Abstract
Aberrations of Notch signaling have been implicated in a variety of human cancers. Oncogenic mutations in NOTCH1 are common in human T-cell leukemia and lymphomas. However, loss-of-function somatic mutations in NOTCH1 arising in solid tumors imply a tumor suppressor function, which highlights the need to understand Notch signaling more completely. Here we describe the small GTPase RhoE/Rnd3 as a downstream mediator of Notch signaling in squamous cell carcinomas (SCCs) that arise in skin epithelia. RhoE is a transcriptional target of activated Notch1, which is attenuated broadly in SCC cells. RhoE depletion suppresses Notch1-mediated signaling in vitro, rendering primary keratinocytes resistant to Notch1-mediated differentiation and thereby favoring a proliferative cell fate. Mechanistic investigations indicated that RhoE controls a key step in Notch1 signaling by mediating nuclear translocation of the activated portion of Notch1 (N1IC) through interaction with importins. Our results define RhoE as a Notch1 target that is essential for recruitment of N1IC to the promoters of Notch1 target genes, establishing a regulatory feedback loop in Notch1 signaling. This molecular circuitry may inform distinct cell fate decisions to Notch1 in epithelial tissues where carcinomas such as SCC arise.
Introduction
Squamous cell carcinomas (SCCs) are the most common cancers worldwide with more then 700 000 new cases diagnosed each year. A major regulator of squamous cell differentiation is the Notch signaling pathway (1–3). It has been previously recognized that NOTCH1 gene expression and activity are substantially down modulated in keratinocyte cancer cell lines and tumors and suppression of Notch signaling in this system promotes aggressive tumor growth (4, 5). These findings are likely of clinical significance since recent studies identified loss-of-function mutation in NOTCH1 in squamous cell carcinomas (SCCs) (6–8). This is in contrast to previously described oncogenic gain-of-function aberrations in Notch in T-cell leukemia and lymphomas suggesting that this signaling pathway may function as a tissue-specific tumor suppressor in squamous epithelia (3). While in the majority of mammalian systems, Notch activation is generally thought to maintain stem cell potential, promote proliferation and inhibit differentiation (9–12), in squamous cells, increased Notch signaling results in cell cycle arrest and initiation of a terminal differentiation program (1–3). Another major pathway that has been linked to control of squamous cell fate determination, is that triggered by the small GTP-ases of the Rho family (13–15). Particularly, a new member of the small GTP-ase family of proteins, RhoE/Rnd3, was identified as a potential regulator of keratinocyte withdrawal from the cell cycle and commitment to differentiation (16). GTP-ases are regulatory proteins that function as molecular switches cycling between the active GDP-bound, and inactive, GTP-bound states (17). In contrast to typical Rho family proteins, Rnd proteins, including RhoE/Rnd3 remain in the constitutively active, GTP-bound state without GTP hydrolytic regulation (18–21). Recently, key effectors of small Rho GTP-ases like ROCK1/2 and MRCKa (5) were found to be transcriptional targets of the tumor suppressor p53/Notch1 signaling in the epidermis and to counteract the Notch mediated commitment to differentiation in keratinocytes.
Materials and Methods
Cell Culture Experiments
Primary and immortalized HKC were cultured in SFM Medium (Invitrogen). U2OS cells and all SCC cell lines were grown in DMEM supplemented with 10% bovine serum.
Quantitative real time RT PCR, chromatin immunoprecipitation and immunodetection techniques
The list of relevant antibodies is provided in the Supplemental Information. Conditions for real time and conventional PCR analysis, chromatin immunoprecipitation ChIP, immunoblotting and immunofluorescence were as previously described (5). Significant increase or decrease of mRNA levels or %bound Chromatin throughout the experiments was considered when p< 0.05.
RhoE loxp/loxp mice
Mutant mice were generated at inGenious Targeting Laboratory, USA (detailed strategy for generating the animals is described in the Supplemental Information).
The genotyping PCR primers for the RhoE-loxp mutant allele were as follows:
P1-F : TGCTGGTGGTGAAATTCAAGTCGC
P2-R: ACTCCAGTCATTCCAAGTCTCCCT
Promoter activity assays
RhoE-luc, Hey2-luc, HES1-AB-luc, HES1-ΔAB-luc, and CSL-responsive luciferase reporter constructs were previously described (2, 5, 22).
In vitro differentiation assay
Primary human keratinocytes were brought into suspension and plated on Petri dishes coated with poly-HEMA (10 mg/ml in ethanol, Sigma). At indicated time-points cells were collected by centrifugation and processed for total RNA preparation (RNeasy, Qiagen).
In vivo cysts formation assays
For in vivo cyst formation assays, control and RhoE siRNA transfected cells were brought into suspension and injected (1.5×106 cells/injection) intradermaly in 8 weeks old female athymic nude mice. Seven days later animals were sacrificed and formed cysts were excised and frozen in OCT. Sections were analyzed by H&E staining and immunolabeling with a Keratin 5 and Keratin 10 antibody.
Results
RhoE/RND3 and Notch1 levels correlate in human squamous cell carcinoma
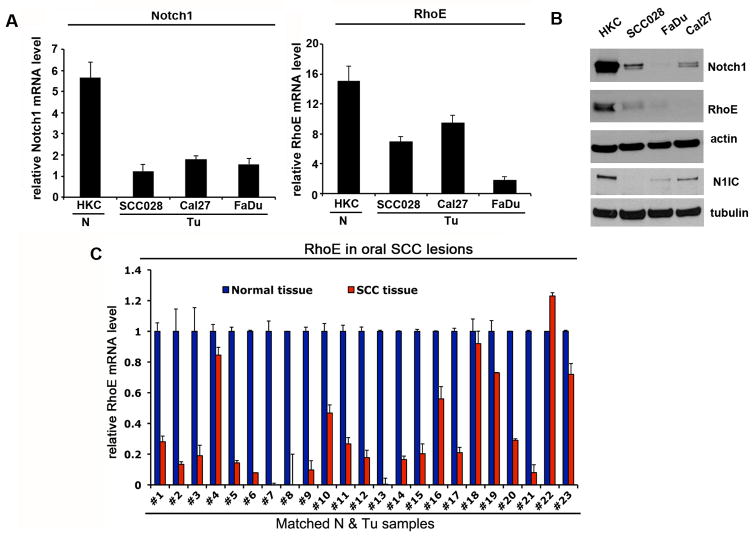
Since exome sequencing data of oral and skin SCCs did not identify mutations in RhoE (6–8) we sought to determine its expression pattern in patient derived SCC samples as well as SCC cell lines. We observed down-modulated Notch1 expression levels, lower levels of activated Notch1 and decreased RhoE expression in SCC samples as compared to controls (Fig. 1A and B). A similar analysis of a panel of patient-derived oral or skin SCC samples and normal controls (32 total number of samples) showed a significant suppression of RhoE expression in the SCC samples as compared to the matched controls (Fig. 1C and Supplementary Fig. S1).
Figure 1. RhoE expression is down-regulated in SCC cell lines and tumors in parallel with Notch1 levels.
A, Primary HKC were analyzed in parallel with SCC cell lines (SCCO28, Cal27 and FaDu) by real time RT-PCR for Notch1 and RhoE mRNA levels using 36B4 as internal control. B, The same set of cells as in A, were analyzed for protein levels of Notch1 and RhoE using β-actin as a loading control. In addition protein levels of the cleaved part of Notch1 were detected with Val1744 specific antibody (lower panel). C, Total RNA from 23 surgically excised oral SCC subjected to laser dissection microscopy (as in (5) were analyzed along with normal epithelium for RhoE mRNA expression by real time RT-PCR using 36B4 as control. The results are expressed as fold changes of RhoE expression in the tumors versus normal epithelium. Error bars represent Standard Deviations, calculated based on triplicate values.
RhoE is a direct transcriptional target gene of Notch signaling in squamous epithelium
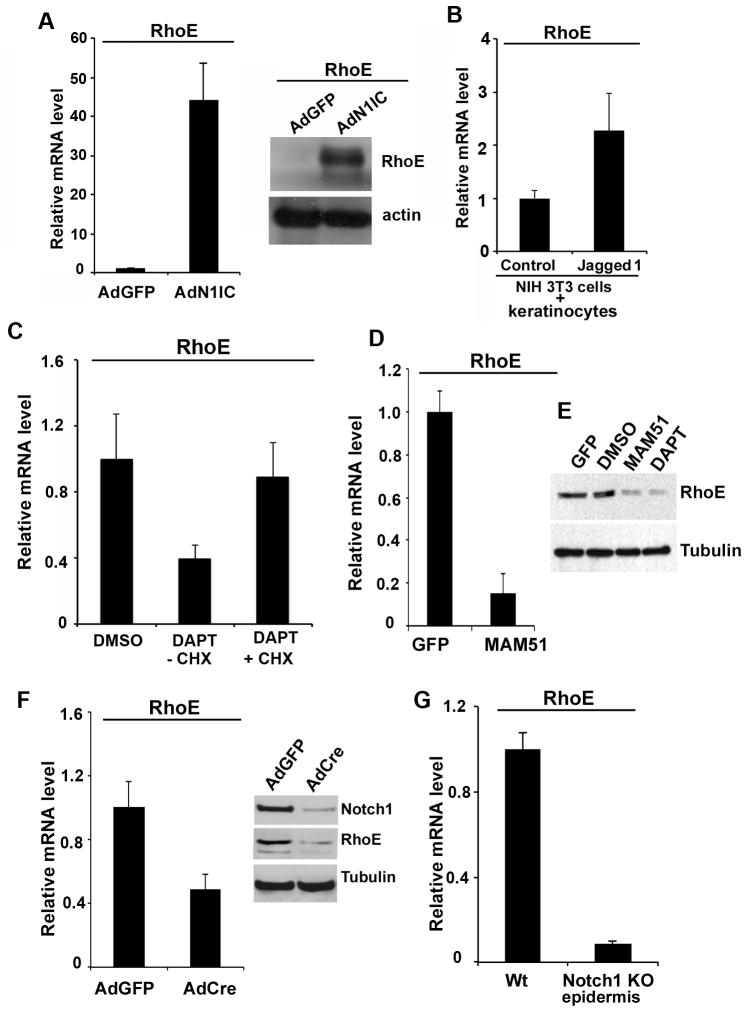
To investigate the possibility that RhoE is a downstream effector of Notch signaling in squamous epithelial cells, we checked its expression in primary human keratinocytes (HKC) infected with an adenovirus expressing activated form of Notch1 (AdN1IC) and detected a significant induction of RhoE mRNA and protein levels upon ectopic expression of activated Notch1 (N1IC) (Fig. 2A). This effect was specific for RhoE only, since other members of the same family of proteins remained unchanged under similar condition (Supplementary Fig. S2A, B). In addition, Notch1 seems to be a specific activator of RhoE expression in keratinocytes since down modulation of Notch2 was not sufficient to modulate RhoE levels (Supplementary Fig. S2C). Alternatively, to mimic endogenous activation of Notch1 by ligand interaction with neighboring cells, HKC were co-cultured for 48 hrs with NIH3T3 fibroblasts expressing the full-length Jagged 1 ligand (2, 23). Real-time RT-PCR analysis with human-specific primers showed that RhoE mRNA was increased significantly (Fig. 2B) correlating to increased mRNA levels of the Notch target gene Hey2 (Supplementary Fig. S3). Conversely, to assess the effects of suppression of endogenous Notch signaling in HKC on RhoE expression, we relied on two complementary approaches. Treatment with γ-secretase inhibitors, for example DAPT, blocks activation of endogenous Notch receptors (24, 25), whereas retroviral mediated expression of a 51-amino-acid peptide corresponding to the amino-terminus of the MAML1 protein (MAM51) provides an effective method to suppress canonical Notch/CSL/MAML-dependent transcription (26). Both conditions resulted in suppression of RhoE mRNA (Fig. 2C left panel, D) and protein levels (Fig. 2E). To determine whether de novo protein synthesis is required for suppression of the RhoE expression by DAPT, human primary keratinocytes were pre-treated with the protein synthesis inhibitor cycloheximide for 2 hrs and RhoE levels in the presence or absence of DAPT were assessed by real time RT-PCR. We observed a decrease of RhoE mRNA expression in the presence of DAPT. When cycloheximide treatment was applied after DAPT wash-out, RhoE mRNA expression returned to control levels (Fig. 2C, right panel). We next investigated whether RhoE expression is under Notch control also in the mouse epidermis both in vitro and in vivo. Primary mouse keratinocytes from mice with the Notch1 gene flanked by loxP (Notch1loxp/loxp) sites were infected with a Cre-expressing adenovirus (AdCre) for deletion of the Notch1 gene. Efficient loss of Notch1 expression upon infection with AdCre was confirmed by immunoblot analysis with an anti-Notch1 antibody (Fig. 2F). Relative to parallel cultures infected with a control GFP-expressing adenovirus (AdGFP), keratinocytes with deletion of the Notch1 gene exhibited a significantly decreased RhoE mRNA (Fig. 2F). In vivo, conditional keratinocyte specific deletion of the Notch1 gene in the epidermis upon crossing of the Notch1loxp/loxp mice with mice expressing the Cre recombinase from the Keratin 14 promoter, also resulted in markedly decreased RhoE mRNA levels (Fig. 2G).
Figure 2. RhoE expression is regulated by Notch1 in keratinocytes.
A, Notch activation induces RhoE expression. HKC were infected with AdN1IC or control AdGFP, followed by assessment of RhoE mRNA levels by real time RT-PCR (left panel) and protein levels after Western blot. B, HKC were co-cultured with control mouse NIH 3T3 fibroblasts or NIH 3T3 fibroblasts stably expressing full-length Jagged1 for 48 hours, followed by real-time RT-PCR analysis of RhoE mRNA levels. C, HKC were treated with 10 μM DAPT or DMSO vehicle control for 24 hrs, after that DAPT was washed out with PBS and cell culture medium and cells were treated with cycloheximide for 2. RhoE mRNA levels were then assessed by real time RT-PCR analysis. D, MAM51-mediated inhibition of Notch transcription suppresses RhoE expression. (E) Western blot analysis of RhoE protein levels in MAM51 overexpressing or DAPT treated primary HKC with tubulin as a loading control. F, Primary mouse keratinocytes from homozygous Notchloxp/loxp were infected with AdCre or AdGFP control. At 72 hours after infection, cells were analyzed either by real time RT-PCR analysis and immunoblotting for RhoE expression levels using 36B4 mRNA levels or γ-tubulin protein levels for normalization, respectively. The deletion of the Notch1 gene was confirmed by immunoblotting with a Notch specific antibody. G, Real time RT-PCR analysis was used to analyze RhoE mRNA levels, using mouse GAPDH for internal normalization. Relative mRNA levels are presented as a fold change as compared to the control condition. Error bars represent Standard Deviations, calculated based on three independent measurements.
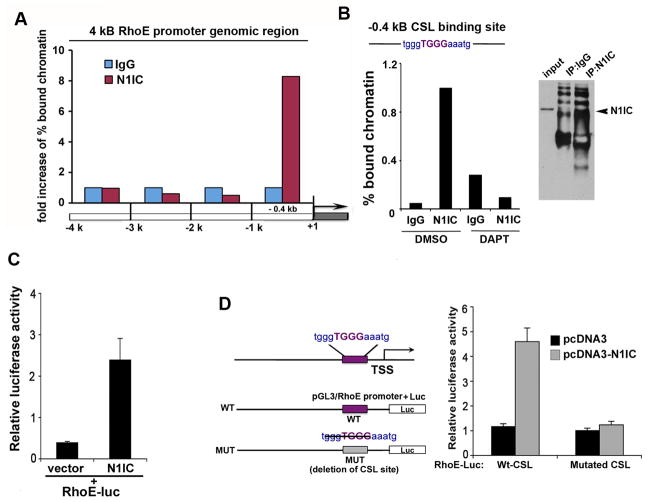
We next hypothesized that RhoE acts as a direct Notch1/CSL transcriptional target. To determine if Notch1 transcriptional complex occupies genomic segments of the RhoE promoter in vivo we used chromatin immunoprecipitation (ChIP) scanning assay and surveyed ~4kb of genomic DNA upstream of the transcription starting site (TSS) of human RhoE. The scanning ChIP was performed in primary HKC after IP with an antibody against the endogenous N1IC and real-time PCR was used for chromatin binding quantification. We detected a significant enrichment of Notch1 containing complex in the −1kB region of the RhoE promoter (Fig. 3A). Nucleotide sequence analysis of the human RhoE promoter revealed the presence of a consensus CSL binding site at ~0.4kB upstream of the TSS. Quantitative ChIP assay by real time PCR using primers targeting the −0.4kB region containing the CSL binding site revealed a significant amplification of this genomic fragment. Importantly, binding od activated Notch1 to the RhoE promoter was abrogated upon DAPT treatment (Fig. 3B, left panel). Cotransfection of an N1IC expression plasmid with a luciferase-linked RhoE promoter construct containing around 2kb of the genomic region upstream of the RhoE TSS into SCC028 cells resulted in an increased luciferase activity, whereas co-transfection with vector alone failed to do so (Fig. 3C). We next generated a deletion mutation in the −0.4 kb CSL-binding (Fig. 3D). The mutated promoter significantly decreased luciferase activity in response to activated Notch1 when compared to the wild type RhoE promoter (Fig. 3D). These results demonstrate that RhoE is a transcriptional target of Notch1 and that the −0.4 kb CSL-binding site plays a role in Notch-mediated RhoE transcription.
Figure 3. Identification of RhoE as a transcriptional target of Notch1.
A, Endogenous Notch1 is enriched at −1kb genomic region from the transcription start site (TSS) of human RhoE. Primary HKC were forced to differentiate in suspension and processed for scanning ChIP of the −5kb genomic region upstream of the human RhoE TSS, as described in the Supplementary Information, using an antibody against the activated form of Notch1 or purified rabbit IgGs. B, Differentiated primary HKC, teraetd with DMSO control or DAPT were processed for ChIP as in (A) followed by real time PCR analysis (upper panel) or conventional PCR (lower pane) using specific primers (described in Supplementary Information) to amplify the −0.4kb region of the human RhoE promoter containing the CSL binding site. Efficiency of the immunoprecipitation is shown in the right panel: Western blot analysis of immunoprecipitates performed either with non-specific IgG or with the antibody against the activated form of Notch1. C, SCC028 cells were transfected with a N1IC construct or empty vector together with the RhoE reporter (RhoE-luc) and phRL-TK Renilla reporter. D, The promoter regions were analyzed using the rVista software for identification of transcription factor binding sites. The CSL binding site at position −0.4 kb was deleted, and SCC028 keratinocyte cells were transfected with the mutated or with the wild type luciferase reporter constructs together with the NICD construct and phRL-TK Renilla reporter. Error bars represent Standard Deviations, calculated based on three independent measurements.
RhoE potentiates Notch1 activity in SCC cells and normal keratinocytes in vitro and in vivo
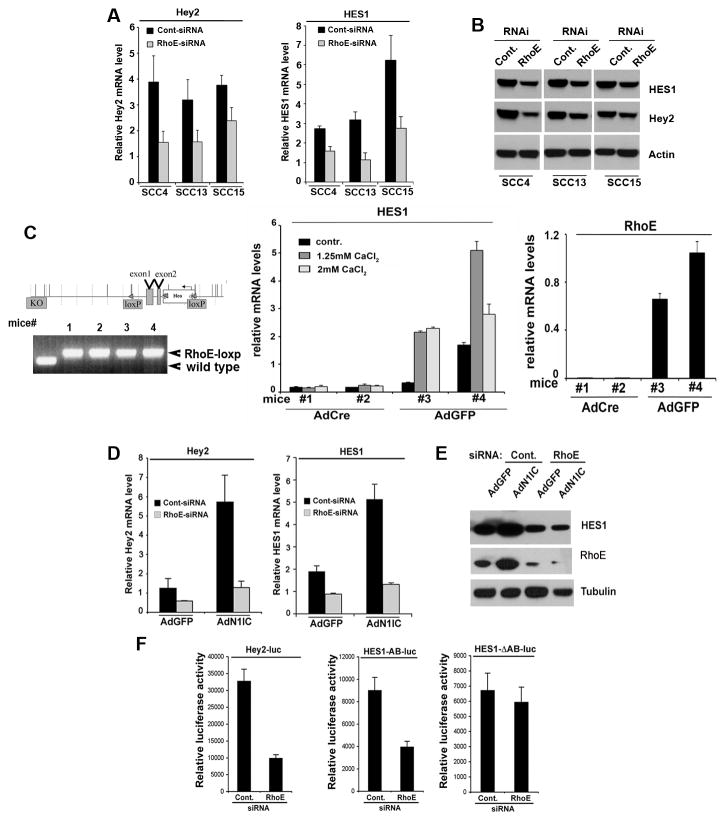
Next we examined the biological significance of RhoE for downstream Notch signaling in SCC cells and observed a suppression of Notch target genes expression upon depletion of RhoE levels with two unique siRNA oligonucleotides (Fig. 4A, B; Supplementary Fig. 4SA). While the effects of RhoE depletion for the SCC cells were not very striking, they were comparable in magnitude with pharmacological inhibition of Notch signaling using DAPT (Supplementary Fig. S4B). In order to explore the effects of a complete suppression of RhoE on Notch1 signaling, we used primary mouse keratinocytes derived from mutant animals, where the first 2 exons of the RhoE gene are surrounded by loxP sites and transduced them with control AdGFP or AdCre constructs. In agreement with our previous findings, we were able to show that lack of functional RhoE in the mouse epidermis results in decrease of mRNA levels of the Notch target gene Hes1 (Fig. 4C). Notably, the expression of other Rho family members was not affected upon RhoE depletion (Supplementary Fig. S5A, B). These results suggest that RhoE is not only a new direct target of Notch1, but RhoE may be required for the ability of Notch to induce its downstream targets. To further address this question, we transfected primary HKC with RhoE siRNA and subsequently infected them with AdN1IC or control AdGFP. Real time RT-PCR analysis showed that the basal levels of Notch1 targets Hey2 and Hes1 were reduced by RhoE knockdown (Fig. 4D). In order to avoid off-target effects of the siRNA, a second RhoE siRNA was used for transfection (Supplementary Fig. S4C). Moreover, the induction of these transcripts by activated Notch1 was also blocked in the RhoE siRNA transfected cells (Fig. 4D). Similar results were obtained for Hes1 protein levels after immunoblot analysis (Fig. 4E). In addition, silencing RhoE expression in SCC028 cells resulted in a significant suppression of Notch1 target Hey2 promoter activity when compared to cells transfected with a control siRNA (Fig. 4F). Applying identical conditions, we observed inhibition of luciferase activity from a reporter construct carrying the promoter of another Notch1 target gene, Hes1 (Hes1-AB). The specificity of the assay was tested with a mutated Hes1 promoter construct devoid of CSL-binding sites in its sequence (Hes1-ΔAB-luc), which didn’t respond to RhoE depletion (Fig. 4F).
Figure 4. Depletion of RhoE inhibits downstream Notch signaling.
A, SCC4, SCC13 and SCC15 cancer cell lines were transfected with control and RhoE siRNA and the levels of the Notch target genes Hey2 and HES1 were analyzed by real time RT-PCR analysis using 36B4 as an internal control. B, Protein levels of Hey2, HES1 and RhoE in the same cells were analyzed by Western blot using actin levels as an internal loading control. C, Inducible keratinocyte-specific deletion of the RhoE gene and resulting effects on Notch signaling and keratinocyte differentiation. Left upper panel: Schematic representation of the murine RhoE gene and of the expected recombination products leading to its inducible tissue-specific deletion. After induction of Cre recombinase activity (the loxP sites are indicated with red arrows), the genomic portion harboring the first two exons are deleted. Left lower panel: PCR analysis of genomic DNA of offspring from the RhoE loxp/loxp homozygous mice: the primers amplify a larger fragment including the loxp site and wild type genomic DNA is presented as a control. Middle panel: Real-time RT-PCR analysis of mRNA isolated from primary keratinocytes derived from mutant RhoE loxp/loxp mice and infected with an adenovirus expressing the Cre recombinase or an Ad-GFP control. At 2 days after infection levels of Hes1 are decreased in cells with Cre activity upon calcium mediated induction of differentiation. Depletion of RhoE mRNA levels shown on the right. D, Primary HKC under growing adherent conditions were transfected with control (scrambled siRNA) or RhoE siRNA, and 48 hours later were infected with adenoviruses expressing either GFP (AdGFP) or the activated form of Notch1 (AdN1IC). 24 hours later expression of Notch1 target genes, Hey2 and HES1 was analyzed by real time RT-PCR. E, RhoE and HES1 expression in the same cells was analyzed by Western blot. F, SCC028 cells transfected with control or RhoE siRNA were transfected with luciferase reporters for Hey2 (Hey2-luc, 0.5mg), wild type HES1 containing both CSL binding sites in the promoter region (HES1-AB-luc, 0.5mg) or HES1-ΔAB-luc with mutated CSL binding sites together with a Renilla minimal reporter (0.05 mg) for internal normalization. 24 hrs after transfection, luciferase activity was measured and normalized with Renilla activity. Error bars represent Standard Deviations, calculated based on three independent measurements.
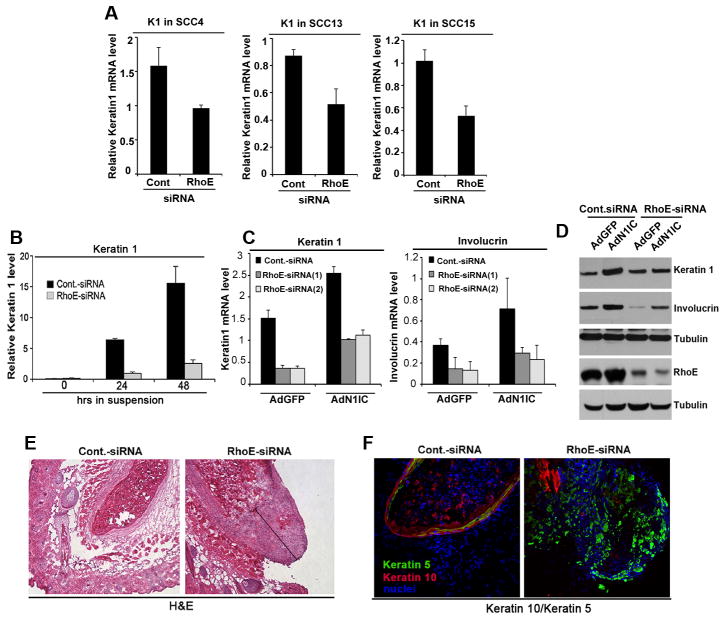
Because activated Notch1 is a key inducer of growth arrest and commitment to differentiation in both SCC cells and normal HKC (1, 2, 27) and RhoE was previously implicated in control of HKC differentiation (16), we depleted RhoE using siRNA in SCC cells and looked at their differentiation profile. We observed a down-modulation of the differentiation marker Keratin 1 (Fig. 5A and Supplementary Fig. S5A with a second independent RhoE siRNA). To dissect the effect of RhoE on the differentiation program of squamous epithelium and ask the question if it is part of a disrupted Notch signaling in the system, we used primary HKC and looked at the “suspension” model of Notch-mediated kerarinocyte differentiation induced upon loss of attachment to the surface (5, 28). We observed a strong increase of RhoE expression upon keratinocytes differentiation mirroring the expression level of Keratin 1 (Supplementary Fig. S6A, B). When RhoE expression was knocked-down by siRNA approach, the suspension conditions failed to induce Keratin 1 expression (Fig. 5B). In agreement with these findings, overexpression of N1IC was not able to induce expression levels of Keratin1 and Involucrin in keratinocytes with RhoE depletion (Fig. 5C, D).
Figure 5. RhoE ablation suppresses growth and commitment to differentiation in vitro and enhances the proliferative phenotype in vivo.
A, SCC4, SCC13 and SCC15 cells were depleted of RhoE expression using siRNA and Keratin 1 mRNA was analyzed 48 hrs later using real time RT-PCR. B, Primary HKC transfected with control or RhoE siRNA were kept under growing conditions (time point 0) or induced to differentiate by suspension or the indicated times. Keratin1 mRNA levels were analyzed by real time RT-PCR. C, Knockdown of RhoE inhibits the Notch1-mediated induction of Keratin 1 and Involucrin. Primary HKC under growing adherent conditions were transfected with control or two different RhoE siRNAs, and 48 hours later infected with adenoviruses AdGFP or AdN1IC. 24 hours later commitment of keratinocytes to differentiation was analyzed by Keratin 1 or Involucrin mRNA levels. D, HKC transfected with control or RhoE siRNA were treated as in C, and analyzed for RhoE, Keratin 1 and Involucrin protein levels by Western blot using tubulin as a loading control. E, Human HKC transfected with control or RhoE siRNA were injected into immunodeficient nude mice at the dermal-epidermal junction. To minimize individual animal variations, the same mice were injected in parallel with control siRNA transfected keratinocytes. 7 days later cysts were harvested, and tissues were processed. F, The same cysts from above experiments were stained with a Keratin 5 specific antibody (green staining in the right panel). Shown are multilayer cysts with well-differentiated keratinocytes building a cornified envelope in the control samples and a large “basal” layer of proliferating keratinocytes with increased Keratin 5 expression (green staining) and decreased Keratin 10 expression (red staining) in the RhoE siRNA tissues. Nuclei were stained in blue (Hoechst staining). Error bars represent Standard Deviations, calculated based on three independent measurements.
We next determined the relevance of RhoE in this context in vivo. Primary HKC transfected with control or RhoE siRNA were injected into immunodeficient nude mice at the dermal-epidermal junction, i.e. a location allowing the formation of multilayer cysts closely resembling the differentiation profile of the normal epidermis. In control mice, as expected, the injected cells gave rise to cornified epithelial cysts with well distinguishable keratinocyte layers starting from a proliferating basal layer and finishing with one or two cornified layers (Fig. 5E, left panel). However, RhoE siRNA transfected keratinocytes formed lesions with a significantly expanded basal proliferating layer and missing cornified envelope (Fig. 5E, right panel). The knock-down of RhoE in the cysts was confirmed by real time RT-PCR as well as Western blot (Supplementary Fig. S7B) and the effect of RhoE knock-down on cyst formation was reproduced using a second RhoE siRNA (Supplementary Fig. S7C). Immunofluorescence analysis of these cysts further confirmed the expansion of the basal layer of keratinocytes as showed by the increase of Keratin 5 and decrease of Keratin 10 expression in the RhoE siRNA transfected cells versus control (Fig. 5F). Therefore, our results strongly suggest that RhoE counteracts the “basal cell-like” phenotype in the epidermis, and suppression of RhoE expression results in an increased proliferation and the failure to commit to differentiation in keratinocytes both in vitro and in vivo.
RhoE binds to Notch1 and mediates its translocation to the nucleus
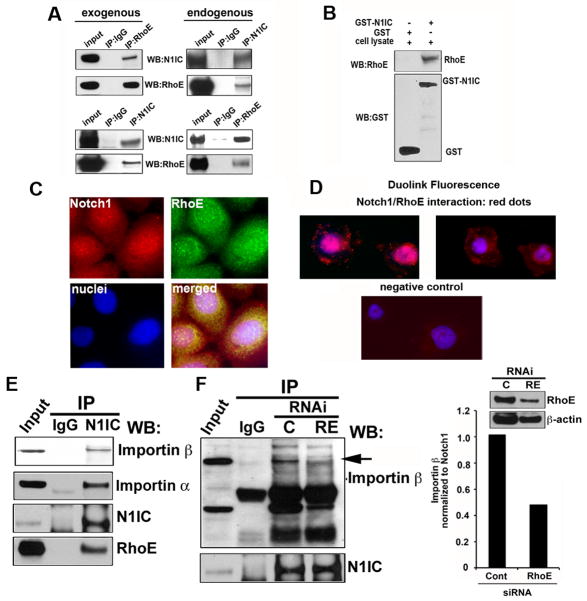
Our findings suggest that RhoE and Notch1 are linked by a molecular circuitry, whereby activated Notch1 signaling increases the transcription of RhoE, while suppression of RhoE levels abbrogates downstream Notch1 signaling. Given the trafficking function of several other small GTP-ases (29–32), we hypothesized that RhoE may influence Notch1 transport into the nucleus. Indeed, in U2OS osteosarcoma cells stably expressing Flag-RhoE and infected with AdN1IC, N1IC was found in a complex with RhoE upon reciprocal immunoprecipitation and Western blot (Fig. 6A). The interaction in vivo was additionally confirmed by reciprocal immunoprecipitaion of endogenous N1IC or RhoE followed by Western blotting for endogenous RhoE or N1IC respectively in SCC4 cells (Fig. 6A). GST-pull down assay using recombinant N1IC (aa1758-aa2556)-GST fusion protein and Western blot for endogenous RhoE confirmed the direct nature of this interaction (Fig. 6B). These biochemical results are consistent with the immunofluorescence results, in which co-localization of endogenous N1IC and RhoE in the cytoplasm and the nucleus was detected in SCC4 cells (Fig. 6C) by confocal microscopy. The specificity of the labeling was tested on the same cells using only secondary antibodies and double labeling of cells treated with DAPT or RhoE siRNA (Supplementary Fig. 8A-C). In addition the co-localization of RhoE and Notch1 in SCC4 cells was visualized using a Duolink assay for RhoE/Notch1 interaction in situ. In this assay the secondary antibodies are conjugated to oligonucleotides that comprise one-half of a closed circle that can be ligated together only when the antibodies are in close proximity (<40 nm) (Fig. 6D). Next we addressed the question of how the activated portion of Notch1 translocates to the nucleus. Since Notch1 contains a classical nuclear localization signal (33), its nuclear import might depend on the association with transport receptors of the importin α and β family (reviewed in (34)). Indeed, the interaction between activated Notch1 and importin α / β1 was revealed in extracts from SCC4 cells, immunoprecipitated with an anti N1IC antibody and analyzed by Western blot for importin α / β1 (Fig. 6E). Importantly, this N1IC/ importin β1interaction required the presence of RhoE since we observed a decrease of N1IC binding to importin β1 when RhoE expression was down-modulated (Fig. 6F). Similar findings were obtained using a second set of RhoE siRNA oligos (Supplementary Fig. 9A, B).
Figure 6. RhoE binds to N1IC and promotes its importin β1-mediated nuclear import.
A, Coimmunoprecipitation of N1IC by RhoE. Cellular extracts from U2OS cells stably expressing Flag-tagged RhoE and infected with AdN1IC were immunoprecipitated using Flag antibody followed by Western blotting for N1CD and RhoE. Endogenous association between N1IC and RhoE in SCC4 cells was analyzed be coimunoprecipitation of N1IC or RhoE followed by Western blot for the indicated proteins. B, GST-pull down assay was also performed: RhoE binds to recombinant N1IC fused to GST but not to GST alone. C, N1IC and RhoE colocalize in the cytoplasm and the nucleus of SCC4 cells by immunofluorescence staining using antibodies for endogenous RhoE and N1IC. The images were then analyzed by confocal microscopy. D, Representative images of SCC4 cell with Duolink fluorescence (red spots) generated by the interaction of the anti RhoE and anti N1IC antibodies (DAPI was used to stain the nucleus). Negative control images were generated by omitting the RhoE antibody. E, N1IC interacts with Importin family members. Cellular lysates of SCC4 cells were tested for the endogenous association between N1IC and importin β1 or α by immunoprecipitation with anti N1IC versus control IgG followed by Western blotting for importin α and β1. F. Depletion of RhoE abrogates the binding between NIIC and importin β1. SCC4 cells were transfected with either scrambled or RhoE siRNA and 48 hours later subjected to immunoprecipitation with an anti N1IC antibody followed by Western blotting using anti importin β1 and Notch1 antibodies. Rabbit IgG was used as a control (left panel). The decrease of immunoprecipitaed importin β1 was measured using UNISCANIT software. RhoE knock-down was measured by immunoblotting (right panel).
RhoE is essential for recruitment of activated Notch1 to its target gene promoters in SCC cells
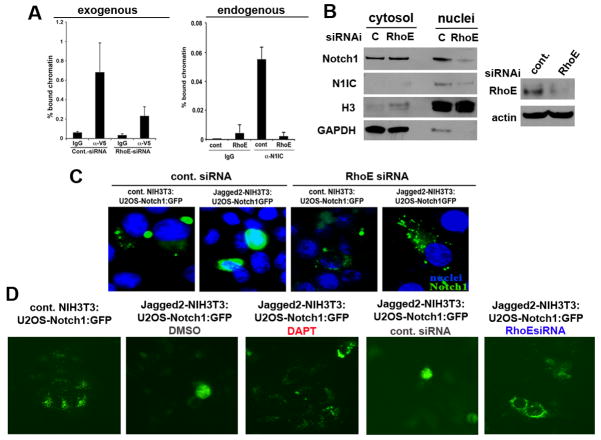
We next hypothesized that by mediating the translocation of N1IC to the nucleus, RhoE regulates the ability of N1IC to affect transcription of its target genes. First, we investigated the binding of NICD to the Hes1 promoter after infection with V5 tagged AdN1IC in primary HKC previously transfected with control siRNA versus cells with RhoE siRNA. Knock-down of RhoE expression inhibited the binding of activated Notch1 to the CSL binding site of the Hes1 promoter as assessed by quantitative ChIP/real-time PCR analysis using a primer pair able to amplify the CSL-binding site of the human Hes1 promoter (Fig. 7A right panel). Similarly, when immortalized HKC were forced to differentiate, ChIP using an antibody against endogenous N1IC detected decrease of binding upon depletion of RhoE expression (Fig. 7A right panel). To assess whether this is reflection of a block of cytoplasmic-nuclear translocation of the N1IC, we transfected SCC4 cells with RhoE or control siRNA. RhoE down-modulation (using two different sets of RNAi oligos) prevented accumulation of activated Notch1 in the nuclear fraction of the cells (Fig. 7B). To further confirm the defect of Notch1 nuclear translocation upon RhoE depletion, we developed a co-culture system in which U2OS cells stably expressing full length Notch1 with a GFP tag at the intracellular part were transfected with control or RhoE siRNA and 24 hrs later seeded on NIH3T3 cells expressing a control or Jagged2 plasmid to initiate ligand dependent cleavage of the GFP-Notch1 construct. Down-modulation of RhoE expression by siRNA prevented the translocation of intracellular Notch to the nucleus (Fig. 7C). Importantly, the block in N1IC nuclear translocation upon RhoE depletion was comparable to the effect of DAPT in the same cells (Fig. 7D). Together, theses data suggest that RhoE is essential for the transduction of downstream Notch signaling in keratinocytes and the binding of activated Notch1 on the promoter regions of its direct target genes.
Figure 7. RhoE promotes N1IC transcriptional activity and importin β1-mediated nuclear import.
A, RhoE knock-down inhibits Notch1-induced CSL-transcription in normal keratinocytes. Human primary keratinocytes under growing conditions were transfected with control siRNA, or RhoE siRNA followed by infection with an adenoviral construct encoding the activated form of Notch1 with a V5 C-terminal tag. 24 hours later cells were processed for ChIP with V5 specific antibodies or non-immune IgG control followed by PCR amplification of the CSL/Notch binding site-containing region of the human HES1 promoter (left panel). Differentiated primary HKC were processed for ChIP as in Figure 3 using endogenous N1IC for immunoprecipitation (right panel). The results from the ChIP assay were quantified by real time PCR analysis. Error bars represent Standard Deviations, calculated based on three independent measurements. B, RhoE inhibition prevents accumulation of activated Notch1 in the nucleus. Primary HKC transfected with RhoE siRNA were induced to differentiate by suspension culture, followed by nuclear and cytoplasmic fractionation and immunoblot analysis for the full length and activated Notch1 proteins using H3 and GAPDH as loading controls for the nuclear and cytosolic fractions, respectively. RhoE knockdown was assessed by Western blot analysis as shown in the right panel. C, U2OS cells stably expressing the full length Notch1-GFP (the GFP tag is at the intracellular end of the protein) transfected with control or RhoE siRNA and seeded over NIH3T3 cells expressing control empty vector or full length Jagged2 were analyzed for ligand mediated translocation of cleaved Notch1-GFP to the nucleus 24hrs later by fluorescent microscopy. D, Same co-culture system as in C but imaging was performed on live cells and the effects of RhoE siRNA were compared to DAPT treatment.
Discussion
We describe here a novel role for RhoE in the control of Notch mediated differentiation of SCC cells. RhoE expression is suppressed in human SCCs and down-modulation of RhoE levels leads to suppressed differentiation potential. Our results demonstrate that RhoE is a novel Notch1 effector, which is essential for binding of activated Notch to its downstream targets and ability to modulate the differentiation program in SCC cells. Recently, it became evident that loss-of-function mutations in NOTCH1 are very frequent in skin and oral SCCs (6–8). These studies also demonstrated that aberrations in Notch signaling in SCCs are important for tumor progression rather than initiation and predict that loss-of-function of the Notch pathway serves as a second “hit” to allow cells with “initiating” oncogenic mutations to progress and develop into tumors. In this context, the role of Notch as a major driver of squamous differentiation becomes critical since the key feature of squamous neoplasia of all types is disrupted commitment to differentiation. Interestingly, the majority of the identified NOTCH1 mutations in SCCs are heterozygous. This observation suggests that although loss of a single copy is probably sufficient to functionally decrease Notch activity, the remaining level of intact signaling may be essential to prevent excessive growth, as SCCs, specifically those in the skin, are known for lack of aggressiveness and slow progression. Hence, downstream effectors of the Notch pathway involved in the differentiation program of squamous cells become essential for controlling growth of SCC cells. From our data, it appears that depletion of RhoE in SCC cells causes inappropriate differentiation phenotype probably through altered Notch downstream signaling. While several studies described earlier how ligand binding activates Notch function through the proteolytically mediated generation of an active intracellular domain of Notch (35), the direct translocation of the intracellular domain to the nucleus remains elusive. Here we identify RhoE as an essential player in this process. RhoE enables the binding of NICD to importin β1 and thus ensures the proper translocation of NICD to the nucleus. We argue that the Notch1 target RhoE may influence Notch regulation through a positive feed-back loop, which regulates the translocation of the activated form of Notch to the nucleus. Alterations in this mode of action may result in growth-related disorders such as cancer. It was reported that RhoE expression is reduced in some cancer cells (36, 37) and this is linked to its anti-proliferative function (38) given the fact that RhoE function, due to its lack of GTPase activity, is primarily regulated on transcriptional level. Our finding that in human squamous cell carcinomas, down-modulation of Notch1 expression correlates with deregulated expression of RhoE, underscores the relevance of these observations for the development and growth of keratinocyte derived cancers. As recently reported, Notch signaling in HKC doesn’t have a general growth inhibitory effect, but it rather inhibits the expansion of “stem cell” like populations partially through inhibition of ROCK1/2 and MRCKa expression (5). Since RhoE functions as an inhibitor of ROCK signaling in various cell types, the inhibition of the expansion of “stem cell” like population by Notch signaling in SCC cells may be mediated through Notch1→RhoE pathway.
Supplementary Material
Acknowledgments
Grant Support:
This work was supported by NIH Grants CA127247, CA97216, CA80058, AR39190, CA16038 and CA73796, the Swiss National Foundation, a grant from the European Union (Epistem, Sixth Framework Program, LSHB-CT-2005-019067) and Shiseido Research Core funding).
Footnotes
The authors declare no competing interests.
References
- 1.Devgan V, Mammucari C, Millar SE, Brisken C, Dotto GP. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes & development. 2005;19:1485–95. doi: 10.1101/gad.341405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen BC, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, et al. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes & development. 2006;20:1028–42. doi: 10.1101/gad.1406006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. The EMBO journal. 2001;20:3427–36. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolev V, Mandinova A, Guinea-Viniegra J, Hu B, Lefort K, Lambertini C, et al. EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol. 2008;10:902–11. doi: 10.1038/ncb1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes & development. 2007;21:562–77. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science (New York, NY. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science (New York, NY. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang NJ, Sanborn Z, Arnett KL, Bayston LJ, Liao W, Proby CM, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17761–6. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, Lefort K, et al. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3799–804. doi: 10.1073/pnas.0600065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–61. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 11.Hopfer O, Zwahlen D, Fey MF, Aebi S. The Notch pathway in ovarian carcinomas and adenomas. British journal of cancer. 2005;93:709–18. doi: 10.1038/sj.bjc.6602719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nature medicine. 2002;8:979–86. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 13.Benitah SA, Frye M, Glogauer M, Watt FM. Stem cell depletion through epidermal deletion of Rac1. Science (New York, NY. 2005;309:933–5. doi: 10.1126/science.1113579. [DOI] [PubMed] [Google Scholar]
- 14.Dotto GP, Cotsarelis G. Developmental biology. Rac1 up for epidermal stem cells. Science (New York, NY. 2005;309:890–1. doi: 10.1126/science.1117192. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Lyle S, Liu Y, Solky B, Cotsarelis G. Differential expression of cyclin D1 in the human hair follicle. Am J Pathol. 2003;163:969–78. doi: 10.1016/S0002-9440(10)63456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liebig T, Erasmus J, Kalaji R, Davies D, Loirand G, Ridley A, et al. RhoE Is Required for Keratinocyte Differentiation and Stratification. Molecular biology of the cell. 2008 doi: 10.1091/mbc.E07-11-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 18.Chardin P. Function and regulation of Rnd proteins. Nat Rev Mol Cell Biol. 2006;7:54–62. doi: 10.1038/nrm1788. [DOI] [PubMed] [Google Scholar]
- 19.Foster R, Hu KQ, Lu Y, Nolan KM, Thissen J, Settleman J. Identification of a novel human Rho protein with unusual properties: GTPase deficiency and in vivo farnesylation. Molecular and cellular biology. 1996;16:2689–99. doi: 10.1128/mcb.16.6.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guasch RM, Scambler P, Jones GE, Ridley AJ. RhoE regulates actin cytoskeleton organization and cell migration. Molecular and cellular biology. 1998;18:4761–71. doi: 10.1128/mcb.18.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nobes CD, Lauritzen I, Mattei MG, Paris S, Hall A, Chardin P. A new member of the Rho family, Rnd1, promotes disassembly of actin filament structures and loss of cell adhesion. J Cell Biol. 1998;141:187–97. doi: 10.1083/jcb.141.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ongusaha PP, Kim HG, Boswell SA, Ridley AJ, Der CJ, Dotto GP, et al. RhoE is a pro-survival p53 target gene that inhibits ROCK I-mediated apoptosis in response to genotoxic stress. Curr Biol. 2006;16:2466–72. doi: 10.1016/j.cub.2006.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Mandinova A, Lefort K, Tommasi di Vignano A, Stonely W, Ostano P, Chiorino G, et al. The FoxO3a gene is a key negative target of canonical Notch signalling in the keratinocyte UVB response. The EMBO journal. 2008;27:1243–54. doi: 10.1038/emboj.2008.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3:688–94. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morohashi Y, Kan T, Tominari Y, Fuwa H, Okamura Y, Watanabe N, et al. C-terminal fragment of presenilin is the molecular target of a dipeptidic gamma-secretase-specific inhibitor DAPT (N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester) The Journal of biological chemistry. 2006;281:14670–6. doi: 10.1074/jbc.M513012200. [DOI] [PubMed] [Google Scholar]
- 26.Weng AP, Nam Y, Wolfe MS, Pear WS, Griffin JD, Blacklow SC, et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Molecular and cellular biology. 2003;23:655–64. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rengarajan J, Mittelstadt PR, Mages HW, Gerth AJ, Kroczek RA, Ashwell JD, et al. Sequential involvement of NFAT and Egr transcription factors in FasL regulation. Immunity. 2000;12:293–300. doi: 10.1016/s1074-7613(00)80182-x. [DOI] [PubMed] [Google Scholar]
- 28.Watt FM, Jordan PW, O’Neill CH. Cell shape controls terminal differentiation of human epidermal keratinocytes. Proc Natl Acad Sci USA. 1988;85:5576–80. doi: 10.1073/pnas.85.15.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 30.Molendijk AJ, Ruperti B, Palme K. Small GTPases in vesicle trafficking. Curr Opin Plant Biol. 2004;7:694–700. doi: 10.1016/j.pbi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 31.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Sandilands E, Frame MC. Endosomal trafficking of Src tyrosine kinase. Trends Cell Biol. 2008;18:322–9. doi: 10.1016/j.tcb.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Kopan R, Nye JS, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development (Cambridge, England) 1994;120:2385–96. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 34.Strom AC, Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2:REVIEWS3008. doi: 10.1186/gb-2001-2-6-reviews3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–47. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Bektic J, Pfeil K, Berger AP, Ramoner R, Pelzer A, Schafer G, et al. Small G-protein RhoE is underexpressed in prostate cancer and induces cell cycle arrest and apoptosis. The Prostate. 2005;64:332–40. doi: 10.1002/pros.20243. [DOI] [PubMed] [Google Scholar]
- 37.Trojan L, Schaaf A, Steidler A, Haak M, Thalmann G, Knoll T, et al. Identification of metastasis-associated genes in prostate cancer by genetic profiling of human prostate cancer cell lines. Anticancer research. 2005;25:183–91. [PubMed] [Google Scholar]
- 38.Villalonga P, Guasch RM, Riento K, Ridley AJ. RhoE inhibits cell cycle progression and Ras-induced transformation. Molecular and cellular biology. 2004;24:7829–40. doi: 10.1128/MCB.24.18.7829-7840.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boswell SA, Ongusaha PP, Nghiem P, Lee SW. The protective role of a small GTPase RhoE against UVB-induced DNA damage in keratinocytes. The Journal of biological chemistry. 2007;282:4850–8. doi: 10.1074/jbc.M610532200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.