Abstract
DNA damage occurs during DNA replication, spontaneous chemical reactions, and assaults by external or metabolism-derived agents. Therefore, all living cells must constantly contend with DNA damage. Cells protect themselves from these genotoxic stresses by activating the DNA damage checkpoint and DNA repair pathways. Coordination of these pathways requires tight regulation in order to prevent genomic instability. The checkpoint clamp complex consists of Rad9, Rad1 and Hus1 proteins, and is often called the 9-1-1 complex. This PCNA (proliferating cell nuclear antigen)-like donut-shaped protein complex is a checkpoint sensor protein that is recruited to DNA damage sites during the early stage of the response, and is required for checkpoint activation. As PCNA is required for multiple pathways of DNA metabolism, the checkpoint clamp has also been implicated in direct roles in DNA repair, as well as in coordination of the pathways. Here we discuss roles of the checkpoint clamp in DNA damage response (DDR).
Keywords: DNA damage checkpoint, checkpoint clamp, DNA repair, Rad9
1. Introduction
The homologs of Rad9, Rad1 and Hus1 in fission yeast were originally discovered by genetic screens to identify genes that affect cell survival upon genotoxic stresses. On the basis of computational modeling, electron microscopy (EM), and biochemical analyses, it is known that Rad9, Rad1 and Hus1 form a heterotrimeric ring structure that resembles PCNA [1,2,3,4,5]. The crystal structure of the checkpoint clamp complex was determined in 2009, and the structure indeed shows a closed-ring architecture of the checkpoint clamp, formed through a head-to-tail association of its subunits similar to that in PCNA [6,7,8]. The homotrimeric complex PCNA is recruited to DNA by the replication clamp loader RFC1–5 complex, and stimulates DNA polymerases and other enzymes for DNA repair [9,10,11,12,13,14,15,16,17,18,19]. Analogously, the checkpoint clamp is recruited to DNA damage sites by the checkpoint clamp loader complex that consists of Rad17 and RFC 2–5 [2]. Although the PCNA and the checkpoint clamp function in different pathways of DNA metabolism, several DNA repair enzymes interact with both clamps. One big difference between PCNA and the checkpoint clamp is that the checkpoint clamp contains an unstructured C-terminal tail on Rad9. This tail is required for checkpoint activation through interaction with TopBP1 [20,21,22]. We are beginning to understand mechanisms of the checkpoint activation through the checkpoint clamp complex. However, roles of the checkpoint clamp in DNA repair regulation are largely unknown. This review describes current understanding of checkpoint activation and repair regulations by the checkpoint clamp.
2. Role of the Checkpoint Clamp in Checkpoint Activation
Depending on the nature of genomic perturbation, different components of the checkpoint factors are employed to deal with DNA damage. The phosphoinositide kinase-related kinases, ATM and ATR, operate near the apex of checkpoint pathways [20,23]. ATM-dependent pathways are initiated primarily by double-strand breaks (DSBs), while ATR responds to a broad spectrum of DNA damage and replication disruption, especially during the S-phase. Critical functions of ATM and ATR involve activation of the downstream checkpoint effector kinases Chk2 and Chk1, respectively. These downstream effector kinases regulate cell cycle progression by phosphorylating cell cycle proteins such as Cdc25 [24].
It has been thought that the checkpoint clamp complex functions in the ATR-dependent pathway, because it is required for activation of Chk1 but not for Chk2 [25,26,27]. The role of the checkpoint clamp in Chk1 activation is to bind TopBP1, which stimulates ATR-mediated Chk1 phosphorylation via TopBP1’s activation domain (AD), a domain that binds and activates ATR [20,21,22]. Similar results were obtained by studies of a budding yeast homolog of TopBP1, Dpb11 [28,29]. Interaction between the checkpoint clamp and TopBP1 requires the C-terminal tail of Rad9. Human Rad9 contains several phosphorylation sites, most of which are constitutively phosphorylated throughout the cell cycle [22]. This interaction is indeed required for Chk1 activation, because “tailless” Rad9 fused with TopBP1 is able to activate Chk1. Importantly, this complex must be recruited to DNA by the Rad17-RFC checkpoint-clamp loader for Chk1 activation [22]. The C-terminal tail, however, is not required for clamp formation or clamp loading [6,7]. Interestingly, two distinct mechanisms of ATR activation through the checkpoint clamp are proposed in budding yeast. Biochemical studies identified two factors, the checkpoint clamp and the Dpb11/TopBP1 replication protein, as potential activators of Mec1/ATR. The author showed that G1 phase activation of Mec1/ATR is achieved by the Ddc1/Rad9 subunit of the checkpoint clamp, while Dpb11/TopBP1 is dispensable. In G2, however, the checkpoint clamp activates Mec1/ATR by two distinct mechanisms. One mechanism involves direct activation of Mec1/ATR by Ddc1/Rad9, while the other proceeds by Dpb11/TopBP1 recruitment mediated through Ddc1/Rad9 phosphorylation [30].
Activation of the checkpoint requires recruitment of the checkpoint clamp to chromatin by the checkpoint clamp loader upon replication stresses. Unlike the PCNA homotrimeric complex that three identical subunits of PCNA impart symmetry to its structure and provide the same interaction surface to the clamp loader RFC, the checkpoint heterotrimeric clamp complex loading requires stricter control (because the interaction sites with RFC should be particular sites of one of the checkpoint clamp components). Rad17 has been shown to interact directly with Rad1 and Rad9 [2,8,31], implicating these two subunits in clamp opening. In fact, it has been suggested that Rad1–Rad9 interface is the weakest and the most likely opening site as evidenced by computational analysis, as well as an examination of interface buried surface areas [6,8]. Further studies are needed to determine the mechanism of the checkpoint clamp opening that is required for the clamp loading and also unloading.
3. Role of the Checkpoint Clamp in DSB Repair
Biochemical analyses have shown that the checkpoint clamp preferentially binds to 5’ recessed DNA [32], and single-strand DNA areas on double-strand DNA seem to be required for checkpoint activation [20]. These 5’ recessed structures could be generated in many biological processes in response to many types of genotoxic stresses, and the checkpoint clamp is recruited to chromatin in response to these stresses, including DNA replication inhibition, ultraviolet light, alkylation, and ionizing radiation (IR) [32]. Rad9−/− and Rad9 knockdown cells are sensitive to these genotoxic treatments [27,33]. Therefore, the checkpoint clamp plays a role in response to DSBs, as well as to replication perturbation. Interestingly, however, Rad9−/− cells are not defective in the Chk2 phosphorylation that is activated in response to DSBs. Furthermore, the C-terminal tail is not required for resistance to IR [27], implying that the “tailless” clamp might play a direct role in DSB repair. Indeed, the checkpoint clamp proteins are recruited to DSB sites, and the foci co-localize with γH2AX foci [34].
The sequence of the DDR protein-loading to chromatin has been shown in response to DSBs in yeast. The MRE11 and the ATM-related Tel1 kinase are the first proteins detected at DSBs. Next, the replication protein A (RPA) single-strand DNA (ssDNA) binding protein relocalizes to resected ssDNA areas and recruits checkpoint proteins. Later, and only in the S and G2 phase, the homologous recombination proteins assemble at the site. Although the checkpoint clamp loader and clamp are recruited to the site before the homologous recombination proteins, they are not required for the recruitment of a homologous recombination protein Rad52 [35]. It is also shown in mammalian cells that ATM and MRE11 nuclease generate RPA-coated ssDNA, and ATR/ATRIP are subsequently recruited to the sites. MRE11 is required for ATM activation [36,37,38,39], and RPA coated ssDNA is required for loading of the checkpoint clamp and ATRIP [40,41]. CtIP is recruited to the DSB sites in an MRN (MRE11-RAD50-NBS1)-complex and ATM-activation-dependent manner, and is required for DSB-end resection. However, the timing of the CtIP recruitment is significantly delayed compared to that of NBS1 [42]. The CtIP protein interacts with BRCA1 protein, and the BRCA1-CtIP complex is thought to mediate 5’ end resection of DSBs, which is abrogated by three independent tumor-associated mutations in the BRCT domain of BRCA1 [43,44]. A CtIP mutant that fails to interact with BRCA1 was shown to have a significant defect in HR in chicken DT40 cells [45]. However, another group showed that the same CtIP mutant is competent in HR, and that BRCA1 and CtIP might play distinct roles in DSB repair pathways by genetic analyses [46]. Further studies are required to verify functions of BRCA1 and CtIP in the HR process.
A two-step model has been proposed for DSB-end resection. In this model, the MRN complex and CtIP initiate the resection process to remove nucleotides. After this first step, the EXO1 exonuclease and BLM helicase (a RecQ-like helicase)-DNA2 helicase/endonuclease function redundantly to carry out further generation of long 3’ ssDNA tails [33,47,48,49,50,51,52]. Another RecQ-like helicase WRN is also shown to be involved in the DSB resection process in the Xenopus egg extract system [53,54]. However, the WRN protein fails to support reconstitution of DSB resection with human proteins in vitro [55]. It has been shown that the loading of a checkpoint clamp component Rad9 requires resection of DSB ends by CtIP [56]. However, the precise timing of the checkpoint clamp loading to chromatin upon damage remains elusive in mammalian cells.
The author’s group has shown that Rad9Ser272 becomes phosphorylated during unperturbed S-G2 phase as well, and this phosphorylation occurs in an ATM-dependent manner. Our data indicate that Rad9Ser272 phosphorylation governs repair pathways [57]. ATM-dependent Rad9Ser272 phosphorylation is not required for survival or checkpoint activation after DNA damage. This phenotype is reminiscent of BRCA1-S988 cells. BRCA1-deficient human cells expressing BRCA1-S988A have defects in homologous recombination (HR). However, the mutant cells retain normal checkpoint function and are resistant to IR, implying that the HR function of BRCA1 is distinct from its other functions in the DNA damage response [58]. Importantly, knock-in mice expressing an S971A mutant (human S988) develop mammary and endometrial tumors after treatment with DNA-damaging agents [59].
This Rad9 phosphorylation requires the MRN complex, confirming requirement of ATM kinase activation for Rad9Ser272 phosphorylation. Furthermore, the mutant cells show a defect in homologous recombination (HR), and induce GCRs. Interestingly GCRs are suppressed in a p53−/− background. These results indicate that the checkpoint clamp is involved in HR repair regulation. Further investigation is required to define the function of the checkpoint clamp and ATM-dependent phosphorylation of Rad9 in control of DSB repair.
4. Role of the Checkpoint Clamp in Other Repairs
Biochemical and genetic studies have revealed functional interactions between the checkpoint clamp and repair proteins that include translesion polymerases and base excision repair enzymes [60,61].
A checkpoint clamp-loader component Rad17 is required for replication perturbation-induced mutagenesis by translation polymerases, polymerase κ and ζ, in fission yeast. Checkpoint activation is required for transcriptional induction of polymerase κ, and the checkpoint clamp components Hus1 and Rad1 physically interact with polymerase κ [60,61]. Similar results were obtained in budding yeast. The checkpoint clamp physically interacts with polymerase ζ, and is partially required for pol ζ-dependent mutagenesis [62].
Biochemical analyses have shown interaction between the checkpoint clamp and the long-patch base excision repair machinery, MutY DNA glycosylase, DNA polymerase β, and Flap endonuclease I. The checkpoint clamp complex enhanced enzymatic activities of these proteins [63,64,65,66]. Nevertheless, confirmation of the biochemical results at the cellular level is required. Some of these repair proteins, including translesion polymerases and Flap endonuclease I, have also been shown to interact with PCNA, and PCNA can stimulate the long-patch base excision repair, as well as translesion synthesis [67,68,69]. Why are two clamps needed for repair pathways? This is still an important, yet unanswered, question.
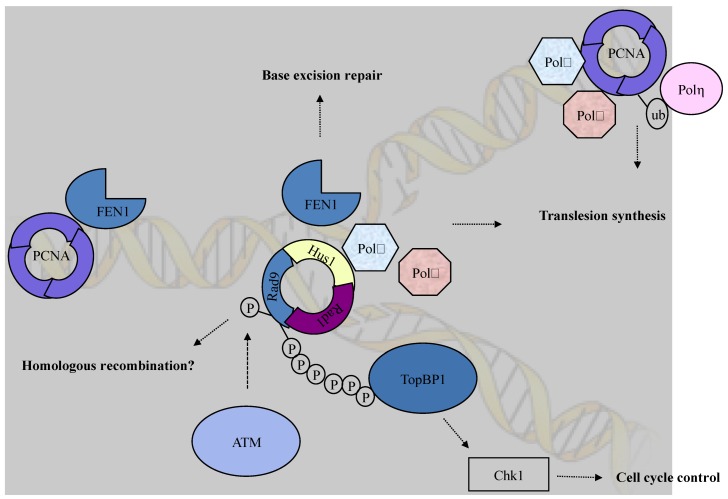
Figure 1.
The checkpoint clamp (Rad9-Husl-Radl) complex plays various roles in DNA damage response pathways. The C-terminal tail of Rad9 is required for Chk1 activation through interaction with TopBP1. In addition to the role in checkpoint activation, the checkpoint clamp functions in multiple DNA repair pathways such as translesion synthesis and base excision repair.
5. Conclusions
Since it was realized that Rad9, Rad1 and Hus1 form a PCNA-like ring structure, significant progress has been made in understanding the function of the checkpoint clamp. Despite much progress, however, the function of the checkpoint clamp, especially in regulation of DNA repair pathways, remains largely unknown.
References
- 1.Venclovas C., Thelen M.P. Structure-based predictions of rad1, rad9, hus1 and rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 2000;28:2481–2493. doi: 10.1093/nar/28.13.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bermudez V.P., Lindsey-Boltz L.A., Cesare A.J., Maniwa Y., Griffith J.D., Hurwitz J., Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hrad17-replication factor c complex in vitro. Proc. Natl. Acad. Sci. USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiomi Y., Shinozaki A., Nakada D., Sugimoto K., Usukura J., Obuse C., Tsurimoto T. Clamp and clamp loader structures of the human checkpoint protein complexes, rad9-1-1 and rad17-rfc. Genes Cells. 2002;7:861–868. doi: 10.1046/j.1365-2443.2002.00566.x. [DOI] [PubMed] [Google Scholar]
- 4.Burtelow M.A., Roos-Mattjus P.M., Rauen M., Babendure J.R., Karnitz L.M. Reconstitution and molecular analysis of the hrad9-hhus1-hrad1 (9-1-1) DNA damage responsive checkpoint complex. J. Bio. Chem. 2001;276:25903–25909. doi: 10.1074/jbc.M102946200. [DOI] [PubMed] [Google Scholar]
- 5.Lindsey-Boltz L.A., Bermudez V.P., Hurwitz J., Sancar A. Purification and characterization of human DNA damage checkpoint rad complexes. Proc. Natl. Acad. Sci. USA. 2001;98:11236–11241. doi: 10.1073/pnas.201373498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dore A.S., Kilkenny M.L., Rzechorzek N.J., Pearl L.H. Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex--implications for clamp loading and regulation. Mol. Cell. 2009;34:735–745. doi: 10.1016/j.molcel.2009.04.027. [DOI] [PubMed] [Google Scholar]
- 7.Sohn S.Y., Cho Y. Crystal structure of the human rad9-hus1-rad1 clamp. J. Mol. Biol. 2009;390:490–502. doi: 10.1016/j.jmb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 8.Xu M., Bai L., Gong Y., Xie W., Hang H., Jiang T. Structure and functional implications of the human rad9-hus1-rad1 cell cycle checkpoint complex. J. Bio. Chem. 2009;284:20457–20461. doi: 10.1074/jbc.C109.022384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S.H., Hurwitz J. Mechanism of elongation of primed DNA by DNA polymerase delta, proliferating cell nuclear antigen, and activator 1. Proc. Natl. Acad. Sci. USA. 1990;87:5672–5676. doi: 10.1073/pnas.87.15.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng L., McConnell M., Tan C.K., Downey K.M., Fisher P.A. Interaction of DNA polymerase delta, proliferating cell nuclear antigen, and synthetic oligonucleotide template-primers. Analysis by polyacrylamide gel electrophoresis-band mobility shift assay. J. Bio. Chem. 1993;268:13571–13576. [PubMed] [Google Scholar]
- 11.Jonsson Z.O., Hindges R., Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mozzherin D.J., Tan C.K., Downey K.M., Fisher P.A. Architecture of the active DNA polymerase delta.Proliferating cell nuclear antigen.Template-primer complex. J. Bio. Chem. 1999;274:19862–19867. doi: 10.1074/jbc.274.28.19862. [DOI] [PubMed] [Google Scholar]
- 13.Matsumoto Y. Molecular mechanism of pcna-dependent base excision repair. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:129–138. doi: 10.1016/S0079-6603(01)68095-4. [DOI] [PubMed] [Google Scholar]
- 14.Maga G., Villani G., Ramadan K., Shevelev I., Tanguy Le Gac N., Blanco L., Blanca G., Spadari S., Hubscher U. Human DNA polymerase lambda functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J. Bio. Chem. 2002;277:48434–48440. doi: 10.1074/jbc.M206889200. [DOI] [PubMed] [Google Scholar]
- 15.Umar A., Buermeyer A.B., Simon J.A., Thomas D.C., Clark A.B., Liskay R.M., Kunkel T.A. Requirement for pcna in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/S0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 16.Gary R., Ludwig D.L., Cornelius H.L., MacInnes M.A., Park M.S. The DNA repair endonuclease xpg binds to proliferating cell nuclear antigen (pcna) and shares sequence elements with the pcna-binding regions of fen-1 and cyclin-dependent kinase inhibitor p21. J. Bio. Chem. 1997;272:24522–24529. doi: 10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- 17.Hosfield D.J., Mol C.D., Shen B., Tainer J.A. Structure of the DNA repair and replication endonuclease and exonuclease fen-1: Coupling DNA and pcna binding to fen-1 activity. Cell. 1998;95:135–146. doi: 10.1016/S0092-8674(00)81789-4. [DOI] [PubMed] [Google Scholar]
- 18.Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. Rad6-dependent DNA repair is linked to modification of pcna by ubiquitin and sumo. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 19.Essers J., Theil A.F., Baldeyron C., van Cappellen W.A., Houtsmuller A.B., Kanaar R., Vermeulen W. Nuclear dynamics of pcna in DNA replication and repair. Mol. Cell. Biol. 2005;25:9350–9359. doi: 10.1128/MCB.25.21.9350-9359.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J., Kumagai A., Dunphy W.G. The rad9-hus1-rad1 checkpoint clamp regulates interaction of topbp1 with atr. J. Bio. Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- 21.Yan S., Michael W.M. Topbp1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. J. Cell. Biol. 2009;184:793–804. doi: 10.1083/jcb.200810185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delacroix S., Wagner J.M., Kobayashi M., Yamamoto K., Karnitz L.M. The rad9-hus1-rad1 (9-1-1) clamp activates checkpoint signaling via topbp1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abraham R.T. Cell cycle checkpoint signaling through the atm and atr kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 24.Perry J.A., Kornbluth S. Cdc25 and wee1: Analogous opposites? Cell Div. 2007;2:12. doi: 10.1186/1747-1028-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bao S., Lu T., Wang X., Zheng H., Wang L.E., Wei Q., Hittelman W.N., Li L. Disruption of the rad9/rad1/hus1 (9-1-1) complex leads to checkpoint signaling and replication defects. Oncogene. 2004;23:5586–5593. doi: 10.1038/sj.onc.1207753. [DOI] [PubMed] [Google Scholar]
- 26.Wang X., Zou L., Lu T., Bao S., Hurov K.E., Hittelman W.N., Elledge S.J., Li L. Rad17 phosphorylation is required for claspin recruitment and chk1 activation in response to replication stress. Mol. Cell. 2006;23:331–341. doi: 10.1016/j.molcel.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 27.Roos-Mattjus P., Hopkins K.M., Oestreich A.J., Vroman B.T., Johnson K.L., Naylor S., Lieberman H.B., Karnitz L.M. Phosphorylation of human rad9 is required for genotoxin-activated checkpoint signaling. J. Bio. Chem. 2003;278:24428–24437. doi: 10.1074/jbc.M301544200. [DOI] [PubMed] [Google Scholar]
- 28.Puddu F., Granata M., Di Nola L., Balestrini A., Piergiovanni G., Lazzaro F., Giannattasio M., Plevani P., Muzi-Falconi M. Phosphorylation of the budding yeast 9-1-1 complex is required for dpb11 function in the full activation of the uv-induced DNA damage checkpoint. Mol. Cell. Biol. 2008;28:4782–4793. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puddu F., Piergiovanni G., Plevani P., Muzi-Falconi M. Sensing of replication stress and mec1 activation act through two independent pathways involving the 9-1-1 complex and DNA polymerase epsilon. PLoS Genetics. 2011;7:e1002022. doi: 10.1371/journal.pgen.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navadgi-Patil V.M., Burgers P.M. The unstructured C-terminal tail of the 9-1-1 clamp subunit ddc1 activates mec1/atr via two distinct mechanisms. Mol. Cell. 2009;36:743–753. doi: 10.1016/j.molcel.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker A.E., Van de Weyer I., Laus M.C., Verhasselt P., Luyten W.H. Identification of a human homologue of the schizosaccharomyces pombe rad17+ checkpoint gene. J. Bio. Chem. 1998;273:18340–18346. doi: 10.1074/jbc.273.29.18340. [DOI] [PubMed] [Google Scholar]
- 32.Ellison V., Stillman B. Biochemical characterization of DNA damage checkpoint complexes: Clamp loader and clamp complexes with specificity for 5' recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin M.H., Yuan M., Zhang H., Margolick J.B., Kai M. Atm-dependent phosphorylation of the checkpoint clamp regulates repair pathways and maintains genomic stability. Cell Cycle. 2012;11:1796–1803. doi: 10.4161/cc.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kai M., Tsui E. USA Unpublished Data 2013. Department of Radiation Oncology, Johns Hopkins University; School of Medicine, Baltimore MD 21231, USA : Role of the checkpoint clamp in double-strand break repair. [Google Scholar]
- 35.Lisby M., Barlow J.H., Burgess R.C., Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 36.Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y. Requirement of the mrn complex for atm activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carson C.T., Schwartz R.A., Stracker T.H., Lilley C.E., Lee D.V., Weitzman M.D. The mre11 complex is required for atm activation and the g2/m checkpoint. EMBO J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J.H., Paull T.T. Direct activation of the atm protein kinase by the mre11/rad50/nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 39.Lee J.H., Paull T.T. Atm activation by DNA double-strand breaks through the mre11-rad50-nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 40.Zou L., Liu D., Elledge S.J. Replication protein a-mediated recruitment and activation of rad17 complexes. Proc. Natl. Acad. Sci. USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou L., Elledge S.J. Sensing DNA damage through atrip recognition of rpa-ssdna complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 42.You Z., Shi L.Z., Zhu Q., Wu P., Zhang Y.W., Basilio A., Tonnu N., Verma I.M., Berns M.W., Hunter T. Ctip links DNA double-strand break sensing to resection. Mol. Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu X., Wu L.C., Bowcock A.M., Aronheim A., Baer R. The C-terminal (brct) domains of brca1 interact in vivo with ctip, a protein implicated in the ctbp pathway of transcriptional repression. J. Bio. Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 44.Chen L., Nievera C.J., Lee A.Y., Wu X. Cell cycle-dependent complex formation of brca1.Ctip.Mrn is important for DNA double-strand break repair. J. Bio. Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- 45.Yun M.H., Hiom K. Ctip-brca1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K.M., Tsutsui K., Hartsuiker E., Ogi T., Takeda S., Taniguchi Y. Collaborative action of brca1 and ctip in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genetics. 2010;6:e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mimitou E.P., Symington L.S. DNA end resection: Many nucleases make light work. DNA Repair. 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Budd M.E., Campbell J.L. Interplay of mre11 nuclease with dna2 plus sgs1 in rad51-dependent recombinational repair. PLoS One. 2009;4:e4267. doi: 10.1371/journal.pone.0004267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mimitou E.P., Symington L.S. Sae2, exo1 and sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. Sgs1 helicase and two nucleases dna2 and exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nimonkar A.V., Ozsoy A.Z., Genschel J., Modrich P., Kowalczykowski S.C. Human exonuclease 1 and blm helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. USA. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gravel S., Chapman J.R., Magill C., Jackson S.P. DNA helicases sgs1 and blm promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan H., McCane J., Toczylowski T., Chen C. Analysis of the xenopus werner syndrome protein in DNA double-strand break repair. J. Cell. Biol. 2005;171:217–227. doi: 10.1083/jcb.200502077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toczylowski T., Yan H. Mechanistic analysis of a DNA end processing pathway mediated by the xenopus werner syndrome protein. J. Bio. Chem. 2006;281:33198–33205. doi: 10.1074/jbc.M605044200. [DOI] [PubMed] [Google Scholar]
- 55.Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C., Modrich P., Kowalczykowski S.C. Blm-dna2-rpa-mrn and exo1-blm-rpa-mrn constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warmerdam D.O., Freire R., Kanaar R., Smits V.A. Cell cycle-dependent processing of DNA lesions controls localization of rad9 to sites of genotoxic stress. Cell Cycle. 2009;8:1765–1774. doi: 10.4161/cc.8.11.8721. [DOI] [PubMed] [Google Scholar]
- 57.Shin M.H., Yuan M., Zhang H., Margolick J.B., Kai M. Atm-dependent phosphorylation of the checkpoint clamp regulates repair pathways and maintains genomic stability. Cell Cycle. 2012;11:1796–1803. doi: 10.4161/cc.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J., Willers H., Feng Z., Ghosh J.C., Kim S., Weaver D.T., Chung J.H., Powell S.N., Xia F. Chk2 phosphorylation of brca1 regulates DNA double-strand break repair. Mol. Cell. Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim S.S., Cao L., Li C., Xu X., Huber L.J., Chodosh L.A., Deng C.X. Uterus hyperplasia and increased carcinogen-induced tumorigenesis in mice carrying a targeted mutation of the chk2 phosphorylation site in brca1. Mol. Cell. Biol. 2004;24:9498–9507. doi: 10.1128/MCB.24.21.9498-9507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kai M., Wang T.S. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 2003;17:64–76. doi: 10.1101/gad.1043203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kai M., Wang T.S. Checkpoint responses to replication stalling: Inducing tolerance and preventing mutagenesis. Mutation Res. 2003;532:59–73. doi: 10.1016/j.mrfmmm.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 62.Sabbioneda S., Minesinger B.K., Giannattasio M., Plevani P., Muzi-Falconi M., Jinks-Robertson S. The 9-1-1 checkpoint clamp physically interacts with polzeta and is partially required for spontaneous polzeta-dependent mutagenesis in saccharomyces cerevisiae. J. Bio. Chem. 2005;280:38657–38665. doi: 10.1074/jbc.M507638200. [DOI] [PubMed] [Google Scholar]
- 63.Smirnova E., Toueille M., Markkanen E., Hubscher U. The human checkpoint sensor and alternative DNA clamp rad9-rad1-hus1 modulates the activity of DNA ligase i, a component of the long-patch base excision repair machinery. Biochem. J. 2005;389:13–17. doi: 10.1042/BJ20050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toueille M., El-Andaloussi N., Frouin I., Freire R., Funk D., Shevelev I., Friedrich-Heineken E., Villani G., Hottiger M.O., Hubscher U. The human rad9/rad1/hus1 damage sensor clamp interacts with DNA polymerase beta and increases its DNA substrate utilisation efficiency: Implications for DNA repair. Nucleic Acids Res. 2004;32:3316–3324. doi: 10.1093/nar/gkh652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Friedrich-Heineken E., Toueille M., Tannler B., Burki C., Ferrari E., Hottiger M.O., Hubscher U. The two DNA clamps rad9/rad1/hus1 complex and proliferating cell nuclear antigen differentially regulate flap endonuclease 1 activity. J. Mol. Biol. 2005;353:980–989. doi: 10.1016/j.jmb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 66.Wang W., Brandt P., Rossi M.L., Lindsey-Boltz L., Podust V., Fanning E., Sancar A., Bambara R.A. The human rad9-rad1-hus1 checkpoint complex stimulates flap endonuclease 1. Proc. Natl. Acad. Sci. USA. 2004;101:16762–16767. doi: 10.1073/pnas.0407686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gary R., Kim K., Cornelius H.L., Park M.S., Matsumoto Y. Proliferating cell nuclear antigen facilitates excision in long-patch base excision repair. J. Bio. Chem. 1999;274:4354–4363. doi: 10.1074/jbc.274.7.4354. [DOI] [PubMed] [Google Scholar]
- 68.Kedar P.S., Kim S.J., Robertson A., Hou E., Prasad R., Horton J.K., Wilson S.H. Direct interaction between mammalian DNA polymerase beta and proliferating cell nuclear antigen. J. Bio. Chem. 2002;277:31115–31123. doi: 10.1074/jbc.M201497200. [DOI] [PubMed] [Google Scholar]
- 69.Bunting K.A., Roe S.M., Pearl L.H. Structural basis for recruitment of translesion DNA polymerase pol iv/dinb to the beta-clamp. EMBO J. 2003;22:5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]