Abstract
Background and Aims
The ability of plant lineages to reach all continents contributes substantially to their evolutionary success. This is exemplified by the Poaceae, one of the most successful angiosperm families, in which most higher taxa (tribes, subfamilies) have global distributions. Due to the old age of the ocean basins relative to the major angiosperm radiations, this is only possible by means of long-distance dispersal (LDD), yet the attributes of lineages with successful LDD remain obscure. Polyploid species are over-represented in invasive floras and in the previously glaciated Arctic regions, and often have wider ecological tolerances than diploids; thus polyploidy is a candidate attribute of successful LDD.
Methods
The link between polyploidy and LDD was explored in the globally distributed grass subfamily Danthonioideae. An almost completely sampled and well-resolved species-level phylogeny of the danthonioids was used, and the available cytological information was assembled. The cytological evolution in the clade was inferred using maximum likelihood (ML) as implemented in ChromEvol. The biogeographical evolution in the clade was reconstructed using ML and Bayesian approaches.
Key Results
Numerous increases in ploidy level are demonstrated. A Late Miocene–Pliocene cycle of polyploidy is associated with LDD, and in two cases (the Australian Rytidosperma and the American Danthonia) led to secondary polyploidy. While it is demonstrated that successful LDD is more likely in polyploid than in diploid lineages, a link between polyploidization events and LDD is not demonstrated.
Conclusions
The results suggest that polyploids are more successful at LDD than diploids, and that the frequent polyploidy in the grasses might have facilitated the extensive dispersal among continents in the family, thus contributing to their evolutionary success.
Keywords: Long-distance dispersal, LDD, biogeography, cytology, Danthonioideae, evolution, diversity, Poaceae, polyploidy
INTRODUCTION
During the last two decades, the central importance of long-distance dispersal (LDD) in the generation of biodiversity has become obvious. Not only has it been the only process by which isolated archipelagos such as Hawaii and the Canary Islands have become vegetated (Wagner and Funk, 1995), but it has also led to the transoceanic expansion of numerous genera [e.g. Hordeum (Blattner, 2006), Festuca (Inda et al., 2008), Cardamine (Carlsen et al., 2009)] and families [e.g. Proteaceae (Barker et al., 2007), Fabaceae (Lavin et al., 2005)] where they often become important contributors to the diversity of the newly colonized region. Dispersal to new continents, isolated islands or new habitats is also often associated with the dramatic and rapid evolution of structural diversity or the generation of great species richness. Such radiations are well documented from archipelagos such as Hawaii (Givnish et al., 1995, 2009 Baldwin, 1997), but are also known from large islands, such as New Zealand (Pirie et al., 2010; Wagstaff et al., 2010), or isolated habitats on continents (Klak et al., 2004; Hughes and Eastwood, 2006). There has been substantial recent research seeking predictors for these LDD events, with a strong focus on extrinsic environmental explanations. In particular, it has been suggested that habitat similarity (Crisp et al., 2009), dominant wind directions (Wright et al., 2000; Muñoz et al., 2004; Sanmartín et al., 2007) or bird migration routes (Coleman et al., 2003; Blattner, 2006) might determine dispersal routes and events. Although intrinsic attributes such as seed morphological structures were explored early in the last century (Ridley, 1930), in the past decade these intrinsic factors have not received much attention. Indeed, a review of whether morphology relates to LDD concluded ‘that the relationship between morphologically defined dispersal syndrome and long-distance dispersal is poor’ (Higgins et al., 2003).
Polyploidy (the possession of at least three complements of chromosomes) or whole-genome duplication (WGD), followed by diploidization, is an important factor in plant evolution. The origins of seed plants and angiosperms (Jiao et al., 2011) and the diversification of angiosperms have been linked to a series of WGD events (Soltis et al., 2009). Polyploidy may impact plant–animal interactions (Thompson et al., 2004) due to differences in the secondary chemicals among the cytotypes (Levin, 2002), and may extend the ecological range available to species, resulting in range expansions into often hostile habitats (Brochmann et al., 2004). Polyploids are linked to adaptive radiations, as exemplified by the silversword radiation on Hawaii (Barrier et al., 1999). Invasive species (Ellstrand and Schierenbeck, 2000) are also more likely to be polyploid than diploid, as shown in a meta-analysis of 81 invasive species and their congeners (Pandit et al., 2011). Much of this success has been hypothesized to be due to allopolyploids combining the genetic information of two parent species (Mummenhoff and Franzke, 2007; Doyle et al., 2008), where the larger genetic variance might enable a wider adaptive range (Lee, 2002).
An association between polyploidy and LDD (interpreted here in the historical biogeographical sense, i.e. crossing ocean basins) has been reported from diverse groups, from several continents and scattered over the angiosperm phylogeny, illustrating that the association is widespread (Coleman et al., 2003; Kadereit et al., 2006; Moore and Donoghue, 2007; Mummenhoff and Franzke, 2007; Inda et al., 2008; Marcussen et al., 2012). There are a number of cases where it is clear that the polyploidy happened after dispersal, e.g. in the amphitropical American disjunctions in the Polemoniaceae (Johnson et al., 2012) or the Australian Lepidium (Brassicaceae) (Mummenhoff et al., 2004), corroborating the interpretation that polyploidy has more to do with post-dispersal establishment, or maybe even persistence, than with the actual dispersal. There are also numerous cases where polyploids within a species complex are dispersed much more widely than diploids [e.g. the soybean Glycine tabacina, where the polyploids have reached the Pacific islands, while the diploid populations are restricted to the ancestral area in eastern Australia (Doyle et al., 1990)].
The association between polyploidy and LDD can operate at two levels. The more general hypothesis is that LDD is more frequent in polyploid than diploid lineages. Thus the polyploidization event can precede LDD, and one polyploidization event can facilitate a series of LDD events. A more restrictive hypothesis is that LDD events are associated with polyploidization events, thus ploidy changes and LDD co-occur more frequently than expected, and a polyploidization event can be associated with only one LDD event.
Here we explore the correlation between polyploidy and LDD in the danthonioid grasses. The 281 species of the C3, temperate, grass subfamily Danthonioideae (Linder et al., 2010) are found on all continents excluding Antarctica (Fig. 1), but are both ecologically more common and more species rich on the southern continents (Linder et al., 2013). Well known austral danthonioids include the Australian wallaby grasses (Rytidosperma), New Zealand snowgrasses (Chionochloa) and South American pampas grasses (Cortaderia). In the northern continents, the danthonioids are less well known, and include the oat grasses (Danthonia) from North America. By correlating LDD events with the ploidy level change in the danthonioids we show that successful LDD is significantly more frequent in polyploid than diploid clades. However, we find no support for the more restrictive hypothesis, that LDD and polyploidization events occur simultaneously.
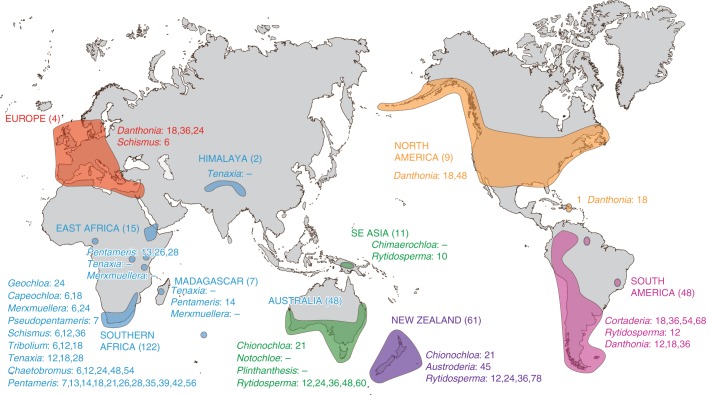
Fig. 1.
Geographical distribution of the danthonioid grasses. The number of species in each region is given, as well as the genera and the haploid chromosome numbers recorded. The largest diversity is in the Southern Hemisphere, particularly in southern Africa, with only three genera (non-endemic) and 15 species on the northern continents.
MATERIALS AND METHODS
Phylogeny
The phylogeny of the grass subfamily Danthonioideae was obtained from Antonelli et al. (2011) and Linder et al. (2013). This is based on 270 accessions (representing approx. 81 % of the 281 described species) and eight plastid and two nuclear DNA sequence regions: trnL-trnF, rpl16, rbcL, ndhF, matK, atpB-rbcL, trnT-trnL, trnC-trnD, ITS (internal transcribed spacer) and 26S, resulting in a matrix with 14 425 characters. The cladistic topology was inferred using both Bayesian and maximum likelihood (ML) methods, implemented in MrBayes (Huelsenbeck and Ronquist, 2001) and Garli (Zwickl, 2006), respectively. The trees were rate corrected with BEAST (Drummond and Rambaut, 2007), using two independent runs of 107 generations each, sampling every 1000th generation and assuming a Yule tree prior, and calibrated against the median crown node age of the subfamily as established by Bouchenak-Khelladi et al. (2010). We removed all taxa lacking ploidy data, and among the duplicated taxa (where the plastid and nuclear partitions were incongruent) retained only those based on nuclear data (as the chromosome number is nuclear inherited) using Drop-tip implemented in the R package APE (Paradis et al., 2004).
There is evidence for hybridization in the subfamily (Pirie et al., 2009); consequently, the taxon phylogeny should be seen as a network rather than a strictly dichotomously branching tree. Such a network could be inferred from the nuclear gene trees. However, our focus here is on the number of chromosomes rather than the patterns of hybridization; consequently, the simplified ‘tree-like’ approximation of the phylogeny should be adequate.
Ploidy data
The chromosome numbers for the danthonioid species were obtained from a literature survey, and were all derived from counts either from root tips or from pollen mother cells. We checked the data against the IPCN database, accessed 16–17 August 2012 (http://www.tropicos.org/Project/IPCN). For the analyses, we used the minimum chromosome number in species with several ploidy levels, on the assumption that the lowest number is ancestral in the species and that higher numbers are the result of secondary duplications within the species. Consequently the lowest number is relevant to understanding the supra-specific evolution of chromosome numbers. Randomly sampling counts from the set reported per species (Pandit et al., 2011) assumes that intraspecific gains and losses are equally likely. We assume that, despite some evidence of potential diploidization, the number of chromosomes can be used to infer the number of polyploidization events, while completely aware that chromosome rearrangements could reduce these. Where available, the reported counts were compared with published photographs and camera lucida drawings, and the vouchers checked.
Dispersal events
The Danthonioideae date to the Oligocene (Bouchenak-Khelladi et al., 2010), too recent for vicariance explanations for the disjunctions across the Indian and Atlantic Oceans. Consequently we did not use analytical methods that allowed for vicariance, such as DIVA (Ronquist, 1996; Yan Yu et al., 2011) or DEC (Ree and Smith, 2008). Instead we obtained an initial mapping of the distribution ranges over the set of Bayesian trees using stochastic mapping, implemented in ‘Multistate’ in BayesTraits (Pagel and Meade, 2010). In order to retain compatibility with the ploidy analysis, we used the set of reduced trees. The analysis was iterated 5 million times, the first 200 000 replicates were removed as burn-in and subsequent sampling density was every 1000 replicates. In order to find the best priors, we implemented a hyperprior. The Rate Dev was set to 0·001 in order to obtain a 20 % acceptance rate. The nodes of interest were defined with the ‘AddMRCA’ command. The areas defined are Africa (including East Africa, the only count for Madagascar is of a species widespread in southern Africa), Australia (including Tasmania and New Guinea), New Zealand (including the islands), South America, North America (including the Caribbean islands) and Europe. Biogeographical areas ‘missing’ due to the absence of chromosome counts of endemic species are Madagascar, Amsterdam Island (in the southern Indian Ocean) and the Himalayas.
Mapping ploidy changes
We used ChromEvol (Mayrose et al., 2010) to estimate the number of genome polyploidizations (duplication of the genomes), demi-polyploidizations (combination of, for example, diploid and tetraploid genomes to form a hexaploid genome) and ascending or descending dysploidy (the gains or loss of single or, more rarely, several chromosomes). This tests a set of eight models (chromosome gains, losses, duplication events and demi-polyploidization events) and obtains the likelihood for each model. The optimal model is then used to map the events over a tree. This method uses only one tree, so cannot take account of topological uncertainty. We set the initial parameters as ‘gainConstR’ = 0·5, ‘lossConstR’ = 0·5, ‘duplConstR’ = 0·5 and ‘DemiPloidyR’ = 0·5. In order to obtain a random model, 1 000 000 simulations were performed.
Hypothesis 1: ploidy increase and dispersal
In order to determine whether LDD was more likely in a polyploid than a diploid clade, we tested whether dispersed clades are polyploid, and undispersed clades diploid. We scored each species as either diploid (2n = 12 or 14) or polyploid (2n > 14). Species from southern Africa were scored as ‘undispersed’, consistent with our findings that the ancestral area is southern Africa (Linder et al., 2013) and our results here. We consequently scored all species from outside southern Africa as ‘dispersed’. This test was conducted using a Bayesian approach as implemented in BayesTraits (Pagel and Meade, 2010), as this takes into account branch lengths as well as topological uncertainty. Reversible jump was used to select between models; these could be classified as ‘dependent’ (dispersal constrained to occur only in polyploid taxa) or ‘independent’ (dispersal and polyploidy allowed to vary independently on the trees) models. The analyses were run over 2 million generations, with the first 500 000 discarded as burn-in. The acceptance rate was used as an indicator to show when the results stabilized. One tree in every 500 replicates was sampled. The priors were unrestricted (reversible jump to a uniform prior with very wide margins). Rate dev was set to 0·001, in order to obtain an acceptance rate between 20 and 40 %.
Hypothesis 2: LDD linked to polyploidization events
We tested whether polyploidization events predict LDD by calculating a logistic regression with LDD as the response variable and polyploidization events as the predictor variable. Because both LDD and polyploidization events are more likely on longer branches (both were mapped over the topology assuming a response to branch length), we included branch length as a predictor variable. As the distribution of branch lengths was strongly left-skewed, we normalized the distribution by log-transforming branch length. These analyses were conducted in R (R Development Core Team, 2012), using the packages (aod) (Lesnoff and Lancelot, 2012).
RESULTS
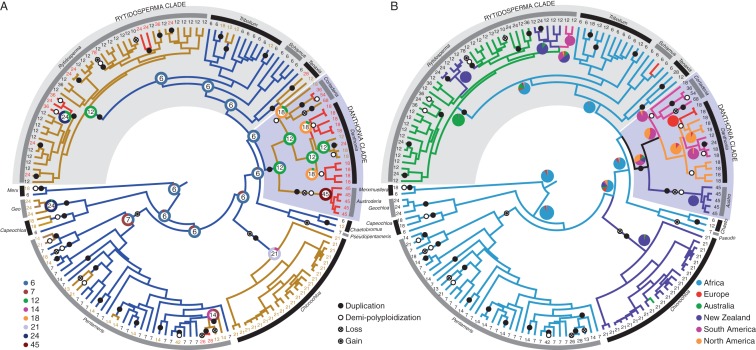
Chromosome numbers are available for 180 of the 315 species and subspecies of danthonioids (Supplementary Data Table S1), and, of these, 154 are included in the phylogeny (Supplementary Data Fig. S1). The reconstruction of the evolution of ploidy numbers using ChromEvol (Mayrose et al., 2010) suggests that the most complicated evolutionary model applies, which allows for separate rates of polyploidizations and demi-polyploidizations, as well as separate rates of individual chromosome losses and gains (ascending or descending dysploidy) scaled to the total number of chromosomes (Table 1). According to this model, there should be 40·3 genome duplications, 19·9 demi-polyploidizations, 3·6 chromosome gains and 11·2 chromosome losses. The ancestral haploid number is probably six (Fig. 2A; Supplementary Data Table S2).
Table 1.
Comparison of the evolutionary models
| Model | Variables in the model | Log-likelihood | AIC |
|---|---|---|---|
| LINEAR_RATE_DEMI_EST | Dups/demi/linear loss/linear gain | –259·5 | 531·1 |
| CONST_RATE_DEMI_EST | Dups/demi/loss/gain | –262 | 531·9 |
| LINEAR_RATE_DEMI | Dups = demi/linear loss/linear gain | –262·7 | 535·3 |
| CONST_RATE_DEMI | Dups = demi/loss/gain | –264·9 | 535·9 |
| LINEAR_RATE | Dups/linear loss/linear gain | –392·2 | 794·5 |
| CONST_RATE | Dups/loss/gain | –398·2 | 802·5 |
| LINEAR_RATE_NO_DUPL | Linear gain/linear loss | –589·8 | 1188 |
| CONST_RATE_NO_DUPL | Gain/loss | –699·8 | 1404 |
Note that the models are ranked by their AIC values
Dups, polyploidization; demi, demi-polyploidization; loss, loss of a chromosome; gain, gain of a chromosome; linear, loss and gain rates scaled to the total chromosome number.
Fig. 2.
Maximum clade credibility (MCC) tree topology of 154 species of Danthonioideae for which ploidy data are available, based on 14 425 aligned DNA base pairs. The branches are scaled to age. Whole-genome duplications are indicated as filled circles (either one or more duplications), demi-polyploidizations by open circles, losses of one or more chromosomes by circles with a plus, and gains of one or more chromosome by a circle with a central dot; species names have been replaced by the haploid chromosome number. (A) The most likely haploid chromosome numbers are indicated at the critical nodes, inside a doughnut indicating the posterior probabilities of all haploid numbers at that node. Branch colours reflect the ploidy level: the ancestral haploid number is six; nodes that retain this ploidy level are coloured blue. A first increase in ploidy is coloured brown, and a second increase (either duplication or demi-polyploidization) red. (B) Branch colours indicate the geographical regions as inferred from ML, and the probability distribution (as determined by stochastic mapping) at critical nodes is given as pie-charts. Abbreviations of generic names: Austro, Austroderia; Chaeto, Chaetobromus; Geo, Geochloa; Merx, Merxmuellera; Pseudo, Pseudopentameris.
According to the area optimizations, the ancestral area of the danthonioids was, with a high probability, southern Africa (Fig. 2B). Subsequent areas occupied were New Zealand, South America and Australia. According to the phylogenetic reconstruction, many of these range expansions occurred repeatedly. The ancestral area of the Danthonia clade is unclear, and is with almost equal probability South or North America, with much lower probability New Zealand or Australia, or even Europe, but clearly not Africa. Subsequent areas occupied are North America, Europe and Malaysia. Thus we have range expansion from southern Africa to all other continents. These range expansions are most probably by LDD, as the dates estimated for the danthonioid disjunctions are much younger than the age of the ocean basins. The first Indian Ocean crossing is dated to 4·35–18·66 million years ago (Ma) (Chionochloa), while the ocean floor is dated to 105–120 Ma (McLoughlin, 2001); for the Atlantic ocean it is 3·76–16·79 Ma (Cortaderia) and the ocean floor dates to 105–120 Ma; the first crossing of the Pacific is 1·04–5·1 Ma (Rytidosperma) against an ocean floor age of 30–50 Ma; the earliest trans-Tasman dispersals are 1·93–9·55 Ma (Notochloe–Plinthanthesis) over an ocean floor dated 76–95 Ma.
Bayesian analysis confirmed that LDD occurs significantly more frequently in polyploid than diploid lineages. Indeed, inspection of Fig. 2 shows that LDD events occur only in polyploid lineages or in association with genome duplications. This is supported by Pentameris natalensis, with diploid and tetraploid populations in Africa, and only tetraploids known from Madagascar (Tateoka, 1965; Davidse et al., 1986; Du Plessis and Spies, 1992).
The logistic regression model of LDD as the response variable and polyploidization events and branch lengths as predictor variables is weakly better than an empty model (P < 0·05), and retained log-branch length in the model, at P < 0·05, but not polyploidization events.
DISCUSSION
Our results, based on a well sampled phylogeny and substantial cytological data, find support for the hypothesis that successful LDD is more likely in polyploid than diploid lineages, but finds no support for the more restrictive hypothesis that ploidy changes and LDD co-occur significantly in the subfamily Danthonioideae. The optimizations indicate an ancestral haploid number of six for the Danthonioideae, the same as that estimated for the PACCAD clade in which the Danthonioideae are nested (Grass Phylogeny Working Group, 2001). As is common in the grasses, polyploids are numerous in the clade, and in some instances form nested series. Forty-two of the 180 taxa (23 %) have more than one ploidy level, and this number is probably an underestimate due to the small number of species which have been studied more than once (for 52 of the 180 species only a single report on the ploidy level is available). Only 40 of the 180 species are diploid, thus 78 % of the species are polyploid, a value which agrees closely with the approx. 80 % polyploidy suggested for the grasses (Hunziker and Stebbins, 1986). Danthonioids have among the highest ploidy levels recorded for the grasses (Rytidosperma exiguum is 2n = 156, and thus 26-ploid). The reconstruction suggests 38 duplication events and 19 demi-polyploidization events with a probability of >0·5. These events are evenly scattered across the phylogeny (Fig. 2). Twenty-six of these duplications affect only a single terminal species, and more sampling might reveal that these species contain polyploid series. The earliest, Miocene, duplications are inherited by several species, and these species may be considered mesopolyploids (in the sense of Mandakova et al., 2010). In two instances these provide the basis of a secondary round of polyploidization, a pattern that appears to be typical of the grasses (Stebbins, 1985) and other families such as Brassicaceae (Mandakova et al., 2010).
Dysploid evolution has received little attention in the literature, but three of the ten instances in the danthonoids are associated with dramatic changes. In Pentameris ascending dysploidy is associated with a major diversification: Pentameris is the most species-rich grass clade in the otherwise quite grass-poor Cape flora, and members of the genus are also important components of the grass flora of some African tropic–alpine habitats (P. mannii on Mt Cameroon and P. minor on Mt Kilimanjaro). Its ancestral haploid count of n = 7 is, presumably, derived by the dysploid gain of a chromosome. This gain occurred at least three times in the danthonioids: at the base of the Pentameris clade, in the lineage leading to Chionochloa, and also in Pseudopentameris. In the New Zealand Chionochloa, this led to the radiation of the snowgrasses (Pirie et al., 2010), one of the important elements of the southern New Zealand tussock grasslands (Wardle, 1991). The unusual n = 13 appears to have evolved twice in Pentameris, probably by a duplication and descending dysploidy from n = 7, or, less likely, by hybridization with Merxmuellera giving 6 + 7 (Du Plessis and Spies, 1992). The one origin is in the widespread and rather unusual Cape grass P. eriostoma, and the second is in the tropical African clade of P. borussica, P. mannii and P. minor. The latter clade is associated with an LDD, first from southern to eastern Africa (where the lineage is found in the montane zone of the Rift Valley mountains), then to Mt Cameroon, where P. mannii is a co-dominant grass in the upper alpine reaches of the peak.
The Danthonia and the Rytidosperma clades are examples of nested polyploidizations (Stebbins, 1985). The Danthonia clade, which includes Cortaderia, Austroderia, Danthonia, Notochloe, Plinthanthesis and Chimaerochloa, originated with a chromosome doubling event associated with dispersal from Africa to the Americas (Fig. 2). This laid the foundation for numerous subsequent increases in ploidy in the genera in this clade. Most Danthonia species are hexaploids (n = 18), resulting from demi-polyploidizations. The current phylogenetic resolution suggests that this evolved twice, but it also remains possible that this demi-polyploidization occurred only once, as the phylogeny of Danthonia is still not robustly resolved. Cortaderia shows a plethora of numbers, including very high ploidy (to 25-ploid C. peruvianus), building up from a possible ancestral number of n = 18, as found in C. pilosa. However, additional chromosome counts are needed from all species in this genus to understand the chromosomal evolution within Cortaderia. Cortaderia and Austroderia both show extensive gynodioecism, an unusual breeding system in the grasses (Connor, 1970, 1979). Austroderia in New Zealand (n = 45) is an example of repeated secondary duplications. The Rytidosperma clade is an excellent example of the evolution of a new ‘base’ number, from which numerous polyploid complexes evolved. The ancestral condition of n = 6 is retained by all African members of the clade (Schismus, Tribolium and Tenaxia). At the base of the genus Rytidosperma is a chromosome duplication which co-occurs with dispersal from Africa to Australia. These tetraploids are reported to behave as diploids (Brock and Brown, 1961). Although the substantial variations in chromosome numbers in some widespread Australian species of Rytidosperma are linked to ecological and geographical differentiation (Waters et al., 2010), subsequent dispersal to New Zealand and South America is not linked to further ploidy increases, and the dispersal of R. oreoboloides from Australia to New Guinea is correlated with descending dysploidy. We have no further information on the other ten Malesian species, so do not know whether the whole group have n = 10.
Many reasons have been advanced to explain why polyploids may be successful as long-distance dispersers; these are all associated with the biology of establishment, rather than of dispersal. Polyploidy (and particularly allopolyploidy) allows the immigrants to survive genetic bottlenecks resulting from single-seed immigrants and the subsequent inbreeding in small, newly established populations (Barrett, 1988). This also includes the masking of deleterious alleles, thus reducing the impact of inbreeding depression and fixed heterozygosity (Soltis and Soltis, 2000; te Beest et al., 2012). Adaptation to the different environmental and ecological opportunities in the new continent could be enhanced by the wider range of gene expression found in heterozygote allopolyploids, leading to novel trait expression and new ecological opportunity (Doyle et al., 2008; te Beest et al., 2012). It is usually assumed that most grass polyploids are allopolyploids (De Wet, 1986) resulting from the hybridization of two species. The situation is unclear in the danthonioids. In Pentameris airoides, meiotic studies revealed chromosome pairing consistent with an allopolyploid, thus hybrid, origin (Spies et al., 1994). In all Rytidosperma species investigated, the meiotic chromosomes formed bivalents (Brock and Brown, 1961), but this could also result from diploidization. Another line of evidence comes from incongruent nuclear and plastid phylogenies, which suggest the hybrid origin of the hexaploid core Cortaderia. However, other similarly demonstrated hybrids (Pirie et al., 2008) show no increase in ploidy (e.g. Capeochloa arundinacea and Danthonia alpina) or are chromosomally unknown (Notochloe). It is therefore unknown how many of the polyploid events, and particularly the palaeopolyploid events, are associated with hybridization.
Polyploid grasses may have a further advantage in LDD, as they have an unusual, two-locus, gametophytic incompatibility system, the S–Z system (Pandey, 1979). If the allelles of both loci of pollen and stigma match, then they are incompatible: the pollen still germinates, but the pollen tube does not successfully penetrate the stigma (Hayman, 1992; Langridge and Baumann, 2008). The S–Z system differs in several significant ways from the S system, which is common in the rest of the angiosperms. First, even selfed flowers have a low seed set. Thus the system is ‘leaky’; this low seed set is referred to as ‘pseudocompatible’. This means that a single seed establishing after LDD is still capable of producing offspring, albeit in small numbers. The clonal growth form of grasses may allow the plants to persist, thus generating more opportunity for occasional seed set. The second important attribute is that the S–Z system does not break down in polyploids, as incompatibility occurs if either or any of the S–Z sets are identical. This guarantees an outcrossing advantage in small populations, and reduces inbreeding. However, the sparse current evidence is that both Australian and New Zealand danthonioids are self-compatible (Cashmore, 1932; Brock and Brown, 1961), suggesting that this mechanism may not apply. A more detailed survey of the breeding systems of the danthonioids, and the grasses in general, is needed to test this model.
How general is the association between LDD and polyploidy? We know of seven well-demonstrated cases (Table 2); there are no doubt many more. If the association with the S–Z incompatability system is valid, then they should be particularly common in the grasses, and, if the association with diversification holds, then also in the Asteraceae and Poaceae (both with simple flowers, and recent radiations). This is corroborated by the very high percentage of polyploidy in the New Zealand grasses (Murray et al., 2005), all of which arrived by LDD. A total of 186 of the 203 species have been counted; of these only eight (4 %) are diploid, compared with a global average of 20 %.
Table 2.
Instances of polyploid long-distance dispersal in angiosperms
| Clade | Family | Ancestral area | Derived area | Reference |
|---|---|---|---|---|
| Loliinae (fescues) | Poaceae | Eurasia, diploid | Global, polyploid | Inda et al. (2008) |
| Santalum (sandalwoods) | Santalaceae | Australia, all diploid | Pacific islands, mostly polyploid | Harbaugh (2008) |
| Nicotiana (tobacco) | Solanaceae | South America, most diploid | Africa, Australia, polyploid | Aoki and Ito (2000); Chase et al. (2003) |
| Microseris | Asteraceae | Americas, diploid | Australia, allotetraploid | Vijverberg et al. (1999) |
| Argyroxiphium (silverswords) | Asteraceae | Americas, diploid | Hawaii, tetraploid | Baldwin and Wagner (2010) |
| Lepidium | Brassicaceae | Americas, diploid | Australia, allotetraploid | Mummenhoff et al. (2006) |
| Collomia, Naverretia | Polemoniaceae | North America, diploid | Patagonia, allotetraploid | Johnson et al. (2012) |
Polyploidy was inferred based on the chromosome numbers, with allotratraploidy inferred from the formation of bivalents at meiosis.
Stebbins (1985) proposed that there are cycles of polyploidization, followed by diploidization, in the grasses, associated with the occupation of new habitats (e.g. the result of mountain building, the Miocene evolution of pyrophytic grasslands and the climatic turbulence of the Plio-Pleistocene). We show that in the danthonioids these cycles can indeed be detected, but that they may be associated with the colonization of new continents. From an ancestral diploid lineage in Africa, colonization of South America by the tetraploid Danthonia clade, New Zealand by the hexaploid Chionochloa and Australia by the tetraploid Rytidosperma followed. In these three regions, these lineages became diploidized, and in two (Danthonia and Rytidosperma clades) provided the base from which further cycles of polyploidization occurred. These led to the hexadecaploid New Zealand Austroderia and the hexaploid North American and European Danthonia. The polyploid cycles in the grass subfamily Danthonioideae may be linked to successive waves of dispersal and subsequent diversification. Either way, the remarkable success of this austral grass clade may be intimately linked to the chromosome evolution in the clade. The high species richness of the Poaceae, with >10 000 species the fifth largest angiosperm family, and by far the largest family of wind-pollinated plants, may thus be due to polyploidy facilitating LDD, resulting in parallel radiations on new continents.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
This work was supported by Swiss National Science Foundation grant no. 3100A0-107927 to H.P.L. We would like to thank Itay Mayrose for assistance with ChromEvol, Richard Carter for R-scripts, and several anonymous reviewers and Susanne Renner for numerous insightful comments.
LITERATURE CITED
- Antonelli A, Humphreys AM, Lee WG, Linder HP. Absence of mammals and the evolution of New Zealand grasses. Proceedings of the Royal Society B: Biological Sciences. 2011;278:695–701. doi: 10.1098/rspb.2010.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki S, Ito M. Molecular phylogeny of Nicotiana (Solanaceae) based on the nucleotide sequence of the matK gene. Plant Biology. 2000;2:316–324. [Google Scholar]
- Baldwin BG. Adaptive radiation of the Hawaiian Silversword Alliance: congruence and conflict of phylogenetic evidence from molecular and non-molecular investigations. In: Givnish TJ, Systma KJ, editors. Molecular evolution and adaptive radiation. Cambridge: Cambridge University Press; 1997. pp. 103–128. [Google Scholar]
- Baldwin BG, Wagner WL. Hawaiian angiosperm radiations of North American origin. Annals of Botany. 2010;105:849–879. doi: 10.1093/aob/mcq052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker NP, Weston PH, Rutschmann F, Sauquet H. Molecular dating of the ‘Gondwanan’ plant family Proteaceae is only partially congruent with the timing of the break-up of Gondwana. Journal of Biogeography. 2007;34:2012–2027. [Google Scholar]
- Barrett SCH. Genetics and evolution of agricultural weeds. In: Altieri MA, Liebman M, editors. Weed management in agroecosystems: ecological approaches. Boca Raton, FL: CRC Press; 1988. pp. 57–75. [Google Scholar]
- Barrier M, Baldwin BG, Robichaux RH, Purugganan MD. Interspecific hybrid ancestry of a plant adaptive radiation: allopolyploidy of the Hawaiian silversword alliance (Asteraceae) inferred from floral homeotic gene duplications. Molecular Biology and Evolution. 1999;16:1105–1113. doi: 10.1093/oxfordjournals.molbev.a026200. [DOI] [PubMed] [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, et al. The more the better? The role of polyploidy in facilitating plant invasions. Annals of Botany. 2012;109:19–45. doi: 10.1093/aob/mcr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR. Multiple intercontinental dispersals shaped the distribution area of Hordeum (Poaceae) New Phytologist. 2006;169:603–614. doi: 10.1111/j.1469-8137.2005.01610.x. [DOI] [PubMed] [Google Scholar]
- Bouchenak-Khelladi Y, Verboom GA, Savolainen V, Hodkinson TR. Biogeography of the grasses (Poaceae): a phylogenetic approach to reveal evolutionary history in geographical space and geological time. Botanical Journal of the Linnean Society. 2010;162:543–557. [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG, et al. Polyploidy in arctic plants. Biological Journal of the Linnean Society. 2004;82:521–536. [Google Scholar]
- Brock RD, Brown JAM. Cytotaxonomy of Australian Danthonia. Australian Journal of Botany. 1961;9:62–91. [Google Scholar]
- Carlsen T, Bleeker W, Hurka H, Elven R, Brochmann C. Biogeography and phylogeny of Cardamine (Brassicaceae) Annals of the Missouri Botanical Garden. 2009;96:215–236. [Google Scholar]
- Cashmore AB. 1932. An investigation of the taxonomic and agricultural characters of the Danthonia group. Commonwealth of Australia. Council for Scientific and Industrial Research Bulletin no. 69. [Google Scholar]
- Chase MW, Knapp S, Cox AV, et al. Molecular systematics, GISH and the origin of hybrid taxa in Nicotiana (Solanaceae) Annals of Botany. 2003;92:107–127. doi: 10.1093/aob/mcg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman M, Liston A, Kadereit JW, Abbott RJ. Repeat intercontinental dispersal and Pleistocene speciation in disjunct Mediterranean and desert Senecio (Asteraceae) American Journal of Botany. 2003;90:1446–1454. doi: 10.3732/ajb.90.10.1446. [DOI] [PubMed] [Google Scholar]
- Connor HE. Gynodioecism in Danthonia archboldii. Australian Journal of Botany. 1970;18:233–236. [Google Scholar]
- Connor HE. Breeding systems in the grasses: a survey. New Zealand Journal of Botany. 1979;17:547–574. [Google Scholar]
- Crisp MD, Arroyo MTK, Cook LG, et al. Phylogenetic biome conservatism on a global scale. Nature. 2009;458:754–756. doi: 10.1038/nature07764. [DOI] [PubMed] [Google Scholar]
- Davidse G, Hoshino T, Simon BK. Chromosome counts of Zimbabwean grasses (Poaceae) and an analysis of polyploidy in the grass flora of Zimbabwe. South African Journal of Botany. 1986;52:521–528. [Google Scholar]
- De Wet JMJ. Hybridization and polyploidy in the Poaceae. In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME, editors. Grass systematics and evolution. Washington, DC: Smithsonian Institution Press; 1986. pp. 188–194. [Google Scholar]
- Doyle JJ, Doyle JL, Brown AH, Grace JP. Multiple origins of polyploids in the Glycine tabacina complex inferred from chloroplast DNA polymorphism. Proceedings of the National Academy of Sciences; USA. 1990. pp. 714–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, et al. Evolutionary genetics of genome merger and doubling in plants. Annual Review of Genetics. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. 2007 doi: 10.1186/1471-2148-7-214. BMC Evolutionary Biology 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Plessis H, Spies JJ. Chromosome numbers in the genus Pentaschistis (Poaceae, Danthonieae) Taxon. 1992;41:709–721. [Google Scholar]
- Ellstrand NC, Schierenbeck KA. Hybridization as a stimulus for the evolution of invasiveness in plants. In: Ayala FJ, Fitch WM, Clegg MT, editors. Variation and evolution in plants and microorganisms. Washington, DC: National Academy Press; 2000. pp. 289–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish TJ, Millam KC, Mast AR, et al. Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proceedings of the Royal Society B: Biological Sciences; 2009. pp. 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givnish TJ, Sytsma KJ, Smith JF, Hahn WJ. Molecular evolution, adaptive radiation, and geographical speciation in Cyanea (Campanulaceae, Lobelioideae) In: Wagner WL, Funk VA, editors. Hawaiian biogeography. Washington, DC: Smithsonian Institution Press; 1995. pp. 288–337. Evolution on a hot spot archipelago. [Google Scholar]
- Grass Phylogeny Working Group. Phylogeny and subfamilial classification of the grasses (Poaceae) Annals of the Missouri Botanical Garden. 2001;88:373–457. [Google Scholar]
- Harbaugh DT. Polyploid and hybrid origins of pacific island sandalwoods (Santalum, Santalaceae) inferred from low-copy nuclear and flow cytometry data. International Journal of Plant Sciences. 2008;169:677–685. [Google Scholar]
- Hayman DL. The S–Z incompatibility system. In: Chapman GP, editor. Grass evolution and domestication. Cambridge: Cambridge University Press; 1992. pp. 117–137. [Google Scholar]
- Higgins SI, Nathan R, Cain ML. Are long-distance dispersal events in plants usually caused by nonstandard means of dispersal? Ecology. 2003;84:1945–1956. [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MrBayes. Bayesian analysis of phylogeny. Version 3.1.2. [DOI] [PubMed] [Google Scholar]
- Hughes C, Eastwood R. Island radiation on a continental scale: exception rates of plant diversification after uplift of the Andes. Proceedings of the National Academy of Sciences; USA. 2006. pp. 10334–10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker JH, Stebbins GL. Chromosomal evolution in the Gramineae. In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME, editors. Grass systematics and evolution. Washington, DC: Smithsonian Institution Press; 1986. pp. 179–187. [Google Scholar]
- Inda LA, Segarra-Moragues JG, Muller J, Peterson PM, Catalan P. Dated historical biogeography of the temperate Loliinae (Poaceae, Pooideae) grasses in the northern and southern hemispheres. Molecular Phylogenetics and Evolution. 2008;46:932–957. doi: 10.1016/j.ympev.2007.11.022. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Johnson LA, Chan LM, Pozner R, Glazier LD. Allotetraploids in Patagonia with affinities to western North American diploids: did dispersal or genome doubling occur first? Botanical Review. 2012;78:288–306. [Google Scholar]
- Kadereit G, Mucina L, Freitag H. Phylogeny of Salicornioideae (Chenopodiaceae): diversification, biogeography, and evolutionary trends in leaf and flower morphology. Taxon. 2006;55:617–642. [Google Scholar]
- Klak C, Reeves G, Hedderson TA. Unmatched tempo of evolution in Southern African semi-desert ice plants. Nature. 2004;427:63–65. doi: 10.1038/nature02243. [DOI] [PubMed] [Google Scholar]
- Langridge P, Baumann U. Self-incompatibility in the grasses. In: Franklin-Tong VE, editor. Self-incompatibility in flowering plants. Berlin: Springer; 2008. pp. 275–287. Evolution, diversity, and mechanisms. [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Systematic Biology. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Lee CE. Evolutionary genetics of invasive species. Trends in Ecology and Evolution. 2002;17:386–391. [Google Scholar]
- Lesnoff M, Lancelot R. aod: analysis of overdispersed data. 2012. Version 1.3. http://cran.r-project.org/package=aod . [Google Scholar]
- Levin DA. The role of chromosomal change in plant evolution. Oxford: Oxford University Press; 2002. [Google Scholar]
- Linder HP, Antonelli A, Humphreys AM, Pirie MD, Wüest RO. What determines biogeographical ranges? Historical wanderings and ecological constraints in the danthonioid grasses. Journal of Biogeography. 2013;40:821–834. [Google Scholar]
- Linder HP, Baeza P. CM, Barker NP, et al. A generic classification of the Danthonioideae (Poaceae) Annals of the Missouri Botanical Garden. 2010;97:306–364. [Google Scholar]
- Mandakova T, Joly S, Krzywinski M, Mummenhoff K, Lysak MA. Fast diploidization in close mesopolyploid relatives of Arabidopsis. The Plant Cell. 2010;22:2277–2290. doi: 10.1105/tpc.110.074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcussen T, Jakobsen KS, Danihelka J, et al. Inferring species networks from gene trees in high-polyploid North American and Hawaiian violets (Viola, Violaceae) Systematic Biology. 2012;61:107–126. doi: 10.1093/sysbio/syr096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose I, Barker MS, Otto SP. Probabilistic models of chromosome number evolution and the inference of polyploidy. Systematic Biology. 2010;59:132–144. doi: 10.1093/sysbio/syp083. [DOI] [PubMed] [Google Scholar]
- McLoughlin S. The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Australian Journal of Botany. 2001;49:271–300. [Google Scholar]
- Moore BR, Donoghue MJ. Correlates of diversification in the plant clade Dipsacales: geographic movement and evolutionary innovations. American Naturalist. 2007;170:S28–S55. doi: 10.1086/519460. [DOI] [PubMed] [Google Scholar]
- Mummenhoff K, Franzke A. Gone with the bird: late Tertiary and Quaternary intercontinental long-distance dispersal and allopolyploidization in plants. Systematics and Biodiversity. 2007;5:255–260. [Google Scholar]
- Mummenhoff K, Linder HP, Friesen N, Bowman JL, Lee J-Y, Franzke A. Molecular evidence for bicontinental hybridogenous genomic constitution in Lepidium sensu stricto (Brassicaceae) species from Australia and New Zealand. American Journal of Botany. 2004;91:254–261. doi: 10.3732/ajb.91.2.254. [DOI] [PubMed] [Google Scholar]
- Mummenhoff K, Linder HP, Friesen N, Bowman JL, Lee J-Y, Franzke A. African species of Lepidium (Brassicaceae) contributed via hybridization to the origin of Australian/New Zealand species. In: Ghanzafar SA, Beentje HJ, editors. Taxonomy and ecology of African plants, their conservation and sustainable use. Kew: Royal Botanic Gardens; 2006. pp. 291–308. [Google Scholar]
- Muñoz J, Felicisimo AM, Cabezas F, Burgaz AR, Martinez I. Wind as a long-distance dispersal vehicle in the Southern Hemisphere. Science. 2004;304:1144–1147. doi: 10.1126/science.1095210. [DOI] [PubMed] [Google Scholar]
- Murray BG, De Lange PJ, Ferguson AR. Nuclear DNA variation, chromosome numbers and polyploidy in the endemic and indigenous grass flora of New Zealand. Annals of Botany. 2005;96:1293–1305. doi: 10.1093/aob/mci281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M, Meade A. 2010. BayesTraits manual. http://www.evolution.rdg.ac.uk/BayesTraits.html , accessed 26 January 2014. [Google Scholar]
- Pandey KK. Long-distance dispersal and self-incompatibility. New Zealand Journal of Botany. 1979;17:225–226. [Google Scholar]
- Pandit MK, Pocock MJO, Kunin WE. Ploidy influences rarity and invasiveness in plants. Journal of Ecology. 2011;99:1108–1115. [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pirie MD, Humphreys AM, Galley C, et al. A novel supermatrix approach improves resolution of phylogenetic relationships in a comprehensive sample of danthonioid grasses. Molecular Phylogenetics and Evolution. 2008;48:1106–1119. doi: 10.1016/j.ympev.2008.05.030. [DOI] [PubMed] [Google Scholar]
- Pirie MD, Humphreys AM, Barker NP, Linder HP. Reticulation, data combination, and inferring evolutionary history: an example from Danthonioideae (Poaceae) Systematic Biology. 2009;58:612–628. doi: 10.1093/sysbio/syp068. [DOI] [PubMed] [Google Scholar]
- Pirie MD, Lloyd K, Lee W, Linder HP. Diversification of Chionochloa (Poaceae) and biogeography of the New Zealand Southern Alps. Journal of Biogeography. 2010;37:379–392. [Google Scholar]
- R Development Core Team. 2012. R: a language and environment for statistical computing. Version 2.15.2 GUI 1.53 Vienna, www.r-project.org . [Google Scholar]
- Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- Ridley HN. The dispersal of plants throughout the world. Ashford, Kent: Reeve; 1930. [Google Scholar]
- Ronquist F. DIVA. 1996 Version 1.1. http://www.ebc.uu.se/systzoo/research/diva/diva.html . [Google Scholar]
- Sanmartín I, Wanntorp L, Winkworth RC. West Wind Drift revisited: testing for directional dispersal in the Southern Hemisphere using event-based tree fitting. Journal of Biogeography. 2007;34:398–416. [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, et al. Polyploidy and Angiosperm diversification. American Journal of Botany. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. The role of genetic and genomic attributes in the success of polyploids. Proceedings of the National Academy of Sciences; USA. 2000. pp. 7051–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies JJ, Linder HP, Labuschagne IF, Du Plessis H . Cytogenetic evidence for the species delimitation of Pentaschistis airoides and P. patula (Poaceae: Arundineae) In: Seyani JH, Chikuni AC, editors. Proceedings of the XIII Plenary Meeting of AETFAT, Zomba, Malawi, 2–11 April 1991. Zomba: National Herbarium and Botanic Garden of Malawi; 1994. pp. 373–383. [Google Scholar]
- Stebbins GL. Polyploidy, hybridization, and the invasion of new habitats. Annals of the Missouri Botanical Garden. 1985;72:824–832. [Google Scholar]
- Tateoka T. Chromosome numbers of some grasses from Madagascar. Botanical Magazine (Tokyo) 1965;78:306–311. [Google Scholar]
- Thompson JN, Nuismer SL, Merg K. Plant polyploidy and the evolutionary ecology of plant/animal interactions. Biological Journal of the Linnean Society. 2004;82:511–519. [Google Scholar]
- Vijverberg K, Mes THM, Bachmann K. Chloroplast DNA evidence for the evolution of Microseris (Asteraceae) in Australia and New Zealand after long-distance dispersal from western North America. American Journal of Botany. 1999;86:1448–1463. [PubMed] [Google Scholar]
- Wagner WL, Funk VA. Hawaiian biogeography. Washington, DC: Smithsonian Institution Press; 1995. Evolution of a hotspot archipelago. [Google Scholar]
- Wagstaff SJ, Dawson MI, Venter S, et al. Origin, diversification, and classification of the Australasian genus Dracophyllum (Richeeae, Ericaceae) Annals of the Missouri Botanical Garden. 2010;97:235–258. [Google Scholar]
- Wardle P. Vegetation of New Zealand. Cambridge: Cambrige University Press; 1991. [Google Scholar]
- Waters C, Murray BG, Melville G, Coates D, Young A, Virgona J. Polyploidy and possible implications for the evolutionary history of some Australian Danthonieae. Australian Journal of Botany. 2010;58:23–34. [Google Scholar]
- Wright SD, Yong CG, Dawson JW, Whittaker DJ, Gardner RC. Riding the ice age El Nino? Pacific biogeography and evolution of Metrosideros subg. Metrosideros (Myrtaceae) inferred from nuclear ribosomal DNA. Proceedings of the National Academy of Sciences; USA. 2000. pp. 4118–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Yu, Harris AJ, He X. RASP (Reconstruct Ancestral State in Phylogenies) 2011. Version 2.1b. http://mnh.scu.edu.cn/soft/blog/RASP . [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. 2006. The University of Texas at Austin. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.