Abstract
Water deficit imposed by either drought or salinity brings about severe growth retardation and yield loss of crops. Since Brassica crops are important contributors to total oilseed production, it is urgently needed to develop tolerant cultivars to ensure yields under such adverse conditions. There are various physiochemical mechanisms for dealing with drought and salinity in plants at different developmental stages. Accordingly, different indicators of tolerance to drought or salinity at the germination, seedling, flowering and mature stages have been developed and used for germplasm screening and selection in breeding practices. Classical genetic and modern genomic approaches coupled with precise phenotyping have boosted the unravelling of genes and metabolic pathways conferring drought or salt tolerance in crops. QTL mapping of drought and salt tolerance has provided several dozen target QTLs in Brassica and the closely related Arabidopsis. Many drought- or salt-tolerant genes have also been isolated, some of which have been confirmed to have great potential for genetic improvement of plant tolerance. It has been suggested that molecular breeding approaches, such as marker-assisted selection and gene transformation, that will enhance oil product security under a changing climate be integrated in the development of drought- and salt-tolerant Brassica crops.
Keywords: Brassica, drought, salinity, tolerance, genetic study
Introduction
Plants growing under field conditions are subjected to various environmental stresses, such as high or low temperature, drought and salinity. At any given point of time, plants may have to face two or more stresses (e.g., drought and salinity). Among these stresses, drought is the most serious problem for global agriculture, approximately affecting 40% of the world’s land area (UN Environment Management Group 2011; http://www.unep-wcmc.org/medialibrary/2011/10/19/3faf1880/Global-Drylands-FINAL-LR.pdf). Even worse, global climate change is predicted to lead to extreme temperatures and severer prolonged drought in some parts of the world, which will have a dramatic impact on crop growth and productivity (Trenberth et al. 2014). Apart from drought, salinity has emerged as another major factor limiting productivity of crops. It is estimated that approximately 7% of the world’s land area is affected by salinity (Rozema and Flowers 2008). In total, drought- and salt-affected soils cover areas of 60 and 10.5 million km2, respectively. These areas are not necessarily arable, but include all global drought- and salt-affected lands. The loss of farmable land due to either salinization or drought has posed a major challenge for maintaining world food supplies for the growing population. Thus, there is an urgent need to develop varieties that can maintain optimum yield levels under abiotic stresses. This objective has been highlighted by the United Nations Secretary General Kofi Annan: “We need a Blue Revolution in agriculture that focuses on increasing productivity per unit of water—more crop per drop” (Pennisi 2008). However, due to the multigenic and quantitative nature of stress tolerance in plants, efforts to improve crop performance under drought and salinity have been elusive. This challenge has given plant scientists an impetus to improve drought and salt tolerance.
Among the major food crops, Brassica crops are the most affected by drought and salinity, due to the fact that they are mainly grown in arid and semiarid areas. The genus Brassica belongs to the family Brassicaceae, which comprises some 100 species, including rapeseed (Brassica napus L.), mustard (Brassica juncea L.), cabbage (Brassica oleracea L.) and turnip rape (Brassica rapa L.) that are mainly grown for oil, condiments, vegetables or fodder (Ashraf and McNeilly 2004). There are also many wild relatives which possess useful agronomic traits, such as tolerance for cold, salty and drought conditions, which could be incorporated into breeding programs (Warwick 1993). Rapeseed accounts for most of the oilseed production in Europe and North America, whereas mustard is mainly grown in India and North Africa. The annual production of rapeseed is more than 60 million tons (http://faostat.fao.org, 2011). However, its growth and seed yield production have greatly decreased owing to drought and salinity. This situation can be alleviated by an approach combining water storage and irrigation, crop management and plant breeding. There is great interest in breeding stress-tolerant varieties, since significant inter- and intraspecific variation for drought and salinity tolerance exists within Brassica, which needs to be exploited through selection and breeding.
In the context of agriculture, drought can be defined as the situation in which the amount of water available to the plant in the root zone is less than that required to sustain maximum growth and productivity (Deikman et al. 2012). Adaptation to drought is definitely one of the most complex biological processes, which include numerous alterations: reduced growth, up- or down-regulation of specific genes, transient increase in ABA levels, build-up of compatible solutes and protective enzymes, enhanced levels of antioxidants and inhibition of energy-consuming pathways (Salekdeh et al. 2009). These biological changes can help plants improve their water-use efficiency, either by dehydration avoidance or through dehydration tolerance (Blum 2005). Drought avoidance strategies include deep rooting, conservative use of available water and adjustment of life cycle to match rainfall (Touchette et al. 2007). Dehydration tolerance ensures plants remain viable after partial dehydration and grow again when moisture recovers. Drought may also lead to stomatal closure, reduction of CO2 intake and ultimately impairment of photosynthesis (Lawlor and Tezara 2009). Alteration of the leaf angle (wildering) will help plants protect against excess solar radiation and decrease evaporation.
Salinity is a soil condition featured by a high concentration of soluble salts (electrical conductivity ≥ 4 dS/m), which is equivalent to ~40 mM NaCl and generates an osmotic pressure of ~0.2 MPa (Rhoades and Loveday 1990). Salinity-affected soils, arising either from long-term natural accumulation of salts or improper land irrigation, are generally occupied by excessive Na+ ion at exchange sites and a high concentration of carbonate/bicarbonate anions. They have a higher pH (>7.5) and poor soil structure (Horneck et al. 2007), and thus significantly reduce the yield of most crops. Soil salinity stresses plants in two ways: dehydration and toxicity. Salts on the outside of roots make it harder to extract water and have an immediate effect on cell growth and related metabolic processes; toxicity only takes place tardily when accumulation of salts inside plants is beyond a certain threshold. During the long process of biological evolution and natural selection, plants have developed three strategies for the maintenance of a low cytosolic sodium concentration: sodium exclusion, compartmentation, and secretion (Purty et al. 2008). As an example, one mechanism for sodium transport out of the cell is through operation of plasma membrane-bound Na+/H+ antiports, putative plasma membranes encoded by SOS1 (Salt Overly Sensitive 1) in Arabidopsis (Shi et al. 2000).
Obviously, there are some common features between drought and salinity stresses in plants. Both stresses impose cellular dehydration, which causes osmotic stress and removal of water from the cytoplasm into the intercellular space. Early responses to drought and salt stress are also similar to each other, except for the ionic component in the cells of plants under salt stress. Thus, it seems that a salinity-tolerant species could also be drought-tolerant, and has largely identical mechanisms to deal with these stresses (Ashraf and O’Leary 1996, Farooq and Azam 2001). In the recent decade, great progresses have been made in the understanding of genetic control of drought and salt tolerance in major crops, such as maize (Tollefson 2011), rice (Fukao and Xiong 2013) and the model plant Arabidopsis (Nakashima et al. 2009), which can certainly shed some light on the mechanisms underlining drought and salt tolerance in Brassica crops and also directly contribute to agronomic-trait improvement (Flowers 2004, Rozema and Flowers 2008). Since the 1980s, with the aid of newly developed genetic experimental tools (Ashraf and McNeilly 2004, El Sharkawy 1989), a number of physiological and genetic studies on drought and salt tolerance have been reported.
Although studies on the molecular mechanism of stress tolerance in Brassica crops have not advanced much, there is an advantage to Brassica studies in that information obtained on Arabidopsis thaliana, which belongs to the family Brassicaceae, may be directly applied to the breeding of Brassica crops. In this review, we first highlight the recent achievements in physiological mechanisms and genetic improvement for drought stress in Brassica crops. Second, a new understanding of salt tolerance in these crops is presented. Third, the challenges and perspectives of breeding drought- and/or salt-tolerant crop varieties are also discussed.
Physiological and genetic studies on drought tolerance in Brassica
Indices for drought tolerance
Drought tolerance is a complex trait, and a crucial aspect is assessment of the degree of drought tolerance of different genotypes. Since physiological responses of plants to drought stress may vary at different developmental stages, it is considered that different indicators should be used for the phenotyping of drought tolerance (Tuberosa 2012). Currently, a number of indicators, such as water-use efficiency (WUE), drought susceptibility index (DSI), relative vigor index (RVI) and leaf wilting index (LWI), are widely used in research and breeding practices. The technical details of these indices and their applications in Brassica crops are summarized in Table 1.
Table 1.
Reported methods and indices for drought tolerance in Brassica species
| Method or indicator | Species | Developmental stage | Traits | Drought regime | Literature |
|---|---|---|---|---|---|
| Biomass and water potential | B. napus, B. juncea, B. campestris, B. carinata | Seedling | Biomass, water content, osmotic potentials | Greenhouse | Ashraf and Mehmood 1990 |
| DSIa | B. juncea | Mature | Seed yield and component traits | Experimental field | Chauhan et al. 2007 |
| LS, PSS, RVIb | B. napus | Germination | Biological and biophysical traits | Petri dish, 10% PEG6000 solution |
Yang et al. 2007 |
| Principal component, clustering, subordinate function analysis | B. napus | Flowering | Morphological and agronomic traits | Rain shelter | Zhu et al. 2011 |
| Leaf wilting index (LWI) c | B. napus | Seedling | Biological and physiological traits | Pots in rain shelter | Li et al. 2012 |
| TDM, LAI, RGR, CGRd | B. napus | Whole stages | Physiological growth indices | Field | Moaveni et al. 2010. |
DSI, drought susceptibility index, which is calculated as (1 − As/Ai)/(1 − Bs/Bi), where As and Ai are traits for a given genotype measured under drought-stressed and irrigated conditions, respectively, and Bs and Bi are traits averaged across all genotypes measured under drought and irrigated conditions, respectively.
RVI, relative vigor index, which is calculated as (Cs × Ds)/(Ci × Di), where Cs and Ci are lengths of seedlings under drought-stressed and irrigated conditions, respectively, and Ds and Di are the percentage of surviving seedlings under drought and irrigated conditions, respectively. LS, length of seedlings (from root to leaf). PSS, percentage of seedlings surviving after drought stress.
LWI = (1 − E/F) × 100, where E is the number of wilting leaves and F is total number of leaves.
TDM, total dry matter of 10 plants. LAI, leaf area index, which is defined as the one-sided green leaf area per unit ground surface area in broad leaf canopies. RGR, relative growth rate. CGR, crop growth rate. Crop growth rate is a measure of the increase in mass over a period of time.
The increase can be plotted as a logarithmic or exponential curve. The CGR is the slope of the curve, while RGR is the slope of the curve that represents logarithmic growth over a period of time.
In a study of Indian mustard (B. juncea L.), DSI values for seed yield and component characteristics were calculated to characterize the relative tolerance of 14 genotypes under watered and drought conditions (Chaunhan et al. 2007). Biomass and water potential have also been used as indices to evaluate the relative drought tolerance of different Brassica species (Ashraf and Mehmood 1990). Biomass production was found to be closely related with water content and water relations. B. carinata showed lower water content and yielded significantly less biomass. However, it had a higher leaf water potential at wilting than the other species. Conversely, B. napus had an increased value for biomass and water content, but decreased leaf water and osmotic potentials. It was concluded that B. napus is the most drought-tolerant, followed by B. juncea and B. rapa and that B. carinata is the most sensitive. Zhu et al. (2011) used principal component, regression and clustering analyses to evaluate the drought tolerance of 49 rapeseed genotypes at the flowering stage. Plants were grown under watered and drought conditions, and morphological and agronomic traits (e.g., lateral root dry weight, stem diameter, number of green leaves, leaf area, plant height, number of primary branches, number of pods per plant, number of seeds per plant, number of seeds per pod, 1000-seed weight and yield per plant) were recorded. As a result, 20 lines were classified as drought-tolerant. However, evaluation of drought tolerance of genotypes at the late developmental stage (i.e., reproductive stage) is laborious and time-consuming. It also requires a greenhouse or rain shelter with a large area, which is not available in most cases. Therefore, a rapid, simple and high-throughput method is needed.
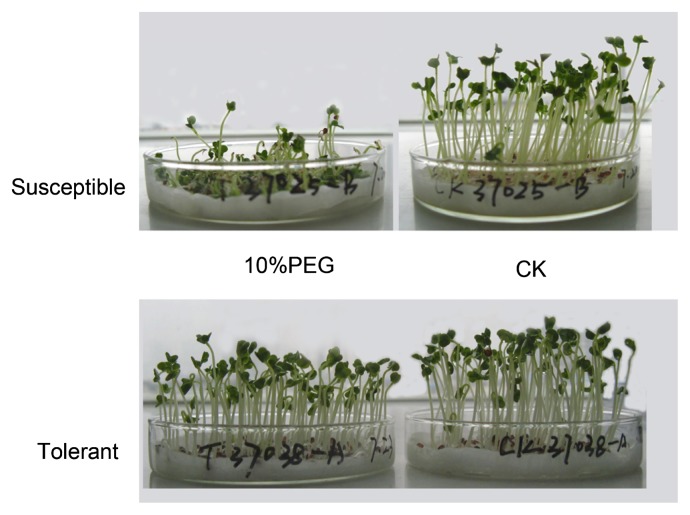
Evaluation of drought tolerance at the germination or seedling stage is now an available method, and it allows a high-throughput screening of germplasm in a laboratory and can greatly reduce the time and labour for field testing at a later developmental stage (Li et al. 2012, Yang et al. 2007). Yang et al. (2007) grew uniformly germinated seedlings under conditions of drought stress simulated by 10% polyethylene glycol 6000 (PEG6000) solution or moistened condition as a control (Fig. 1). Biological traits such as length of seedlings (LS), percentage of surviving seedlings (PSS) and RVI were recorded. The results showed that RVI varied from 0.32 to 0.79, with an average of 0.49, which could be used as an index for drought tolerance at the germination stage. In another study, plants of 17 rapeseed genotypes were grown in pots until the 3–5 leaf stage and then subjected to a drought regime in a rain shelter for one week. LWI had the largest C.V. (coefficient of variation) and was closely associated with eight other physiological indices, indicating that it can be used as a key index for drought tolerance at the seedling stage (Li et al. 2012).
Fig. 1.
Evaluation of drought tolerance for Brassica napus at germination stage. Uniformly germinated seeds (2 d) were transferred into petri dishes, which contained 3 layers of filter paper soaked with 10 ml of 10% polyethylene glycol 6000 (PEG6000) solution to simulate drought stress (10% PEG) or water as a control (CK), and allowed for additional 7 d of growth. Biological traits were then recorded to determine the tolerance level of each genotype (Yang et al. 2007).
The above studies have demonstrated that drought tolerance in plants is a very complex trait and can be evaluated by many indicators. However, the existing large numbers of indicators make it difficult to choose and apply in breeding practice. In the context of crop production, seed yield is the final target. Taking this concept in mind, some drought-related phenotypic traits can be incorporated into the equation: yield = water use (WU) × WUE × harvest index (HI) (Passioura 1977). This equation allows us to genetically depict the drought tolerance mechanism and postulate a trait-based breeding scheme. Traits associated with WU include deep rooting, osmatic adjustment (OA), seedling vigor and ground cover (Salekdeh et al. 2009). Deep rooting allows plants to find more water in the lower part of the soil, which can be indirectly reflected by other measurements, such as canopy temperature (Blum 1988).
Drought impact on agronomical traits
Drought can severely affect seed germination, plant growth, flowering, seed yield and quality as well. The degree of these impacts depends on the plant physiological, biochemical and molecular biological processes, as well as the ability of the plant to adapt to drought stress (Massonnet et al. 2007). It has been reported that the performance of most agro-morphological characteristics, such as plant height, primary branches per plant, secondary branches per plant, and seed yield of mustard, is greatly reduced under drought conditions (Chauhan et al. 2007). Seed fatty acid (FA) composition, oil content and protein content of rapeseed are also affected by drought stress, as reported by Aslam et al. (2009). When moderate drought conditions further developed into severe drought conditions at the growing season, decreases were found in oleic acid (by 3.8%) and saturated FA (by 0.4%), while increases occurred in linoleic acid (by 2.0%) and linolenic acid (by 1.7%). Furthermore, seed oil decreased by 3.2% and protein in meal increased by 3.9% under drought stress. The impact of drought on the root system has also been addressed. Wang, L. et al. (2005) found that roots were able to forage for fixed water patches by selectively placing themselves in the wet part of the pot. Under alternate watering, roots were as effective as in normal watering in terms of water-use efficiency. Compared with uniform watering, up to a 10% increase in shoot biomass was observed both in patchy and alternate watering regimes.
Physiochemical responses to drought
The physiologically relevant indicators of drought effects are the water content and the water potential of plants (Jones 2007). A wide range of physiological processes can be triggered by a decrease of water potential. Some of these responses are either directly attributed to the changing water status or to the flux of phytohormones caused by water stress (Chaves et al. 2003). Adaptive responses are observed as a consequence of such changes, including early flowering and growth inhibition. The accumulation of certain new metabolites is frequently observed in acclimated plants to improve plant functions under drought stress (Pinhero et al. 1997). In rapeseed, reduced relative water contents, osmotic potential and potassium contents, as well as increased total greenness and proline contents, were observed under various levels of water stresses (Alikhan et al. 2010). Total dry matter (TDM), leaf area index (LAI), relative growth rate (RGR) and crop growth rate (CGR) were overwhelmingly varied among the rapeseed cultivars and were greatly affected by drought stress (Moaveni et al. 2010). In mustard, the drought-tolerant cultivar was observed to have a higher OA value, higher proline contents and activity of proline biosynthetic enzyme P5CS (pyrroline-5-carboxylate synthetase), but lower activity of the proline degrading enzyme PO (proline oxidase) (Phutela et al. 2000).
The most potent and active member of Brassinosteroids (BRs), i.e., 28-homobrassinolide (HBL), counteracts abiotic stress in plants (Sharma et al. 2008). Plant growth and photosynthesis were greatly inhibited when exposed to drought stress but could be recovered by the subsequent HBL treatment, which brought about an increase of activity of antioxidant enzymes and proline content in leaves. Amelioration of drought stress has been suggested to be attributable to the elevated antioxidant system (Fariduddin et al. 2009). Water stress could expedite leaf senescence and impede leaf development as N depletes, although the symptom of wilting in N-deprived plants was not so overt as that in drought-affected plants (Albert et al. 2012). It was estimated that 25–85% of the total free amino acid in the pool resulted from the accumulation of proline. A great deal of proline was reallocated to emerging leaves from senescing leaves since the emerging leaves cannot accumulate proline effectively.
From the above studies, it is clear that proline contents are increased, stomata close and photosynthesis is inhibited under drought stress. Such responses are dominantly mediated by plant hormone, i.e. abscisic acid (ABA) (Wilkinson and Davies 2010). ABA is a well-characterized hormone that can serve as messenger for environmental adversity (Raghavendra et al. 2010). In Arabidopsis seedlings, Huang et al. (2008) has illustrated drought-enhanced ABA biosynthesis, which led to an increase in ABA contents. Interestingly, the drought-tolerant cultivars tended to have more ABA than susceptible ones (Thameur et al. 2011, Veselov et al. 2008). These findings have provided a simple approach of enhancing plant drought tolerance. Indeed, exogenous application of ABA can really enhance the tolerance of plants or plant cells to drought (Lu et al. 2009). This may be due to the fact that a high level of endogenous ABA can induce stomatal closure and consequently minimize the water loss by transpiration.
Classical genetics and QTL mapping of traits related to drought tolerance
Due to the complexity of drought tolerance, few genetic studies on Brassica crops have been reported. A pilot study was presented by Richards and Thurling (1979a). They showed that leaf proline accumulation, leaf chlorophyll stability, and germination at low osmotic potentials were largely controlled by genetic factors and were responsive to breeding selection. Broad-sense heritability of proline accumulation was 40% in both B. napus and B. rapa, while those of germination rate in B. rapa and chlorophyll stability in B. napus were 55% and 64%, respectively. However, narrow-sense heritabilities of both proline accumulation and germination rate were markedly lower than the broad-sense heritabilities of these traits. Cheema and Sadaqat (2004) analyzed the yield and its components of six populations (i.e., P1, P2, F1, F2, BC1, and BC2) derived from each of the three crosses under watered and drought conditions. For all plant traits studied, the genotype × environment interaction was significant. Most of the traits were under non-additive control, although both additive and non-additive gene actions were involved. A duplicate or complementary type of epistasis was also observed. Oil content is dominantly inherited, and plant height, days to first bud and days to maturity are controlled by more than two genes acting in a complex fashion. Yang et al. (2008) estimated genetic parameters of drought tolerance during seed germination using a complete diallel crossing design. General combining ability (GCA) was significantly different in the six germination traits. Apart from RVI, special combining ability (SCA) values for the other five traits were also significantly different. The highest GCA effects for the six traits were found in ‘Zhongshuang9’.
Plant morphological responses to water stress are controlled by many genes and known as quantitative traits. In B. oleracea, Hall et al. (2005) carried out quantitative trait loci (QTL) mapping of WUE (indicated by carbon isotope ratios) and photosynthetic traits. As a result, thirty significant QTLs were identified on seven linkage groups, explaining 3.4–36.6% of the phenotypic variance in the measured traits. QTLs for several traits (e.g., carbon isotope discrimination, photosynthetic capacity, nitrogen content, leaf thickness and stomatal density) were found to share the same location in a physiologically meaningful way, indicating that gene(s) at these loci may have pleiotropical effects on water-use- and photosynthesis-related traits.
Molecular basis of drought tolerance in Brassica crops
Drought tolerance in plants is recognised as a quantitative trait conditioned by many genes through various pathways. When drought stress is perceived by the plant, expression patterns are changed in genes, including those involved in water transport, osmotic balance, oxidative stress and damage repair (Table 2). In recent years, differentially expressed genes have been highlighted by new technologies, such as RNA-Seq, and bioinformatic software tools have been also developed to accelerate the discovery of new stress response genes from publicly available databases.
Table 2.
Summary of drought- and/or salt-tolerant-associated genes mentioned in this review
| Species | Genes | Encoding protein | Tolerant to | Literature |
|---|---|---|---|---|
| B. rapa | BrERF4 | Ethylene-responsive factors | Drought and salt | Seo et al. 2010 |
| Arabidopsis | DREB1A | Dehydration response element binding protein | Drought, salt and freezing | Kasuga et al. 1999 |
| B. napus | BnLAS | Transcriptional regulator members in GRAS family | Drought | Yang et al. 2011 |
| B. napus | AnnBn1 | Membrane-binding proteins for Ca2+ | Drought | Xiao et al. 2012 |
| Arabidopsis | FTA | Farnesyl transferase | Drought | Wang, Y. et al. 2005 |
| Arabidopsis | AtFTB | β-subunit of Farnesyl transferase | Drought | Wang et al. 2009 |
| Arabidopsis | SOS1 | Plasma membrane-bound Na+/H+ antiports | Salt | Martinez-Atienza et al. 2007, Shi et al. 2000 |
| Arabidopsis | AtHKT1 | Na+ transporter | Salt | Berthomieu et al. 2003 |
| Arabidopsis | AtNHX1 | Vacuolar Na+/H+ antiporter | Salt | Zhang and Blumwald 2001, Zhang et al. 2001 |
| Arthrobacter globiformis | codA | Choline oxidase | Salt | Wang et al. 2010 |
| B. napus | BnLEA4-1 | Late-embryogenesis abundant proteins in group 4 | Salt | Dalal et al. 2009 |
| B. juncea | BrECS | Glutamylcysteine synthetase | Salt | Bae et al. 2013 |
Many studies have attempted to elucidate the molecular mechanism of drought tolerance in plants. With the help of next-generation sequencing technology, Yu et al. (2012) conducted a pilot genome-wide analysis of gene expression in leaves during a drought treatment in B. rapa using a Solexa Illumina array. A total of 1,092 drought-responsive genes were identified, among which 37 were transcription factors. Twenty-eight were involved in signal transduction and 61 were involved in water- and osmo-sensing-responsive pathways. Similarly, Dong et al. (2012) identified 308 down-regulated genes and 248 up-regulated genes under drought in Sinapis alba. Differentially expressed genes were found to be involved in cell division and catalytic and metabolic processes, as revealed by gene ontology (GO) analysis.
Roots are believed to play an important role in drought response, because in plants they are the major organs for water uptake and thus can first experience and sense water deficit. To investigate the transcriptional changes associated with root hydrotropism, Moriwaki et al. (2010) performed a whole-genome microarray analysis of Arabidopsis to monitor the transcription levels of 22,810 genes during the early phase of the hydrotropic response. One or two hours after hydrotropic stimulation, 793 genes were identified with substantially changed transcript levels. Among these genes, many were involved in response to abscisic acid (ABA) or water stress, indicating that ABA and water-stress-mediated signal transductions are possible mechanisms for the root hydrotropic response. Such studies have provided us a large number of candidate drought-tolerant genes, which can be manipulated (mainly via overexpression) to achieve enhanced drought performance in target crops.
With the availability of massive gene expression data in the public domain, a new bioinformatic tool was developed by improving the SVM-RFE (Support Vector Machine— Recursive Feature Elimination) method to predict key genes involved in water stress (Liang et al. 2011). The SVM-RFE is a simple and efficient algorithm that can be used to conduct gene selection in a backward elimination procedure (Guyon et al. 2002). Using 22 sets of Arabidopsis gene expression data from the Gene Expression Omnibus (GEO, a public functional genomics data repository at http://www.ncbi.nlm.nih.gov/geo/), Liang et al. (2011) analyzed the top 10 genes predicted by the SVM-RFE to be involved in water tolerance. Seven of them were confirmed to be connected to known biological processes in drought resistance, exhibiting the power of this method in analyzing plant microarray data for studying genotype-phenotype relationships. Similarly, Li et al. (2008) reported a cis-regulatory element-based computational approach to genome-wide identification of genes putatively responding to various osmotic stresses in Arabidopsis. With this approach, the 500 top-scoring genes were selected, 91.3% of which were related to the stress response and ABA response in GO enrichment analysis. These predicted results can be further confirmed by RT-PCR analysis. Among the 41 top-ranked genes, 27 (65.8%) were found to have altered transcript levels under various osmotic stress treatments.
In recent years, the functions of some drought-related genes have been well characterized. Seo et al. (2010) reported that overexpression of an ethylene-responsive factor (ERF) from B. rapa (BrERF4) led to improved tolerance to salt and drought stresses in Arabidopsis. Expression of BrERF4 was induced by ethylene or methyl jasmonate, but not by ABA or NaCl. Thus, BrERF4 seems to be activated through a network of signalling pathways in response to salinity and drought. In another study, an Arabidopsis lateral suppressor (LAS) homologous gene (BnLAS) was cloned from B. napus, which was highly expressed in the roots, shoot tips, lateral meristems and flower organs (Yang et al. 2011). Constitutive overexpression of BnLAS in Arabidopsis plants exhibited enhanced drought tolerance and increased recovery after drought treatment. It is expected that genetic improvement of drought tolerance in rapeseed can also be achieved by overexpressing this gene, since Arabidopsis is closely related to rapeseed.
Annexins are a multifunctional family of membrane-binding proteins that have an important role in Ca2+ signalling pathways in both animal and plant cells (Mortimer et al. 2008). It is known that genes encoding Annexins in B. juncea play an important role in abiotic stress responses (Jami et al. 2008). Xiao et al. (2012) cloned an Annexin gene (AnnBn1) from drought-tolerant rapeseed genotype ‘Q2’. Four repeated annexin superfamily domains and a calcium-binding motif were found in the deduced amino acid sequence of AnnBn1. AnnBn1 was expressed in leaves, roots, stems and shoots at a similar level under normal condition, but the expression of AnnBn1 increased 2–4 fold in stems and shoots and 10 fold in leaves and roots within 30 h after drought stress treatment, suggesting that this is a drought-stress-responsive gene. Although AnnBn1 is up-regulated (especially in roots and leaves) under drought stress, it is not so straightforward to infer that enhanced drought tolerance can be achieved by overexpressing this gene in rapeseed, and thus further investigation is needed.
Breeding of drought-tolerant Brassica crops
Breeding and utilization of drought-resistant varieties are ideal ways to deal with drought problems in agriculture. Due to the complexity of drought as a stress signal, elucidating drought tolerance mechanisms has been a major challenge for plant biologists. Nevertheless, some efforts have been made to genetically improve drought tolerance in Brassica crops.
One of the earliest attempts was made by Richards and Thurling (1979b). B. rapa grown in a drought environment was assessed for grain yield improvement response to joint selection for yield, harvest index, 1000-seed weight and seeds per pod. It was shown that the joint selection was 20% more effective than direct selection for yield only under drought. In the B. napus population, a genetic advancement could be achieved by direct selection for flowering time or for harvest index, which was equal to or even better than by direct selection for yield. Interestingly, joint selection for flowering time and yield resulted in a 16% greater increase in yield.
The wild relatives of Brassica crops, such as Moricandia arvensis and S. alba, exhibit greater tolerance to drought (Warwick 1993). To transfer this valuable trait into Brassica crops, intergeneric crosses were made between B. oleracea and M. arvensis utilizing embryo rescue. By backcrossing F1 with B. oleracea, some sesquidiploid BC1 plants with 2n = 32 chromosomes were developed. In the BC2 population, some monosomic addition lines of B. oleracea carrying a single chromosome from M. arvensis were generated, which could be used in future genetic and breeding research (Bang et al. 2007). Similarly, S. alba plants were pollinated with B. oleracea, and the fertile F1 plants were pollinated with B. oleracea to obtain BC1 plants. BC1 plants had 18 chromosomes of B. oleracea and one, five or six additional S. alba chromosomes. These monosomic alien addition lines can be incorporated into the breeding program of B. oleracea (Wei et al. 2007).
In the past decade, a promising breakthrough has been made in understanding phytohormone ABA biosynthesis and the signal transduction pathway (Century et al. 2008). To avoid potential water deficit damage, plants can rapidly accumulate ABA which induces stomatal closure to reduce transpiration. Farnesyl-transferase (FTA) is a key negative regulator controlling ABA sensitivity in the guard cells. Wang, Y. et al. (2005) showed that the plant response to ABA and drought tolerance in Arabidopsis could be enhanced by down-regulation of either the α- or β-subunit of FTA. Interestingly, down-regulation of FTA in rapeseed also brought about significant reduction in stomatal conductance and water transpiration under drought stress. Furthermore, transgenic rapeseed plants having an rd29A:anti-AtFTB construct were more resistant to water-deficiency-induced seed abortion during flowering (Wang et al. 2009). Seed yields of transgenic rapeseed plants were as high as the wild-type controls under watered conditions in field experiments, but significantly higher than controls under moderate drought stress conditions at the flowering stage. Later, they found that conditional and specific down-regulation of FTA in rapeseed with a drought-induced promoter resulted in much better yield protection against drought stress in the field. Using this molecular strategy, they have made significant progress in engineering drought tolerance in B. napus.
Physiological and genetic studies on salt tolerance in Brassica
Methods for evaluation of salt tolerance
Salt tolerance is a complex trait to study for the following reasons: (a) salt tolerance can only be evaluated under stress conditions, which can affect multiple physiological responses of the plants; (b) salt tolerance is a quantitative trait which requires an efficient and effective means to quantify the tolerance level; (c) the “salt” in “salt stress” is often ambiguous as it can be different mineral salts, such as NaCl, MgCl2, and CaCl2; despite the high-frequency use of NaCl for salinity, we cannot ignore the harm of other ions; and (d) other physiological stresses (e.g., drought, extreme acidity and alkalinity) are often associated with salt-stressed plants, which also makes this trait more difficult to study. Therefore, efficient and effective methods, including plant culture under salt conditions, trait detection and grading to evaluate the salt tolerance degree, should be employed in the primary stage of study.
Growing plants under some controllable conditions (e.g., hydroponics) is commonly used for salt tolerance studies because very few natural salty soils can provide a consistent and representative condition (Flowers 2004). The model plant Arabidopsis is usually germinated and grown either on an agar medium and/or in pots, where it can complete its whole life cycle. For Brassica crops and radish, big-volume pots or containers in controlled conditions (potted soils or hydroponics) are necessary for growth. A few experiments for yield evaluation have been conducted in salty land.
It is notable that salt tolerance of Brassicaceae species is likely controlled by different genetic components, which have been revealed by salt tolerance performances at different developmental stages (Almodares et al. 2007, Ashraf and Sharif 1998, El Madidi et al. 2004, Islam and Karim 2010, Quesada et al. 2002). For example, Quesada et al. (2002) found that Arabidopsis accessions with good germination rates showed more reductions in fresh weight and dry weight at the vegetative stage under salt stress than the poorly germinating ones. Therefore, salt tolerance tests in the entire life cycle or at the most salt-sensitive stages would be required for comparison of salt tolerance of different lines.
Artificial salt stress methods, such as gradual stress and salt shock, may lead to outcomes different from those by field tests (Shavrukov 2013). Imposition of salt stress by gradual exposure to NaCl rather than salt shock has been recommended for genetic and molecular studies because it reflects more closely natural incidences of salinity stress (Katori et al. 2010). However, the ‘ideal’ type of gradual salt application is technically difficult. Katori et al. (2010) developed a method named salt acclimation (SA) assay, in which plants were transferred to a high-salinity medium following placement on a moderate-salinity medium for 7 d. There was no significant difference in salt tolerance between Arabidopsis ecotypes ‘Col-0’ and ‘Bu-5’ without SA, while Bu-5 showed higher salt tolerance than ‘Col-0’ plants with SA (Katori et al. 2010).
Researchers are seeking an easy method or an ideal trait value to forecast the salt tolerance to enable more efficient selection of tolerant crop types or tolerant genotypes. Munir et al. (2013) found that the photosynthetic rate could be an effective selection criterion under a salt regime. Photosynthetic capacity, proline and glycinebetaine accumulation ability, and ion discrimination can be used as potential biochemical or physiological selection criteria for salt tolerance in canola (Ulfat et al. 2007). Kumar et al. (2009) revealed that the transcript accumulation pattern for various salt overly sensitive (SOS) members after 24 h of salt stress in various cultivars showed a strong positive correlation with salt tolerance among Brassica species. Khayat et al. (2010) considered shoot Na+ content to be a useful criterion. Ashrasf and Ali (2008) suggested that relative cell membrane permeability and activities of antioxidant enzymes (superoxide dismutase, catalase and peroxidase) could be very effective in identifying canola cultivars with high salt tolerance. So far, there are no uniform criteria applied to the assessment of salt tolerance. A polygon method summarizing various recorded phenotypes can be applied to compare the salinity tolerance of different plants (Orsini et al. 2010). In such a method, each apex of a polygon points to the detected phenotype. The comparative value of a certain phenotype is displayed on the line from the centre to the apex proportionally. Therefore, every phenotype has a point on the corresponding line and an inside polygon is formed by connecting the points. The area of the inner polygon clearly represents the salinity tolerance of a certain species.
Germplasm screening of Brassicaceae
There is significant interspecific and intraspecific variation for salt tolerance. Thellungiella salsuginea grows in a salt environment with a life cycle and genome background similar to its close relative Arabidopsis, and therefore, this species can be used as a model plant for conducting salt tolerance research (Wu et al. 2012). Orsini et al. (2010) collected 11 wild relatives of Arabidopsis from different environments and identified four categories of tolerance: (i) halophytic (T. salsuginea and Thellungiella parvula), (ii) highly tolerant (Lepidium densiflorum and Lepidium virginicum), (iii) moderately tolerant (Malcolmia triloba, Hirschfeldia incana, Descuraina pinnata), and (iv) marginally better than Arabidopsis (Thlaspi arvense, Sisymbrium officinale, Barbarea verna) based on the overall growth performance. The authors suggested that T. parvula can be a new model species for comparative analyses of halophytes.
Arabidopsis itself is also a good model species for salt tolerance studies. Quesada et al. (2002) first observed the natural variation of 102 wild-type races of Arabidopsis in salt tolerance during germination and vegetative growth, and screened the most salt-tolerant accessions, i.e., ‘Ak-1’ and ‘La-1’, and the most salt-sensitive ones, i.e., ‘Bu-18’ and ‘Hodja-Obi-Garm’. Galpaz and Reymond (2010) tested 87 lines of Arabidopsis from Asian and European countries and identified ‘Sha’, ‘Neo-2’ and ‘Neo-3’ as the most salt-tolerant ones. According to the vegetative growth of 350 accessions of Arabidopsis under salt stress, Katori et al. (2010) selected salt-tolerant accessions, i.e., ‘Bu-5’, ‘Bur- 0’, ‘Ll-1’, ‘Wl-0’, and ‘Zu-0’; and salt-sensitive accessions, i.e., ‘Bch-4’, ‘Cvi-0’, ‘Sg-2’, ‘Sh-0’, and ‘Col-0’. Variation in salt tolerance appears to have a geographic pattern with Western European and Swedish samples having the highest salt tolerance and Northern and Southern Europe samples having the lowest salt tolerance (DeRose-Wilson and Gaut 2011).
Assessment of salt tolerance has also been performed in Brassica and its closely related species. Ashraf et al. (2001) and Nazir et al. (2001) compared salt tolerance of amphidiploid and diploid Brassica species and found that under salt stress, shoot and root weights and seed yield of the three amphidiploid species were significantly greater than those of their ancestral diploids. They suggested that the salt tolerance of these species originated from A and C genomes. Kumar et al. (2009) explored the variation of salt tolerance within six common Brassica species plus Eruca sativa and Brassica tournefortii by assessing the morphological, physiological and biochemical parameters. B. juncea, which had been thought to be the most salt-tolerant species, showed the least decrease of shoot length and root length, and lesser electrolyte leakage, higher proline content and higher K+/Na+ ratio than the other species. Munir et al. (2013) assessed the physiological and biochemical responses of winter vegetable radish (Raphanus sativus L.) to salt stress and found that salt stress significantly reduced chlorophyll contents in ‘Deci White’ and ‘Lal Pari’. Higher contents of chlorophyll a and chlorophyll b and a higher chlorophyll a/b ratio were found in variety ‘Deci White’ while such contents were lower in ‘Lal Pari’ under different levels of sodium chloride. Noreen et al. (2012) studied salt-induced changes in growth, various gas exchange characteristics, and ion accumulation during a greenhouse experiment on six radish cultivars and found that ‘Mannu Early’ and ‘Desi’ were higher in shoot and root dry weights than the other cultivars, and thus, they were ranked as highly salt-tolerant. Salt-tolerant lines in Brassica crops have been screened by various methods. Some varieties (lines) were approved as salt-tolerant genotypes and used in some studies (Ashraf and McNeilly 2004). It is important that the performances of the screened germplasms in real salinity fields be used to confirm their salt tolerance ability.
Classical genetics of salt tolerance in Brassicaceae
Exploration of the heritable potential of a certain trait within the existing germplasm for a given crop would supply information on factors such as salt tolerance for breeders. Rezai and Saeidi (2005) estimated the genetic parameters for different characteristics related to salt tolerance of rapeseed (B. napus L.) by using 8 × 8 diallel crosses. The results indicated the involvement of both additive and non-additive gene actions in the inheritance of characteristics. High narrow-sense heritability estimates were observed for Ca2+, K+, Na+, K+/Na+, Ca2+/Na+ and stress tolerance index, indicating the prime importance of additive effects in their genetic control. Qiu and Li (2009) investigated genetics of salt tolerance in B. rapa by an eight-parent complete diallel cross design and showed that salt tolerance was mainly controlled by dominant genes with an additive effect. The dominant effect played a major role and over-dominance might have existed in salt tolerance. Long et al. (2013) analyzed multi-family generations (P1, F1, P2, B1, B2 and F2) derived from a cross of ‘DT16’ (a salt-tolerant B. napus line) × ‘NY16’ (a salt-sensitive B. napus line) and found that salt tolerance was dominated by at least three major genes with additive-dominance effects. On the whole, all the outcomes were in accordance with former findings by Thakral and Prakash (1998) in B. juncea, Kumar and Yadav (1985) in B. rapa and Thakral and Singh (1994) in B. carinata.
QTL mapping of salt tolerance
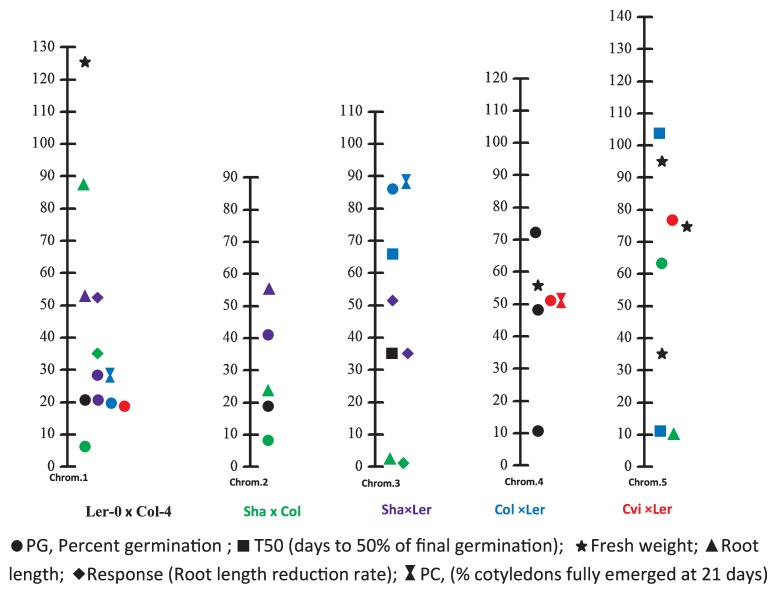
Interesting results have been obtained by independent studies on salt tolerance in Brassicaceae, especially in Arabidopsis. Most of the mapped QTLs controlling the salt tolerance were different from each other, since the mapping populations were different and the investigated traits were not all the same. Herein, we summarize the QTLs for salt tolerance in A. thaliana in three studies (DeRose-Wilson and Gaut 2011, Galpaz and Reymond 2010, Quesada et al. 2002) in Fig. 2. In these studies, the assessments of salt tolerance were all conducted using an agar medium. A common QTL for percent germination (PC) was detected at 20 cM on chromosome 1 across four RIL populations. This QTL was co-localized with the gene RAS1, a negative regulator of salt tolerance during seed germination and early growth, which was cloned from the Sha × Ler population (Ren 2010). Another QTL for PC located at 50 cM on chromosome 4 was detected in two RIL populations, Ler- 0 × Col-4 and Col × Ler. In this QTL, the candidate gene AT4G19030 (Lee et al. 2006), the expression level of which is reduced by ABA and NaCl, was predicted (DeRose-Wilson and Gaut 2011). The other eight QTLs of PC distributed in different positions. QTL mapping of T50 (days to 50% of final germination), root length, response (root length reduction rate), and PC (% cotyledons fully emerged at 21 days) was performed in two or three RIL populations, whereas no consistent QTLs of each trait were detected. These results suggest a complicated genetic work controlling salt tolerance and that the genetic determinants are different in different accessions. Some QTLs for different traits were overlapped: for example, QTLs for root length and response on chromosome 1 and 3, indicating that these two loci may contain genes controlling root length and those for salt tolerance exhibited by root growth. QTLs for PC and PG were also overlapped on chromosome 1, 3, and 4, in consistence with the results of phenotype assessments, in which PC and PG were highly correlated across 96 accessions (r2 = 0.69) of A. thaliana (DeRose-Wilson and Gaut 2011).
Fig. 2.
Comparison of QTLs mapped using different populations for salt tolerance in Arabidopsis. The QTLs in RIL populations Ler-0 × Col-4 (black) were detected by Quesada et al. (2002), those of Sha × Col-0 (green) and Sha × Ler (purple) were by Galpaz and Reymond (2010), and those of Col × Ler(blue) and Cvi × Ler(red) were by DeRose-Wilson and Gaut (2011).
Although QTL mapping remains the best method for identifying causal genes, it is quite laborious and time-consuming. Association mapping, which utilizes a higher number of historical recombination events that have occurred throughout the entire evolutionary process of the population, enables mapping of the genes in smaller genomic regions (Nordborg and Tavaré 2002). Using an association population of 96 accessions, DeRose-Wilson and Gaut (2011) identified ten genomic regions associated with phenotypic variation in salinity tolerance, and six locations were overlapped with the results of QTL analysis of two RIL populations. However, due to the small size of the association population, the results of the association mapping might be underpowered (Zhao et al. 2007). Genome-wide association studies with larger samples are considered to be more reliable and fruitful.
Molecular basis of salt tolerance in Brassicaceae
In Arabidopsis, some key genes related to salt tolerance have been identified. The SOS signalling pathway governing salt tolerance in Arabidopsis well clarifies the mechanisms of salt tolerance. The SOS pathway comprises three family members, i.e., the plasma membrane Na+/H+ antiporter SOS1, the protein kinase SOS2, and the Ca2+- binding protein SOS3. An increase of Na+ concentration leads to an elevation of intracellular Ca2+, and SOS3 binds Ca2+ and activates SOS2 to form a compound which phosphorylates the plasma membrane-localized SOS1. Finally, overexpression of SOS1 results in an efflux of more Na+ (Martinez-Atienza et al. 2007). SOS1 and SOS3 are constitutively expressed in Brassica crops, while the expression pattern for SOS2 amongst Brassica species was found to be very unique (Kumer et al. 2009). SOS2 may be upregulated by salinity stress in the roots of all the Brassica species except for B. juncea, which maintains high SOS2 transcripts even under non-stress conditions, indicating a very unique feature of B. juncea (Kumer et al. 2009). Strong correlation between transcript abundance for SOS pathway-related genes and salinity stress tolerance was observed in Brassica crops (Chakraborty et al. 2012, Kumer et al. 2009). In addition, Berthomieu et al. (2003) found that AtHKT1 is involved in the recirculation of Na+ from shoots to roots, presumably by promoting Na+ movement into phloems in shoots and translocation into roots. Apse et al. (1999) confirmed the function of AtNHX1 in salt tolerance through increased Na+ compartmentation in the vacuoles, which was also confirmed in tomato and B. napus by Zhang and Blumwald (2001) and Zhang et al. (2001).
Molecular mechanism of salt tolerance revealed in the model plants will facilitate identification of candidate genes and development of transgenic plants with salt tolerance in Brassica crops. Overexpression of genes encoding enzymes related to abiotic stresses enhanced crop salt tolerance. Transgenic B. rapa spp. chinensis plants expressing a choline oxidase (codA) gene from Arthrobacter globiformis showed a significantly higher net photosynthetic rate and a higher photosynthetic rate under high salinity conditions than wild-type plants (Wang et al. 2010). Dalal et al. (2009) proved that LEA4-1 plays a crucial role in salt stress tolerance during the vegetative stage of B. napus and that transgenic Arabidopsis plants overexpressing BnLEA4-1 have enhanced tolerance to salt stress. Glutathione (GSH) plays an important role in cell function and metabolism as an antioxidant. Bae et al. (2013) developed transgenic plants by introducing the γ-ECS ( Glutamylcysteine synthetase) gene from B. juncea (BrECS) into rice. Overexpression of BrECS confers plants with significantly enhanced tolerance to salinity by sustaining a cellular GSH redox state to avoid attacks from reactive oxygen species produced by salt. Furthermore, the transgenic rice plants also exhibited a 15–18% increase in grain yield under general paddy field conditions.
Breeding of salt-tolerant Brassica crops
Recently, studies of salt tolerance in plants have covered genetic mapping to molecular characterization of salt-induced genes. Increasing the understanding of biochemical pathways and mechanisms that participate in plant stress responses has made it possible to genetically improve the salinity performance of new varieties through various routes. Currently, transgenic plants have been used to test the effect of overexpression of specific plant genes that are known to be up-regulated by salt stress (Table 2). Great progress of salt tolerance has been made in major crops, such as rice, wheat and tomatoes. A number of QTLs have been mapped (Thomson et al. 2010) and some important genes have been cloned (Gaxiola et al. 1999, Ishitani et al. 2000, Liu et al. 2000, Ren et al. 2005, Rus et al. 2001, Shi et al. 2002, Zhu et al. 1998). However, studies on QTLs or genes controlling salt tolerance in Brassica oil crops are still very limited. To date, the breeding practice of salt tolerance in Brassica crops has been largely unsuccessful, although some salt-tolerant cultivars of B. juncea have been developed in India, e.g., ‘CS56’. Researchers and breeders endeavor to understand the mechanisms of salt tolerance and screen for stable salt-tolerant genotypes to use in breeding programs. Attempts have also been made to develop salt-tolerant transgenic Brassica crops with candidate genes with proven roles in ion homeostasis and osmolytes accumulation (Zhang et al. 2004).
Conclusion and future prospective
A series of studies using the phenotyping method have clearly revealed the physiological responses to drought and salinity in Arabidopsis as well the involved metabolic pathways. Through broad application of forward and reverse genetic approaches in model plants with powerful genomics and proteomics tools, great progress has been achieved in understanding the mechanisms of drought and salt tolerance, although we are still far from having a clear picture. However, since the Brassica A genome sequences from B. rapa have been published (Wang et al. 2011) and sequencing of the Brassica C genome from B. oleracea has also been completed and will be published soon (http://www.ocri-genomics.org/bolbase/). These data together with the known genes involved in drought and salt tolerance in model plants will serve as a fine platform for identification of “candidate genes”.
Phenotypic traits associated with drought-tolerant or salt-tolerant crops serve as important breeding tools in identifying stress-tolerant genotypes and in introducing such tolerance traits into cultivated genotypes. However, most modern cultivars of Brassica crops are sensitive to drought and salt stress and thus do not perform well under field stress conditions. Fortunately, genetic resources of drought and salt tolerance have been identified in their wild relatives. Advances in molecular markers and mapping technology promises opportunities to identify genes or QTLs for drought and salt tolerance. The identified drought- and salt-tolerant germplasms as well as DNA markers delineating QTLs for these traits can be used in marker-assisted selection (MAS) breeding programs of Brassica crops for areas threatened by drought and salt stresses. To develop reliable marker information for quantitative traits, more efforts, such as precise phenotyping in the field and minimizing environmental effects on the traits, should be made. Moreover, QTLs are required to be identified for individual developmental stages as well as for individual physiological parameters. Pyramiding of such QTLs may lead to the development of plants with improved drought or salt tolerance.
Although QTL mapping and MAS in Brassica crops has great potential for developing varieties with high yield and quality, very few drought- or salt-tolerant cultivars and lines have been developed, which is mainly due to the quantitative nature of stress tolerance, difficulty of transferring stress-tolerant traits from interspecific and intergeneric sources and the linkage drag between desirable and undesirable genes. Moreover, Brassica plants tend to be exposed to multiple stresses (i.e., drought and salinity), which makes the breeding of stress-tolerant lines more difficult using conversional strategies. Fortunately, the transgenic approach promises a substantial improvement in desired traits. Understanding the molecular mechanisms of drought and salt tolerance may lead to a generalized master mechanism for stress tolerance. This will be possible because there were many common features between the two stresses. For example, there is cross-talk between drought and salt stress, since both stresses will finally result in dehydration of the cell and osmotic imbalance. Engineered genes encoding organic osmolytes, plant growth regulators, antioxidants, late embryo-genesis abundant proteins, and transcription factors have been introduced into transgenic lines which performed well under controlled stress conditions (Table 2). In the future, it is desirable to use multiple tolerance mechanisms of drought and salt stresses to achieve high levels of tolerance for commercial exploitation in Brassica crops. There already exists an example for this target. The cis-acting dehydration response element (DRE) plays an important role in regulating gene expression in response to these stresses. Kasuga et al. (1999) showed that overexpression of cDNA encoding DREB1A (DRE binding protein) in Arabidopsis transgenic plants activated the expression of many stress-tolerant genes and resulted in improved tolerance to drought, salt and freezing. Under field conditions, plants are often subjected to multiple stresses. Transgenic approaches should be integrated with conventional breeding and molecular breeding as well as more recent innovative strategies. It is also imperative to note that most drought- and salt-tolerant transgenic lines have been developed using a single gene transformation, which may not be as productive as using transformation of many genes. Thus, it is considered to be a more logical approach to enhance crop stress tolerance by transferring a number of target genes.
Finally, drought and salinity stress is always accompanied by high temperature stress, especially in the tropics, which increases the water requirements of crops. The responses of crops to a combination of these stress factors appear to differ from the responses to a single stress (Pinto et al. 2010). Therefore, more attention should be paid to the joint impact of a combination of stresses of drought, salinity and high temperature. Exploiting the genetic variability in crop species may be a useful strategy to improve climate-change-related stress tolerance.
Acknowledgement
Cooperation of the authors was supported by the China-Japan Joint Research Program of the Chinese government (2012GR0080) and Japan Science and Technology Agency (J120000331).
Literature Cited
- Albert, B., Cahérec, F.L., Niogret, M.F., Faes, P., Avice, J.C., Leport, L. and Bouchereau, A. (2012) Nitrogen availability impacts oilseed rape (Brassica napus L.) plant water status and proline production efficiency under water-limited conditions. Planta 236: 659–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan, M., Ashraf, M.Y., Mujtaba, S.M., Shirazi, M.U., Khan, M.A., Shereen, A., Mumtaz, S., Siddiqui, M.A. and Kaleri, G.M. (2010) Evaluation of high yielding canola type Brassica genotypes/mutants for drought tolerance using physiological indices as screening tool. Pak. J. Bot. 42: 3807–3816 [Google Scholar]
- Almodares, A., Hadi, M.R. and Dosti, B. (2007) Effects of salt stress on germination percentage and seedling growth in sweet sorghum cultivars. J. Biol. Sci. 7: 1492–1495 [Google Scholar]
- Apse, M.P., Aharon, G.S., Snedden, W.A. and Blumwald, E. (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Ashraf, M. and Mehmood, S. (1990) Response of four Brassica species to drought stress. Envir. Exp. Bot. 30: 93–100 [Google Scholar]
- Ashraf, M. and O’Leary, J.W. (1996) Effect of drought stress on growth, water relations, and gas exchange of two lines of sunflower differing in degree of salt tolerance. Int. J. Plant Sci. 157: 729–732 [Google Scholar]
- Ashraf, M. and Sharif, R. (1998) Does salt tolerance varies in a potential oilseed Brassica carinata at different growth stages? J. Agron. Crop Sci. 181: 103–115 [Google Scholar]
- Ashraf, M., Nazir, N. and McNeilly, T. (2001) Comparative salt tolerance of amphidiploid and diploid Brassica species. Plant Sci. 160: 683–689 [DOI] [PubMed] [Google Scholar]
- Ashraf, M. and McNeilly, T. (2004) Salinity tolerance in Brassica oilseeds. Crit. Rev. Plant Sci. 23: 157–174 [Google Scholar]
- Ashraf, M. and Ali, Q. (2008) Relative membrane permeability and activities of some antioxidant enzymes as the key determinants of salt tolerance in canola (Brassica napus L.). Environ. Exp. Bot. 63: 266–273 [Google Scholar]
- Aslam, M.N., Nelson, M.N., Kailis, S.G., Bayliss, K.L., Speijers, J. and Cowling, W.A. (2009) Canola oil increases in polyunsaturated fatty acids and decreases in oleic acid in drought-stressed Mediterranean-type environments. Plant Breed. 128: 348–355 [Google Scholar]
- Bae, M.J., Kim, Y.S., Kim, I.S., Choe, Y.H., Lee, E.J., Kim, Y.H., Park, H.M. and Yoon, H.S. (2013) Transgenic rice overexpressing the Brassica juncea gamma-glutamylcysteine synthetase gene enhances tolerance to abiotic stress and improves grain yield under paddy field conditions. Mol. Breed. 31: 931–945 [Google Scholar]
- Bang, S.W., Sugihara, K., Jeung, B.H., Kaneko, R., Satake, E., Kaneko, Y. and Matsuzawa, Y. (2007) Production and characterization of intergeneric hybrids between Brassica oleracea and a wild relative Moricandia arvensis. Plant Breed. 126: 101–103 [Google Scholar]
- Berthomieu, P., Conéjéro, G., Nublat, A., Brackenbury, W.J., Lambert, C., Savio, C., Uozumi, N., Oiki, S., Yamada, K., Cellier, F.et al. (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+recirculation by the phloem is crucial for salt tolerance. EMBO J. 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, A. (1988) Plant Breeding for Stress Environments, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- Blum, A. (2005) Drought resistance, water-use efficiency, and yield potential are they compatible, dissonant, or mutually exclusive? Aust. J. Agric. Res. 56: 1159–1168 [Google Scholar]
- Century, K., Reuber, T.L. and Ratcliffe, O.J. (2008) Regulating the regulators: the future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol. 147: 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, K., Sairam, R.K. and Bhattacharya, R.C. (2012) Differential expression of salt overly sensitive pathway genes determines salinity stress tolerance in Brassica genotypes. Plant Physiol. Biochem. 51: 90–101 [DOI] [PubMed] [Google Scholar]
- Chauhan, J.S., Tyagi, M.K., Kumar, A., Nashaat, N.I., Singh, M., Singh, N.B., Jakhar, M.L. and Welham, S.J. (2007) Drought effects on yield and its components in Indian mustard (Brassica juncea L.). Plant Breed. 126: 399–402 [Google Scholar]
- Chaves, M.M., Maroco, J.P. and Pereira, J.S. (2003) Understanding plant responses to drought from genes to the whole plant. Funct. Plant Biol. 30: 239–264 [DOI] [PubMed] [Google Scholar]
- Cheema, K.L. and Sadaqat, H.A. (2004) Potential and genetic basis of drought tolerance in canola (Brassica napus): I. Generation mean analysis for some phenological and yield components. Int. J. Agric. Biol. 6: 1560–8530 [Google Scholar]
- Dalal, M., Tayal, D., Chinnusamy, V. and Bansal, K.C. (2009) Abiotic stress and ABA-inducible Group 4 LEA from Brassica napus play a key role in salt and drought tolerance. J. Biotechnol. 139: 137–145 [DOI] [PubMed] [Google Scholar]
- Deikman, J., Petracek, M. and Heard, J.E. (2012) Drought tolerance through biotechnology: improving translation from the laboratory to farmers’ fields. Curr. Opin. Biotechnol. 23: 243–250 [DOI] [PubMed] [Google Scholar]
- DeRose-Wilson, L. and Gaut, B.S. (2011) Mapping salinity tolerance during Arabidopsis thaliana germination and seedling growth. PLoS ONE 6: e22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C.H., Li, C., Yan, X.H., Huang, S.M., Huang, J.Y., Wang, L.J., Guo, R.X., Lu, G.Y., Zhang, X.K., Fang, X.P.et al. (2012) Gene expression profiling of Sinapis alba leaves under drought stress and rewatering growth conditions with Illumina deep sequencing. Mol. Biol. Rep. 39: 5851–5857 [DOI] [PubMed] [Google Scholar]
- El Madidi, S.E., Baroudi, B.E. and Aameur, F.B. (2004) Effects of salinity on germination and early growth of barley cultivars. Int. J. Agri. Biol. 6: 767–770 [Google Scholar]
- El Sharkawy, H. (1989) A review of genetic advances on breeding salt tolerant crops. In: Bouchet, R. (ed.) Reuse of low quality water for irrigation, CIHEAM, Bari, pp. 183–190 [Google Scholar]
- Fariduddin, Q., Khanam, S., Hasan, S.A., Ali, B., Hayat, S. and Ahmad, A. (2009) Effect of 28-homobrassinolide on the drought stress-induced changes in photosynthesis and antioxidant system of Brassica juncea L. Acta Physiol. Plant. 31: 889–897 [Google Scholar]
- Farooq, S. and Azam, F. (2001) Co-existence of salt and drought tolerance in Triticeae. Hereditas 135: 205–210 [DOI] [PubMed] [Google Scholar]
- Flowers, T.J. (2004) Improving crop salt tolerance. J. Exp. Bot. 55: 307–319 [DOI] [PubMed] [Google Scholar]
- Fukao, T. and Xiong, L. (2013) Genetic mechanisms conferring adaptation to submergence and drought in rice: simple or complex? Curr. Opin. Plant Biol. 16: 196–204 [DOI] [PubMed] [Google Scholar]
- Galpaz, N. and Reymond, M. (2010) Natural variation in Arabidopsis thaliana revealed a genetic network controlling germination under salt stress. PLoS One 5: e15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola, R.A., Rao, R., Sherman, A., Grisafi, P., Alper, S.L. and Fink, G.R. (1999) The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. USA 96: 1480–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon, I., Weston, J., Barnhill, S. and Vapnik, V. (2002) Gene selection for cancer classification using support vector machines. Mach. Learn. 46: 389–422 [Google Scholar]
- Hall, N.M., Griffiths, H., Corlett, J.A., Jones, H.G., Lynn, J. and King, G.J. (2005) Relationships between water-use traits and photosynthesis in Brassica oleracea resolved by quantitative genetic analysis. Plant Breed. 124: 557–564 [Google Scholar]
- Horneck, D.A., Ellsworth, J.W., Hopkins, B.G., Sullivan, D.M. and Stevens, R.G. (2007) Managing salt-affected soils for crop production. Pacific Northwest Extension publication, p. 24 http://extension.oregonstate.edu/catalog/pdf/pnw/pnw601-e.pdf [Google Scholar]
- Huang, D., Wu, W., Abrams, S.R. and Cutler, A.J. (2008) The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 59: 2991–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani, M., Liu, J., Halfter, U., Kim, C.S., Shi, W. and Zhu, J.K. (2000) SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M.M. and Karim, M.A. (2010) Evaluation of rice genotypes at germination and early seedling stage for their tolerance to salinity. Agriculturists 8: 57–65 [Google Scholar]
- Jami, S.K., Clark, G.B., Turlapati, S.A., Handley, C., Roux, S.J. and Kirti, P.B. (2008) Ectopic expression of an annexin from Brassica juncea confers tolerance to abiotic and biotic stress treatments in transgenic tobacco. Plant Physiol. Biochem. 46: 1019–1030 [DOI] [PubMed] [Google Scholar]
- Jones, H.G. (2007) Monitoring plant and soil water status: established and novel methods revisited and their relevance to studies of drought tolerance. J. Exp. Bot. 58: 119–130 [DOI] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K. and Shinozaki, K. (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Katori, T., Ikeda, A., Iuchi, S., Kobayashi, M., Shinozaki, K., Maehashi, K., Sakata, Y., Tanaka, S. and Taji, T. (2010) Dissecting the genetic control of natural variation in salt tolerance of Arabidopsis thaliana accessions. J. Exp. Bot. 61: 1125–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayat, P.N., Jamaati-e-Somarin, S., Zabihi-e-Mahmoodabad, R., Yari, A., Khayatnezhad, M. and Gholamin, R. (2010) Screening of salt tolerance canola cultivars (Brassica napus L.) World Appl. Sci. J. 10: 817–820 [Google Scholar]
- Kumar, D. and Yadav, I.S. (1985) Line × tester analysis for certain quantitative traits in brown sarson (B. campestris var. brown sarson) on normal and alkaline soils. Soc. Adv. Breed. Res. Asia Ocea. J. 17: 21–28 [Google Scholar]
- Kumar, G., Purty, R.S., Sharma, M.P., Singla-Pareek, S.L. and Pareek, A. (2009) Physiological responses among Brassica species under salinity stress show strong correlation with transcript abundance for SOS pathway-related genes. J. Plant Physiol. 166: 507–520 [DOI] [PubMed] [Google Scholar]
- Lawlor, D.W. and Tezara, W. (2009) Causes of decreased photosynthetic rate and metabolic capacity in water-deficient leaf cells: a critical evaluation of mechanisms and integration of processes. Ann. Bot. 103: 561–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, B.H., Kapoor, A., Zhu, J. and Zhu, J.K. (2006) STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 18: 1736–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Zhu, Y.M., Liu, Y., Shu, Y.J., Meng, F.J., Lu, Y.M., Bai, X., Liu, B. and Guo, D.J. (2008) Genome-wide identification of osmotic stress response gene in Arabidopsis thaliana. Genomics 92: 488–493 [DOI] [PubMed] [Google Scholar]
- Li, Z., Wu, B.J., Lu, G.Y., Chen, Y., Zou, C.S. and Zhang, X.K. (2012) Differences in physiological responses of Brassica napus genotypes under water stress during seedling stage. Chin. J. Oil Crops Sci. 34: 033–039 [Google Scholar]
- Liang, Y., Zhang, F., Wang, J., Joshi, T., Wang, Y. and Xu, D. (2011) Prediction of drought-resistant genes in Arabidopsis thaliana using SVM-RFE. PLoS One 6: e21750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Ishitani, M., Halfter, U., Kim, C.S. and Zhu, J.K. (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97: 3730–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, W.H., Pu, H.M., Zhang, J.F., Qi, C.K. and Zhang, X.K. (2013) Screening of Brassica napus for salinity tolerance at germination stage. Chin. J. Oil Crop Sci. 35: 271–275 [Google Scholar]
- Lu, S., Su, W., Li, H. and Guo, Z. (2009) Abscisic acid improves drought tolerance of triploid bermudagrass and involves H2O2- and NO-induced antioxidant enzyme activities. Plant Physiol. Biochem. 47: 132–138 [DOI] [PubMed] [Google Scholar]
- Martinez-Atienza, J., Jiang, X., Garciadeblas, B., Mendoza, I., Zhu, J.K., Pardo, J.M. and Quintero, F.J. (2007) Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 143: 1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet, C., Costes, E., Rambal, S., Dreyer, E. and Regnard, J.L. (2007) Stomatal regulation of photosynthesis in apple leaves: evidence for different water-use strategies between two cultivars Catherine. Ann. Bot. 100: 1347–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moaveni, P., Ebrahimi, A. and Farahani, H.A. (2010) Physiological growth indices in winter rapeseed (Brassica napus L.) cultivars as affected by drought stress at Iran. J. Cereals Oilseeds 1: 11–16 [Google Scholar]
- Moriwaki, T., Miyazawa, Y. and Takahashi, H. (2010) Transcriptome analysis of gene expression during the hydrotropic response in Arabidopsis seedlings. Envir. Exp. Bot. 69: 148–157 [Google Scholar]
- Mortimer, J.C., Laohavisit, A., Macpherson, N., Webb, A., Brownlee, C., Battey, N.H. and Davies, J.M. (2008) Annexins: multifunctional components of growth and adaptation. J. Exp. Bot. 59: 533–544 [DOI] [PubMed] [Google Scholar]
- Munir, S., Hussain, E., Bhatti, K.H., Nawaz, K., Hussain, K., Rashid, R. and Hussain, I. (2013) Assessment of inter-cultivar variations for salinity tolerance in winter radish (Raphanus sativus L.) using photosynthetic attributes as effective selection criteria. World Appl. Sci. J. 21: 384–388 [Google Scholar]
- Nakashima, K., Ito, Y. and Yamaguchi-Shinozaki, K. (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149: 88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazir, N., Ashraf, M. and Rasul, E. (2001) Genomic relationships in oilseeds Brassicas with respect to salt tolerance-photosynthetic capacity and ion relations. Pak. J. Bot. 33: 483–501 [Google Scholar]
- Nordborg, M. and Tavaré, S. (2002) Linkage disequilibrium: what history has to tell us. Trends Genet. 18: 83–90 [DOI] [PubMed] [Google Scholar]
- Noreen, Z., Ashraf, M. and Akram, N.A. (2012) Salt-induced regulation of photosynthetic capacity and ion accumulation in some genetically diverse cultivars of radish (Raphanus sativus L.). J. Appl. Bot. Food Qual. 85: 91–96 [Google Scholar]
- Orsini, F., D’Urzo, M.P., Inan, G., Serra, S., Oh, D.H., Mickelbart, M.V., Consiglio, F., Li, X., Jeong, J.C., Yun, D.J.et al. (2010) A comparative study of salt tolerance parameters in 11 wild relatives of Arabidopsis thaliana. J. Exp. Bot. 61: 3787–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passioura, J.B. (1977) Grain yield, harvest index and water use of wheat. J. Aust. Inst. Agric. Sci. 43: 117–120 [Google Scholar]
- Pennisi, E. (2008) Plant genetics. The blue revolution, drop by drop, gene by gene. Science 320: 171–173 [DOI] [PubMed] [Google Scholar]
- Phutela, A., Jain, V., Dhawan, K. and Nainawatee, H.S. (2000) Proline metabolism under water stress in the leaves and roots of Brassica juncea cultivars differing in drought tolerance. J. Plant Biochem. Biotechnol. 9: 35–39 [Google Scholar]
- Pinhero, R.G., Rao, M.V., Palyath, G., Murr, D.P. and Fletcher, R.A. (1997) Changes in activities of antioxidant enzymes and their relationship to genetic and paclobutrazol-induced chilling tolerance of maize seedlings. Plant Physiol. 114: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, R.S., Reynolds, M.P., Mathews, K.L., McIntyre, C.L., Olivares-Villegas, J.J. and Chapman, S.C. (2010) Heat and drought adaptive QTL in a wheat population designed to minimise confounding agronomic effects. Theor. Appl. Genet. 121: 1001–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purty, R.S., Kumar, G., Singla-Pareek, S.L. and Pareek, A. (2008) Towards salinity tolerance in Brassica: an overview. Physiol. Mol. Biol. Plants 14: 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Y. and Li, X.X. (2009) Genetic analysis of salinity tolerance in Brassica campestris L. China Vegetab. 1: 21–25 [Google Scholar]
- Quesada, V., García-Martínez, S., Piqueras, P., Ponce, M.R. and Micol, J.L. (2002) Genetic architecture of NaCl tolerance in Arabidopsis. Plant Physiol. 130: 951–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra, A.S., Gonugunta, V.K., Christmann, A. and Grill, E. (2010) ABA perception and signalling. Trends Plant Sci. 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Ren, Z.H., Gao, J.P., Li, L.G., Cai, X.L., Huang, W., Chao, D.Y., Zhu, M.Z., Wang, Z.Y., Luan, S. and Lin, H.X. (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Ren, Z., Zheng, Z., Chinnusamy, V., Zhu, J., Cui, X., Iida, K. and Zhu, J.K. (2010) RAS1, a quantitative trait locus for salt tolerance and ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 5669–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezai, A.M. and Saeidi, G. (2005) Genetic analysis of salt tolerance in early growth stages of rapeseed (Brassica napus L.) genotypes. Ind. J. Genet. Plant Breed. 65: 269–273 [Google Scholar]
- Rhoades, J.D. and Loveday, J. (1990) Salinity in irrigated agriculture. In: Stewart, B.A.et al. (eds.) Irrigation of Agricultural Crops, American Society of Agronomy, Madison, WI: pp. 1089–1142 [Google Scholar]
- Richards, R.A. and Thurling, N. (1979a) Genetic analysis of drought stress response in rapeseed (Brassica campestris and B. napus). III. physiological characters. Euphytica 28: 755–759 [Google Scholar]
- Richards, R.A. and Thurling, N. (1979b) Genetic analysis of drought stress response in rapeseed (Brassica campestris and B. napus). II. Yield improvement and the application of selection indices. Euphytica 28: 169–177 [Google Scholar]
- Rozema, J. and Flowers, T. (2008) Crops for a salinized world. Science 322: 1478–1480 [DOI] [PubMed] [Google Scholar]
- Rus, A., Yokoi, S., Sharkhuu, A., Reddy, M., Lee, B.H., Matsumoto, T.K., Koiwa, H., Zhu, J.K., Bressan, R.A. and Hasegawa, P.M. (2001) AtHKT1 is a salt tolerance determinant that controls Naentry into plant roots. Proc. Natl. Acad. Sci. USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salekdeh, G.H., Reynolds, M., Bennett, J. and Boyer, J. (2009) Conceptual framework for drought phenotyping during molecular breeding. Trends Plant Sci. 14: 488–496 [DOI] [PubMed] [Google Scholar]
- Seo, Y.J., Park, J.B., Cho, Y.J., Jung, C., Seo, H.S., Park, S.K., Nahm, B.H. and Song, J.T. (2010) Overexpression of the ethylene-responsive factor gene BrERF4 from Brassica rapa increases tolerance to salt and drought in Arabidopsis plants. Mol. Cells 30: 271–277 [DOI] [PubMed] [Google Scholar]
- Sharma, P., Bhardwaj, R., Arora, H.K., Arora, N. and Kumar, A. (2008) Effects of 28-homobrassinolide on nickel uptake, protein content and antioxidative defence system in Brassica juncea. Biol. Plant 52: 767–770 [Google Scholar]
- Shavrukov, Y. (2013) Salt stress or salt shock: which genes are we studying? J. Exp. Bot. 64: 119–127 [DOI] [PubMed] [Google Scholar]
- Shi, H., Ishitani, M., Kim, C. and Zhu, J.K. (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Quintero, F.J., Pardo, J.M. and Zhu, J.K. (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakral, N.K. and Singh, H. (1994) Combining ability for vigour character sunder normal and saline environment in Ethiopian mustard. Ann. Arid Zone 33: 245–247 [Google Scholar]
- Thakral, N.K. and Prakash, K. (1998) Combining ability for vigour characters under normal and saline environments in Indian mustard [Brassica juncea (L.) Czern. & Coss.]. J. Oilseed Res. 15: 234–237 [Google Scholar]
- Thameur, A., Ferchichi, A. and López-Carbonell, M. (2011) Quantification of free and conjugated abscisic acid in five genotypes of barley (Hordeum vulgare L.) under water stress conditions. S. Afr. J. Bot. 77: 222–228 [Google Scholar]
- Thomson, M.J., de Ocampo, M., Egdane, J., Rahman, M.A., Sajise, A.G., Adorada, D.L., Tumimbang-Raiz, E., Blumwald, E., Seraj, Z.I., Singh, R.K.et al. (2010) Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 3: 148–160 [Google Scholar]
- Tollefson, J. (2011) Drought-tolerant maize gets US debut. Nature 469: 144. [DOI] [PubMed] [Google Scholar]
- Touchette, B.W., Iannacone, L.R., Turner, G.E. and Frank, A.R. (2007) Drought tolerance versus drought avoidance: A comparison of plant-water relations in herbaceous wetland plants subjected to water withdrawal and repletion. Wetlands 27: 656–667 [Google Scholar]
- Trenberth, K.E., Dai, A., van der Schrier, G., Jones, P.D., Barichivich, J., Briffa, K.R. and Sheffield, J. (2014) Global warming and changes in drought. Nat. Climate Change 4: 17–22 [Google Scholar]
- Tuberosa, R. (2012) Phenotyping for drought tolerance of crops in the genomics era. Front. Physiol. 3: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulfat, M., Athar, H.R., Ashraf, M., Akram, N.A. and Jamil, A. (2007) Appraisal of physiological and biochemical selection criteria for evaluation of salt tolerance in canola (Brassica napus L.). Pak. J. Bot. 39: 1593–1608 [Google Scholar]
- Veselov, D.S., Sharipova, G.V., Veselov, S.U. and Kudoyarova, G.R. (2008) The effects of NaCl treatment on water relations, growth and ABA content in barley cultivars differing in drought tolerance. J. Plant Growth Regul. 27: 380–386 [Google Scholar]
- Wang, L., de Kroon, H., Bögemann, G.M. and Smits, A.J.M. (2005) Partial root drying effects on biomass production in Brassica napus and the significance of root responses. Plant Soil 276: 313–326 [Google Scholar]
- Wang, Q.B., Xu, W., Xue, Q.Z. and Su, W.A. (2010) Transgenic Brassica chinensis plants expressing a bacterial codA gene exhibit enhanced tolerance to extreme temperature and high salinity. J. Zhejiang Univ. Sci. B. 11: 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., Bai, Y., Mun, J.H., Bancroft, I., Cheng, F.et al. (2011) Brassica rapa Genome Sequencing Project Consortium. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Ying, J., Kuzma, M., Chalifoux, M., Sample, A., McArthur, C., Uchacz, T., Sarvas, C., Wan, J., Dennis, D.T.et al. (2005) Molecular tailoring of farnesylation for plant drought tolerance and yield protection. Plant J. 43: 413–424 [DOI] [PubMed] [Google Scholar]
- Wang, Y., Beaith, M., Chalifoux, M., Ying, J., Uchacz, T., Sarvas, C., Griffiths, R., Kuzma, M., Wan, J. and Huang, Y. (2009) Shoot-specific down-regulation of protein farnesyltransferase (α-subunit) for yield protection against drought in canola. Mol. Plant 2: 191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick, S.I. (1993) Wild species in the tribe Brassicaceae (Cruciferae) as sources of agronomic traits. In: Guide to the wild germplasm of Brassica and allied crops. Technical Bulletin 17E, 1–19.Center for Land and Biological Resources Research Branch, Agriculture Canada, Ottawa, Ontario, Canada [Google Scholar]
- Wei, W.H., Zhang, S.F., Wang, L.J., Li, J., Chen, B., Wang, Z., Luo, L.X. and Fang, X.P. (2007) Cytogenetic analysis of F1, F2 and BC1 plants from intergeneric sexual hybridization between Sinapis alba and Brassica oleracea by genomic in situ hybridization. Plant Breed. 126: 392–398 [Google Scholar]
- Wilkinson, S. and Davies, W.J. (2010) Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant Cell Environ. 33: 510–525 [DOI] [PubMed] [Google Scholar]
- Wu, H.J., Zhang, Z., Wang, J.Y., Oh, D.H., Dassanayake, M., Liu, B., Huang, Q., Sun, H.X., Xia, R., Wu, Y.et al. (2012) Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. USA 109: 12219–12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Q.S., Zhang, X.K., Xu, B.B., Cheng, Y., Zheng, P.Y. and Lu, G.Y. (2012) Cloning and expression pattern of AnnBn1 gene in Brassica napus. Chin. J. Oil Crops Sci. 34: 123–128 [Google Scholar]
- Yang, C.J., Zhang, X.K., Zou, C.S., Cheng, Y., Zheng, P.Y. and Li, G.Y. (2007) Effects of drought simulated by PEG-6000 on germination and seedling growth of rapeseed (Brassica napus L.). Chin. J. Oil Crops Sci. 29: 425–430 [Google Scholar]
- Yang, C.J., Cheng, Y., Zou, C.S., Zhang, X.K., Zheng, P.Y., Li, G.Y. and Yang, C. (2008) Combining ability and genetic effects of germination ability in different Brassica napus L. cultivar under simulated drought stress. Acta Agron. Sin. 34: 1744–1749 [Google Scholar]
- Yang, M., Yang, Q., Fu, T. and Zhou, Y. (2011) Overexpression of the Brassica napus BnLAS gene in Arabidopsis affects plant development and increases drought tolerance. Plant Cell Rep. 30: 373–388 [DOI] [PubMed] [Google Scholar]
- Yu, S., Zhang, F., Yu, Y., Zhang, D., Zhao, X. and Wang, W. (2012) Transcriptome profiling of dehydration stress in the Chinese cabbage (Brassica rapa L. ssp. pekinensis) by tag sequencing. Plant Mol. Biol. Rep. 30: 17–28 [Google Scholar]
- Zhang, H.X. and Blumwald, E. (2001) Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 19: 765–768 [DOI] [PubMed] [Google Scholar]
- Zhang, H.X., Hodson, J.N., Williams, J.P. and Blumwald, E. (2001) Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl. Acad. Sci. USA 98: 12832–12836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.Z., Creelman, R.A. and Zhu, J.K. (2004) From laboratory to field. Using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol. 135: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, K., Aranzana, M.J., Kim, S., Lister, C., Shindo, C., Tang, C., Toomajian, C., Zheng, H., Dean, C., Marjoram, P.et al. (2007) An Arabidopsis example of Association Mapping in structured samples. PLoS Genet. 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z.H., Zheng, W.Y. and Zhang, X.K. (2011) Principal component analysis and comprehensive evaluation on morphological and agronomic traits of drought tolerance in rapeseed (Brassica napus L.). Sci. Agri. Sin. 44: 1775–1787 [Google Scholar]
- Zhu, J.K., Liu, J. and Xiong, L. (1998) Genetic analysis of salt tolerance in Arabidopsis: Evidence for a critical role of potassium nutrition. Plant Cell 10: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]