Abstract
Background
PhyC levels have been observed to be markedly lower in phyB mutants than in Arabidopsis or rice wild type etiolated seedlings, but the mechanism of this phenomenon has not been fully elucidated.
Results
In the present study, we investigated the mechanism by which phyB affects the protein concentration and photo-sensing abilities of phyC and demonstrated that rice phyC exists predominantly as phyB/phyC heterodimers in etiolated seedlings. PHYC-GFP protein was detected when expressed in phyA phyC mutants, but not in phyA phyB mutants, suggesting that phyC requires phyB for its photo-sensing abilities. Interestingly, when a mutant PHYB gene that has no chromophore binding site, PHYB(C364A), was introduced into phyB mutants, the phyC level was restored. Moreover, when PHYB(C364A) was introduced into phyA phyB mutants, the seedlings exhibited de-etiolation under both far-red light (FR) and red light (R) conditions, while the phyA phyB mutants were blind to both FR and R. These results are the first direct evidence that phyC is responsible for regulating seedling de-etiolation under both FR and R. These findings also suggest that phyB is indispensable for the expression and function of phyC, which depends on the formation of phyB/phyC heterodimers.
Significance
The present report clearly demonstrates the similarities and differences in the properties of phyC between Arabidopsis and rice and will advance our understanding of phytochrome functions in monocots and dicots.
Introduction
Plants sense diverse light signals from the environment via a family of plant photoreceptors, including phytochromes, cryptochromes, and phototropins. Phytochromes are chromoproteins that regulate the expression of a large number of light-responsive genes and thus influence many photomorphogenic events [1]–[5].
The phytochrome monomer is an approximately 120 kDa protein attached to a linear tetrapyrrole chromophore, and the phytochrome molecule is thought to exist as a dimer in two stable photointerconvertible forms, Pr and Pfr. Photochromicity between inactive, red light (R)-absorbing Pr and active, far-red light (FR)-absorbing Pfr endows phytochromes with the capacity to sense the relative ratio of R and FR [6]. Phytochromes in higher plants are encoded by small gene families [7], [8]. Molecular phylogenetic analyses indicate that the angiosperm phytochrome gene family is composed of four subfamilies, PHYTOCHROME A (PHYA), PHYB, PHYC/F, and PHYE [9]. In Arabidopsis, PHYD is further derived from an ancestral PHYB gene by a recent gene duplication event [7], and as a result, Arabidopsis has five PHY genes, PHYA to PHYE [7], [10].
phyA and phyB exist as homodimers in wild type (WT) and phytochrome-overexpressing lines of Arabidopsis [11], [12], and the native complement of phytochromes in plants has often been assumed to consist of only homodimeric forms. However, in Arabidopsis, heterodimers of type II phytochromes have been observed in vivo [13]. Furthermore, there is no evidence for the homodimerization of phyC or phyE, indicating that these two phytochromes are present in cells only as heterodimers [14]. The formation of such heterodimeric phytochromes increases the potential complexity of R/FR light-sensing and signaling mechanisms in plants.
In rice (Oryza sativa), the phytochrome gene family is composed of three members, PHYA, PHYB, and PHYC [15]–[18]. Recently, the generation and characterization of rice phytochrome single, double, and triple mutants have revealed that individual members of the rice phytochrome family have synergistic, as well as overlapping, functions in the control of the responses to R and FR in terms of de-etiolation and flowering processes [19]–[22]. Under continuous FR, phyA mutants exhibited partially impaired de-etiolation, and phyA phyC double mutants exhibited no significant residual phytochrome responses, indicating that both phyA and phyC are involved in the photo-sensing of FR in rice [20]. However, phyB phyC double mutants did not show any apparent decrease in their sensitivity to FR compared with phyB mutants, indicating that the mutation of phyC in the phyB-deficient background did not have any additive effect. Moreover, the responses to FR were completely canceled in phyA phyB double mutants. It has also been reported that seedlings of phyA phyB double mutants are blind to R and that phyB phyC and phyB mutants showed similar sensitivity to R regarding de-etiolation responses [20]. These observations implied that phyB somehow affects phyC in the photo-sensing of FR or R in rice.
The dependence of phyC on phyB has been reported in Arabidopsis. Monte et al. found that phyB is necessary for the function of phyC in mediating the responses to R in Arabidopsis and that the functional dependency of phyC on phyB correlates with constitutively lower levels of phyC in phyB mutants compared with in WT [23], which was observed in several reports [24], [25]. These observations could result from the reduced stability of phyC when phyB is absent, preventing heterodimer formation [13], [14].
In rice, it has also been reported that phyC levels are lower in phyB mutant than WT etiolated seedlings and that phyB affects the photo-sensing ability of phyC [20]. In this study, we demonstrated that phyB and phyC form heterodimers in rice, consistent with observations in Arabidopsis. To reveal the properties of phyB/phyC heterodimers in rice, two types of transgenic plants overexpressing phyC-GFP or chromophore-less PHYB were generated and examined for their responsiveness to light. Our results revealed that phyB is indispensable for phyC stability and for its ability to sense both R and FR in the control of de-etiolation in rice seedlings. This study provides a new molecular mechanism for the interaction among phytochromes in the photoregulation of diverse developmental processes.
Materials and Methods
Plant Materials
Plant materials used in this study were as follows: WT, Oryza sativa L. cv. Nipponbare; phyA mutants, phyA-4; phyB mutants, phyB-1; phyC mutants, phyC-1; phyA phyC mutants, phyA-4 phyC-1; and phyA phyB mutants, phyA-4 phyB-1 [19], [20]. The background of these mutants is Nipponbare. Arabidopsis is in the gl-1 genetic background.
Protein Expression in Escherichia coli and Protein Purification
For quantifying the relative levels of phyB and phyC proteins, phyB-His and phyC-His were expressed in Escherichia coli as described by Takano et al. [20]. Dilutions of purified proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue [26]. Signal intensities of purified proteins were analyzed using NIH image 1.62.
To facilitate phyA or phyC apoprotein binding to chromophores in E. coli, the protein was expressed with a calmodulin binding domain at its N-terminus and a 6-histidine tag at its C-terminus in phytochromobilin-expressing Rosetta 2 cells (Novagen, Merck KGaA, Darmstadt, Germany) and purified, as previously described [27].
Immunoblotting and Co-immunoprecipitation
Soluble protein was extracted from above-ground parts of etiolated or light-grown seedlings as described by Takano et al. [19]. Protein extracts were separated on SDS-PAGE and blotted onto PVDF membranes (Immobilon-P, Millipore, MA). phyA, phyB, and phyC proteins were detected immunochemically using a colorimetric detection method (NBT/BCIP stock solution, Roche Applied Science, Basel, Switzerland) or ECL chemiluminescence kits (GE Healthcare, Uppsala, Sweden). For immunoblot analysis of Arabidopsis phyA protein, monoclonal antibody, AA01, was used.
For co-immunoprecipitation (co-IP), 200 µg of protein extract from dark-grown seedlings or 500 µg from continuous white light (cW)-grown seedlings were precleared by adding 50 µL of γProteinA-Sepharose Fast Flow (GE Healthcare), incubating for 30 min at 4°C, and centrifuging at 15,000 rpm for 10 min. The γProteinA-Sepharose-anti-PHYC conjugates were prepared by mixing 20 µL of γProteinA-Sepharose with anti-PHYC antibody or preimmune serum from the same rabbit, gently rocking for 1 h at 4°C, and washing twice with PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4). Then, precleared protein extracts were added to the Sepharose-anti-PHYC conjugates and incubated for 3–4 h at 4°C. The Sepharose-anti-PHYC-protein complex was washed six times with NETN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris-Cl pH 8.0, 0.5% (v/v) Nonidet P-40) containing 900 mM NaCl and twice with NETN buffer without additional NaCl. Proteins bound to the Sepharose beads were eluted by boiling for 5 min in 2× SDS sample buffer. The eluted proteins were analyzed by immunoblotting as described in Takano et al. [19]. Immunochemical detection was performed using an immunoblot kit (alkaline phosphatase system, using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) stock solution; Roche Applied Science).
Size Exclusion Chromatography
Protein extracts from WT, phyB, and phyA phyC rice seedlings or WT Arabidopsis seedlings grown in the dark for 8 days were prepared, and 0.25-mL samples containing 1 mg of total soluble protein were applied to a Superdex 200 HR 10/30 (GE Healthcare). The column was eluted with buffer (50 mM Na-phosphate, pH 7.0, 150 mM NaCl) at 4°C at a rate of 0.25 mL/min, and 0.5 mL fractions were collected. Co-IP and immunoblot analyses of each fraction were performed as described by Takano et al. [19]. The column was calibrated with protein molecular weight standards (GE Healthcare). The elution volume-molecular weight curve was plotted. The plotted data were fitted with a regression line, and the molecular weight of each fraction was estimated based on the linear regression line.
Plasmid Construction and Rice Transformation
For the construction of the mutant PHYB(C364A), the codon (TGC) encoding the chromophore-binding site (cysteine) was substituted with GCC encoding alanine by site-directed PCR. Rice PHYB full-length cDNA or mutated PHYB(C364A)cDNA was subcloned into the pPZP2Ha3 vector [28] in the sense orientation. These plasmids were introduced into Agrobacterium tumefaciens strain EHA101 by electroporation.
To construct the PHYC-GFP (PCG) overexpressing lines, rice PHYC cDNA was amplified by PCR with a nucleotide substitution that replaced the PHYC translation termination codon (TAG) with an oligonucleotide sequence containing a KpnI site. The PHYB moiety of Arabidopsis phyB-GFP fusion construct [29] was replaced with PHYC to obtain the PCG vector, in which the PHYC-GFP fusion sequence was inserted between the constitutive cauliflower mosaic virus 35S promoter and the Nos terminator of pPZP211/35S-nosT26 [30]. This plasmid was introduced into Agrobacterium tumefaciens strain EHA105 [31] by electroporation.
Rice (Oryza sativa L. Cv. Nipponbare) was transformed via the agroinfection methods of Hiei et al. [32]. The PHYA allele is heterozygous in the host phyB mutants, permitting the creation of phyA phyB double mutants in the progeny of the overexpressor.
Determination of Mutant Alleles in the Transgenic Lines
DNA was extracted from seedlings of transgenic lines and used as templates for PCR. For the phyA-4 lines, the primer pair phyA BF (5′ -CCAACATCATGGACCTTGTG-3′)/phyA HR (5′ -CCATTGACCAATCCATTGCT-3′) was used to detect the WT allele, and phyA BF/T17 R1 (5′ -CAGCAACGATGTAGATGGTCAAGC-3′) was used to detect the mutant allele (insertion of Tos17). To detect insertion of PHYB(C364A) and PHYB in transgenic lines, the primer pair phyB Fw2032 (5′ -GAGACAGCAACAGTACCCATCTTTG- 3′)/phyB R1 (5′-CTTCCCCTCTTGACCATCCT-3′) was used to amplify an approximately 600-bp fragment of PHYB. To detect insertion of PHYC-GFP, the primer pair 35S Fw (5′ -TGACGTAAGGGATGACGCACAATC-3′)/phyC Rv444 (5′ -GGGGTTGAGCAGGTTGACG-3′) was used. PCR reagents were obtained from the Qiagen Taq system (QIAGEN GmbH, Germany).
RNA Analysis
For analyzing light-induced gene expression, total RNAs were extracted from the seedlings using an RNeasy Plant Mini Kit (QIAGEN, GmbH). Expression of ribrose-1,5-bisphosphate carboxylase small subunit (RbcS) was analyzed by RNA blot analysis as described by Takano et al. [20]. RT-PCR was used to examine the expression levels of two light-harvesting chlorophyll a/b binding protein (Lhcb) genes, Os03g0592500 and Os09g0346500. For Os03g0592500, the primer pair used was F1 (5′ -TGAGCACAAC GACACGAT-3′)/R1 (5′ -TCTCCTCGATCGATCACA-3′). For Os09g0346500, the primer pair used was F1 (5′ -GTAGCTTAGCAGTGGTTAATTGT-3′)/R1 (5′ -TCTTCATCTTCTTAGTfGTACACAAC-3′). The rice ubiquitin gene (UBQ) was used as an internal control, and its primer pair was 237F (5′ -GAGCCTCTGTTCGTCAAGTA-3′)/304R (5′ -ACTCGATGGTCCATTAAACC-3′). The PCR conditions were 94°C for 3 min and 20 cycles of 94°C for 1 min, 50°C, 45°C, or 55°C for 1 min for Os03g0592500, Os09g0346500, or UBQ, respectively, and 72°C for 1 min. PCR reagents were obtained from the Qiagen Taq system.
For analyzing gene expression in PCG/aabb lines, PCG/Aabb transgenic lines with homozygous PHYC-GFP were harvested and rapidly frozen in liquid nitrogen. Then, the individual seedlings were numbered. A small portion from each seedling was used for analyzing the genotype to distinguish the phyA-4 mutant allele from the WT allele by PCR, as described above. The left portion of PCG/aabb seedlings were used for RNA analysis.
Spectrophotometric Assays
Nine-day-old dark grown seedlings of rice were harvested and frozen with liquid nitrogen. The frozen samples were powdered by a mortar and pestle and suspended in 10 volumes of extraction buffer (100 mM Tris-HCl pH 7.5, 5 mM EDTA pH 8.0, 0.2% 2-mercaptoethanol, and protease inhibitor cocktail (Complete, Roche Diagnostics GmbH, Mannheim, Germany)). After being filtered with two layers of Miracloth (Calbiochem, Merck KGaA, Darmstadt, Germany), the suspension was centrifuged for 10 min at 15,000 g and 4°C. The supernatant was combined with solid ammonium sulfate (230 g L−1) to concentrate the phytochrome [33] and centrifuged for 30 min at 15,000 g after incubation on ice for at least 30 min. The precipitate was re-suspended with re-suspension buffer (100 mM Tris-HCl pH 7.5, 5 mM EDTA). The re-suspension was clarified with centrifugation for 10 min at 15,000 g, and the supernatant was used for the measurement.
Spectrophotometric measurements of phytochrome were performed with a photometer (Biospec 1650, Shimazu, Japan). Red and far-red illumination was obtained using red (660 nm; SLP-838A-37, Sanyo Semiconductor Corp., Japan) and far-red (771 nm; HE7601SG, Hitachi, Japan) light-emitting diodes.
Measurement of Coleoptile Length
Sterilized seeds of WT, mutant, or transgenic lines were sown in 0.6% (w/v) agar and then grown in the dark, under FR, R, or white light (W) at 28°C for eight days. Images of the seedlings were captured using a scanner (EPSON GP9600, SEIKO EPSON, Suwa, Japan), and coleoptile lengths were measured by scale.
For analyzing the coleoptile length of PCG/aabb lines, we sowed the seeds of PCG/Aabb transgenic lines. After seedlings had grown for eight days, we labeled and measured the coleoptile length of individual seedlings. Then, we analyzed the genotype of individual seedlings to distinguish the phyA-4 mutant allele from the WT allele by PCR, as described above.
Growth Conditions for the Flowering Time Measurements
Nipponbare, phyB, and PHYB(C364A)/Aabb lines were grown in a growth chamber in LD conditions (light cycle, 14.5 h of light/9.5 h of dark; 28°C in the day/23°C at night). The light sources were metal halide lamps (MLBOC400C-U, MITSUBISHI/OSRAM; 390 µmol m−2 s−1).
Treatment with MG132
The stock solution of 10 mM MG132 was prepared by dissolving MG132 in dimethyl sulfoxide (DMSO; Sigma-Aldrich, MO). Four-day-old dark-grown seedlings were pretreated with either 0.5% DMSO or 50 µM MG132 (Calbiochem). After 1.5 h of pretreatment, seedlings were transferred to R light for 6 h. The soluble protein was extracted from seedlings and analyzed by western blotting using anti-PHYA or anti-PHYC antibodies [20].
Light Sources and Light Intensities
Monochromatic light sources and W in this study were the same as those described by Takano et al. [20]. The fluence rates were 15 µmol m−2 s−1 for FR, 15 µmol m−2 s−1 for R, and 50 µmol m−2 s−1 for W.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB109892 (PHYB), AB018442 (PHYC), X07515 (RbcS), and AY072820 (UBQ).
Results
Quantification of the Relative Levels of phyB and phyC Proteins in Rice Seedlings
To analyze the interaction between phyB and phyC, we initially quantified the relative levels of phyB and phyC in WT rice seedlings grown under either continuous darkness or cW by comparative western blotting by using purified proteins as standards (Fig. S1).
Protein extracts from 5-day-old etiolated seedlings and dilution series of standard proteins (PHYB-His or PHYC-His) were separated by SDS-PAGE on the same gel, and phyB and phyC were detected by anti-PHYB and anti-PHYC antibodies, respectively. For each blot, signal intensities of phyB or phyC for the same protein extracts were compared with those of standard proteins (Figs. S1B and S1C). The phyB to phyC ratio was estimated to be 1.3∶1 in the etiolated seedlings (for calculation details, see supporting information online).
The same methods were applied to quantify the relative level of phyB:phyC in 7-day-old seedlings grown under W. However, the immunoblot signals of phyC protein were too low to estimate the phyC level because rice phyC is light-labile. The protein concentration of phyC was reduced to about one-sixteenth of the level in etiolated seedlings after 24-h growth under W, whereas phyB was light-stable, only less than a one-fourth reduction of levels in etiolated seedlings after growth for 24 h under W (Fig. S2).
phyB and phyC Form Heterodimers in Rice Seedlings
It has been reported that phyB and phyC form heterodimers in Arabidopsis [13], [14]. To examine the physical interaction between phyB and phyC in rice, co-IP experiments were performed using protein extracts from rice seedlings grown under either dark or W conditions (Fig. 1). When protein extracts from WT etiolated seedlings were used, phyB was co-immunoprecipitated with anti-PHYC antibody (Fig. 1A, anti-PHYC/WT). No phyB was co-immunoprecipitated in the control experiments using protein extracts from phyC seedlings (Fig. 1A, anti-PHYC/phyC) or preimmune serum as the precipitating antibody (Fig. 1A, Pre/WT). The interaction between phyC and phyB was also detected in W-grown WT seedlings (Fig. 1B), although the co-IP signals were weaker than those of etiolated seedlings. These results indicate that a potential physical interaction exists between phyB and phyC in both etiolated and light-grown rice seedlings.
Figure 1. phyB and phyC proteins form complexes in both etiolated and light-grown rice seedlings.

A. Protein extracts (200 µg) from 7-day-old etiolated seedlings of WT or phyC mutants (phyC) were immunoprecipitated with an anti-PHYC antibody (Anti-PHYC/WT; Anti-PHYC/phyC) or with preimmune serum from the same rabbit (Pre/WT). phyC or phyB was immunodetected in the precipitation. Thirty micrograms of protein extracts from WT were loaded as the positive control (Ext). B. Protein extracts (500 µg) from WT seedlings grown under W for 7 days were immunoprecipitated with an anti-PHYC antibody (Anti-PHYC/WT) or with preimmune serum from the same rabbit (Pre/WT). phyC or phyB was immunodetected in the precipitation. Fifty micrograms of protein extracts from WT were loaded as the positive control (Ext).
However, the question remains whether phyB and phyC form heterodimers in rice seedlings as observed in Arabidopsis [13], [14]. To address this question, protein extracts from etiolated seedlings of WT or phytochrome mutants were fractionated by SEC. The column was calibrated with protein molecular weight standards, and the molecular size of each fraction was calculated based on the calibration line.
Using the same column with the same conditions, protein extracts from etiolated seedlings of WT rice and various phytochrome mutants were fractionated. Aliquots of each eluted fraction were separated by SDS-PAGE, and phyA, phyB, and phyC were immunochemically detected. As shown in Figures 2A-1, 2B-1, and 2C-1, phyA, phyB, and phyC from WT rice were mainly detected in Fraction #20 with a calculated molecular mass in the range of 316 to 404 kDa, similar to observations in Arabidopsis where all five native phytochrome proteins migrate at apparent masses in the range of 300 to 380 kDa on SEC [13]. We characterized the migration of phyA from etiolated Arabidopsis seedlings using the same assay conditions, and confirmed that Arabidopsis phyA was mainly detected in Fractions #20 and #21 (Fig. 2A-5). Since Arabidopsis phyA is a homodimer [13], we concluded that rice phyA, phyB, and phyC predominantly exist in dimeric forms. In WT and phyA-mutant extracts, phyB was detected in the monomer fractions as a second peak (Fig. 2B-1 and 2B-2). In phyC-deficient mutants, phyB was only detected in dimeric forms (Fig. 2B-3 and 2B-4), suggesting that the absence of phyC prevents monomerization of phyB. As dimerization is necessary for full activity of phyB [12], [34], it is hard to imagine that bioactive phyB exists as monomers within cells. It remains possible that phyB/phyC dimers are not as stable as phyB homodimers and a small proportion of heterodimers may dissociate during size exclusion chromatography (SEC). To dissect the dimer composition of phyC in WT rice, protein extracts from etiolated seedlings of various phytochrome mutants were fractionated by SEC and phyC was detected. In the phyA mutant, phyC migrated in a position equivalent to a dimer, the same as in WT (Fig. 2C-2). However, in the phyB-deficient mutants (phyB and phyA phyB mutants), phyC was mainly detected as a putative monomer in Fraction #22 with a molecular mass of 192 to 246 kDa (Figs. 2C-3 and 2C-4). Thus, phyB is necessary for forming a phyB/phyC heterodimer and phyC exists in a monomeric form in the absence of phyB.
Figure 2. Immunoblot analyses of phyA, phyB, and phyC in the column fractions of protein extracts.
Protein was extracted from 7-day-old etiolated seedlings of WT and all phytochrome single and double mutants and fractionated by SEC. phyA, phyB, and phyC were immunochemically detected with anti-PHYA (A), anti-PHYB (B), and anti-PHYC antibodies (C), respectively, in the individual fractions (#17–#24). Small numbers above the fraction numbers are the molecular sizes which were calculated based on the calibration line of standard proteins. Arabidopsis seedlings were used to characterize the migration of homodimer phyA. D. Physical interactions between phyB and phyC in the fractions (#17–#26) of protein extracts from 7-day-old etiolated seedlings of WT. The individual fractions were immunoprecipitated with an anti-PHYC antibody. phyB and phyC in the precipitates were detected by immunoblotting. Thirty micrograms of protein extracts from WT were loaded as the positive control (Ext).
Figure 5. phyC is biologically active in PHYB/aabb and PHYB(C364A)/aabb transgenic lines.
A. Diagram of PHYB and mutant PHYB(C364A) constructs introduced into phyB mutants. B. Immunoblot analyses of phyB and phyC levels in the dark-grown seedlings of PHYB/Aabb and PHYB(C364A)/Aabb transgenic lines. Protein extracts from 7-day-old etiolated seedlings of different transgenic lines of PHYB(C364A)/Aabb (#27, #69, #71, and #72) and PHYB/aabb (#7 and #11) were probed with anti-PHYC and anti-PHYB antibodies. WT and phyB were used as positive and negative controls, respectively. Each lane was loaded with 50 µg of protein extracts. C. FR inhibited coleoptile growth in PHYB/aabb transgenic lines. Mean coleoptile lengths were shown for the seedlings of WT, phyA, phyB, phyA phyB, and two lines of PHYB/aabb (#7-2- and #11-3-) grown under FR (15 µmol m−2 s−1, open bars) or in the dark (filled bars) for eight days. The mean ± SE obtained from at least 12 seedlings is plotted. D and E. FR (D) or R (E) inhibited coleoptile growth in the PHYB(C364A)/Aabb and PHYB(C364A)/aabb transgenic lines. Mean coleoptile lengths are shown for WT, phyA (only in D), phyB, phyA phyB, PHYB(C364A)/Aabb, and PHYB(C364A)/aabb seedlings grown under FR (15 µmol m−2 s−1, D), R (15 µmol m−2 s−1, E), or in the dark (filled bars) for eight days. Four independent lines (#27-6-, #69-2-, #71-1-, and #72-1-) of PHYB(C364A) were used, and results from different genetic backgrounds (Aabb or aabb) were depicted with separate bars. The mean ± SE obtained from at least 12 seedlings is plotted.
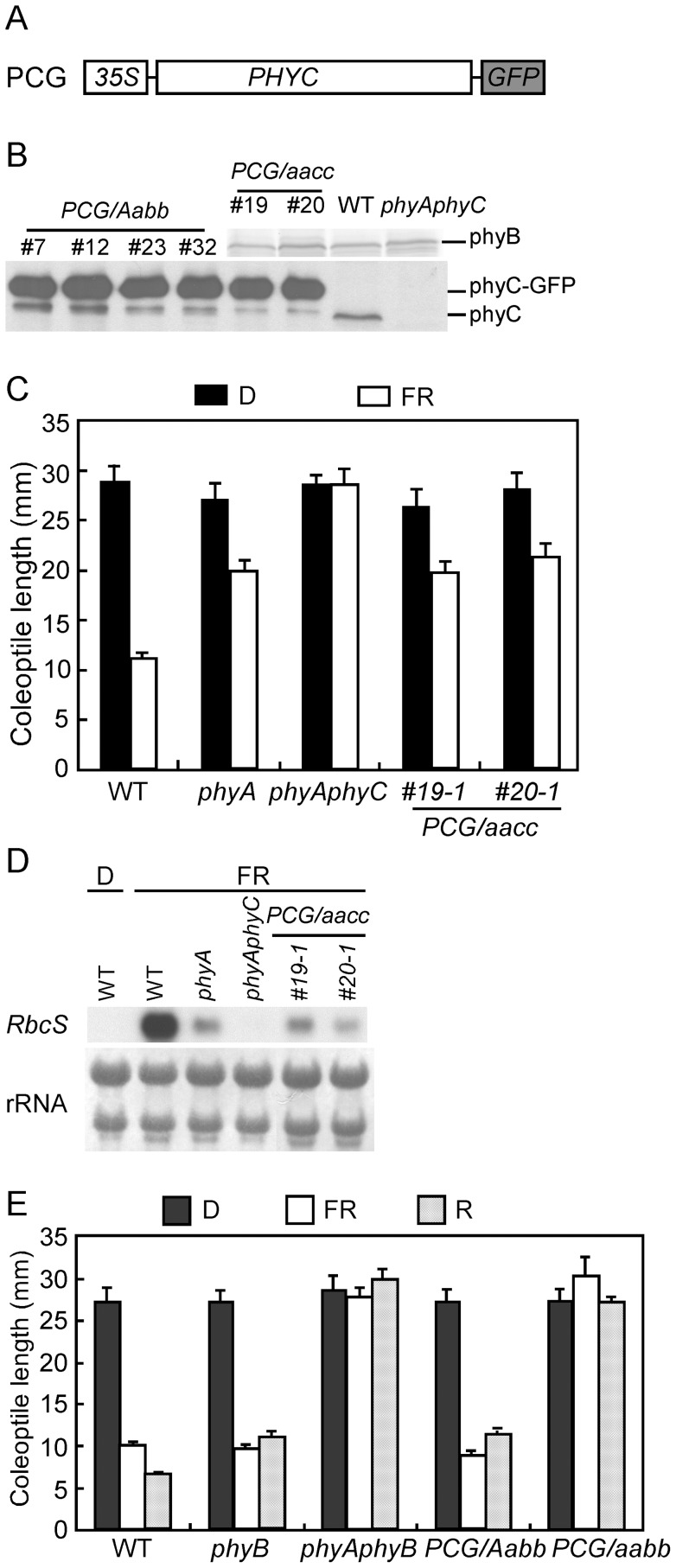
Figure 3. phyC-GFP is biologically active in phyA phyC backgrounds but inactive in phyB-mutant backgrounds.
A. Diagram of the PHYC-GFP fusion construct introduced into phyA phyC and phyB mutants. B. Immunoblot analyses of phyB and the phyC-GFP fusion protein in WT, phyA phyC, and PCG/aacc transgenic lines using anti-PHYB and anti-PHYC antibodies. Protein extracts (25 µg from PCG/aacc lines and PCG/Aabb lines; 50 µg from WT and phyA phyC seedlings) were loaded to detect phyC-GFP and phyC. Fifty micrograms of protein were loaded to detect phyB. Aabb is the phyB mutant where the PHYA mutant allele is heterozygous. C. The phyC-GFP fusion protein caused inhibition of coleoptile growth by FR irradiation in PCG/aacc transgenic lines. Mean coleoptile lengths are shown for WT, phyA, phyA phyC, and PCG/aacc seedlings (#19 and #20) grown under FR (15 µmol m−2 s−1, open bars) or in the dark (filled bars) for 8 days. The mean ± SE (standard error) obtained from at least 12 seedlings is plotted. D. phyC-GFP fusion protein exerted the phyC function in the expression of RbcS. The expression of RbcS induced by FR was comparatively analyzed by RNA blotting in the seedlings of WT, phyA, phyA phyC, and PCG/aacc (#19 and #20). RbcS was used as a probe. rRNA was stained by methylene blue as a quantity control. E. phyC-GFP did not participate in the photoinhibitory responses of coleoptile growth under FR or R in the phyB mutant background. Mean coleoptile lengths are shown for WT, phyB, phyA phyB, PCG/Aabb, and PCG/aabb seedlings grown under FR (15 µmol m−2 s−1, open bars) or under R (15 µmol m−2 s−1, hatched bars) or in the dark (filled bars). The mean ± SE obtained from at least 12 seedlings is plotted, excluding the PCG/aabb genotype, for which only six seedlings were grown.
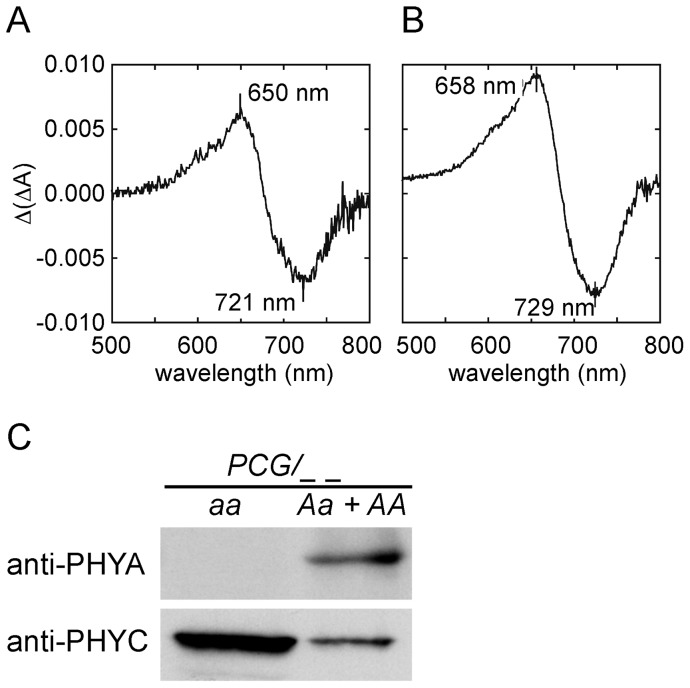
Figure 4. Absorption difference spectra of phyC-GFP.
Difference spectrum characteristics of phytochromes in protein extracts from 7-day-old etiolated seedlings of PCG/aabb (A) or PCG/Aabb (B). Difference spectra were measured using a double-beam spectrophotometer and normalized by 1 mg total protein per mL. C. Immunoblot analyses of phyA and the phyC-GFP fusion protein in PCG/Aabb or PCG/aabb transgenic lines using anti-PHYA and anti-PHYC antibodies. Each lane was loaded with 5 µg of total protein.
The physical interaction between phyB and phyC was also supported by co-IP assays in the eluted fractions. When precipitated with an anti-PHYC antibody, phyC was detected in Fractions #18 to #22 (Fig. 2D, anti-PHYC), and phyB was co-immunoprecipitated in the fractions corresponding to the dimeric complexes (Fig. 2D, anti-PHYB). These results further confirm that phyB and phyC form heterodimers, like those observed in Arabidopsis [14].
These deductions were supported by in vitro experiments using recombinant proteins. Recombinant phyC with phytochromobilins as a chromophore was expressed in E. coli (Fig. S3A). As a control, recombinant phyA was also expressed using the same experimental procedure. The recombinant phyA and phyC showed typical R/FR difference spectra with maximum peaks of 667 nm and 650 nm, and minimum peaks of 724 nm and 725 nm, respectively (Figs. S3B and S3C), indicating that recombinant phyA and phyC are spectrophotometrically active. When recombinant phyA or phyC was fractionated by SEC using the same column, recombinant phyA was mainly detected in the dimeric Fraction #20, whereas recombinant phyC was mainly detected in the monomeric Fraction #24 (Fig. S3D) as detected using both anti-His and phytochrome-specific antibodies. These results suggest that rice phyC exists as a monomer, while phyA exists as a homodimer.
phyC-GFP is biologically active in phyA phyC mutants, but inactive in phyB mutants
Studies on phytochrome single and double mutants have revealed that phyC is involved in FR photo-sensing for de-etiolation (e.g. inhibition of coleoptile growth) as well as induction of light-regulated genes, such as Lhcb or RbcS, in rice [20]. However, in the mutants deficient in phyB, these responses mediated by phyC were negligible, as indicated by the findings that phyB and phyB phyC showed similar responses to both R and FR and that the phyA phyB and phyA phyB phyC mutants always exhibited the same phenotype under either FR or R [20], [21]. These observations suggest that phyC function depends on the presence of phyB. phyC protein concentration is greatly reduced in phyB mutants [20], so it is possible that reduced phyC levels are responsible for the functional inactivity of phyC in phyB mutants. An alternate possibility is that phyC functions only in the phyB/phyC heterodimeric form. To test the first possibility, we produced transgenic plants overexpressing PHYC-GFP and examined their responses to light.
To increase the phyC levels in phyB mutants, 35S:PHYC-GFP (Fig. 3A) was introduced into phyB mutants, where the PHYA allele is heterozygous in the host phyB mutant line (PCG/Aabb) to obtain progenies of PHYC overexpressors in phyA phyB double mutants (PCG/aabb). The same construct was also introduced into the phyA phyC double mutant as a positive control (PCG/aacc). Accumulation of phyC-GFP in transgenic seedlings was examined by immunoblot analysis. Protein extracts from the 7-day-old etiolated T2 seedlings of PCG/Aabb lines (#7, #12, #23, and #32) and PCG/aacc lines (#19 and #20) were probed with anti-PHYB and anti-PHYC antibodies (Fig. 3B). Strong signals corresponding to the phyC-GFP fusion protein were detected around 140 kDa in the transgenic lines (Fig. 3B). Signal intensities of the phyC-GFP in the mutants were significantly higher than those of the intrinsic phyC in WT seedlings. On the other hand, the abundance of phyB protein in the transgenic seedlings were comparable to that of WT (Fig. 3B). These results indicate that phyC-GFP overexpression did not increase phyB levels in the PCG/aacc transgenic lines.
First, T2 seedlings of PCG/aacc transgenic lines were grown under FR for 8 days to examine the biological activity of the phyC-GFP fusion protein in the phyA phyC double mutant background. The phyA phyC double mutant did not show any responses to FR (Fig. 3C; Fig. S4A) as reported by Takano et al. [20]. In PCG/aacc transgenic seedlings (#19 and #20), however, coleoptile growth was inhibited (Fig. 3C; Fig. S4A), and RbcS or Lhcb was induced by FR to levels similar to those observed in phyA mutants (Fig. 3D; Fig. S4B). These results indicate that overexpression of PHYC-GFP complemented the loss of phyC function in inhibiting coleoptile growth and inducing light-regulated genes under FR. Therefore, we can conclude that phyC-GFP is biologically active in the phyA phyC seedlings.
Second, the biological activities of phyC-GFP were examined in the phyA phyB mutant background. Coleoptile growth and induction of Lhcb genes were analyzed in the progeny of PCG/Aabb plants grown under FR or R for 8 days. Although phyC-GFP fusion protein concentrations in several PCG/Aabb transgenic lines (#7, #12, #23, and #32) were as high as those in PCG/aacc lines (#19 and #20) in etiolated seedlings (Fig. 3B), the coleoptile lengths of the segregated PCG/aabb seedlings grown under either FR or R were the same as the lengths of phyA phyB seedlings (Fig. 3E; Figs. S4C and S4E). In addition, coleoptile lengths of the segregated PCG/Aabb seedlings were not significantly different from those of phyB seedlings under FR or R (Fig. 3E; Figs. S4C and S4E). Furthermore, two Lhcb genes, Os03g0592500 and Os09g0346500, were not induced by FR or R in the segregated PCG/aabb transgenic seedlings (Figs. S4D and S4F), although phyC-GFP levels in these two lines were comparable with those in PCG/aacc seedlings (Fig. 3B). These results indicate that phyC-GFP participated neither in the photoinhibitory responses of coleoptile growth nor in the induction of Lhcb genes under FR or R in the phyB-deficient backgrounds (phyB or phyA phyB). Therefore, the phyC-GFP fusion protein is biologically inactive in the phyA phyB and phyB transgenic lines. Taken together with the findings that phyC-GFP fusion proteins are biologically active in the phyA phyC double mutant (Figs. 3B and 3C), phyB protein is indispensable for the phyC-mediated responses to FR and R. These findings also suggest that reduced phyC levels are not the major factor in its functional loss in phyB-deficient mutants.
The lack of biological responses to FR and R in PCG/aabb seedlings might be due to the photochemical inability of expressed phyC-GFP. To examine this possibility, an absorption difference spectrum was obtained. As shown in Figure 4A, a difference spectrum characteristic for the phytochrome was obtained for the protein extract from 7-day-old etiolated seedlings of PCG/aabb. The absorption difference spectrum (Fig. 4A) is believed to be that of phyC-GFP because western blotting confirmed that phyC-GFP was the only phytochrome present in PCG/aabb seedlings (Fig. 4C). As a reference, a difference spectrum was also obtained from protein extracts of PCG/Aabb seedlings (Fig. 4B), where both phyA and phyC are expressed (Fig. 4C). It was also noted by comparing the results of Figures 4A and 4B that maximum and minimum peaks were blue-shifted by 8 nm in phyC-GFP-expressing seedlings compared with those in seedlings expressing a mixture of phyA and phyC-GFP. Such a blue-shift in the difference spectrum of phyC was also observed in our experiments using recombinant phyA and phyC (Fig. S3C), and has been reported in Arabidopsis as well [35]. Therefore, phyC-GFP in PCG/aabb seedlings is spectrophotometrically active but biologically inactive. These results suggest that phyC can absorb light but not initiate downstream signaling without the presence of phyB. Therefore, we examined the mechanism by which phyB is involved in phyC function.
Because phyB and phyC form heterodimers in the etiolated seedlings, it is assumed that the function of phyC-GFP in PCG/aacc seedlings is probably attributed to the formation of phyB/phyC-GFP complexes. Co-IP assays revealed the physical interaction between phyC-GFP and phyB (Fig. S5A). Even so, the mechanism by which phyB affects phyC-mediated responses in the phyB/phyC heterodimer remains elusive. It is unclear whether the binding of phyB to phyC alone is sufficient or whether photo-sensing by phyB is required.
Expression of Chromophore-less phyB Restores phyC Levels as well as phyC Function in phyB Mutants
To address this question in detail, the PHYB derivative, PHYB(C364A) was used, where the chromophore attachment site, cysteine 364, was converted to alanine (Fig. 5A). The resulting molecule is expected to be inactive, as observed for PHYB(C357S) derivatives in Arabidopsis [12]. PHYB(C364A) or intact PHYB cDNA was introduced into phyB mutants, in which the PHYA allele is heterozygous to obtain PHYB(C364A)/phyA phyB (PHYB(C364A)/aabb) or PHYB/phyA phyB (PHYB/aabb) as segregated progenies. Immunoblot analyses showed that expression levels of PHYB(C364A) in the PHYB(C364A) lines and those of phyB in the PHYB-transgenic lines were comparable with those of WT in the etiolated seedlings (Fig. 5B). Notably, phyC levels were also increased in these transgenic lines corresponding to the increased levels of phyB, even if phyB is inactive. The resultant phyC levels were similar to those of WT (Fig. 5B). These results suggest that the expression of chromophore-less phyB restored phyC protein concentrations in the phyB mutant to levels comparable to WT.
To examine the biological activities of recovered phyC in the PHYB and PHYB(C364A) transgenic lines, coleoptile growth and Lhcb expression were analyzed in these transgenic lines. When grown in darkness, coleoptile lengths were not significantly different between the genotypes tested (Fig. 5C; Fig. S6A). Under FR, phyA phyB double mutants exhibited long coleoptiles similar to etiolated seedlings, but the expression of PHYB in the phyA phyB double mutants (PHYB/aabb) resulted in coleoptile lengths as short as those of phyA mutants (Fig. 5C; Fig. S6A). Lhcb genes (Os03g0592500 and Os09g0346500) were also induced by FR in these PHYB/aabb transgenic seedlings (Fig. S6B). It is known that FR-mediated responses are attributed to only phyA or phyC in rice [20]. Therefore, recovered phyC, not introduced phyB, was responsible for de-etiolation of seedlings under FR in the PHYB/aabb lines, indicating that the phyC in PHYB/aabb transgenic lines is biologically active.
Before analyzing the biological activities of phyC in PHYB(C364A) transgenic lines, we first examined the activity of PHYB(C364A). Takano et al. [20] reported that seedlings of phyB mutants exhibited a pale green phenotype under R. When seedlings of WT, phyB, PHYB(C364A)/aabb, and PHYB/aabb lines were grown under R for 8 days, WT and PHYB/aabb seedlings exhibited a dark green phenotype, while the PHYB(C364A) seedlings (#27-6 and #71-1) exhibited a pale green phenotype similar to that of phyB mutants (Fig. S6C). These observations indicate that PHYB(C364A) is insensitive to R in the transgenic lines. Thus, the PHYB(C364A)/aabb lines made it possible to analyze the real function of phyC in rice photomorphogenesis without the functional interference of phyA or phyB.
To examine the phyC-mediated responses to FR, seedlings from PHYB(C364A)/Aabb lines (#27-6) were grown under FR for 8 days (Fig. S6D). The coleoptile growth of PHYB(C364A)/aabb seedlings was significantly inhibited by FR, exhibiting dramatically shorter coleoptile lengths than those of phyA phyB double mutants (Fig. S6D, PHYB(C364A)/aabb). As shown in Figure 5D, the coleoptile growth was obviously inhibited by FR in all lines examined excluding the phyA phyB double mutant seedlings. Interestingly, the inhibition of coleoptile growth was more severe in PHYB(C364A)/aabb mutants than in phyA mutants, in both of which phyC is the only functional photoreceptor for FR (Fig. 5D). These results indicate that phyC is less sensitive to FR in the presence of functional phyB than in the presence of nonfunctional phyB. In addition, PHYB(C364A)/Aabb mutants had shorter coleoptiles compared with those of phyB mutants, which could be interpreted as the additive response of phyA and phyC to FR (Fig. 5D). Furthermore, expression of Lhcb genes (Os03g0592500 and Os09g0346500) was also induced by FR in PHYB(C364A)/aabb seedlings (Fig. S6B, #27-6- and #71-1-). Thus, phyC can initiate downstream signaling only in the presence of phyB, even if phyB is spectrophotometrically inactive.
phyC-mediated responses to R in the inhibition of coleoptile growth and induction of light-regulated genes were also examined using PHYB(C364A) seedlings. When WT, phyB, phyA phyB, PHYB(C364A)/aabb, and PHYB(C364A)/Aabb seedlings were grown under R for 8 days, all seedlings, excluding those of phyA phyB double mutants, exhibited short coleoptiles (Fig. S6E). Moreover, the coleoptile lengths of PHYB(C364A)/aabb seedlings were as short as those of PHYB(C364A)/Aabb under R, and the coleoptile lengths of PHYB(C364A)/aabb seedlings were shorter than those of phyB. The differences were small but statistically significant (Fig. 5E). These results indicate that phyA and phyC respond to R in a highly redundant manner and that the phyC-mediated response to R is larger in magnitude than that of phyA, at least for the inhibition of coleoptile growth. In addition, PHYB(C364A)/aabb seedlings exhibited a pale green phenotype (Fig. S6E), indicating that phyC has a similar role to phyA in chlorophyll accumulation. Moreover, Lhcb genes were induced in PHYB(C364A)/aabb transgenic lines by R, and the expression levels of Lhcb genes were higher in PHYB(C364A)/aabb mutants than in phyB mutants (Fig. S6F, #27-6-, #69-2-, and #71-1-), which is consistent with the stronger inhibition of coleoptile growth in PHYB(C364A)/aabb mutants than in phyB mutants. These results indicate that phyC can sense R to control the de-etiolation of rice seedlings in the presence of phyB.
Collectively, phyC is biologically active in the presence of both functional and nonfunctional phyB protein, suggesting that the exertion of phyC function is not related to either the photo-sensing of phyB or the phyB-mediated responses but depends on complex formation with phyB. The co-IP assay revealed that phyC had a direct physical interaction with phyB in both PHYB and PHYB(C364A) transgenic seedlings (Fig. S5B).
These findings are different from the results obtained in Arabidopsis, in which expression of chromophore-less phyB restored phyC levels, but did not restore phyC activity [14]. It was concluded that phyC requires dimerization with chromophore-bearing phyB to have activity in Arabidopsis.
Flowering Times of the Phytochrome Mutants
Under LD conditions (14L/10D), phyB and phyC mutants showed similar early flowering phenotypes, and phyB phyC double mutants flowered as early as phyB or phyC single mutants [20]. These observations suggest that phyB and phyC are probably equally involved in the control of flowering time for delaying response to LD conditions. However, we have not been able to exclude the possibility that phyB is not involved in the control of flowering times under the LD conditions because phyC is also dysfunctional in the phyB mutants. However, we can now overcome this limitation via phyB(C364A) mutants, which have functional phyA and phyC but dysfunctional phyB.
Three independent phyB(C364A) lines were grown under LD (14.5L/9.5D) conditions, and their flowering times were compared with those of WT and phyB mutants (Fig. 6). All PHYB(C364A)/Aabb lines flowered earlier than the WT by the same extent as the phyB mutant. These observations clearly confirmed that phyB and phyC are redundantly involved in the control of flowering time in response to LD conditions.
Figure 6. PHYB(C364A)/Aabb transgenic lines flower earlier than WT under LD conditions.
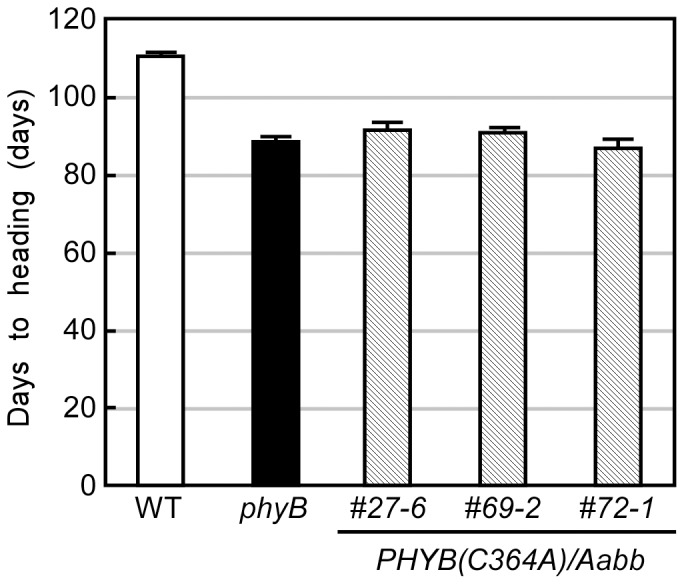
Nipponbare (WT), phyB-1 mutant (phyB), and PHYB(C364A)/Aabb transgenic lines were grown in a growth chamber set with LD (14.5L/9.5D) condition. The mean ± SE obtained from 20 plants is shown.
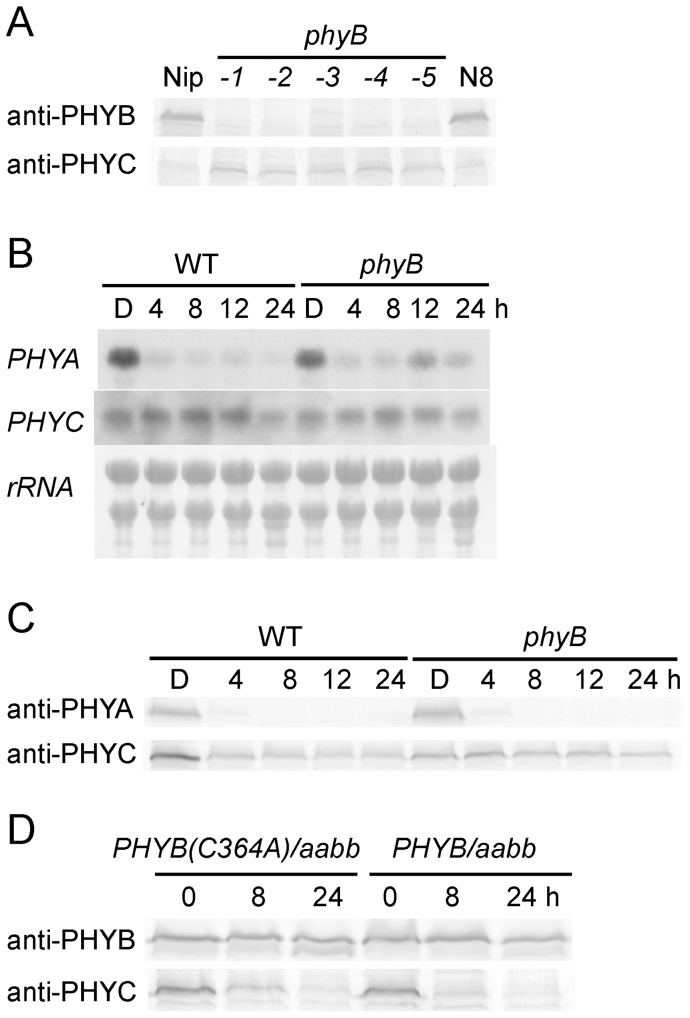
phyB Is Involved in the Light-Induced Degradation of phyC in Rice
In Arabidopsis, phyC concentrations have been shown to be lower in phyB mutants than in WT seedlings grown under dark as well as R conditions [23]–[25]. A similar phenomenon was observed in rice etiolated seedlings [20]. In the seedlings grown under W, however, phyC levels were similar in WT and phyB mutants (Fig. 7A). To address this issue more comprehensively, we examined the effect of W on both transcript and protein abundance of phyC at different time points after W irradiation in WT and phyB mutants. As shown in Figure 7B, PHYA mRNA levels were significantly down-regulated under these conditions, whereas transcript levels of PHYC remained relatively constant in WT seedlings as well as in phyB mutants, in accordance with the observation of Basu et al. [18]. These results suggest that the effect of W on the transcript level of PHYC is indistinguishable between WT and phyB mutants. Next, the light-stabilities of phyA and phyC proteins were examined in these seedlings. As shown in Figure 7C, protein abundance of phyA was rapidly reduced by W exposure in both WT and phyB mutants. On the other hand, phyC levels were reduced in WT but not in phyB mutants. As a result, the residual phyC levels in the phyB mutant were slightly higher than those in WT after being exposed to W for 24 h. These results imply that phyB protein is somehow involved in the light-induced degradation of phyC, but not phyA, in rice.
Figure 7. phyB is involved in light-induced degradation of phyC in rice.
A. Levels of phyC are not reduced in phyB mutant seedlings grown under W. Protein extracts were prepared from WT and phyB seedlings grown under W for 7 days. Each lane was loaded with 50 µg of protein extracts for the detection of phyB and phyC using anti-PHYB and anti-PHYC antibodies, respectively. Nipponbare (Nip) and Norin8 (N8) were used as controls. phyB-1, -2, -3, -4, and -5 are five different mutant alleles of PHYB. B. Effect of W on the transcript levels of PHYA and PHYC genes in WT and the phyB mutant. The seedlings of WT and phyB-1 mutant (phyB) were grown for 5 days in the dark (D) and then exposed to W for 4, 8, 12, or 24 h before harvesting. For detecting the transcripts of PHYA and PHYC, each lane was loaded with 10 µg of total RNA. As a quantity control, rRNA was stained with methylene blue. C. Effect of W on phyA and phyC protein concentrations in WT and the phyB mutant. Growth conditions of the seedlings were the same as those in (B). Fifty micrograms of protein extract were loaded in each lane. D. PHYB(C364A) protein is necessary for the R-induced degradation of phyC. PHYB(C364A)/aabb (#27) and PHYB/aabb (#11) seedlings were grown in the dark (D) or in the D and then exposed to R for 8 or 24 h before harvesting. phyB and phyC proteins in 50 µg of protein extracts were detected with anti-PHYB and anti-PHYC antibodies, respectively.
Next, we investigated the light-stability of restored phyC in PHYB/aabb and PHYB(C364A)/aabb transgenic lines. As shown in Figure 7D, the levels of phyC were obviously reduced by R irradiation in the seedlings of both PHYB and PHYB(C364A) mutants, while PHYB protein concentrations were nearly constant under these conditions in both seedlings. These results indicate that the restored phyC undergoes light-induced degradation. The formation of heterodimers between phyB and phyC is considered necessary for the light-induced selective degradation of phyC in rice.
It has been well established that the ubiquitin/26S proteasome pathway is involved in the Pfr-specific degradation of phyA in Arabidopsis [36]–[38] because a proteasome inhibitor, MG132, repressed the R-induced degradation of phyA in Arabidopsis [38]. Therefore, we tested the effect of MG132 on the R-induced degradation of phyC. Treatment with 50 µM MG132 significantly repressed the reduction of phyC and phyA in rice seedlings grown under R for 6 h (Fig. S7). These results suggest that the ubiqutin/26S proteasome pathway is involved in the light-induced degradation of phyC in rice seedlings, as is the case for Arabidopsis phyA.
Discussion
phyC Forms Heterodimers with phyB in Rice
In this study, it was revealed that all three native phytochrome proteins predominantly migrated at masses in the range of 316 to 404 kDa in rice etiolated seedlings (Fig. 2). These apparent masses are larger than the calculated 250 kDa for the dimeric forms of the phytochromes. However, similar observations were reported in Arabidopsis and oat. All five native Arabidopsis phytochrome proteins migrated at masses in the range of 300 to 380 kDa on SEC, and purified oat phyA had an apparent mass of 350 to 360 kDa on SEC, all of which are dimers [11], [13], [39]. Therefore, we conclude that native rice phytochromes also predominantly exist as dimers.
PhyB and phyC form heterodimeric complexes in Arabidopsis [13]. Native rice phyC was detected in dimer fractions on SEC using protein extracts from WT and phyA mutants and in monomer fractions in the absence of phyB both in vivo and in vitro (Fig. 2; Fig. S3D). Given the direct interaction between phyC and phyB (Figs. 1 and 2D), we deduced that phyC forms heterodimers with phyB in rice, similar to Arabidopsis. However, if phyB is absent, then phyC exists as a monomer. Even in its monomeric form, phyC is spectrophotometrically active (Fig. S3C), but biologically inactive.
The phyB to phyC ratio was estimated to be 1.3∶1 in etiolated seedlings (Fig. S1 and Supporting Information S1). Therefore, phyB exists as both phyB/phyC heterodimers and phyB/phyB homodimers in WT seedlings, and phyB homodimers and phyB/phyC heterodimers might have different roles in response to light. Microarray experiments using rice etiolated seedlings revealed that there is a cluster of genes that shows early and transient expression after an R-pulse in WT and phyA mutants, but not in phyA phyC mutants [40]. Because only phyB exists in the phyA phyC mutants, these results suggest that the early and transient gene expression induced by R is mediated by phyB/phyC heterodimers, but not by phyB homodimers.
PHYB Protein is Essential for phyC-Mediated Responses in Rice
In this study, PCG/aabb seedlings exhibited the same phenotypes as phyA phyB seedlings, which do not respond to R and FR. In contrast, PCG/aacc seedlings responded to FR and showed de-etiolated phenotypes (Figs. 3C and 3D; Figs. S4A and S4B). These results indicate that phyC-GFP fusion proteins are biologically inactive in the phyA phyB background, but biologically active in the phyA phyC background, although in both cases higher phyC-GFP fusion protein concentrations were accumulated in the mutants compared to WT (Fig. 3B; Fig. S4C). Therefore, different phenotypes were observed depending on the absence or presence of phyB.
The molecular mechanism of the functional dependency of phyC on phyB was revealed by the experiments using PHYB(C364A)/aabb transgenic lines, in which chromophore-less PHYB was expressed, and phyC levels were recovered to those observed in the WT. These seedlings responded to both R and FR, which indicates that the functional rescue of phyC in PHYB(C364A)/aabb is attributed to the presence of phyB, even when it is non-functional, but not to the increased levels of phyC because phyC levels are significantly lower in PHYB(C364A)/Aabb transgenic lines than phyC-GFP levels in PCG/aabb mutants (Figs. 3B and 5B). Furthermore, it became clear for the first time that phyC is involved in the responses to both FR and R for the inhibition of coleoptile growth as well as the induction of Lhcb genes in rice (Fig. 5; Fig. S6).
It is worth noting that PCG/aacc mutants exhibited the same phenotypes as phyA mutants under FR despite the significantly higher abundance of the phyC-GFP fusion protein in these transgenic seedlings compared to those in phyA seedlings (Fig. 3). This suggests that the limiting factor is the level of phyB, which contributes to the formation of phyB/phyC heterodimers in PCG/aacc seedlings. In turn, this observation provides indirect evidence that the heterodimeric complex of phyB/phyC is the only functional form for the phyC-mediated responses to light.
Photosensory Specificities of phyC in Rice
As Clack et al. [14] mentioned, heterodimerization of phyB and phyC is likely fundamental throughout plants. However, there is a difference between Arabidopsis and rice. Arabidopsis phyC requires dimerization with functional phyB to have activity [14], while rice phyC is active even while forming heterodimers with chromophore-less PHYB. This difference seems to be correlated with the functional differentiation of phyC during evolution. Rice phyC is involved in the responses to both R and FR in the presence of phyB (Fig. 5; Fig. S6). Thus, the photosensory specificity of phyC is similar to that of phyA and different from that of phyB in rice. In contrast, Arabidopsis phyC does not participate in the control of seedling de-etiolation under FR, but does participate in controlling responses to R [23], [41].
Regarding the inhibition of coleoptile growth under R, the inhibitory effect mediated by phyC was similar to that mediated by phyA, as demonstrated by the fact that PHYB(C364A)/aabb seedlings exhibited the same phenotype as PHYB(C364A)/Aabb seedlings (Fig. 5E). In addition, the seedlings of phyA and phyC mutants did not show any different phenotypes from those of WT under R [20]. These observations indicate that phyA, phyB, and phyC act in a highly redundant manner to control coleoptile growth under R.
phyA is the principal photoreceptor for FR, while phyC plays a minor role, because phyA mutants show more pronounced phenotypes than phyC mutants under FR [20]. Unexpectedly, however, PHYB(C364A)/aabb seedlings exhibited significantly shorter coleoptiles than phyA seedlings under FR, although phyC is the only photoreceptor for FR in these two genotypes (Fig. 5D). These findings may indicate that phyB has an antagonistic function against the phyC-mediated responses to FR. When phytochrome molecules are activated to their Pfr forms, a “Z” to “E” isomerization occurs in the C-15 double bond between the C and D rings of the linear tetrapyrrole [42]. The Pfr chromophore is in a distorted, high-energy C15-E anticonfiguration. Chromophore-apoprotein interactions (including the structural relaxation at the chromophore-binding site) and subsequent conformational changes in the protein backbone are required to maintain the high-energy state [42]–[45]. In this context, when phyB/phyC heterodimers are exposed to FR, phyC subunits, similar to Arabidopsis phyA, absorb FR and adopt the phyB Pr/phyC Pfr configuration because phyB Pr cannot sense FR [20]. Pr-conformation of phyB subunits may prevent phyC subunits from maintaining the high-energy Pfr form, whereas phyB(C364A) subunits lacking the chromophore may not affect the Pfr conformation of phyC subunits.
Hening and Schäfer [46] prepared Arabidopsis phyA dimers that incorporate the essential chromophore only in one subunit by using a coexpression system in yeast and demonstrated that such mixed dimers showed unaltered difference spectra. Their results, together with our findings, indicate that the absence of a chromophore from one subunit of a phytochrome dimer does not affect the photoreversible change of the functional phytochrome.
phyB Is Involved in the Flowering Time Determination under LD
For the involvement of phytochromes in the regulation of flowering time, phyB has been considered responsible for delaying flowering in response to LD conditions because phyB mutants flower earlier than WT under LD conditions [20]. However, because phyB, phyC, and phyB phyC mutants all flowered earlier than WT in LD [20], and because phyC is also dysfunctional in the phyB mutants, the results for the phyB mutants must be carefully reconsidered. In this report, we created PHYB(C364A)/Aabb transgenic rice, in which only phyB is dysfunctional, and demonstrated that phyB is surely involved in the control of flowering time by suppressing the floral initiation under LD conditions in rice (Fig. 6). Moreover, the observations that phyB, phyC, and phyB phyC mutants similarly flowered earlier than WT in LD can be easily explained if phyB/phyC heterodimers are responsible for the suppression of flowering under LD.
phyB Affects the Stability of phyC in Both Etiolated and Light-grown Rice Seedlings
PhyC levels are decreased in phyB mutants in rice, which is attributed to a posttranslational event because RNA blot analyses of PHYC did not show any differences between phyB mutants and WT [20]. In this study, chromophore-less phyB recovered phyC to WT levels (Fig. 5B), suggesting that phyB is indispensable for stabilizing phyC by forming phyB/phyC heterodimers in etiolated seedlings.
RNA blot and immunoblot analyses suggest that phyB is necessary for light-induced degradation of phyC by formation of phyB/phyC heterodimers (Fig. 7). Recently, several studies about the degradation of Arabidopsis phyA have revealed that light-induced degradation of phyA is in part controlled by light-regulated import into the nucleus where the turnover rate is significantly faster than in the cytoplasm [47], [48]. Based on these observations, we speculate that the rice phyC monomer is probably unable to undergo nuclear translocation, which imparts stability.
Treatment of etiolated seedlings with MG132 delayed light-induced degradation of phyC (Fig. S7), suggesting that the ubiquitin/26S proteasome pathway is involved in phyC degradation steps as observed for Arabidopsis phyA. For light-induced degradation of Arabidopsis phyA, multiple ubiquitins are covalently attached to the phyA Pfr; these ubiquitin-protein conjugates then serve as substrates for the 26S proteasome complex [36]–[38], [49]. Regarding rice phyC, we hypothesize that formation of the phyB/phyC heterodimer is necessary for nuclear translocation to transduce the signal and allow phyC to undergo ubiquitination (Fig. 8). It should also be noted that light irradiation in etiolated seedlings caused concentrations of phyC, but not phyB, to decrease. We speculate that the phyC subunit is a selective target of ubiquitination in the phyB/phyC heterodimer and that after degradation of phyC, the remaining phyB subunits likely form homodimers.
Figure 8. State models of phyC and phyB in rice seedlings.
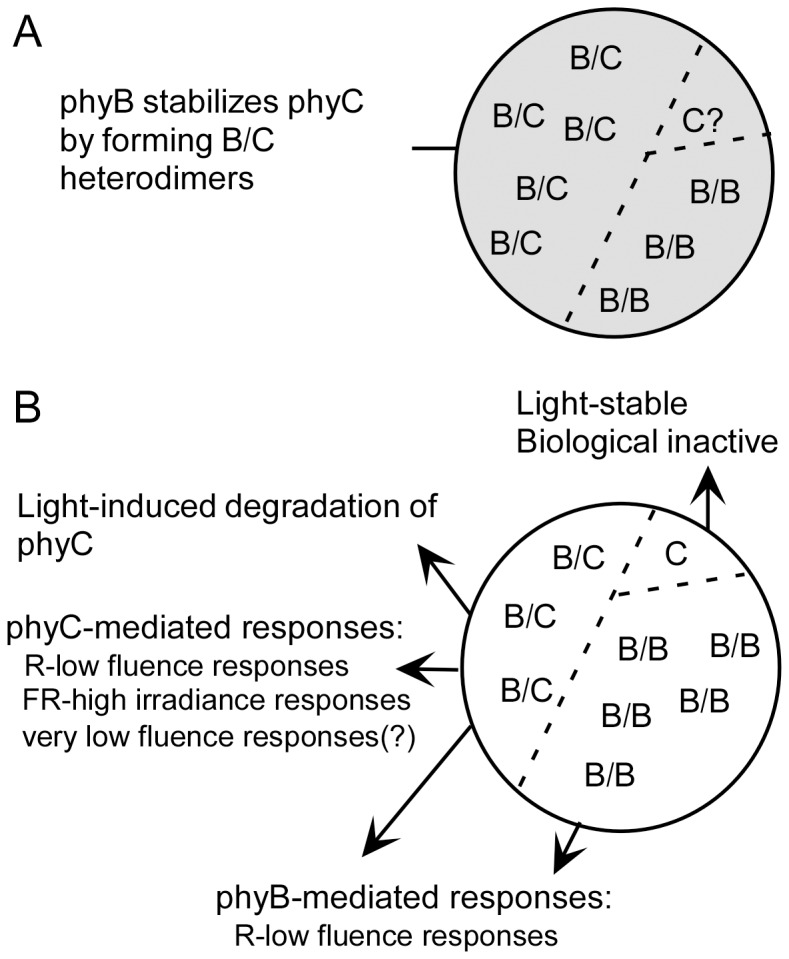
A. In the cells of WT seedlings grown in the dark, most of the phyC exists as phyB/phyC heterodimers (B/C) and, probably, to a smaller degree as phyC monomers (C?), while phyB exist as phyB/phyC heterodimers (B/C) and phyB/phyB homodimers (B/B). In etiolated seedlings, phyB stabilizes phyC in the B/C conformation. Consequently, phyC levels are quite low in the phyB-deficient mutants. B. When seedlings are exposed to light, including W, R, and FR, phyC subunits in phyB/phyC heterodimers (B/C) are light-labile and biologically active to participate in the multiple processes of rice development (inhibition of coleoptile growth, induction of light-regulated genes, and chlorophyll accumulation). By contrast, phyC monomers (C) are light-stable but do not participate in the de-etiolation of rice seedlings. phyB in its heterodimeric (B/C) and homodimeric (B/B) forms probably play different roles in the responses to light.
In summary, our data indicate that phyC predominantly exists as phyB/phyC heterodimers in etiolated seedlings (Fig. 8A). As such, the amount of phyC in phyA phyB mutants is greatly reduced, and the remaining phyC exists in monomeric form and has no function (Fig. 8A). Homodimer formation of phyC has not been observed in Arabidopsis [13], [14]. Therefore, formation of the phyB/phyC heterodimer stabilizes phyC and is indispensable for the light-induced function of phyC in the control of the de-etiolation of rice seedlings and for the light-induced degradation of phyC (Fig. 8B). Physical interaction between phyB and phyC has been conserved in both Arabidopsis and rice, but the function of phyC has been differentiated during evolution. Recent progress in the structure–function relationship of phytochromes, which extensively revealed the function of the various domains of phytochrome molecules in light-dependent signaling [50]–[52], will provide clues for explaining the functional differentiations of phyC in future.
Supporting Information
Quantification of relative phyB and phyC concentrations in the protein extracts of rice seedlings. A. CBB staining of PHYB-His and PHYC-His proteins. The purified PHYB-His and PHYC-His proteins were separated by 12% SDS-PAGE and stained by CBB R-250. The signal intensities were analyzed using NIH image 1.62. The loaded amounts of proteins were 15, 10, 7.5, 5.0, and 2.5 µl for PHYB-His and PHYC-His. B. Immunoblots of phyB and PHYB-His proteins. Protein extracts from 5-day-old etiolated WT seedlings and dilution series of PHYB-His standard proteins were separated by SDS-PAGE in the same gel. PhyB was detected using anti-PHYB antibody. The loaded amounts of proteins were 50, 25, 12.5, and 6.3 µg for detecting phyB proteins in the protein extracts. The loaded amounts of proteins were 13, 6.5, and 3.2 µl of 1000× diluted purified PHYB-His protein for quantifying standard PHYB-His protein. The signal intensities were analyzed using NIH image 1.62. C. Immunoblots of phyC and PHYC-His proteins. Protein extracts from 5-day-old etiolated WT seedlings and dilution series of PHYB-His standard proteins were separated by SDS-PAGE in the same gel. PhyC was detected using anti-PHYC antibody. The loaded amounts of proteins were 50, 25, 12.5, and 6.3 µg for detecting phyC protein in the protein extracts. The loaded amounts of proteins were 13, 6.5, and 3.2 µl of 1000× diluted purified PHYC-His protein for quantifying standard PHYC-His proteins. The signal intensities were analyzed using NIH image 1.62.
(TIF)
Immunoblot analyses of phyB and phyC light-stabilities in rice seedlings. A. Effect of W on phyB and phyC levels in the WT seedlings. The WT seedlings were grown in the dark (D) for 6 days or in the dark for 6 days and then exposed to W for 0.5, 1, 2, 4, 8, 12, or 24 h before harvesting. Protein extracts were prepared from these seedlings. Fifty micrograms of protein extract were loaded for detecting phyB and phyC with anti-PHYB and anti-PHYC antibodies, respectively. Relative signal intensities of protein bands were analyzed using Gel-Pro Analyzer 4.0 software (Media Cybernetics, USA). B. Dilution series of protein extracts from the seedlings grown in the dark (D) were compared with the protein extracts from the seedlings exposed to W for 24 h.
(TIF)
Difference spectra and SEC profiles of recombinant phyA and phyC proteins expressed in E. coli . A. Schematic drawing of a phytochrome molecule and a construction for expression of full length phytochromes (for both phyA and phyC). For the purification, CBP (calmodulin-binding peptide) is attached at N-terminal and 6× His at C-terminal. Native chromophore, phytochromobilin is used. B and C. Absorbance spectra (left) and R/FR difference spectrum (right) of recombinant rice phyA (B) and phyC (C). D. Recombinant proteins expressed in E. coli were fractionated by SEC and phyA and phyC proteins were immunochemically detected with anti-His tag (upper) or anti-phytochrome antibodies (lower) in the individual fractions (#17–#24). Small numbers above the fraction numbers are the molecular sizes which were calculated based on the calibration line of standard proteins.
(TIF)
phyC-GFP is biologically active in phyA phyC backgrounds and inactive in phyB- deficient backgrounds. A. Visual phenotypes of WT, phyA, phyA phyC, and PCG/aacc seedlings (#19-1) grown under D or FR for 8 days. White arrow heads indicate apices of coleoptiles in the seedlings grown under FR. Bar = 10 mm. B. FR-induced expression of Lhcb genes in PCG/aacc transgenic seedlings. WT, phyA, phyA phyC, and PCG/aacc seedlings (#19-1 and #20-1) grown under D or FR for 7 days. Transcript levels of two Lhcb genes (Os03g0592500 and Os09g034650) were analyzed by RT-PCR. Ubiquitin (UBQ) was used as an internal control. C. Visual phenotypes of WT, phyB, phyA phyB, and PCG/Aabb (#7-4-) seedlings grown under D or FR for 8 days. Segregated PCG/aabb genotypes were identified by genotyping PCR. The abundance of phyC-GFP fusion proteins was compared between PCG/Aabb and PCG/aabb mutants by immunoblot analysis. White arrow heads indicate apices of coleoptiles in the PCG transgenic seedlings grown under FR. Bar = 10 mm. D. FR could not induce the expression of Lhcb genes in PCG/aabb transgenic seedlings. WT, phyB, phyA phyB and PCG/aabb seedlings (#23 and #32) grown under D or FR for 7 days. Transcript levels of two Lhcb genes (Os03g0592500 and Os09g034650) were analyzed by RT-PCR. Ubiquitin (UBQ) was used as an internal control. E. Visual phenotypes of WT, phyB, phyA phyB, and PCG/Aabb (#12-5-) and segregated PCG/aabb seedlings grown under D or R for 8 days. White arrow heads indicate apices of coleoptiles in the PCG transgenic seedlings grown under R. Bar = 10 mm. F. R could not induce the expression of Lhcb genes in PCG/aabb transgenic seedlings (#7-4 and #12-5).
(TIF)
A physical interaction exists between phyB and phyC in overexpresser lines of PHYC-GFP , PHYB , and PHYB ( C364A ). A. Co-IP assay of phyC-GFP and phyB in PCG/aacc seedlings. The protein extracts from 7-day-old etiolated seedlings of PCG/aacc (#5, #15, #19, #7, and #8) were immunoprecipitated with anti-PHYC antibody. PhyB and phyC-GFP were detected by immunoblot analyses. Thirty micrograms of protein extracts from PCG/aacc #7 were loaded as the positive control (Ext). B. Co-IP assay of phyC and phyB in PHYB and PHYB(C364A) transgenic seedlings. The protein extracts from 7-day-old etiolated seedlings of PHYB/aabb (#5 and #7) and PHYB(C364A)/aabb (#27 and #62) mutants were immunoprecipitated with anti-PHYC antibody. PhyB and phyC were detected by immunoblot analyses. Thirty micrograms of protein extracts from WT seedlings were loaded as the positive control (Ext).
(TIF)
phyC is biologically active in PHYB and PHYB(C364A) transgenic lines. A. Visual phenotypes of WT, phyA, phyA phyB, and PHYB/aabb (#7-2- and #11-3-) seedlings grown in the dark (D) or under FR (FR) for 8 days. White arrow heads indicate the apices of coleoptiles. Bar = 10 mm. B. FR-induced expression of Lhcb genes in PHYB/aabb and PHYB(C364A)/aabb transgenic seedlings. WT, phyB, phyA phyB, PHYB/aabb (#7-2- and #11-3-), and PHYB(C364A)/aabb (#27-6- and #71-1-) seedlings grown for 7 days. Transcript levels of two Lhcb genes (Os03g0592500 and Os09g034650) were analyzed by RT-PCR. Ubiquitin (UBQ) was used as an internal control. C. PHYB(C364A) transgenic lines and phyB mutants exhibited a pale green phenotype under R. Visual phenotypes of WT, phyB, phyA phyB, two lines of PHYB(C364A)/aabb (#27-6- and #71-2-), and two lines of PHYB/aabb (#7-1- and #11-3-) seedlings grown under R for 7 days. Bar = 10 mm. D and E. Visual phenotypes of WT, phyB, phyA phyB, and PHYB(C364A)/Aabb (#27-6-) seedlings grown under FR (D) or R (E) for 8 days. Mutated and wild-type PHYA alleles are indicated by a and A, respectively. White arrow heads in (D) and (E) indicate the apices of coleoptiles of 8-day old seedlings. Bar = 10 mm. F. R-induced expression of Lhcb genes in the PHYB(C364A)/aabb seedlings. Transcript levels of two Lhcb genes (Os03g0592500 and Os09g034650) were analyzed by RT-PCR in the PHYB(C364A)/aabb seedlings (#27-6- #69-2-, and #71-1-). Ubiquitin (UBQ) was used as an internal control.
(TIF)
Treatment with MG132 delays light-induced degradation of phyA and phyC in rice seedlings. Four-day-old etiolated WT seedlings (D) were treated with 50 µM MG132 (+) or 0.5% DMSO (−) for 1.5 h and then exposed to R for 0 or 6 h before harvesting. Protein extracts (50 µg) from these seedlings were used to detect phyA and phyC with monoclonal anti-rye PHYA (mAR08) and anti-PHYC antibodies, respectively.
(TIF)
Quantification of relative phyB and phyC protein concentrations in rice seedlings. To establish the calibration curves of purified proteins, the purified proteins were separated by SDS-PAGE and stained with CBB R-250. Signal intensities of CBB-stained purified protein bands were analyzed using NIH image 1.62 software for PHYB-His and PHYC-His (Fig. S1A). The graph (A) was produced using the loaded amounts of proteins as X-axes and the signal intensities of CBB-stained PHYB-His or PHYC-His standard proteins as Y-axes. The regression lines were fitted as Y = 61.9X for PHYB-His and Y = 78X for PHYC-His. Therefore, given the loaded amounts of the standard proteins, the corresponding intensities of CBB-stained PHYB-His or PHYC-His were calculated according to the regression lines. For quantifying the relative levels of phyB and phyC proteins, protein extracts from 5-day-old etiolated seedlings and dilution series of standard proteins (PHYB-His or PHYC-His, they were diluted by 1000-fold for immunodetection) were separated by SDS-PAGE in the same gel. phyB and phyC were immunochemically detected with anti-PHYB and anti-PHYC antibodies, respectively. Signals intensities of the representative immunoblot for phyB (or phyC) and PHYB-His (or PHYC-His) were scanned and analyzed using NIH image 1.62 software (Figs. S1B and S1C). Because the signal intensities of immunoblots with different antibodies cannot be compared directly. They were converted into the intensities of CBB staining using the data from standard proteins (PHYB-His or PHYC-His). The graph (B) was plotted with the signal intensities of CBB-stained PHYB-His or PHYC-His proteins as the X-axis and signal intensities of immunoblots as the Y-axis. The regression line was fitted. Thus, the corresponding CBB-stained intensity per immunoblot signal for phyB and phyC in the protein extracts were calculated. Then, the graph (C) was plotted with the loaded amount of protein extracts as the X-axis and the calculated CBB signals as the Y-axis. The linear regression lines of Y = 16.7X and Y = 13.2X were obtained for phyB and phyC proteins, respectively. Therefore, the ratio of phyB:phyC was 1.3∶1 in the dark-grown WT seedlings.
(PDF)
Acknowledgments
We thank Hiroko Hanzawa and Dr. Akira Nagatani for providing monoclonal antibodies against rye phyA and Arabidopsis phyA (AA01), respectively. We also thank Kazuko Yagi and Yumiko Iguchi for their technical assistance.
Funding Statement
This work was supported by grants from the Ministry of Agriculture, Forestry, and Fisheries, the Rice Genome Programs IP1006 and SY1108, and the Genomics for Agricultural Innovation GPN0003. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Neff MM, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14: 257–271. [PubMed] [Google Scholar]
- 2. Quail PH (2002a) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93. [DOI] [PubMed] [Google Scholar]
- 3. Quail PH (2002b) Photosensory perception and signalling in plant cells: new paradigms? Curr Opin Cell Biol 14: 180–188. [DOI] [PubMed] [Google Scholar]
- 4. Franklin KA, Quail PH (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen M, Chory J (2011) Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21: 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smith H (1994) Phytochrome transgenics: functional, ecological and biotechnological applications. Semin Cell Biol 5: 315–325. [DOI] [PubMed] [Google Scholar]
- 7. Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE . Plant Mol Biol 25: 413–427. [DOI] [PubMed] [Google Scholar]
- 8. Mathews S, Sharrock RA (1996) The phytochrome gene family in grasses (Poaceae): a phylogeny and evidence that grasses have a subset of the loci found in dicot angiosperms. Mol Biol Evol 13: 1141–1150. [DOI] [PubMed] [Google Scholar]
- 9. Alba R, Kelmenson PM, Cordonnier-Pratt MM, Pratt LH (2000) The phytochrome gene family in tomato and the rapid differential evolution of this family in angiosperms. Mol Biol Evol 17: 362–373. [DOI] [PubMed] [Google Scholar]
- 10. Sharrock RA, Quail PH (1989) Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev 3: 1745–1757. [DOI] [PubMed] [Google Scholar]
- 11. Jones AM, Quail PH (1986) Quaternary structure of 124-kilodalton phytochrome from Avena sativa L. Biochem. 25: 2987–2995. [Google Scholar]
- 12. Wagner D, Koloszvari M, Quail PH (1996) Two small spatially distinct regions of phytochrome B are required for efficient signaling rates. Plant Cell 8: 859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharrock RA, Clack T (2004) Heterodimerization of type II phytochromes in Arabidopsis . Proc Natl Acad Sci USA 101: 11500–11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clack T, Shokry A, Moffet M, Liu P, Faul M, et al. (2009) Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21: 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kay SA, Keith B, Shinozaki K, Chua NH (1989) The sequence of the rice phytochrome gene. Nucleic Acids Res 17: 2865–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dehesh K, Tepperman J, Christensen AH, Quail PH (1991) phyB is evolutionarily conserved and constitutively expressed in rice seedling shoots. Mol Gen Genet 225: 305–313. [DOI] [PubMed] [Google Scholar]
- 17. Tahir M, Kanegae H, Takano M (1998) Phytochrome C (PHYC) gene in rice: Isolation and characterization of a complete coding sequence. Plant Physiol 118: 1535. [Google Scholar]
- 18. Basu D, Dehesh K, Schneider-Poetsch HJ, Harrington SE, McCouch SR, et al. (2000) Rice PHYC gene: structure, expression, map position and evolution. Plant Mol Biol 44: 27–42. [DOI] [PubMed] [Google Scholar]
- 19. Takano M, Kanegae H, Shinomura T, Miyao A, Hirochika H, et al. (2001) Isolation and characterization of rice phytochrome A mutants. Plant Cell 13: 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, et al. (2005) Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell 17: 3311–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takano M, Inagaki N, Xie X, Kiyota S, Baba-Kasai A, et al. (2009) Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice. Proc Natl Acad Sci USA 106: 14705–14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Osugi A, Itoh H, Ikeda-Kawakatsu K, Takano M, Izawa T (2011) Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice. Plant Physiol 157: 1128–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Monte E, Alonso JM, Ecker JR, Zhang Y, Li X, et al. (2003) Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell 15: 1962–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hirschfeld M, Tepperman JM, Clack T, Quail PH, Sharrock RA (1998) Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics 149: 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Palecanda L, Sharrock RA (2001) Molecular and phenotypic specificity of an antisense PHYB gene in Arabidopsis . Plant Mol Biol 46: 89–97. [DOI] [PubMed] [Google Scholar]
- 26. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 27. Kagawa T, Suetsugu N (2007) Photometrical analysis with photosensory domains of photoreceptors in green algae. FEBS Lett 581: 368–374. [DOI] [PubMed] [Google Scholar]
- 28. Fuse T, Sasaki T, Yano M (2001) Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotech 18: 219–222. [Google Scholar]
- 29. Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A (1999) Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis . J Cell Biol 145: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994. [DOI] [PubMed] [Google Scholar]
- 31. Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2: 208–218. [Google Scholar]
- 32. Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282. [DOI] [PubMed] [Google Scholar]
- 33. Nakazawa M, Yoshida Y, Manabe K (1991) Differences between the surface properties of the PR and PFR forms of native pea phytochrome, and their application to a simplified procedure for purification of the phytochrome. Plant Cell Physiol 32: 1187–1194. [Google Scholar]
- 34. Matsushita T, Mochizuki N, Nagatani A (2003) Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424: 571–574. [DOI] [PubMed] [Google Scholar]
- 35. Eichenberg K, Bäurle I, Paulo N, Sharrock RA, Rüdiger W, et al. (2000) Arabidopsis phytochromes C and E have different spectral characteristics from those of phytochromes A and B. FEBS Lett. 470: 107–112. [DOI] [PubMed] [Google Scholar]
- 36. Clough RC, Vierstra RD (1997) Phytochrome degradation. Plant Cell Environ 20: 713–721. [Google Scholar]
- 37. Clough RC, Jordan-Beebe ET, Lohman KN, Marita JM, Walker JM, et al. (1999) Sequences within both the N- and C-terminal domains of phytochrome A are required for PFR ubiquitination and degradation. Plant J 17: 155–167. [DOI] [PubMed] [Google Scholar]
- 38. Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18: 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lagarias JC, Mercurio FM (1985) Structure function studies on phytochrome. Identification of light-induced conformational changes in 124-kDa Avena phytochrome in vitro . J Biol Chem 260: 2415–2423. [PubMed] [Google Scholar]
- 40. Kiyota S, Xie X, Takano M (2012) Phytochromes A and C cooperatively regulate early and transient gene expression after red-light irradiation in rice seedlings. Plant Physiol Biochem 51: 10–17. [DOI] [PubMed] [Google Scholar]
- 41. Franklin KA, Davis SJ, Stoddart WM, Vierstra RD, Whitelam GC (2003) Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell 15: 1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Andel FR, Lagarias JC, Mathies RA (1996) Resonance raman analysis of chromophore structure in the lumi-R photoproduct of phytochrome. Biochemisty 35: 15997–16008. [DOI] [PubMed] [Google Scholar]
- 43.Kim JI, Song PS (2005) A structure-function model based on inter-domain crosstalks in phytochromes. In: Wada M, Shimazaki K, Iino M, editors. Light Sensing in Plants, Springer-Verlag, Tokyo. pp 57–67.
- 44. Wang H (2005) Signaling mechanisms of higher plant photoreceptors: a structure-function perspective. Curr Top Dev Biol 68: 227–261. [DOI] [PubMed] [Google Scholar]
- 45. Mroginski MA, Murgida DH, Hildebrandt P (2007) The chromophore structural changes during the photocycle of phytochrome: a combined resonance Raman and quantum chemical approach. Acc Chem Res 40: 258–266. [DOI] [PubMed] [Google Scholar]
- 46. Hennig L, Schäfer E (2001) Both subunits of the dimeric plant photoreceptor phytochrome require chromophore for stability of the far-red light-absorbing form. J Biol Chem 276: 7913–7918. [DOI] [PubMed] [Google Scholar]
- 47. Debrieux D, Fankhauser C (2010) Light-induced degradation of phyA is promoted by transfer of the photoreceptor into the nucleus. Plant Mol Biol 73: 687–695. [DOI] [PubMed] [Google Scholar]
- 48. Wolf I, Kircher S, Fejes E, Kozma-Bognár L, Schäfer E, et al. (2011) Light-regulated nuclear import and degradation of Arabidopsis phytochrome-A N-terminal fragments. Plant Cell Physiol 52: 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trupkin SA, Debrieux D, Hiltbrunner A, Fankhauser C, Casal JJ (2007) The serine-rich N-terminal region of Arabidopsis phytochrome A is required for protein stability. Plant Mol Biol 63: 669–678. [DOI] [PubMed] [Google Scholar]
- 50. Rockwell NC, Su YS, Lagarias JC (2006) Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 57: 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nagatani A (2010) Phytochrome: structural basis for its functions. Curr Opin Plant Biol 13: 565–570. [DOI] [PubMed] [Google Scholar]
- 52. Ulijasz AT, Vierstra RD (2011) Phytochrome structure and photochemistry: recent advances toward a complete molecular picture. Curr Opin Plant Biol 14: 498–506. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification of relative phyB and phyC concentrations in the protein extracts of rice seedlings. A. CBB staining of PHYB-His and PHYC-His proteins. The purified PHYB-His and PHYC-His proteins were separated by 12% SDS-PAGE and stained by CBB R-250. The signal intensities were analyzed using NIH image 1.62. The loaded amounts of proteins were 15, 10, 7.5, 5.0, and 2.5 µl for PHYB-His and PHYC-His. B. Immunoblots of phyB and PHYB-His proteins. Protein extracts from 5-day-old etiolated WT seedlings and dilution series of PHYB-His standard proteins were separated by SDS-PAGE in the same gel. PhyB was detected using anti-PHYB antibody. The loaded amounts of proteins were 50, 25, 12.5, and 6.3 µg for detecting phyB proteins in the protein extracts. The loaded amounts of proteins were 13, 6.5, and 3.2 µl of 1000× diluted purified PHYB-His protein for quantifying standard PHYB-His protein. The signal intensities were analyzed using NIH image 1.62. C. Immunoblots of phyC and PHYC-His proteins. Protein extracts from 5-day-old etiolated WT seedlings and dilution series of PHYB-His standard proteins were separated by SDS-PAGE in the same gel. PhyC was detected using anti-PHYC antibody. The loaded amounts of proteins were 50, 25, 12.5, and 6.3 µg for detecting phyC protein in the protein extracts. The loaded amounts of proteins were 13, 6.5, and 3.2 µl of 1000× diluted purified PHYC-His protein for quantifying standard PHYC-His proteins. The signal intensities were analyzed using NIH image 1.62.
(TIF)
Immunoblot analyses of phyB and phyC light-stabilities in rice seedlings. A. Effect of W on phyB and phyC levels in the WT seedlings. The WT seedlings were grown in the dark (D) for 6 days or in the dark for 6 days and then exposed to W for 0.5, 1, 2, 4, 8, 12, or 24 h before harvesting. Protein extracts were prepared from these seedlings. Fifty micrograms of protein extract were loaded for detecting phyB and phyC with anti-PHYB and anti-PHYC antibodies, respectively. Relative signal intensities of protein bands were analyzed using Gel-Pro Analyzer 4.0 software (Media Cybernetics, USA). B. Dilution series of protein extracts from the seedlings grown in the dark (D) were compared with the protein extracts from the seedlings exposed to W for 24 h.
(TIF)
Difference spectra and SEC profiles of recombinant phyA and phyC proteins expressed in E. coli . A. Schematic drawing of a phytochrome molecule and a construction for expression of full length phytochromes (for both phyA and phyC). For the purification, CBP (calmodulin-binding peptide) is attached at N-terminal and 6× His at C-terminal. Native chromophore, phytochromobilin is used. B and C. Absorbance spectra (left) and R/FR difference spectrum (right) of recombinant rice phyA (B) and phyC (C). D. Recombinant proteins expressed in E. coli were fractionated by SEC and phyA and phyC proteins were immunochemically detected with anti-His tag (upper) or anti-phytochrome antibodies (lower) in the individual fractions (#17–#24). Small numbers above the fraction numbers are the molecular sizes which were calculated based on the calibration line of standard proteins.
(TIF)
phyC-GFP is biologically active in phyA phyC backgrounds and inactive in phyB- deficient backgrounds. A. Visual phenotypes of WT, phyA, phyA phyC, and PCG/aacc seedlings (#19-1) grown under D or FR for 8 days. White arrow heads indicate apices of coleoptiles in the seedlings grown under FR. Bar = 10 mm. B. FR-induced expression of Lhcb genes in PCG/aacc transgenic seedlings. WT, phyA, phyA phyC, and PCG/aacc seedlings (#19-1 and #20-1) grown under D or FR for 7 days. Transcript levels of two Lhcb genes (Os03g0592500 and Os09g034650) were analyzed by RT-PCR. Ubiquitin (UBQ) was used as an internal control. C. Visual phenotypes of WT, phyB, phyA phyB, and PCG/Aabb (#7-4-) seedlings grown under D or FR for 8 days. Segregated PCG/aabb genotypes were identified by genotyping PCR. The abundance of phyC-GFP fusion proteins was compared between PCG/Aabb and PCG/aabb mutants by immunoblot analysis. White arrow heads indicate apices of coleoptiles in the PCG transgenic seedlings grown under FR. Bar = 10 mm. D. FR could not induce the expression of Lhcb genes in PCG/aabb transgenic seedlings. WT, phyB, phyA phyB and PCG/aabb seedlings (#23 and #32) grown under D or FR for 7 days. Transcript levels of two Lhcb genes (Os03g0592500 and Os09g034650) were analyzed by RT-PCR. Ubiquitin (UBQ) was used as an internal control. E. Visual phenotypes of WT, phyB, phyA phyB, and PCG/Aabb (#12-5-) and segregated PCG/aabb seedlings grown under D or R for 8 days. White arrow heads indicate apices of coleoptiles in the PCG transgenic seedlings grown under R. Bar = 10 mm. F. R could not induce the expression of Lhcb genes in PCG/aabb transgenic seedlings (#7-4 and #12-5).
(TIF)
A physical interaction exists between phyB and phyC in overexpresser lines of PHYC-GFP , PHYB , and PHYB ( C364A ). A. Co-IP assay of phyC-GFP and phyB in PCG/aacc seedlings. The protein extracts from 7-day-old etiolated seedlings of PCG/aacc (#5, #15, #19, #7, and #8) were immunoprecipitated with anti-PHYC antibody. PhyB and phyC-GFP were detected by immunoblot analyses. Thirty micrograms of protein extracts from PCG/aacc #7 were loaded as the positive control (Ext). B. Co-IP assay of phyC and phyB in PHYB and PHYB(C364A) transgenic seedlings. The protein extracts from 7-day-old etiolated seedlings of PHYB/aabb (#5 and #7) and PHYB(C364A)/aabb (#27 and #62) mutants were immunoprecipitated with anti-PHYC antibody. PhyB and phyC were detected by immunoblot analyses. Thirty micrograms of protein extracts from WT seedlings were loaded as the positive control (Ext).
(TIF)
phyC is biologically active in PHYB and PHYB(C364A) transgenic lines. A. Visual phenotypes of WT, phyA, phyA phyB, and PHYB/aabb (#7-2- and #11-3-) seedlings grown in the dark (D) or under FR (FR) for 8 days. White arrow heads indicate the apices of coleoptiles. Bar = 10 mm. B. FR-induced expression of Lhcb genes in PHYB/aabb and PHYB(C364A)/aabb transgenic seedlings. WT, phyB, phyA phyB, PHYB/aabb (#7-2- and #11-3-), and PHYB(C364A)/aabb (#27-6- and #71-1-) seedlings grown for 7 days. Transcript levels of two Lhcb genes (Os03g0592500 and Os09g034650) were analyzed by RT-PCR. Ubiquitin (UBQ) was used as an internal control. C. PHYB(C364A) transgenic lines and phyB mutants exhibited a pale green phenotype under R. Visual phenotypes of WT, phyB, phyA phyB, two lines of PHYB(C364A)/aabb (#27-6- and #71-2-), and two lines of PHYB/aabb (#7-1- and #11-3-) seedlings grown under R for 7 days. Bar = 10 mm. D and E. Visual phenotypes of WT, phyB, phyA phyB, and PHYB(C364A)/Aabb (#27-6-) seedlings grown under FR (D) or R (E) for 8 days. Mutated and wild-type PHYA alleles are indicated by a and A, respectively. White arrow heads in (D) and (E) indicate the apices of coleoptiles of 8-day old seedlings. Bar = 10 mm. F. R-induced expression of Lhcb genes in the PHYB(C364A)/aabb seedlings. Transcript levels of two Lhcb genes (Os03g0592500 and Os09g034650) were analyzed by RT-PCR in the PHYB(C364A)/aabb seedlings (#27-6- #69-2-, and #71-1-). Ubiquitin (UBQ) was used as an internal control.
(TIF)
Treatment with MG132 delays light-induced degradation of phyA and phyC in rice seedlings. Four-day-old etiolated WT seedlings (D) were treated with 50 µM MG132 (+) or 0.5% DMSO (−) for 1.5 h and then exposed to R for 0 or 6 h before harvesting. Protein extracts (50 µg) from these seedlings were used to detect phyA and phyC with monoclonal anti-rye PHYA (mAR08) and anti-PHYC antibodies, respectively.
(TIF)
Quantification of relative phyB and phyC protein concentrations in rice seedlings. To establish the calibration curves of purified proteins, the purified proteins were separated by SDS-PAGE and stained with CBB R-250. Signal intensities of CBB-stained purified protein bands were analyzed using NIH image 1.62 software for PHYB-His and PHYC-His (Fig. S1A). The graph (A) was produced using the loaded amounts of proteins as X-axes and the signal intensities of CBB-stained PHYB-His or PHYC-His standard proteins as Y-axes. The regression lines were fitted as Y = 61.9X for PHYB-His and Y = 78X for PHYC-His. Therefore, given the loaded amounts of the standard proteins, the corresponding intensities of CBB-stained PHYB-His or PHYC-His were calculated according to the regression lines. For quantifying the relative levels of phyB and phyC proteins, protein extracts from 5-day-old etiolated seedlings and dilution series of standard proteins (PHYB-His or PHYC-His, they were diluted by 1000-fold for immunodetection) were separated by SDS-PAGE in the same gel. phyB and phyC were immunochemically detected with anti-PHYB and anti-PHYC antibodies, respectively. Signals intensities of the representative immunoblot for phyB (or phyC) and PHYB-His (or PHYC-His) were scanned and analyzed using NIH image 1.62 software (Figs. S1B and S1C). Because the signal intensities of immunoblots with different antibodies cannot be compared directly. They were converted into the intensities of CBB staining using the data from standard proteins (PHYB-His or PHYC-His). The graph (B) was plotted with the signal intensities of CBB-stained PHYB-His or PHYC-His proteins as the X-axis and signal intensities of immunoblots as the Y-axis. The regression line was fitted. Thus, the corresponding CBB-stained intensity per immunoblot signal for phyB and phyC in the protein extracts were calculated. Then, the graph (C) was plotted with the loaded amount of protein extracts as the X-axis and the calculated CBB signals as the Y-axis. The linear regression lines of Y = 16.7X and Y = 13.2X were obtained for phyB and phyC proteins, respectively. Therefore, the ratio of phyB:phyC was 1.3∶1 in the dark-grown WT seedlings.
(PDF)