Summary
Background
In the fly’s visual motion pathways, two cell types - T4 and T5 - are the first known relay neurons to signal small-field direction-selective motion responses [1]. These cells then feed into large tangential cells that signal wide-field motion. Recent studies identified two types of columnar neurons in the second neuropil, or medulla, that relay input to T4 from L1, the ON-channel neuron in the first neuropil, or lamina, thus providing a candidate substrate for the elementary motion detector (EMD) [2]. Interneurons relaying the OFF-channel from L1’s partner, L2, to T5 are so far not known however.
Results
Here we report that multiple types of transmedulla (Tm) neurons provide unexpectedly complex inputs to T5 at their terminals in the third neuropil, or lobula. From the L2 pathway, single-column input comes from Tm1 and Tm2 and multiple-column input from Tm4 cells. Additional input to T5 comes from Tm9, the medulla target of a third lamina interneuron L3, providing a candidate substrate for L3’s combinatorial action with L2 [3]. Most numerous, Tm2 and Tm9’s input synapses are spatially segregated on T5’s dendritic arbor, providing candidate anatomical substrates for the two arms of a T5 EMD circuit; Tm1 and Tm2 provide a second. Transcript profiling indicates that T5 expresses both nicotinic and muscarinic cholinoceptors, qualifying T5 to receive cholinergic inputs from Tm9 and Tm2, which both express ChAT.
Conclusions
We hypothesize that T5 computes small-field motion signals by integrating multiple cholinergic Tm inputs using nicotinic and muscarinic cholinoceptors.
Indexing terms: fly, photoreceptors, visual pathway, column, motion sensitivity, directional selectivity, ‘off’ edge response, elementary motion detector
Introduction
More than 50 years ago, quantitative behavioral studies on insects gave rise to an influential model of visual motion detection, the Hassenstein-Reichardt elementary motion detector (EMD) [4, 5]. In this mathematical model, two visual signals with a spatial offset, one instantaneous and the other delayed by a temporal filter, are combined at a “multiplication” unit to generate direction-selective motion signals. Electrophysiological investigation on direction-selective motion-sensitive neurons in the rabbit’s retina led to the Barlow-Levick model, which uses a similar delay-and-compare strategy but employs an inhibitory mechanism to suppress motion signals in the null direction [6]. Despite the widespread influence exerted for the last half century on those working on both insect and vertebrate visual systems, the Hassenstein-Reichardt EMD has remained a purely biocybernetic model. Detailed knowledge of its implementation by actual neurons to generate local motion detection in the insect visual system is only now being unravelled. Still unresolved however is how multiplication, a mathematical necessity, might be generated physiologically by purely synaptic means.
Differing pathways from L1 and L2 are required to detect motion
It is now well established that L1 and L2, a pair of columnar neurons in each column or cartridge in the first optic neuropil, the lamina, together are needed for a fly to sense motion. L1 and L2 are required to detect moving ON- and OFF-edges, respectively [7], providing inputs to circuits specialized to detect moving light and dark edges [8] that serve analogous functions to ON- and OFF-bipolar neurons of the vertebrate retina [9, 10]. At their inputs, L1 and L2 are postsynaptic at each of the tetrad synapses from the lamina terminals of photoreceptor neurons R1-R6 [11–13]. These express a single rhodopsin, Rh1 [14], and respond to a broad spectrum of light [15], providing a monochromatic input, like vertebrate rods. Because every tetrad invariably incorporates L1 and L2 as a pair at two of its four postsynaptic sites, the overall synaptic input to these two cells is carefully matched. Independently, L2 also provides input from lamina collaterals of L4. That input is closely reciprocated.
By contrast, L1 and L2 have completely different outputs. Their terminals innervate specific strata of the distal medulla, L1 in strata M1 and 5, and L2 between these, in stratum M2 [16]. The cells provide inputs to two anti-parallel motion pathways previously identified anatomically mostly in large fly species (review: [17]). Silencing both pathways blocks motion sensing [3, 18, 19], as does silencing their eventual outputs, T4 and T5 [20]. In turn, T4 and T5 cells both exhibit direction-selective responses [1], and in turn converge upon the dendrites of giant lobula plate tangential cells (LPTCs) [17], that signal wide-field motion in either a horizontal (HS cells) or vertical (VS cells) direction [21]. Each T4 and T5 cell type comprises four subtypes, a–d [2, 16], that terminate one each in four strata of the lobula plate (Figure 1A). Each stratum exhibits direction-specific activity, for example in the pattern of [3H]-2-deoxyglucose uptake in response to wide-field motion stimuli [22, 23]. A direct synaptic contact has been reported from T4 to an HS-cell dendrite [24], but pathway details are otherwise sparse.
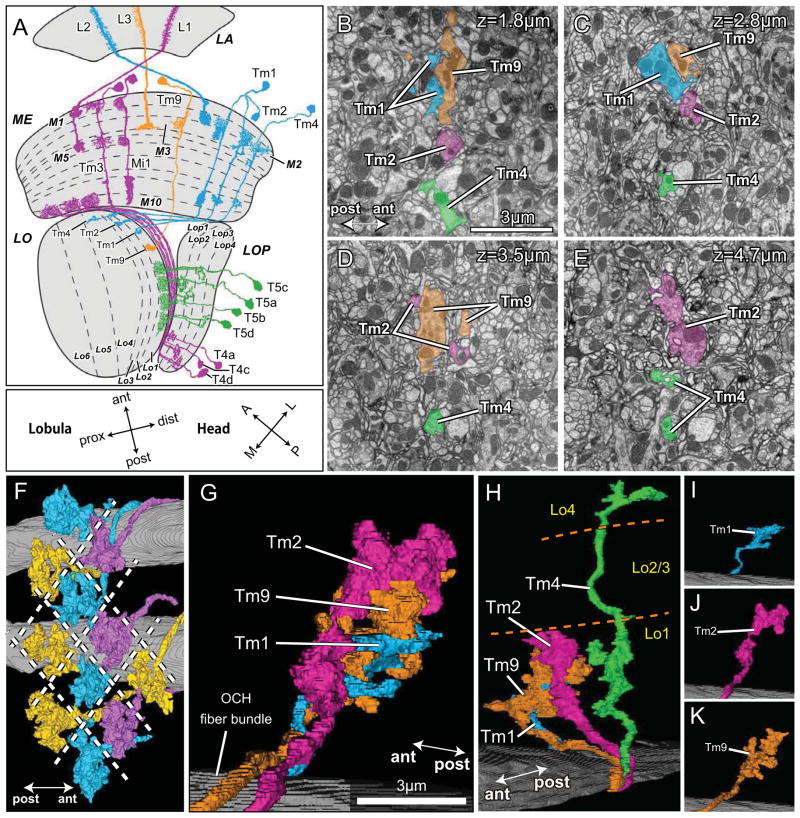
Figure 1.
T4 and T5 provide two independent pathways to four different lobula plate strata. (A) T4 (magenta) and T5 (green) inputs to four strata of the lobula plate, LOP (Lop1-Lop4). Tm cell inputs to T5 come via L2 (Tm1, Tm2, Tm4: cyan) and L3 (Tm9: orange) pathways from the lamina (LA). L1 pathway inputs to T4 in the proximal medulla (ME) are shown only in part for clarity, omitting the terminals of Tm3 in the lobula (LO). Orientation markers: the lobula is rotated 90° with respect to the medulla so that its anterior (ant) direction points towards the medulla and its posterior (post), towards the head’s midline (M). The lobula’s Tm cell inputs enter at its distal strata (dist), roughly towards the eye’s lateral edge (L), and extend proximally (prox). The lobula’s anterior edge receives input from frontally directed ommatidia and lamina cartridges, towards the head’s anterior (A), after two inversions of the pathways through the chiasmata between lamina and medulla, and between medulla and lobula. (B–E) Tm cell terminals innervate T5 cells in the lobula. Terminals of Tm1, Tm2, Tm4, and Tm9 at four depths in lobula stratum Lo1: 1.8μm (B); 2.8μm (C); 3.5μm (D); and 4.7μm (E) from the border with the inner chiasma. Lo1 completely contains the terminals of Tm9, Tm2 and the dendrites of T5, with Tm9 falling more superficially within this stratum [16]. The terminals of Tm2 extend deeper than those of Tm9 (ED), and Tm4 extends down to stratum Lo4. The profiles of terminals are often divided (Tm2, Tm9 in D). (F–K) Terminals of four modular Tm cell types. (F) Viewed from distal to proximal within the lobula neuropil, the terminals of 11 Tm9 cells tile the lobula, one per column, without touching; overlap is apparent, an artifact of the reconstruction angle. (G) Terminals of Tm1, Tm2 and Tm9 overlap each other as a fascicle in a single column. A bundle of horizontally directed axons in the optic chiasma (OCH) reveals the entry path for Tm axons to the neuropil. (H) Tm4’s terminal extends more deeply and is separate from the bundled terminals of the other three Tm cells, and lacks presynaptic sites in Lo2 and Lo3. (I–K) The terminal of Tm1 (I) is smallest, while the terminal of Tm9 is larger (K), more complex and compact than that of Tm2 (J), which extends deeper.
The medulla targets of L1 and L2
Recent reports from Drosophila do much to identify the medulla targets of L1 and L2 that connect to T4 and T5 [2, 25]. L1 provides input to a single-column medulla intrinsic (Mi) neuron Mi1 that contacts T4 cells in the proximal medulla, while L2’s chief targets are not one but two single-column transmedulla (Tm) cells, Tm1 and Tm2 (Figure 1A), that penetrate the medulla and terminate in the lobula [2, 25]. Many of these two cells’ dendrites receive input at the same synapses [2], much as L1 and L2 duplicate the R1–R6 pathway in the lamina. Tm2 also receives input from the terminals of L4 [2, 25].
Even though a major single-column target of L1 is indeed Mi1, previously undetected inputs are also made to a group of Tm3 neurons with dendrites that spread between columns [2]. Neurotransmitter candidates have yet to be identified for these two inputs. Likewise, L2 provides inputs to a corresponding group of Tm4 cells [2] (Figure 1A), so that both L1 and L2 provide respective input to single-column (Mi1, and Tm1/Tm2) and multiple-column (Tm3, Tm4) medulla target neurons. Dense reconstruction of a single medulla column [2] reveals that Mi1 and Tm3 neurons alone provide strong connections, having many synapses, spanning between the terminal of L1 and the dendrites of T4 cells in stratum M10 of the proximal medulla. The strength and exclusivity of anatomical connection between L1 and T4, and the correspondence between the anatomical vector of T4’s dendrite and that of the lobula plate, provide a strong basis to propose that vector comparisons between the Mi1 and combined Tm3 inputs from L1 to T4 could constitute two arms of an EMD. These same features also provide a basis against which to compare L2’s corresponding pathways to T5 cells that we now report. Such a correspondence first required documentation from serial-section electron microscopy (ssEM) of which Tm cells provide input to T5 in the lobula, the interpretation of which is aided by identifying the transmitter systems employed by cells in the L2 pathway.
T5 receives input from the lobula terminals of four specific types of Tm cells
T5 cells have an axon that divides in the inner chiasma, doubles back, and then innervates the lobula plate. Large fly species have up to four T5 cells per column [24] and Drosophila has four anatomical subtypes overall [16], each morphologically similar to those of T4 but with dendritic arbors in the lobula that resemble those of T4 in the proximal medulla (Figures 1A, 2). These four exhibit direction-selective responses to moving stimuli [1, 26].
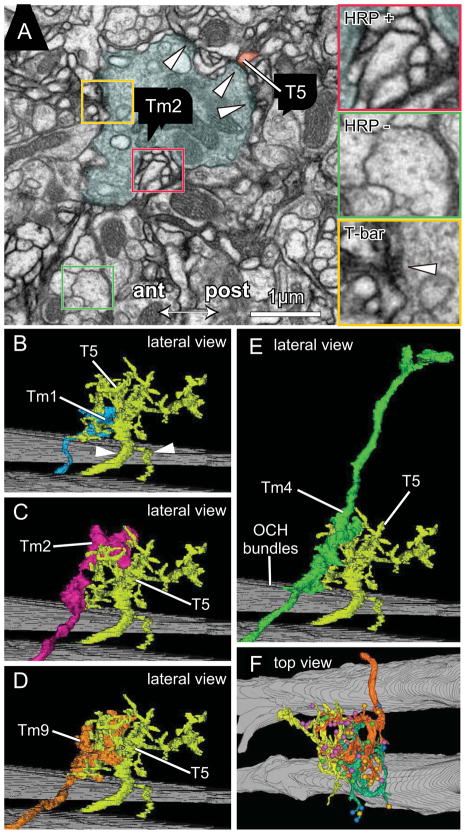
Figure 2.
Tm cell synaptic inputs to T5 dendrites. (A) EM of lobula stratum Lo1 showing a terminal of Tm2 surrounded by T5 dendrites expressing HRP over their membranes. Insets: HRP + (red) and HRP − (green) profiles; with presynaptic T-bar ribbon visible in Tm2 (yellow) providing input to HRP labeled T5 dendrites. (B–F) Representative pairs of Tm terminals overlapping the dendritic arbor of cell T5-1. Confirming its identity the latter has a bifurcated axon with the more slender branch going to the lobula plate, and its stouter partner connecting to the cell body (arrowheads in B). With T5-1 are lateral views of: (B) Tm1 (same terminal as Figure 1I); (C) Tm2 (same terminal as Figure 1J); (D) Tm9 (same terminal as Figure 1K); and (E) Tm4 (same terminal as Figure 1H). Background: bundles of horizontally directed chiasmal axons. (F) In a tangential plane, the overlap between neighboring T5 cells: T5-02 (orange), T5-01 (yellow) and T5-11 (green) with their color-coded synaptic inputs from four types of Tm input cells, Tm1, Tm2, Tm4 and Tm9 (between 1 and 3 cells per type, color coded as in Figure 3A–L).
Results
Synaptic studies on the lobula have so far to be reported, and inputs to T5 have been those surmised solely by overlap between Tm terminals and T5 dendrites. Using EM we now reveal that these inputs differ in unexpected ways from those to the T4 cells. We could identify the terminals of Tm cells from their shapes and depths of termination, by comparison with those previously reported from Golgi impregnation [16]. Given the complex branching patterns of T5’s dendrites however we used T4/T5-Gal4 driven UAS-HRP::CD2 to target expression of horseradish peroxidase (HRP) to the cell membrane, and render it osmiophilic after treatment with diaminobenzidine [27] (Figure 2A). This method, which successfully marks medulla neurons (e.g. [28]), labels T5’s tiny dendrites that would be difficult to trace in unlabeled preparations. The resulting reconstructions (Figure 2B–E) are as complete as those reported for T4 cells using semi-automated reconstruction methods ([2], their Figure 6), but are obtained much more efficiently. The HRP label also enabled us to identify any small, orphaned neurites that we could not trace back to a parent T5 cell, but which their HRP expression revealed had such an origin. We could clearly see the presence of T-bar ribbons, sites of neurotransmitter release at active zones [29], in terminals that abutted HRP labeled T5 profiles. The HRP label was sufficiently fine so as not to obscure the T5 cytoplasm, enabling us to ascertain that T5 dendrites entirely lacked T-bar ribbons themselves and were thus nowhere presynaptic. Use of targeted HRP expression is of course limited to individual neurons and does not circumvent the need to trace the unlabeled input terminals of Tm neurons, which also required considerable labor, especially in the case of the deep terminals of Tm4.
We concentrated on those Tm neurons that Golgi impregnation revealed had greatest overlap with the dendrites of T5, in lobula stratum Lo1 [16]. Most obvious were inputs from the L2 pathway [2, 25]: Tm1, Tm2, and Tm4. In addition, we could readily identify the terminals of a fourth Tm neuron, Tm9, from the size of its profiles in all columns, and previously identified to converge upon Lo1 in large fly species [30]. The axons from Tm1 and Tm2 entered the lobula with Tm9 as a bundle, which we presumed to correspond to a medulla column, whereas Tm4 was separate and its terminal penetrated deeper. Because this was more slender and needed to be traced a considerable depth, reconstructed Tm4 neurons were fewer, as were their identified synaptic inputs to T5.
In addition to the input they provide to T5, Tm4 also formed synapses in Lo4, a deeper stratum of the lobula. So, unlike their L2 pathway partners Tm1 and Tm2, which formed output synapses exclusively onto T5 cells, Tm4 terminals presumably provide a substrate for additional deeper circuits. For the five most completely reconstructed Tm4 terminals, the average total number of synapses was 29.2 ± 4.66 (mean ± SD), of which 20.2 ± 3.56 were in Lo1 and 9.0 ± 2.45 in Lo4. Thus, about 70% of Tm4’s synapses were distributed as inputs to T5 cells, within Lo1. We could not reconstruct or identify any of the targets of Tm4 in Lo4, but many visual projection neurons, the likely targets of Tm4, project from this proximal stratum to the central brain [16, 31], where they are anatomically qualified to convey L2 pathway information.
Inputs to T5 incorporate numerically strong and weak pathways
We found that Tm1 and Tm2, two major targets of L2 [2, 25], provide strong inputs to T5 neurons. We also reveal that a third L2 target, Tm4, provides input to T5 that was never previously identified. Since these three cell types receive most of the input from L2, our findings are consistent with the notion that T5 computes motion signals based on the OFF-channel L2 pathway. For example, in a reference L2 that provided identified input to a total of 483 postsynaptic sites, 69% of contacts were to dendrites of Tm1, Tm2 and Tm4 cells, allocated to 31.6%, 21.9% and 15.7% of those contacts respectively [2]. In sharp contrast to the L1 pathway that provides input to T4 in the medulla, however, T5 in the lobula also receives strong input from the L3 pathway, via its main medulla target cell, Tm9. That input also differs from those arriving from L2. L3 provides input to multiple medulla cell types, of which Tm9 is only the third most populated by synaptic contacts. Thus, a reference L3 provided input to a total of 137 identified postsynaptic sites, of which only 12% were to dendrites of Tm9[2].
Together these four Tm cell inputs constitute 87% of the total input to eight reconstructed T5 cells. Thus in aggregate they are the only numerically strong pathways between L2 and T5, and the sole pathway from L3. The numbers of presynaptic contacts from Tm1, Tm2, Tm4 and Tm9 differed significantly however, both for different T5 cell targets and between the input terminals themselves. Thus, the average number of synapses per Tm terminal providing input to the four most completely reconstructed T5 cells (T5-01, T5-02, T5-08, T5-11) ranged from 12.25 synapses for Tm2 to 4.25 synapses for Tm4 (Table S1), and for other or unidentified inputs only ~3 synapses. The dendrite: terminal ratios for the numbers of input synapses (from [2]) to output (Table S1), synapses therefore ranged from about 20 for both Tm1 and Tm4 (for example Tm1 has >220 medulla inputs to ~7 T5 outputs, for a ratio of ~30), less for Tm2 and about 2 for Tm9. Tm9 thus differed from the other Tm cells in having many T5 outputs for its few L3 input synapses. The final validity of such numerical comparisons relies of course upon knowledge of the synaptic gain at each class of synapse. For this, some knowledge of neurotransmission at each class of synapse is at a minimum necessary.
T5 dendrites receive spatially segregated inputs from Tm cells
Inputs to T5 dendrites arrive from three prominent Tm cells (Figure 1B–E, G): Tm9, with a large conspicuous terminal in Lo1 readily identified in all columns from its size (Figure 1K); Tm1 with a terminal in lobula stratum Lo1 (Figure 1I); and Tm2 with a terminal also in stratum Lo1 (Figure 1J). The Tm9 terminals tile the lobula’s stratum Lo1, overlapped by the terminals of Tm1 and Tm2, leaving small spaces between (Figure 1F), through which pass the axons of Tm4, with a terminal that extends deeper, from Lo1 to Lo4 (Figure 1H). Because this needed to be traced a considerable depth, which for technical reasons was sometimes difficult, we reconstructed fewer Tm4 neurons.
T5 cells exhibit directionally selective responses to moving dark edges, just as do T4 cells for moving bright edges [1]. The medulla dendrites of T4 have two subcomponents to their anatomical receptive field that arise from the two strong pathways, Mi1 and Tm3. Plotted as the corresponding weighted centers of synaptic input for each cell type, there is a small spatial offset between these subcomponents, and this provides a vector corresponding to the direction selectivity of the T4 cell, as defined by the lobula plate stratum in which this terminates [2]. For the dendrites of T5, inputs from Tm1 and Tm2, and of Tm2 and Tm9 are both spatially segregated over the dendritic arbor (Figure 3), leading us to examine whether the members of these two pairs of inputs may likewise signal from divergent fields of view, so as to provide a possible substrate for a T5 EMD circuit, but in this case two EMDs rather than one. Our analysis of this possibility is limited because we cannot identify the medulla columns of origin for the Tm neurons we trace, for which we would have to trace axons through the orthogonal inflection in their trajectory through the internal chiasma between medulla and lobula. Nevertheless, if we reasonably assign each axon bundle to a single medulla column, and neighboring bundles to neighboring columns, each signaling a neighboring point in visual space, we could plot the relative numbers of inputs to single T5 cell dendrites that arose from different bundles (Figure 3M).
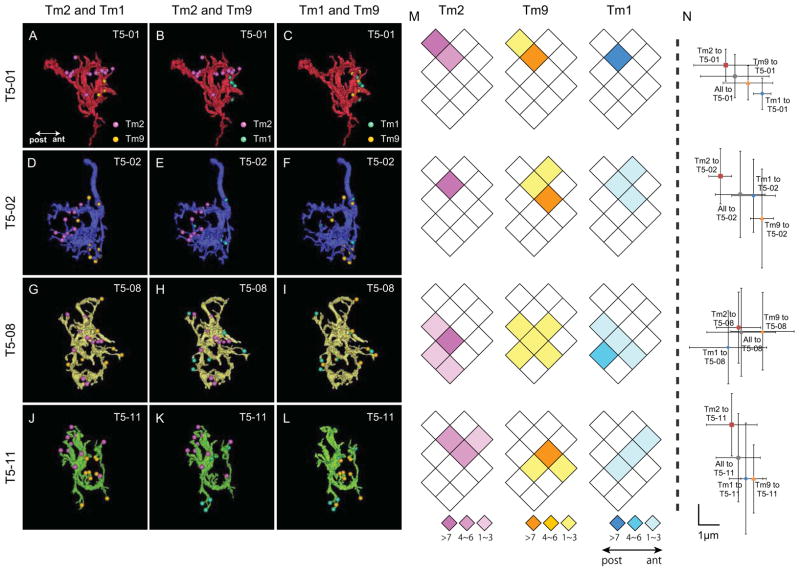
Figure 3.
Anatomical receptive fields of four T5 dendritic arbors. (A–L) Four reconstructed T5 cells (T5-01 (A–C), T5-02 (D–F), T5-08 (G–I) and T5-11 (J–L) as seen from the proximal to distal face of the lobula. Synaptic input sites from three types of Tm cells (Tm1, Tm2 and Tm9) are color coded (cyan, magenta and orange, respectively) in paired combinations. Synaptic contacts (T-bar ribbons) are from Tm2 and Tm9 (A, D, G, J); Tm1 and Tm2 (B, E, H, K); or from Tm1 and Tm9 (C, F, I, L). Each Tm cell’s synaptic input is restricted to a limited zone of the T5 dendritic arbor, and input areas for different Tm cells (Tm2 and Tm1, Tm2 and Tm9) to each single T5 cell are generally segregated, suggesting that the T5 receptive field incorporates at least two pairs of anatomical subcomponent fields: Tm1/Tm2, and Tm2/Tm9. M) Diagram of the array of lobula columns, rhomboids (each assumed to correspond to a bundle of axon terminals, and to an overlying medulla column), within which are plotted the distribution of Tm cell synapses corresponding to the same T5 cells shown in the horizontal row to the left. Color-coded rhomboids indicate the number of input synapses in each column (from 1 to >7) from specific Tm cells (Tm2: magenta, Tm9: orange, Tm1: cyan). One T5 cell may receive inputs from multiple Tm cells in multiple columns, the receptive field encompassing more than a single column. For each T5 cell, inputs come from 2–6 columns overall, contributed by all three types of Tm input. These arise for each Tm input from between 1 and 5 of the columns. So, for example, comparing inputs from Tm1 with those from Tm9, most inputs arise from a corresponding set of columns, but the overlap is not perfect. Thus for T5-08, Tm9 inputs come from five columns, whereas Tm1 inputs come from only four. N) The summed numbers of synapses distributed over several columns, shown as the corresponding x, y coordinates (μm) for Tm1 (cyan), Tm2 (magenta) and Tm9 (orange) inputs, relative to their sum (grey). Error bars: SD of the x, y coordinates for each synapse, provide a measure of the spread of inputs from each Tm input. Tm2 and Tm9, and Tm1 and Tm2 provide two anatomical receptive field subcomponents with significant angular separation. The angular offset between each Tm1, Tm2 and Tm2, Tm9 pair falls in the same direction for each T5 cell, indicating congruence of the displacement vector for the T5 subtype. For T5-08 the offset between Tm2 and Tm9 is ambiguous. No significant difference is seen between the summed distributions of Tm1 and Tm9 inputs. Tm4 inputs are too few to plot reliably.
We reconstructed eight T5 cells and plotted their four numerically strong synaptic inputs from Tm1, Tm2, Tm4 and Tm9. Because the number of inputs to T5 cells varied considerably, we selected four with fewer than 8.6% of their inputs unidentified or not from Tm1, Tm2, Tm4 or Tm9. All had more than 22% of their inputs from Tm9. Four other T5 cells had more unidentified inputs, up to 16.7% of the total, and two had fewer Tm9 inputs (one cell in fact having only a single such input). The distributions of these numbers suggested considerable heterogeneity among the T5 cells, but with some consistencies. For example all had more inputs from Tm2 than Tm1, in some cases up to twice as many. We made plots for the four selected T5 cells. These revealed that inputs from Tm2 and Tm9 and from Tm2 and Tm1 have weighted distributions with centers that diverged from each other. That divergence constituted a vector for each pair and each T5 target (Figure 3N). The divergence between the weighted centers for Tm1 and Tm9 inputs was, by contrast, less than those for the other two pairs.
Our data do not yet let us say whether the vector angles form four subtypes, compatible with the four subtypes a–d of their T5 targets. The vector angles for the four T5 cells fell along different directions (Figure 3M, N), at least two of the four quadrants of a polar plot, but we lack information on the depth in the lobula plate at which each T5 cell terminated, and thus on the subtype to which each cell belonged. This would have required us to trace the T5 cells across the internal chiasma, which was not possible in our EM series, which was cut in a direction from the distal surface of the lobula into the depth of the lobula neuropil to identify the Tm terminals, whereas to trace the terminals of T4 and T5 would have required us to cut ultrathin series in the opposite direction, into the lobula plate.
T4 and T5 provide cholinergic input to the lobula plate
Neurotransmitters have not been well documented in Drosophila neither for T4 nor T5, although at least one subtype (b) of T4 is labeled by a Cha-Gal4 line [32]. In different larger species of flies some T4 and T5 cells are reported GABA [33] or aspartate (T5) immunoreactive [30]. To determine the neurotransmitter genotype of Drosophila T4 and T5 neurons, we therefore manually isolated GFP-labeled T4 and T5 neurons and used RT-PCR to assess mRNA expression of diagnostic transporters or biosynthetic enzymes for known neurotransmitters [34, 35]. While RT-PCR detected the presence of transcripts for all diagnostic genes in the optic lobe (data not shown), only transcripts of choline acetyltransferase (ChAT) were identified in T4 and T5 (Figures S1A, S1A′), suggesting that these two cell types are likely to be cholinergic. We quantified ChAT transcript level using real-time PCR and found that T5 expressed high levels of ChAT transcript (5952 ± 617 copies per cell), comparable to those of Rp49, a ribosomal protein (8513 ± 412 copies per cell; Figures S1D, F). The expression of ChAT but not VGlut in T4 and T5 neurons was further confirmed by immunohistochemistry against the products of these two genes, using anti-ChAT and anti-VGlut antibodies (Figures 4A, B), strongly suggesting that all T-cells of both types, T4 and T5, express a cholinergic phenotype. In agreement, intersectional genetics (split Gal4:[36]) and pharmacologic approaches together indicate that the LPTCs receive excitatory cholinergic input from both T4 and T5 [37]. We cannot exclude that either or both cell types, or their four respective subtypes, may additionally express another fast neurotransmitter. Insofar as nicotinic Dα7 cholinoceptors are expressed on the fine dendrites of HS cells [38], it is moreover likely that the cholinergic inputs of T4 and T5 to the LPTCs are excitatory [32], as also concluded in a parallel study [37].
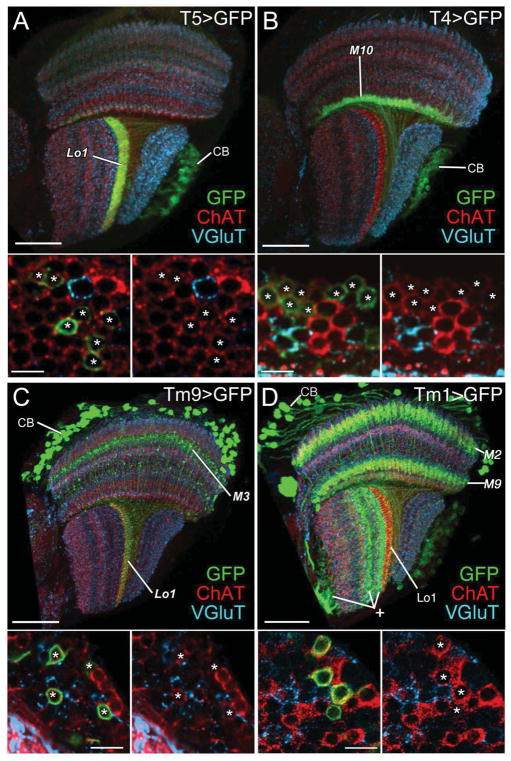
Figure 4.
T4, T5, Tm9, and Tm1 neurons express immunoreactivity to choline acetyltransferase (ChAT, red) but not vesicular glutamate transporter (VGlut, cyan). (A–D) T5, T4, Tm9, and Tm1 neurons are labeled with the mCD8::GFP membrane marker (green) using T5-Gal4, T4-Gal4, Tm9-Gal4, and Tm1-Gal4 drivers, respectively. (A) T5-GFP expression highlights a band of dendrites in lobula stratum Lo1 and cell bodies (CB) in the lobula plate cortex and their axons in the internal chiasma between lobula and lobula plate. (B) T4-GFP expression highlights a band of dendrites concentrated in medulla stratum M10 and cell bodies (CB) in the lobula plate cortex and their axons penetrating the lobula plate and entering the internal chiasma between medulla and lobula plate. (C) Tm9-GFP expression reveals somata (CB) in the medulla cortex, axons that penetrate the medulla neuropil, an arborization in stratum M3, and a band of terminals in lobula stratum Lo1. (D) Tm1-GFP expression reveals somata (CB) in the medulla cortex, dendrites that arborize in medulla strata M2 and M9, and axons that terminate in lobula stratum Lo1. The Gal4 line also labels uncharacterized neurons in the lobula (plus sign) and lobula plate. Lower panels: high-magnification views of the corresponding cell bodies (asterisks) in the cortex of the lobula plate (A, B) and medulla (C, D), distributed amongst cell bodies of other neurons (no asterisk). The left hand image of each pair shows all three labels; for clarity the green channel is omitted in the right hand panel, to show that all GFP expressing somata are also ChAT-positive. Scale bars: 30 μm in A–D; and 5 μm in lower panels.
L2 and L3 pathways provide cholinergic inputs to T5
Although reported to converge upon the T5 layer of the lobula in larger fly species [30], Tm9 was initially an unexpected synaptic input to T5 in Drosophila because its own major input in the distal medulla comes from neither of the L1 nor L2 pathways, but from a third type of lamina input neuron, L3 [2], not previously recognized for its involvement in motion sensing. L3 has been implicated in landmark orientation [18] but in parallel with our identification of Tm9’s input to T5, imaging and silencing experiments revealed that L3 also cooperates with L1 and L2 in the circuits that detect moving light and dark edges [3]. Tm9’s inputs to T5 thus provide us with an anatomical substrate for that cooperation in the lobula.
We next sought to identify candidate neurotransmitters in these pathways. The L2 pathway contains cells with cholinergic phenotypes. Thus L2, L4 and their common target Tm2 all express transcripts for ChAT [25] so that at least one input to T5 should be cholinergic. To identify the neurotransmitter credentials of Tm9, we used RT-PCR to access mRNA expression of diagnostic transporters or biosynthetic enzymes for known neurotransmitters in GFP-labeled Tm9 cells [39]. We detected only ChAT mRNA in Tm9, suggesting that Tm9 is cholinergic (Figure S1A″), even though this morphological cell type in large fly species is reported to be GABA-positive [30]. Quantitative real-time PCR revealed that Tm9 expressed high levels of ChAT transcript (6272 ± 510 copies per cell), comparable to those of Rp49, a ribosomal protein (8397 ± 707 copies per cell; Figures S1D, F). The cholinergic phenotype of Tm9 was further confirmed using anti-ChAT immunolabeling (Figure 4C). Based on the expression pattern of ChAT-Gal4, Tm1 and Tm4 have also previously been suggested to be cholinergic [32]. We confirmed this possibility for Tm1 by immunolabeling a Tm1-Gal4 line [40] with anti-ChAT, which revealed that Tm1 expresses ChAT but is not immunoreactive to VGluT and therefore is indeed likely to be cholinergic (Figure 4D). However, we were not able to confirm a cholinergic phenotype for Tm4 cells using single-cell transcript profiling or immunohistochemistry because we lack a specific Gal4 line for this cell type. In summary, T5 receives inputs from Tm1, Tm2 and Tm9 that are genetically qualified to be cholinergic, and also from Tm4, which is also reported to be cholinergic, thus receiving acetylcholine from all its anatomically identified synaptic inputs.
To further characterize the nature of transmission at Tm/T5 connections, we extended our transcript profile analyses of T5 to include all known Drosophila acetylcholine receptors. The Drosophila genome encodes 7 nicotinic alpha (nAcRα1-7) and 3 beta (nAcRβ1-3) subunits as well as two muscarinic (mAchR-A and mAchR-B) cholinoceptors (Figures S1B, S1B′; [25, 34, 41]). Interestingly, we found that T5 expresses the transcripts of nAcRα3 (CG2303) and nAcRβ3 (CG11822) nicotinic cholinoceptors as well as those of an A-type muscarinic cholinoceptor (mAchR-A [CG4356]), suggesting that T5 signals cholinergic Tm inputs by means of both ionotropic and metabotropic cholinoceptors (Figure S1B′). The Drosophila mAchR-A is homologous to the vertebrate m1/3-type muscarinic cholinoceptors, which couple to Gq/11 and the IP3 (inositol 1,4,5-trisphosphate) second messenger pathway to mobilize intracellular calcium (reviewed in [42]). Quantitative PCR further revealed that T5 expresses mAchR-A but at a rather more modest level (2914 ± 725 copies per cell: Figure S1E), compared with that of ChAT or Rp49 (Figures S1D, F). For comparison with T5, and in view of the similar functions and common targets of these two cell types, we also determined the expression patterns of cholinoceptors in T4 cells by means of single-cell transcript profiling. We found that T4 likewise expresses both nicotinic and muscarinic cholinoceptors but that these differ somewhat from those expressed in T5: T4 expresses nAcRα7 (CG8109) and nAcRβ3 (shared with T5) as well as mAchR-B (CG7918) (Figure S1B). The significance of cholinergic receptor expression in T4 must await resolution of the transmitters used by both Mi1 and Tm3.
Discussion
Does T5 use its nicotinic and muscarinic cholinoceptors to integrate multiple Tm inputs for motion detection?
The L2 pathway has been identified as the substrate for detecting moving dark edges [7, 43]. L2’s partner cells, L4, and their common medulla targets Tm2 are both essential components of this dark edge pathway, prior to the computation of directionality, which occurs first in the dendrites of T5 [1]. Tm2 and L4 cells both respond with a non-directional increase in activity to moving dark (OFF) edges, and silencing both, singly or in combination also abolishes the response to moving dark edges in downstream LTPCs [44]. The anatomical receptive fields we report for the input terminals to T5 cell dendrites are compatible with Tm2 being wired as one of the two arms of an EMD circuit, and either or both Tm1 and Tm9 as the other (Figure 5A, B). The existence of two EMD circuits would require that the inputs from Tm1/Tm2 and Tm9/Tm2 have aligned vector angles, since each T5 receives both circuits but must respond to dark edge motion in only one of the four cardinal directions. This alignment is clear for three of the T5 cells plotted in detail, but less so for T5-08 (Figure 3N). Clearly, additional detailed plots of T5 anatomical receptive fields are needed.
Figure 5.
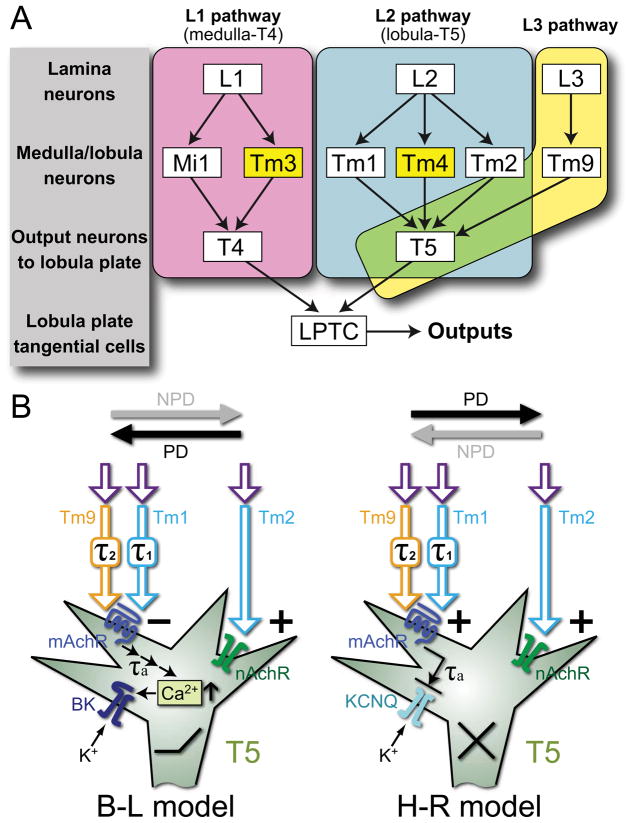
Models of motion detection pathways and proposed molecular mechanisms. A) Input pathways to T4 and T5 comprise single-column and multi-column (yellow) Tm cells. L1 pathways (magenta) via single-column Mi1 and multi-column Tm3 cells converge upon T4 in the proximal medulla [2]. L2 pathways (cyan) via single-column Tm1 and Tm2 [25] and multi-column Tm4 [2] cells converge upon T5 in the distal lobula, along with Tm9 [2] from the L3 pathway (yellow). T4 and T5 cells in turn provide direction-selective inputs to LPTCs. B) Hypothetical implementation of Barlow-Levick (B–L) and Hassenstein-Reichardt (H–R) models. In both, T5 is proposed to use muscarinic cholinoceptors (mAchR) to receive a delayed signal from Tm1 and Tm9, and nicotinic receptors (nAchR) to receive an instantaneous signal from Tm2. In the B–L model, these two signals converge antagonistically: the activation of nAchR increases sodium conductance and depolarizes the membrane while the activation of mAchR leads to calcium release from internal stores, the activation of a high-conductance calcium-dependent potassium (BK) channel, and eventual membrane hyperpolarization. In the H–R model, the instantaneous and delayed signals interact synergistically: activated mAchR inhibits a Kv type potassium channel, leading to membrane depolarization while activating nAchR depolarizes the membrane. PD: preferred, and NPD: non-preferred directions of motion; τ1, τ2, τa: time delays in Tm1, Tm9, and T5, respectively.
Even though this scheme is still highly speculative, nonlinear interaction between two input arms of the EMD circuits is the computational basis of local motion-detection models while specific computational models favor different types of interactions: either multiplication/facilitation for the Hassenstein-Reichardt model [4] or inhibition for the Barlow-Levick model [6] (Figure 5B).
A parallel may be seen in the vertebrate retina. There, turtle B10 bipolar cells use ionotropic and metabotropic glutamate receptors to signal, respectively, L- and M-cone inputs, a dual deployment that has been suggested to form the basis for B10 red-blue/green color opponency [45]. The expression of both nicotinic and muscarinic cholinoceptors in T5 may provide a similar means to integrate multiple cholinergic Tm inputs and so compute small-field motion signals. Vertebrate muscarinic receptors are coupled to G-proteins and various downstream signaling pathways to regulate a broad spectrum of cellular functions, including neuronal excitability [46]. In Drosophila, agonist activation of mAchR-A acts via the IP3 pathway to increase calcium release from internal stores [47], which elsewhere is reported in turn to activate high-conductance calcium-dependent potassium (BK) channels (the slowpoke channel), leading to membrane hyperpolarization [48]. It was therefore interesting that, using single-cell RT-PCR, we detected slowpoke transcripts in both T4 and T5 (Figure S1C), consistent with a previous immunohistochemical study that slowpoke is expressed in the optic lobe, including the lobula [49]. We propose that these two postsynaptic events, mAchR-A mediated increased intracellular calcium and mAchR activation of slowpoke channels, occur at postsynaptic sites activated by different inputs distributed over the T5 dendritic arbor (Figure 5B). Given the relative temporal inflexibility of an excitatory nicotinic cholinoceptor synaptic response, and the fact that their anatomy qualifies Tm2/Tm9 and Tm2/Tm1 input pairs as two independent pairs of EMD input arms to T5 that share Tm2, it seems most reasonable to us that T5 uses the nicotinic receptor inputs for fast excitation from Tm2 (as the instantaneous signal) and compares this with a slow inhibitory input from either Tm9 or Tm1 (as the delayed signal). This arrangement could allow T5 to inherit motion information with two temporal characteristics provided that Tm1 and Tm9 have their own time delay (τ1 and τ2, Figure 5B). The coupling of the inhibitory inputs to secondary messenger system also provides a potential mechanism to adapt the temporal delay filter (τa in Figure 5B; [50–52]). In addition, the activation of Drosophila mAchR-A in cultured cells also induces a secondary calcium influx, potentially originating from an extracellular calcium pool [47], while the activation of vertebrate muscarinic receptors has been shown to inhibit potassium channels, including those of the Kv7 (KCNQ/M) type, and to lower the excitability threshold (Figure 5B; [53, 54]). It is therefore possible that an interaction between the nicotinic and muscarinic inputs provides some form of multiplication, as suggested in the Hassenstein-Reichardt model (Figure 5B). Further definition of the roles of the Tm1, Tm2 and Tm9 input cells as input arms to different EMD circuits, and determination of the synaptic mechanisms for detecting moving dark edges, must obviously await additional genetic silencing experiments and electrophysiological recordings from all these cells, which this study now clearly identifies as synaptic inputs to T5 in Drosophila.
Experimental Procedures
Fly stocks
Fly stocks were maintained on standard medium at 23–25°C. The genotypes of the flies are listed in Supplemental Data Table 2.
Cell dissociation, single-cell isolation, and RT-PCR
Single Tm2, T4, and T5 neurons labeled with GFP were dissociated from adult medulla or lobula plate cortices and single GFP-positive cells were collected using a custom-made capillary aspiration system as previously described [25]. Briefly, total RNA from lysed single cells was reverse-transcribed to cDNA. PCR analyses were used to determine the presence of specific transcripts for neurotransmitter transporters, biosynthesis enzymes, and for acetylcholine receptors, and the Slowpoke gene. Detailed procedures for cell dissociation, single-cell isolation, PCR primers, and single-cell PCR assay were carried out as described previously [25, 35]. PCR primers for amplifying slowpoke transcript are as follows: First-round PCR primer sequence (5′-3′) Forward: TATACTCAGCATTGCATCCCTC Reverse: GATGTATAATGCCCGCTGCTG Nested PCR primer sequence (5′-3′) Forward: AGCGAAGAAGTCGAAAGATGC Reverse: GTTGAGCCAAGCGTATGGAG
Immunohistochemistry
Brain dissection, fixation, immunohistochemistry, and confocal imaging were undertaken as described previously [55]. The following primary antibodies were used: 1) mouse anti-GFP (3E6, Life Technologies, 1:200); 2) mouse anti-ChAT (4B1, DSHB, 1:200); and 3) rabbit anti-DVGlut ([56], 1:10000). The secondary antibodies used were goat anti-mouse, or rabbit conjugated with Alexa Fluro 488, 568, and 647 (Life Technologies) at a 1:400 dilution.
Electron microscopy
Specimens were prepared for EM, and reconstructions made from series of 50 nm sections. Ultrathin series were prepared as previously reported [57]; those with targeted expression of HRP were processed also as previously reported [58]. Images were captured using a Gatan Orius 832 11MB camera with a Philips Tecnai 12 electron microscope operated at 80kV. Image series were aligned using the TrakEM2 plug-in [59] for Fiji (http://fiji.sc/) and cells reconstructed. Procedures to locate synaptic inputs and their distributions over T5 arbors are summarized in the Supplemental Experimental Procedures.
Supplementary Material
Figure S1: Transcript profiles of T4, T5 and Tm9 neurons reveal their expression of transcripts for neurotransmitter transporters and biosynthesis enzymes, as well as cholinoceptors and the potassium channel Slowpoke.
(A) T4 cells. (A′) T5 cells. (A″) Tm9 cells. PCR-amplified products of the transcripts from single Gal4-driven GFP-expressing neurons. Choline acetyl transferase (ChAT) transcript appears in T4, T5 and Tm9. Lane M: 100bp DNA ladder marker; lane C: negative control (without reverse transcription). 1: ChAT; 2: VGlut; 3: VGAT; 4: Hdc; 5: DAT; 6: SerT; 7: Tβh; and 8: Tdc2
(B, B′) PCR-amplified products of cholinoceptor transcripts from single T4 (B) and T5 (B′) cells. T4 neurons express two nicotinic receptor subunits, Dα7 and Dβ3, as well as the muscarinic receptor mAcR-B (CG7918). T5 neurons express two nicotinic receptor subunits, Dα3 and Dβ3, as well as a different muscarinic receptor, mAcR-A (CG4356). Lane M: 100bp DNA ladder marker; 1: nAcRα-96Aα (Dα1); 2: nAcRα-96Ab (Dα2); 3: nAcRα-7E (Dα3); 4: nAcRα-80B (Dα4); 5: nAcRa-34E (Dα5); 6: nAcRa-30D (Dα6); 7: nAcRα-7 (Dα7); 8: nAcRβ-64B (Dβ1); 9: nAcRβ-96A (Dβ2); 10: nAcRβ-21C (Dβ3); 11: mAcR-A (CG4356); 12: mAcR-B (CG7918).
(C) PCR-amplified products of the potassium channel slowpoke transcripts from T4 (lane i) and T5 (lane ii). Lane M: 100bp DNA ladder marker.
(D–F) Standard curves for T5 cells generated by plotting CT values against the known initial DNA copy number. Abscissa is the logarithm of the input cDNA; ordinate is the number of PCR cycles required to reach a given fluorescence signal level (Ct). Threshold cycle, CT, values (ordinate) were obtained using a controlled amount of input cDNA for ChAT (D); mAcR-A (E); and Rp49 (F). The final PCR product was analyzed by DNA gel electrophoresis (D′, E′, F′). Ct and the logarithm of the input cDNA varied in inverse linear proportion over a large concentration range. Standard linear regression analysis was used to calculate the standard curve, the amplification factor (AmF), PCR efficiency (Eff), and the coefficient of determination (R2) for each curve. The standard curves were used to calculate the ChAT [D] and mAcR-A transcript level in Tm9 (red arrows) and T5 (blue arrows) neurons. All qRT-PCR amplification reactions were carried out in three biological replicates for each cell type. (D′) Lane M: 100bp DNA ladder marker; lane C: negative control (without reverse transcription). Lanes 1-9: ChAT amplicon using different amounts of input DNA (D). (E′) as in (D′) but for mAcR-A. (F′) as in (D′) but for Rp49.
Table S1: The numbers of input synapses from four classes of Tm cell input (Tm1, Tm2, Tm9, Tm4) to their T5 target cells
Table S2: Fly stocks used in this study.
Highlights.
EM reveals synaptic inputs to T5 cells that signal motion direction in Drosophila
Four classes of transmedulla (Tm) interneurons relay the L2 and L3 pathways to T5
T5 expresses nicotinic and muscarinic receptors to integrate Tm cholinergic inputs
Fast nicotinic excitation and delayed muscarinic inhibition mediate two arms of EMD
Acknowledgments
This work was supported by NIH grant EY-03592 (to I.A.M.) and the Intramural Research Program of the NIH, Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant Z01-HD008776 to C.-H. L.). During part of his work, K.S. was in receipt of a Government of Canada Post-Doctoral Research Fellowship from Foreign Affairs and International Trade Canada (DFAIT). We thank Jane Anne Horne (Dalhousie) for assistance in making the EM reconstructions, Chun-Yuan Ting (NIH) for assistance in analyzing synaptic distribution and immunohistochemistry, Mark Stopfer (NIH) for helpful discussions, and Axel Borst (Martinsried) for reagents and communicated intelligence.
Footnotes
Supplemental Information includes one figure and two tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maisak MS, Haag J, Ammer G, Serbe E, Meier M, Leonhardt A, Schilling T, Bahl A, Rubin GM, Nern A, et al. A directional tuning map of Drosophila elementary motion detectors. Nature. 2013;500:212–216. doi: 10.1038/nature12320. [DOI] [PubMed] [Google Scholar]
- 2.Takemura S, Bharioke A, Lu Z, Nern A, Vitaladevuni S, Rivlin PK, Katz WT, Olbris DJ, Plaza SM, Winston P, et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature. 2013;500:175–181. doi: 10.1038/nature12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silies M, Gohl DM, Fisher YE, Freifeld L, Clark DA, Clandinin TR. Modular use of peripheral input channels tunes motion-detecting circuitry. Neuron. 2013;79:111–127. doi: 10.1016/j.neuron.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassenstein B, Reichardt W. Systemtheoretische Analyse der Zeit-, Reihenfolgen- und Vorzeichenauswertung bei der Bewegungsperzeption des Rüsselkäfers Chlorophanus. Z Naturforsch. 1956;11:513–524. [Google Scholar]
- 5.Reichardt W. Autocorrelation, a principle for the evaluation of sensory information by the central nervous system. In: Rosenblith WA, editor. In Sensory communication. New York/London: MIT Press; 1961. pp. 303–317. [Google Scholar]
- 6.Barlow HB, Levick WR. The mechanism of directionally selective units in rabbit’s retina. J Physiol. 1965;178:477–504. doi: 10.1113/jphysiol.1965.sp007638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joesch M, Schnell B, Raghu SV, Reiff DF, Borst A. ON and OFF pathways in Drosophila motion vision. Nature. 2010;468:300–304. doi: 10.1038/nature09545. [DOI] [PubMed] [Google Scholar]
- 8.Eichner H, Joesch M, Schnell B, Reiff DF, Borst A. Internal structure of the fly elementary motion detector. Neuron. 2011;70:1155–1164. doi: 10.1016/j.neuron.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 9.Dowling JE. The Retina: an Approachable Part of the Brain (revised edition) Cambridge, MA: Harvard University Press; 2012. [Google Scholar]
- 10.Wässle H. Parallel processing in the mammalian retina. Nat Rev Neurosci. 2004;5:747–757. doi: 10.1038/nrn1497. [DOI] [PubMed] [Google Scholar]
- 11.Meinertzhagen IA, O’Neil SD. Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J Comp Neurol. 1991;305:232–263. doi: 10.1002/cne.903050206. [DOI] [PubMed] [Google Scholar]
- 12.Meinertzhagen IA, Sorra KE. Synaptic organization in the fly’s optic lamina: few cells, many synapses and divergent microcircuits. Prog Brain Res. 2001;131:53–69. doi: 10.1016/s0079-6123(01)31007-5. [DOI] [PubMed] [Google Scholar]
- 13.Rivera-Alba M, Vitaladevuni SN, Mishchenko Y, Lu Z, Takemura S, Scheffer L, Meinertzhagen IA, Chklovskii DB, de Polavieja GG. Wiring economy and volume exclusion determine neuronal placement in the Drosophila brain. Curr Biol. 2011;21:2000–2005. doi: 10.1016/j.cub.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Tousa JE, Baehr W, Martin RL, Hirsh J, Pak WL, Applebury ML. The Drosophila ninaE gene encodes an opsin. Cell. 1985;40:839–850. doi: 10.1016/0092-8674(85)90343-5. [DOI] [PubMed] [Google Scholar]
- 15.Hardie RC. Electrophysiological analysis of fly retina. I: Comparative properties of R1-6 and R7 and 8. J Comp Physiol A. 1979;129:19–33. [Google Scholar]
- 16.Fischbach KF, Dittrich APM. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 1989;258:441–475. [Google Scholar]
- 17.Douglass JK, Strausfeld NJ. Anatomical organization of retinotopic motion-sensitive pathways in the optic lobes of flies. Microsc Res Tech. 2003;62:132–150. doi: 10.1002/jemt.10367. [DOI] [PubMed] [Google Scholar]
- 18.Rister J, Pauls D, Schnell B, Ting CY, Lee CH, Sinakevitch I, Morante J, Strausfeld NJ, Ito K, Heisenberg M. Dissection of the peripheral motion channel in the visual system of Drosophila melanogaster. Neuron. 2007;56:155–170. doi: 10.1016/j.neuron.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Tuthill JC, Nern A, Holtz SL, Rubin GM, Reiser MB. Contributions of the 12 neuron classes in the fly lamina to motion vision. Neuron. 2013;79:128–140. doi: 10.1016/j.neuron.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schnell B, Raghu SV, Nern A, Borst A. Columnar cells necessary for motion responses of wide-field visual interneurons in Drosophila. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2012;198:389–395. doi: 10.1007/s00359-012-0716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borst A, Haag J, Reiff DF. Fly motion vision. Annu Rev Neurosci. 2010;33:49–70. doi: 10.1146/annurev-neuro-060909-153155. [DOI] [PubMed] [Google Scholar]
- 22.Buchner E, Buchner S. Mapping stimulus-induced nervous activity in small brains by [3H]2-deoxy-D-glucose. Cell Tissue Res. 1980;211:51–64. doi: 10.1007/BF00233722. [DOI] [PubMed] [Google Scholar]
- 23.Bausenwein B, Fischbach KF. Activity labeling patterns in the medulla of Drosophila melanogaster caused by motion stimuli. Cell Tissue Res. 1992;270:25–35. doi: 10.1007/BF00381876. [DOI] [PubMed] [Google Scholar]
- 24.Strausfeld NJ, Lee JK. Neuronal basis for parallel visual processing in the fly. Vis Neurosci. 1991;7:13–33. doi: 10.1017/s0952523800010919. [DOI] [PubMed] [Google Scholar]
- 25.Takemura S, Karuppudurai T, Ting CY, Lu Z, Lee CH, Meinertzhagen IA. Cholinergic circuits integrate neighboring visual signals in a Drosophila motion detection pathway. Curr Biol. 2011;21:2077–2084. doi: 10.1016/j.cub.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Douglass JK, Strausfeld NJ. Visual motion-detection circuits in flies: parallel direction- and non-direction-sensitive pathways between the medulla and lobula plate. J Neurosci. 1996;16:4551–4562. doi: 10.1523/JNEUROSCI.16-15-04551.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graham RC, Jr, Karnovsky MJ. The early stages of absorption of injected horseradish peroxidase in the proximal tubules of mouse kidney: ultrastructural cytochemistry by a new technique. J Histochem Cytochem. 1966;14:291–302. doi: 10.1177/14.4.291. [DOI] [PubMed] [Google Scholar]
- 28.Gao S, Takemura S, Ting CY, Huang S, Lu Z, Luan H, Rister J, Thum AS, Yang M, Hong ST, et al. The neural substrate of spectral preference in Drosophila. Neuron. 2008;60:328–342. doi: 10.1016/j.neuron.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saint Marie RL, Carlson SD. Synaptic vesicle activity in stimulated and unstimulated photoreceptor axons in the housefly. A freeze-fracture study. J Neurocytol. 1982;11:747–761. doi: 10.1007/BF01153517. [DOI] [PubMed] [Google Scholar]
- 30.Sinakevitch I, Strausfeld NJ. Chemical neuroanatomy of the fly’s movement detection pathway. J Comp Neurol. 2004;468:6–23. doi: 10.1002/cne.10929. [DOI] [PubMed] [Google Scholar]
- 31.Otsuna H, Ito K. Systematic analysis of the visual projection neurons of Drosophila melanogaster. I. Lobula-specific pathways. J Comp Neurol. 2006;497:928–958. doi: 10.1002/cne.21015. [DOI] [PubMed] [Google Scholar]
- 32.Raghu SV, Reiff DF, Borst A. Neurons with cholinergic phenotype in the visual system of Drosophila. J Comp Neurol. 2011;519:162–176. doi: 10.1002/cne.22512. [DOI] [PubMed] [Google Scholar]
- 33.Strausfeld NJ, Kong A, Milde JJ, Gilbert C, Ramaiah L. Oculomotor control in calliphorid flies: GABAergic organization in heterolateral inhibitory pathways. J Comp Neurol. 1995;361:298–320. doi: 10.1002/cne.903610208. [DOI] [PubMed] [Google Scholar]
- 34.Meinertzhagen IA, Lee CH. The genetic analysis of functional connectomics in Drosophila. Adv Genet. 2012;80:99–151. doi: 10.1016/B978-0-12-404742-6.00003-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karuppudurai T, Lin TY, Ting CY, Pursley R, Melnattur KV, Diao F, White BH, Macpherson LJ, Gallio M, Pohida T, et al. A Hard-Wired Glutamatergic Circuit Pools and Relays UV Signals to Mediate Spectral Preference in Drosophila. Neuron. 2014;81:603–615. doi: 10.1016/j.neuron.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mauss AS, Meier M, Serbe E, Borst A. Optogenetic and pharmacologic dissection of feedforward inhibition in Drosophila motion vision. J Neurosci. 2014;34:2254–2263. doi: 10.1523/JNEUROSCI.3938-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raghu SV, Joesch M, Sigrist SJ, Borst A, Reiff DF. Synaptic organization of lobula plate tangential cells in Drosophila: Dα7 cholinergic receptors. J Neurogenet. 2009;23:200–209. doi: 10.1080/01677060802471684. [DOI] [PubMed] [Google Scholar]
- 39.Ting CY, McQueen PG, Pandya N, Lin TY, Yang M, Reddy OV, O’Connor MB, McAuliffe M, Lee CH. Photoreceptor-derived activin promotes dendritic termination and restricts the receptive fields of first-order interneurons in Drosophila. Neuron. 2014;81:830–846. doi: 10.1016/j.neuron.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morante J, Desplan C. The color-vision circuit in the medulla of Drosophila. Curr Biol. 2008;18:553–565. doi: 10.1016/j.cub.2008.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collin C, Hauser F, Gonzalez de Valdivia E, Li S, Reisenberger J, Carlsen EM, Khan Z, Hansen NO, Puhm F, Sondergaard L, et al. Two types of muscarinic acetylcholine receptors in Drosophila and other arthropods. Cell Mol Life Sci. 2013;70:3231–3242. doi: 10.1007/s00018-013-1334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishii M, Kurachi Y. Muscarinic acetylcholine receptors. Curr Pharm Des. 2006;12:3573–3581. doi: 10.2174/138161206778522056. [DOI] [PubMed] [Google Scholar]
- 43.Clark DA, Bursztyn L, Horowitz MA, Schnitzer MJ, Clandinin TR. Defining the computational structure of the motion detector in Drosophila. Neuron. 2011;70:1165–1177. doi: 10.1016/j.neuron.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meier M, Serbe E, Maisak MS, Haag J, Dickson BJ, Borst A. Neural Circuit Components of the Drosophila OFF Motion Vision Pathway. Curr Biol. 2014;24:385–392. doi: 10.1016/j.cub.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Haverkamp S, Mockel W, Ammermuller J. Different types of synapses with different spectral types of cones underlie color opponency in a bipolar cell of the turtle retina. Vis Neurosci. 1999;16:801–809. doi: 10.1017/s0952523899164186. [DOI] [PubMed] [Google Scholar]
- 46.Wess J, Eglen RM, Gautam D. Muscarinic acetylcholine receptors: mutant mice provide new insights for drug development. Nat Rev Drug Discov. 2007;6:721–733. doi: 10.1038/nrd2379. [DOI] [PubMed] [Google Scholar]
- 47.Cordova D, Delpech VR, Sattelle DB, Rauh JJ. Spatiotemporal calcium signaling in a Drosophila melanogaster cell line stably expressing a Drosophila muscarinic acetylcholine receptor. Invert Neurosci. 2003;5:19–28. doi: 10.1007/s10158-003-0024-2. [DOI] [PubMed] [Google Scholar]
- 48.Lagrutta A, Shen KZ, North RA, Adelman JP. Functional differences among alternatively spliced variants of Slowpoke, a Drosophila calcium-activated potassium channel. J Biol Chem. 1994;269:20347–20351. [PubMed] [Google Scholar]
- 49.Becker MN, Brenner R, Atkinson NS. Tissue-specific expression of a Drosophila calcium-activated potassium channel. J Neurosci. 1995;15:6250–6259. doi: 10.1523/JNEUROSCI.15-09-06250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maddess T, Laughlin SB. Adaptation of the motion-sensitive neuron H1 is generated locally and governed by contrast frequency. Proc Roy Soc Lond B. 1985;225:251–275. [Google Scholar]
- 51.de Ruyter van Steveninck RR, Zaagman WH, Mastebroek HAK. Adaptation of transient responses of a movement-sensitive neuron in the visual system of the blowfly Calliphora erythrocephala. Biol Cybern. 1986;54:223–236. [Google Scholar]
- 52.Borst A, Egelhaaf M. Temporal modulation of luminance adapts time constant of fly movement detectors. Biol Cybern. 1987;56:209–215. [Google Scholar]
- 53.Delmas P, Brown DA. Pathways modulating neural KCNQ/M (Kv7) potassium channels. Nat Rev Neurosci. 2005;6:850–862. doi: 10.1038/nrn1785. [DOI] [PubMed] [Google Scholar]
- 54.Shen W, Hamilton SE, Nathanson NM, Surmeier DJ. Cholinergic suppression of KCNQ channel currents enhances excitability of striatal medium spiny neurons. J Neurosci. 2005;25:7449–7458. doi: 10.1523/JNEUROSCI.1381-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ting CY, Yonekura S, Chung P, Hsu SN, Robertson HM, Chiba A, Lee CH. Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development. 2005;132:953–963. doi: 10.1242/dev.01661. [DOI] [PubMed] [Google Scholar]
- 56.Daniels RW, Gelfand MV, Collins CA, DiAntonio A. Visualizing glutamatergic cell bodies and synapses in Drosophila larval and adult CNS. J Comp Neurol. 2008;508:131–152. doi: 10.1002/cne.21670. [DOI] [PubMed] [Google Scholar]
- 57.Meinertzhagen IA. Ultrastructure and quantification of synapses in the insect nervous system. J Neurosci Methods. 1996;69:59–73. doi: 10.1016/S0165-0270(96)00021-0. [DOI] [PubMed] [Google Scholar]
- 58.Edwards TN, Meinertzhagen IA. Photoreceptor neurons find new synaptic targets when misdirected by overexpressing runt in Drosophila. J Neurosci. 2009;29:828–841. doi: 10.1523/JNEUROSCI.1022-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, Douglas RJ. TrakEM2 software for neural circuit reconstruction. PLoS One. 2012;7:e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Transcript profiles of T4, T5 and Tm9 neurons reveal their expression of transcripts for neurotransmitter transporters and biosynthesis enzymes, as well as cholinoceptors and the potassium channel Slowpoke.
(A) T4 cells. (A′) T5 cells. (A″) Tm9 cells. PCR-amplified products of the transcripts from single Gal4-driven GFP-expressing neurons. Choline acetyl transferase (ChAT) transcript appears in T4, T5 and Tm9. Lane M: 100bp DNA ladder marker; lane C: negative control (without reverse transcription). 1: ChAT; 2: VGlut; 3: VGAT; 4: Hdc; 5: DAT; 6: SerT; 7: Tβh; and 8: Tdc2
(B, B′) PCR-amplified products of cholinoceptor transcripts from single T4 (B) and T5 (B′) cells. T4 neurons express two nicotinic receptor subunits, Dα7 and Dβ3, as well as the muscarinic receptor mAcR-B (CG7918). T5 neurons express two nicotinic receptor subunits, Dα3 and Dβ3, as well as a different muscarinic receptor, mAcR-A (CG4356). Lane M: 100bp DNA ladder marker; 1: nAcRα-96Aα (Dα1); 2: nAcRα-96Ab (Dα2); 3: nAcRα-7E (Dα3); 4: nAcRα-80B (Dα4); 5: nAcRa-34E (Dα5); 6: nAcRa-30D (Dα6); 7: nAcRα-7 (Dα7); 8: nAcRβ-64B (Dβ1); 9: nAcRβ-96A (Dβ2); 10: nAcRβ-21C (Dβ3); 11: mAcR-A (CG4356); 12: mAcR-B (CG7918).
(C) PCR-amplified products of the potassium channel slowpoke transcripts from T4 (lane i) and T5 (lane ii). Lane M: 100bp DNA ladder marker.
(D–F) Standard curves for T5 cells generated by plotting CT values against the known initial DNA copy number. Abscissa is the logarithm of the input cDNA; ordinate is the number of PCR cycles required to reach a given fluorescence signal level (Ct). Threshold cycle, CT, values (ordinate) were obtained using a controlled amount of input cDNA for ChAT (D); mAcR-A (E); and Rp49 (F). The final PCR product was analyzed by DNA gel electrophoresis (D′, E′, F′). Ct and the logarithm of the input cDNA varied in inverse linear proportion over a large concentration range. Standard linear regression analysis was used to calculate the standard curve, the amplification factor (AmF), PCR efficiency (Eff), and the coefficient of determination (R2) for each curve. The standard curves were used to calculate the ChAT [D] and mAcR-A transcript level in Tm9 (red arrows) and T5 (blue arrows) neurons. All qRT-PCR amplification reactions were carried out in three biological replicates for each cell type. (D′) Lane M: 100bp DNA ladder marker; lane C: negative control (without reverse transcription). Lanes 1-9: ChAT amplicon using different amounts of input DNA (D). (E′) as in (D′) but for mAcR-A. (F′) as in (D′) but for Rp49.
Table S1: The numbers of input synapses from four classes of Tm cell input (Tm1, Tm2, Tm9, Tm4) to their T5 target cells
Table S2: Fly stocks used in this study.