Abstract
Dysfunction of the RNA-binding protein, TDP-43, is strongly implicated as a causative event in many neurodegenerative diseases including amyotrophic lateral sclerosis (ALS). TDP-43 is normally found in the nucleus and pathological hallmarks of ALS include the presence of cytoplasmic protein aggregates containing TDP-43 and an associated loss of TDP-43 from the nucleus. Loss of nuclear TDP-43 likely contributes to neurodegeneration. Using Drosophila melanogaster to model TDP-43 loss of function, we show that reduced levels of the voltage-gated calcium channel, cacophony, mediate some of the physiological effects of TDP-43 loss. Null mutations in the Drosophila orthologue of TDP-43, named TBPH, resulted in defective larval locomotion and reduced levels of cacophony protein in whole animals and at the neuromuscular junction. Restoring the levels of cacophony in all neurons or selectively in motor neurons rescued these locomotion defects. Using TBPH immunoprecipitation, we showed that TBPH associates with cacophony transcript, indicating that it is likely to be a direct target for TBPH. Loss of TBPH leads to reduced levels of cacophony transcript, possibly due to increased degradation. In addition, TBPH also appears to regulate the inclusion of some alternatively spliced exons of cacophony. If similar effects of cacophony or related calcium channels are found in human ALS patients, these could be targets for the development of pharmacological therapies for ALS.
1. Introduction
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disease that leads to the selective death of motor neurons and has no cure or treatment (Turner et al, 2013). A significant breakthrough in understanding the etiology of ALS has been the identification of the TAR DNA-binding protein (TDP-43) as a major component of the cytoplasmic aggregates in motor neurons that are a classical pathological symptom of the disease (Arai et al, 2006; Neumann et al, 2006). TDP-43 is the major component of the ubiquitinated inclusions found in ALS and in frontotemporal lobar degeneration (FTLD) (Arai et al, 2006; Neumann et al, 2006). TDP-43 containing aggregates are recognized as an important feature in other neurological diseases, such as Alzheimer disease, Parkinson disease, Huntington disease, as well as some other rare diseases (Geser et al, 2009). In addition, numerous dominant mutations in the TARDBP gene have been characterized in either familial or sporadic cases of ALS and FTLD (Gitcho et al, 2008; Kabashi et al, 2008; Sreedharan et al, 2008; Yokoseki et al, 2008), indicating that the pathogenic causes of these diseases are due to the dysfunction of TDP-43. TDP-43 is a predominantly nuclear protein and diseased neurons containing cytoplasmic TDP-43 aggregates also exhibit reduced levels of TDP-43 in the nucleus (Neumann et al, 2006). It is currently believed that the loss of normal TDP-43 function plays a critical role in neurodegenerative diseases (Lee et al, 2012).
TDP-43 is a member of the heterogeneous nuclear ribonucleoprotein family (Krecic and Swanson, 1999) and was first observed as binding the polypyrimidine region of HIV TAR DNA (Ou et al, 1995). Subsequently, studies have demonstrated that TDP-43 participates in many aspects of RNA metabolism, including RNA alternative splicing and stability (Buratti and Baralle, 2001; Volkening et al, 2009; Ayala et al, 2011; Fiesel and Kahle, 2011), transcriptional regulation (Ou et al, 1995), mRNA transport and translation (Wang et al, 2008; Fiesel et al, 2012; Buratti and Baralle, 2012). We have been using Drosophila to explore the role of the fly TDP-43 homologue, named TBPH, in a TDP-43 loss of function model for ALS (Hazelett et al, 2012). Loss of TBPH is late pupal lethal and results in severe deficits in larval locomotion (Feiguin et al, 2009; Hazelett et al, 2012; Diaper et al, 2013). We examined the transcriptome of the CNS from third instar larvae and identified about 1,000 genes that showed differential expression or splicing in TBPH loss of function larvae compared to wild type animals (Hazelett et al, 2012). Of these genes, it was notable that a gene encoding a CaV2 calcium channel, named cacophony, showed the highest score for splicing changes. Cacophony is responsible for the majority of neuronal calcium current (Peng and Wu, 2007), is localized at the active zone of the neuromuscular junction (NMJ) (Kawasaki et al 2004), where it is required for neurotransmitter release and synaptic growth (Rieckhof et al, 2003). In the current study, we show that loss of TBPH causes reduced levels of cacophony and restoring this expression either pan-neuronally or selectively in motor neurons rescues the locomotion defects caused by the loss of TBPH.
2. Results
2.1. Loss of TBPH leads to reduced cacophony protein
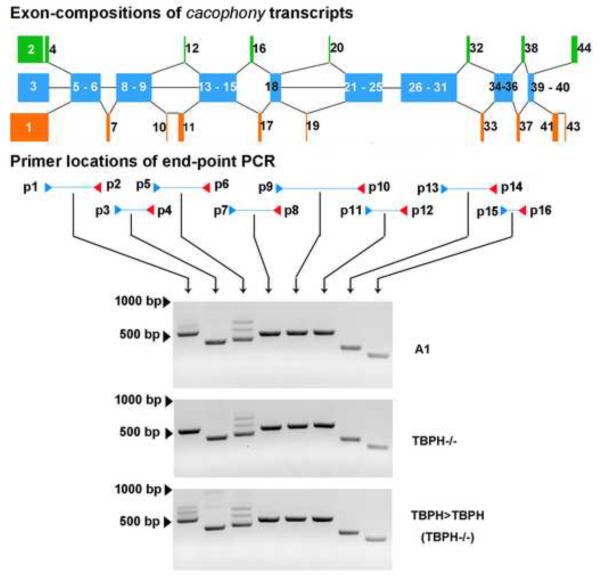
Our previous studies examining the transcriptome of the central nervous system in TBPH null mutants showed that loss of TBPH led to altered splicing of cacophony transcripts with no overall change in total cacophony transcript levels (Hazelett et al 2012). The cacophony gene generates multiple transcripts through alternative splicing of several exons (Figure 1) generating at least 15 different predicted transcripts (FlyBase). Also shown in Figure 1 are the predicted TBPH binding sites located in cacophony introns based on the sequences of the mammalian TDP-43 binding sites (see Hazelett et al, 2012).
Figure 1.
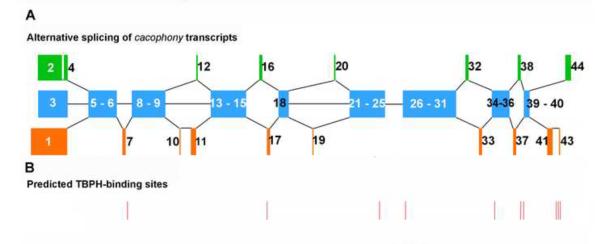
Schematic diagram of cacophony alternatively spliced exons and the positions of predicted TBPH binding sites. A. Alternatively spliced exons. Each colored block represents individual or groups of exons, which are numbered according to their designations on FlyBase. Blue blocks represent conserved exons, common to each transcript whereas orange or green blocks represent alternatively spliced exons. B. Positions of the predicted TBPH binding sites identified previously (Hazelett et al, 2012).
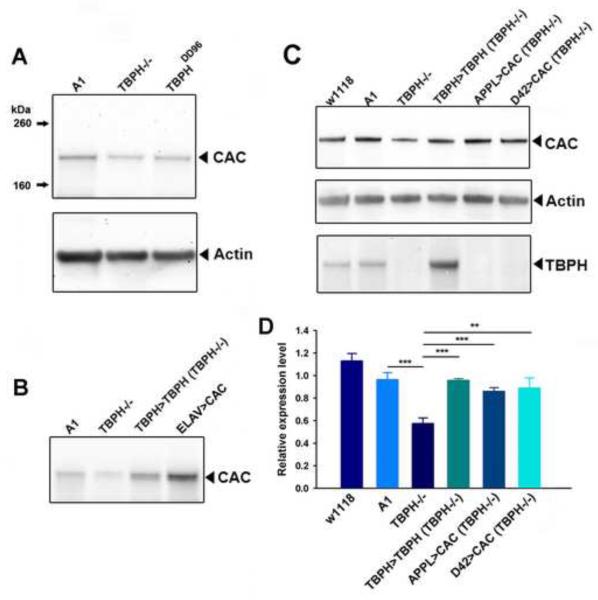
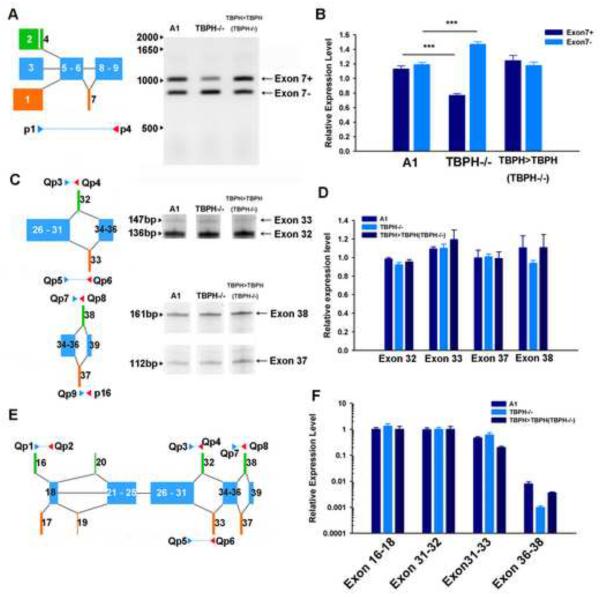
To determine whether the altered splicing of cacophony led to changes in the levels of cacophony protein, we generated an antibody that would recognize all predicted protein products and used immunoblots to measure the levels of cacophony in third instar larvae with a variety of different genotypes. We first compared the levels of cacophony in our TBPH mutant (Hazelett et al., 2012) and the control line (A1) generated in parallel and also to a separately isolated TBPH null (TBPHDD96, Diaper et al, 2013). Both of these null mutants showed reduced levels of cacophony protein (Figure 2A), which were restored when TBPH was expressed under the control of the TBPH-GAL4 promoter (Hazelett et al, 2012) (Figure 2B). Figures 2C & D show that the reduced levels of cacophony were also restored when the expression of a cacophony cDNA transgene (Kawasaki et al, 2002) was driven using either a pan-neuronal or motor neuron-specific GAL4 driver.
Figure 2.
The levels of cacophony are reduced in TBPH null mutants. A. Representative immunoblot from 3rd instar larvae showing the protein levels of cacophony (CAC) and actin as a loading control. The levels of cacophony were reduced in TBPH null mutants (TBPH −/−) compared to the A1 control line (line generated by precise excision of transposon – see Hazelett et al, 2012). Cacophony is also reduced in another independently generated TBPH mutant line, TBPHDD96 (Diaper et al, 2013). B. Cacophony levels were restored in TBPH rescue (TBPH>TBPH) lines and enhanced when cacophony was expressed under the control of a pan-neuronal driver (ELAV>CAC). C. Cacophony levels were also restored when cacophony was expressed either in all neurons (APPL>CAC) or only in motor neurons (D42>CAC). This blot was also re-probed for TBPH. D. Quantification of immunoblots. The intensity of the cacophony immunoreactive band was quantified, normalized to the intensity of the actin band and compared to the ratio obtained for the A1 control. ANOVA analysis shows that the relative expression of cacophony in TBPH mutants is significantly reduced (** p<0.01, *** p<0.001, n=6) compared to all other genotypes.
2.2. Restoring the levels of cacophony protein rescues the locomotion defects in TBPH mutant larvae
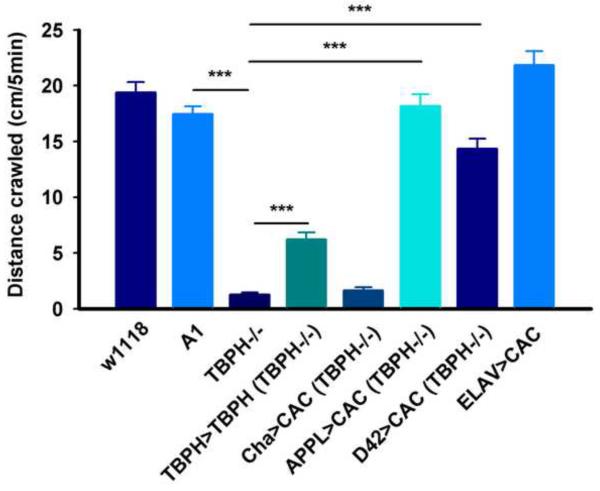
Loss of TBPH leads to a dramatic reduction in the crawling behavior of third instar larvae (Feguin et al, 2009; Hazelett et al, 2012; Diaper et al. 2013). As cacophony is required for neurotransmitter release (Kawasaki et al, 2002) we reasoned that at least part of these defects could be due to the loss of cacophony expression. To test this hypothesis we used larvae of the genotypes shown in Figure 2 and measured the distance they crawled in 5 minutes (Figure 3). These results showed that, as previously reported (Hazelett et al, 2012), loss of TBPH resulted in greatly reduced distance crawled compared to w1118 and A1 larvae. Also as previously reported (Hazelett et al, 2012), rescue of TBPH expression using the TBPH-GAL4 driver only partially rescued the crawling defects, probably due to the deleterious effects of over expression of TBPH (Hazelett et al, 2012). By contrast, expressing cacophony cDNA using either the pan-neuronal or motor neuron-specific GAL4 driver almost completely rescued the larval crawling deficits. Interestingly, rescue of cacophony levels with either of these drivers failed to rescue the pupal lethality also associated with loss of TBPH (data not shown). The rescue of the larval crawling defects appeared to be motor neuron specific as expression of cacophony using a cholinergic (Cha) GAL4 driver did not show increased crawling compared to the TBPH null mutants (Figure 3).
Figure 3.
Restoration of cacophony levels rescues locomotion defects in TBPH null larvae. Third instar larvae were placed on agar plates, allowed to crawl for 5 minutes and the total distance crawled quantified. As described previously, TBPH null mutants (TBPH−/−) crawled dramatically less distance than both control larvae (w1118 and A1) and this was partially restored in TBPH rescue (TBPH>TBPH) larvae (Hazelett et al, 2012). By contrast the locomotion behavior was significantly restored by expressing cacophony either in all neurons (APPL>CAC) or only in motor neurons (D42>CAC) but no change in crawling distance was observed when cacophony was expressed in cholinergic neurons (Cha>CAC). Over-expression of cacophony in all neurons (ELAV>CAC) in a wild type background had no effect on crawling distance. ** p<0.01, *** p<0.001, ANOVA, n=12–25 larvae.
2.3. Loss of TBPH leads to reduced levels of cacophony at the larval neuromuscular junction
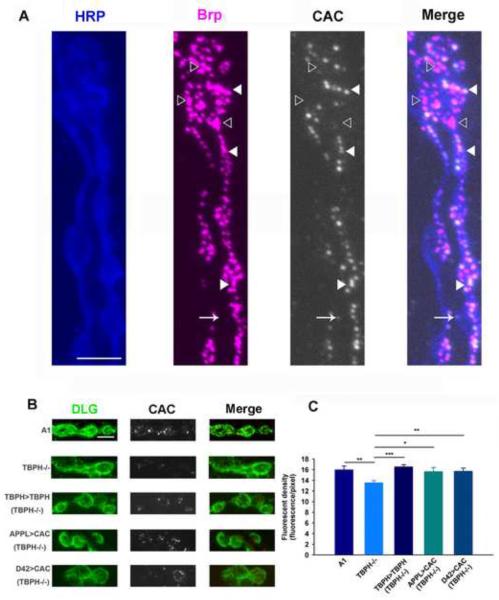
As the rescue of the larval crawling defects appeared to require expression of cacophony in motor neurons, we examined the distribution of cacophony in the motor neuron terminals at the larval neuromuscular junction (NMJ). Previous studies using exogenously expressed cacophony show that it co-localizes to active zone markers such as bruchpilot at the larval NMJ (Kittel et al, 2006). To confirm that endogenous cacophony is also co-localized with bruchpilot, we double labeled the larval NMJ preparation with our cacophony antibody and an antibody to bruchpilot. As expected cacophony immunofluorescence frequently co-localized with bruchpilot (Figure 4A), although there were also examples where we detected bruchpilot staining in the absence of cacophony. Occasionally we also detected cacophony outside of boutons, where it did not co-localize with bruchpilot. We next examined the levels of expression of cacophony at the NMJ of TBPH mutant larvae, using the post-synaptic marker, discs-large (Dlg) to define the synaptic terminals. This revealed fewer immunofluorescent puncta within the boutons compared to wild-type animals (Figure 4B). We then quantified this fluorescence by measuring the number of pixels representing cacophony immunofluorescence that were located within boutons defined by Dlg staining. This analysis showed that there was a significant reduction in the cacophony immunofluorescence in TBPH null animals compared to wild-type controls (Figure 4C). This reduction was restored in the TBPH rescue animals and when cacophony was expressed in all neurons or when selectively expressed in motor neurons (Figure 4B&C). There are two anatomically distinct types of terminals at the larval NMJ that can be distinguished based on their size and the intensity of Dlg staining, named 1B and 1S (Kurdyak et al, 1994). The data shown in Figure 4C is the pooled data from both types of terminals as there appeared to be a reduction in both 1s and 1b terminals, although this reduction was smaller in the 1s terminals.
Figure 4.
Localization and quantification of cacophony at the NMJ. A. Cacophony co-localizes with a pre-synaptic active site marker. Triple labeled image of the NMJ of muscle 6 showing motor neuron axons and terminals labeled with HRP (blue), the pre-synaptic active site marker bruchpilot (Brp, magenta) and cacophony (CAC, white). The fourth panel (Merge) is a merged image showing that cacophony is frequently co-localized with bruchpilot (closed arrow head) although occasionally we observed cacophony in the absence of bruchpilot (arrow) and bruchpilot in the absence of cacophony (open arrow head). B & C. Quantification of cacophony within the pre-synaptic terminals of motor neurons. B. Representative images of 1B terminals from larvae of the same genotypes shown in figures 2 and 3. The terminals were defined by staining with discs large (DLG, green). C. The cacophony fluorescence intensity within terminals was measured and showed a significant reduction in TBPH null mutants compared to the other genotypes (ANOVA, * p<0.05, ** p<0.01, *** p<0.001, n=3 animals each with at least 6 terminals). Scale bar is 5 μm in A and B.
There have been several previous studies that have described the anatomy of the larval NMJ in response to changes in TBPH levels. Initially, Feguin et al. (2009) showed that loss of TBPH led to reduced bouton number and terminal length. A more recent report, however, showed that there were no significant differences in the anatomy of the NMJ in third instar larvae comparing TBPH null mutants and control animals (Diaper et al, 2013). Although our quantification of cacophony immunofluorescence at the NMJ measured the level of immunoreactivity within boutons and thus shouldn't be affected by differences in the number of boutons or terminal branch length, it is possible that anatomical differences could affect cacophony distribution. We measured the number and area of boutons, motor neuron terminal branch length and number of branch points for both motor neurons innervating two different muscles in both A1 and TBPH mutant larvae and saw no significant differences in any of these parameters between these two genotypes (data not shown), confirming the findings of Diaper et al. (2013) that loss of TBPH has no effect on the anatomy of the larval NMJ, at least at the resolution of light microscopy.
2.4. TBPH associates with cacophony transcripts
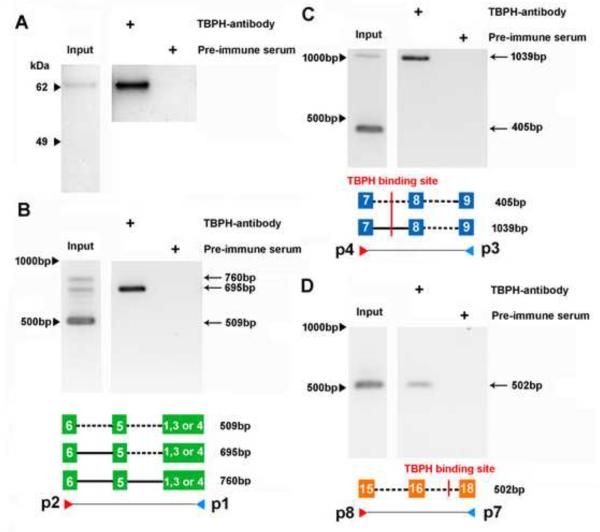
Our previous studies had identified genes with predicted TBPH binding sites based on the mammalian TDP-43 binding site sequences (Hazelett et al, 2012). One of these genes is cacophony, which has 10 predicted binding sites located in introns (Figure 1A). To determine if these predicted binding sites were authentic, we used TBPH immunoprecipitation followed by PCR to detect cacophony transcripts. These results clearly show that cacophony transcript can be detected in immunoprecipitates from adult flies using a TBPH antibody compared to using the pre-immune control serum (Figure 5). Because the predicted TBPH binding sites are in introns, we expected that the PCR fragments that we isolated would represent pre-spliced RNA rather than the mature transcript. This was true for some of the primer pairs that we tested, but not all. Primers p1–p2 amplified a product between a conserved region of alternative exons 1–3 and exon 6 and covers 2 introns. The mature RNA product is predicted to be 509bp and was seen as the major band from the input sample, but was not detected in the immunoprecipitated sample, where we detected a band of 695bp, which is the size predicted if one of the introns is retained in the transcript (Figure 5B). Sequencing of this product confirmed the inclusion of the intron between exons 5 & 6. Similarly, primers p3–p4 amplified across exons 7 and 9 and covered two introns, one of which contained a predicted TBPH binding site (Figure 1 and 5C). The mature RNA product is predicted to be 405bp and the pre-spliced form that includes the intron between exons 7 and 8, which contains the predicted TBPH binding site is 1039bp (Figure 5C). Both products were detected in the input sample, whereas only the pre-spliced product was detected in the immunoprecipitated sample. Again, sequencing confirmed the introns and exons included in this PCR product. By contrast, primers p7–p8 amplified across exons 16–18 and included up to 3 introns. In this case only the product of 502bp from mature RNA was detected in both input and immunoprecipitated samples, whereas the product from pre-spliced RNA (4342bp) was not detected in either sample. It should be noted however, that for these primers the lack of a product for the pre-spliced RNA could just reflect inefficient amplification of this larger product rather than its absence in the complex.
Figure 5.
TBPH is part of a complex that contains the cacophony transcript. Wild type adult flies were homogenized and any complexes present covalently cross linked with UV illumination. TBPH was immunoprecipitated and the pellet assayed for the presence of cacophony transcripts using RT-PCR. A. Immunoblot confirming that immunoprecipitation with the TBPH antibody compared to pre-immune serum specifically pelleted TBPH. B,C&D. RT-PCR showing the presence of cacophony immunoprecipitated with TBPH. The positions of the primers used in each case are shown in the accompanying schematic diagram and also in Figure 6. These diagrams also show the presence (solid lines) or absence (dashed lines) of introns in the different RT-PCR products and the location of predicted TBPH binding sites. In each case the amplified product obtained from the immunoprecipitated pellet was confirmed by sequencing. Primers p1–p2 (B) and p3–p4 (C) amplify both mature (509bp and 405bp) and pre-spliced RNA (760bp, 695bp and 1039bp) from the input sample and only pre-spliced RNA from the immunoprecipitated pellet, whereas primers p7–p8 only amplify mature RNA (502bp) in both samples. The gels shown are representative examples of experiments carried out in triplicate with similar results.
2.5. Loss of TBPH alters cacophony splicing
Our previously reported RNAseq data (Hazelett et al, 2012) indicated that alternative splicing of cacophony was altered in TBPH mutants compared to control animals. To analyze this in more detail, we used RT-PCR to scan cacophony transcripts with primer pairs that tiled groups of adjacent exons. The positions of the primer pairs and the results of the RT-PCR are shown in Figure 6. These results reveal no apparent differences in the sizes of the different PCR products between TBPH mutant and control samples, suggesting that TBPH does not regulate inclusion or exclusion of specific exons. The RNA-seq data suggested that the expression levels of specific exons and exon/exon boundaries were changed in TBPH mutant animals (Hazelett et al, 2012). To more accurately quantify these changes we used a combination of semi-quantitative and real time RT-PCR to measure the expression levels of a variety of the alternatively spliced exons that our RNAseq data suggested were changed in the TBPH mutant larvae. These data are shown in Figure 7 and reveal only subtle changes in the expression of specific exons. Exon 7 is either included or excluded from different transcripts and the RT-PCR data shows that transcripts that include this exon are significantly reduced in TBPH mutants (Figure 7A & B). In parallel with this change, the PCR product that excludes exon 7 is increased in TBPH mutants and both of these changes are reversed in the TBPH rescue animals (Figure 7A & B). Cloning and sequencing of these two bands confirmed the identity of the predicted products. By contrast a variety of additional primers, which were designed to amplify products containing the mutually exclusive alternatively spliced exons 16, 17, 32, 33, 37 and 38, revealed no changes in the expression levels for these products in TBPH mutants (Figure 7C–F).
Figure 6.
RT-PCR using pairs of primers that cover all of cacophony reveal no broad change in splicing patterns in TBPH mutants. The positions of the primer pairs used are shown in the upper portion of the panel and although some primer pairs show more than one alternatively spliced product (p1–p2 and p5–p6) there are no observed differences between TBPH mutants and controls.
Figure 7.
Semi-quantitative and real-time RT-PCR reveal subtle changes in cacophony splicing in TBPH null mutant larvae. A & B. Semi-quantitative RT-PCR shows that loss of TBPH leads to a reduction in inclusion of exon 7. B. Quantification of the intensities of the PCR products shows a significant decrease in the amount of product containing exon 7 (exon7+) and a concomitant increase in the product lacking exon 7 (exon7−) in TBPH mutants, which is reversed in the TBPH rescue animals. The intensity of each band was measured and normalized to the intensity of the band amplified in w1118 control animals. Mean and standard error from four independent samples are shown, *** p<0.001. C & D. Semi-quantitative RT-PCR showed no change in the relative abundance of mutually exclusive alternate exons 32 and 33 and exons 37 and 38. D. Quantification of the relative abundance of these products performed as in B, E & F. Real-time RT-PCR from the CNS of third instar larvae of products covering alternate exons 16, 17, 32, 33 & 38 confirms no effect of TBPH loss on the splicing of these exons. E. Schematic diagram showing positions of primers. F. Relative expression levels of products was quantified using real time RT-PCR and normalized to actin expression levels. Mean and standard error from five independent samples are shown.
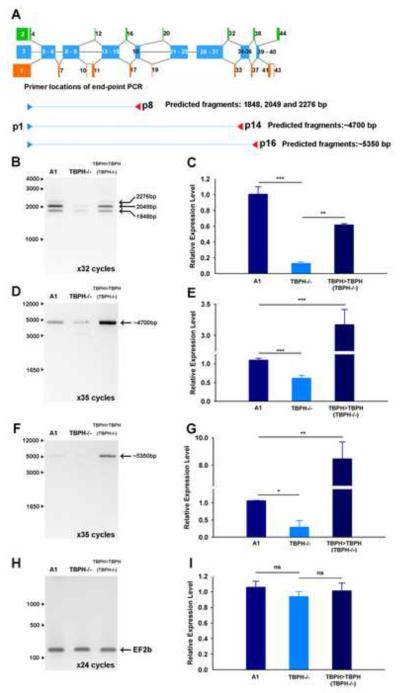
For the data shown in Figure 7, we designed primers that covered relatively short regions (up to about 1kb) of the cacophony transcript. In addition to detecting only subtle differences in the splicing patterns of alternative transcripts, these data showed no significant differences in the overall levels of cacophony RNA. In an attempt to gain a better understanding of the regulation of full length cacophony transcripts we used PCR conditions that favored amplification of longer products and measured the levels of these products in mutant and control animals. These results are shown in Figure 8 and reveal that when longer products are amplified we detected a significant reduction in the levels of cacophony transcript in TBPH mutant animals and importantly, these levels were restored in the TBPH rescue lines.
Figure 8.
Long-range RT-PCR reveals a reduction in the levels of cacophony transcripts in TBPH nulls. A. Schematic diagram showing the positions of the primers used. B–G. Semi-quantitative RT-PCR showed reduced levels of products in TBPH mutants compared to controls. The intensity of each band was measured and normalized to the intensity of the band amplified in w1118 control animals. H & I. Amplification of a control gene, EF2b, showed no change in expression between these genotypes. Mean and standard error from three independent samples are shown, ** p<0.01, *** p<0.001.
3. Discussion
ALS is a devastating neurodegenerative disease with no known cure and with only palliative care currently available to sufferers. Identification of molecular targets for pharmacological intervention is critically needed to provide alternative methods of treating the symptoms of this disease (Turner et al, 2013). An important breakthrough was achieved in 2006 when the RNA-binding protein TDP-43 was identified as a major constituent of the protein aggregates found in the cytoplasm of motor neurons from ALS patients (Arai et al., 2006; Neumann et al, 2006). TDP-43 is normally located predominantly in the nucleus and in these patient samples it was notable that, in parallel to the accumulation of cytoplasmic TDP-43 containing aggregates, there was a loss of nuclear TDP-43 (Neumann et al, 2006). It has recently become apparent that loss of TDP-43 may play a significant role in the development of the symptoms associated with ALS (Lee et al, 2012). TDP-43 has been implicated in a wide variety of RNA metabolic processes (Buratti and Baralle, 2010) and a variety of studies have identified hundreds of potential targets for TDP-43 (e.g. Sephton et al, 2011; Polymenidou et al, 2011). Using Drosophila as a model for TDP-43 loss of function, we have also shown that many different genes are likely to be direct targets for TBPH, the Drosophila orthologue of TDP-43 (Hazelett et al, 2012). It is now important to identify which of these targets are responsible for the phenotypes phenotypes associated with the loss of TDP-43 or TBPH.
In the present study, we identify the voltage-gated calcium channel, cacophony, as a likely direct target for TBPH and also a critical mediator of the effects of TBPH loss. TBPH null mutants are adult lethal with severe larval locomotion deficits (Feguin et al, 2009; Hazelett et al, 2012; Diaper et al, 2013). Loss of TBPH results in an almost 50% reduction of the levels of cacophony protein in larval Drosophila, which can be restored by expressing TBPH under the control of the TBPH promoter. Importantly, when we reversed the reduced levels of cacophony by expressing a cDNA for the gene in all neurons, not only was the protein level of cacophony restored, but larval locomotion was also restored to normal levels. We were also able to fully restore locomotion by selectively expressing cacophony in motor neurons, suggesting that these neurons were particularly susceptible to loss of cacophony. Although cacophony is known to also be expressed in cholinergic neurons (Gu et al, 2009) there was no effect on larval locomotion when cacophony was expressed selectively in cholinergic neurons. Interestingly, only larval locomotion was restored, expression of cacophony had no effect on adult lethality associated with TBPH loss.
We also examined the distribution of cacophony at the larval NMJ, where it is known to be critical for transmitter release (Kawasaki et al, 2002). We showed that endogenous cacophony was localized to pre-synaptic active zones, co-localizing with bruchpilot, as has been previously described for exogenously expressed cacophony (Kittel et al, 2006). When we quantified the expression at the NMJ, we again found the levels of cacophony reduced in TBPH mutants, but interestingly this effect was primarily seen in the 1B or tonic motor terminals (Kurdyak et al., 1994).
Our previous studies identified predicted TBPH binding sites based on the sequences of mammalian TDP-43 binding sites and cacophony was identified as a gene with multiple predicted sites (Hazelett et al, 2012). Using TBPH immunoprecipitation followed by RT-PCR, we showed that cacophony transcripts and TBPH associate together suggesting that cacophony is a direct target of TBPH. Nevertheless, the precise molecular mechanisms that mediate the TBPH-regulated levels of cacophony are unclear. Our previous RNA-seq data (Hazelett et al, 2012) and the real-time RT-PCR data from the present study suggested that the overall levels of cacophony transcript are unaffected in TBPH null larvae. These methods both detect relatively short lengths of RNA rather than full-length transcripts. When we measured the levels of longer PCR products we observed a significant reduction in the levels of cacophony transcript, which would be sufficient to account for the reduction in protein levels. This dependence on the length of the PCR product is intriguing and suggests that TBPH is required for the stability of the full length transcript rather than directly regulating the rate of transcription. Interestingly, studies in mammals have shown that long pre-RNA species are particularly susceptible to loss of TDP-43 (Polymenidou et al, 2011).
One of the initial functional roles for TDP-43 was alternative splicing regulation (Buratti and Baralle, 2010) and our initial RNA-seq data showed that alternative splicing of cacophony was most dramatically affected by loss of TBPH (Hazelett et al, 2012). It was therefore surprising that a more detailed analysis of the splicing patterns of cacophony revealed only subtle changes in splicing as a result of TBPH loss. Only the inclusion and exclusion of exon 7 was significantly affected, with TBPH mutants having a higher level of transcripts lacking exon 7 compared to controls. Exon 7 codes for a region of the cytoplasmic C-terminal tail of cacophony. Although the specific function for the region of protein coded for by exon 7 is unknown, the C-terminal cytoplasmic tail is known to affect inactivation and calcium regulation of cacophony and interactions with other proteins such as Ca2+/calmodulin-dependent serine kinase and munc-18 interacting protein (Macleod et al, 2006). It is also interesting to note that of the 15 predicted cacophony transcripts on FlyBase only one, cac-RM, lacks exon 7 suggesting that it plays a significant physiological role.
Alternative splicing is ubiquitous in mammalian neuronal circuits, controlling neuron development, axon targeting and the synthesis of neuron-specific components (Lipscombe et al, 2013). As a major factor of conveying signals from cell body to the terminus in myriad forms of neurons, cell- or district-specific voltage gated calcium channels are normally derived from the alternative splicing of conserved exons of pre-mature transcripts (Lipscombe et al, 2013). Recently, the connection between alternative exons and behavior has been reported on the CaV2.2 gene cacna1b. In contrast to pan-neuronal expression of exon 37b containing transcripts, exon 37a possessing CaV2.2 channels are enriched in nociceptors of dorsal root ganglia (Andrade et al, 2010). Furthermore, exon 37a was demonstrated to contribute to μ-opioid receptor-mediated inhibition of native N-type calcium channels in nociceptors and the behavioral sensitivity to spinal morphine-induced analgesia (Andrade et al, 2010). Similarly, mutations in exon 8a or exon8 of cacna1c determine the symptoms in Type I and Type II Timothy Syndrome (Splawski et al, 2004), as well as the association between Cacna1a (CaV2.1) and familial hemiplegic migraine on the short (D47) and long (+47) splice isoforms of CaV2.1 (Adams et al, 2009), are additional examples of alternative splicing of voltage-gated calcium in neural disease.
In summary, results presented here show that loss of TBPH results in reduced levels of cacophony in larval Drosophila and restoration of the levels of cacophony, selectively in motor neurons rescues the locomotion defects caused by loss of TBPH. TBPH appears to regulate the levels of cacophony by directly binding to the transcript and reducing its susceptibility to degradation. As cacophony mediates some of the physiological effects of TBPH loss, it is a good candidate to explore as a pharmacological target for ALS therapies. Supporting this idea is the recent finding that motor defects in a zebra fish TDP-43 over-expression model for ALS are rescued by calcium channel agonists (Armstrong and Drapeau, 2013).
4. Experimental Procedures
4.1. Fly Stocks
All Drosophila stocks were reared at 25°C using standard procedures (Greenspan, 2004). The following fly strains were obtained from the Bloomington stock center (http://flystocks.bio.indiana.edu/): APPL-GAL4 driver (P{Appl-GAL4.G1a}1, y1 w*), D42-GAL4 motor neuron driver (w*; P{GawB}D42), ELAV-GAL4 (P{GawB}elavC155), and UAS-CACOPHONY (w*; P{UAS-cac1-EGFP}422A). The cac1 isoform of cacophony (Kawasaki et al., 2002) codes for a protein that is equivalent to the cac-PE and cac-PO splice forms listed in FlyBase. The loss of function, UAS-TBPH and TBPH-GAL4 driver lines were described previously (Hazelett et al, 2012). The additional TBPH null mutant used, TBPHDD96 was generously provided by Drs. Diaper and Hirth, Kings College, London, UK (Diaper et al, 2013). In most experiments, two control fly lines were used, w1118 and A1. The A1 line was generated at the same time as the TBPH null mutant line by precise excision of a transposon located just upstream of the TBPH gene (Hazelett et al, 2012).
4.2. Immunoblotting
Anti-peptide antisera specific for the voltage-gated calcium channel cacophony and TBPH were produced in rabbits (Yao-Hong Biotechnology Inc., New Taipei City, Taiwan). The antigens were synthesized by coupling with keyhole limpet hemocyanin to a peptide corresponding to the 18 C-terminal amino acids of cacophony (SSRERERDRERLRDRDRD) or the 20 N-terminal amino acids of TBPH (VSEEEGDEPIELPAEEDGT). The resulting antisera were then affinity purified using affinity chromatography on peptide-conjugated columns.
The third instar larvae were collected, washed with PBS followed by 70% ethanol and lysed in lysis buffer (50 mM Tris-HCl to pH 7.5, 150 mM NaCl, 1% TritonX100, 1mM EDTA, 5% glycerol, 2 mM AEBSF, 0.3 μM aprotinin, 130 μM bestatin, 14 μM E-64, and 1 μM leupeptin) and centrifuged at 10,000 × g for 15 min at 4°C. The supernatants were denatured in 1X Laemmli sample buffer at 85°C and proteins fractionated by SDS-PAGE. Following transfer to polyvinylidine difluoride membranes, the membranes were blocked with 0.1% gelatin in PBST (PBS containing 0.05% tween-20) and incubated with either 1:10,000 anti-cacophony combining 1:200 anti-actin antisera (Developmental Studies Hybridoma Bank, Iowa City, Iowa, USA) followed by 1:10,000 HRP-conjugated goat-anti rabbit antisera (Jackson Immunoresearch, West Grove, PA, USA) and 1:10,000 HRP-conjugated goat-anti mouse antisera (Jackson Immunoresearch). The proteins were then detected with enhanced chemiluminescence (Invitrogen, Carlsbad, CA, USA). The proteins were then detected with enhanced chemiluminescence (Invitrogen). The same blot were stripped by use of stripping buffer (Thermo, Rockford, USA) at room temperature for 5 mins, followed by blocking, probed with 1:5,000 anti-TBPH antisera, incubation of 1:10,000 HRP-conjugated goat-anti rabbit antisera (Jackson Immunoresearch) and the detection with enhanced chemiluminescence (Invitrogen). The relative cacophony levels were normalized to actin levels by use of Gel plugin of ImageJ (http://imagej.nih.gov/ij/).
4.3. Locomotion assays
Third instar larvae were rinsed with PBS and placed on the heat-sterilized 2% agarose plate. The crawled paths of the larvae were then traced for 5 min. the plates photographed and the distances traveled determined using ImageJ software (http://imagej.nih.gov/ij/).
4.4. Immunofluorescence and quantification of the larval neuromuscular junction
For localization of cacophony at the NMJ, third instar larvae were dissected in ice-cold PBS and fixed in Bouin's fixative (3ml saturated picric acid, 1ml 36% formaldehyde, and 200 μl acetic acid) for 20 min at room temp. The dissected tissues were blocked with 5% normal goat serum (NGS) in PBS+0.2% Triton X100 (PBSTX) overnight, followed by the incubation in anti-cacophony antibody (1:2000) overnight at 4°C PBSTX, 5X washes with PBSTX, incubation in 1:500 biotinylated goat anti rabbit IgG (Jackson Immunoresearch), 5X washes with PBSTX and then incubation in 1:500 AlexaFluor 647 streptavidin (Life Technologies, Grand Island, NY). Tissues were then incubated with 1:200 discs-large (Dlg) antibody (MAb 4f3; Developmental Studies Hybridoma Bank) for 24 h at 4°C, 5X washes with PBSTX followed by 1:500 AlexaFlour 488 goat anti-mouse secondary antibodies (Life Technologies) for 4 h at room temperature. Tissues were rinsed in a series of concentrated glycerol and mounted in 50%Vectashield (Vector Laboratories, INC., Burlingame, CA). Images were acquired with an Olympus Fluoview FV1000 confocal microscope using the same setting of Olympus Fluoviewer3.1 for experimental and control samples. For the punctate cacophony signal, the fluorescence of the pixels situated in boutons (delineated by Dlg staining) were quantified by use of histogram plugin of FIJI, following subtraction of the background signal, and then normalized to the total pixels within the boutons.
To examine anatomical differences at the NMJ, third instar larvae were cut along their dorsal midline, pinned open and immediately fixed with Histochoice (Amresco, Solon, OH) for 30 mins at RT. After blocking for 1 hr in 10% NGS in PBSTx (PBS plus 0.3% triton X-100) the tissue was incubated in 1:1,000 rabbit anti-synaptotagmin (dsyt2; kindly provided by Dr. Karla Kent) in PBSTx with 10% NGS overnight at 4°C. After 4 × 30 min washes in PBST samples were incubated with 1:100 Cy5-labeled goat anti-horseradish peroxidase (Jackson Immuno Research) and 1:100 anti-Dlg in PBSTx with 10% NGS overnight at 4°C. Following 4 × 30 min washes in PBST samples were incubated with 1:1,000 Alexa Fluor 488-conjugated goat anti-mouse and 1:1,000 Alexa Fluor 546-conjugated goat anti-rabbit (Life Technologies) in PBSTx with 10% NGS for 2 hrs at room temperature. After 4 × 15 min washes in PBSTx and 4 × 15 min washes in PBS, the samples were mounted in Histochoice mounting medium (Amresco) and imaged by confocal microscope using 488 nm, 543 nm and 637 nm excitation wavelengths and visualized using exclusion filters of 500–530 nm, 570–635 nm and 660 nm-long pass respectively. The images were quantified using ImageJ after microscope calibration and the 1B or 1S terminals identified by the more intense Dlg staining and more irregular shape of the 1B terminals.
4.5. RNA immunoprecipitation
RNA immunoprecipitation assays were modified from previously described procedures (Polymenidou et al, 2011). Drosophila heads were partially fragmented with sharp forceps, and then passed through a 100 μm cell strainer (Becton Dickinson Falcon, Franklin Lakes, NJ, USA) in iced-cold PBS to yield dissociated cells. The cells were exposed to 200 mJ of UV irradiation three times. UV-treated or control cells were spun down by 2,500 RPM for 3min at 4°C and the supernatants removed. The cell pellets were re-suspended with PBS containing RNaseOut (0.6 U/μl; Invitrogen) and DNAse I (0.1U/μl) for 25 min at room temperature to remove the genomic DNA. Subsequently, DNAse was removed by centrifugation and the pellets were re-suspended with Tris-lysis buffer (50 mM Tris-HCl to pH 7.5, 150 mM NaCl, 1% TritonX100, 1mM EDTA, 5% glycerol, 2 mM AEBSF, 0.3 μM aprotinin, 130 μM bestatin, 14 μM E-64, 1 μM leupeptin and 0.6 U/μl RNaseOut). The re-suspended lysates were incubated with 1:150 anti-TBPH antisera and Protein-A Sepharose (40 μg) for 2h at 4 °C, followed by lysis buffer washing, and used for reverse-transcription or immunoblot detection. Samples for reverse-transcription were treated with DNAse I (0.1U/μl) and protease K (100μg/ml; Invitrogen) for 30 min at 37°C and RNA isolated with Trizol-chloroform extraction. The purified RNAs were reverse-transcribed for PCR detection as described below. Additional immuno-precipitated pellets were visualized by immunoblot.
4.6. Reverse transcription (RT)-PCR
To systematically examine different possible splicing patterns of cacophony we designed primers that spanned groups of nearby exons and carried out RT-PCR on cDNA synthesized from total RNA extracted from third instar larvae. Total RNA was extracted using Trizol (Life Technologies), treated with DNAse I and cDNA synthesized using SuperScript III (Life Technologies). The sets of PCR primers are shown in table 1 and the reaction conditions were 94°C for 2 min, followed by 32 cycles of 30 sec at 94°C, 20 sec at 60°C, and 60 sec at 72°C and a final incubation at 68°C for 7 min. In some cases we amplified much larger regions of cacophony and to increase the yield of large DNA fragments, Platinum® Taq High Fidelity DNA Polymerase (Life Technologies) was used for PCR synthesis. These reactions were 94°C for 30 sec, followed by 35 cycles of 30 sec at 94°C, 20 sec at 60°C, and 6 min 30 sec at 68°C and a final incubation at 72°C for 7 min. To gain a more quantitative evaluation of the splicing regulation, we also performed real time PCR using Power SYBR® Green PCR Master Mix (Applied Biosystems, Warrington, UK) in a MX3000P thermocycler (Stratagene, La Jolla, CA, USA). Following cDNA synthesis the PCR reactions were 95°C for 5 min, followed by 40 cycles of 15 sec at 95°C, 60 sec at 60°C.
Table 1.
Primer pairs for RT-PCR reactions of cacophony. All primers are listed 5′–3′. Their locations are shown in Figures 5 – 8.
| Exons amplified | Size | Primer pairs | |||
|---|---|---|---|---|---|
| 1–6 | 509 | P1 | CCCAGATCTGATCCTTTCGAT | p2 | ACCGTCAACACTGCAGCTC |
| 7–9 | 405 | p3 | GTCTCGTTCACGCATCTCCT | p4 | GATGCTGGCCATTTACATCC |
| 9–14 | 661 | p5 | AGGATGTAAATGGCCAGCAT | p6 | GGGTTTTGGCAACAAGTGTC |
| 15–18 | 502 | P7 | GCACCCAATATGCTGGAGTC | p8 | ACTCAATCAACGTCGGCTTC |
| 18–23 | 518 | p9 | GCCGATAATGGCGTATATGAA | p10 | CACGACCTCTCGAACGCTAT |
| 24–26 | 523 | p11 | GTAACTCAGCTTCGCCTTGC | p12 | CTACGCCCATTGAAGACCAT |
| 30–34 | 361 | p13 | CCGCTTCCTCTTCCTCTGTA | p14 | AACGACGCATTAGGTTCAGC |
| 36–39 | 297 | p15 | CCGAATAAAACTCGAGTCCA | p16 | CCTTAGGGCTTGTTCTGCAT |
| 16–18 | 129 | Qp1 | TGGAATTTTCCACCGTACCT | Qp2 | TGGACATTTGTGCAATCCTT |
| 31–32 | 136 | Qp3 | GCCAGGATTACCTCCTCAGC | Qp4 | TGAATTCGCAAAAGAACGAG |
| 31–33 | 147 | Qp5 | CCTCCATTATGTGCATTTTCTC | Qp6 | AACGTCGCATGGAGTTTCA |
| 36–38 | 112 | Qp7 | ACACCAAGAGACCGATTTGC | Qp8 | TTTCGTGTTTTACGCCCACT |
| Actin | CAGAGCAAGCGTGGTATCCT | GGTGTGGTGCCAGATCTTCT | |||
| EF2b | GCGTTCACCCTCAAGCAGTTCT | AGCGTTTGTTGTCAGCCTCTTTCT | |||
Acknowledgments
We wish to thank Ms. Kayly Lembke, Drs. Scott Vanderwerf and Baotong Xie for helpful comments on this manuscript and during the course of this study. We also wish to thank Drs. Diaper and Hirth for providing us with their TBPH mutant alleles.This work was supported by grants 23VU14 from the ALS Association and NS071186 from NINDS to DBM. The imaging carried out during this study was performed at the OHSU Advanced Imaging Facility, supported by NINDS grant P30-NS061800 to Dr. Sue Aicher.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PJ, Garcia E, David LS, Mulatz KJ, Spacey SD, Snutch TP. Ca(V)2.1 P/Q-type calcium channel alternative splicing affects the functional impact of familial hemiplegic migraine mutations: implications for calcium channelopathies. Channels. 2009;3:110–121. doi: 10.4161/chan.3.2.7932. [DOI] [PubMed] [Google Scholar]
- Andrade A, Denome S, Jiang Y-Q, Marangoudakis S, Lipscombe D. Opioid inhibition of N-type Ca2+ channels and spinal analgesia couple to alternative splicing. Nat. Neurosci. 2010;13:1249–1256. doi: 10.1038/nn.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Comm. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- Armstrong GAB, Drapeau P. Calcium channel agonists protect against neuromuscular dysfunction in a genetic model of TDP-43 mutation in ALS. J Neurosci. 2013;33:1741–1752. doi: 10.1523/JNEUROSCI.4003-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala YM, De Conti L, Avendaño-Vázquez SE, Dhir A, Romano M, D'Ambrogio A, Tollervey J, Ule J, Baralle M, Buratti E, Baralle FE. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, novel splicing regulator of CFTR exon 9. J. Biol. Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. The multiple roles of TDP-43 in pre-mRNA processing and gene expression regulation. RNA Biology. 2010;7:420–429. doi: 10.4161/rna.7.4.12205. [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. TDP-43: gumming up neurons through protein-protein and protein-RNA interactions. Trends Biochem. Sci. 2012;37:237–247. doi: 10.1016/j.tibs.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Diaper DC, Adachi Y, Sutcliffe B, Humphrey BM, Elliott CJH, Stepto A, Ludlow ZN, Vanden Broeck L, Callaerts P, Dermaut B, Al-Chalabi A, Shaw CE, Robinson IM, Hirth F. Loss and gain of Drosophila TDP-43 impair synaptic efficacy and motor control leading to age-related neurodegeneration by loss-of-function phenotypes. Human Mol. Genet. 2013;22:1539–1557. doi: 10.1093/hmg/ddt005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguin F, Godena VK, Romano G, D'Ambrogio A, Klima R, Baralle FE. Depletion of TDP-43 affects Drosophila motor neurons terminals synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Fiesel FC, Kahle PJ. TDP-43 and FUS/TLS: cellular functions and implications for neurodegeneration. FEBS J. 2011;278:3550–3568. doi: 10.1111/j.1742-4658.2011.08258.x. [DOI] [PubMed] [Google Scholar]
- Fiesel FC, Weber SS, Supper J, Zell A, Kahle PJ. TDP-43 regulates global translational yield by splicing of exon junction complex component SKAR. Nucleic Acids Res. 2012;40:2668–2682. doi: 10.1093/nar/gkr1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geser F, Martinez-Lage M, Robinson J, Uryu K, Neumann M, Brandmeir NJ, Xie SX, Kwong LK, Elman L, McCluskey L, Clark CM, Malunda J, Miller BL, Zimmerman EA, Qian J, Van Deerlin V, Grossman M, Lee VM-Y, Trojanowski JQ. Clinical and pathological continuum of multisystem TDP-43 proteinopathies. Arch. Neurol. 2009;66:180–189. doi: 10.1001/archneurol.2008.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, Hatanpaa KJ, White CL, Bigio EH, Caselli R, Baker M, Al-Lozi MT, Morris JC, Pestronk A, Rademakers R, Goate AM, Cairns NJ. TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan RJ. Fly Pushing. Ed 2 Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2004. [Google Scholar]
- Gu H, Jiang SA, Campusano JM, Iniguez J, Su H, Hoang AA, Lavian M, Sun X, O'Dowd DK. Cav2-type calcium channels encoded by cac regulate AP-independent neurotransmitter release at cholinergic synapses in adult Drosophila brain. J Neurophysiol. 2009;101:42–53. doi: 10.1152/jn.91103.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelett DJ, Chang J-C, Lakeland DL, Morton DB. Comparison of parallel hi-throughput RNA-sequencing between knockout of TDP-43 and its overexpression reveals primarily non-reciprocal and non-overlapping gene expression changes in the central nervous system of Drosophila. G3:Genes, Genomes, Genetics. 2012;2:789–802. doi: 10.1534/g3.112.002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande Velde C, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nature Genetics. 2008;40(5):572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kawasaki F, Collins SC, Ordway RW. Synaptic calcium-channel function in Drosophila: analysis and transformation rescue of temperature-sensitive paralytic and lethal mutations of cacophony. J Neurosci. 2002;22:5856–5864. doi: 10.1523/JNEUROSCI.22-14-05856.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki F, Zou B, Xu X, Ordway RW. Active zone localization of presynaptic calcium channels encoded by the cacophony locus of Drosophila. J. Neurosci. 2004;24:282–285. doi: 10.1523/JNEUROSCI.3553-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, Schmid A, Wagh DA, Pawlu C, Kellner RR, Willig KI, Hell SW, Buchner E, Heckmann M, Sigrist SJ. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 2006;312:1051–1054. doi: 10.1126/science.1126308. [DOI] [PubMed] [Google Scholar]
- Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- Kurdyak P, Atwood HL, Stewart BA, Wu C-F. Differential physiology and morphology of motor axons to ventral longtitudinal muscles in larval Drosophila. J. Comp. Neurol. 1994;350:463–472. doi: 10.1002/cne.903500310. [DOI] [PubMed] [Google Scholar]
- Lee EB, Lee VM-Y, Trojanowski JQ. Gains and losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat. Rev. Neurosci. 2012;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D, Allen SE, Toro CP. Control of neuronal voltage-gated calcium ion channels from RNA to protein. Trends Neurosci. 2013 doi: 10.1016/j.tins.2013.06.008. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D, Andrade A, Allen SE. Alternative splicing: functional diversity among voltage-gated calcium channels and behavioral consequences. Biochim Biophys Acta. 2013;1828:1522–1529. doi: 10.1016/j.bbamem.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod GT, Chen L, Karunanithi S, Peloquin JB, Atwood HL, McRory JE, Zamponi GW, Charlton MP. The Drosophila cacts2 mutation reduces presynaptic Ca2+ entry and defines an important element in CaV2.1 channel inactivation. Eur. J. Neurosci. 2006;23:3230–3244. doi: 10.1111/j.1460-9568.2006.04873.x. [DOI] [PubMed] [Google Scholar]
- Miller TM, Pestronk A, David W, Rothstein J, Simpson E, Appel SH, Andres PL, Mahoney K, Allred P, Alexander K, Ostrow LW, Schoenfeld D, Macklin EA, Norris DA, Manousakis G, Crisp M, Smith R, Bennett CF, Bishop KM, Cudkowicz ME. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12:435–442. doi: 10.1016/S1474-4422(13)70061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Samputhu DM, Kwong LK, Truax AK, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM-Y. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng IF, Wu CF. Drosophila cacophony channels: a major mediator of neuronal Ca2+ currents and a trigger for K+ channel homeostatic regulation. J Neurosci. 2007;27:1072–1081. doi: 10.1523/JNEUROSCI.4746-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, Lagieer-Tourenne C, Hutt K, Huelga SC, Moran J, Liang TY, Ling SC, Sun E, Wancewicz E, Kordasiewicz H, Sedaghat Y, Donohue JP, Shiue L, Bennett CF, Yeo GW, Cleveland DW. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckhof GE, Yoshihara M, Guan Z, Littleton JT. Presynaptic N-type calcium channels regulate synaptic growth. J Biol. Chem. 2003;278:41099–41108. doi: 10.1074/jbc.M306417200. [DOI] [PubMed] [Google Scholar]
- Sephton CF, Cenik C, Kucukural A, Dammer EB, Cenik B, et al. Identification of Neuronal RNA Targets of TDP-43-containing Ribonucleoprotein Complexes. J Biol Chem. 2011;286:1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, Tager-Flusberg H, Priori SG, Sanguinetti MC, Keating MT. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MR, Hardiman O, Benatar M, Brooks BR, Chio, de Carvalho AM, Ince PG, Lin C, Miller RG, Mitsumoto H, Nicholson G, Ravits J, Shaw PJ, Swash M, Talbot K, Traynor BJ, Van den Berg LH, Veldink JH, Vucic S, Kiernan MC. Controversies and priorities in amyotrophic lateral sclerosis. Lancet Neurol. 2013;12:310–322. doi: 10.1016/S1474-4422(13)70036-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkening K, Leystra-Lantz C, Yang W, Jaffee H, Strong MJ. Tar DNA binding protein of 43 kDa (TDP-43), 14-3-3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS) Brain Res. 2009;1305:168–182. doi: 10.1016/j.brainres.2009.09.105. [DOI] [PubMed] [Google Scholar]
- Wang I-F, Wu L-S, Shen C-KJ. TDP-43: and emerging new player in neurodegenerative diseases. Trends Mol Med. 2008;14:479–485. doi: 10.1016/j.molmed.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Yokoseki A, Shiga A, Tan CF, Tagawa A, Kaneko H, Koyama A, Eguchi H, Tsujino A, Ikeuchi T, Kakita A, Okamoto K, Nishizawa M, Takahashi H, Onodera O. TDP-43 mutation in amyotrophic lateral sclerosis. Ann Neurol. 2008;63:538–542. doi: 10.1002/ana.21392. [DOI] [PubMed] [Google Scholar]