Abstract
Signal peptide-driven secretion of precursor proteins directs polypeptides across the plasma membrane of bacteria. Two pathways, Sec- and SRP-dependent, converge at the SecYEG translocon to thread unfolded precursor proteins across the membrane, whereas folded preproteins are routed via the Tat secretion pathway. Gram-positive bacteria lack an outer membrane and are surrounded by a rigid layer of peptidoglycan. Interactions with their environment are mediated by proteins that are retained in the cell wall, often through covalent attachment to the peptidoglycan. In this review, we describe the mechanisms for both Sec-dependent secretion and sortase-dependent assembly of proteins in the envelope of Gram-positive bacteria.
Keywords: Sec, leader peptide, LPXTG, sortase, cell wall, peptidoglycan
1. Introduction
Bacterial cells can be visualized by bright-field microscopy, a technique that takes advantage of the contrast difference between the specimen and its surrounding medium. Often, dyes with cationic properties are used to increase this contrast. The Gram stain procedure, developed by Christian Gram in Denmark in 1884 [1], represents such a differential staining procedure that has been used to classify bacteria for over 100 years. Most, but not all, bacteria take up the basic dye crystal violet. Gram-positive bacteria retain the purple dye, whereas Gram-negative bacteria are destained during fixation and washes and must be counter-stained with safranin. Acid-fast bacteria such as mycobacteria cannot be stained with the Gram procedure but will retain carbolfuchsin efficiently following a procedure known as Ziehl-Neelsen (acid-fast) staining [2]. These staining attributes of bacteria can be explained by fundamental differences in their respective cell envelope. Gram-positive bacteria have a relatively thick (~ 20–80 nm) cell wall also called murein sacculus that surrounds the plasma membrane. This thickness accounts for retention of crystal violet. The cell wall is largely composed of peptidoglycan, also called glycopeptide (muropeptide) and in Gram-positive bacteria, it serves as the matrix for the covalent attachement of wall teichoic acid, surface proteins and polysaccharide capsule [3–6]. In contrast, the peptidoglycan layer of Gram-negative bacteria is thin (~5–10 nm) and surrounded by an outer membrane structure (~7.5–10 nm thick) that itself is tethered to peptidoglycan by Braun’s lipoprotein [7]. Mycobacterium tuberculosis and other mycobacteria contain a layer of peptidoglycan linked to arabinogalactan that is in turn linked to mycolic acids forming an outer membrane. The high mycolic acid content of the envelope is responsible for the poor absorption of some dyes. The Corynebacterium, Mycobacterium and Nocardia species produce a complex envelope containing lipid species and porins and this outer layer is reminiscent of the function of the outer membrane (OM) of Gram-negative bacteria [8–10].
Bacterial peptidoglycan is responsible for the shape of bacteria and for protection against osmotic lysis [11]. Because they lack an outer membrane, Gram-positive bacteria cannot embed proteins in a lipid bilayer for surface display, yet these bacteria engage in molecular interactions that are mediated by proteins at the cell surface and thus have evolved several mechanisms for the trafficking and retention of proteins in the envelope. In Gram-positive bacteria, most secreted proteins are transported across the plasma membrane via the universally conserved and essential Sec pathway. Proteins carrying a cleavable Sec-dependent signal sequence, but lacking any other type of topogenic information, are released into the extra-cellular milieu. Additional sequence motifs within secreted substrates are necessary to target proteins to discrete sites within the envelope. Dedicated factors are responsible for deciphering such signals and implementing these protein-targeting mechanisms. Here, we will review the molecular events leading to the display of proteins known as cell wall-anchored proteins (CWP) in the envelope, beginning with their secretion across the plasma membrane mediated by the Sec system and followed by the covalent attachment to peptidoglycan by transpeptidase enzymes known as sortases. Surface display of proteins in the envelope of Gram-positive bacteria can also be achieved by noncovalent interactions with peptidolgycan or wall polymers. These interations are often mediated by repeated segments such as the GW module, LysM motif or Surface Layer Homology domain (see references [12–14] for reviews of these mechanisms).
2. The Sec system in Gram-positive bacteria
Sec-machinery mediated secretion is an essential pathway that provides for the transport of most proteins into and across the plasma membrane. Sec-mediated protein secretion has been best studied in Escherichia coli and serves as a paradigm for all other bacteria [15–17]. Genes involved in Sec-dependent secretion are largely conserved leading to the general assumption that the mechanism of protein translocation is also conserved.
2.1. Sec-mediated protein translocation in E. coli: a paragdim
This section is meant to provide a brief overview of the Sec pathway of E. coli as it represents the starting point for all in silico predictions of conserved elements in Gram-positive bacteria. Readers should refer to Chapters 2, 3, 5 and 6 for details.
In E. coli, three membrane proteins SecYEG assemble into the translocation pore for the secretion of protein substrates referred as preproteins or precursor proteins. The signal (leader) peptide at the N-terminus of the preprotein is the critical feature for recognition by the SecYEG translocon. The SecA ATPase pushes precuror proteins through the hydrophilic channel of the SecYEG translocon forming the so-called translocase [18–22] and is assisted by three other membrane proteins SecD, SecF and YajC [23–26]. Precursor proteins are maintained in a secretion competent state by one of two separate pathways [27, 28]. In one pathway, the signal peptide of nascent chains is bound by the signal recognition particle (SRP) and translation is temporarily arrested [29, 30]. E. coli SRP is a ribonucleoprotein particle consisting of Ffh and 4.5S RNA [31–34]. Upon docking of the SRP-ribosome complex on the membrane receptor FtsY, translation resumes and the nascent polypeptide is transferred to the SecYEG translocon [28, 35]. In the second pathway, fully synthesized precursor proteins are bound by a secretion chaperone such as SecB that maintains precursors in an unfolded, translocation-competent state in the cytoplasm [36–38]. Although both pathways, SRP-mediated co-translational secretion and chaperone-implemented post-translational secretion, ultimately converge at the translocon [28, 39], depletion analyses demonstrated that the SRP pathway is required for the secretion of polytopic membrane proteins in E. coli [40]. Precursors secreted in a post-translational manner are substrate of signal (leader) peptidase encoded by lepB in E. coli. This enzyme removes the signal peptide of newly translocated proteins as they emerge into the periplasm making the process unidirectional [41, 42].
2.2. Sec encoding genes in Gram-positive bacteria: a reverse genetic approach
The discovery of the sec genes and the elucidation of their biochemical activity in E. coli are the results of several genetic approaches and experimental validation of clever predictive models. In contrast, sec genes of other bacteria were mostly identified by homology searches. Investigations that have focused on low GC-content Gram-positive bacteria with an emphasis on Firmicutes are described herein. Secretion systems for only a few high GC-content Gram-positive bacteria have been examined and the readers are directed to the following articles and reviews describing these in Actinobacteria including Corynebacterineae and Streptomycetaceae [43–45]. Genome analyses have revealed homologs of SecA, SecD, SecE, SecF, SecG, SecY, Ffh, FtsY, YajC and LepB in organisms such as B. subtilis, B. anthracis, Staphylococcus aureus, Listeria monocytogenes and various Streptococcal species [46]. In particular, the SecA, and to a lesser extend SecY, are highly conserved with their E. coli homologues. SecE and SecG are less well conserved and are much shorter in B. subtilis, S. aureus and S. epidermidis. Most Gram-positive bacteria carry a secDF fusion gene, instead of single secD and secF genes [47]. The secB gene is conspicuously missing. In B. subtilis, the general chaperone CsaA has been proposed to serve a similar purpose as SecB in E. coli; CsaA interacts tightly with both SecA and preprotein [48]. Nevertheless, the csaA gene is missing in the genome of many Gram-positive bacteria and it seems likely that other cytosolic chaperones are responsible for maintaining precursors in an export-competent form. In E. coli, heat shock protein chaperones such as DnaK and DnaJ have been shown participate in this process, independently of SecB [49]. Genes encoding Ffh, 4.5S RNA, and FtsY are also conserved in Gram-positive bacteria. Ffh is essential for growth in B. subtilis but dispensible in Streptococcus mutans and S. pyogenes [50–52]. In S. pyogenes, SRP is responsible for secretion of extracellular virulence factors and an ffh mutant is attenuated for virulence [51, 52]. These observations suggest that the biogenesis of plasma membrane proteins is not restricted to the SRP pathway in Gram-positive bacteria and importantly that SRP may contribute to protein translocation across the plasma membrane.
Second sets of secA and/or secY genes (paralogues) can be found in the genomes of Staphylococci and Streptococci and these genes are referred to as secA-2, and secY-2 (see Chapter 25). Although this has not been rigorously tested, the secA and secY genes are considered to be essential in Gram-positive bacteria, whereas the secA2 and secY2 genes are dispensable, at least in Staphylococci and Streptococci [14]. Exceptions to this rule include C. difficile, for which both sets of genes have been found to be essential for growth [53] as well as the high GC-content organism C. glutamicum [44]. In S. aureus, expression of antisense secY RNA inhibits colony formation on agar plates [54], suggesting it represents the functional homologue of E. coli secY. The secA-2 and secY-2 genes may be found immediately adjacent to or in close proximity of the gene(s) whose product enters the secretion pathway defined by SecA2/SecY2; often these secreted proteins are glycosylated and promote bacterial virulence functions (see Chapter 25). Thus, for some species, SecA-2 and SecY-2 are highly specialized transporters dedicated to the secretion of specific substrates that cannot travel the canonical Sec pathway. Detailed information regarding SecA-2 and SecY-2 and secretion substrate determinants for SecA-2/SecY-2 translocase can be found in Chapter 25.
2.3. Translocation of cell wall anchored protein
Most cell wall anchored proteins are synthesized as precursor proteins and targeted for export to the canonical SecYEG translocon by the presence of a signal sequence at their extreme N-terminus. Signal peptides of wall anchored proteins encompass the canonical tripartite structure where a short, basic n-region precedes a longer hydrophobic stretch of amino acids (h-region), followed by the c-region with the recognition sequence for the enzyme signal peptidase (see Chapter 9) [41, 55]. Sec-dependent secretion of cell wall anchored proteins has been confirmed in S. aureus by several proteomic analyses including comparing secretomes of wild-type and secDF mutant strains [56] or by chemical inhibition of type I signal peptidase [57]. Further, by examining the secretion profile of a tatC mutant, the involvment of the Tat pathway for secretion of sortase A-anchored surface proteins in S. aureus has been ruled out [58]. Although the signal sequence of preproteins is not conserved, the site of cleavage (c-region) determines which enzyme, signal peptidase I (SPI) or signal peptidase II (SPII) will cleave the precursor (see Chapter 9). SPI often cleaves after an alanine residues whereas SPII cleaves proteins immediately before the conserved cysteine residue of the lipobox, a feature of lipoproteins that are modified with thioether linked diacylglycerol at their N-terminal cysteinyl [43, 59–61]. While E. coli encodes only one SPI enzyme, LepB [62], that is essential for growth, many Gram-positive bacteria express multiple SPI type genes. B. subtilis encodes five lepB-like genes with the products SipS, SipT, SipU, SipV, and SipW [63]. Together, SipS and SipT process preproteins and are essential for growth [63]. The SipW protein is unusual in that it is required for the expression of genes that elaborate biofilm matrix components when bacilli are grown on a solid surface [64]. However, the enzymatic signal peptidase activity appears to be dispensable for this process [64].
S. aureus encodes two SPI, SpsA and SpsB [65]. SpsB, but not SpsA (presumably because it lacks the critical catalytic residues), can substitute for E. coli LepB and is required for growth of S. aureus [65, 66]. S. epidermidis contains two SpsB homologues, most related to SipS and SipU of B. subtilis. Thus, the contribution of SpsA (if any) for preprotein secretion remains unknown. Finally, in B. subtilis the ABC transporter EcsAB has been implicated in the secretion of proteins. While the transport substrate(s) of EcsAB is still unknown, an ecsA mutant was shown to disrupt the secretion of the AmiQ precursor in a manner whereby preAmyQ remained associated with the membrane, suggesting a defect in its processing by signal peptidases [67]. Further, the activity of the intramembrane cleaving protease RasP (YluC) was also altered in the ecsA mutant [67]. RasP belongs to a family of proteases that cleaves proteins and peptides spaning the plasma membrane and the first enzyme of this family, Eep, was characterized for its role in the processing and secretion of an 8 amino acid peptide pheromone called cAD1; cAD1 induces a mating response in enterococci that harbor plasmid pAD1 [68, 69]. It was later demonstrated that pheromone peptides are the degradation products of lipoprotein signal sequences [70] and also require an ABC transporter to be dislodged from the lipid bilayer. This mechanism of degradation appears to be conserved between bacteria and mammals and has been designated Regulated Intramembrane Proteolysis (RIP)[71]. It is thus interesting that mutations in the ABC transporter EscA block processing mediated by the RIP protein RasP [67] and that escA mutants affects the overall secretion of proteins in B. subtilis and S. aureus (including secretion of cell wall proteins in this organism) [72, 73]. Together, these findings suggest that RIP proteases and the Esc ABC transporter may be generally required for the degradation or recycling of signal peptides leftover in the plasma membrabe following processing by LepA and LepB.
It seems important to note that signal peptides of Gram-positive bacteria, in particular signal peptides of cell wall anchored proteins, differ from those of Gram-negative bacteria. They are generally longer, more hydrophobic and with more charged residues at the N-terminal end [60]. While signal peptides of Gram-positive bacteria appear to be processed in Gram-negative bacteria, the converse is not always true [74]. This leaves open the possibility that additional factors contribute to the recognition of signal peptides. However, biochemical approaches using membrane extracts with or without bound ribosomes failed to find such candidates [75–78]. Many cell wall anchored proteins like staphylococcal protein A (S. aureus) carry a long signal peptide with the YSIRK-G/S motif (Fig. 1) [79, 80]. Removal of the YSIRK-G/S motif slows but does not abrogate the secretion and cell wall anchoring of protein A [81]. Interestingly, when examined by immunofluorescence microscopy, protein A was found to assemble in a ring-like structure encircling the spherical cells, with increased abundance at cell division sites [58, 82]. Staphylococci divide perpendicular to their previous cell division plane and appear to be born unit size during logarithmic growth [83, 84]. Peptidoglycan synthesis occurs at the cross wall (cell division site) and allows for the bulk of bacterial envelope expansion [84]. Streptococcus pyogenes M protein also harbors the YSIRK-G/S motif within its signal peptide and is also deposited into peptidoglycan at the division plane of the cell [85]. Conversely, it was observed that surface proteins bearing a conventional signal peptide (non-YSIRK), for example SasF in S. aureus and PrtF in S. pyogenes, are deposited at the cell poles [58, 85]. Replacement of the signal peptide of SasF (non-YSIRK) with that of ClfA (+YSIRK) leads to the deposition of SasF at the cross-wall of staphylococci [58]. Incubation of staphylococci with sub-inhibitory concentrations of penicillin or moenomycin triggers the accumulation of cell wall-anchored preproteins at the cross wall, irrespective of the presence of the YSIRK motif [86]. Together, these observations suggest that distinct sequence motifs such as the YSIRK-G/S motif of some preproteins may represent new topogenic sequences for secretion at the cross wall. It seems unlikely that the SecA/SecYEG translocase is solely responsible for the observed differences in the trafficking of precursors with discrete signal sequence motifs. Using various microscopy approaches, several patterns of Sec translocase distribution have been reported and invoked as mechanism for the selection of preproteins. For example, the Sec machine has been reported to localize to the so-called ExPortal microdomain in S. pyogenes [87, 88] and Enterococcus faecalis [89], a localization that nonetheless does not seem compatible with the secretion of preproteins at both cross wall and cell poles. Indeed, other work reported a more random distribution for membrane associated SecA in S. pyogenes [85, 90]. In B. subtilis, the Sec machine is distributed along the cell in a spiral pattern [91] wherease in S. pneumoniae, its localization is subject to dynamic changes and relocates from the cell division plane to variable sites at mid-cell, perhaps co-localizing with active peptidoglycan synthesis [92].
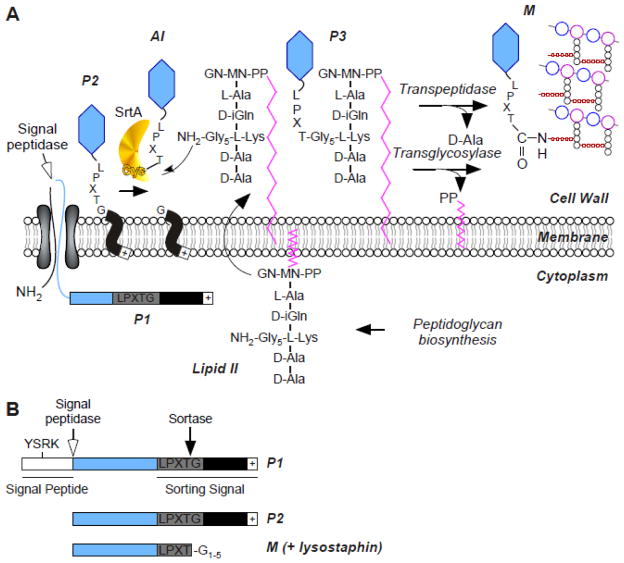
FIG. 1.
Sortase A-dependent surface display of proteins. (A) Model for preprotein secretion and sorting in S. aureus. Surface proteins are first synthesized in the bacterial cytoplasm as full-length precursors (P1) containing an N-terminal signal sequence and a C-terminal sorting signal. The signal sequence directs the cellular export of the polypeptide through the Sec system and, upon translocation, is cleaved by signal peptidase. The product of this reaction, P2 precursor harboring only the C-terminal sorting signal, is retained within the secretory pathway via its C-terminal hydrophobic domain (black box) and positively charged tail (+). Sortase, a membrane anchored transpeptidase with active site cysteine, cleaves the peptide bond between the threonine (T) and the glycine (G) of the LPXTG motif, generating an acyl intermediate (AI). Lipid II, the peptidoglycan biosynthesis precursor, and its pentaglycine crossbridge (Gly5) amino group attacks the acyl intermediate, linking the C-terminal threonine of surface protein to lipid II (P3 precursor) and regenerating the active site of sortase. P3 precursor functions as a substrate for penicillin binding proteins and is incorporated into the cell wall envelope to generate mature anchored surface protein (M) that is also displayed on the bacterial surface. Note that the pentaglycine cell wall crossbridge is not commonly found in other bacteria nonetheless this pathway is conserved in many Gram-positive bacteria where the functional elements of cell wall crossbridges, LPXTG motif, sortase and penicillin binding proteins are conserved (adapted from [188]). (B) Schematic representation of protein precursors identified by using metabolic labeling of staphylococcal cultures with radiolabeled methionine. Immunoprecipitation of labeled polypeptides identifies the precursor P2 and P1. The short-lived AI and P3 intermediates are not identified. Treatment of the staphylococcal peptidoglycan with lysostaphin (glycyl-glycine endopeptidase) releases the final product, i.e. M, the surface protein anchored to the cell wall.
A genetic search for mutants displaying altered secretion of staphylococcal protein A identified three transmembrane proteins, SpdA, SpdB and SpdC (named for surface protein display) each one carrying an ABI (abortive infectivity) domain. Spd factors were shown to be required for the abundant deposition of protein A at the cross wall, albeit that a precise mechanism for their activity could not be assigned [93].
3. Sorting reactions
3.1. Processing and attachment to peptidoglycan
Covalent anchoring of secreted proteins was first appreciated in Group A streptococci and S. aureus. These bacteria are endowed with two very abundant surface proteins, the streptococcal M protein [94, 95] and staphylococcal protein A [96, 97]. Early investigations used electron microscopy to demonstrate that the M protein forms a long fiber (~500 Å long) that can be visualized as a fuzzy coat on the surface of bacteria [98, 99]. Cicrular dichroïsm studies suggested that the protein is formed of repeats primarily arranged in an alpha-helical coiled-coil allowing the fiber to span the thick peptidoglycan and protrude on the cell surface [98, 99], which was recently corrborated by X-ray crystallography studies [100]. Attempts at solubilizing streptococcal M protein or staphylococcal protein A proved remarkably difficult. Because, protein A could be esaliy purified, owing to its ability of binding to the constant region of immunoglobulins [97], it became obvious that treatment of bacteria with acids, bases, detergents, proteases or boiling, were insufficient to release these surface proteins [99, 101]. Nonetheless, treatment of bacteria with cell wall-hydrolytic enzymes led to successful solubilzation of protein A [99, 102]. Importantly, treatment of staphylococci with lysostaphin, a bacteriolytic enzyme secreted by Staphylococcus simulans, released protein A as a homogeneous population, judged by its migration profile on sodium dodecyl sulfate-polyacrylamide gel electrophoresis [103]. Treatment with egg white lysozyme released small amounts of protein A that migrated more slowly than the lysostaphin-released counterpart, suggesting an increase in molecular mass [103]. Lysosyme does not effectively degrade the staphylococcal cell wall, and the mass difference of released protein A could be attributed to the amino sugars N-acetylglucosamine and N-acetylmuramic acid that form the repeating dissacharide units in peptidoglycan. Thus, these finding suggested that protein A must be linked to the staphylococcal cell wall [103].
In addition to the N-terminus signal sequence, protein A contains a second hydrophobic domain at its C-terminus (Fig. 1A). When compared with the C-termini of other surface proteins of Gram-positive bacteria, the conservation of the LPXTG motif can be discerned. X represents here any amino acid and the LPXTG motif is always followed by a C-terminal hydrophobic domain and a tail of mostly positively charged residues [104]. Deletion experiments revealed that all three sequence features, LPXTG motif, charged tail and hydrophobic domain are necessary for cell wall anchoring [74]. Fusing the C-terminal domain of protein A to otherwise secreted proteins is sufficient to promote attachment of reporter proteins to the cell wall [105]. Swapping of C-terminal sorting domains from different bacteria demonstrated the universality and conservation of the cell wall anchoring motif [105]. In S. aureus, the cytoplasmic precursor (P1) of protein A is first cleaved by leader peptidase following translocation across the membrane, yielding P2, and processed a second time at the C-terminal LPXTG motif that serves a sorting signal for retention in the envelope (Fig. 1A). Processing includes cleavage between the threonine (T) and the glycine (G) residues [106] followed by transfer of the protein from the carboxyl of threonine onto the free amino group of pentaglycine cross-bridges in the staphylococcal cell wall, a reaction referred to as transpeptidation [5] (Fig. 1A). Together, this series of processing events has been designated “sorting reaction” and is catalyzed by the sortase A enzyme [106, 107]. Methanethiosulfonates and p-hydroxymercuribenzoic acid were shown to inhibit the sorting reaction in vivo [108] suggesting that the enzyme proposed to cleave surface proteins at the LPXTG motif, must be a sulfhydryl-containing enzyme. It was observed that vancomycin and moenomycin, inhibitors of cell wall polymerization, slowed the sorting reaction but addition of the transpeptidation inhibitor, penicillin G, had no effect on surface protein anchoring [108]. However, cleavage of surface protein precursors could be observed in protoplasts, suggesting that the mature assembled cell wall is not required for attachment. Together, these findings suggested that lipid II, the peptidoglycan biosynthesis precursor may serve as the substrate of the sorting reaction [108]. In S. aureus lipid II is is comprised of C55-PP-MurNAc-(D-Ala-D-iGln-(NH2-Gly5)-L-Lys-D-Ala-D-Ala)-GlcNAc, i.e. a murein disaccharide-pentapeptide subunit linked to the membrane carrier bactoprenol (C55) [109]. By using metabolic labeling of staphylococcal cultures with radiolabeled phosphoric acid, it was later revealed that the last step of the sorting reaction involves a P3 intermediate that could be immunoprecipitated along with nisin, an antibiotic that forms a complex with lipid II; following the discovery of sortase, it was further confirmed that the P3 intermediate is not observed in a strain where the sortase A gene is disrupted [110]. P3 is eventually incorporated into peptidolgycan strands yielding mature protein A (M) (Fig. 1A) [110, 111].
In S. aureus, the peptidoglycan layer is 30 to 100-nm thick and composed of the repeating disaccharide N-acetylmuramic acid-(β1–4)-N-acetylglucosamine (MurNAc-GlcNAc) [112]. MurNAc is amide-bonded to the first alanine of the tetrapeptide [L-Ala-D-isoGln-L-Lys(NH2-Gly5)-D-Ala]. Crosslinking between tetrapeptide through the pentaglycine (NH2-Gly5) of one strand to the terminal D-Ala of an adjacent tetrapeptide, effectively links strands together yielding a single large macromolecule, the murein sacculus, that encloses the cell (Fig. 1A) [113–116]. Thus, in S. aureus cell wall anchored proteins appear to be linked to lipid II and ultimately incorporated into the cell wall envelope via the transpeptidation and transglycosylation reactions of peptidoglycan synthesis [117–119] along with other secondary polymers such as carbohydrates and teichoic acids [113, 120]. It is interesting that in L. monocytogenes, some SrtA-substrates produced upon stress are anchored to the cell pole [121]. It has been proposed that either lipid II is present at the pole of low-dividing bacteria or that sortase A directly anchors such proteins to cross-linked peptidoglycan [122]. As noted by Bierne and Dramsi [122], lipid II is most abundantly found at sites of peptidoglycan synthesis and during exponential growth, sites that also serve to recruit the secretion and sorting machineries and help shape bacterial cells. Thus, cellular shape and sites of peptidoglycan biogenesis may account for polar, septal and lateral distribution of cell wall anchored proteins on the bacterial surface [122].
3.2. Discovery of S. aureus sortase A
To identify mutants unable to anchor protein A to the cell wall, a collection of temperature-sensitive variants was screened for defects in protein processing using a reporter protein, secreted staphylococcal enterotoxin B (SEB) fused to the LPXTG sorting signal of protein A (SEB-SPA) [123]. Cultures of bacterial variants were grown in non-permissive conditions in the presence of radioactive methionine, lysed and SEB was immunoprecipitated with specific antibody. Radiolabeled SEB products were separated on a denaturing gel and visualized by autoradiography to reveal the three expected species: SEB-SPA P1 precursor synthesized in the cytoplasm, the P2 intermediate processed by leader peptidase and the mature, surface-anchored protein (M) (Fig. 1B). The transient P3 precursor is not observed in this assay. In wild-type bacteria, the P1, P2 and M species represented 5%, 19% and 76% of total SEB-SPA and a pulse-chase analysis showed that P2 was cleaved and anchored within 2 min [123]. Several hundred mutants were screened using this assay. One of them displayed aberrant accumulation of P2 and a P2 to M conversion time > 10 min, suggesting a defect in cell wall sorting [123]. A complementation analysis was performed in a similar manner by screening a plasmid library of 2,000 random DNA fragments from S. aureus for gene candidates that could reduce the concentration of P2 in the mutant [123]. Two clones were identified that shared the critical S. aureus sequences for a gene that was named srtA for surface protein sorting A [123]. Analysis of mutants lacking srtA revealed that sortase A is responsible for anchoring most cell wall proteins to the cell wall, at least all of those carrying an LPXTG motif [124].
3.3. Sortase A-Catalyzed Transpeptidation
S. aureus sortase A was the first sortase to be characterized biochemically. It is a 206 amino acid protein with an N-terminal membrane anchor. Removing the membrane anchor yields a soluble protein that could be purified in E. coli. The catalytic activity of SrtA could be recapitulated in vitro by using the substrate abz-LPETG-dnp that harbors the fluorophore, 2-amino-benzyl, at the N-terminus and the quencher, 2,4-dinitrophenyl, at the C-terminus. Due to the close proximity between fluorophore and quencher, the substrate does not emit a fluorescence signal when excited at the proper wavelength. However, sortase A-mediated cleavage separates fluorophore and quencher, abz -LPET and G-dnp, causing an increase in fluorescence [119, 125]. Recombinant sortase A was found to cleave the peptide bond between the threonine and the glycine of the LPET peptide and to form a thioester-linked intermediate between the carboxyl group of threonine and the active site cysteine residue of sortase [106, 123, 126]. Hydroxylamine or water could serve as nucleophiles in vitro, resolving the acyl-enzyme intermediate. Addition of hydroxylamine yielded LPET-hydroxamate products, indicating that hydroxylamine attacks the acyl-enzyme intermediate [123, 125]. Petidoglycan fragments or surrogates, including Gly, Gly2, Gly3, Gly4, or Gly5 (i.e., mimicking the pentaglycine cross-bridge), also stimulated sortase cleavage of abz-LPETG-dnp in vitro [119, 125]. Analysis of the reaction products (abz -LPET-GlyN, where N=1–5) demonstrated that the amino group of the cell wall crossbridge performs the nucleophilic attack at the acyl-enzyme intermediate of sortase A [119, 123, 125].
In agreement with the finding that the sorting reaction could be inhibited by methanethiosulfonates and p-hydroxymercuribenzoic acid [108], sortase A was found to contain a single cysteine, Cys184. Substitution of Cys184 with alanine abolishes sortase A activity both in vivo and in vitro [119, 125]. Bioinformatic analysis revealed that the active site cysteine is a conserved feature of sortases, in agreement with the general hypothesis that mechanisms of surface protein anchoring to the cell wall envelope are conserved in Gram-positive bacteria. The structure of of S. aureus sortase A was solved by NMR and X-ray crystallography and revealed a unique eight-stranded β-barrel, with several short helices and loops [127, 128]. The active site of the enzyme was found to be located in a hydrophobic depression formed by two β-stands. Two conserved residues, His120 and Arg197, are positioned in close proximity to the active site sulfhydryl Cys184 [128, 129] and were shown to be involved in catalysis by forming a catalytic triad [130–133].
In summary, cell wall-anchored surface proteins of Gram-positive bacteria encode two topogenic sequences, an N-terminal signal peptide for secretion of precursor proteins via the Sec pathway and a C-terminal cell wall sorting signal with a conserved LPXTG motif. In staphylococci, sortase A anchors proteins to pentaglycine cross-bridges that connect peptidoglycan strands. These pentaglycine cross-bridges are otherwise covalently linked to the ε-amino group of lysine residues. Not all gram-positive bacteria share the same cross-bridge structure. In S. pyogenes, two alanine residues are found in place of the pentaglycine and it is assumed that the LPXT substrate is linked to the di-alanyl bridge. In many bacteria, meso-diaminopimelic acid (m-Dpm) replaces the lysine residue in peptidoglycan and a direct covalent bond is formed between the free ε-amino group of m-Dpm and the terminal D-Ala of a neighboring strands. In L. monocytogenes, the T of LPXTG proteins is directly attached to the free ε-amino group of m-Dpm [134].
3.4. Class A Sortases: housekeeping functions
A search for proteins with a predicted signal sequence and a C-terminal LPXTG motif can identify putative sortase A-anchored surface proteins in the sequenced genomes of Gram-positive bacteria. For example, 18–22 cell wall proteins were identified in the genomes of S. aureus isolates, and similar results were obtained with related Gram-positive pathogens. Many of these cell wall anchored proteins function as microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) and represent bacterial elements of tissue adhesion and immune evasion [135].
When injected into the bloodstream of mice, most staphylococci survive innate immune response, disseminate to peripheral tissues and establish abscesses in multiple organ systems over 3–4 days. Abscess lesions continue to develop over weeks and cannot be cleared by the host. Staphylococci grow as communities at the center of these lesions and are enclosed by pseudocapsules, separating the pathogen from immune cells. S. aureus srtA mutants display a large virulence defect in this model of infection [124] and are in fact unable to form abscesses [136]. By testing insertional variants in genes for cell wall-anchored surface proteins, the stage at which some of the sortase A-anchored products function could be inferred [136]. For example, fibrinogen-binding proteins ClfA and ClfB were found to be required during the early phase of staphylococcal dissemination. The heme scavenging factors IsdA and IsdB, as well as SdrD and protein A, were found to be necessary for abscess formation. Other types of infection have also been tested, including septic arthritis and endocarditis. In both models, srtA mutants displayed large reductions in pathogenesis [137–139]. Virulence attributes of sortase and their protein substrates extend well beyond staphylococci, and defects in the pathogenesis of sortase mutants have been reported for many bacterial species, including actinomyces, enterococci, streptococci, bacilli, or listeria [140–150]. Finally, cell wall anchored proteins have been shown to raise immunity following vaccination with purified components against Group B streptococcal, pneumococcal, and staphylococcal infections [151, 152]. Collectively, these studies illustrate the many important and diverse roles that surface proteins play during the pathogenesis of infections caused by Gram-positive pathogens.
Genome sequencing of Gram-positive bacteria resulted in the discovery of thousands of genes encoding proteins with LPXTG or LPXTG-like sorting signals, as well as a plethora of sortase genes [153–155], suggesting the existence of distinct classes of substrates and cognate sortases [156, 157]. A complete classification of sortase enzymes, classes A-D, has been reported by Dramsi and colleagues [158] (Table 1) and is used in the remaining sections of this review. However, it is important to note that the function of sortases are explored only in few organisms and that some sortase genes have been named in the order of their discovery, which does not match the aforementioned nomenclature [158].
Table 1.
Classes of Sortases along with substrates and nucleophiles.
| Sortase Class | Substrates | Substrate Motif | Nucleophile |
|---|---|---|---|
| A (housekeeping) | Surface proteins | LPXTG | Lipid II |
| B | Heme transporter | N(P/A)(Q/K)(T/S)(N/S) | Peptidoglycan crossbridge |
| C | Pilin subunits | (I/L)(P/A)XTG | Lys of Pilin |
| D | Mother cell and endospore envelope protein | LPNTA | Lipid II |
Class A sortase represents the housekeeping enzyme described above. Sortases described in the subsequent sections are accessory and appear to anchor dedicate factors. Often, the surface protein gene is found in the same transcriptional unit with a sortase gene. The general assumption is that such genes encode enzyme-substrate pairs.
3.5. Class B Sortase: iron acquisition
While the great majority of sorting signals carry the LPXTG motif, others harbor variations of this sequence (Table 1) and often appear to be encoded by genes adjacent to a sortase-encoding gene. This was first observed and tested for S. aureus and B. anthracis isd-srtB [159, 160] as well as L. monocytogenes svpA-srtB [161]. srtB encodes the enzyme sortase B that is required for anchoring of one or more proteins that promote heme-iron uptake [159]. Enzymes belonging to the sortase B subgroup recognize substrates that often contain an NPQTN motif rather than the canonical LPXTG [158, 159, 162]. SrtB of L. monocytogenes has two subsrates with NAKTN and NPKSS sorting motifs, respectively [163]. Nonetheless, sortase B enzymes are structurally conserved with sortase A [132, 164]. In S. aureus, srtB is located in the so-called isd locus that encodes the IsdA, IsdB and IsdC cell wall-anchored heme-binding proteins, a membrane-based heme transport system (IsdD, IsdE, and IsdF), a heme degrading monooxygenase (IsdG), and SrtB [159]. IsdC is the only substrate anchored by SrtB, and is cleaved at its NPQTN motif-sorting signal and immobilized at cell wall crossbridges [162] (Fig. 2). IsdA and IsdB are anchored to the cell wall by sortase A [165]. In contrast to IsdA and IsdB, IsdC remains buried within the staphylococcal cell wall envelope, because it is linked to mature assembled peptidoglycan but not to lipid II [159, 162] (Fig. 2).
FIG. 2.
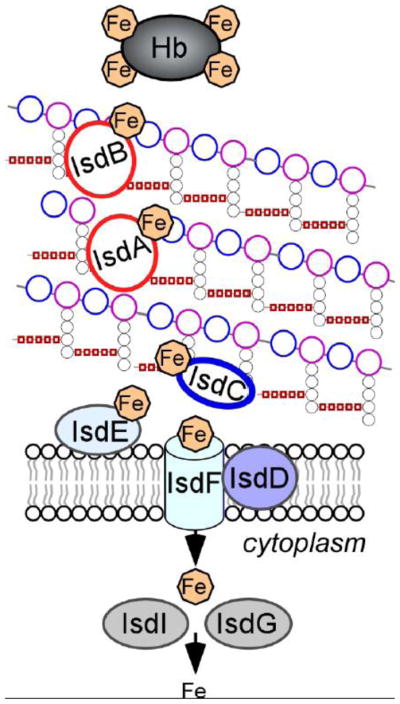
Isd-mediated heme-iron uptake across the cell wall of S. aureus. IsdA, IsdB, and IsdH are anchored to the cell wall by sortase A and function as receptors for hemoprotein ligands, such as hemoglobin (Hb). Upon binding to Isd receptors, heme is released from the hemoproteins and passaged through the cell wall in an IsdC-dependent manner. IsdC carries the NPQTN motif and is the substrate of Sortase B. When S. aureus are incubated with proteinase K, IsdB is completely degraded, IsdA is partially degraded but IsdC remains intact, suggesting that the three proteins are displayed at varying distances from the surface of the envelope. The heme molecule is taken up by the membrane transport system composed of IsdDEF and upon entry into the cytoplasm, is degraded by the heme oxygenases IsdG and IsdI to the release free iron (adapted from [189]).
3.6. Class C Sortase: assembly of pili
A subgroup of sortases was found to assemble pili on the surface of Gram-positive bacteria (Table 1), a finding first documented for Corynebacterium diphtheriae [166, 167]. While in Gram-negative bacteria, pili are formed by non-covalent interactions between pilin subunits, C. diphtheriae pili are formed by covalent polymerization of pilin subunits [168, 169]. Pili are macromolecular structures that consist of repeating protein subunits (pilin), extending from the bacterial surface into the surrounding milieu. These fiber-like structures (fimbriae) are often capped with a protein with adhesive properties that promote bacterial binding and virulence [168]. Owing to their small diameter, pili were not detected in studies that examined the morphology of Gram-positive bacteria by electron microscopy or rather were mistaken as artifacts [170]. The first evidence of pili was revealed in actinomyces species responsible for tooth decay, where an operon encoding a pilin-like gene and a gene later characterized as sortase were identified [171, 172]. While the function of sortase was still unknown at the time of this observation, mutagenesis of the corresponding gene abolished actinomyces adherence and led to the loss of pili that could otherwise be visualized by immuno-gold electron microscopy [171, 172].
Experimental evidence for the contribution of sortases in polymerization of pili was gained following the characterization of six sortase-like genes in C. diphtheriae [166]. C. diphtheriae expresses at least three gene operons specifying a sortase and surface proteins now recognized as pilin subunits [166]. For example, one of these loci encodes the SpaA protein (sortase-mediated pilin assembly A) that can be stained by immunogold to reveal fimbrae-like structures distributed uniformly on the cell surface. Immunogold labeling of the other two proteins encoded by the locus, SpaB and SpaC, revealed that SpaB is distributed along the fiber shaft whereas SpaC can be stained at the tip of pilus fibers [166]. Deletion of the sortase gene encoded in the spaABC locus abolished the formation of SpaA pili [166]. This sortase is now classified as a class C sortase (Table 1). Importantly, SpaA subunits within the polymer were crosslinked to each other and ultimately attached to peptidoglycan [166]. SpaA assembly was also dependent on a conserved lysine residue of SpaA, located in the so-called pilin motif, together with the canonical sorting signal, the pilin motif was found to be required for the formation of pili in a sortase-dependent manner [166, 173]. It was thus hypothesized that class C sortases may polymerize pili by forming covalent bonds between individual pilin subunits. Indeed, it was demonstrated that the transpeptidation reaction between pilin subunits involved formation of an acyl-enzyme intermediate between sortase and pilin precursors at the canonical sorting signal (LPXTG) [173–175] and resolution by nucleophilic attack of the incoming pilin subunit through a conserved YPKN pilin motif (Fig. 3) [176]. Examination of B. cereus pili established that the major pilin subunit is the substrate for both class C sortase and class A (housekeeping) sortase [177]. The housekeeping sortase transfers the entire pilus structure onto lipid II and ultimately, it becomes incorporated into peptidoglycan [177, 178] (Fig. 3). Class C sortases and their corresponding pili have been discovered in several organisms and similar mechanisms for their assembly have been reported [169, 179].
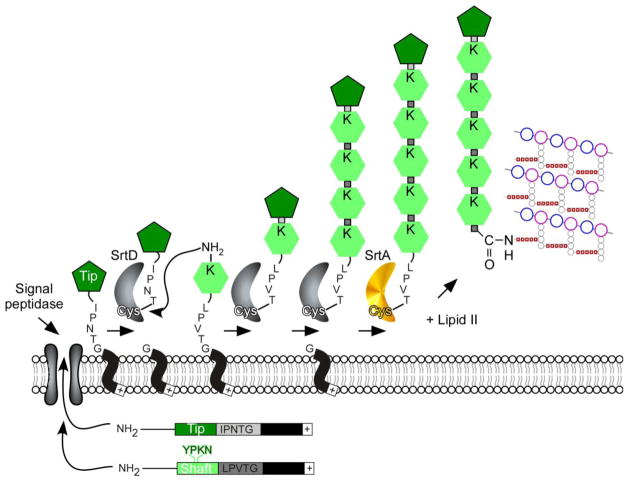
FIG. 3.
Model for sortase-mediated polymerization of heterodimeric pili. Pilin subunits are typical sortase substrates, containing an N-terminal signal peptide that promotes secretion through the Sec system, and a C-terminal cell wall sorting signal. The pilin-specific sortase (SrtC) cleaves the sorting signals of the Tip and Shaft proteins such as the BcpA and BcpB proteins of Bacillus cereus at the Thr of the LPXTG motif, generating an acyl–enzyme intermediate that is resolved by the free amino group of a conserved lysine residue (K) present in the YPKN pilin motif of an incoming pilin subunit. Nucleophilic attack by the side chain of Lys in the YPKN motif results in intermolecular isopeptide bond between tip and shaft. The remainder of the filament assembles by a sequence of similar transpeptidation reactions. Nucleophilic attack of lipid II on the acyl intermediate formed by the housekeeping sortase (SrtA) and the polymerized pilus results in transfer to the cell wall envelope and terminates pilus assembly. Note: a similar mechanism occurs with heterotrimeric pili for example SpaCAB pili of Corynebacterium diphtheriae. SpaC is the pilus tip and SpA, the shaft protein. A third subunit SpaC serves to anchor the polymerized pilus to the cell wall in SrtA-dependent manner (adapted from [169]).
Pilin subunits of Gram-positive bacteria have been shown to fold into two adjacent immunoglobulin (Ig)-like domains with seven anti-parallel β-strands as first demonstrated for the minor pilin GBS52 of Streptococcus agalactiae [180]. Interestingly, an intramolecular isopeptide bond between the side chains of Lys and Asn residues was observed in the crystal structure of the major pilin subunit from S. pyogenes in the so-called Cna domain [181]. The prototypical Cna domain is found in the surface collagen adhesin protein of S. aureus anchored by sortase A [182]. As a result, Ig-like folds containing an intramolecular isopeptide bond are referred as Cna protein B-type domain (Cna-B; pfam05738).
3.7. Class D Sortase
Class D sortases can be found in the genome of many bacilli, clostridia and actinomycetales. A single member of this family has been characterized, the class D enzyme from B. anthracis named SrtC [183, 184]. B. anthracis harbors three sortase genes, srtA, srtB and srtC. SrtA and SrtB share properties similar to those described for S. aureus SrtA and SrtB and thus act as housekeeping sortase (SrtA) [185] or as an iron-scavenging system (SrtB)[160], respectively. Interestingly, B. anthracis encodes a minimal number of SrtA-substrates and instead displays the majority of surface proteins by means of an S-layer, a process that does not require covalent attachment to the cell wall peptidoglycan [14]. The class D SrtC enzyme is encoded in a gene locus that carries a two-component regulatory system (sctR and sctS) as well as basI, that specifies a secreted protein with an LPNTA motif (Table 1) [183]. BasH, a second LPNTA-motif containing protein, is encoded elsewhere on the B. anthracis chromosome. Recombinant SrtC was shown to cleave the LPNTA peptide between the threonine and alanine residues but not LPATG and NPKTG peptides, the SrtA and SrtB substrates, respectively [183]. Both BasI and BasH were shown to be the substrates of SrtC in vivo [183]. Interestingly, SrtC was found to be expressed only during sporulation in a manner that required the response regulator SctR [183]. BasH is expressed in the developing forespore. Analysis of cell wall fragments demonstrated that BasH and BasI are anchored to the forespore and mother cell peptidoglycan, respectively [183, 184]. The exact function of BasH and BasI is unknown and deletion of srtC did not result in a virulence defect in B. anthracis [183]. To complete its infectious cycle, B. anthracis must sporulate in animals that have succumbed to anthrax disease. Sporulation ensures survival of this bacterial species in the environment until it is taken up, presumably by ingestion, by another predator or grazing animal. Unlike wild-type bacilli, srtC mutants cannot readily form spores in guinea pig carcass tissue or sheep blood unless their vegetative forms were exposed to air. A general model derived from these observations suggests that sortase C-mediated anchoring of BasI and BasH is critical for the generation of spores in host tissues [183]. As noted by Dramsi et al. [158], it is interesting that many of the organisms endowed with class D sortases display unique morphological differentiation cycles, perhaps pointing to discrete spatial-temporal distribution for these enzymes and their cognate substrates.
4. Outlook
Protein anchoring to the cell wall is a common and conserved mechanism of Gram-positive bacteria for the display of proteins in an envelope devoid of an outer lipid bilayer. Cell wall anchored proteins carry two topogenic signals, one for translocation across the membrane and one for recognition by transpeptidase enzyme, designated sortases. Sec-dependent secretion in Gram-positive bacteria uses conserved elements of Gram-negative bacteria including the translocase and signal sequence processing enzymes, albeit that some of these genes have been duplicated (secY, secA genes) or amplified (lepB-like genes). Importantly, signal sequences of some cell wall anchored proteins appear to encompass additional topogenic information that may target subsets of preproteins to distinct locations in the cell envelope. Nonetheless, the ultimate reason for this trafficking requirement remains unclear and trans-acting factors responsible for the travels of proteins remain to be identified. Obvisouly, SecB the dedicated preprotein chaperone of Gram-negative bacteria cannot be involved in this process since it is altogether missing in many Gram-positive microbes. Finally, the spatial and temporal localization of secretons, sortase and cytoskeletal proteins must also be interpreted in relation to cell shape and adjustements in gene expression [122].
All sortases, class A through D, share similar structures, catalytic attributes and substrates, albeit that in the case of pili, sortase-mediated transpeptidation reaction occurs between two pilin subunits. In all other cases examined, the nucleophile appears to be the peptidoglycan crossbidge of the lipid II precursor or assembled, but not crosslinked peptidoglycan. Once attached to the cell wall, it is still unclear how proteins are released. During infection, the abundance and content of the cell wall proteome changes and cannot be solely regulated at the level of gene expression. Presumably, extension of the cell wall in dividing bacteria may provide a dilution effect. It is also likely that enzymes that remodel the peptidoglycan will also contribute to the turnover of cell wall anchored proteins. Extracellular proteases and chaperones such as HtrA, rotamases and disulfide isomerases provide some quality control for the folding of secreted proteins or when secretion of proteins is placed under stress [61, 186, 187]. However, it is unclear whether anchored proteins can be repaired, degraded or recycled through such pathways. It is thus noteworthy that pilin subunits and many anchored proteins contain highly protease-resistant immunoglobulin folds with stabilizing isopeptide bonds in their Cna-B domains. A considerable challenge for the field is to decipher the function of all cell wall anchored proteins that are encoded by human pathogens.
Highlights.
Cell wall anchored proteins are displayed on the surface of Gram-positive bacteria.
We describe the processes involved in secretion and processing of these proteins.
We describe how sortase catalyzes the covalent attachment of these proteins to peptidoglycan
We describe the physiological contributions of the four classes of known sortase enzymes.
Acknowledgments
Research on protein secretion in the laboratories of D.M. and O.S. is supported by grants from the National Institute of Allergy and Infectious Diseases, Infectious Diseases Branch AI038897 (O.S.), AI052747 (O.S.) and AI075258 (D.M.), The authors acknowledge membership of and support from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (NIH Award 1-U54-AI-057153).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gram HC. Über die isolierte Färbung der Schizomyceten in Schnitt- und Trockenpräparaten. Fortschritte der Medizin. 1884;2:185–189. [Google Scholar]
- 2.Bishop PJ, Neumann G. The history of the Ziehl-Neelsen stain. Tubercle. 1970;51:196–206. doi: 10.1016/0041-3879(70)90073-5. [DOI] [PubMed] [Google Scholar]
- 3.Munoz E, Ghuysen JM, Heymann H. Cell walls of Streptococcus pyogenes, type 14. C polysaccharide-peptidoglycan and G polysaccharide-peptidoglycan complexes. Biochemistry. 1967;6:3659–3670. doi: 10.1021/bi00864a007. [DOI] [PubMed] [Google Scholar]
- 4.Coley J, Archibald AR, Baddiley J. A linkage unit joining peptidoglycan to teichoic acid in Staphylococcus aureus H. FEBS Lett. 1976;61:240–242. doi: 10.1016/0014-5793(76)81047-2. [DOI] [PubMed] [Google Scholar]
- 5.Schneewind O, Fowler A, Faull KF. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 6.Richter S, Anderson VJ, Garufi G, Lu L, Budzik JM, Joachimiak A, He C, Schneewind O, Missiakas D. Capsule anchoring in Bacillus anthracis occurs by a transpeptidation reaction that is inhibited by capsidin. Mol Microbiol. 2009;71:404–420. doi: 10.1111/j.1365-2958.2008.06533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun V, Rehn K. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem. 1969;10:426–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 8.Minnikin D. Lipids: complex lipids, their chemistry, biosynthesis and roles. In: Ratledge C, Stanford J, editors. The Biology of Mycobacteria. Vol. 1. Academic Press; London: 1982. pp. 95–184. [Google Scholar]
- 9.Niederweis M, Danilchanka O, Huff J, Hoffmann C, Engelhardt H. Mycobacterial outer membranes: in search of proteins. Trends Microbiol. 2010;18:109–116. doi: 10.1016/j.tim.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bansal-Mutalik R, Nikaido H. Quantitative lipid composition of cell envelopes of Corynebacterium glutamicum elucidated through reverse micelle extraction. Proc Natl Acad Sci U S A. 2011;108:15360–15365. doi: 10.1073/pnas.1112572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giesbrecht P, Wecke J, Reinicke B. On the morphogenesis of the cell wall of staphylococci. Int Rev Cytol. 1976;44:225–318. doi: 10.1016/s0074-7696(08)61651-4. [DOI] [PubMed] [Google Scholar]
- 12.Desvaux M, Dumas E, Chafsey I, Hebraud M. Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiol Lett. 2006;256:1–15. doi: 10.1111/j.1574-6968.2006.00122.x. [DOI] [PubMed] [Google Scholar]
- 13.Scott JR, Barnett TC. Surface proteins of gram-positive bacteria and how they get there. Annu Rev Microbiol. 2006;60:397–423. doi: 10.1146/annurev.micro.60.080805.142256. [DOI] [PubMed] [Google Scholar]
- 14.Schneewind O, Missiakas DM. Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond B Biol Sci. 2012;367:1123–1139. doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duong F, Eichler J, Price A, Leonard MR, Wickner W. Biogenesis of the gram-negative bacterial envelope. Cell. 1997;91:567–573. doi: 10.1016/s0092-8674(00)80444-4. [DOI] [PubMed] [Google Scholar]
- 16.Pohlschröder M, Prinz WA, Hartmann E, Beckwith J. Protein translocation in the three domains of life: variations on a theme. Cell. 1997;91:563–566. doi: 10.1016/s0092-8674(00)80443-2. [DOI] [PubMed] [Google Scholar]
- 17.Pugsley AP. The complete general secretory pathway in Gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver DB, Beckwith J. E. coli mutant pleiotropically defective in the export of secreted proteins. Cell. 1981;25:765–772. doi: 10.1016/0092-8674(81)90184-7. [DOI] [PubMed] [Google Scholar]
- 19.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 20.Duong F, Eichler J, Price A, Leonard MR, Wickner W. Biogenesis of the gram-negative bacterial envelope. Cell. 1997;91:567–573. doi: 10.1016/s0092-8674(00)80444-4. [DOI] [PubMed] [Google Scholar]
- 21.Eichler J, Wickner W. Both an N-terminal 65-kDa domain and a C-terminal 30-kDa domain of SecA cycle into the membrane at SecYEG during translocation. Proc Natl Acad Sci U S A. 1997;94:5574–5581. doi: 10.1073/pnas.94.11.5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Wolk JP, de Wit JG, Driessen AJ. The catalytic cycle of the escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 1997;16:7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 24.Matsuyama S, Fujita Y, Mizushima S. SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:265–270. doi: 10.1002/j.1460-2075.1993.tb05652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duong F, Wickner W. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 1997;16:4871–4879. doi: 10.1093/emboj/16.16.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nouwen N, Piwowarek M, Berrelkamp G, Driessen AJ. The large first periplasmic loop of SecD and SecF plays an important role in SecDF functioning. J Bacteriol. 2005;187:5857–5860. doi: 10.1128/JB.187.16.5857-5860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Gier JW, Valent QA, Von Heijne G, Luirink J. The E. coli SRP: preferences of a targeting factor. FEBS Lett. 1997;408:1–4. doi: 10.1016/s0014-5793(97)00402-x. [DOI] [PubMed] [Google Scholar]
- 28.Valent QA, Scotti PA, High S, de Gier JW, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter P, Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci USA. 1980;77:7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poritz MA, Strub K, Walter P. Human SRP RNA and E. coli 4.5S RNA contain a highly homologous structural domain. Cell. 1988;55:4–6. doi: 10.1016/0092-8674(88)90003-7. [DOI] [PubMed] [Google Scholar]
- 32.Struck JC, Toschka HY, Specht T, Erdmann VA. Common structural features between eukaryotic 7SL RNAs, eubacterial 4.5S RNA and scRNA and archaebacterial 7S RNA. Nucleic Acids Res. 1988;16:7740. doi: 10.1093/nar/16.15.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- 34.Bernstein HD, Rapoport TA, Walter P. Cytosolic protein translocation factors. Is SRP still unique? Cell. 1989;58:1017–1019. doi: 10.1016/0092-8674(89)90497-2. [DOI] [PubMed] [Google Scholar]
- 35.Miller JD, Bernstein HD, Walter P. Interaction of E. coli Ffh/4.5S ribonucleoprotein and FtsY mimics that of mammalian signal recognition particle and its receptor. Nature. 1994;367:657–659. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- 36.Muller M, Blobel G. In vitro translocation of bacterial proteins across the plasma membrane of Escherichia coli. Proc Natl Acad Sci U S A. 1984;81:7421–7425. doi: 10.1073/pnas.81.23.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Randall LL. Peptide binding by chaperone SecB: implications for recognition of nonnative structure. Science. 1992;257:241–245. doi: 10.1126/science.1631545. [DOI] [PubMed] [Google Scholar]
- 38.Kumamoto CA, Francetic O. Highly selective binding of nascent polypeptides by an Escherichia coli chaperone protein in vivo. J Bacteriol. 1993;175:2184–2188. doi: 10.1128/jb.175.8.2184-2188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valent QA, Kendall DA, High S, Kusters R, Oudega B, Luirink J. Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 1995;14:5494–5505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulbrandt ND, Newitt JA, Bernstein HD. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 41.Chang CN, Blobel G, Model P. Detection of prokaryotic signal peptidase in an Escherichia coli membrane fraction: endoproteolytic cleavage of nascent f1 pre-coat protein. Proc Natl Acad Sci USA. 1978;75:361–365. doi: 10.1073/pnas.75.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalbey RE, Wickner W. Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coli plasma membrane. J Biol Chem. 1985;260:15925–15931. [PubMed] [Google Scholar]
- 43.van Roosmalen ML, Geukens N, Jongbloed JD, Tjalsma H, Dubois JY, Bron S, van Dijl JM, Anne J. Type I signal peptidases of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:279–297. doi: 10.1016/j.bbamcr.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 44.Caspers M, Freudl R. Corynebacterium glutamicum possesses two secA homologous genes that are essential for viability. Arch Microbiol. 2008;189:605–610. doi: 10.1007/s00203-008-0351-0. [DOI] [PubMed] [Google Scholar]
- 45.Anne J, Maldonado B, Van Impe J, Van Mellaert L, Bernaerts K. Recombinant protein production and streptomycetes. J Biotechnol. 2012;158:159–167. doi: 10.1016/j.jbiotec.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 46.Lee VT, Schneewind O. Protein secretion and the pathogenesis of bacterial infections. Genes Dev. 2001;15:1725–1752. doi: 10.1101/gad.896801. [DOI] [PubMed] [Google Scholar]
- 47.Bolhuis A, Broekhuizen CP, Sorokin A, van Roosmalen ML, Venema G, Bron S, Quax WJ, van Dijl J. SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins. J Biol Chem. 1998;273:21217–21224. doi: 10.1074/jbc.273.33.21217. [DOI] [PubMed] [Google Scholar]
- 48.Muller JP, Ozegowski J, Vettermann S, Swaving J, Van Wely KH, Driessen AJ. Interaction of Bacillus subtilis CsaA with SecA and precursor proteins. Biochem J. 2000;348(Pt 2):367–373. [PMC free article] [PubMed] [Google Scholar]
- 49.Wild J, Altman E, Yura T, Gross CA. DnaK and DnaJ heat shock proteins participate in protein export in Escherichia coli. Genes Dev. 1992;6:1165–1172. doi: 10.1101/gad.6.7.1165. [DOI] [PubMed] [Google Scholar]
- 50.Crowley PJ, Svensäter G, Snoep JL, Bleiweis AS, Brady LJ. An ffh mutant of Streptococcus mutans is viable and able to physiologically adapt to low pH in continuous culture. FEMS Microbiol Lett. 2004;234:315–324. doi: 10.1016/j.femsle.2004.03.043. [DOI] [PubMed] [Google Scholar]
- 51.Rosch JW, Vega LA, Beyer JM, Lin A, Caparon MG. The signal recognition particle pathway is required for virulence in Streptococcus pyogenes. Infect Immun. 2008;76:2612–2619. doi: 10.1128/IAI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trevino J, Perez N, Sumby P. The 4.5S RNA component of the signal recognition particle is required for group A Streptococcus virulence. Microbiology. 2010;156:1342–1350. doi: 10.1099/mic.0.036558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fagan RP, Fairweather NF. Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem. 2011;286:27483–27493. doi: 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji Y, Zhang B, Van SF, Horn, Warren P, Woodnutt G, Burnham MK, Rosenberg M. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- 55.Chang CN, Nielsen JBK, Izui K, Blobel G, Lampen JO. Idenification of the signal peptidase cleavage site in Bacillus licheniformis prepenicillinase. J Biol Chem. 1982;257:4340–4344. [PubMed] [Google Scholar]
- 56.Quiblier C, Seidl K, Roschitzki B, Zinkernagel AS, Berger-Bachi B, Senn MM. Secretome analysis defines the major role of SecDF in Staphylococcus aureus virulence. PLoS One. 2013;8:e63513. doi: 10.1371/journal.pone.0063513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schallenberger MA, Niessen S, Shao C, Fowler BJ, Romesberg FE. Type I Signal Peptidase and Protein Secretion in Staphylococcus aureus. J Bacteriol. 2012;194:2677–2686. doi: 10.1128/JB.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeDent A, Bae T, Missiakas DM, Schneewind O. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 2008;27:2656–2668. doi: 10.1038/emboj.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 60.von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 1989;2:531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- 61.Tjalsma H, Antelmann H, Jongbloed JD, Braun PG, Darmon E, Dorenbos R, Dubois JY, Westers H, Zanen G, Quax WJ, Kuipers OP, Bron S, Hecker M, van Dijl JM. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol Mol Biol Rev. 2004;68:207–233. doi: 10.1128/MMBR.68.2.207-233.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolfe PB, Silver P, Wickner W. The isolation of homogeneous leader peptidase from a strain of Escherichia coli which overproduces the enzyme. J Biol Chem. 1982;257:7898–7902. [PubMed] [Google Scholar]
- 63.Tjalsma H, Bolhuis A, van Roosmalen ML, Wiegert T, Schumann W, Broekhuizen CP, Quax WJ, Venema G, Bron S, van Dijl JM. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 1998;12:2318–2331. doi: 10.1101/gad.12.15.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Terra R, Stanley-Wall NR, Cao G, Lazazzera BA. Identification of Bacillus subtilis SipW as a bifunctional signal peptidase that controls surface-adhered biofilm formation. J Bacteriol. 2012;194:2781–2790. doi: 10.1128/JB.06780-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cregg KM, Wilding I, Black MT. Molecular cloning and expression of the spsB gene encoding an essential type I signal peptidase from Staphylococcus aureus. J Bacteriol. 1996;178:5712–5718. doi: 10.1128/jb.178.19.5712-5718.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuhn A, Wickner W. Conserved residues of the leader peptide are essential for cleavage by leader peptidase. J Biol Chem. 1985;260:15914–15918. [PubMed] [Google Scholar]
- 67.Heinrich J, Lunden T, Kontinen VP, Wiegert T. The Bacillus subtilis ABC transporter EcsAB influences intramembrane proteolysis through RasP. Microbiology. 2008;154:1989–1997. doi: 10.1099/mic.0.2008/018648-0. [DOI] [PubMed] [Google Scholar]
- 68.Dunny GM, Leonard BA. Cell-cell communication in gram-positive bacteria. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 69.An FY, Sulavik MC, Clewell DB. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J Bacteriol. 1999;181:5915–5921. doi: 10.1128/jb.181.19.5915-5921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clewell DB, An FY, Flannagan SE, Antiporta M, Dunny GM. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol Microbiol. 2000;35:246–247. doi: 10.1046/j.1365-2958.2000.01687.x. [DOI] [PubMed] [Google Scholar]
- 71.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 72.Leskela S, Wahlstrom E, Hyyrylainen HL, Jacobs M, Palva A, Sarvas M, Kontinen VP. Ecs, an ABC transporter of Bacillus subtilis: dual signal transduction functions affecting expression of secreted proteins as well as their secretion. Mol Microbiol. 1999;31:533–543. doi: 10.1046/j.1365-2958.1999.01194.x. [DOI] [PubMed] [Google Scholar]
- 73.Jonsson IM, Juuti JT, Francois P, AlMajidi R, Pietiainen M, Girard M, Lindholm C, Saller MJ, Driessen AJ, Kuusela P, Bokarewa M, Schrenzel J, Kontinen VP. Inactivation of the Ecs ABC transporter of Staphylococcus aureus attenuates virulence by altering composition and function of bacterial wall. PLoS One. 2010;5:e14209. doi: 10.1371/journal.pone.0014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 75.Adler LA, Arvidson S. Correlation between the rate of exoprotein synthesis and the amount of the multiprotein complex on membrane-bound ribosomes (MBRP-complex) in Staphylococcus aureus. J Gen Microbiol. 1987;133:803–813. doi: 10.1099/00221287-133-3-803. [DOI] [PubMed] [Google Scholar]
- 76.Adler LA, Arvidson S. Cloning and expression in Escherichia coli of genes encoding a multiprotein complex involved in secretion of proteins from Staphylococcus aureus. J Bacteriol. 1988;170:5337–5343. doi: 10.1128/jb.170.11.5337-5343.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hemila H, Palva A, Paulin L, Arvidson S, Palva I. Secretory S complex of Bacillus subtilis: sequence analysis and identity to pyruvate dehydrogenase. J Bacteriol. 1990;172:5052–5063. doi: 10.1128/jb.172.9.5052-5063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hemila H, Palva A, Paulin L, Adler L, Arvidson S, Palva I. The secretory S complex in Bacillus subtilis is identified as pyruvate dehydrogenase. Res Microbiol. 1991;142:779–785. doi: 10.1016/0923-2508(91)90055-f. [DOI] [PubMed] [Google Scholar]
- 79.Rosenstein R, Gotz F. Staphylococcal lipases:biochemical and molecular characterization. Biochimie. 2000;82:1005–1014. doi: 10.1016/s0300-9084(00)01180-9. [DOI] [PubMed] [Google Scholar]
- 80.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 81.Bae T, Schneewind O. The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J Bacteriol. 2003;185:2910–2919. doi: 10.1128/JB.185.9.2910-2919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.DeDent AC, McAdow M, Schneewind O. Distribution of protein A on the surface of Staphylococcus aureus. J Bacteriol. 2007;189:4473–4484. doi: 10.1128/JB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tzagoloff H, Novick R. Geometry of cell division in Staphylococcus aureus. J Bacteriol. 1977;129:343–350. doi: 10.1128/jb.129.1.343-350.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giesbrecht P, Kersten T, Maidhof H, Wecke J. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol Mol Biol Rev. 1998;62:1371–1414. doi: 10.1128/mmbr.62.4.1371-1414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carlsson F, Stalhammar-Carlemalm M, Flardh K, Sandin C, Carlemalm E, Lindahl G. Signal sequence directs localized secretion of bacterial surface proteins. Nature. 2006;442:943–946. doi: 10.1038/nature05021. [DOI] [PubMed] [Google Scholar]
- 86.Yu W, Gotz F. Cell wall antibiotics provoke accumulation of anchored mCherry in the cross wall of Staphylococcus aureus. PLoS One. 2012;7:e30076. doi: 10.1371/journal.pone.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rosch J, Caparon M. A microdomain for protein secretion in Gram-positive bacteria. Science. 2004;304:1513–1515. doi: 10.1126/science.1097404. [DOI] [PubMed] [Google Scholar]
- 88.Rosch JW, Caparon MG. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol. 2005;58:959–968. doi: 10.1111/j.1365-2958.2005.04887.x. [DOI] [PubMed] [Google Scholar]
- 89.Kline KA, Kau AL, Chen SL, Lim A, Pinkner JS, Rosch J, Nallapareddy SR, Murray BE, Henriques-Normark B, Beatty W, Caparon MG, Hultgren SJ. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol. 2009;191:3237–3247. doi: 10.1128/JB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Raz A, Fischetti VA. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc Natl Acad Sci U S A. 2008;105:18549–18554. doi: 10.1073/pnas.0808301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, Westermann M, Muller JP, Bron S, Kok J, Kuipers OP, Jongbloed JD. Subcellular sites for bacterial protein export. Mol Microbiol. 2004;53:1583–1599. doi: 10.1111/j.1365-2958.2004.04278.x. [DOI] [PubMed] [Google Scholar]
- 92.Tsui HC, Keen SK, Sham LT, Wayne KJ, Winkler ME. Dynamic distribution of the SecA and SecY translocase subunits and septal localization of the HtrA surface chaperone/protease during Streptococcus pneumoniae D39 cell division. MBio. 2011;2 doi: 10.1128/mBio.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Frankel MB, Wojcik BM, DeDent AC, Missiakas DM, Schneewind O. ABI domain-containing proteins contribute to surface protein display and cell division in Staphylococcus aureus. Mol Microbiol. 2010;78:238–252. doi: 10.1111/j.1365-2958.2010.07334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuttner AG, Lenert TF. The Occurrence of Bacteriostatic Properties in the Blood of Patients after Recovery from Streptococcal Pharyngitis. J Clin Invest. 1944;23:151–161. doi: 10.1172/JCI101478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lancefield RC. Current knowledge of type-specific M antigens of group A streptococci. J Immunol. 1962;89:307–313. [PubMed] [Google Scholar]
- 96.Jensen K. A normally occurring Staphylococcus antibody in human serum. Acta path microbiol scand. 1958;44:421–428. doi: 10.1111/j.1600-0463.2007.apm_731a.x. [DOI] [PubMed] [Google Scholar]
- 97.Forsgren A, Sjoquist J. “Protein A” from S. aureus. I. Pseudo-immune reaction with human gamma-globulin. J Immunol. 1966;97:822–827. [PubMed] [Google Scholar]
- 98.Phillips GN, Jr, Flicker PF, Cohen C, Manjula BN, Fischetti VA. Streptococcal M protein: alpha-helical coiled-coil structure and arrangement on the cell surface. Proc Natl Acad Sci U S A. 1981;78:4689–4693. doi: 10.1073/pnas.78.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fischetti VA. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McNamara C, Zinkernagel AS, Macheboeuf P, Cunningham MW, Nizet V, Ghosh P. Coiled-coil irregularities and instabilities in group A Streptococcus M1 are required for virulence. Science. 2008;319:1405–1408. doi: 10.1126/science.1154470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lancefield R. Current knowledge of type-specific M antigens of group A streptococci. J Immunol. 1962;89:307–313. [PubMed] [Google Scholar]
- 102.Hjelm H, Hjelm K, Sjoquist J. Protein A from Staphylococcus aureus. Its isolation by affinity chromatography and its use as an immunosorbent for isolation of immunoglobulins. FEBS Lett. 1972;28:73–76. doi: 10.1016/0014-5793(72)80680-x. [DOI] [PubMed] [Google Scholar]
- 103.Sjöquist J, Meloun B, Hjelm H. Protein A isolated from Staphylococcus aureus after digestion with lysostaphin. Eur J Biochem. 1972;29:572–578. doi: 10.1111/j.1432-1033.1972.tb02023.x. [DOI] [PubMed] [Google Scholar]
- 104.Fischetti VA, Pancholi V, Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol Microbiol. 1990;4:1603–1605. doi: 10.1111/j.1365-2958.1990.tb02072.x. [DOI] [PubMed] [Google Scholar]
- 105.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface protein of Gram-positive bacteria. EMBO. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Navarre WW, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 107.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ton-That H, Schneewind O. Anchor structure of staphylococcal surface proteins. IV. Inhibitors of the cell wall sorting reaction. J Biol Chem. 1999;274:24316–24320. doi: 10.1074/jbc.274.34.24316. [DOI] [PubMed] [Google Scholar]
- 109.Higashi Y, Strominger JL, Sweeley CC. Structure of a lipid intermediate in cell wall peptidoglycan synthesis: a derivative of C55 isoprenoid alcohol. Proc Natl Acad Sci USA. 1967;57:1878–1884. doi: 10.1073/pnas.57.6.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem. 2002;277:16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- 111.Ruzin A, Severin A, Ritacco F, Tabei K, Ingh GS, Bradford PA, Siegel MM, Projan SJ, Shlaes DM. Further evidence that a cell wall precursor [C(55)-MurNAc-(peptide)-GlcNAc] serves as an acceptor in a sorting reaction. J Bacteriol. 2002;184:2141–2147. doi: 10.1128/JB.184.8.2141-2147.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ghuysen JM, Strominger JL. Structure of the Cell Wall of Staphylococcus Aureus, Strain Copenhagen. Ii. Separation and Structure of Disaccharides. Biochemistry. 1963;2:1119–1125. doi: 10.1021/bi00905a036. [DOI] [PubMed] [Google Scholar]
- 113.Ghuysen JM, Tipper DJ, Strominger JL. Structure of the Cell Wall of Staphylococcus Aureus, Strain Copenhagen. Iv. The Teichoic Acid-Glycopeptide Complex. Biochemistry. 1965;4:474–485. doi: 10.1021/bi00879a016. [DOI] [PubMed] [Google Scholar]
- 114.Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-alanine. Proc Natl Acad Sci USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Archibald AR, Stafford GH. A polymer of N-acetylglucosamine 1-phosphate in the wall of Staphylococcus lactis 2102. Biochem J. 1972;130:681–690. doi: 10.1042/bj1300681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Beveridge TJ. Ultrastructure, chemistry, and function of the bacterial wall. Int Rev Cytol. 1981;72:229–317. doi: 10.1016/s0074-7696(08)61198-5. [DOI] [PubMed] [Google Scholar]
- 117.Strominger JL. Penicillin-sensitive enzymatic reactions in bacterial cell wall synthesis. Harvey Lectures. 1968;64:179–213. [PubMed] [Google Scholar]
- 118.Ton-That H, Faull KF, Schneewind O. Anchor structure of staphylococcal surface proteins. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J Biol Chem. 1997;272:22285–22292. doi: 10.1074/jbc.272.35.22285. [DOI] [PubMed] [Google Scholar]
- 119.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dmitriev BA, Toukach FV, Holst O, Rietschel ET, Ehlers S. Tertiary structure of Staphylococcus aureus cell wall murein. J Bacteriol. 2004;186:7141–7148. doi: 10.1128/JB.186.21.7141-7148.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bruck S, Personnic N, Prevost MC, Cossart P, Bierne H. Regulated shift from helical to polar localization of Listeria monocytogenes cell wall-anchored proteins. J Bacteriol. 2011;193:4425–4437. doi: 10.1128/JB.01154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bierne H, Dramsi S. Spatial positioning of cell wall-anchored virulence factors in Gram-positive bacteria. Curr Opin Microbiol. 2012;15:715–723. doi: 10.1016/j.mib.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 123.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 124.Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci USA. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ton-That H, Mazmanian SK, Faull KF, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J Biol Chem. 2000;275:9876–9881. doi: 10.1074/jbc.275.13.9876. [DOI] [PubMed] [Google Scholar]
- 126.Huang X, Aulabaugh A, Ding W, Kapoor B, Alksne L, Tabei K, Ellestad G. Kinetic mechanism of Staphylococcus aureus sortase SrtA. Biochemistry. 2003;42:11307–11315. doi: 10.1021/bi034391g. [DOI] [PubMed] [Google Scholar]
- 127.Ilangovan U, Ton-That H, Iwahara J, Schneewind O, Clubb RT. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2001;98:6056–6061. doi: 10.1073/pnas.101064198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zong Y, Bice TW, Ton-That H, Schneewind O, Narayana SV. Crystal structures of Staphylococcus aureus sortase A and its substrate complex. J Biol Chem. 2004;279:31383–31389. doi: 10.1074/jbc.M401374200. [DOI] [PubMed] [Google Scholar]
- 129.Ilangovan U, Iwahara J, Ton-That H, Schneewind O, Clubb RT. Assignment of the 1H, 13C and 15N signals of Sortase. J Biomol NMR. 2001;19:379–380. doi: 10.1023/a:1011299500628. [DOI] [PubMed] [Google Scholar]
- 130.Ton-That H, Mazmanian SK, Alksne L, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Cysteine 184 and histidine 120 of sortase form a thiolate-imidazolium ion pair for catalysis. J Biol Chem. 2002;277:7447–7452. doi: 10.1074/jbc.M109945200. [DOI] [PubMed] [Google Scholar]
- 131.Dessen A. A new catalytic dyad regulates anchoring of molecules to the Gram-positive cell wall by sortases. Structure. 2004;12:6–7. doi: 10.1016/j.str.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 132.Zhang R, Wu R, Joachimiak G, Mazmanian SK, Missiakas DM, Gornicki P, Schneewind O, Joachimiak A. Structures of sortase B from Staphylococcus aureus and Bacillus anthracis reveal catalytic amino acid triad in the active site. Structure. 2004;12:1147–1156. doi: 10.1016/j.str.2004.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Race PR, Bentley ML, Melvin JA, Crow A, Hughes RK, Smith WD, Sessions RB, Kehoe MA, McCafferty DG, Banfield MJ. Crystal structure of Streptococcus pyogenes sortase A: implications for sortase mechanism. J Biol Chem. 2009;284:6924–6933. doi: 10.1074/jbc.M805406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dhar G, Faull KF, Schneewind O. Anchor structure of cell wall surface proteins in Listeria monocytogenes. Biochemistry. 2000;39:3725–3733. doi: 10.1021/bi992347o. [DOI] [PubMed] [Google Scholar]
- 135.Foster TJ, Höök M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6:484–488. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 136.Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jonsson IM, Mazmanian SK, Schneewind O, Verdrengh M, Bremell T, Tarkowski A. On the role of Staphylococcus aureus sortase and sortase-catalyzed surface protein anchoring in murine septic arthritis. J Infect Dis. 2002;185:1417–1424. doi: 10.1086/340503. [DOI] [PubMed] [Google Scholar]
- 138.Jonsson IM, Mazmanian SK, Schneewind O, Bremell T, Tarkowski A. The role of Staphylococcus aureus sortase A and sortase B in murine arthritis. Microbes Infect. 2003;5:775–780. doi: 10.1016/s1286-4579(03)00143-6. [DOI] [PubMed] [Google Scholar]
- 139.Weiss WJ, Lenoy E, Murphy T, Tardio L, Burgio P, Projan SJ, Schneewind O, Alksne L. Effect of srtA and srtB gene expression on the virulence of Staphylococcus aureus in animal models of infection. J Antimicrob Chemother. 2004;53:480–486. doi: 10.1093/jac/dkh078. [DOI] [PubMed] [Google Scholar]
- 140.Bierne H, Mazmanian SK, Trost M, Pucciarelli MG, Liu G, Dehoux P, Jansch L, Garcia-del Portillo F, Schneewind O, Cossart P. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol Microbiol. 2002;43:869–881. doi: 10.1046/j.1365-2958.2002.02798.x. [DOI] [PubMed] [Google Scholar]
- 141.Garandeau C, Reglier-Poupet H, Dubail I, Beretti JL, Berche P, Charbit A. The sortase SrtA of Listeria monocytogenes is involved in processing of internalin and in virulence. Infect Immun. 2002;70:1382–1390. doi: 10.1128/IAI.70.3.1382-1390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kharat AS, Tomasz A. Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect Immun. 2003;71:2758–2765. doi: 10.1128/IAI.71.5.2758-2765.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lee SF, Boran TL. Roles of sortase in surface expression of the major protein adhesin P1, saliva-induced aggregation and adherence, and cariogenicity of Streptococcus mutans. Infect Immun. 2003;71:676–681. doi: 10.1128/IAI.71.2.676-681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen S, Paterson GK, Tong HH, Mitchell TJ, DeMaria TF. Sortase A contributes to pneumococcal nasopharyngeal colonization in the chinchilla model. FEMS Microbiol Lett. 2005;253:151–154. doi: 10.1016/j.femsle.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 145.Lalioui L, Pellegrini E, Dramsi S, Baptista M, Bourgeois N, Doucet-Populaire F, Rusniok C, Zouine M, Glaser P, Kunst F, Poyart C, Trieu-Cuot P. The SrtA Sortase of Streptococcus agalactiae is required for cell wall anchoring of proteins containing the LPXTG motif, for adhesion to epithelial cells, and for colonization of the mouse intestine. Infect Immun. 2005;73:3342–3350. doi: 10.1128/IAI.73.6.3342-3350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Levesque CM, Voronejskaia E, Huang YC, Mair RW, Ellen RP, Cvitkovitch DG. Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infect Immun. 2005;73:3773–3777. doi: 10.1128/IAI.73.6.3773-3777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Paterson GK, Mitchell TJ. The role of Streptococcus pneumoniae sortase A in colonisation and pathogenesis. Microbes Infect. 2006;8:145–153. doi: 10.1016/j.micinf.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 148.Sabet C, Lecuit M, Cabanes D, Cossart P, Bierne H. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect Immun. 2005;73:6912–6922. doi: 10.1128/IAI.73.10.6912-6922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Leigh JA, Egan SA, Ward PN, Field TR, Coffey TJ. Sortase anchored proteins of Streptococcus uberis play major roles in the pathogenesis of bovine mastitis in dairy cattle. Vet Res. 2010;41:63. doi: 10.1051/vetres/2010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Turner LS, Kanamoto T, Unoki T, Munro CL, Wu H, Kitten T. Comprehensive evaluation of Streptococcus sanguinis cell wall-anchored proteins in early infective endocarditis. Infect Immun. 2009;77:4966–4975. doi: 10.1128/IAI.00760-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stranger-Jones YK, Bae T, Schneewind O. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc Natl Acad Sci U S A. 2006;103:16942–16947. doi: 10.1073/pnas.0606863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Arrecubieta C, Matsunaga I, Asai T, Naka Y, Deng MC, Lowy FD. Vaccination with clumping factor A and fibronectin binding protein A to prevent Staphylococcus aureus infection of an aortic patch in mice. J Infect Dis. 2008;198:571–575. doi: 10.1086/590210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hendrickx AP, Willems RJ, Bonten MJ, van Schaik W. LPxTG surface proteins of enterococci. Trends Microbiol. 2009;17:423–430. doi: 10.1016/j.tim.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 154.Soriani M, Telford JL. Relevance of pili in pathogenic streptococci pathogenesis and vaccine development. Future Microbiol. 2010;5:735–747. doi: 10.2217/fmb.10.37. [DOI] [PubMed] [Google Scholar]
- 155.Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pallen MJ, Lam AC, Antonio M, Dunbar K. An embarrassment of sortases - a richness of substrates? Trends Microbiol. 2001;9:97–102. doi: 10.1016/s0966-842x(01)01956-4. [DOI] [PubMed] [Google Scholar]
- 157.Comfort D, Clubb RT. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect Immun. 2004;72:2710–2722. doi: 10.1128/IAI.72.5.2710-2722.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Dramsi S, Trieu-Cuot P, Bierne H. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol. 2005;156:289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 159.Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, Joachmiak A, Missiakas DM, Schneewind O. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 160.Maresso AW, Chapa TJ, Schneewind O. Surface protein IsdC and Sortase B are required for heme-iron scavenging of Bacillus anthracis. J Bacteriol. 2006;188:8145–8152. doi: 10.1128/JB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Newton SM, Klebba PE, Raynaud C, Shao Y, Jiang X, Dubail I, Archer C, Frehel C, Charbit A. The svpA-srtB locus of Listeria monocytogenes: fur-mediated iron regulation and effect on virulence. Mol Microbiol. 2005;55:927–940. doi: 10.1111/j.1365-2958.2004.04436.x. [DOI] [PubMed] [Google Scholar]
- 162.Marraffini LA, Schneewind O. Anchor structure of staphylococcal surface proteins. V. Anchor structure of the sortase B substrate IsdC. J Biol Chem. 2005;280:16263–16271. doi: 10.1074/jbc.M500071200. [DOI] [PubMed] [Google Scholar]
- 163.Mariscotti JF, Garcia-del Portillo F, Pucciarelli MG. The Listeria monocytogenes sortase-B recognizes varied amino acids at position 2 of the sorting motif. J Biol Chem. 2009;284:6140–6146. doi: 10.1074/jbc.M807989200. [DOI] [PubMed] [Google Scholar]
- 164.Zong Y, Mazmanian SK, Schneewind O, Narayana SV. The structure of sortase B, a cysteine transpeptidase that tethers surface protein to the Staphylococcus aureus cell wall. Structure. 2004;12:105–112. doi: 10.1016/j.str.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 165.Mazmanian SK, Ton-That H, Su K, Schneewind O. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc Natl Acad Sci U S A. 2002;99:2293–2298. doi: 10.1073/pnas.032523999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 167.Skaar EP, Gaspar AH, Schneewind O. Bacillus anthracis IsdG, a heme degrading monooxygenase. J Bacteriol. 2006;188:1071–1080. doi: 10.1128/JB.188.3.1071-1080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kline KA, Dodson KW, Caparon MG, Hultgren SJ. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 2010;18:224–232. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hendrickx AP, Budzik JM, Oh SY, Schneewind O. Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol. 2011;9:166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- 170.Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, Telford JL. Genome analysis reveals pili in Group B streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 171.Yeung MK, Donkersloot JA, Cisar JO, Ragsdale PA. Identification of a gene involved in assembly of Actinomyces naeslundii T14V type 2 fimbriae. Infect Immun. 1998;66:1482–1491. doi: 10.1128/iai.66.4.1482-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yeung MK, Ragsdale PA. Synthesis and function of Actinomyces naeslundii T14V type 1 fimbriae require the expression of additional fimbria-associated genes. Infect Immun. 1997;65:2629–2639. doi: 10.1128/iai.65.7.2629-2639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 174.Ton-That H, Schneewind O. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 2004;12:228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 175.Swaminathan A, Mandlik A, Swierczynski A, Gaspar A, Das A, Ton-That H. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol. 2007;66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Budzik JM, Marraffini LA, Souda P, Whitelegge JP, Faull KF, Schneewind O. Amide bonds assemble pili on the surface of bacilli. Proc Natl Acad Sci U S A. 2008;105:10215–10220. doi: 10.1073/pnas.0803565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Budzik JM, Oh SY, Schneewind O. Cell wall anchor structure of BcpA pili in Bacillus anthracis. J Biol Chem. 2008;283:36676–36686. doi: 10.1074/jbc.M806796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Oh SY, Budzik JM, Schneewind O. Sortases make pili from three ingredients. Proc Natl Acad Sci U S A. 2008;105:13703–13704. doi: 10.1073/pnas.0807334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Telford JL, Barocchi M, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 180.Krishnan V, Gaspar AH, Ye N, Mandlik A, Ton-That H, Narayana SV. An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure. 2007;15:893–903. doi: 10.1016/j.str.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 182.Deivanayagam CC, Rich RL, Carson M, Owens RT, Danthuluri S, Bice T, Hook M, Narayana SV. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure. 2000;8:67–78. doi: 10.1016/s0969-2126(00)00081-2. [DOI] [PubMed] [Google Scholar]
- 183.Marraffini LA, Schneewind O. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol Microbiol. 2006;62:1402–1417. doi: 10.1111/j.1365-2958.2006.05469.x. [DOI] [PubMed] [Google Scholar]
- 184.Marraffini LA, Schneewind O. Sortase C-mediated anchoring of BasI to the cell wall envelope of Bacillus anthracis. J Bacteriol. 2007;189:6425–6436. doi: 10.1128/JB.00702-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gaspar AH, Marraffini LA, Glass EM, Debord KL, Ton-That H, Schneewind O. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J Bacteriol. 2005;187:4646–4655. doi: 10.1128/JB.187.13.4646-4655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Antelmann H, Darmon E, Noone D, Veening JW, Westers H, Bron S, Kuipers OP, Devine KM, Hecker M, van Dijl JM. The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol Microbiol. 2003;49:143–156. doi: 10.1046/j.1365-2958.2003.03565.x. [DOI] [PubMed] [Google Scholar]
- 187.Vitikainen M, Hyyrylainen HL, Kivimaki A, Kontinen VP, Sarvas M. Secretion of heterologous proteins in Bacillus subtilis can be improved by engineering cell components affecting post-translocational protein folding and degradation. J Appl Microbiol. 2005;99:363–375. doi: 10.1111/j.1365-2672.2005.02572.x. [DOI] [PubMed] [Google Scholar]
- 188.Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Skaar EP, Schneewind O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 2004;6:390–397. doi: 10.1016/j.micinf.2003.12.008. [DOI] [PubMed] [Google Scholar]