Ptx3 increases with age. In particular, demonstrated by immunohistochemistry, both miR-224 and Ptx3 are expressed in hair and supporting cells. Yet, Ptx3 localization is restricted to the periphery of the cell, possibly due to its function to be secreted upon induction of inflammation. As Ptx3 recruits immune system factors, both Ptx3 and miR-224 are required to be present in the same cells at the same time point.
The fact that both Ptx3 and miR-224 promoters have a binding site for NF-κB (14,16) is intriguing, since it implies they are co-expressed. To understand how NF-κB is both responsible for the transcription of Ptx3 and down-regulates it through the induction of miR-224, we should look into the role of miRNA molecules. miRNAs tend to operate by fine tuning their mRNA targets (4). Therefore, a powerful protein that acts upstream of the cascade of the complement immune response, such as Ptx3, has to be subjected to close regulation, hereby offered by miR-224. We propose a functional mechanism of the inner ear inflammatory response, orchestrated by NF-κB, Ptx3 and miR-224. NF-κB is induced by IL-1β and TNFα upon inflammation, and then up-regulates the expression of Ptx3 and miR-224. In this manner, NF-κB activates the complement immune cascade and regulates it by fine-tuning Ptx3. As a result, miR-224 and Ptx3 both act in a regulatory loop in the NF-κB pathway. It is expected there are other players here as well. TargetScan predicted many potential targets for miR-224, and other miRNAs that complement the Ptx3 3′UTR, implicating these two in a larger and more complex system.
We described a similar direction of change for miR-224 and Ptx3 in the LPS-treated ears. Since both miR-224 and Ptx3 are induced by the transcription factor NF-κB, their expression may be induced simultaneously during inflammation, due to the expression of NF-κB in this course of action. Our model involves a short scale, acute inflammatory response. There may be changes in the miR-224 and Ptx3 expression levels or in the directions of change during the progression of the immune response, since there are more factors other than NF-κB that regulate expression of both miR-224 and Ptx3. Although inner ear inflammation caused by chronic otitis media also involves NF-κB, we speculate that the intensity of these immune factors might change (26). This scenario might be different in autoimmune inner ear inflammation, which is non-LPS mediated (27).
A previous study showed that miR-224 directly targets Smad4 (28). In the inner ear, Smad4 has an important role during development. Chondrocyte-specific Smad4 knockout mice had abnormalities in cochlear structure, hair cell development, vestibular organ structure and exhibited sensorineural hearing loss (29). Smad4 is a major mediator in the TGFβ anti-inflammatory signaling pathway (30). Upon TGFβ induction, Smad4 (Co-Smad), creates a complex with other Smad family proteins, and acts in the recruitment of transcription factors that induce anti-inflammatory TGFβ target genes (31). These finding suggest that miR-224, by down-regulating Smad4, might be involved in the anti-inflammatory pathway of the inner ear.
In conclusion, we found that miR-224 has a key presence in the inner ear. We propose a miR-224 - Ptx3 regulatory axis that ablates inflammation and blunts the cytokine storm, thereby protecting the inner ear from damage. This regulatory mechanism is biologically relevant as hair cells are mitotically quiescent and given the importance of the hearing phenotype, we speculate that complex regulatory systems have evolved to protect the inner ear from damage during an individual's lifetime. This finding sheds light on the role of miRNAs in the complex regulation of the inflammatory pathway in the inner ear. Given the influence of inflammation in inner ear disease, a better understanding of the molecular factors controlling inner ear inflammation may lead to optimal care and treatment. miR-224 activation in the inner ear could therefore improve therapeutic treatments aimed at reducing inflammation, by further influencing Ptx3.
MATERIALS AND METHODS
Target prediction
MicroRNA targets were predicted using the overlap of three bioinformatics programs: targetScanMouse, Release 5.2 (http://www.targetscan.org/mmu_50/), miRanda (http://www.microrna.org/microrna/home.do) and Diana (http://diana.imis.athena-innovation.gr).
To search for immune response-related targets, predicted targets were submitted to the PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System (http://www.pantherdb.org/), and immune response-related targets were further considered.
Animal handling and dissections
Procedures involving animals met the guidelines defined by the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Animal Care and Use Committees of Tel Aviv University (M-12-046) and the University of Michigan Institutional Committee on the Care and Use of Animals (UCUCA). Cochlear and vestibular sensory epithelia were dissected from P0 wild-type C57Bl/6J mice and collected separately. The vestibular epithelia consisted of the saccule, utricle and the lateral and anterior cristae. Both the cochlear and vestibular sensory epithelia were dissected with their underlying mesenchyme and attached neurons.
miRNA overexpression and protein analysis
NIH3T3 cells, grown under blasticidin selection, were transfected with the miR-224 miR-VEC expressing vector, or an empty miR-VEC vector, which served as a control. Proteins were extracted from harvested cells using NP40 with a phosphatase inhibitor and quantified using the Quick Start™ Bradford Protein Assay (BioRad). Lysates were separated by 8% SDS polyacrylamide gel, and transferred to PVDF membrane by electrophoresis. Western blots were analyzed using the rabbit-anti-Ptx3 human antibody (Abcam) and mouse anti-HSC70 antibody (Santa Cruz Biotechnology) as a loading control. Ptx3 relative quantities were calculated using the Image J software (32).
Luciferase assays
A portion of the 3′UTR of each target was cloned into the pGL3 vector, downstream to the firefly luciferase gene (Promega). To produce the Ptx3 luciferase construct, a segment of 140 bp upstream to the first miR-224 binding site and 65 bp downstream to the second binding site of the 3′-UTR was cloned. To produce the Eya4 luciferase construct, a segment of 300 bp upstream to the miR-224 first binding site and 350 bp downstream to the second binding site of the 3′-UTR was cloned. For Ptx3 and Eya4, both of the predicted sites were cloned. The first three nucleotides of both miR-224 binding sites in the 3′UTR of each target were mutated using the QuikChange® Site-Directed Mutagenesis Kit (Stratagene). The miR-224 expression vector was created by cloning the pre-miR-224 region into miR-VEC (gift from Reuven Agami). HEK293T cells were transfected with the wild-type or mutant 3′UTR, or empty vector, for each of the targets, together with the miR-224 expression vector and Renilla expressing vector. Lysates were collected after 48 h after transfection and Renilla and firefly luciferase activities were measured with the Dual-Luciferase Reporter System (Promega). The relative firefly luciferase/Renilla activity ratios were calculated for each sample and normalized. Three biological repeats were performed.
In situ hybridization
C57BL/6 P0 mice inner ears were dissected and fixed in 4% paraformaldehyde (PFA). Whole mount in situ hybridization was performed on whole mount inner ears using miRCURY LNA™ microRNA Detection Probes following the manufacturer's protocol (Exiqon). Digoxygenin (DIG)-labeled probes were incubated with the samples at 20–22°C below the melting temperature of the probe. Probes were detected with an alkaline phosphatase-conjugated anti-DIG antibody (Roche). Color reaction was developed by adding NTB/BCIP (Sigma). The ears were cryo-sectioned and mounted. Images were taken using the Zeiss Axiovert200M microscope. Three independent ISH experiments were performed; n = 3 inner ears for each.
RNA preparation and qRT–PCR
Whole inner ear or sensory epithelium of the cochlea and vestibule were dissected, frozen in liquid nitrogen and stored at −80°C. Total RNA containing small RNA was extracted by either the RNeasy Plus Mini Kit (Qiagen) for whole inner ear or RNeasy Micro Kit (Qiagen) for sensory epithelium. cDNA was prepared with the Taqman® Reverse Transcription Kit (Applied Biosystems). miRNA and mRNA expression was evaluated using the TaqMan® microRNA Assays and TaqMan® Gene Expression Assay (Applied Biosystems) in the StepOne™ Real-Time PCR System (Applied Biosystems). miRNA expression was normalized to U6B and mRNA expression was normalized to TBP. Three biological repeats were performed, in triplicate (n = 3 ears).
Immunohistochemistry
Whole mount and section immunohistochemistry was prepared as previously described (33). Primary antibodies and dilutions were used as follows: rabbit-anti-Ptx3 (Abcam) antibody 1:50 for whole mount, 1:200 for sections, goat-anti-active C3 (34). The DRAQ5 nuclear marker was used to label nuclei (Abcam).
LPS in vivo injections
P30 mice were anesthetized using ketamine (120 mg/kg) and xylazine (7 mg/kg) IP. The inner ear of the left ear was exposed by an incision behind the pinna and the muscles covering the ear were cut to expose the bulla and avoid damage to the facial nerve. The bulla was then opened by first piercing using a 26-gauge needle and then expanded using forceps, allowing for easy access to the round window. A small polyethylene tube was connected on one side to a fine polyimide tube and on the other to a 30-gauge 10 µl Hamilton syringe filled with sterile water and placed in a micropump, the tube was filled with 2 µl of LPS (1 mg/ml). The tube was positioned in the window and advanced towards the ST in order to make sure the round window membrane was broken and the LPS was injected directly into the ST and did not leak out (1 µl per min). The tube was left in place for an extra 2 min and then covered with a small piece of muscle from adjacent tissue. The wound was closed using 5-0 suture. After the procedure, mice were treated with ketoprofen (5 mg/kg) to control the pain and were left under a heating lamp until full recovery from the anesthesia. The mice were sacrificed 24 h or 72 h later, treated and control inner ears were extracted and either processed for RNA extraction or for paraffin sections using 4% PFA and 5% EDTA overnight, then washed and placed in EDTA for 1 day.
Statistics
Student's two-tailed t-test P-values of <0.05 were considered to be statistically significant and those of <0.005 were considered to be highly statistically significant. The data in the figures are presented by means ± SEM.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Israel Science Foundation to K.B.A (grant number 1320/11); Israeli Centers of Research Excellence (I-CORE) and ISF Gene Regulation in Complex Human Disease Center to K.B.A (grant number 41/11), National Institutes of Health/National Institutes on Deafness and Other Communicative Disorders (NIDCD) grant to Y.R. (P30-DC05188) and the Berte and Alan Hirschfield Foundation to K.B.A. and Y.R.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Eithan Galun, Carmit Levy, Ram-Shankar Mani and Zvi Fishelson for helpful discussions and critical review of the manuscript.
Conflict of Interest statement: None declared.
REFERENCES
- 1.Friedman L.M., Dror A.A., Mor E., Tenne T., Toren G., Satoh T., Biesemeier D.J., Shomron N., Fekete D.M., Hornstein E., et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc. Natl Acad. Sci. USA. 2009;106:7915–7920. doi: 10.1073/pnas.0812446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mencia A., Modamio-Hoybjor S., Redshaw N., Morin M., Mayo-Merino F., Olavarrieta L., Aguirre L.A., del Castillo I., Steel K.P., Dalmay T., et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 3.Lewis M.A., Quint E., Glazier A.M., Fuchs H., De Angelis M.H., Langford C., van Dongen S., Abreu-Goodger C., Piipari M., Redshaw N., et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat. Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo H., Ingolia N.T., Weissman J.S., Bartel D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan A.F., Harris J.P., Keithley E.M. Immune-mediated hearing loss: basic mechanisms and options for therapy. Acta Otolaryngol. Suppl. 2002;548:38–43. doi: 10.1080/00016480260094965. [DOI] [PubMed] [Google Scholar]
- 6.Cureoglu S., Schachern P.A., Rinaldo A., Tsuprun V., Ferlito A., Paparella M.M. Round window membrane and labyrinthine pathological changes: an overview. Acta Otolaryngol. 2005;125:9–15. doi: 10.1080/00016480410022534. [DOI] [PubMed] [Google Scholar]
- 7.Nadol J.B., Jr. Hearing loss. N. Engl. J. Med. 1993;329:1092–1102. doi: 10.1056/NEJM199310073291507. [DOI] [PubMed] [Google Scholar]
- 8.Elkan-Miller T., Ulitsky I., Hertzano R., Rudnicki A., Dror A.A., Lenz D.R., Elkon R., Irmler M., Beckers J., Shamir R., et al. Integration of transcriptomics, proteomics, and microRNA analyses reveals novel microRNA regulation of targets in the mammalian inner ear. PLoS One. 2011;6:e18195. doi: 10.1371/journal.pone.0018195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mees S.T., Mardin W.A., Sielker S., Willscher E., Senninger N., Schleicher C., Colombo-Benkmann M., Haier J. Involvement of CD40 targeting miR-224 and miR-486 on the progression of pancreatic ductal adenocarcinomas. Ann. Surg. Oncol. 2009;16:2339–2350. doi: 10.1245/s10434-009-0531-4. [DOI] [PubMed] [Google Scholar]
- 10.Arndt G.M., Dossey L., Cullen L.M., Lai A., Druker R., Eisbacher M., Zhang C., Tran N., Fan H., Retzlaff K., et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer. 2009;9:374. doi: 10.1186/1471-2407-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prueitt R.L., Yi M., Hudson R.S., Wallace T.A., Howe T.M., Yfantis H.G., Lee D.H., Stephens R.M., Liu C.G., Calin G.A., et al. Expression of microRNAs and protein-coding genes associated with perineural invasion in prostate cancer. Prostate. 2008;68:1152–1164. doi: 10.1002/pros.20786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu S., Sachdeva M., Wu F., Lu Z., Mo Y.Y. Ubc9 promotes breast cell invasion and metastasis in a sumoylation-independent manner. Oncogene. 2010;29:1763–1772. doi: 10.1038/onc.2009.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Lee A.T., Ma J.Z., Wang J., Ren J., Yang Y., Tantoso E., Li K.B., Ooi L.L., Tan P., et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J. Biol. Chem. 2008;283:13205–13215. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 14.Scisciani C., Vossio S., Guerrieri F., Schinzari V., De Iaco R., D'Onorio de Meo P., Cervello M., Montalto G., Pollicino T., Raimondo G., et al. Transcriptional regulation of miR-224 upregulated in human HCCs by NFκB inflammatory pathways. J. Hepatol. 2012;56:855–861. doi: 10.1016/j.jhep.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Bottazzi B., Doni A., Garlanda C., Mantovani A. An integrated view of humoral innate immunity: Pentraxins as a paradigm. Annu. Rev. Immunol. 2010;28:157–183. doi: 10.1146/annurev-immunol-030409-101305. [DOI] [PubMed] [Google Scholar]
- 16.Basile A., Sica A., d'Aniello E., Breviario F., Garrido G., Castellano M., Mantovani A., Introna M. Characterization of the promoter for the human long pentraxin PTX3. Role of NF-κB in tumor necrosis factor-α and interleukin-1β regulation. J. Biol. Chem. 1997;272:8172–8178. doi: 10.1074/jbc.272.13.8172. [DOI] [PubMed] [Google Scholar]
- 17.Wayne S., Robertson N.G., DeClau F., Chen N., Verhoeven K., Prasad S., Tranebjarg L., Morton C.C., Ryan A.F., Van Camp G., et al. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum. Mol. Genet. 2001;10:195–200. doi: 10.1093/hmg/10.3.195. [DOI] [PubMed] [Google Scholar]
- 18.Mondal M., Schilling B., Folger J., Steibel J.P., Buchnick H., Zalman Y., Ireland J.J., Meidan R., Smith G.W. Deciphering the luteal transcriptome: potential mechanisms mediating stage-specific luteolytic response of the corpus luteum to prostaglandin F2α. Physiol. Genomics. 2011;43:447–456. doi: 10.1152/physiolgenomics.00155.2010. [DOI] [PubMed] [Google Scholar]
- 19.Kanda R., Hamada C., Kaneko K., Nakano T., Wakabayashi K., Io H., Horikoshi S., Tomino Y. Pentraxin 3 as a new biomarker of peritoneal injury in peritoneal dialysis patients. J. Artif. Organs. 2013;16:66–73. doi: 10.1007/s10047-012-0663-3. [DOI] [PubMed] [Google Scholar]
- 20.O'Connell R.M., Taganov K.D., Boldin M.P., Cheng G., Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl Acad. Sci. USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng H.S., Sivachandran N., Lau A., Boudreau E., Zhao J.L., Baltimore D., Delgado-Olguin P., Cybulsky M.I., Fish J.E. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol. Med. 2013;5:949–966. doi: 10.1002/emmm.201202318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebert M.S., Sharp P.A. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oeckinghaus A., Ghosh S. The NF-kB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009;1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantovani A., Garlanda C., Doni A., Bottazzi B. Pentraxins in innate immunity: from C-reactive protein to the long pentraxin PTX3. J. Clin. Immunol. 2008;28:1–13. doi: 10.1007/s10875-007-9126-7. [DOI] [PubMed] [Google Scholar]
- 25.Wan G., Corfas G., Stone J.S. Inner ear supporting cells: Rethinking the silent majority. Semin. Cell Dev. Biol. 2013;24:448–459. doi: 10.1016/j.semcdb.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghaheri B.A., Kempton J.B., Pillers D.A., Trune D.R. Cochlear cytokine gene expression in murine acute otitis media. Laryngoscope. 2007;117:22–29. doi: 10.1097/01.mlg.0000240170.48584.73. [DOI] [PubMed] [Google Scholar]
- 27.Bovo R., Ciorba A., Martini A. Vertigo and autoimmunity. Eur. Arch. Otorhinolaryngol. 2010;267:13–19. doi: 10.1007/s00405-009-1122-5. [DOI] [PubMed] [Google Scholar]
- 28.Yao G., Yin M., Lian J., Tian H., Liu L., Li X., Sun F. MicroRNA-224 is involved in transforming growth factor-β-mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Mol. Endocrinol. 2010;24:540–551. doi: 10.1210/me.2009-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang S.M., Hou Z.H., Yang G., Zhang J.S., Hu Y.Y., Sun J.H., Guo W.W., He D., Han D.Y., Young W.Y., et al. Chondrocyte-specific Smad4 gene conditional knockout results in hearing loss and inner ear malformation in mice. Dev. Dyn. 2009;238:1897–1908. doi: 10.1002/dvdy.22014. [DOI] [PubMed] [Google Scholar]
- 30.Letterio J.J., Roberts A.B. Regulation of immune responses by TGF-β. Annu. Rev. Immunol. 1998;16:137–161. doi: 10.1146/annurev.immunol.16.1.137. [DOI] [PubMed] [Google Scholar]
- 31.Hanada T., Yoshimura A. Regulation of cytokine signaling and inflammation. Cytokine Growth Factor Rev. 2002;13:413–421. doi: 10.1016/s1359-6101(02)00026-6. [DOI] [PubMed] [Google Scholar]
- 32.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dror A.A., Politi Y., Shahin H., Lenz D.R., Dossena S., Nofziger C., Fuchs H., Hrabe de Angelis M., Paulmichl M., Weiner S., et al. Calcium oxalate stone formation in the inner ear as a result of an Slc26a4 mutation. J. Biol. Chem. 2010;285:21724–21735. doi: 10.1074/jbc.M110.120188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Attali G., Gancz D., Fishelson Z. Increased sensitivity of early apoptotic cells to complement-mediated lysis. Eur. J. Immunol. 2004;34:3236–3245. doi: 10.1002/eji.200425011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.