Abstract
Large expansions of a CGG-repeat element (>200 repeats; full mutation) in the fragile X mental retardation 1 (FMR1) gene cause fragile X syndrome (FXS), the leading single-gene form of intellectual disability and of autism spectrum disorder. Smaller expansions (55–200 CGG repeats; premutation) result in the neurodegenerative disorder, fragile X-associated tremor/ataxia syndrome (FXTAS). Whereas FXS is caused by gene silencing and insufficient FMR1 protein (FMRP), FXTAS is thought to be caused by ‘toxicity’ of expanded-CGG-repeat mRNA. However, as FMRP expression levels decrease with increasing CGG-repeat length, lowered protein may contribute to premutation-associated clinical involvement. To address this issue, we measured brain Fmr1 mRNA and FMRP levels as a function of CGG-repeat length in a congenic (CGG-repeat knock-in) mouse model using 57 wild-type and 97 expanded-CGG-repeat mice carrying up to ∼250 CGG repeats. While Fmr1 message levels increased with repeat length, FMRP levels trended downward over the same range, subject to significant inter-subject variation. Human comparisons of protein levels in the frontal cortex of 7 normal and 17 FXTAS individuals revealed that the mild FMRP decrease in mice mirrored the more limited data for FMRP expression in the human samples. In addition, FMRP expression levels varied in a subset of mice across the cerebellum, frontal cortex, and hippocampus, as well as at different ages. These results provide a foundation for understanding both the CGG-repeat-dependence of FMRP expression and for interpreting clinical phenotypes in premutation carriers in terms of the balance between elevated mRNA and lowered FMRP expression levels.
INTRODUCTION
The FMR1 gene, located on the X chromosome, harbors a non-coding CGG-repeat element that, when expanded beyond ∼40 repeats, gives rise to several distinct disorders in a manner dependent on the size of the repeat (NCBI gene 2332). Full-mutation alleles (>200 CGG repeats) are generally transcriptionally silenced, with the consequent diminishment/absence of FMR1 protein (FMRP) giving rise to fragile X syndrome (FXS) (OMIM #300624) (1–5). Smaller expansions (55–200 CGG repeats; premutation range) are common in the general population with prevalence estimates of 1:130–250 in females and 1:260–800 in males (6,7). Approximately 70% of premutation alleles in the general population possess <70 CGG repeats, and in a newborn screening study (n ∼20 900 alleles), only 1 subject, out of 50 premutations ascertained, had an allele that exceeded 120 CGG repeats (130 repeats) (6,8).
Carriers of a premutation allele can present with a spectrum of neurodevelopmental and adult-onset phenotypes that are thought to arise through a ‘toxic’ gain-of-function of elevated CGG-repeat-containing FMR1 mRNA levels (9). One such phenotype is fragile X-associated tremor/ataxia syndrome (FXTAS), a late-onset neurodegenerative disorder characterized by intention tremor, gait ataxia, cognitive decline and peripheral neuropathy, as well as ancillary features reflecting both CNS and non-CNS dysfunction (OMIM #300623) (9–17). In addition, women with premutation expansions have an increased risk of primary ovarian insufficiency (18,19), which involves impaired ovarian function and/or menstrual cessation prior to the age of 40.
In carriers of premutation alleles, while FMR1 message levels are markedly elevated (20–23), FMRP levels remain unchanged or moderately decrease with increasing CGG-repeat length (20,24), because CGG repeats progressively impede the translation of FMR1 mRNA (25–27). Thus, though RNA toxicity is thought to be sufficient to cause the neuropathology of FXTAS (28–31), it is possible that lowered FMRP levels, particularly in the upper premutation range, could exacerbate clinical involvement for either/both neurodevelopmental and neurodegenerative disorders, particularly for clinical domains involving mood, behavior and cognition, which are shared features in individuals with either a premutation or a full-mutation allele. Confounding this issue, disparate FMRP levels in CGG-knock-in (KI) premutation mice have been reported, although for very small numbers of mice in each study (32–38).
To assess the potential for FMRP deficiency as an additional contributing factor for clinical involvement in premutation-associated disorders, we have determined the relative FMRP levels in the brains of a large cohort of mice in which the native murine Fmr1 CGG-repeat element (∼9–11 CGG) has been replaced with an expanded (∼70–250 CGG) repeat in the premutation KI mouse (39). As in humans, the KI mouse exhibits repeat-length instabilities upon transmission of the repeat. Importantly, the mouse also displays physical and behavioral phenotypes similar to those seen in human premutation individuals. These issues include motor deficits and anxiety (40–42), as well as impairments in exploration (41), memory, learning and spatial organization (41,43,44). Neuronal and astrocytic intranuclear inclusions are also evident in older KI mice (34,38, review: 45).
In addition, we have also measured FMRP expression levels in postmortem brain tissue (frontal cortex) from 24 human cases (CGG ranged from 56 to 118 CGG repeats). In both mice and humans, we observed that there is only a modest reduction in mean FMRP level with increasing CGG-repeat size in a background of relatively large inter-individual variation in FMRP level, albeit with human CGG repeats restricted toward the lower end of the premutation range. These data suggest that any contribution to the premutation clinical phenotype would likely be relatively modest, on average, but could be quite variable within a given population because of the high degree of FMRP variability.
RESULTS
Murine brain Fmr1 mRNA levels increase substantially with increasing CGG-repeat size throughout the premutation range
To assess the degree to which mouse Fmr1 mRNA levels parallel the increase observed in humans with the premutation, we utilized a KI mouse model in which the native murine CGG-repeat element was replaced (originally) by a ∼98-CGG-repeat element (39), from which a broad range of allele sizes (<70 to >300 CGG repeats) have since been generated through breeding owing to contraction and expansion of the meiotically unstable repeat element (32,38,45,46).
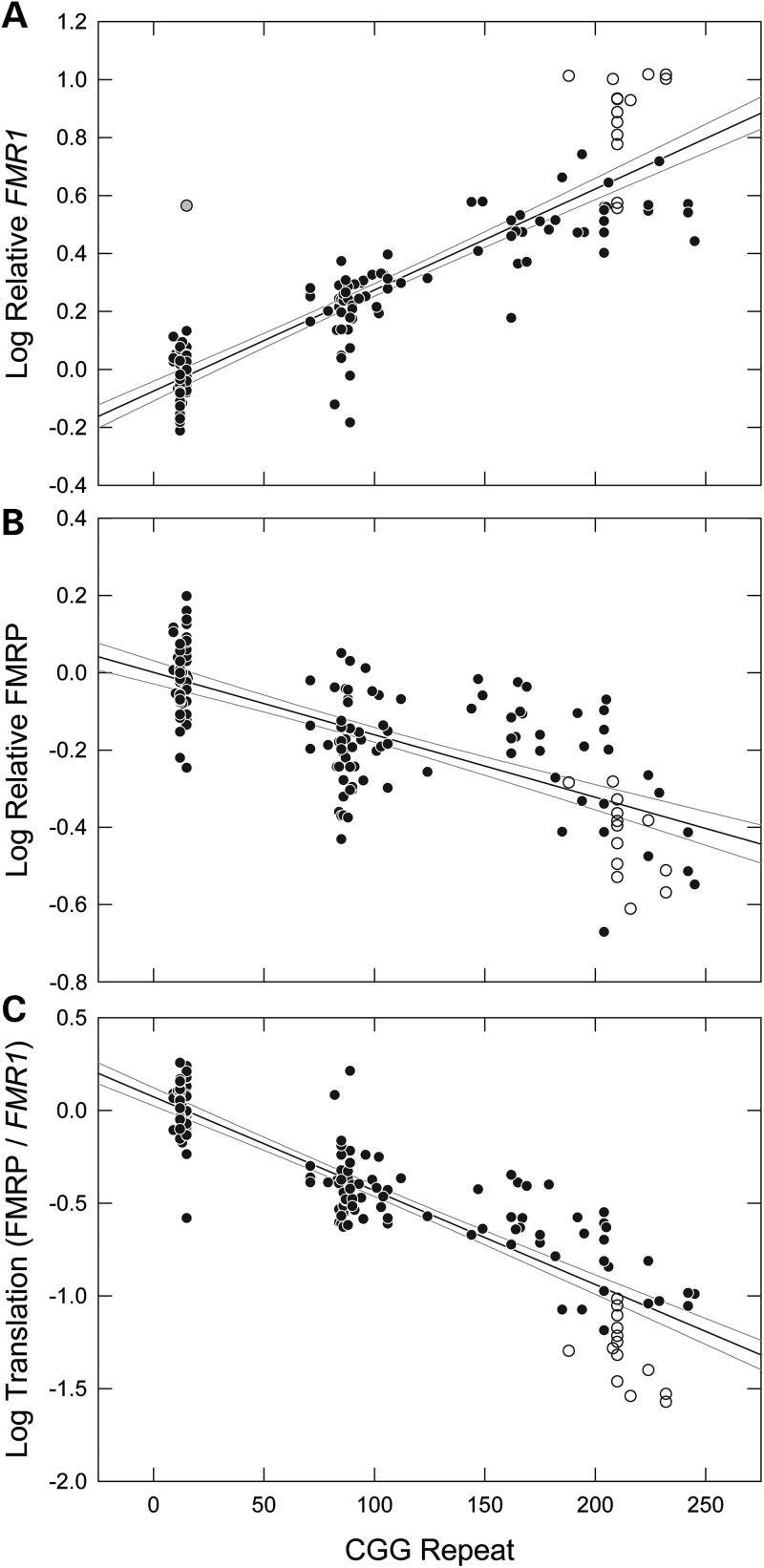
We bred female mice (C57BL/6J background) heterozygous for the expanded (KI) repeat with male wild-type (wt) or KI mice to obtain male offspring that were either wt or hemizygous for the expanded allele. Fmr1 mRNA and FMRP levels were determined for whole right and left hemispheres in all male offspring at 3 weeks of age. In this manner, wt and CGG-repeat males were littermates and their in utero, parenting, and external environments were the same. The cohort for these studies comprised 57 wt (∼10 CGG repeats) and 97 KI (71–245 CGG repeats) animals at postnatal day 21 (P21) (Table 1). We found that Fmr1 mRNA levels increase substantially with increasing CGG-repeat length (Fig. 1A), as anticipated from previous studies with smaller cohorts (33,34,38), with levels for ∼200 CGG-repeat alleles (180–220 CGG) approaching 10-fold higher than the mean for wt mice (excluding one extreme outlier among wt mice; range = 2.5 to 10.3). The observed range among the wt animals (∼1.9-fold between the 5% lower and 95% upper CI bounds; 0.67 to 1.3) is similar to the range for normal FMR1 alleles in humans (∼3-fold, 22, 47). The highest Fmr1 mRNA levels were found in several litters from breedings of KI males (238 and 243 CGG) to KI females (214 and 204 CGG) (Fig. 1A, open circles). These offspring had Fmr1 mRNA levels higher than other animals within the same repeat range, as well as animals with even larger repeats. It is not apparent why this breeding pattern would lead to increased Fmr1 transcription in the offspring, because male pups receive the Fmr1 allele from their mothers and are expected to be unaffected by the paternal genotype. The female mice giving rise to these litters did not have the largest CGG repeats among the breeding females.
Table 1.
Molecular and repeat-expansion properties of the mice measured in Figure 1
| Dataset feature | Count | SEM |
|---|---|---|
| Number wt mice | 57 | — |
| Number exp-CGG mice | 97 | — |
| Largest repeat contraction | 161 | — |
| Largest repeat expansion | 143 | — |
| Avg repeat contraction | 17 | ±5.0 |
| Avg repeat expansion | 31 | ±5.3 |
| Avg exp-CGG-repeat length | 143 | ±5.8 |
| Avg exp-CGG Fmr1 mRNA level | 3.2 | ±0.24 |
| Avg exp-CGG FMRP level | 0.62 | ±0.021 |
| Avg exp-CGG translation | 0.29 | ±0.023 |
Molecular measurements are all versus wt, the average of which was set to 1. exp, expanded; avg, average.
Figure 1.
Scatter plots of mouse CGG repeat versus: (A) Fmr1 mRNA level (normalized to the endogenous control Gusb), (B) FMRP protein level (normalized to Gapdh) and (C) the ratio of FMRP to Fmr1 mRNA, an indication of Fmr1 translation efficiency. Each graph includes solid black regression lines and gray 95% confidence interval lines. Open circles indicate animals that were bred as F2 crosses of animals from the initial breading. One outlier is shaded gray. FMR1 mRNA measurements were performed in triplicate, and FMRP measurements were in duplicate. R-squared values are as follows: Fmr1 = 0.80, FMRP = 0.52 and Fmr1/FMRP = 0.80.
FMRP levels decrease gradually throughout the premutation range
FMRP levels had been reported to be normal in mice with moderate expansions (∼70–110 CGG) or reduced in KI mice with larger expansions (up to ∼230 CGG); however, these observations were based on small numbers of animals and specific CGG-repeat ranges (33,34,38). To more precisely estimate the dependence of FMRP level on CGG-repeat length in this mouse line, we measured whole-brain protein levels relative to Gapdh protein for both wt and KI mice, using much larger cohorts (wt, n = 57; KI, n = 97) than had been used previously (Fig. 1B). Large errors of replicate FMRP measurements, along with a visible protein precipitation in frozen brain lysates, led us to re-exam our lysis procedure (Supplementary Material, Methods and Fig. S1). Therefore, we adapted a new method for protein extraction that was reported to solubilize 98% of mouse brain tissue (48), and, using this method, we found that sample replicates were consistent from blot to blot, a drastic improvement over the initial method (Supplementary Material, Figs S1 and S2).
As can be seen in Figure 1B, FMRP levels clearly decrease with increasing CGG repeat throughout the premutation range, with an average 38% reduction in FMRP levels within that range. However, it is also evident that there is broad variation in FMRP levels for any given CGG-repeat size, even within the wt group (95% CIs of FMRP are indicated by curved lines on the graph), likely due at least in part to variation in the western blot method. This latter observation suggests that previous differences in FMRP levels with respect to CGG-repeat size (33–38) could be accounted for by sampling differences for small cohorts (∼1–10 animals; Supplementary Material, Fig. S3 andTable S2).
Reductions and variation in FMRP levels in premutation postmortem human cerebral cortex are similar to those observed in mice
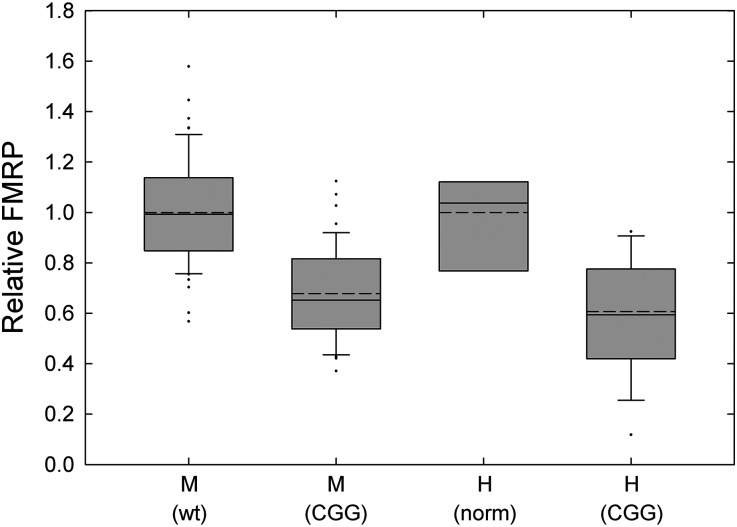
To determine whether the characteristics of FMRP expression in human postmortem brain tissue exhibit a similar repeat-dependence to that observed in the KI mouse, we measured FMRP levels in mid-frontal cortex from 7 individuals with normal-CGG-repeat alleles (controls), and from 17 cases with CGG repeats in the premutation range (56–118 CGG repeats; Table 2, Fig. 2), all of whom had features of FXTAS prior to death. We compared FMRP levels in human frontal cortex to FMRP levels in a subset of 47 mice that were within the same repeat range and observed that the reduction in FMRP in this range is comparable. Setting the normal human and wt mouse groups to 1, the average FMRP in expanded alleles is 0.59 in humans and 0.67 in mice (P = 0.19 using t-test). As the human samples were subject to a range of postmortem intervals (from 1.5 to 25 h), some decrease in FMRP is expected due to protein degradation. Regardless of postmortem interval, FMRP reductions are moderate (∼60% of normal) in the premutation range of 56–118 CGG repeats.
Table 2.
Age, CGG repeat and FXTAS stage of the human patients from which FMRP levels were measured in frontal cortex, as shown in Figure 2
| CGG repeat | Age at death | FXTAS stage |
|---|---|---|
| 25 | 53 | NA |
| 31 | 81 | NA |
| 34 | 81 | NA |
| 34, 42 | 69 | NA |
| 37 | NS | NA |
| 43 | NS | NA |
| 49 | NS | NA |
| 56 | 72 | |
| 62, 71, 83 | 72 | 3 |
| 66 | 82 | |
| 71 | 70 | |
| 77 | 82 | 0 |
| 78 | 87 | |
| 82 | 77 | |
| 82 | 79 | 4 |
| 90 | NS | |
| 96 | 75 | 3 |
| 96 | 82 | |
| 100 | 81 | 5 |
| 102 | 76 | |
| 105 | 67 | 6 |
| 109 | 77 | 6 |
| 112 | 78 | |
| 118 | 87 | 0 |
NS, not specified; NA, not applicable.
Figure 2.
Box-plot comparison of human and mouse brain FMRP levels. FMRP levels in the frontal cortex of normal (n = 7) and premutation (n = 17) human individuals and wt (n = 57) and expanded-CGG (n = 48) mice. Solid bisecting lines are medians, dotted lines are means and individual points represent outliers. Error bars are standard errors of the mean (SEM). Significant differences were seen between the following groups: P < 0.001 for mouse wt versus mouse expanded; mouse wt versus human expanded; human normal versus human expanded; and human normal versus mouse expanded, using one-way ANOVA. The P-value between mouse expanded and human expanded is 0.26.
The efficiency of Fmr1 translation decreases dramatically over the premutation range
Although there is only a modest decrease in FMRP levels with increasing CGG repeat, in both mouse and human brain tissue and in human blood lymphocytes (22,24,50), expanded-CGG-repeat alleles are well known to impede FMRP production owing to secondary structure formation within the repeat tract and, consequently, to inhibit scanning of the 43S ribosomal pre-initiation complex through the structured 5′ UTR (25,51,52). Thus, to get an estimate of the efficiency of FMRP production within the premutation range, we determined the ratio of FMRP to Fmr1 mRNA for each tissue sample. When FMRP levels (Fig. 1B) were corrected for Fmr1 mRNA levels (Fig. 1A) for each animal, the decrease in translation efficiency is striking (Fig. 1C), with relative efficiencies of ∼10% of wt levels for the highest repeats (180–245 CGG; 95% CI Fmr1/FMRP for difference of means, −0.90 ± 0.10).
Dependency of FMRP levels on mouse age
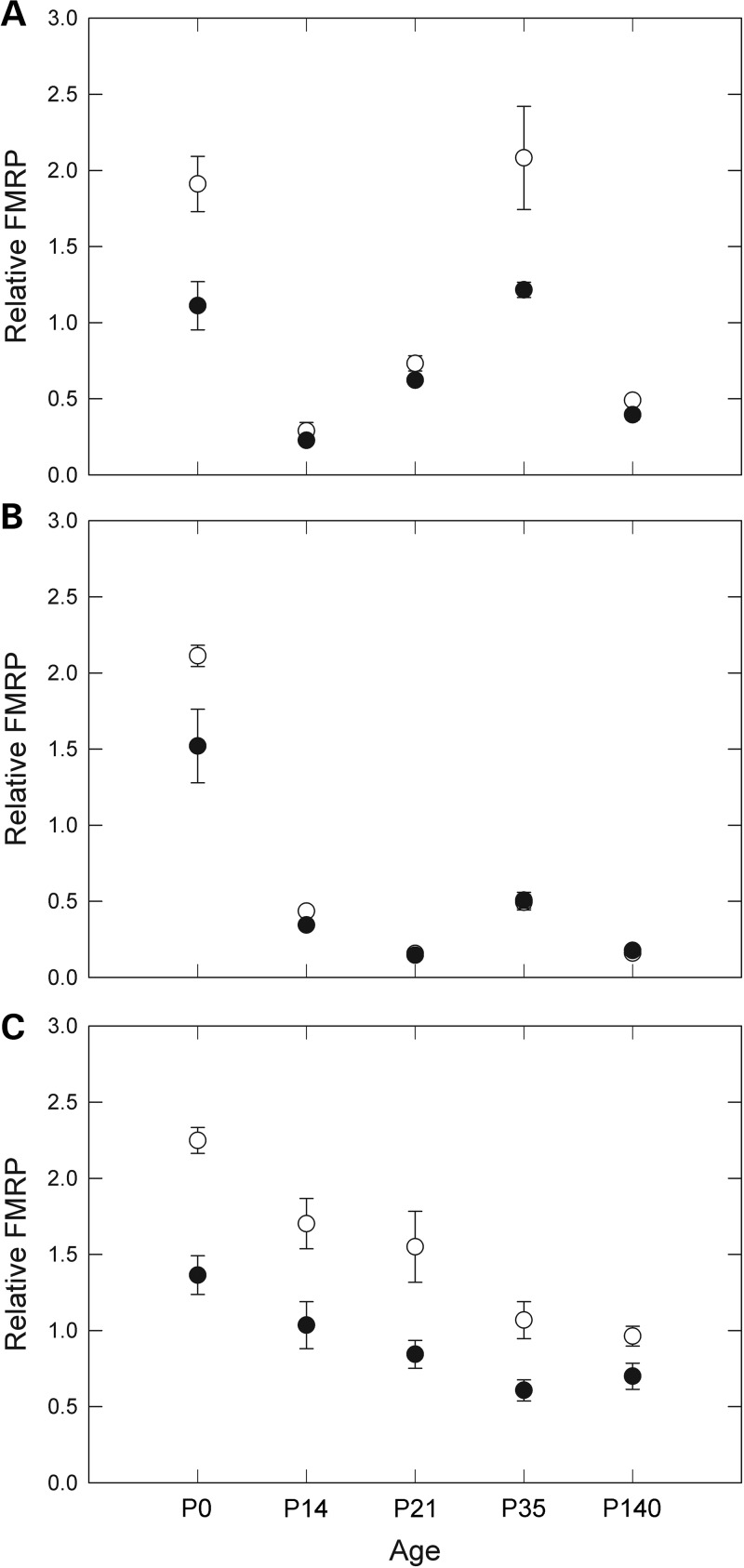
We next sought to determine what effect age has on FMRP levels in mice. FMRP levels may change throughout development and into adulthood; for example, it has been shown that FMRP levels are highest in mice during development in utero (53–56). We measured levels of FMRP in frontal cortex, hippocampus and cerebellum in age-matched control (n = 24) and KI mice (n = 29) at P0, P14, P21, P35 and P140 (Fig. 3); the CGG repeat of each mouse is listed in Table 3. FMRP expression in the cerebellum was highest at birth and decreased throughout the time measured. In the frontal cortex and hippocampus, expression was also high at P0 with a general decrease with age; however, time point P35 showed increased expression in both of these tissues. As mice reach sexual maturation at 5–8 weeks, the spike in FMRP around that time may be associated with reproductive development. While the P35 cohort of expanded-repeat mice also had the smallest CGG repeats, on average, of all the age groups, smaller repeat sizes do not account for the increases in FMRP at P35; those repeat lengths were only significantly different from the P14 age group repeats, which had the overall largest repeats, and cerebellum FMRP expression was actually lowest at P35.
Figure 3.
Dependence of FMRP expression on mouse age. FMRP levels in (A) frontal cortex, (B) hippocampus and (C) cerebellum of wt (open circles) and expanded-repeat (solid circles) mice at five different ages. Each point represents the average of three to seven mice, with the exception of P35 expanded-CGG hippocampus, which included two mice. Error bars correspond to SEMs for each age group.
Table 3.
CGG-repeat lengths of mice used for mouse-age experiments
| Age | CGG-repeat lengths | Average |
|---|---|---|
| P0 | 85, 101, 181, 184, 214 | 153 |
| P14 | 191, 193, 226, 226, 259 | 219 |
| P21 | 155, 155, 158, 160, 204 | 166 |
| P35 | 83, 100, 135, 145, 159, 184, 197 | 143 |
| P140 | 139, 141, 142, 146, 164, 166, 168 | 152 |
CGG repeats for all age groups do not significantly differ except for P14 versus P35 (P = 0.011), P14 versus P140 (P = 0.029) and P14 versus P0 (P = 0.049), by ANOVA.
Our data indicate that FMRP levels are highest at the earliest stages of postnatal development, which may be the critical period in which FMRP is needed in humans. However, the small number of animals at each time point does not provide us with enough power to separately assess the dependencies of FMRP level on age, brain region and genotype. Therefore, we performed an ANOVA for the main effects (single variable) and two-variable effects of these groups (Supplementary Material, Table S1). Combining, for example, samples from all ages and genotypes for one brain region, or FMRP levels from all brain regions for one genotype, provides groups with large enough Ns for statistical analysis. Overall, FMRP levels are highest in wt mice, in the cerebellum, and at time P0. When brain region data are combined, wt and CGG levels are significantly different at every age except for P140, where FMRP levels have dropped to their lowest levels for both groups.
Instability of the CGG-repeat during maternal transmission
Transmission of the CGG repeat from heterozygous female mice to their offspring yielded both expansions and contractions (Supplementary Material, Fig. S4), with the average expansion (+31 CGG repeats) nearly twice the average contraction (-17 CGG repeats) (Table 1). Moreover, more progeny had expanded alleles (39 of 95) than had contractions (29 of 95). For these estimates, only animals whose repeat lengths differed by at least 5% from their mothers' genotypes were included, to avoid biasing for sizing inaccuracies; 27 animals fell below this threshold, whereas 2 animals had an unknown maternal genotype. The largest expansions occurred in the low premutation range, whereas larger contractions occurred only for the largest parental repeats. However, this trend is highly influenced by the 6 largest expansions (114, 115, 136, 143, 143 and 143) and 2 largest CGG-repeat contractions (-128, -161), and there is no significant inverse correlation between the CGG repeat of the mother and the repeat expansions in the progeny (R2 = 0.0831).
Immunohistochemistry within specific brain regions reveals reductions of FMRP that are consistent with whole-brain levels
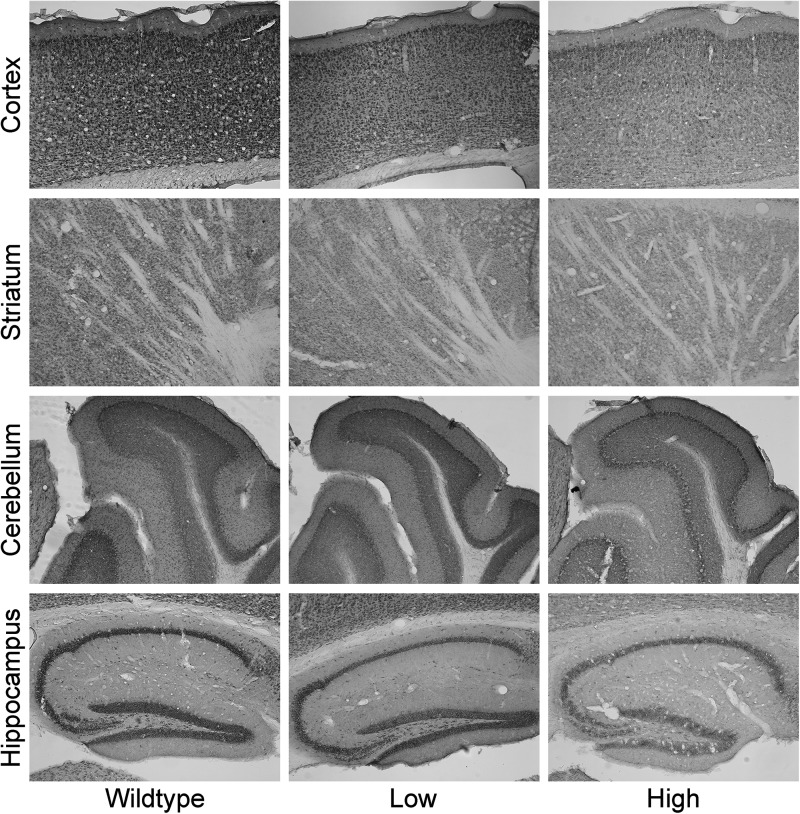
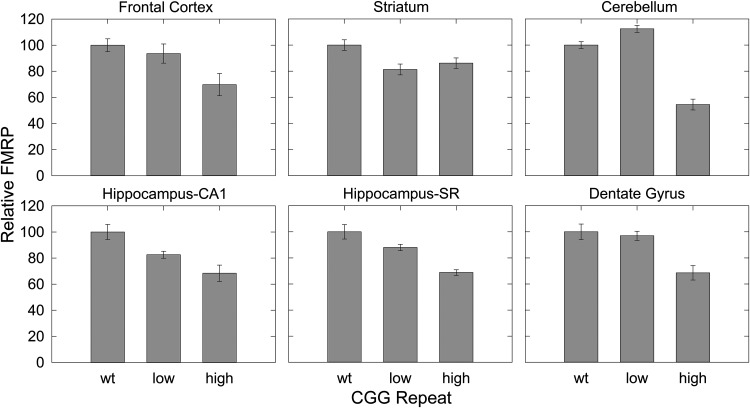
We next immunostained for FMRP expression across several brain regions for wt (n = 3), and for low-CGG (CGG = 82, 89 and 96) and high-CGG repeat (CGG = 175, 181 and 222) KI adult mice. Brains were fixed, parasagittally sectioned and stained for FMRP. As can be seen in Figure 4, expression of FMRP is concentrated across the neocortex, hippocampal CA1 and dentate gyrus regions, and Purkinje and granule layers of the cerebellum, where Purkinje cell bodies and granule cells reside, respectively. These observations are consistent with higher FMRP levels in regions of high neuronal cell-body density. There is a general, mild reduction of FMRP in the KI mice (Figs 4 and 5), with average FMRP levels of 92% (95% CI, 92 ± 6.9) and 69% (95% CI, 69 ± 10) of wt levels for low- and high-repeat KI mice, respectively. Across all regions of interest, the lowest level (54%) was observed in the cerebellum of high-repeat mice. We also compared FMRP levels among different brain regions in the mice, observing that expression (staining) was highest in the CA1 and dentate gyrus of the hippocampus, and lowest in the striatum and hippocampus stratum radiatum (SR) (Table 4).
Figure 4.
FMRP immunohistochemistry in mouse brain. FMRP staining of brain regions of mice with wt (∼10), low (82, 89 and 96) and high (175, 181 and 222) CGG-repeat lengths.
Figure 5.
Quantification of FMRP levels in the mouse tissues shown in Figure 4. Error bars are SEM of four or five separate experiments.
Table 4.
Quantification of FMRP in wt mouse brain from IHC
| Brain region | wt |
Low CGG |
High CGG |
|||
|---|---|---|---|---|---|---|
| % FMRP | SEM (%) | % FMRP | SEM (%) | % FMRP | SEM (%) | |
| Hippocampus CA1 | 100 | ±5.8 | 92.6 | ±3.0 | 100 | ±9.1 |
| Dentate Gyrus | 91.9 | ±5.4 | 100 | ±3.6 | 92.2 | ±7.4 |
| Cortex | 69.6 | ±3.4 | 73.1 | ±5.8 | 71.1 | ±8.6 |
| Cerebellum | 62.2 | ±1.2 | 78.4 | ±1.3 | 49.5 | ±2.7 |
| Striatum | 49.6 | ±2.0 | 45.2 | ±2.3 | 62.4 | ±2.9 |
| Hippocampus SR | 38.5 | ±2.1 | 38.0 | ±1.0 | 38.8 | ±1.2 |
The region of highest expression, hippocampus CA1, is set to 100%.
DISCUSSION
In this study, we demonstrated that both enhanced Fmr1 mRNA production and diminished FMRP production with increasing CGG-repeat size in mice recapitulate the observed trends in humans (20–24). The preponderance of human and mouse data indicates that the trends are smooth and monotonic, with approximate log-linear relationships between the CGG-repeat length and both mRNA and FMRP levels. Despite increased FMR1/Fmr1 mRNA levels, FMRP levels in humans and mice are diminished, particularly in the upper premutation range, likely due to the fact that the 5′ non-coding rCGG-repeat element acts as a structural impediment to translational initiation (21,25–27,51,52). Also consistent with the mouse data, we observed a general diminution in FMRP levels with increasing CGG repeat for human postmortem cerebral cortex, although the data are limited to the lower half of the premutation range and are subject to additional uncertainties associated with the postmortem interval.
It is evident that both FMR1 mRNA and protein (FMRP) measures vary substantially about their mean levels throughout the normal and premutation ranges (47,57; Fig. 1). The basis of this variation is not completely understood, although based on our current analysis, a significant portion of this variation is likely due to technical variation (PCR, western), evident even within a single mouse strain (Supplementary Material, Fig. S2). From a practical standpoint, this variation is important, particularly for FMRP levels, whose mean levels decrease only gradually (and modestly) within the premutation range, because it diminishes the utility of FMRP as a diagnostic tool within that range. However, FMRP levels will remain important as an adjunct to other clinical and molecular measures for those with very low FMRP levels, for both high-premutation and full-mutation alleles, where FMRP levels are likely to be influenced by various epigenetic, environmental and/or metabolic factors.
Previous reports of the current and other premutation (KI) mouse models (33,34,36,38) have described a range of trends in Fmr1 mRNA and FMRP expression levels within the premutation range; though all generally show a positive correlation between CGG-repeat length and Fmr1 mRNA expression, and a negative correlation between the size of the repeat and FMRP levels. In light of the variation in mRNA and FMRP levels (for a given CGG-repeat size) that we observed across the premutation range, the most likely explanation for the widely differing reported levels is that many of these studies used very small cohorts, resulting in sampling differences (Supplementary Material, Fig. S3 and, Table S2). The results of the present study also show that developmental age (Fig. 3) and brain region examined (Table 4, Figs 3 and 4) are important variables influencing levels of FMRP in the KI mouse model. Previous studies have not systematically examined the effect of age on FMRP levels or Fmr1 mRNA levels (33,34,37). The present study has found that levels were highest early in development (i.e. P0), consistent with the critical role of FMRP in brain development (49,58), followed by what appears to be a general pattern of decline with age. There may also be additional developmental periods when levels may change (i.e. P35); this interesting observation should be pursued in future studies.
Substantial regional brain differences were also found in relative FMRP expression (Table 4, Fig. 4). The highest total levels were found in the CA1 subregion of the hippocampus, with relatively lower levels in the dentate gyrus, cortex, cerebellum, striatum and stratum lacunosum of the hippocampus, respectively. Regional differences may be related to function, such that high levels in the hippocampus in the premutation could reflect an important role for FMRP in memory processes, with reduced levels contributing to cognitive impairment in premutation mice. However, regional variations may be due to differences in cell density, with higher levels of FMRP in regions of high cell-body density (e.g. CA1 and striatum radiatum) and lower levels in areas of sparse cell density, including the dendritic fields of the hippocampal striatum radiatum and cerebellar molecular layer (Fig. 4). Additional studies on regional transport and distribution of FMRP within cells will be needed in order to clarify the importance of brain regional differences in the function of FMRP, as well as the reduced levels found in the premutation.
Unlike in humans, full-mutation expansions (>200 CGG repeats) of the mouse Fmr1 gene have not been found to undergo DNA hypermethylation and transcriptional silencing (34,36). As a consequence, full-mutation murine alleles continue to express high levels of Fmr1 mRNA, providing an opportunity to gauge the efficiency of FMRP production from mRNAs possessing full-mutation CGG-repeat expansions (Fig. 1C). This issue is of some importance from a therapeutic standpoint, because any consideration of gene reactivation (i.e. induction of mRNA production) must still account for the markedly reduced translational efficiency in that range. Interestingly, in both humans, in the absence of methylation (59), and in mice, transcription rates remain quite high well into the full-mutation range. Moreover, more than half of males with full-mutation alleles still produce some FMR1 mRNA. Therefore, an alternative strategy to increasing FMR1 transcript expression would be to increase the efficiency of translation, even if only by a factor of 2- to 3-fold (60). Attempts to increase FMR1 mRNA levels alone in FXS may in fact be detrimental later in life, as intranuclear inclusions have been found, albeit at low levels, in the postmortem brain tissue of three individuals with full-mutation FMR1 alleles and FXS (61), suggesting that efforts at further mRNA production should be avoided.
Concluding remarks
Evidence to date indicates that FMRP levels decrease throughout the premutation range in a manner that is both monotonic and relatively mild, with an approximate 2-fold decrease from normal (population mean) levels upon reaching the upper end of the premutation range (21,22,24). The decreased FMRP levels observed in human and mouse appear to be comparable, indicating that the mouse model is a reasonable one for the study of FMRP effects in the premutation range. However, there is an important caveat to the above-mentioned trend. We and others consistently observed substantial variation in FMRP levels for any given CGG repeat, a variation that is not accounted for by variation in age or protein source (e.g. blood versus brain). This dispersion of protein levels, even within an individual mouse line, indicates both that individual-specific metabolic (and perhaps epigenetic) effects are regulating protein level and that simple measurement of FMRP level is unlikely to be a solid indicator of clinical involvement—except perhaps for the low-protein outliers in the premutation range. However, this diagnostic caveat is also a potential opportunity in that identification of those factors that modulate FMRP level may lead to effective therapeutic agents for treatment of individuals who have clinical involvement owing to low protein levels, in both the premutation and full-mutation ranges.
MATERIALS AND METHODS
Premutation knock-in mice
A CGG KI mouse model (C57BL/6J) that harbors premutation-length CGG expansions has been described and characterized (38,39,44,62). For the present study, we bred female mice heterozygous for the CGG-repeat expansion (71–245 CGG repeats on the expanded allele) to wt or expanded-repeat males to produce wt and expanded-CGG-repeat animals. Male offspring were used for molecular and immunohistochemical investigations. The dams and their pups (wt mice were littermates with CGG KI mice) were housed under standard conditions, approved by the University of California, Davis, Institutional Animal Care and Use Committee, and all effort was taken to reduce stress in the mice.
Tissue preparation for molecular measurements
Mice were sacrificed by cervical dislocation, and whole brains were either hemisected (one half each for Fmr1 mRNA and FMRP measurements) or dissected, separating cerebellum, frontal cortex and hippocampus. A total of 97 CGG KI and 57 wt 3-week-old (p21) male mice were used for Fmr1 mRNA and FMRP molecular measurements using brain hemispheres. For longitudinal studies across brain regions, 29 expanded-repeat (85–259 CGG repeats) and 25 wt (9–11 CGG repeats) male mice were used to measure FMRP at P0, P14, P21, P35 and p140. Tail snips of ∼5 mm in length were taken for immediate extraction of genomic DNA and genotyping from all animals.
Human brain tissue samples
Postmortem brain tissue from 7 normal-CGG-repeat and 17 premutation male individuals was previously acquired and sectioned in accordance with University of California, Davis, IRB-approved protocols as described (63), and brain sections of 1 cm thickness were stored long-term at −80°C. Small tissue samples (∼2 × 2 cm) were taken from (approximately) superolateral mid-frontal cortex, in order to provide tissue from an equivalent region across all cases and controls, and each were ground to powder in liquid nitrogen and stored at −80°C until use.
Molecular measurements
DNA isolation and CGG-repeat analysis
Genomic DNA was isolated from mouse tail snips and human brain tissue using the Gentra Puregene genomic DNA purification kit (Qiagen, Valencia, CA, USA), which routinely yields 5–15 µg of genomic DNA per sample. Mouse DNAs—40 to 80 ng of mouse DNA was used to PCR-amplify the CGG-repeat region of each sample using AmplideX FMR1 PCR kit (Asuragen, Austin, TX, USA), as per manufacturer's instructions, after which sizing was performed using the Agilent DNA 7500 Kit on a 2100 Bionalalyzer (Agilent Technologies, Santa Clara, CA, USA) (Supplementary Material, Fig. S5A). Human DNAs—Repeat lengths in human DNA samples were sized following PCR amplification of the repeat region using a standard protocol (64) with 100 ng of genomic DNA, utilizing the Expand Long Template PCR System (Roche Diagnostics Corporation, Indianapolis, IN, USA) and primers C3 (5′ TGTTTACACCCGCAGCGGGCCGGGGGTTC) and F (5′ AGCCCCGCACTTCCACCACCAGCTCCTCCA). PCR reactions were resolved by agarose gel electrophoresis and then sized by regression relative to the HiLo DNA Marker (Bionexus, Inc., Oakland, CA, USA) using the Alpha Innotech Fluorchem 8900 BioImaging System running AlphaEaseFC software (ProteinSimple, Santa Clara, CA, USA)(Supplementary Material, Fig. S5B).
Total RNA isolation and cDNA synthesis
The right hemisphere of each brain was homogenized in 1 ml HEPES Buffer (10 mm HEPES, 300 mm KCl, 3 mm MgCl2, 100 µm CaCl2, 0.45% Triton X-100, 0.05% Tween-20 and 3 mm Dithiothreitol) using the Tissuelyzer LT system (Qiagen). 100 µl of homogenate plus 500 µl QIAzol (Qiagen) were used for extraction of total RNA using the RNeasy Lipid Tissue Mini Kit (Qiagen), which routinely extracted 5 µg of RNA per extraction. We performed first-strand synthesis with the QuantiTect Reverse Transcription kit (Qiagen) according to the manufacturer's protocol (1 µg of total RNA in 100 µl).
Analysis of mRNA expression
Quantitative PCR reactions were performed using the TaqMan Universal Master Mix with Fmr1 (Mm00484415_m1) and Gusb (Mm00446953_m1) primer/probe sets (Life Technologies, Carlsbad, CA, USA). We serially diluted cDNA to 5.0, 2.5 and 1.25 ng/µl and then performed qPCR on 5 µl of cDNA in 12 µl reactions on a 7900 HT Real-Time PCR System (Life Technologies), in 384-well format. Temperature cycling was as follows: 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Three qPCR reactions were performed for each cDNA dilution, for a total of nine measurements per sample. As a fiducial control, six to eight unrelated wt C57BL/6J mouse brains were pooled and then homogenized and extracted for RNA, as described earlier. To account for inter-plate variability of Fmr1 mRNA expression, this pooled, wt fiducial was included in every qPCR experiment. We determined relative Fmr1 mRNA levels for all samples (including the fiducial) compared with the endogenous control, Gusb, for each sample using the following formula (20): log2(Fmr1/Gusb) = CT(Fmr1) − CT(Gusb), where CT values are the cycles at which fluorescence reached a defined threshold value. These corrected Fmr1 levels of expanded-CGG and wt mice were normalized to total brain Fmr1 expression of the fiducial for each qPCR experiment, and the wt group was set to 1 by dividing all mRNAs by the average of wt, and reported as relative FMRP expression levels. At least three separate qPCR experiments were performed for each sample.
Protein extraction, western blotting and FMRP analysis
Mouse protein extraction was performed by a variation of the protocol described by Ericsson and Nister (48) after extensive trouble-shooting (see Supplementary Material, Methods and Fig. S6A). The left hemisphere of each brain was ground to powder in liquid nitrogen, and ∼30 mg of sample was weighed out for protein extraction. Tissue powder was resuspended in 20 µl protein extraction buffer (PEB) per mg tissue (65) [0.125 m Tris–HCl (pH 6.8), 2% sodium dodecyl sulfate (SDS), 10% glycerol and 5% 2-mercaptoethanol] followed by manual grinding with a pestle for 1 min. After incubating at 70°C in a Thermomixer (Eppendorf, Hauppauge, NY, USA) for 20 min at max speed, samples were cooled to room temperature and spun at 16.1×g in a tabletop centrifuge (Eppendorf). Protein concentrations of lysate supernatants were then immediately measured using the reducing agent and detergent-compatible (RC DC) protein assay (Bio-Rad Laboratories, Hercules, CA, USA) with 1:25 dilutions of lysates in PEB according to the manufacturer's microfuge tube instructions. One wt sample was chosen as a fiducial and was run on all western blots to account for blot-to-blot variations. Mouse FMRP measurements were performed in duplicate from fresh, frozen tissue. On a 10.5–14% Criterion Tris–HCl gel (Bio-Rad) in running buffer (25 mm Tris, 192 mm glycine, 0.1% SDS, pH 8.3), 10 µg of each sample was then separated by electrophoresis. Following transfer of the proteins onto nitrocellulose (30 mA overnight at 4°C in 5 mm Tris, 192 mm glycine and 20% methanol), the membrane was blocked in Odyssey Blocking Buffer (LI-COR Biosciences, Lincoln, NE, USA) for 1 h. The membrane was incubated with both mouse anti-FMRP (1:5000 v/v; #MAB2160; EMD Millipore, Billerica, MA, USA) and chicken anti-Gapdh (1:10 000 v/v; #AB2302; Millipore) antibodies in Blocking Buffer plus 0.1% Tween-20 overnight at 4°C. After washing the membranes five times with PBST (PBS, 0.1% Tween-20), they were incubated in secondary goat anti-mouse IRDye 800 nm (1:20 000 v/v) and donkey anti-chicken IRDye 700 nm (1:25 000 v/v) antibodies (LI-COR) in Blocking Buffer for 1 h. The separate excitation frequencies (680 and 780 nm) of the two infrared-fluor-conjugated antibodies allow for FMRP (70 kDa with multiple isoforms) and Gapdh (36 kDa) to be independently detected on the same membrane. Membranes were washed five times with PBST, plus two final washes with PBS, and then scanned on the Odyssey Infrared Imager (LI-COR) with laser excitations at 680 and 780 nm and emission measurements at ∼720 and 820 nm. For each western blot lane, the sum of all FMRP isoforms was measured by integrating the background-subtracted intensities of all bands of 60–70 kDa (Supplementary Material, Fig. S6B). We calculated the relative expression of FMRP by background-subtracting the integrated intensities of FMRP and Gapdh and then dividing FMRP values by Gapdh values using the Odyssey Software Version 3.0.21 (LI-COR). Each relative FMRP value was then normalized first to the fiducial sample on that blot and then to the average of wt.
Human brain protein extraction and western blotting were performed essentially as mentioned earlier, in duplicates from fresh, frozen tissue (Supplementary Material, Fig. S6C). About 30 mg of powdered frontal cortex tissue was lysed by adding 20 µl PEB per 1 mg of tissue and proceeding with mechanical and thermal disruption. A normal fiducial sample was chosen and included on all western blots, FMRP (75 kDa) and GAPDH (38 kDa) levels in each blot were normalized to the local fiducial, and the mean of the normal group was set to 1, as per the mouse samples.
Protein extraction and western blotting of samples for age studies were performed with the following alterations. Samples were lysed in T-PER (Thermo Scientific), concentrations of samples were determined using the BCA protein assay kit (Pierce, Rockford, IL, USA) according to manufacturer's microplate instructions, and western blots were analyzed by horseradish peroxidase development on film using chicken anti-FMRP (1:2500 v/v; 66) and mouse anti-Gapdh (1:80 000 v/v; #MAB374; EMD Millipore) primary antibodies and donkey anti-chicken (1:5000 w/v; #703-035-155; Jackson ImmunoResearch Laboratories, Inc.) or goat anti-mouse (1:5000 v/v; #70-6516; Bio-Rad) secondary antibodies. Expression of FMRP was calculated by densitometry using background subtraction of the integrated intensities of FMRP and Gapdh bands and then normalizing FMRP values to Gapdh values using ImageJ software (67).
Tissue preparation for immunohistochemistry
Male mice with wt, low (82, 89 and 96 CGG) or high (175, 181 and 222 CGG) CGG-repeat lengths, between 3 and 8 weeks of age, were deeply anesthetized with sodium pentobarbital (100 mg/kg, i.p.; Euthasol; Virbac AH, Inc., Fort Worth, TX, USA) and then perfused intracardially with 12 ml of isotonic saline with heparin (1000 U heparin per 1 ml of saline) for >1 min, followed by 60 ml of a chilled, freshly prepared 4% paraformaldehyde solution in 0.1 m sodium phosphate buffer, pH 7.4 (PB) administered by gravity feed over 20 min. We immediately removed the brains from the skull and placed them in the same fixative for 1 h at 4°C with gentle agitation on a table shaker. After post-fixation, the brains were rinsed in 0.1 m PB, cryoprotected in 10% sucrose in 0.1 m PB for 1 h, followed by 30% sucrose in 0.1 m PB for 24 h at 4°C, then flash-frozen and stored at −80°C until use. Parasagittal serial sections were cut at 35 µm on a sliding microtome equipped with a freezing stage.
Immunohistochemistry
FMRP was quantified in situ by immunohistochemistry (IHC) in several brain regions, including CA1, dentate gyrus and SR of the hippocampus; neocortex; striatum; and granule cell layer of the cerebellum. We processed sections for IHC using a modification of the avidin–biotin complex (ABC)-peroxidase technique as previously described (44,62). Briefly, free-floating sections were rinsed in 10% sucrose in 0.1 m PB, followed by 0.1 m PB and pretreated with 0.1% sodium borohydride for antigen retrieval for 15 min. Sections underwent treatment with 0.5–2% H2O2 in PB for 90 min to inactivate endogenous peroxidases and then were rinsed in 0.1 m PB for 15 min, followed by 0.01 m PB, 0.15 m NaCl, pH 7.4 (PBS) for 15 min. Sections were then treated with 3% goat serum (Sigma, St. Louis, MO, USA; DAKO, Inc., Carpinteria, CA, USA), 3% BSA and 0.3% Triton X-100 (TX) in 0.01 m PBS for 1 h to reduce nonspecific staining. Sections were incubated for 48–72 h at 4°C in a chicken polyclonal anti-FMRP primary antibody (Aves Labs, Tigard, OR, USA; 66) diluted 1:20 000 in PBS containing 1% goat serum, 2% BSA and 0.3% TX. Following rinses for 1 h in PBS, sections were incubated in biotinylated goat anti-chicken IgG (DAKO, Inc.; Vector Laboratories, Burlingame, CA, USA), diluted 1:500 in 1% goat serum, 2% BSA and 0.3% TX for 24 h at 4°C. After 1 h of rinses in PBS, sections were incubated in ABC reagent (Elite ABC Kit, Vector Laboratories) and diluted 1:2000 in 1% goat serum, 2% BSA, 0.3% TX and PBS for 24 h at 4°C. Sections were transferred to Tris–HCl buffers (pH 7.4, 7.6) and then incubated for 15 min in 0.025% 3,3′-diaminobenzidine (DAB, Sigma) in TB (pH 7.6). Staining was initiated by addition of 0.003% H2O2 until development was optimized. Sections were then rinsed in TB, then PB and mounted on gelatin-coated slides. Sections were air-dried overnight at room temperature, dehydrated, cleared and cover-slipped with Permount. For all experiments, sections from wt-, low- and high-repeat mice were run in parallel.
Analysis of immunostained sections
Stained sections were visualized at 100× and images captured on a Nikon ECLIPSE E600 microscope (Nikon Instruments, Melville, NY, USA) equipped with a digital camera using Pictureframe software (MicroFire; Olympus America, Inc., Melville, NY, USA). The images were converted to gray scale using Adobe Photoshop (Adobe Systems, Inc., San Jose, CA, USA), and ImageJ software (http://rsbweb.nih.gov/ij/; 67) was used to measure optical densities of immunostained sections in the following brain regions of interest (ROI): CA1, dentate gyrus and SR of the hippocampus; neocortex; striatum and the granular layer of the cerebellum. We analyzed three animals for each of the three (wt-, low- and high-repeat) groups, and each animal was analyzed for all brain ROI. ImageJ was calibrated in optical density units using a 600-dpi gray-scale density wedge. Briefly, gray-scale images of each brain section were opened in Image J, and optical densities were measured at 4–5 circular areas for each ROI using the ellipsoid selection tool. The size of the circle was determined by the size and shape of the ROI. For example, a circle with a diameter of 40 µm was used for the granular cell layer of the cerebellum, whereas a smaller oval with a diameter of ∼7 µm was used for the pyramidal cell layer of the CA1 subregion of the hippocampus. The optical density values were then normalized to wt values and reported as a percentage of wt.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (grant numbers R01HD040661 to P.J.H. and R01NS079775 to R.F.B.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Danh V. Nguyen for discussions on plotting the mouse age data, and the families whose support has made this work possible.
Conflicts of Interest statement. Dr Hagerman holds patents for quantification of CGG-repeat number and for measurement of FMRP levels. He has submitted, with Pacific Biosciences, a patent application for SMRT-sequencing methodology. He collaborates with Pacific Biosciences without compensation; he and Pacific Biosciences are co-recipients of an STTR grant from the NICHD. Dr Tassone holds a patent for quantification of CGG-repeat number. She and Pacific Biosciences are co-recipients of an STTR grant from the NICHD. Dr Tassone has received funds from Roche Molecular Systems.
REFERENCES
- 1.Loesch D.Z., Huggins R.M., Hagerman R.J. Phenotypic variation and FMRP levels in fragile X. Ment. Retard. Dev. Disabil. Res. Rev. 2004;10:31–41. doi: 10.1002/mrdd.20006. [DOI] [PubMed] [Google Scholar]
- 2.Hagerman R.J., Berry-Kravis E., Kaufmann W.E., Ono M.Y., Tartaglia N., Lachiewicz A., Kronk R., Delahunty C., Hessl D., Visootsak J., et al. Advances in the treatment of fragile X syndrome. Pediatrics. 2009;123:378–390. doi: 10.1542/peds.2008-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider A., Hagerman R.J., Hessl D. Fragile X syndrome -- from genes to cognition. Dev. Disabil. Res. Rev. 2009;15:333–342. doi: 10.1002/ddrr.80. [DOI] [PubMed] [Google Scholar]
- 4.Pieretti M., Zhang F.P., Fu Y.H., Warren S.T., Oostra B.A., Caskey C.T., Nelson D.L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 5.Fu Y.H., Kuhl D.P., Pizzuti A., Pieretti M., Sutcliffe J.S., Richards S., Verkerk A.J., Holden J.J., Fenwick R.G., Jr, Warren S.T., et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 6.Tassone F., Iong K.P., Tong T.H., Lo J., Gane L.W., Berry-Kravis E., Nguyen D., Mu L.Y., Laffin J., Bailey D.B., et al. FMR1 CGG allele size and prevalence ascertained through newborn screening in the United States. Genome Med. 2012;4:100. doi: 10.1186/gm401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maenner M.J., Baker M.W., Broman K.W., Tian J., Barnes J.K., Atkins A., McPherson E., Hong J., Brilliant M.H., Mailick M.R. FMR1 CGG expansions: prevalence and sex ratios. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2013;162B:466–473. doi: 10.1002/ajmg.b.32176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hantash F.M., Goos D.M., Crossley B., Anderson B., Zhang K., Sun W., Strom C.M. FMR1 premutation carrier frequency in patients undergoing routine population-based carrier screening: insights into the prevalence of fragile X syndrome, fragile X-associated tremor/ataxia syndrome, and fragile X-associated primary ovarian insufficiency in the United States. Genet. Med. 2011;13:39–45. doi: 10.1097/GIM.0b013e3181fa9fad. [DOI] [PubMed] [Google Scholar]
- 9.Hagerman P. Fragile X-associated tremor/ataxia syndrome (FXTAS): pathology and mechanisms. Acta Neuropathol. 2013;126:1–19. doi: 10.1007/s00401-013-1138-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagerman R.J., Leehey M., Heinrichs W., Tassone F., Wilson R., Hills J., Grigsby J., Gage B., Hagerman P.J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- 11.Hagerman P.J., Hagerman R.J. Fragile X-associated tremor/ataxia syndrome (FXTAS) Ment. Retard. Dev. Disabil. Res. Rev. 2004;10:25–30. doi: 10.1002/mrdd.20005. [DOI] [PubMed] [Google Scholar]
- 12.Leehey M.A., Hagerman P.J. Fragile X-associated tremor/ataxia syndrome. Handb. Clin. Neurol. 2012;103:373–386. doi: 10.1016/B978-0-444-51892-7.00023-1. [DOI] [PubMed] [Google Scholar]
- 13.Apartis E., Blancher A., Meissner W.G., Guyant-Marechal L., Maltete D., De Broucker T., Legrand A.P., Bouzenada H., Thanh H.T., Sallansonnet-Froment M., et al. FXTAS: new insights and the need for revised diagnostic criteria. Neurology. 2012;79:1898–1907. doi: 10.1212/WNL.0b013e318271f7ff. [DOI] [PubMed] [Google Scholar]
- 14.Juncos J.L., Lazarus J.T., Graves-Allen E., Shubeck L., Rusin M., Novak G., Hamilton D., Rohr J., Sherman S.L. New clinical findings in the fragile X-associated tremor ataxia syndrome (FXTAS) Neurogenetics. 2011;12:123–135. doi: 10.1007/s10048-010-0270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacquemont S., Hagerman R.J., Leehey M., Grigsby J., Zhang L., Brunberg J.A., Greco C., Des Portes V., Jardini T., Levine R., et al. Fragile X premutation tremor/ataxia syndrome: molecular, clinical, and neuroimaging correlates. Am. J. Hum. Genet. 2003;72:869–878. doi: 10.1086/374321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leehey M.A., Legg W., Tassone F., Hagerman R. Fibromyalgia in fragile X mental retardation 1 gene premutation carriers. Rheumatology. 2011;50:2233–2236. doi: 10.1093/rheumatology/ker273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunsaker M.R., Greco C.M., Spath M.A., Smits A.P., Navarro C.S., Tassone F., Kros J.M., Severijnen L.A., Berry-Kravis E.M., Berman R.F., et al. Widespread non-central nervous system organ pathology in fragile X premutation carriers with fragile X-associated tremor/ataxia syndrome and CGG knock-in mice. Acta Neuropathol. 2011;122:467–479. doi: 10.1007/s00401-011-0860-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz C.E., Dean J., Howard-Peebles P.N., Bugge M., Mikkelsen M., Tommerup N., Hull C., Hagerman R., Holden J.J., Stevenson R.E. Obstetrical and gynecological complications in fragile X carriers: a multicenter study. Am. J. Med. Genet. 1994;51:400–402. doi: 10.1002/ajmg.1320510419. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan S.D., Welt C., Sherman S. FMR1 and the continuum of primary ovarian insufficiency. Semin. Reprod. Med. 2011;29:299–307. doi: 10.1055/s-0031-1280915. [DOI] [PubMed] [Google Scholar]
- 20.Tassone F., Hagerman R.J., Taylor A.K., Gane L.W., Godfrey T.E., Hagerman P.J. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenneson A., Zhang F., Hagedorn C.H., Warren S.T. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum. Mol. Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- 22.Tassone F., Beilina A., Carosi C., Albertosi S., Bagni C., Li L., Glover K., Bentley D., Hagerman P.J. Elevated FMR1 mRNA in premutation carriers is due to increased transcription. RNA. 2007;13:555–562. doi: 10.1261/rna.280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen E.G., He W., Yadav-Shah M., Sherman S.L. A study of the distributional characteristics of FMR1 transcript levels in 238 individuals. Hum. Genet. 2004;114:439–447. doi: 10.1007/s00439-004-1086-x. [DOI] [PubMed] [Google Scholar]
- 24.Peprah E., He W., Allen E., Oliver T., Boyne A., Sherman S.L. Examination of FMR1 transcript and protein levels among 74 premutation carriers. J. Hum. Genet. 2010;55:66–68. doi: 10.1038/jhg.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Primerano B., Tassone F., Hagerman R.J., Hagerman P., Amaldi F., Bagni C. Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. RNA. 2002;8:1482–1488. [PMC free article] [PubMed] [Google Scholar]
- 26.Napierala M., Michalowski D., de Mezer M., Krzyzosiak W.J. Facile FMR1 mRNA structure regulation by interruptions in CGG repeats. Nucleic Acids Res. 2005;33:451–463. doi: 10.1093/nar/gki186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zumwalt M., Ludwig A., Hagerman P.J., Dieckmann T. Secondary structure and dynamics of the r(CGG) repeat in the mRNA of the fragile X mental retardation 1 (FMR1) gene. RNA Biol. 2007;4:93–100. doi: 10.4161/rna.4.2.5039. [DOI] [PubMed] [Google Scholar]
- 28.Jin P., Zarnescu D.C., Zhang F., Pearson C.E., Lucchesi J.C., Moses K., Warren S.T. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 29.Arocena D.G., Iwahashi C.K., Won N., Beilina A., Ludwig A.L., Tassone F., Schwartz P.H., Hagerman P.J. Induction of inclusion formation and disruption of lamin A/C structure by premutation CGG-repeat RNA in human cultured neural cells. Hum. Mol. Genet. 2005;14:3661–3671. doi: 10.1093/hmg/ddi394. [DOI] [PubMed] [Google Scholar]
- 30.Hashem V., Galloway J.N., Mori M., Willemsen R., Oostra B.A., Paylor R., Nelson D.L. Ectopic expression of CGG containing mRNA is neurotoxic in mammals. Hum. Mol. Genet. 2009;18:2443–2451. doi: 10.1093/hmg/ddp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Arocena D., Yang J.E., Brouwer J.R., Tassone F., Iwahashi C., Berry-Kravis E.M., Goetz C.G., Sumis A.M., Zhou L., Nguyen D.V., et al. Fibroblast phenotype in male carriers of FMR1 premutation alleles. Hum. Mol. Genet. 2010;19:299–312. doi: 10.1093/hmg/ddp497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berman R.F., Murray K.D., Arque G., Hunsaker M.R., Wenzel H.J. Abnormal dendrite and spine morphology in primary visual cortex in the CGG knock-in mouse model of the fragile X premutation. Epilepsia. 2012;53(Suppl 1):150–160. doi: 10.1111/j.1528-1167.2012.03486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brouwer J.R., Huizer K., Severijnen L.A., Hukema R.K., Berman R.F., Oostra B.A., Willemsen R. CGG-repeat length and neuropathological and molecular correlates in a mouse model for fragile X-associated tremor/ataxia syndrome. J. Neurochem. 2008;107:1671–1682. doi: 10.1111/j.1471-4159.2008.05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brouwer J.R., Mientjes E.J., Bakker C.E., Nieuwenhuizen I.M., Severijnen L.A., Van der Linde H.C., Nelson D.L., Oostra B.A., Willemsen R. Elevated Fmr1 mRNA levels and reduced protein expression in a mouse model with an unmethylated Fragile X full mutation. Exp. Cell Res. 2007;313:244–253. doi: 10.1016/j.yexcr.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brouwer J.R., Severijnen E., de Jong F.H., Hessl D., Hagerman R.J., Oostra B.A., Willemsen R. Altered hypothalamus-pituitary-adrenal gland axis regulation in the expanded CGG-repeat mouse model for fragile X-associated tremor/ataxia syndrome. Psychoneuroendocrinology. 2008;33:863–873. doi: 10.1016/j.psyneuen.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Entezam A., Biacsi R., Orrison B., Saha T., Hoffman G.E., Grabczyk E., Nussbaum R.L., Usdin K. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395:125–134. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin M., Entezam A., Usdin K., Huang T., Liu Z.H., Hoffman G.E., Smith C.B. A mouse model of the fragile X premutation: effects on behavior, dendrite morphology, and regional rates of cerebral protein synthesis. Neurobiol. Dis. 2011;42:85–98. doi: 10.1016/j.nbd.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willemsen R., Hoogeveen-Westerveld M., Reis S., Holstege J., Severijnen L.A., Nieuwenhuizen I.M., Schrier M., van Unen L., Tassone F., Hoogeveen A.T., et al. The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum. Mol. Genet. 2003;12:949–959. doi: 10.1093/hmg/ddg114. [DOI] [PubMed] [Google Scholar]
- 39.Bontekoe C.J., Bakker C.E., Nieuwenhuizen I.M., van der Linde H., Lans H., de Lange D., Hirst M.C., Oostra B.A. Instability of a (CGG)98 repeat in the Fmr1 promoter. Hum. Mol. Genet. 2001;10:1693–1699. doi: 10.1093/hmg/10.16.1693. [DOI] [PubMed] [Google Scholar]
- 40.Hunsaker M.R., von Leden R.E., Ta B.T., Goodrich-Hunsaker N.J., Arque G., Kim K., Willemsen R., Berman R.F. Motor deficits on a ladder rung task in male and female adolescent and adult CGG knock-in mice. Behav. Brain Res. 2011;222:117–121. doi: 10.1016/j.bbr.2011.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Dam D., Errijgers V., Kooy R.F., Willemsen R., Mientjes E., Oostra B.A., De Deyn P.P. Cognitive decline, neuromotor and behavioural disturbances in a mouse model for fragile-X-associated tremor/ataxia syndrome (FXTAS) Behav. Brain Res. 2005;162:233–239. doi: 10.1016/j.bbr.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 42.Diep A.A., Hunsaker M.R., Kwock R., Kim K., Willemsen R., Berman R.F. Female CGG knock-in mice modeling the fragile X premutation are impaired on a skilled forelimb reaching task. Neurobiol. Learn. Mem. 2012;97:229–234. doi: 10.1016/j.nlm.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunsaker M.R., Goodrich-Hunsaker N.J., Willemsen R., Berman R.F. Temporal ordering deficits in female CGG KI mice heterozygous for the fragile X premutation. Behav. Brain Res. 2010;213:263–268. doi: 10.1016/j.bbr.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunsaker M.R., Wenzel H.J., Willemsen R., Berman R.F. Progressive spatial processing deficits in a mouse model of the fragile X premutation. Behav. Neurosci. 2009;123:1315–1324. doi: 10.1037/a0017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berman R.F., Willemsen R. Mouse models of fragile X-associated tremor ataxia. J. Investig. Med. 2009;57:837–841. doi: 10.231/JIM.0b013e3181af59d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y., Tassone F., Berman R.F., Hagerman P.J., Hagerman R.J., Willemsen R., Pessah I.N. Murine hippocampal neurons expressing Fmr1 gene premutations show early developmental deficits and late degeneration. Hum. Mol. Genet. 2010;19:196–208. doi: 10.1093/hmg/ddp479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lessard M., Chouiali A., Drouin R., Sebire G., Corbin F. Quantitative measurement of FMRP in blood platelets as a new screening test for fragile X syndrome. Clin. Genet. 2012;82:472–477. doi: 10.1111/j.1399-0004.2011.01798.x. [DOI] [PubMed] [Google Scholar]
- 48.Ericsson C., Nister M. Protein extraction from solid tissue. Methods Mol. Biol. 2011;675:307–312. doi: 10.1007/978-1-59745-423-0_17. [DOI] [PubMed] [Google Scholar]
- 49.Saffary R., Xie Z. FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J. Neurosci. 2011;31:1427–1439. doi: 10.1523/JNEUROSCI.4854-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tassone F., Hagerman R.J., Garcia-Arocena D., Khandjian E.W., Greco C.M., Hagerman P.J. Intranuclear inclusions in neural cells with premutation alleles in fragile X associated tremor/ataxia syndrome. J. Med. Genet. 2004;41:e43. doi: 10.1136/jmg.2003.012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen L.S., Tassone F., Sahota P., Hagerman P.J. The (CGG)n repeat element within the 5′ untranslated region of the FMR1 message provides both positive and negative cis effects on in vivo translation of a downstream reporter. Hum. Mol. Genet. 2003;12:3067–3074. doi: 10.1093/hmg/ddg331. [DOI] [PubMed] [Google Scholar]
- 52.Ludwig A.L., Hershey J.W., Hagerman P.J. Initiation of translation of the FMR1 mRNA Occurs predominantly through 5′-end-dependent ribosomal scanning. J. Mol. Biol. 2011;407:21–34. doi: 10.1016/j.jmb.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abitbol M., Menini C., Delezoide A.L., Rhyner T., Vekemans M., Mallet J. Nucleus basalis magnocellularis and hippocampus are the major sites of FMR-1 expression in the human fetal brain. Nat. Genet. 1993;4:147–153. doi: 10.1038/ng0693-147. [DOI] [PubMed] [Google Scholar]
- 54.Hinds H.L., Ashley C.T., Sutcliffe J.S., Nelson D.L., Warren S.T., Housman D.E., Schalling M. Tissue specific expression of FMR-1 provides evidence for a functional role in fragile X syndrome. Nat. Genet. 1993;3:36–43. doi: 10.1038/ng0193-36. [DOI] [PubMed] [Google Scholar]
- 55.Bachner D., Manca A., Steinbach P., Wohrle D., Just W., Vogel W., Hameister H., Poustka A. Enhanced expression of the murine FMR1 gene during germ cell proliferation suggests a special function in both the male and the female gonad. Hum. Mol. Genet. 1993;2:2043–2050. doi: 10.1093/hmg/2.12.2043. [DOI] [PubMed] [Google Scholar]
- 56.Khandjian E.W., Fortin A., Thibodeau A., Tremblay S., Cote F., Devys D., Mandel J.L., Rousseau F. A heterogeneous set of FMR1 proteins is widely distributed in mouse tissues and is modulated in cell culture. Hum. Mol. Genet. 1995;4:783–789. doi: 10.1093/hmg/4.5.783. [DOI] [PubMed] [Google Scholar]
- 57.Wang J.Y., Hessl D., Iwahashi C., Cheung K., Schneider A., Hagerman R.J., Hagerman P.J., Rivera S.M. Influence of the fragile X mental retardation (FMR1) gene on the brain and working memory in men with normal FMR1 alleles. Neuroimage. 2013;65:288–298. doi: 10.1016/j.neuroimage.2012.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo Y., Shan G., Guo W., Smrt R.D., Johnson E.B., Li X., Pfeiffer R.L., Szulwach K.E., Duan R., Barkho B.Z., et al. Fragile X mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 2010;6:e1000898. doi: 10.1371/journal.pgen.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tassone F., Hagerman R.J., Loesch D.Z., Lachiewicz A., Taylor A.K., Hagerman P.J. Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA. Am. J. Med. Genet. 2000;94:232–236. doi: 10.1002/1096-8628(20000918)94:3<232::aid-ajmg9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 60.Tassone F., Hagerman P.J. Expression of the FMR1 gene. Cytogenet. Genome Res. 2003;100:124–128. doi: 10.1159/000072846. [DOI] [PubMed] [Google Scholar]
- 61.Hunsaker M.R., Greco C.M., Tassone F., Berman R.F., Willemsen R., Hagerman R.J., Hagerman P.J. Rare intranuclear inclusions in the brains of 3 older adult males with fragile X syndrome: implications for the spectrum of fragile X-associated disorders. J. Neuropathol. Exp. Neurol. 2011;70:462–469. doi: 10.1097/NEN.0b013e31821d3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wenzel H.J., Hunsaker M.R., Greco C.M., Willemsen R., Berman R.F. Ubiquitin-positive intranuclear inclusions in neuronal and glial cells in a mouse model of the fragile X premutation. Brain Res. 2010;1318:155–166. doi: 10.1016/j.brainres.2009.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greco C.M., Berman R.F., Martin R.M., Tassone F., Schwartz P.H., Chang A., Trapp B.D., Iwahashi C., Brunberg J., Grigsby J., et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- 64.Saluto A., Brussino A., Tassone F., Arduino C., Cagnoli C., Pappi P., Hagerman P., Migone N., Brusco A. An enhanced polymerase chain reaction assay to detect pre- and full mutation alleles of the fragile X mental retardation 1 gene. J. Mol. Diagn. 2005;7:605–612. doi: 10.1016/S1525-1578(10)60594-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laemmli U.K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 1973;80:575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- 66.Iwahashi C., Tassone F., Hagerman R.J., Yasui D., Parrott G., Nguyen D., Mayeur G., Hagerman P.J. A quantitative ELISA assay for the fragile X mental retardation 1 protein. J. Mol. Diagn. 2009;11:281–289. doi: 10.2353/jmoldx.2009.080118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rasband W.S. ImageJ. U. S. National Institutes of Health. Bethesda, Maryland, USA: 1997–2012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.