Abstract
Nutritional metabolomics is rapidly maturing to use small molecule chemical profiling to support integration of diet and nutrition in complex biosystems research. These developments are critical to facilitate transition of nutritional sciences from population-based to individual-based criteria for nutritional research, assessment and management. This review addresses progress in making these approaches manageable for nutrition research. Important concept developments concerning the exposome, predictive health and complex pathobiology, serve to emphasize the central role of diet and nutrition in integrated biosystems models of health and disease. Improved analytic tools and databases for targeted and non-targeted metabolic profiling, along with bioinformatics, pathway mapping and computational modeling, are now used for nutrition research on diet, metabolism, microbiome and health associations. These new developments enable metabolome-wide association studies (MWAS) and provide a foundation for nutritional metabolomics, along with genomics, epigenomics and health phenotyping, to support integrated models required for personalized diet and nutrition forecasting.
Keywords: Systems biology, microbiome, prevention, mass spectrometry, personalized medicine, exposome
INTRODUCTION
Each person has a unique genome and is an ongoing nutritional experiment from conception to death. A large number of functional redundancies and adaptive mechanisms serve to provide homeostasis despite a considerable range of developmental, dietary, infectious and other environmental challenges. Differences in nutrient intake cause epigenomic changes (9; 56) so that an n = 1 nutritional experiment may not be reproducible even within the context of an individual over time. Application of scientific method to define nutritional requirements relevant to an individual therefore requires substantial simplification and many assumptions. Despite this, modern nutrition has provided a manageable system for nutrition scientists to understand the mechanistic basis for requirements and clinical nutritionists and dietitians to develop dietary and nutritional plans to minimize and remediate nutrition-related disease. This was achieved using a central assumption that health and disease among populations and in individuals may be determined by variations of the same biochemical system with common biochemical requirements. To a first approximation, this assumption is true; normative descriptions, such as Dietary Reference Intake (DRI) values, accurately describe the collective needs of individuals; DRI along with Food Guides are useful to promote health and reduce disease collectively for individuals within the population. From the standpoint of public health, this approach is effective and successful.
During the past decade, many distinguished scientists have emphasized (26–29; 78; 97; 100; 106) that new technologies in metabolomics, along with genomics, epigenomics, transcriptomics and proteomics, enable approaches that have the potential to expand this effective public health strategy with a potentially improved approach for personalized nutrition, i.e., to address nutrition at n = 1. The full implications of this are only beginning to be discussed; the transition may not be incremental improvements of current nutritional science, such as having better ways to study pathways of iron metabolism or more precise ways to measure phytochemicals in green tea. Instead, personalized nutrition in the future may require a considerable expansion of the conceptual framework for nutrition science that has emerged over the past century. Diet and nutrition are inherently complex: hundreds of foods are derived from heterogeneous plant - and animal-based nutrients and nutrient substrates; food storage, processing and preparation methods vary; consumption, digestion and absorption are heterogeneous over the lifespan within individuals; specific nutrients, such as zinc or vitamin D, impact hundreds of molecular systems; molecular systems are highly regulated and adaptive. To date, the framework in nutrition science has been largely reductionist in nature (e.g. by focusing on deviations from the norm, one nutrient at a time). Examples include single nutrition deficiencies or excesses, and single gene variants (with variable penetrance), that are useful to predict clinical responses in homogeneous groups of individuals. However, emerging technologies, including metabolomics approaches now allow investigation on the complexity of interactions of all of the nutrients within a complex individual having a unique genome and history of dietary, environmental and behavioral exposures.
In this review, we define nutritional metabolomics as “use of small molecule chemical profiling to integrate diet and nutrition in complex biosystems”. This explicitly defines nutritional metabolomics as an experimental approach that uses chemical profiling in a global manner, i.e., as a component of a complex systems approach to diet and health. With this definition, profiling of chemicals, whether targeted or non-targeted, is necessary but insufficient for the transformation to personalized nutrition. Progress in nutritional metabolomics is measured by steps to support facile use of chemical profiles to enhance practice at the level of an individual. For instance, a manageable system for an average practitioner may be a series of nutrition forecasting models, much like weather forecasting systems used to predict occurrence, paths, time-course and severity of hurricanes. Such nutritional models would use metabolic profiles along with genomic, epigenomic and health phenotyping to predict health outcomes and relevant time frames from dietary and nutritional practices. The practitioner of the future will evaluate results of computer-based models and develop interventional plans based upon these results.
Many reviews and commentaries address the challenges and limitations in application of metabolomics to nutrition, diet and health (26–29; 78; 97; 100; 106). Much of this has been focused on use of nutritional metabolomics to discover new biomarkers of nutritional exposure, nutritional status and nutritional impacts on disease (75). The present review is focused on recent progress in making nutritional metabolomics tractable for nutritional scientists to transform nutritional evaluation and management from a normative, population-based approach to one utilizing individual characteristics in an integrated, complex systems approach. Progress is ongoing in many scientific disciplines that are peripheral to nutrition but central to integrated systems biology (88; 90). By necessity, we only briefly summarized these, focusing on conceptual developments in integrative biology, analytic methods that support hybrid approaches for targeted (e.g. in identification of potential direct or surrogate biomarkers of health, disease and mechanistic pathways) and non-targeted metabolic profiling, and bioinformatic and computational approaches to facilitate interpretation and use of complex data. While routine use of such approaches in applied nutrition must await development and testing prior to implementation, an extensive array of metabolic profiling and computational capabilities are now available to support basic research to enable this transition. Progress in these areas suggests that nutritional metabolomics has advanced toward establishing a critical foundation to define the cumulative dietary and nutritional exposures of an individual impacting personal health.
CONCEPTUAL DEVELOPMENTS
Three important conceptual developments have occurred during recent years that impact the transition from targeted nutritional biochemical studies describing population averages to nutritional metabolomics studies describing personalized nutritional needs. These include the concept of 1) the exposome, in which cumulative exposures throughout life are incorporated into models of health (99); 2) predictive health, in which nutritional guidance to prevent disease is replaced by nutrition designed to optimize vitality and well-being (60); and 3) individual complexity in which models include multiple interacting functional networks rather than unrealistic “reductionist” cause-effect models (17).
The exposome and nutritional metabolomics
Christopher Wild introduced a bold and visionary concept in 2005 advocating the need for research to define exposures that complement the genome in defining health risks (99). He used the term “exposome” to encompass life-course environmental exposures (including lifestyle factors), from the prenatal period onwards, and proposed the development of a conceptual grid for exposure research to complement the Human Genome Project. While relevant for environmental science (74), this also provides a basis for systematic integration of life-cycle research. Varying nutritional needs during the life cycle are central to nutritional sciences and provide a foundation for public health policy (59; 69). Thus, application of nutritional metabolomics to improved life-cycle nutrition research can yield a central foundation to operationalize the conceptual grid of the exposome (Figure 1).
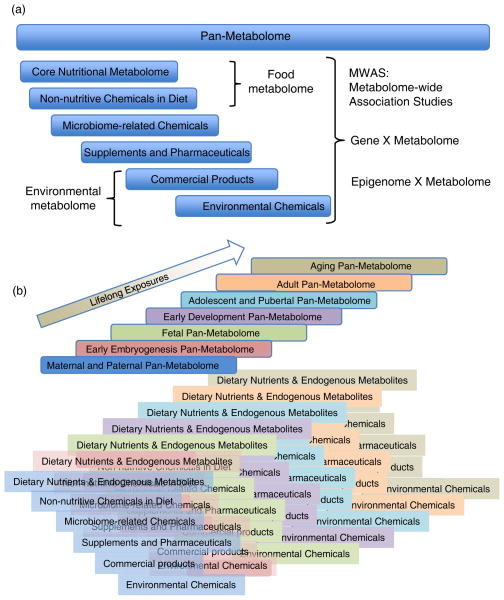
Figure 1. The nutritional metabolome as a component of the exposome.
A. The summation of all chemicals found in an organism can be considered a “pan-metabolome”. Although no method is available to measure all chemicals, the pan-metabolome can be conceptualized to contain a core nutritional metabolome derived from required nutrients and related biochemicals derived from these nutrients in reactions catalyzed by enzymes encoded in the organism. The food metabolome contains many components of this core nutritional metabolome and also a large number of other non-nutritive chemicals. The pan-metabolome also contains microbiome-related chemicals derived from food metabolites, drugs and other environmental agents acted upon by the intestinal microbes. Other components of the pan-metabolome are derived from dietary supplements and pharmaceuticals, commercial products such as sun screen and face creams, and environmental chemicals. B. The exposome is defined as the cumulative exposures from conception onwards. Life cycle nutritional requirements can be viewed within the conceptual grid of the exposome, including the core nutritional metabolome and food metabolome as in Panel A. Together, Panels A and B provide a conceptual grid for the exposome. Consequently, nutritional metabolomics represents a central and critical component of exposome research, impacting expression of the genome and modification of the epigenome through the lifecycle.
Food metabolome
In the exposome, diet is probably the greatest single source of chemical exposures, including nutrients, non-nutritive chemicals, pesticides and others. About 40 required nutrients are consumed daily in variable amounts by individuals. Among these, the organic chemicals are converted through intermediary metabolism to more than 1500 chemicals, which are now included in curated databases such as the KEGG human metabolic pathways (Kyoto Encyclopedia of Genomes and Genomics) (48; 49), Madison Metabolomics Consortium Database (MMCD) (60) and Metlin database (81). Lipidomics research shows that the full spectrum of endogenous metabolites is much larger, perhaps hundreds of thousands of chemical species (39). These nutrients and related products can be considered operationally as a core nutritional metabolome, which contributes to the pan-metabolome (Fig 1A). Systematic study of the effects of under-nutrition and over-nutrition for each of the required nutrients on the core nutritional metabolome under controlled conditions is feasible with current technologies. Such studies are central to a conceptual grid for the exposome created by combining exposures of the life cycle (Fig 1B) with the exposures contributing to the pan-metabolome (Fig 1A).
Most chemicals in food are not nutrients (Fig 1A). Some phytochemicals are biologically active and important to health, such as inducers of detoxification systems that protect against cancer-causing reactive electrophiles (52) and antioxidants that protect against damage from free radical reactions (24). However, these phytochemicals are highly variable within individual food sources and thus in the diet, and the complexity of diet and overlapping biologic activities of chemicals has prevented clear understanding of their importance, as evidenced by the failure of interventional trials based upon extensive experimental and observational research (34; 46). Goodacre et al (33) estimated that there are more than 200,000 metabolites within the plant kingdom, and new web-based tools to facilitate research on plant metabolomics have become available (3; 67). Advances relevant to nutritional metabolomics have been made in food and agricultural sciences (16; 37). Even though the primary purposes are not nutrition, per se, the use of chemical profiling as a basis to characterize plants and plant-derived products is of great importance to support detailed nutritional intervention and epidemiology studies. For instance, characterization of chemicals in grapes and products such as wine provide an important subset of dietary chemicals that are relevant to diet and health (25; 76). Common and abundant phytochemicals are already included in human metabolomics databases, such as KEGG, Metlin and MMCD, and are readily accessible to nutrition scientists. Other chemicals are included in more specialized resources (23; 30; 53), some with open access and others that are proprietary. The task of learning the tools and their respective strengths and weaknesses for nutrition research remains challenging because food and nutrition are inherently complex and there are many databases. Critical challenges remain to link crop metabolomics (80) with food metabolomics (food storage, processing and cooking) (11; 100) and human nutrition. Considerable scientific information is available concerning loss of nutrient content and formation of chemical carcinogens, but manual curation of such information into readily accessible databases is costly and time-consuming. Additionally, characterization of chemicals formed during storage and handling is mostly limited to highly toxic metabolites and degradation products. Thus, an alternative approach would be to use the advances in analytic methods for nutritional metabolomics to create an integrated pipeline for this complex subject, fostering the critical links between plant science, food science and human nutrition needed for complex systems models of diet and health (78).
Microbiome-related metabolome
Important advances have come from the recognition of the contribution of the enteric microbiome to the mammalian metabolome (Fig 1A). Martin et al (63) used a top-down metabolomics approach, i.e., one examining all chemicals detected rather than only a targeted subset, to study the interaction of symbiotic gut microorganisms with mouse metabolism. Comparisons of germ-free mice colonized by a human baby flora or a normal flora to conventional mice showed a simple microbiome/metabolome correlation network, impacting directly on the host’s ability to metabolize lipids. They concluded that the microbiome modulates absorption, storage and energy harvest from the diet at the systems level. Association between early nutrition, the microbiota and the immune system are being actively studied as contributors to obesity and chronic disease (19; 65). Wang et al (94) used liquid chromatography-mass spectrometry and a combination of human and mouse studies to generate unbiased small-molecule metabolic profiles of plasma. They found that three microbiome-dependent metabolites of diet-derived phosphatidylcholine [choline, trimethylamine N-oxide (TMAO) and betaine] predict risk for cardiovascular disease. Wikoff et al (98) used a mass spectrometry approach to study the effect of the intestinal microbiome on plasma metabolites in germ-free mice compared to conventional mice. Of the chemicals found in both, about 10% significantly differed between the mice, and hundreds of chemicals were only detected in one group. One can infer that greater than 10% of the plasma metabolome is directly dependent upon the microbiome. Specific studies showed that bacteria produced indole-containing metabolites derived from tryptophan such as indoxyl sulfate and indole-3-propionic acid (98). Together, these results illustrate significant interplay between bacterial and mammalian metabolism both at the level of macronutrition and also at the level of specific metabolic pathways linked to disease.
Environmental metabolome
The food metabolome also contains herbicides, insecticides, fungicides and other chemicals of interest for environmental health (Fig 1A). Although not a central component of nutritional metabolomics research, per se, these are relevant because environmental chemicals are present to variable extents in all metabolomics studies, and nutrient-chemical interactions could represent a common determinant of environmental health. High-performance metabolic profiling of human plasma revealed over 100 chemicals apparently of environmental origin (83). Such environmental chemicals, as well as other chemicals derived from commercial products (face creams, soaps, disinfectants, flame retardants, etc) and contaminants in the drinking water (antibiotics, fertilizers, drugs, etc) can potentially interact with nutrients to impact health. Environmental contaminants are present in all of the solvents and materials used for collection and analysis of samples, and precautions and controls for proper interpretation have been developed (1; 51). Thus, progress in environmental metabolomics provides information critical to the elaboration and development of the exposome and, ultimately, to effectively utilize nutritional metabolomics for study of complex interactions that impact personalized nutrition.
Predictive health and healthcare economics
Nutritional and dietary recommendations are focused on disease prevention and management, codified as a set of reference values, DRI, defined as the amounts of essential nutrients considered sufficient to meet the physiologic needs of practically all healthy persons in a specified group and the average amount of food sources of energy needed by the members of the group (58). While maintaining a focus on healthy people, a shift has occurred in recent years to include recommendations concerning non-nutritive food components, such as fiber, and also to include recommendations for overweight people to promote health (87). Although gradual, this shift from a focus on sufficient nutrition to avoid disease to a focus on nutrition to optimize overall health and function is fundamental. It redirects nutrition sciences from individual nutrients to eliminate specific disease symptoms to optimized balances for healthy physical, mental and emotional development. This advanced concept sets the stage for evolution of complex systems models for personalized nutrition.
Concept development was advanced in a commentary on predictive health and personalized medicine (89), in which the authors emphasized that establishing “personalized health profiles” will be a challenging task that is dependent upon integrated systems approaches. They suggest that the required shift in thinking by scientists and practitioners will be a greater challenge than to create mathematical models of complex diseases. They predict, however, that this shift will ultimately occur because of the cost of the current model of healthcare. Progress in conceptualizing this shift for nutritional sciences is summarized in Figure 2, based upon this vision for the transition of medicine from a disease-oriented model to a health-oriented model.
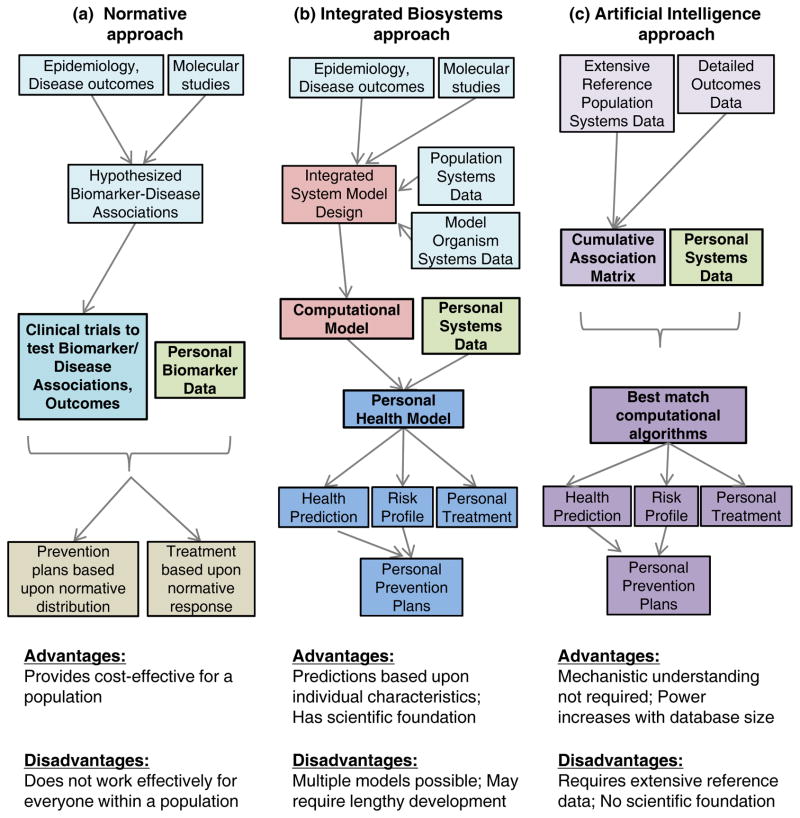
Figure 2. Nutritional metabolomics to support personalized nutrition.
A. Contemporary nutritional recommendations and interventions use a normative approach based upon the characteristics of a healthy population. Hypotheses are based upon experimental and epidemiologic studies, and tested in clinical trials to determine outcomes in individuals meeting certain phenotypic or nutritional criteria. Importantly, the criteria are based upon population-based norms. A person with a predefined deviation from the norm is prescribed an intervention based upon clinical trials which show that this intervention has a significant beneficial effect in at least some of the individuals in the trial. While cost-effective for the population, the approach does not work for all individuals. B. An integrated biosystems approach utilizes the hypotheses of Panel A along with information-rich nutritional metabolomics (systems data) for humans and model organisms to develop computational models. The computational models are tested and refined to correctly describe responses to differences in diet, genetics or other factors. The computational models are used with nutritional metabolomics data for an individual to provide personal health models for health prediction, risk profiling and treatments. The approach takes advantage of the knowledge base as in Panel A but refines this for personalized use. C. An artificial intelligence approach can take advantage of nutritional metabolomics (systems data) as in Panel B but does not require mechanistic development. In this case, artificial intelligence approaches are used to compare personal profiles to profiles and outcomes within a reference population to obtain the best matches for prediction. This has advantages that there is no delay in building models and the power increases with the size of the reference population. On the other hand, due to the correlative nature of these statistical models, there is has no scientific foundation to facilitate development of new interventional strategies. Based upon Voit and Brigham (89).
Normative approach
In this vision, contemporary nutrition uses a normative approach (Fig 2A) in which average characteristics in populations and model systems support development of hypotheses, which are then tested in human studies to determine average response of a selected population to an intervention. Prevention and treatment plans then use this treatment for an individual who meets the selection criteria, i.e., deviates from “normal”, with the expectation that the treatment that showed significant benefit in the test population will be beneficial in the individual.
Integrated biosystems approach
An integrated biosystems approach (Fig 2B) also uses information from population and molecular studies as above, but this information is used as a basis for design of integrated systems models that predict behavior (89). Computational models based upon these integrated designs can then be used to evaluate effects of varying single or multiple elements within the model, as is needed to predict outcome for an individual with a unique set of characteristics. As models are refined and validated with inclusion of information-rich data, such as nutritional metabolomics, they can be used for personalized health prediction, risk evaluation and treatment. This is advantageous in the transition from an average (population) model because it can be individualized as much as the data will allow.
Implementation of integrated systems models in healthcare will face many regulatory hurdles, and the cost of surmounting these barriers limits incremental improvements. Thus, even though gradual improvement would appear possible, substantially improved personalized models are needed to warrant the cost of validation studies. The development of highly precise personalized models is is not likely to be straightforward, and there is a possibility that the interactions of the genome, epigenome, diet and health behaviors, are so complex that useful integrated systems models will not become available for decades.
Artificial intelligence approach
An alternative (Fig 2C) is to use systems data and associated detailed health outcomes data with artificial intelligence. For this, data for a large reference population would be assembled into a cumulative data matrix so that data for an individual can be queried against the matrix to obtain the best matches to directly predict outcomes. Although less attractive from a scientific point of view, this may be more effective in rapidly providing personal predictions for human nutrition because of the extensive variability of human diet, individual genetics, etc. An advantage of this approach is that the power of prediction would increase with the number of individuals in the database and the period of time over which data are collected. Progress with virtual clinical data warehouses (14; 96) and information systems (36; 105) provide capabilities to use metabolomics data to support this approach to personalized nutrition. Although there are many practical issues concerning policy and implementation of such new approaches, the conceptual development has advanced to enable nutrition scientists to effectively use nutritional metabolomics toward the long-term goal of improved individual nutritional assessment, health prediction and therapeutic intervention.
Nutritional metabolomics for complex biosystems research
A third conceptual advance lies in the transition from simple cause-effect models of disease to ones acknowledging that in complex, adaptive systems, a single cause can have multiple effects, and multiple causes can have a single effect. Loscalzo et al (57) addressed the challenging transition from contemporary medical classification of human disease, derived from conventional reductionism, to one that incorporates a non-reductionist approach to systems biomedicine. Contemporary nutrition was derived in a manner similar to contemporary medicine, based upon observed associations between nutrient intake and clinical syndromes, with supportive mechanistic studies to link nutrients to phenotypes. In practice, effectiveness of contemporary nutrition depends upon observational skills of a nutritionist and relatively simple assessment and intervention tools. Although effective, the approach has limitations in sensitivity to detect early or subclinical nutritional insufficiencies, accuracy for different clinical presentations and usefulness in therapeutic interventions for complex phenotypes.
A greater limitation to contemporary approaches, however, is that rational interpretations based upon a system that excessively relies upon reductionism can be mostly or completely wrong. Loscalzo et al (57) used failure of secondary interventional trials with folate to reduce risk of atherothrombotic events with vascular disease and hyperhomocysteinemia to illustrate the failure of this approach. They used sickle cell disease, in which a single DNA mutation can have multiple disease phenotypes, as a second example (57). Other examples are common: cardiovascular disease has multiple nutrition-related components, including high LDL, low HDL, uncontrolled hypertension, physical inactivity, smoking, obesity, and uncontrolled diabetes, but none of these is perfect in prediction; phenylketonuria has multiple phenotypic characteristics that are not uniformly expressed; many diseases have genetic and non-genetic bases. Nutritional metabolomics studies reinforce the conclusion that average population characteristics are not good descriptors of individual characteristics. In a diurnal variation study, an average variation in the plasma metabolic profile was identified that discriminated morning, evening and nighttime metabolic patterns (71). No individual had the average response pattern, i.e., even though an average response was described, this average was not a good descriptor of an individual. A crossover study of sulfur amino acid insufficiency similarly showed that while about half of the individuals showed a common response pattern, no individual had an average response pattern (72). Thus, nutritional metabolomic data underscore the need to transition to complex biosystems approaches to transform nutrition to a personalized level.
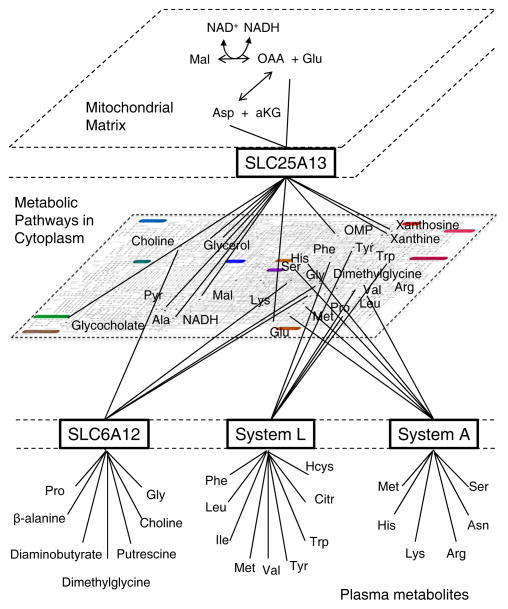
A study by Deo et al (15) illustrates the important contribution of nutritional metabolomics toward the goal of complex biosystems models. Nutritional science with an n=1 is practiced daily with conduct of glucose tolerance tests. Insulin affects many processes in addition to blood glucose levels, and glucose availability impacts many aspects of metabolism in different body compartments. Deo et al (15) showed that considerable additional information about an individual could be obtained by applying nutritional metabolomics methods in association with a challenge test. They analyzed results of targeted metabolic profiling in association with an oral glucose tolerance test and found few chemicals corresponding to those expected from known metabolic pathway metabolites in comparison of normal glucose tolerance and impaired glucose tolerance. However, using unbiased approaches with bioinformatics tools, active modules of metabolites were identified that were naturally grouped according to System A and System L amino acid transporters and the osmolyte transporter SLC6A12 (15). Additional contribution by the mitochondrial glutamate-aspartate tranporter SLC25A13 was indicated by changes associated with pyrimidine biosynthesis (OMP, ribose-1-phosphate), purine biosynthesis (ribose-1-phosphate, xanthine, hypoxanthine, xanthosine), triglyceride biosynthesis (glycerol, glycerol-3-phosphate), urea cycle (citrulline, ornithine), bile salt accumulation (taurochenodeoxycholate, glycocholate, glycochenodeoxycholate), glycolysis (lactate, pyruvate), gluconeogenesis (ala, ser), malate shuttling (glutamate, malate, alpha-ketoglutarate) and aspartate biosynthesis (asparagine). The data illustrate an important conceptual advance in nutritional metabolomics, namely, transitioning from a mono-dimensional view of metabolism involving only substrate clearance or enzymatic conversions to a multi-dimensional view in which biochemical reactions occur in different subcellular compartments linked by transport systems (Figure 3). In such multidimensional models, movement between compartments can compensate for changes within compartments. Because volumes, concentrations and enzyme contents of compartments differ, response of a complex system is often not a simple function. Similarly, differences in gene copy number, alleles, gene expression, epigenomic regulation, microRNAs, cell populations and a number of other mechanisms contribute to the multidimensional character of complex systems. Hence, the use of nutritional metabolomics in integrated biosystems research represents a critical advance in addressing complex issues of diet and health.
Figure 3. Development of multidimensional models for nutritional metabolomics.
Recent use of unbiased pathway models (15) have revealed an important multidimensional character to nutritional metabolomics. Differences in plasma metabolite profiles in individuals with impaired glucose tolerance and normal glucose tolerance were associated with transporters functioning in mitochondrial/cytoplasmic balance of NAD+ and NADH (SLC25A13), and with cell membrane transporters involved in amino acid transport (System A and System L) and osmotic regulation (SLC6A12). In this schematic representation based upon the KEGG human metabolic pathway map, the plasma metabolome (bottom) is linked to KEGG biochemical pathways in the cytoplasm (middle) through System A, System L and SLC6A12, and a subset of metabolites in the cytoplasm is linked to the mitochondrial matrix though the glutamate-aspartate transporter (SCL25A13). Based upon findings of Deo et al (15). Some metabolites are not labeled due to lack of space.
PROFILING METABOLITES FOR NUTRITIONAL METABOLOMICS
The small-molecular weight metabolites within an organism cannot all be measured due to the practical limit of sensitivity, i.e., detection methods are inadequate to measure all individual small molecules. Human metabolic databases include approximately 2,500 metabolic intermediates, hormones and other signaling molecules, over 1000 drugs and over 3500 food components (60; 81; 101). Many analytic methods are available to acquire extensive metabolic information and the strengths and weaknesses of these have been recently reviewed (10; 40). No single method has achieved a level of standardization or widespread use to warrant consideration as a uniform platform for nutritional metabolomics. Although thousands of chemicals can be measured by current technologies, it is humbling to recognize that today’s analytic platforms for targeted measurement of biochemicals cannot quantify more than 10-times the number of chemicals measured by Moore and Stein half a century ago using automated amino acid analysis (64). Nutrition research favors rigorous analytic procedures in which individual analytes are quantified in absolute amounts relative to authentic standards. In amino acid analysis, the amine moiety allowed chemical modification and quantification in terms of an added molecular tag. Because metabolites differ considerably in physical and chemical properties, this approach is inherently limited to capture the entire metabolome. Consequently, a central challenge remaining for nutritional metabolomics is the development of comprehensive profiling capabilities.
Mass spectrometry
All molecules have mass, and advanced mass detectors in mass spectrometry (MS) currently provide the best methods to detect the broad range of chemicals. MS detects chemicals as ions in the gas phase, and this presents practical limitations to obtain absolute quantification of a spectrum of chemicals. Chemicals differ considerably in the conditions required for ionization in the gas phase. The energy required to create ions can result in reactions that convert metabolites to different chemical species. Furthermore, a single chemical can form multiple ionic forms, and relative amounts of these forms can vary due to the presence of other chemicals. These limitations are controlled by standardized separation techniques (gas chromatography, liquid chromatography, capillary electrophoresis), which simplify the mixture of chemicals introduced into the mass spectrometer, and ionization methods (electrospray ionization, ESI; atmospheric pressure chemical ionization, APCI; desorption electrospray ionization, DESI), which ionize a broad range of chemicals (78). Currently, targeted analysis of more than known 300 metabolites in biological samples is widely available with reliable relative quantification within sample sets. Gas and liquid chromatography separations are most commonly used, with ion dissociation using tandem mass spectrometry (MS/MS) to facilitate chemical identification. Ongoing improvements in MS/MS databases (60) (National Institute of Standards http://chemdata.nist.gov/mass-spc/amdis) continue to enhance capabilities, and advances in ultra-high pressure liquid chromatography support separation of thousands of chemicals in relatively short analysis times (95). Absolute quantification by mass spectrometry requires standardization relative to authentic standards and is often readily achieved only for small numbers of metabolites. The limited ability to obtain absolute quantification of large numbers (>2000) of metabolites remains a major obstacle for nutritional metabolomics.
An important practical advance that makes metabolic profiling tractable for nutritional sciences is the availability of the mass spectral analyses in academic cores and commercially (e.g., Metabolon, Research Triangle Park, NC). Although absolute quantification is not achieved, relative quantification is sound for samples collected, stored and analyzed together in sample sets. If one uses the 2500 metabolic intermediates in the Human Metabolome Database (101) as a reference, analytic methods are widely available to gain information on 10% of the metabolites of intermediary metabolism for nutrition studies.
High-resolution mass spectrometry
High-resolution mass spectrometry (62) has been used to facilitate measurement of large numbers of chemicals based upon mass resolution and mass accuracy (Figure 4). These characteristics allow prediction of elemental composition of a chemical using the accurate mass/charge (m/z) values; >90% of the metabolites in the MMCD or KEGG human metabolite databases have unique elemental compositions so that m/z matching to the databases provides a fairly effective approach to map metabolism. Using this approach, a single 10-min analysis by LC-FTMS (83) with data extraction by apLCMS (104) detects about 4000 m/z, which include matches to about 25% of the metabolites in KEGG human metabolic pathways (Figure 5). Targeted quantification can be obtained by stable isotope dilution (44; 45) and by concurrent MS/MS analysis, matched back to authentic standards and MS/MS databases. Greater coverage (about 7000 m/z) is obtained with a dual-chromatography-FTMS approach (83), and data extraction by apLCMS with multiple parameter settings and data merger can further increase the number of m/z features detected in a single sample. Importantly, the coverage can, in principle, be increased to detect over 10,000 chemicals, including representative chemicals of each of the components of the pan-metabolome. Such coverage enables studies similar to genome-wide association studies, in which abundance of individual chemicals are tested for association with health phenotypes. These metabolome-wide association studies (MWAS), will be very important to link dietary chemicals with disease. Related analyses can support metabolome-genome association and metabolome-epigenome association studies. Improved classification of the pan-metabolome as outlined in Figure 1 will further allow more specific analyses of the nutritional metabolome and food metabolome.
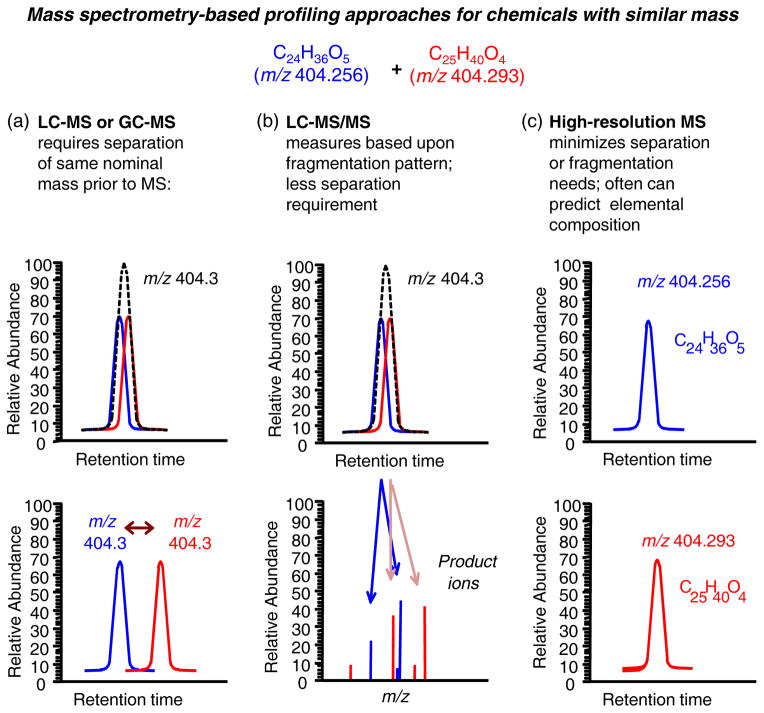
Figure 4. Comparison of mass spectrometry (MS) based metabolic profiling approaches for chemicals have similar but not identical mass.
A. Analysis with gas chromatography (GC) or liquid chromatography (LC) with a single low resolution mass detector requires separation of chemicals prior to detection. B. Analysis with a tandem mass spectrometer using either GC or LC often does not require complete separation because ion dissociation and detection of product ions supports identification without separation. However, quantification typically requires a stable isotopic form of chemicals of interest for internal standardization. C. LC coupled to high-resolution mass spectrometry supports high throughput analysis because chemicals are resolved by mass and have less demand for chromatographic separation. High resolution instruments include Fourier-transform ion cyclotron resonance, Orbitrap (Thermo) and newer time-of-flight (TOF) instruments (62).
Figure 5. Intermediary metabolites detected in 10-min liquid chromatography-Fourier transform mass spectrometry (LC-FTMS) analysis of human plasma.
Matches for ions detected by LC-FTMS with high-resolution mass/charge (m/z) in human plasma to metabolites in the KEGG human metabolic pathways. MS data were extracted using apLCMS (104). Larger dots represent matches for 745 metabolites found in plasma from human, rhesus macaque, common marmoset, rat, mouse, pig and sheep. Metabolites are present for 136 out of 154 pathways in the database.
Other analytic approaches
Many additional approaches are available for nutritional metabolomics, such as capillary electrophoresis as an alternative to chromatography (82) and electrochemical detection as an alternative to mass spectrometry (55). A common approach to improve detection of metabolites that differ in characteristics of ionization or separation is to combine results from multiple analytic platforms. Innovative approaches include biosensor arrays, microfluidics, and alternate spectral methods, such as Fourier-transform infrared spectroscopy (13; 54; 107). An important advance from the commercial sector (Metabolon, Research Triangle Park, NC) is the introduction of a hybrid approach for metabolomics, consisting of targeted analysis of 300 to 500 known chemicals and untargeted analysis of an additional 3000 unidentified m/z. This hybrid approach allows one to test specific hypotheses using targeted analysis while simultaneously performing untargeted testing for other associations among unidentified chemicals.
In summary, progress in development of analytic tools for nutritional metabolomics has made targeted (biomarker or mechanistic discovery) and untargeted metabolite analysis practical for routine inclusion into laboratory, clinical and epidemiological nutrition research. Limitations remain for absolute quantification, but relative quantification of ≈ 10% of the core nutritional metabolome of humans is readily available to support integrated studies of diet and health. Non-targeted analyses using high-resolution mass spectrometry provides relative quantification of thousands of chemicals, over half of which are currently unidentified. These include bioactive phytochemicals and products of the microbiome. Hybrid approaches for targeted and untargeted analyses provide information-rich data sets to enable bionformatic and computational research to address complex interactions of diet and the microbiome as they impact individual nutritional needs.
BIOINFORMATICS AND COMPUTATIONAL SCIENCES
Biostatistical, bioinformatic and computational tools to analyze chemical profiles have been developed over several decades and used extensively for nutritional biomarker discovery (7). In the past several years, important advances have been made in tools and applications to use information-rich metabolomics data to integrate diet and nutrition in complex biosystems. These tools can be used in both “bottom-up” and “top-down” approaches (4; 45; 63; 74). Bottom-up approaches start with known molecular reactions and add known steps with targeted metabolic analyses to create models of complex behavior. Important advances have been made in metabolic flux and contributions of discrete biochemical pathways to complex processes. For example, Wopereis et al (102) studied the effect of the anti-inflammatory drug, diclofenac, on 343 plasma metabolites during an oral glucose tolerance test. Metabolites that change grouped into patterns, one of which included increases in neutral amino acids (Ile, Leu, Thr) over the entire challenge time course and the other including metabolites (5-oxoproline, Gly, Glu) related to glutathione biosynthesis that were increased only after 90–120 min. Diclofenac was found to selectively increase the latter group. Wang et al (93) studied a panel of >60 amino acids, amine and other polar metabolites in a case-control study of normoglycemic individuals followed over 12 years and found branched-chain and aromatic amino acids as predictors of future diabetes. Isotopic tracer methods in nutritional metabolomics have also contributed to a number of important nutritional requirements of disease. Marrero et al (61) used 13C carbon tracer kinetics to study gluconeogenesis in Mycobacterium tuberculosis, which relies on this metabolic process to establish and maintain infection. Beste et al (6) used tracer studies to identify a novel pathway for pyruvate dissimilation requiring CO2 fixation. Ferrara et al (22) combined transcriptomic data for 40,000 probe sets with targeted analysis of 67 intermediary metabolites (amino acids, organic acids, acyl-carnitines) to construct causal networks of metabolic processes in liver. Such examples provide clear evidence of the progress in application of bioinformatics and computational approaches to address complex nutritional questions.
Database tools
Developments for study of specific nutrient effects on metabolism include substantial advances in biosystems databases. The confluence of genome sequencing and bioinformatics has led to the development of thousands of metabolic databases such as the MetaCyc, KEGG, Reactome, Model SEED, and BiGG families (50). These are developed using manual curation of scientific data, often focusing on humans and model organisms, such as Escherichia coli, Saccharomyces cerevisiae, Mus musculus, and Arabidopsis thaliana, and are available through a number of web sites. Software tools for querying and visualizing metabolic networks support description and prediction of metabolic pathways for a wide variety of organisms and constitute an important resource for nutrition research. Such developments have occurred as a consequence of genome sequencing efforts and algorithms for predicting the presence of metabolic pathways in organisms with a sequenced genome. These are mostly non quantitative at the present time, but combination with enzyme databases containing kinetic information (35; 79) is beginning to provide more comprehensive modeling capabilities, such as are required to describe nutrient effects.
As an example, MetaCyc is a database of non-redundant, experimentally elucidated metabolic pathways from >1000 organisms (mostly microorganisms and plants) that has begun to capture enzyme kinetics data. The database includes more than 1500 metabolic pathways and associated 8,600 enzymatic reactions. Such systems can support computational metabolic network prediction, integrate experimental data into metabolic pathways and create metabolic models for simulation (8; 50). An important advance for nutritional metabolomics is the development of a complementary experimental approach to use stable isotopes to elucidate metabolic pathways in a non-targeted manner (41). This method uses the isotopomer distribution and computation analysis for global analysis of flux into all detectable metabolite pools. Such methods remain limited by metabolic cycles, loss of label as CO2, and an assumption that the system is at a steady state. Despite these limitations for precise systems biology descriptions, however, the methods provide means to elucidate new metabolic pathways and could be especially valuable for studies of nutrition and the microbiome.
These database tools support genome-scale models of metabolism as needed to address the effects of diet and nutrition on health of an organism. While review of research on metabolic reconstructions (18; 20) is beyond the scope of the present review, an example of advanced computational methods illustrates the applicability for complex nutritional modeling. Metabolic network reconstruction from genomic and gene expression data is composed of a set of biochemical reactions and provides insight into the hierarchical regulation of metabolic flux at a genome-scale. However, enzyme activity and metabolomics data are needed to provide information on metabolic flux. Yizhak et al (103) used computational modeling to predict metabolic flux distribution for genome-scale models by determining a steady-state flux distribution in which flux through reactions with measured proteomic and metabolomic data is as consistent as possible with kinetically derived flux estimations. Such approaches provide a foundation for nutritional studies where high-throughput metabolomics data are becoming available for nutritional deficiency and excess models. While limited high-throughput flux measurement capabilities exist, such modeling approaches can support understanding of complex nutritional interactions at the level of an entire organism.
Top-down approaches complement bottom-up metabolomics
With time, such approaches will yield an understanding of responses of mammals to changes in the endogenous microbiome. At present, however, the variability of the microbiome introduces a complication in mammalian nutrition that is difficult to address using a bottom-up approach to nutrition at a personalized level. Each microorganism has a metabolome, and the metabolomes of these organisms interact with each other as well as with the host mammalian metabolome. Thus, even though rapid progress is currently being made to understand normative characteristics of the human and mouse microbiomes, individual nutritional modeling becomes unwieldy without a priori knowledge of the contributions of thousands of microbial species within an individual. For such purposes, top-down nutritional metabolomics approaches provide a critical complement to the bottom-up computational frameworks.
Top-down nutritional metabolomics approaches are analogous to a whole body physical examination in medicine, e.g., an elevated blood LDL concentration provides important information about cardiovascular disease risk, but total CVD risk is better evaluated within the context of obesity, hypertension, exercise and other risk factors. While there are no ideal top-down nutritional metabolomics methods currently available, important advances have been made with several analytic platforms. Proton nuclear magnetic resonance (1H-NMR) spectroscopy and phosphorous-31 NMR (31P-NMR) spectroscopy provide spectral measurements that are directly and quantitatively related to abundance of chemicals (12; 92). Many of the signals overlap and are difficult to relate unambiguously to individual chemicals, but the methods are useful to measure abundant chemicals, such as macronutrients in energy metabolism, including fatty acids, sugars and some amino acids (68). Inouye et al (42) used 1H-NMR metabolomic data along with transcriptomic and genomic variation to show concurrent association of metabolites, inflammation and adiposity. Importantly, gene co-expression in circulating leukocytes was dependent upon plasma metabolites, showing networks linkage with lipoproteins, lipids and amino acids. Stringer et al. (86) showed that quantitative metabolomics was potentially useful in sepsis-induced acute lung injury, and Park et al (70) showed a progressive trajectory of the principal component analysis (PCA) of plasma metabolites in albumin-treated patients toward a normal healthy metabolic profile. The most discriminatory regions by PCA consisted of parts of the spectra in which amino acids, glucose and other metabolites are present, suggesting utility to evaluate nutritional imbalance of critical illness and to assess recovery from critical illness (70).
Tracer methods using carbon-13 NMR (13C-NMR) spectroscopy also provide ability to measure flux in pathways containing high abundance metabolites, such as metabolic interactions of pathogenic and commensal bacteria (66). Analogous magnetic resonance spectroscopy (MRS) methods measure metabolites in vivo, and such studies were used many years ago to study nutrient effects on muscle energy metabolism (2) and more recently to show that 3 days of a sulfur amino acid-free diet has no detectable effect on brain GSH, but increases midbrain glutamate and has effects on other high-abundance metabolites in humans (73)
High-resolution mass spectrometry methods provide capabilities for comprehensive top-down analysis by supporting measurement of thousands of metabolites, with relatively high throughput and minimal sample processing. Associated limitations are the capability to only support relative quantification of most species and the inclusion of large numbers of unidentified chemicals. Moreover, statistical methods must be applied with caution because of the large number of comparisons in associated datasets. Methods based upon the principles of false discovery rate (FDR)(5), and positive FDR (85), are valuable for analysis, but these methods are limited in that metabolic adaptation can result in an appearance of no effect when substantial pathway effects are present. For instance, proteolysis and gluconeogenesis maintain blood amino acid and glucose concentrations during starvation, with some, but not all, related metabolites significantly altered (21; 38). Consequently, for nutritional metabolomics, there remains a need for robust statistical methods to complement those for individual metabolites and test for pathway and network effects involving multiple metabolites.
Bioinformatics for top-down metabolomics
Common approaches for top-down nutritional metabolomics rely upon bioinformatics methods, which include both statistical and non-statistical methods. The broader approach of chemometrics, developed over several decades (7), uses multivariate statistics, applied mathematics, and computer science to address chemistry, biology and medicine. Methods are widely available to characterize metabolic patterns associated with nutritional variables and other data-analytic needs in nutritional metabolomics research (78). Clustering algorithms to simplify complexity or select a smaller subset of chemicals or subpopulations for targeted investigation are widely available in biostatistical software packages; hierarchical cluster analysis (HCA) is commonly used in nutrition research in gene expression analysis and can similarly be used to subgroup metabolites or individuals according to metabolic characteristics without a priori knowledge of metabolic pathways or health phenotype. Principal component analysis (PCA) and partial least squares (PLS) are also widely available to reduce complexity by extracting information on factors contributing to differences among the samples. Both HCA and PCA are also widely considered unbiased in the sense that they classify samples without assignment of identifiers to the samples. However, both can be biased in complex biosystems analyses because analytic methods can omit lower abundance metabolites. Furthermore, higher abundance metabolites often contribute more to the total variance so that mean-centering and autoscaling of abundance data can be important to evaluate metabolomic profiles in an unbiased manner. A limit to this approach is that lower abundance chemicals often have greater analytical variability, so scaling and normalization procedures commonly used in statistics can introduce bias in bioinformatics. Critical reviews and commentaries on sample collection, metabolic profiling and data analysis provide important progress in nutritional metabolomics (78).
Advances in probability-based clustering (84) allow co-regulated metabolites to be identified, moving forward capabilities for delineation of the hierarchical structure of metabolic organization. Such approaches are not always reliable because genes of the same pathway can be regulated very differently (91). None-the less, advances in these methods enable sub-classification of the pan-metabolome based upon associations of unidentified metabolites with known metabolites and metabolic pathways (83). In the context of carefully designed nutritional intervention studies, such data will support genome-scale nutritional metabolomics models as needed for personalized nutrition. Toward this goal, important advances have been made in standardization of metabolomics data to support nutrition research (77), but cumulative databases of quantitative human metabolomics data are not yet well developed or widely available.
Artificial intelligence: Fractal analysis
Artificial intelligence methods have been applied to nutritional metabolomics and may be particularly useful to quantify metabolic adaptability. An example is the use of fractal dynamics to measure irregularity and unpredictability in biological systems, as introduced by Goldberger (31). Fractal analysis shows that irregularity of serial physiologic data are important features of health and adaptation; in contrast, characteristics of disease are associated with greater regularity of such data. This is illustrated by fractal analysis of the human heartbeat, where a higher Hurst exponent (H), indicative of greater heartbeat regularity, was associated with cardiovascular disease, while a more irregular heartbeat pattern, with a lower H value, was observed in healthy individuals as an indicator of greater adaptability to ambient conditions (31; 32). Although not developed for nutritional assessment, fractal analysis of diurnal metabolic variation using wavelet transformed 1H NMR spectra of human plasma showed that H was predictive of the plasma concentration of cysteine (47). More powerful multifractal approaches are also available to improve description of metabolic regularity/irregularity by decomposing data into subsets characterized by multifractal spectra with Holder exponent values that quantify local behaviors, i.e., subregions of higher regularity and irregularity (43). Application of such methods to information-rich datasets could be useful to detect metabolic consequences of suboptimal nutrition that may warrant more detailed clinical follow up or to provide quantitative means to monitor clinically-relevant responses to specific nutritional interventions.
CONCLUSIONS AND NEW PERSPECTIVES
Nutrition sciences have a rich tradition of dealing with food and nutrient requirements for heterogeneous populations, simplifying the inherent complexity into manageable recommendations in the form of dietary guidance for the purpose of avoiding disease. While using rigorous experimental designs and sophisticated analytic and biostatistical methods, this approach has inherent limitations due to assumptions that metabolic organizational structure is uniform among individuals and that direct cause-effect relationships exist. In addressing this complexity, Ziesel et al (106) introduced the concept that nutritional phenotype could be defined as an integrated set of genetic, proteomic, metabolomic, functional, and behavioral factors that, when measured, could provide the basis for assessment of human nutritional status. The nutritional phenotype was proposed as a means to integrate the effects of diet on disease/wellness and provide a quantitative indication of the paths by which genes and environment exert their effects on health. As summarized here, advances in available technologies and databases support use of nutritional metabolomics as a central component to define and measure the nutritional phenotype, initially across populations with various states of health and disease, and ultimately at the level of individuals.
Several key advances have been made to transition from cross-sectional descriptions of populations to useful models that are relevant for individuals. Complex biosystems models must accommodate single or multiple nutritional deficiencies and/or excesses as having different health phenotypic consequences among individuals, as well as different nutritional variations among individuals contributing to common health phenotypes. Such characteristics cannot be reliably predicted from normative responses with single cause-effect relationships. Advances that enable use of nutritional metabolomics to support complex biosystems models include the delineation of a conceptual grid for the exposome, a transition from disease-oriented to health-oriented nutrition and transition from a reductionist approach to one that incorporates non-reductionist complex biosystems approaches. These conceptual advances are enhanced by improved analytic techniques allowing relative quantification of thousands of metabolites, encompassing the core nutritional metabolome, the broader food metabolome, a microbiome-associated metabolome, and a range of chemicals derived from pharmaceutics, commercial products and other environmental exposures (Figure 1). Rapid adaptation of chemometric methods to nutritional metabolomics, and evolution of new bioinformatic and computional methods, as well as artificial intelligence, have set the stage to focus nutrition research on requirements of complex biosystems within individuals. These advances in nutritional metabolomics portend a major shift in nutritional assessment and intervention from those based upon normative characteristics of populations to ones utilizing complex biosystems approaches that may better account for individual nutritional needs.
SUMMARY POINTS
Available technologies and databases support use of nutritional metabolomics as a central component to define and measure the nutritional phenotype
The nutritional metabolome is a critical component of the conceptual grid of the exposome, i.e., incorporating nutrition exposures from conception onward
Nutritional metabolomics is rapidly maturing as an experimental approach in the field of nutrition science to support integration of diet and nutrition in complex biosystems research
Personalized computational models can be developed to use nutritional metabolomics to forecast health risks and treatment outcomes, thereby facilitating the transition from population-based to individual-based nutrition.
High-resolution mass spectrometry provides sufficient sensitivity and metabolic coverage to support metabolome-wide association studies (MWAS) of nutrition and disease
An array of chemical and metabolic databases, bioinformatic methods and computational approaches are available to enhance metabolomics use in nutrition research
FUTURE ISSUES
Systematic application of metabolomics to life-cycle nutrition research provides a logical, central foundation for the elaboration of the conceptual grid of the exposome
Mechanistic information is available to use with nutritional metabolomics data to support design and development of integrated systems models for nutrition health prediction
Reference high-performance metabolic profiles and associated health outcomes data will be needed as a resource for nutrition scientists to provide real tests of computational models
Application and utility of nutritional metabolomics in both targeted (e.g. identification of potential direct or surrogate biomarkers of health and disease) and untargeted analysis in nutrition science
Acknowledgments
The authors thank J.F. Gregory, E.O. Voit, Y.-M. Go, J.R. Roede and Q.A. Soltow for helpful comments during manuscript preparation. DPJ is a Professor of Medicine supported in part by NIH Grants ES009047, ES011195, AG038746, NR012021, AA013757, ES016731, DK069322 and DK089369. YP is an Assistant Professor of Medicine supported in part by NIH Grants AG038746, ES016731, DK089369, RR023356 and Children’s Healthcare of Atlanta Center for Developmental Lung Biology. TRZ is a Professor of Medicine supported in part by NIH Grants DK069322, DK089369, HL110044, RR023356 and RR025008 and the Emory Global Health Institute.
Acronyms
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MMCD
Madison Metabolomics Consortium Database (MMCD)
- DRI
Dietary Reference Intakes
- MS
Mass Spectrometry
- FTMS
Fourier-transform mass spectrometry
- apLCMS
adaptive processing of high-resolution liquid chromatography/mass spectrometry data
- NMR
nuclear magnetic resonance spectroscopy
- FDR
false discovery rate
- PCA
principal component analysis
- MWAS
metabolome-wide association studies
Terms/Definitions
- Nutritional metabolomics
use of small molecule chemical profiling to integrate diet and nutrition in complex biosystems
- Exposome
life-course environmental exposures (including lifestyle factors) from the prenatal period onwards
- Reductionist
description of complex system behavior in terms of simple cause-effect relationships
- Pan-metabolome
All small molecular weight chemicals in a biologic system
- Microbiome
Microorganisms associated with a metazoan organism
- Predictive Health
A healthcare concept using lifestyle approaches, including nutrition, to optimize vitality and well-being rather than to serve primarily as a means to prevent disease
- Normative
Description of characteristics and responses in terms of population averages
- Integrated Biosystems
Models incorporating functional relationships of component parts to describe overall behavior of a biological system
- Artificial Intelligence
Use of machine learning for classification or decision-making purposes without regard for underlying mechanisms or relationships of component parts
- Fractal analysis
An approach to quantify the regularity in behavior of a characteristic of a biologic system as a means to discriminate unhealthy (more regular) and healthy (more irregular) individual
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Dean P. Jones, Email: dpjones@emory.edu.
Youngja Park, Email: youngja.park@emory.edu.
Thomas R. Ziegler, Email: tzieg01@emory.edu.
LITERATURE CITED
- 1.Alcock RE, Halsall CJ, Harris CA, Johnston AE, Lead WA, et al. Contamination of Environmental Samples Prepared for PCB Analysis. Environ Sci Technol. 1994;28:1838–42. doi: 10.1021/es00060a013. [DOI] [PubMed] [Google Scholar]
- 2.Argov Z, Chance B. Phosphorus magnetic resonance spectroscopy in nutritional research. Annu Rev Nutr. 1991;11:449–64. doi: 10.1146/annurev.nu.11.070191.002313. [DOI] [PubMed] [Google Scholar]
- 3.Bais P, Moon SM, He K, Leitao R, Dreher K, et al. PlantMetabolomics.org: a web portal for plant metabolomics experiments. Plant Physiol. 2010;152:1807–16. doi: 10.1104/pp.109.151027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bang JW, Crockford DJ, Holmes E, Pazos F, Sternberg MJ, et al. Integrative top-down system metabolic modeling in experimental disease states via data-driven Bayesian methods. J Proteome Res. 2008;7:497–503. doi: 10.1021/pr070350l. [DOI] [PubMed] [Google Scholar]
- 5.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 6.Beste DJ, Bonde B, Hawkins N, Ward JL, Beale MH, et al. (1)(3)C metabolic flux analysis identifies an unusual route for pyruvate dissimilation in mycobacteria which requires isocitrate lyase and carbon dioxide fixation. PLoS Pathog. 2011;7:e1002091. doi: 10.1371/journal.ppat.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown SD, Tauler R, Walczak B, editors. Comprehensive Chemometrics. Chemical and Biochemical Data Analysis. Elsevier; 2009. [Google Scholar]
- 8.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–9. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CDC. Fourth National Report on Human Exposure to Environmental Chemicals: Updated Tables. National Center for Environmental Health, Centers for Disease Control and Prevention; Atlanta, GA: 2011. [Google Scholar]
- 10.Ceglarek U, Leichtle A, Brugel M, Kortz L, Brauer R, et al. Challenges and developments in tandem mass spectrometry based clinical metabolomics. Mol Cell Endocrinol. 2009;301:266–71. doi: 10.1016/j.mce.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Cevallos-Cevallosa JM, Reyes-De-Corcueraa JI, Etxeberriaa E, Danyluka MD, Rodrick GE. Metabolomic analysis in food science: a review. Trends Food Sci Technol. 2009;20:557–66. [Google Scholar]
- 12.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106:14728–33. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corte L, Rellini P, Roscini L, Fatichenti F, Cardinali G. Development of a novel, FTIR (Fourier transform infrared spectroscopy) based, yeast bioassay for toxicity testing and stress response study. Anal Chim Acta. 2010;659:258–65. doi: 10.1016/j.aca.2009.11.035. [DOI] [PubMed] [Google Scholar]
- 14.de Mul M, Alons P, van der Velde P, Konings I, Bakker J, Hazelzet J. Development of a clinical data warehouse from an intensive care clinical information system. Comput Methods Programs Biomed. 2010 doi: 10.1016/j.cmpb.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Deo RC, Hunter L, Lewis GD, Pare G, Vasan RS, et al. Interpreting metabolomic profiles using unbiased pathway models. PLoS Comput Biol. 2010;6:e1000692. doi: 10.1371/journal.pcbi.1000692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon RA, Gang DR, Charlton AJ, Fiehn O, Kuiper HA, et al. Applications of metabolomics in agriculture. J Agric Food Chem. 2006;54:8984–94. doi: 10.1021/jf061218t. [DOI] [PubMed] [Google Scholar]
- 17.Draper J, Enot DP, Parker D, Beckmann M, Snowdon S, et al. Metabolite signal identification in accurate mass metabolomics data with MZedDB, an interactive m/z annotation tool utilising predicted ionisation behaviour ‘rules’. BMC Bioinformatics. 2009;10:227. doi: 10.1186/1471-2105-10-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duarte NC, Herrgard MJ, Palsson BO. Reconstruction and validation of Saccharomyces cerevisiae iND750, a fully compartmentalized genome-scale metabolic model. Genome Res. 2004;14:1298–309. doi: 10.1101/gr.2250904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–4. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feist AM, Palsson BO. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotechnol. 2008;26:659–67. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felig P, Kim YJ, Lynch V, Hendler R. Amino acid metabolism during starvation in human pregnancy. J Clin Invest. 1972;51:1195–202. doi: 10.1172/JCI106913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrara CT, Wang P, Neto EC, Stevens RD, Bain JR, et al. Genetic networks of liver metabolism revealed by integration of metabolic and transcriptional profiling. PLoS genetics. 2008;4:e1000034. doi: 10.1371/journal.pgen.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiehn O, Barupal DK, Kind T. Extending biochemical databases by metabolomic surveys. J Biol Chem. 2011;286:23637–43. doi: 10.1074/jbc.R110.173617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol. 2003;15:247–54. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 25.Flamini R. Mass spectrometry in grape and wine chemistry. Part I: polyphenols. Mass Spectrom Rev. 2003;22:218–50. doi: 10.1002/mas.10052. [DOI] [PubMed] [Google Scholar]
- 26.German JB, Bauman DE, Burrin DG, Failla ML, Freake HC, et al. Metabolomics in the opening decade of the 21st century: building the roads to individualized health. J Nutr. 2004;134:2729–32. doi: 10.1093/jn/134.10.2729. [DOI] [PubMed] [Google Scholar]
- 27.German JB, Roberts MA, Watkins SM. Genomics and metabolomics as markers for the interaction of diet and health: lessons from lipids. J Nutr. 2003;133:2078S–83S. doi: 10.1093/jn/133.6.2078S. [DOI] [PubMed] [Google Scholar]
- 28.German JB, Roberts MA, Watkins SM. Personal metabolomics as a next generation nutritional assessment. J Nutr. 2003;133:4260–6. doi: 10.1093/jn/133.12.4260. [DOI] [PubMed] [Google Scholar]
- 29.Gibney MJ, Walsh M, Brennan L, Roche HM, German B, van Ommen B. Metabolomics in human nutrition: opportunities and challenges. Am J Clin Nutr. 2005;82:497–503. doi: 10.1093/ajcn.82.3.497. [DOI] [PubMed] [Google Scholar]
- 30.Go EP. Database resources in metabolomics: an overview. J Neuroimmune Pharmacol : the official journal of the Society on NeuroImmune Pharmacology. 2010;5:18–30. doi: 10.1007/s11481-009-9157-3. [DOI] [PubMed] [Google Scholar]
- 31.Goldberger AL. Fractal electrodynamics of the heartbeat. Ann N Y Acad Sci. 1990;591:402–9. doi: 10.1111/j.1749-6632.1990.tb15104.x. [DOI] [PubMed] [Google Scholar]
- 32.Goldberger AL, Amaral LA, Hausdorff JM, Ivanov P, Peng CK, Stanley HE. Fractal dynamics in physiology: alterations with disease and aging. Proc Natl Acad Sci U S A. 2002;99(Suppl 1):2466–72. doi: 10.1073/pnas.012579499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–52. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Goodman M, Bostick RM, Kucuk O, Jones DP. Clinical trials of antioxidants as cancer prevention agents: past, present, and future. Free Radic Biol Med. 2011;51:1068–84. doi: 10.1016/j.freeradbiomed.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 35.Gremse M, Chang A, Schomburg I, Grote A, Scheer M, et al. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011;39:D507–13. doi: 10.1093/nar/gkq968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grimaldi D, Claessens YE, Mira JP, Chiche JD. Beyond clinical phenotype: the biologic integratome. Crit Care Med. 2009;37:S38–49. doi: 10.1097/CCM.0b013e3181920cca. [DOI] [PubMed] [Google Scholar]
- 37.Hall RD, Brouwer ID, Fitzgerald MA. Plant metabolomics and its potential application for human nutrition. Physiol Plant. 2008;132:162–75. doi: 10.1111/j.1399-3054.2007.00989.x. [DOI] [PubMed] [Google Scholar]
- 38.Hammarqvist F, Andersson K, Luo JL, Wernerman J. Free amino acid and glutathione concentrations in muscle during short-term starvation and refeeding. Clin Nutr. 2005;24:236–43. doi: 10.1016/j.clnu.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Harkewicz R, Dennis EA. Applications of mass spectrometry to lipids and membranes. Annu Rev Biochem. 2011;80:301–25. doi: 10.1146/annurev-biochem-060409-092612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hertkorn N, Ruecker C, Meringer M, Gugisch R, Frommberger M, et al. High-precision frequency measurements: indispensable tools at the core of the molecular-level analysis of complex systems. Anal Bioanal Chem. 2007;389:1311–27. doi: 10.1007/s00216-007-1577-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hiller K, Metallo CM, Kelleher JK, Stephanopoulos G. Nontargeted elucidation of metabolic pathways using stable-isotope tracers and mass spectrometry. Anal Chem. 2010;82:6621–8. doi: 10.1021/ac1011574. [DOI] [PubMed] [Google Scholar]
- 42.Inouye M, Kettunen J, Soininen P, Silander K, Ripatti S, et al. Metabonomic, transcriptomic, and genomic variation of a population cohort. Mol Syst Biol. 2010;6:441. doi: 10.1038/msb.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ivanov PC, Amaral LA, Goldberger AL, Havlin S, Rosenblum MG, et al. Multifractality in human heartbeat dynamics. Nature. 1999;399:461–5. doi: 10.1038/20924. [DOI] [PubMed] [Google Scholar]
- 44.Johnson JM, Strobel FH, Reed M, Pohl J, Jones DP. A rapid LC-FTMS method for the analysis of cysteine, cystine and cysteine/cystine steady-state redox potential in human plasma. Clin Chim Acta. 2008;396:43–8. doi: 10.1016/j.cca.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson JM, Yu T, Strobel FH, Jones DP. A practical approach to detect unique metabolic patterns for personalized medicine. The Analyst. 2010;135:2864–70. doi: 10.1039/c0an00333f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones DP. Redefining oxidative stress. Antioxid Redox Signal. 2006;8:1865–79. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 47.Jung YY, Park Y, Jones DP, Ziegler TR, Vidakovic B. Self-similarity in NMR spectra: an application in assessing the level of cysteine. J Data Sci. 2010;8:1–19. [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. discussion −3, 19–28, 244–52. [PubMed] [Google Scholar]
- 49.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karp PD, Caspi R. A survey of metabolic databases emphasizing the MetaCyc family. Arch Toxicol. 2011;85:1015–33. doi: 10.1007/s00204-011-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keith LH, Crummett W, Deegan J, Libby RA, Taylor JK, Wentler G. Principles of environmental analysis. Anal Chem. 1983;55:2210–8. [Google Scholar]
- 52.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 53.Kind T, Scholz M, Fiehn O. How large is the metabolome? A critical analysis of data exchange practices in chemistry. PLoS One. 2009;4:e5440. doi: 10.1371/journal.pone.0005440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kraly JR, Holcomb RE, Guan Q, Henry CS. Review: Microfluidic applications in metabolomics and metabolic profiling. Analytica chimica acta. 2009;653:23–35. doi: 10.1016/j.aca.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kristal BS, Shurubor YI, Kaddurah-Daouk R, Matson WR. High-performance liquid chromatography separations coupled with coulometric electrode array detectors: a unique approach to metabolomics. Methods Mol Biol. 2007;358:159–74. doi: 10.1007/978-1-59745-244-1_10. [DOI] [PubMed] [Google Scholar]
- 56.Kwan D, Bartle WR, Walker SE. Abnormal serum transaminases following therapeutic doses of acetaminophen in the absence of known risk factors. Digestive diseases and sciences. 1995;40:1951–5. doi: 10.1007/BF02208663. [DOI] [PubMed] [Google Scholar]
- 57.Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Macronutrients ARotPo. Dietary Reference Intakes for Energy, Carbohydrate, Fiber, Fat, Fatty Acids, Cholesterol, Protein and Amino Acids (Macronutrients) Washington, DC: National Academies Press; 2005. pp. 998–1014. Number of 998–1014 p pp. [Google Scholar]
- 59.Macronutrients ARotPo; SCotSEoDR Intakes, editor. Dietary reference intakes for energy, carbohydrate, fiber, fat, fatty acids, cholesterol, protein, and amino acids (macronutrients) XXV. Washington, D.C: National Academies Press; 2005. p. 1331. Number of 1331 p. pp. [Google Scholar]
- 60.Markley JL, Anderson ME, Cui Q, Eghbalnia HR, Lewis IA, et al. New bioinformatics resources for metabolomics. Pac Symp Biocomput. 2007:157–68. [PubMed] [Google Scholar]
- 61.Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc Natl Acad Sci U S A. 2010;107:9819–24. doi: 10.1073/pnas.1000715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marshall AG, Hendrickson CL. High-resolution mass spectrometers. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:579–99. doi: 10.1146/annurev.anchem.1.031207.112945. [DOI] [PubMed] [Google Scholar]
- 63.Martin FP, Dumas ME, Wang Y, Legido-Quigley C, Yap IK, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore S, Spackman DH, Stein WH. Automatic recording apparatus for use in the chromatography of amino acids. Fed Proc. 1958;17:1107–15. [PubMed] [Google Scholar]
- 65.Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–4. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakanishi Y, Fukuda S, Chikayama E, Kimura Y, Ohno H, Kikuchi J. Dynamic omics approach identifies nutrition-mediated microbial interactions. J Proteome Res. 2011;10:824–36. doi: 10.1021/pr100989c. [DOI] [PubMed] [Google Scholar]
- 67.Neveu V, Perez-Jimenez J, Vos F, Crespy V, du Chaffaut L, et al. Phenol-Explorer: an online comprehensive database on polyphenol contents in foods. Database (Oxford) 2010;2010:bap024. doi: 10.1093/database/bap024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nicholson JK, Foxall PJ, Spraul M, Farrant RD, Lindon JC. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal Chem. 1995;67:793–811. doi: 10.1021/ac00101a004. [DOI] [PubMed] [Google Scholar]
- 69.Otten JJ, Hellwig JP, Meyers LD. DRI, dietary reference intakes : the essential guide to nutrient requirements. Washington, D.C: National Academies Press; 2006. p. 543. [Google Scholar]
- 70.Park Y, Jones DP, Ziegler TR, Lee K, Kotha K, et al. Metabolic effects of albumin therapy in acute lung injury measured by proton nuclear magnetic resonance spectroscopy of plasma: a pilot study. Crit Care Med. 2011;39:2308–13. doi: 10.1097/CCM.0b013e31822571ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Park Y, Kim SB, Wang B, Blanco RA, Le NA, et al. Individual variation in macronutrient regulation measured by proton magnetic resonance spectroscopy of human plasma. Am J Physiol Regul Integr Comp Physiol. 2009;297:R202–9. doi: 10.1152/ajpregu.90757.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park Y, Le NA, Yu T, Strobel F, Gletsu-Miller N, et al. A sulfur amino acid-free meal increases plasma lipids in humans. J Nutr. 2011;141:1424–31. doi: 10.3945/jn.111.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Park Y, Zhao T, Miller NG, Kim SB, Accardi CJ, et al. Sulfur amino acid-free diet results in increased glutamate in human midbrain: A pilot magnetic resonance spectroscopic study. Nutrition. 2011 doi: 10.1016/j.nut.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rappaport SM. Implications of the exposome for exposure science. J Expo Sci Environ Epidemiol. 2010;21:5–9. doi: 10.1038/jes.2010.50. [DOI] [PubMed] [Google Scholar]
- 75.Rezzi S, Ramadan Z, Fay LB, Kochhar S. Nutritional metabonomics: applications and perspectives. Journal of proteome research. 2007;6:513–25. doi: 10.1021/pr060522z. [DOI] [PubMed] [Google Scholar]
- 76.Sandhu AK, Gu L. Antioxidant capacity, phenolic content, and profiling of phenolic compounds in the seeds, skin, and pulp of Vitis rotundifolia (Muscadine Grapes) As determined by HPLC-DAD-ESI-MS(n) J Agric Food Chem. 2010;58:4681–92. doi: 10.1021/jf904211q. [DOI] [PubMed] [Google Scholar]
- 77.Sansone SA, Fan T, Goodacre R, Griffin JL, Hardy NW, et al. The metabolomics standards initiative. Nat Biotechnol. 2007;25:846–8. doi: 10.1038/nbt0807-846b. [DOI] [PubMed] [Google Scholar]
- 78.Scalbert A, Brennan L, Fiehn O, Hankemeier T, Kristal BS, et al. Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics. 2009;5:435–58. doi: 10.1007/s11306-009-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Scheer M, Grote A, Chang A, Schomburg I, Munaretto C, et al. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–6. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schreiber F, Colmsee C, Czauderna T, Grafahrend-Belau E, Hartmann A, et al. MetaCrop 2.0: managing and exploring information about crop plant metabolism. Nucleic Acids Res. 2012;40:D1173–7. doi: 10.1093/nar/gkr1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–51. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 82.Soga T, Igarashi K, Ito C, Mizobuchi K, Zimmermann HP, Tomita M. Metabolomic profiling of anionic metabolites by capillary electrophoresis mass spectrometry. Anal Chem. 2009;81:6165–74. doi: 10.1021/ac900675k. [DOI] [PubMed] [Google Scholar]
- 83.Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics. 2011 doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stone EA, Ayroles JF. Modulated modularity clustering as an exploratory tool for functional genomic inference. PLoS Genet. 2009;5:e1000479. doi: 10.1371/journal.pgen.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Storey JD. 3. The positive false discovery rate: A Bayesian interpretation and the q-value. Ann Statist. 2000;31:2013–35. [Google Scholar]
- 86.Stringer KA, Serkova NJ, Karnovsky A, Guire K, Paine R, 3rd, Standiford TJ. Metabolic consequences of sepsis-induced acute lung injury revealed by plasma (1)H-nuclear magnetic resonance quantitative metabolomics and computational analysis. Am J Physiol Lung Cell Mol Physiol. 2011;300:L4–L11. doi: 10.1152/ajplung.00231.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.USDA. Dietary Guidelines for Americans. U.S. Government Printing Office; Washington, DC: 2010. [Google Scholar]
- 88.Voit E, Neves AR, Santos H. The intricate side of systems biology. Proc Natl Acad Sci U S A. 2006;103:9452–7. doi: 10.1073/pnas.0603337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Voit EO, Brigham KL. The role of systems biology in predictive health and personalized medicine. The Open Pathology Journal. 2008;2:68–70. [Google Scholar]
- 90.Voit EO, Qi Z, Miller GW. Steps of modeling complex biological systems. Pharmacopsychiatry. 2008;41(Suppl 1):S78–84. doi: 10.1055/s-2008-1080911. [DOI] [PubMed] [Google Scholar]
- 91.Voit EO, Radivoyevitch T. Biochemical systems analysis of genome-wide expression data. Bioinformatics. 2000;16:1023–37. doi: 10.1093/bioinformatics/16.11.1023. [DOI] [PubMed] [Google Scholar]
- 92.Walsh MC, Brennan L, Malthouse JP, Roche HM, Gibney MJ. Effect of acute dietary standardization on the urinary, plasma, and salivary metabolomic profiles of healthy humans. Am J Clin Nutr. 2006;84:531–9. doi: 10.1093/ajcn/84.3.531. [DOI] [PubMed] [Google Scholar]
- 93.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Want EJ, Wilson ID, Gika H, Theodoridis G, Plumb RS, et al. Global metabolic profiling procedures for urine using UPLC-MS. Nat Protoc. 2010;5:1005–18. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- 96.Weng C, Bigger JT, Busacca L, Wilcox A, Getaneh A. Comparing the effectiveness of a clinical registry and a clinical data warehouse for supporting clinical trial recruitment: a case study. AMIA Annu Symp Proc. 2010;2010:867–71. [PMC free article] [PubMed] [Google Scholar]
- 97.Whitfield PD, German AJ, Noble PJ. Metabolomics: an emerging post-genomic tool for nutrition. Br J Nutr. 2004;92:549–55. doi: 10.1079/bjn20041243. [DOI] [PubMed] [Google Scholar]
- 98.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14:1847–50. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 100.Wishart DS. Metabolomics: applications to food science and nutrition research. Trends Food Sci Technol. 2008;19:482–93. [Google Scholar]
- 101.Wishart DS, Knox C, Guo AC, Eisner R, Young N, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–10. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wopereis S, Rubingh CM, van Erk MJ, Verheij ER, van Vliet T, et al. Metabolic profiling of the response to an oral glucose tolerance test detects subtlemetabolic changes. PLoS One. 2009;4:e4525. doi: 10.1371/journal.pone.0004525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yizhak K, Benyamini T, Liebermeister W, Ruppin E, Shlomi T. Integrating quantitative proteomics and metabolomics with a genome-scale metabolic network model. Bioinformatics. 2010;26:i255–60. doi: 10.1093/bioinformatics/btq183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu T, Park Y, Johnson JM, Jones DP. apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25:1930–6. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zapletal E, Rodon N, Grabar N, Degoulet P. Methodology of integration of a clinical data warehouse with a clinical information system: the HEGP case. Stud Health Technol Inform. 2010;160:193–7. [PubMed] [Google Scholar]
- 106.Zeisel SH, Freake HC, Bauman DE, Bier DM, Burrin DG, et al. The nutritional phenotype in the age of metabolomics. J Nutr. 2005;135:1613–6. doi: 10.1093/jn/135.7.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao Q, Wang D, Jayawardhana DA, Guan X. Stochastic sensing of biomolecules in a nanopore sensor array. Nanotechnology. 2008;19:505504. doi: 10.1088/0957-4484/19/50/505504. [DOI] [PubMed] [Google Scholar]