Abstract
Phomopsis blight in Lupinus albus is caused by a fungal pathogen, Diaporthe toxica. It can invade all plant parts, leading to plant material becoming toxic to grazing animals, and potentially resulting in lupinosis. Identifying sources of resistance and breeding for resistance remains the best strategy for controlling Phomopsis and reducing lupinosis risks. However, loci associated with resistance to Phomopsis blight have not yet been identified. In this study, quantitative trait locus (QTL) analysis identified genomic regions associated with resistance to Phomopsis pod blight (PPB) using a linkage map of L. albus constructed previously from an F8 recombinant inbred line population derived from a cross between Kiev-Mutant (susceptible to PPB) and P27174 (resistant to PPB). Phenotyping was undertaken using a detached pod assay. In total, we identified eight QTLs for resistance to PPB on linkage group (LG) 3, LG6, LG10, LG12, LG17 and LG27 from different phenotyping environments. However, at least one QTL, QTL-5 on LG10 was consistently detected in both phenotyping environments and accounted for up to 28.2% of the total phenotypic variance. The results of this study showed that the QTL-2 on LG3 interacts epistatically with QTL-5 and QTL-6, which map on LG10 and LG12, respectively.
Keywords: DArT, Rqtl, multi-phase experiments, anamorph Phomopsis leptostromiformis, broad-leaf lupin
Introduction
Lupinus albus L. is grown as a protein crop in numerous Mediterranean-type climatic regions (Baer 2006, Luckett et al. 2008, Noffsinger et al. 2006) and is primarily used as stock feed for sheep and cattle (Hill 2006, May et al. 1993). It has potential for increased inclusion in the human diet (Erbas et al. 2005, Mariotti et al. 2002), although allergies have been reported (Quaresma et al. 2007).
There are several important yield-limiting fungal diseases of L. albus, including anthracnose (Colletotrichum lupini, Thomas et al. 2008), Pleiochaeta root rot (Pleiochaeta setosa, Luckett et al. 2009), and Phomopsis blights caused by Diaporthe toxica (Cowley et al. 2010, 2012b). In commercial crops of L. albus, Phomopsis infection is often not detected until animals exhibit symptoms of lupinosis after grazing on infected stubble and fallen seed after harvest (Cowley et al. 2010). Lupinosis is a degenerative disorder that causes acute liver damage, brain damage and death (Allen et al. 1979, Luduena et al. 1989). It results from the consumption of toxic metabolites produced by the fungus (Peterson et al. 1987). Other Phomopsis species that infect soybean also produce similar toxins (Balducchi and McGee 1987).
In Australia L. albus cultivation has been based on the Kiev-Mutant and Ultra varieties since the late 1970s (Gladstones 1976). These varieties have had adequate resistance to D. toxica over a long period (Wood and Allen 1980, Sweetingham et al. 1998). Epidemics of Phomopsis blights on L. albus have been reported in South Africa (Jaarsveld and Knox-Davies 1974) and Poland (Kochman and Kubicka 1974). This study was prompted by an outbreak of Phomopsis blight on a crop of Kiev-Mutant in southern New South Wales (NSW, Australia) in 2004, which resulted in the death of animals that grazed on crop residues (Cowley et al. 2010). A virulent isolate of D. toxica (isolate DAR80114) capable of invading all plant parts of L. albus was collected from the 2004 outbreak and has been used to identify L. albus genotypes with genetic resistance in stems, leaves and pods (Cowley et al. 2010, 2012b, 2012c). From previous work it appears that resistance to D. toxica in leaves, stems and pods of L. albus is under independent genetic control (Cowley et al. 2010). Principal component analysis showed that resistance to Phomopsis stem and leaf blight was related. Resistance to seed infection and Phomopsis pod blight was also related, but independently of stem and leaf blight. Further work using detached plant part assays demonstrated that the genotypes often displayed varying resistance to Phomopsis leaf and pod blight in L. albus (Cowley et al. 2012b). Shankar et al. (2002a) postulated that stem and pod resistance in the L. angustifolius variety Tanjil is also controlled by different genes. Considerable genetic variation in the resistance to D. toxica exists within L. albus germplasm in leaves, stems and pods (Cowley et al. 2012a, 2012b, 2012c), although knowledge is limited on the underlying genetics.
Ethiopian landrace P27174 is resistant to PPB (Cowley et al. 2012b). This landrace is also a source of resistance to anthracnose in L. albus (Adhikari et al. 2009, Phan et al. 2007). A recombinant inbred line (RIL) population exists between P27174 (anthracnose resistant) and Kiev-Mutant (anthracnose susceptible, Thomas et al. 2008) and has been used to develop the first genetic and comparative map of white lupin (Phan et al. 2007), although the marker density was low. Anthracnose resistance, flowering time and seed alkaloid have been mapped using this population (Phan et al. 2007). Vipin et al. (2013) developed a DArT array of L. albus and increased the marker density in the genetic linkage map of the same RIL population derived from Kiev-Mutant × P27174.
The Kiev-Mutant × P27174 population was phenotyed for resistance to Phomopsis pod and leaf blight (Cowley et al. 2012b) using detached plant part assays. The population is segregated, with resistance in pods but not leaves. The nature of the genetic control of resistance to PPB in L. albus is unknown, but is hypothesised to be polygenic due to the continuous phenotypic variation that exists in structured bi-parental mapping populations assessed for resistance (Cowley et al. 2012b). In this study, QTL mapping was undertaken to understand the genetics underlying resistance and to locate loci associated with resistance to PPB in L. albus.
Materials and Methods
Plant materials
A subset of an F8-derived recombinant inbred line (RIL) population, comprising 93 lines derived from a cross between Kiev-Mutant (susceptible to D. toxica isolate DAR80114) and P27174 (resistant to the same isolate), was used for QTL identification for PPB resistance. Seeds of the mapping population (Phan et al. 2007) were provided by Dr. Huaan Yang (Department of Agriculture and Food, Western Australia).
Experimental design and data analysis using multi-phase experiments
Phenotyping assays in plant pathology using detached plant parts are multi-phase experimental processes (Brien and Bailey 2006, Smith et al. 2006). This involves growing plants in field or controlled-environment trials (Phase 1) and then subjecting a sample removed from these plants to disease assessment, usually under laboratory conditions (Phase 2). Each phase may be subject to non-genetic sources of variation. To be able to separate these sources of variation in both phases from genetic sources requires a multi-phase experiment with an appropriate experimental design and statistical analysis (Cowley et al. 2012b). To achieve this, separate randomization is required for each phase, with additional replication in Phase 2 (Smith et al. 2006).
In each experiment, the spatial arrangement of lupin lines in both phases was optimized using DiGGer design software (available from http://www.austat.gen.org/files/software/downloads). All data were analysed using ASReml-R (Butler et al. 2009) with factors describing the spatial arrangement of both phases in the model (Cowley et al. 2012b).
Inoculum preparation
The single-spore isolate of D. toxica used in this study was isolated from an infected commercial crop of Kiev-Mutant grown in Tarcutta, NSW, Australia, in 2004 (Cowley et al. 2010). The isolate (accession no. DAR80114) was deposited in the Living Culture Collection in the Department of Primary Industries, Orange, NSW, Australia. For this study, inoculum of the isolate was prepared as described previously by Cowley et al. (2010). A spore concentration of 5 × 106 conidia/ml was used in all experiments described below.
Phenotyping for resistance to Phomopsis pod blight
The phenotyping experiments and analysis have been detailed in Cowley et al. (2012b) and are briefly described below.
Screen-house experiment
In phase 1 of the first phenotyping experiment (hereafter referred to as experiment 1), ten seeds from each line from the KievMutant × P27174 RIL population were grown in 50 cm rows with three replicates in a field soil screen-house in Wagga Wagga, NSW, Australia (latitude: 35.05°; longitude: 147.35°) in a randomised complete block. The late-flowering lines (n = 26) were sown on 18 July 2008, with seeds of the remaining mid- to early flowering lines (n = 71) sown 2 weeks later to maximise physiological uniformity among the lines at the time of removing pods.
Glasshouse experiment
In phase 1 of the second set of phenotyping experiments (collectively referred to hereafter as experiment 2), pods were selected from plants grown in sandy-loam soil in 125 mm diameter pots in an evaporative-cooled glasshouse in Wagga Wagga. Nine seeds of each line were sown per pot and later thinned to three plants per pot. The entire population was assessed in three separate experiments, with two pots of both Kiev-Mutant and P27174 sown within each replicate as control lines. Pods were collected at physiological maturity, approximately 24 weeks after sowing.
The experimental design and analysis of Phase 2 of the detached pod assays are described in Cowley et al. (2012b). Briefly, in the first phase of the experiment, plants were grown in a randomised replicated array to produce pods (as described above). The pods were collected when they reached physiological maturity, growth stage 4.4 (Dracup and Kirby 1996), and transported to the laboratory. Additional pods were collected from randomly selected rows to provide the duplication needed for the laboratory phase (Smith et al. 2006). The pods were immersed in a D. toxica spore suspension (5 × 106 spores per ml) before being placed in humidified tubs in a culture room at 20°C with 12-hour fluorescent light. The incubation period was 7 days and then the lids were removed to reduce humidity. The pods were scored for disease symptoms 10 days after inoculation according to a 0 to 9 scale (where, 0 = no symptoms; 9 = total pod rot) as described previously (Cowley et al. 2012b). A score <3 was regarded as resistant to PPB.
The three experiments where Phase 1 occurred in a glasshouse were analysed using meta-analysis performed in ASReml-R. The predicted genotype means were then used in the quantitative genetic analysis for QTL detection.
Quantitative genetic analysis
An integrated genetic linkage map consisting of 441 markers (220 AFLP, 105 genic and 136 DArT) on 38 linkage groups, with a total length of 2,169 cM (Phan et al. 2007, Vipin et al. 2013), was utilised to identify loci associated with PPB resistance. QTL mapping was first conducted on the predicted means for the detached pod assays from screen-house-grown plants. The predicted means from meta-analysis of the glasshouse-grown plants were then analysed separately. We performed both one- and two-dimensional interval mapping analyses using the Rqtl program of the R Statistical package with an error probability of <0.001 (Broman and Sen 2009, Sen and Churchill 2001) [http://www.rqtl.org/]. Multiple QTL models were scanned for epistatically interacting QTLs. The corrected QTL model was optimised by an iterative process. The LOD (logarithm of odds) scores were calculated by Rqtl to provide a measure of the likelihood that the observed data were due to linkage, compared to the alternative that they were due to chance.
Results
Evaluation of Phomopsis pod blight resistance
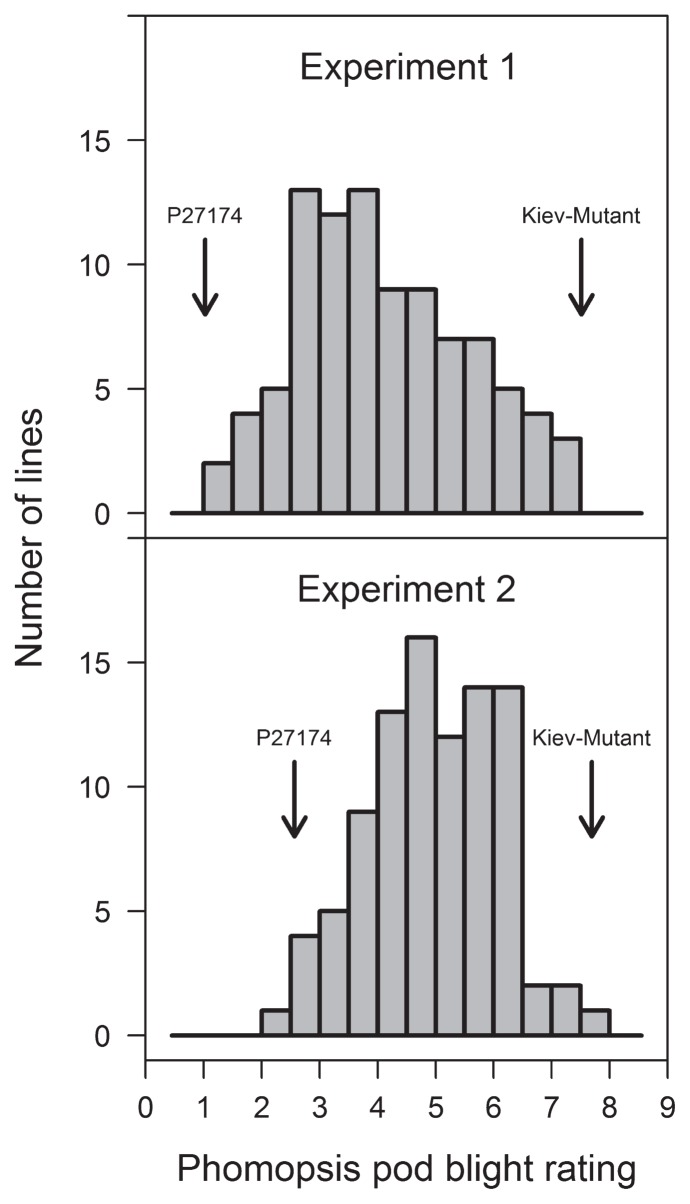
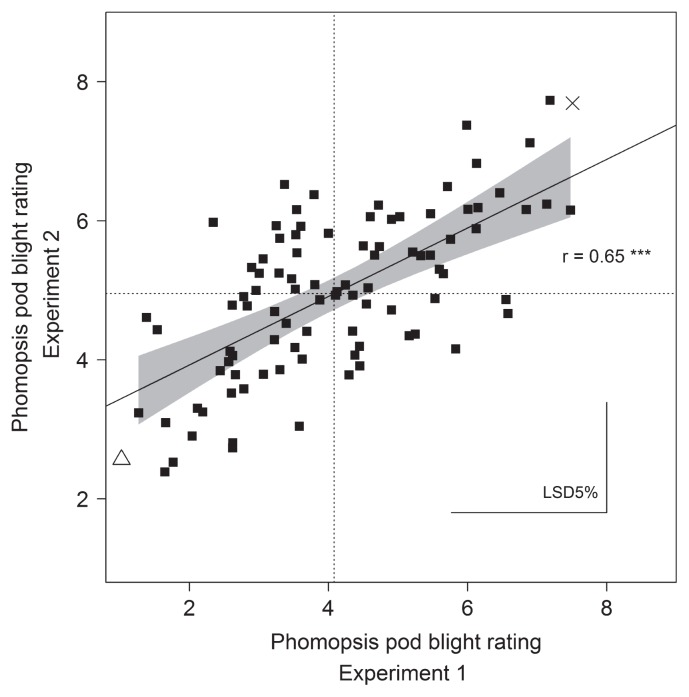
The Kiev-Mutant × P27174 population had a continuous distribution for resistance to PPB in both phenotyping datasets, suggesting that resistance was quantitatively inherited (Fig. 1). In experiment 2 (plants grown in a glasshouse), the grand mean for PPB phenotyping was higher than in experiment 1 (plants grown in screen-house) (4.96 and 4.08, respectively). The predicted mean for P27174 was also higher in experiment 2 than experiment 1, but P27174 was resistant in both experiments (2.56 and 1.04, respectively). There was no difference in the response of the susceptible parent Kiev-Mutant. There was a significant variation in disease scores between the experiments, although the disease scores showed a significant correlation (r = 0.65, P < 0.001, Fig. 2).
Fig. 1.
Frequency distributions for phenotyping response to Phomopsis pod blight in a Lupinus albus population of F8 RILs from Kiev- Mutant × P27174. In experiment 1, plants were grown in a field-soil screen-house. Experiment 2 shows the overall predicted means from meta-analysis of three glasshouse-grown experiments. Phomopsis pod blight was assessed using detached pods in both experiments using a 0 to 9 scale, where 0 = very resistant and 9 = very susceptible.
Fig. 2.
Relationship between phenotyping experiments assessing Phomopsis pod blight severity assessed using a detached pod assay (0 to 9 scale) in a Lupinus albus Kiev-Mutant × P27174 population as F8 RILs. Dashed lines show the grand mean for each data set. The location of the parental lines is shown with a cross for Kiev-Mutant and an open triangle for P27174. The regression line is shown with the 95% confidence interval depicted in grey.
Identification of QTL for Phomopsis pod blight resistance
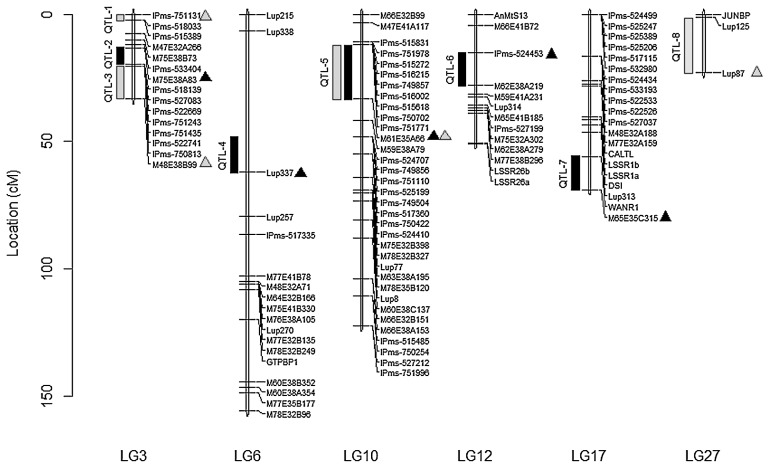
Results of the QTL mapping of the two phenotyping data sets are shown in Table 1. In experiment 1, five QTL explaining 8.3% to 36.9% of the phenotypic variance for resistance to PPB were identified on linkage groups LG3, LG6, LG10, LG12 and LG17 (Fig. 3). Of these QTLs, QTL-2 on LG3 had the maximum LOD score (9.3) and explained 36.9% variance. P27174 contributed alleles for resistance at QTL-2 and QTL-4, whereas Kiev-Mutant contributed alleles for resistance at QTL-5, QTL-6 and QTL-7. QTL-2 on LG3 showed epistatic interaction with QTL-5 (LG10) and QTL-6 (LG12) (Table 1).
Table 1.
Detected QTLs for resistance to Phomopsis pod blight (caused by Diaporthe toxica) using a Lupinus albus F8-RIL population derived from a cross between Kiev-Mutant and P27174
| QTL | LG | Map position (cM) | Pa | LOD | Explained varianceb (%VG) | Additive effectc | Proximal marker |
|---|---|---|---|---|---|---|---|
| Experiment 1 (plants grown in field-soil screen-house) | |||||||
| QTL-2 | LG3 | 17.5 | <0.001 *** | 9.3 | 36.9 | −2.37 | M75E38A83 |
| QTL-4 | LG6 | 61.9 | 0.020 * | 2.1 | 10.0 | −0.39 | Lup337 |
| QTL-5 | LG10 | 30.0 | <0.001 *** | 6.4 | 27.1 | 0.75 | M61E35A66 |
| QTL-6 | LG12 | 10.0 | 0.012 * | 4.3 | 19.4 | 1.31 | lPms-524453 |
| QTL-7 | LG17 | 69.0 | 0.040 * | 1.8 | 8.3 | 0.22 | M65E35C315 |
| QTL-2:QTL-5d | LG3:LG10 | – | <0.001 *** | 5.7 | 38.6 | −0.49 | |
| QTL-2:QTL-6 | LG3:LG12 | – | 0.007 ** | 3.8 | 24.6 | 0.48 | |
| Experiment 2 (plants grown in a glasshouse) | |||||||
| QTL-1 | LG3 | 0.0 | 0.007 ** | 2.5 | 11.5 | 0.58 | lPms-751131 |
| QTL-3 | LG3 | 33.1 | <0.001 *** | 7.1 | 29.8 | −0.37 | M48E38B99 |
| QTL-5 | LG10 | 27.5 | <0.001 *** | 6.7 | 28.2 | 0.45 | M61E35A66 |
| QTL-8 | LG27 | 12.5 | 0.035 * | 1.7 | 8.0 | 0.10 | Lup87 |
| QTL-3:QTL-5d | LG3:LG10 | – | <0.001 *** | 6.0 | 25.8 | −0.38 | |
*: P < 0.05, **: P < 0.01 and ***: P < 0.001.
Proportion of variability explained by the putative QTL. Bold-consistent genomic regions associated with Phomopsis pod blight resistance that was scored under glasshouse and field conditions.
Alleles from QTL with positive additive effect value are from Kiev-Mutant. Alleles from QTL with negative additive effect value are from P27174.
Interaction effect between pairs of QTLs.
Fig. 3.
Five linkage groups from the Lupinus albus Kiev-Mutant × P27174 map showing the location of putative QTLs conditioning resistance to Phomopsis pod blight caused by Diaporthe toxica. Solid rectangles show QTLs identified from phenotyping experiment 1 where Phase 1 was conducted in a screen-house. Grey rectangles show QTLs identified from the glasshouse-grown phenotyping in experiment 2. Triangles indicate proximal markers to the putative QTL locations.
In experiment 2, four QTLs explaining 8.0% to 29.8% phenotypic variance were identified on LG10, LG27, and two on LG3 (Table 1). Kiev-Mutant contributed alleles for resistance in QTL-1, QTL-5 and QTL-8, which were detected on LG3, LG10 and LG 27, respectively, whereas P27174 contributed alleles for resistance on LG3 (QTL-3). The QTLs with the highest LOD scores (>6.7) were located on linkage groups LG3 (QTL-3) and LG10 (QTL-5) and accounted for >28.2% variance. In addition, they showed significant epistatic genetic effects (Table 1). QTL-5 on LG10 was identified in both experiments, was mapped within the same genomic region (2.5 cM) and was likely to be the same QTL (Fig. 3). QTL-3 was mapped within 15.6 cM of QTL-2, and QTL-3 and QTL-2 may be the same gene associated with PPB resistance.
Discussion
The two phenotyping datasets presented here were obtained from two different environments, and the data from each were assessed independently. The overall mean of experiment 2 was higher than experiment 1; however, in both datasets the relative ranking of the lines did not differ greatly and the correlation between the environments was significant (Fig. 2).
Consistent identification of QTLs in similar chromosomal positions from both assessments suggests that they might be the same genomic regions controlling resistance to PPB with isolate DAR80114. QTL-5 on LG10 showed the largest effect in both experiments. On LG3, QTL-2 and QTL-3 detected in experiments 1 and 2 were located at different marker intervals (within 15.6 cM). It is possible that both of these QTLs map to the same genomic region. High resolution mapping of the genomic region is required and has not yet been performed. The results of this study show that QTL-2 and QTL-5 had the largest effect on PPB resistance in this population and showed epistatic interaction.
The positive and negative values representing additive effects of the QTLs indicate that both parents contributed alleles for resistance to the observed variation in the disease scores of the Kiev-Mutant × P27174 RIL population. Kiev-Mutant was commercialised in Australia in the 1970s and had good field resistance to Phomopsis (Sweetingham et al. 1998). This resistance was maintained in Kiev-Mutant for over 30 years until 2004 when an isolate capable of overcoming resistance in Kiev-Mutant was identified (Cowley et al. 2010). Consequently, it is not surprising that Kiev-Mutant donates alleles for resistance to Phomopsis as this variety does have resistance to Phomopsis, but was susceptible to the isolate in this study.
None of the QTLs in this study appeared to be co-located with loci already published for the same Kiev-Mutant × P27174 RIL population for anthracnose resistance (Yang et al. 2010) or flowering time (Phan et al. 2007). This observation suggests that loci controlling resistance to anthracnose and PPB are different and not located in a cluster, as reported in other key crops such as rapeseed and cereals (Delourme et al. 2004, Michelmore and Meyers 1998)
Anthracnose resistance was identified from P27174 and transferred to modern varieties (Adhikari et al. 2009). Two QTLs identified on LG4 and LG17 conditioned resistance to anthracnose (Phan et al. 2007). Using microsatellite-anchored fragment length polymorphism (MFLP), Yang et al. (2010) were able to identify three sequence-specific PCR markers conditioning resistance to anthracnose, which were designated WANR1, WANR2 and WANR3. QTL-7 was detected in the vicinity of WANR1 (~13.2 cM), which has been associated with resistance to anthracnose disease in L. albus. In the first experiment presented here, the marker WANR1 was a flanking marker for QTL-7, located on LG17, at a distance of 13.2 cM from the QTL.
Genetic diversity analysis has shown that P27174 represents a ‘cluster’ of genotypes of Ethiopian origin (Raman et al. 2008). It is possible that other Ethiopian accessions may have the same loci conferring PPB resistance. Further work is required to test linkage disequilibrium for PPB resistance among L. albus accessions. Further alleles (or loci) for Phomopsis resistance in pods, and possibly stems, may exist in other landraces from this geographic region. We have previously identified resistance to PPB in the Ethiopian landrace P28507 (Cowley et al. 2012b), suggesting the merit of further screening of accessions from east Africa. This approach is supported by the numerous Ethiopian accessions that have being identified with resistance to anthracnose (Adhikari et al. 2009), although complementation studies have not been undertaken.
This study established the location of loci and identification of markers closely linked to resistance to PPB in an RIL population of L. albus. Validation of the effects of the QTLs against different genetic backgrounds is required before attempting marker-assisted selection in breeding programs. Phenotyping for resistance to PPB is lengthy as resistance can only be detected towards the end of a growing cycle when large pods are present at the correct physiological maturity. This restricts phenotyping events to one (if using field-based screening) or possibly two events per year if growing plants in controlled conditions per growing cycle. The use of molecular techniques to aid breeding is well understood and currently employed in lupin breeding for anthracnose resistance (Yang et al. 2004, 2010) and Phomopsis stem blight resistance in L. angustifolius (Shankar et al. 2002b, Yang et al. 2002). Breeding efforts to improve PPB resistance in L. albus would be enhanced by marker-assisted selection. However, further research is needed to convert the AFLP and DArT markers located near the QTLs identified in this study into usable PCR-based markers for routine marker-assisted selection in L. albus improvement programs.
Acknowledgements
Richard Oliver and Huyen Phan are thanked for supplying the AFLP and genetic marker data. Huaan Yang is thanked for supplying seed of the Kiev-Mutant × P27174 population. Alison Smith is thanked for analysis of pod data. This research was part-funded by the Grains Research and Development Corporation of Australia (GRDC).
Literature Cited
- Adhikari, K.N., Buirchell, B.J., Thomas, G.J., Sweetingham, M.W. and Yang, H. (2009) Identification of anthracnose resistance in Lupinus albus L. and its transfer from landraces to modern cultivars. Crop Pasture Sci. 60: 472–479 [Google Scholar]
- Allen, J.G., Masters, H.G. and Wallace, S.R. (1979) The effect of lupinosis on liver copper, selenium and zinc concentrations in merino sheep. Vet. Rec. 105: 434–436 [DOI] [PubMed] [Google Scholar]
- Baer, E.v. (2006) Relevant points for the production and use of sweet lupin in Chile. In: Santen, E.v. and Hill, G.D. (eds.) Proceedings of the 11th International Lupin Conference: Mexico, where old and new world lupins meet, International Lupin Association, Canterbury, New Zealand, pp. 116–119 [Google Scholar]
- Balducchi, A.J. and McGee, D.C. (1987) Environmental factors influencing infection of soybean seeds by Phomopsis and Diaporthe species during seed maturation. Plant Dis. 71: 209–212 [Google Scholar]
- Brien, C.J. and Bailey, R.A. (2006) Multiple randomizations. J. Roy. Stat. Soc. B. 68: 571–609 [Google Scholar]
- Broman, K.W. and Sen, Ś. (2009) A Guide to QTL Mapping with R/qtl. Springer, New York [Google Scholar]
- Butler, D.G., Cullis, B.R., Gilmour, A.R. and Gogle, B.J. (2009) ASReml-R reference manual, release 3. Technical report, Queensland Department of Primary Industries [Google Scholar]
- Cowley, R.B., Ash, G., Harper, J.D.I. and Luckett, D.J. (2010) Evidence that Diaporthe toxica infection of Lupinus albus is an emerging concern for the Australian lupin industry. Australas. Plant Pathol. 39: 146–153 [Google Scholar]
- Cowley, R.B., Ash, G.J., Harper, J.D.I. and Luckett, D.J. (2012a) Evaluation of resistance to Phomopsis stem blight (caused by Diaporthe toxica) in Lupinus albus. Eur. J. Plant Pathol. 133: 631–644 [Google Scholar]
- Cowley, R.B., Ash, G.J., Harper, J.D.I., Smith, A.B., Cullis, B.R. and Luckett, D.J. (2012b) Application of multi-phase experiments in plant pathology to identify genetic resistance to Diaporthe toxica in Lupinus albus. Euphytica 186: 655–669 [Google Scholar]
- Cowley, R.B., Luckett, D.J., Harper, J.D.I. and Ash, G.J. (2012c) Development of a reliable and rapid detached leaf assay for Lupinus albus to detect resistance to the fungal disease Phomopsis leaf blight, caused by Diaporthe toxica. Can. J. Plant Pathol. 34: 401–409 [Google Scholar]
- Delourme, R., Pilet-Nayel, M.L., Archipiano, M., Horvais, R., Tanguy, X., Rouxel, T., Brun, H., Renard, M. and Balesdent, M.H. (2004) A cluster of major specific resistance genes to Leptosphaeria maculans in Brassica napus. Phytopathology 94: 578–583 [DOI] [PubMed] [Google Scholar]
- Dracup, M. and Kirby, M.E.J. (1996) Lupin development guide. University of Western Australia [Google Scholar]
- Erbas, M., Certel, M. and Uslu, M.K. (2005) Some chemical properties of white lupin seeds (Lupinus albus L.). Food Chem. 89: 341–345 [Google Scholar]
- Gladstones, J.S. (1976) The Mediterranean white lupin. J. Agric., Western Australia. 17: 70–74 [Google Scholar]
- Hill, G.D. (2006) The utilization of lupins in animal nutrition. In: Santen, E.v. and Hill, G.D. (eds.) Proceedings of the 11th International Lupin Conference: Mexico, where old and new world lupins meet, International Lupin Association, Canterbury, New Zealand, pp. 288–305 [Google Scholar]
- Jaarsveld, A.B.V. and Knox-Davies, P.S. (1974) Resistance of lupins to Phomopsis leptostromiformis. Phytophylactica 6: 55–60 [Google Scholar]
- Kochman, J. and Kubicka, H. (1974) Aggressiveness and pathogenicity of Phomopsis leptostromiformis (Kuhn) Bubak and development of the process of infection caused by the fungus. Acta Agrobot. 27: 5–17 [Google Scholar]
- Luckett, D.J., Cowley, R.B., Richards, M.F. and Roberts, D.M. (2008) Improved methodology for screening for resistance to Pleiochaeta setosa root rot in Lupinus albus. In: Palta, J.A. and Berger, J. (eds.) 12th International Lupin Conference Fremantle, Western Australia, International Lupin Association, Canterbury, New Zealand, pp. 447–451 [Google Scholar]
- Luckett, D.J., Cowley, R.B., Richards, M.F. and Roberts, D.M. (2009) Breeding Lupinus albus for resistance to the root pathogen Pleiochaeta setosa. Eur. J. Plant Pathol. 125: 131–141 [Google Scholar]
- Luduena, R.F., Prasad, V., Roach, M.C. and Lacey, E. (1989) The interaction of phomopsin A with bovine brain tubulin. Arch. Biochem. Biophys. 272: 32–38 [DOI] [PubMed] [Google Scholar]
- Mariotti, F., Pueyo, M.E., Tome, D. and Mahe, S. (2002) The bioavailability and postprandial utilisation of sweet lupin (Lupinus albus)- flour protein is similar to that of purified soybean protein in human subjects: a study using intrinsically N-15-labelled proteins. Br. J. Nutr. 87: 315–323 [DOI] [PubMed] [Google Scholar]
- May, M.G., Otterby, D.E., Linn, J.G., Hansen, W.P., Johnson, D.G. and Putnam, D.H. (1993) Lupins (Lupinus albus) as a protein-supplement for lactating Holstein dairy-cows. J. Dairy Sci. 76: 2682–2691 [DOI] [PubMed] [Google Scholar]
- Michelmore, R.W. and Meyers, B.C. (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8: 1113–1130 [DOI] [PubMed] [Google Scholar]
- Noffsinger, S.L., Bhardwaj, H.L. and Santen, E.v. (2006) An ideotype for winter-type grain white lupin in the Southern USA. In: Santen, E.v. and Hill, G.D. (eds.) Proceedings of the 11th International Lupin Conference: Mexico, where old and new world lupins meet, International Lupin Association, Canterbury, New Zealand, pp. 64–67 [Google Scholar]
- Peterson, J.E., Jago, M.V., Payne, A.L. and Stewart, P.L. (1987) The toxicity of phomopsin for sheep. Aust. Vet. J. 64: 293–298 [DOI] [PubMed] [Google Scholar]
- Phan, H.T.T., Ellwood, S.R., Adhikari, K., Nelson, M.N. and Oliver, R.P. (2007) The first genetic and comparative map of White Lupin (Lupinus albus L.): Identification of QTLs for anthracnose resistance and flowering time, and a locus for alkaloid content. DNA Res. 14: 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaresma, R.R., Viseu, R., Martins, L.M., Tomaz, E. and Inacio, F. (2007) Allergic primary sensitization to lupine seed. Allergy 62: 1473–1474 [DOI] [PubMed] [Google Scholar]
- Raman, R., Luckett, D.J. and Raman, H. (2008) Estimation of genetic diversity in albus lupins (Lupinus albus L.) using DArT and genic markers. In: Palta, J.A. and Berger, J. (eds.) 12th International Lupin Conference Fremantle, Western Australia, International Lupin Association, Canterbury, New Zealand, pp. 236–241 [Google Scholar]
- Sen, S. and Churchill, G.A. (2001) A statistical framework for quantitative trait mapping. Genetics 159: 371–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar, M., Sweetingham, M., Buirchell, B. and Cowling, W. (2002a) Evidence that resistance to phomopsis stem and pod blight in Lupinus angustifolius cv. Tanjil is controlled by different genes. In: McComb, J.A. (ed.) Plant breeding for the 11th millennium: 12th Australasian Plant Breeding Conference, Perth WA Australasian Plant Breeding Association Inc., pp. 429–431 [Google Scholar]
- Shankar, M., Sweetingham, M.W. and Cowling, W.A. (2002b) Identification of alleles at two loci controlling resistance to Phomopsis stem blight in narrow-leafed lupin (Lupinus angustifolius L.). Euphytica 125: 35–44 [Google Scholar]
- Smith, A.B., Lim, P. and Cullis, B.R. (2006) The design and analysis of multi-phase plant breeding experiments. J. Agric. Sci. 144: 393–409 [Google Scholar]
- Sweetingham, M.W., Jones, R.A. and Brown, A.G. (1998) Diseases and pests. In: Gladstones, J.S., Atkins, C. and Hamblin, J. (eds.) Lupins as crop plants: biology, production and utilization, CAB International, Wallingford, UK, pp. 263–289 [Google Scholar]
- Thomas, G.J., Sweetingham, M.W., Yang, H.A. and Speijers, J. (2008) Effect of temperature on growth of Colletotrichum lupini and on anthracnose infection and resistance in lupins. Australas. Plant Pathol. 37: 35–39 [Google Scholar]
- Vipin, C., Luckett, D.J., Harper, J.D.I., Ash, G.J., Kilian, A., Ellwood, S.R., Phan, H.T.T. and Raman, H. (2013) Construction of integrated linkage map of a recombinant inbred line population of white lupin (Lupinus albus L.). Breed. Sci. 63: 292–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, P.M. and Allen, J.G. (1980) Control of ovine lupinosis: use of a resistant cultivar of Lupinus albus - cv. Ultra. Aust. J. Exp. Agr. Ani. Husb. 20: 316–318 [Google Scholar]
- Yang, H., Shankar, M., Buirchell, B.J., Sweetingham, M.W., Caminero, C. and Smith, P.M.C. (2002) Development of molecular markers using MFLP linked to a gene conferring resistance to Diaporthe toxica in narrow-leafed lupin (Lupinus angustifolius L.). Theor. Appl. Genet. 105: 265–270 [DOI] [PubMed] [Google Scholar]
- Yang, H., Boersma, J.G., You, M., Buirchell, B.J. and Sweetingham, M.W. (2004) Development and implementation of a sequence-specific PCR marker linked to a gene conferring resistance to anthracnose disease in narrow-leafed lupin (Lupinus angustifolius L.). Mol. Breed. 14: 145–151 [Google Scholar]
- Yang, H., Lin, R., Renshaw, D., Li, C., Adhikari, K., Thomas, G., Buirchell, B., Sweetingham, M. and Yan, G. (2010) Development of sequence-specific PCR markers associated with a polygenic controlled trait for marker-assisted selection using a modified selective genotyping strategy: a case study on anthracnose disease resistance in white lupin (Lupinus albus L.). Mol. Breed. 25: 239–249 [Google Scholar]