Abstract
Killer cell immunoglobulin-like receptors (KIR) interact with HLA class I ligands to regulate NK cell development and function. These interactions affect the outcome of unrelated donor (URD) hematopoietic cell transplantation (HCT). We have shown previously that donors with KIR B vs. KIR A haplotypes improve the clinical outcome for patients with acute myelogenous leukemia (AML) by reducing the incidence of leukemic relapse and improving leukemia free survival (LFS). Both centromeric and telomeric KIR B genes contribute to the effect, but the centromeric genes are dominant. They include the genes encoding inhibitory KIR that are specific for the C1 and C2 epitopes of HLA-C. We used an expanded cohort of 1532 T-cell replete transplants to examine the interaction between donor KIR B genes and recipient Class I HLA KIR ligands. The relapse protection associated with donor KIR B is enhanced in recipients who have one or two C1-bearing HLA-C allotypes, compared to C2 homozygous recipients, with no effect based on donor HLA. The protective interaction between donors with ≥2 vs. 0–1 KIR B-motifs and recipient C1 was specific to transplants with class I mismatch at HLA-C (RR of LFS 0.57 [0.40–0.79]; P=0.001) irrespective of the KIR ligand mismatch status of the transplant. The survival advantage and relapse protection in C1/x recipients compared to C2/C2 recipients was similar irrespective of the particular donor KIR B genes. Understanding the interactions between donor KIR and recipient HLA class I can be used to inform donor selection to improve outcome of URD HCT for AML.
Introduction
The interactions of variable killer-cell immunoglobulin-like receptors (KIR) with polymorphic HLA class I ligands form an extraordinary immunogenetic system that influences NK cell biology, human susceptibility to disease, and the success of hematopoietic cell transplantation (HCT) as therapy for acute myelogenous leukemia (AML). A key feature of this system is that the KIR and HLA genes are on different chromosomes and thus segregate independently in human populations. This serves to increase the functional diversity of the system and has important consequences for HCT. Unrelated donors (URD) and recipients who are HLA-identical almost never have identical KIR genes. In fact, even in families, only 25% of HLA-identical siblings are also KIR identical (1).
KIR recognize four polymorphic epitopes of HLA-A, B and C molecules. These epitopes, defined by amino-acid substitutions in residues 76–83 of the α1 helix of the HLA class I heavy chain, are also called KIR ligands. The C1 and C2 epitopes are carried by different subsets of HLA-C allotypes, the Bw4 epitope is carried by subsets of HLA-A and –B allotypes and the A3/11 epitope is carried by the HLA-A*03 and –A*11 allotypes. Each of the four epitopes is recognized by different inhibitory KIR which are encoded by polymorphic genes. The C2 epitope is also recognized by the activating receptor encoded by KIR2DS1. Additional members of the KIR gene family encode proteins whose functions are yet to be determined. In addition to the polymorphism of individual genes, the KIR locus exhibits haplotypic gene-content variation. The basis for this component of KIR variation is the presence of two groups of KIR haplotype: KIR-A and KIR-B haplotypes in all human populations. The KIR-A haplotypes have conserved gene content and encode mainly inhibitory receptors, whereas KIR-B haplotypes have varied gene content that includes a variety of activating receptors of unknown function. Further details of HLA and KIR immunogenetics are provided in the Materials and Methods section.
The potential value of NK cell responses in HCT was first demonstrated by Ruggeri et al(2). These investigators observed that certain HLA-B and -C incompatibilities reduce relapse and improve the survival of AML patients receiving a haploidentical, T-cell depleted transplant from a related family members(3, 4). For these transplants, in which donor and recipient share one HLA haplotype but are mismatched for the other haplotype, a beneficial alloreactive response occurs when the donor has a KIR ligand, Bw4, C1 or C2, not present in the recipient. In this situation, subsets of donor-derived NK cells can attack and kill recipient cells because they are missing self-HLA class I. Velardi and colleagues proposed that reduced relapse was due to NK-cell killing of residual leukemia cells that had survived the myeloablative conditioning regimen. They also proposed that the reduced graft-versus-host disease (GVHD) they observed was caused by NK-cell killing of recipient dendritic cells(5). These pioneering observations led to investigations of various other types of transplant examining the effects of alloreactive NK cells and the HLA-A, -B and –C epitopes recognized by KIR(6–10). A general observation emerging from these subsequent studies is that NK cell effects in HCT are principally seen in patients transplanted for AML. A second observation is that the nature of NK cell effects varies considerably and is influenced by factors that include the intensity of the preparative regimen, the extent of HLA match, donor type (sibling or URD) and the source, processing method and T-cell content of the stem cell graft (8, 9). Third, it has been reported that C2/C2 homozygous patients with AML have more relapse (11, 12).
Whereas other studies concentrated on the polymorphic HLA class I ligands that are recognized by KIR, we have studied variation of the KIR gene family and its effect on HCT. For AML patients transplanted with a T-cell replete transplant from an unrelated donor (URD), we found that clinical outcome was better when the donors have one or two KIR-B haplotypes (KIR-B/x donors) than for donors who have two KIR-A haplotypes (KIR-A/A donors). With a KIR-B/x donor, relapse was reduced and leukemia-free survival (LFS) was increased(13). In a subsequent study we sought to determine whether the protective effect of KIR-B could be mapped to either the centomeric or the telomeric region of the KIR locus. The centromeric region contains genes encoding the inhibitory receptors for the C1 and C2 epitopes of HLA-C, whereas the telomeric region contains genes encoding the inhibitory receptors for the Bw4 and A3/11 epitopes and the activating C2 receptor. We found that both the centromeric and telomeric regions of KIR-B correlated with protective effect, but the much stronger association was with the centromeric region(14). The beneficial effects associated with KIR B haplotype donors were consistent in both HLA-matched and HLA-mismatched transplants. Because the genes encoding the inhibitory C1 and C2 receptors are located in the centromeric regions, we have investigated the influence of the C1 and C2 epitopes on the protection provided by donor KIR-B haplotypes in HCT.
Materials and Methods
Patient cohort
We studied 1532 patients with AML, including 1086 previously analyzed(14), who received myeloblative preparation for an unrelated donor (URD) HCT facilitated by the National Marrow Donor Program (NMDP) between 1988 and 2009. DNA samples were obtained from the NMDP Research Sample Repository. Outcome data were obtained from the Center for International Blood and Marrow Transplant Research (CIBMTR). Complete high-resolution HLA-A, HLA-B, HLA-C, HLA-DRB1 and HLA-DQB1 typing data were obtained from the NMDP retrospective typing project. The demographics, KIR genotypes, and multivariate statistical analysis of the clinical data have been described(13, 14). KIR genotyping using MALDI-TOF mass spectroscopy was performed as previously described(15, 16). DNA samples and clinical data were obtained with informed consent and approval from the NMDP and University of Minnesota Institutional Review Boards.
HLA and KIR immunogenetics
Complete high-resolution, allele-level HLA-A, HLA-B, HLA-C, HLA-DRB1 and HLA-DQB1 typing data were obtained from the NMDP retrospective typing project. KIR typing at the level of KIR gene content was performed using MALDI-TOF mass spectroscopy as described previously(15, 16). Four epitopes of HLA-A, -B and –C molecules are recognized by KIR. The epitopes, also called KIR ligands, are situated on the upward face of the HLA class I molecule and involve the amino-terminal part of the α1 helix and the carboxy-terminal parts of the bound peptide and the α2-helix(17). The epitopes are mutually exclusive, such that each HLA-A, -B or –C molecule either carries one of the four epitopes or no epitope at all. Every HLA-C allotype is a KIR ligand whereas only 43% of HLA-A and 35% of HLA-B allotypes are KIR ligands. The KIRs are named according to the number of extracellular Ig-like domains, either 2 or 3, and the length of the cytoplamic tail, either long (L) or short (S), correlating respectively with inhibitory and activating signaling function(18). The C1 and C2 epitopes carried by HLA-C are distinguished by lysine and methionine residues at position 80, respectively(17). The C1 epitope is recognized by the inhibitory KIR2DL2/3 receptor, whereas the C2 epitope is recognized by inhibitory KIR2DL1 and activating KIR2DS1 receptors. The Bw4 epitope, carried by 27% of HLA-A and 35% of HLA-B allotypes, is recognized by inhibitory KIR3DL1(19, 20). The A3/11 epitope, carried by 16% of HLA-A allotypes is recognized by inhibitory KIR3DL2. The C1, C2 and Bw4 epitopes play major roles in NK-cell regulation. Such a role has not been demonstrated for the A3/11 epitope(21) which is unusually dependent upon the sequence of the peptide bound to HLA-A*03 or HLA-A*11(22). For this reason the A3/11 epitope was not included in the analyses of the transplant donors and recipients studied here. In the studies described in this paper, the impact of recipient C1 on transplant outcome dominated C2. In some analyses, recipients with C2/C2 genotype were compared to recipients with either C1/C1 or C1/C2 genotypes. The combined group of C1/C1 or C1/C2 recipients was designated C1/x.
The KIR locus is part of the leukocyte receptor complex on human chromosome 19 and segregates independently of the HLA class I genes in the MHC on chromosome 6. A KIR haplotype is the set of KIR genes that are linked together on the same chromosome. Haplotypes contain 7–15 KIR genes and are 129–215kb in length(23). Every individual has a maternally inherited and a paternally inherited KIR haplotype that together form his or her KIR genotype. Conserved genes in the center of the locus and at the two ends divide the locus into centromeric (Cen) and telomeric (Tel) regions, each of which exhibits variable content of KIR genes. In both regions there are two distinctive types of variable gene-content motif. These are designated Cen-A/Cen–B, and Tel-A/Tel-B. Further variations within these four motifs are differentiated by numbers; e.g. Cen-B1 and Cen-B2. The A motifs are shorter, more conserved and consist mainly of genes for inhibitory KIR that recognize the C1, C2 and Bw4 epitopes. The B motifs are longer, more variable and contain one or more of seven KIR B-specific genes(23). These comprise KIR2DS2 and KIR2DL2 in Cen-B, KIR2DS1 and KIR3DS1 in Tel-B, and KIR2DS3/5 and KIR2DL5 in either Cen-B or Tel-B, or both. Of the KIR encoded by the B-specific genes, only KIR2DL2 (C1-specific) and KIR2DS1 (C2-specific) recognize HLA class I. Haplotypes that consist of a Cen-A motif and a Tel-A motif are called KIR A haplotypes and haplotypes consisting of a Cen-B and a Tel-B motif are called KIR B haplotypes. Recombinant haplotypes, which consist of either Cen-A and Tel-B or Cen-B and Tel-A, are also included in the KIR B haplotypes because of the dominant effect of the B motifs in disease, transplantation and other clinical associations.
In this study we characterized transplant donors according to their KIR B-motif content, a parameter that varies from 0–4 and is simply the sum of the number of Cen-B and Tel-B motifs in the donor’s KIR genotype. Based upon the results of our previous study (2), donors are classified as “Neutral” (0–1 KIR B-motifs),”Better” (≥2 B-motifs without Cen-B/B), or ”Best” (≥ 2 B-motifs with Cen-B/B)(14). In some analyses, the “Better” and “Best” groups were combined to form the KIR-Better/Best donor group (with ≥ 2 B-motifs) (Table I).
Table I.
KIR Haplotype Group Nomenclature
| KIR Genotype | KIR B-Motif Content | Centromeric Haplotypes | Telomeric Haplotypes | KIR Donor Group | |
|---|---|---|---|---|---|
| A/A | 0 | A/A | A/A | “Neutral” | KIR-Neutral (0–1 B-motif) |
| B/x | 1 | A/A | A/B | ||
| A/B | A/A | ||||
| 2 | A/A | B/B | “*Better” | KIR-Better/Best (≥2 B-motifs) | |
| A/B | A/B | ||||
| B/B | A/A | *“Best” | |||
| 3 | A/B | B/B | “Better” | ||
| B/B | A/B | “Best” | |||
| 4 | B/B | B/B | |||
”Better” KIR donors have ≥2 B-motifs without Cen-B/B, and ”Best” KIR donors have ≥2 B-motifs with Cen-B/B
Statistical Analysis
We considered five clinical outcomes of HCT: leukemia free survival (LFS), relapse, treatment-related mortality (TRM), grade II–IV or III–IV acute graft-versus-host disease (aGVHD) and chronic graft-versus-host disease (cGVHD). Kaplan-Meier curves were used to evaluate LFS, whereas cumulative incidence functions were used to evaluate the other outcomes. Unadjusted comparisons between KIR genotypes were made on either the hazard rates for LFS or the crude hazard rates for relapse, TRM, aGVHD and cGVHD. In the cohort we studied, the completeness of follow-up at three years after transplantation was over 98.8%. At this time, 90% of events had occurred. Cox proportional hazard models were used to adjust for important clinical factors. Proportional hazards were checked in a time-dependent covariate model. Factors that violated proportional assumptions were adjusted via stratification. Forward stepwise regression modeling was used to identify clinical and patient factors that needed adjustment using a 5% significance level. Adjusted factors include patient age, cytogenetic risk, sex match, HLA matching, graft source, CMV serostatus, race, Karnofsky score, GVHD prophylaxis, the use of total body irradiation (TBI), time from diagnosis to transplant, disease status and year of transplantation. Cases were excluded from some models if data for the outcome or significant covariates were missing. All analyses were performed using SAS version 9.3 (SAS Institute, Cary, NC).
Results
KIR-Better/Best donors improve recipient survival and reduce relapse in patients transplanted for AML
The cohort of myeloablative, T-cell replete unrelated donor (URD) transplants we studied included adult and pediatric patients with early, intermediate and advanced AML. Fifty-six percent (n=856) of the donor-recipient pairs were 10/10 HLA-allele matched for HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1; the rest (n=676) had varying degrees of HLA mismatch: 407 had one mismatch, 173 had two mismatches, and 85 had three or more mismatches. Whereas 357 (53%) of the HLA-mismatched transplants involved an HLA-C mismatch, only 110 of them were also KIR ligand mismatched in the graft-versus-host direction. Approximately 53% (n=810) of the recipients received bone marrow (BM) grafts, whereas the others received grafts of stem cells present in peripheral blood (PB) mobilized with granulocyte-colony stimulating factor (G-CSF). Additional information describing the transplant characteristics and HLA matching of the cohort is provided in Supplemental Tables I and II.
Transplant donors were typed for the presence or absence of each of the 15 KIR genes. From these KIR genotype data, donors were assigned as either A/A or B/x based upon the absence or presence of KIR B-specific genes(13). Each donor was also assigned to one of three groups based on the number of centromeric and telomeric B-motifs in the KIR genotype: “Neutral” (0 or 1 B-motif), “Better” (≥2 B-motifs without Cen-B/B) or “Best” (≥2 B-motifs with Cen-B/B) (Table I)(23). Analysis of clinical outcome for this cohort of myeloablative transplants confirmed our previous observations that improved LFS and protection from relapse are associated with transplant donors having ≥2 B-motifs. These comprise the combination of the “Better” and “Best” donor groups; KIR-Better/Best(13, 14). Compared to KIR-Neutral donors who have one or no KIR B-motifs, a 30% reduction in the risk of relapse (RR 0.70 [0.57–0.86]; P=0.0005) was associated with KIR-Better/Best donors, which gave improved LFS (RR 0.79 [0.69–0.91]; P=0.001) (Table II: A and B). The magnitude of the protection was similar for HLA-matched and HLA-mismatched transplants. Compared to KIR-neutral donors, the KIR-Better/Best donors improved LFS (RR 0.83 [0.69–1.01]; P=0.063 and RR 0.76 [0.62–0.93]; P=0.0078, for HLA-matched and HLA-mismatched transplants, respectively) and decreased the risk of relapse (RR 0.72 [0.55–0.94]; P=0.016 and RR 0.49 [0.57–0.93]; P=0.016, respectively).
TABLE II.
Effect of Donor KIR Genotype on Outcome
| A. All Transplants | LFS | ||
|---|---|---|---|
| Donor KIR Content Group | N | RR (CI) | P |
| KIR B=0 [i.e. KIR A/A] | 478 | 1.00 | |
| KIR B ≥1 [i.e. KIR B/x] | 964 | 0.89 (0.78–1.01) | 0.075 |
| KIR B=1 | 535 | 0.98 (0.85–1.14) | 0.80 |
| KIR B=2 | 325 | 0.79 (0.66–0.94) | 0.008 |
| KIR B=3+4 | 104 | 0.76 (0.59–0.99) | 0.043 |
| KIR B=0 or 1 [i.e. Neutral Donors] | 1013 | 1.00 | |
| B≥2 (non Cen B/B) [i.e. Better Donors] | 290 | 0.86 (0.73–1.00) | 0.055 |
| B≥2 (Cen B/B) [i.e. Best Donors] | 139 | 0.67 (0.54–0.85) | 0.0007 |
| B≥2 [i.e. Better/Best Donors] | 429 | 0.79 (0.69–0.91) | 0.001 |
| B. All Transplants | Relapse | ||
|---|---|---|---|
| Donor KIR B Content Group | N | RR (CI) | P |
| KIR B=0 [i.e. KIR A/A] | 499 | 1.00 | |
| KIR B ≥1 [i.e. KIR B/x] | 1003 | 0.88 (0.73–1.06) | 0.18 |
| KIR B=1 | 556 | 1.03 (0.84–1.26) | 0.77 |
| KIR B=2 | 339 | 0.73 (0.57–0.94) | 0.014 |
| KIR B=3+4 | 108 | 0.64 (0.43–0.97) | 0.033 |
| KIR B=0 or 1 [i.e. Neutral Donors] | 1055 | 1.00 | |
| B≥2 (non Cen B/B) [i.e. Better Donors] | 302 | 0.82 (0.65–1.02) | 0.078 |
| B≥2 (Cen B/B) [i.e. Best Donors] | 145 | 0.46 (0.31–0.68) | 0.0001 |
| B≥2 [i.e. Better/Best Donors] | 447 | 0.70 (0.57–0.86) | 0.0005 |
Recipient C1 contributes to the benefit provided by a KIR B donor by decreasing the likelihood of relapse
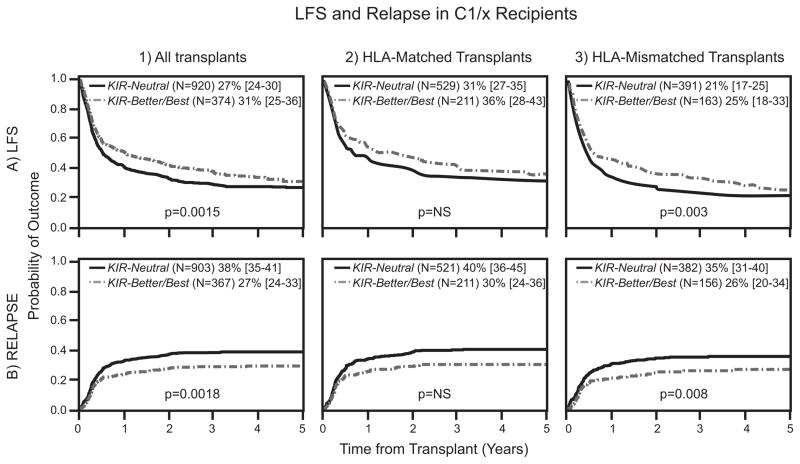
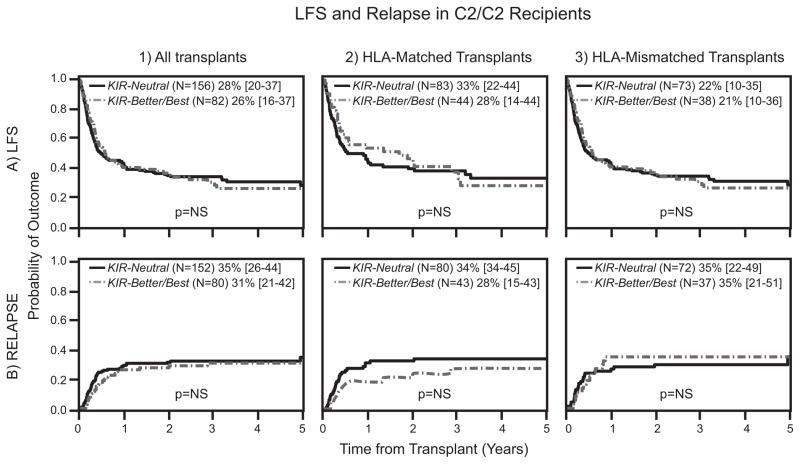
We investigated the mechanism underlying the beneficial effect of KIR B donors in URD transplantation for AML. Multivariate analyses tested the involvement of interactions between donor KIR and the Bw4, C1 and C2 epitopes of donor or recipient HLA class I. In these analyses we distinguished between C2/C2 individuals (N=238), who lack the C1 epitope, and C1/x individuals (N=1294) who are hetrozygous or homozygous for HLA-C bearing the C1 epitope. C1/x recipients had improved LFS when transplanted with grafts from KIR-Better/Best compared to KIR-Neutral donors (RR 0.78 [0.67–0.91]; P=0.0015; Table III, Figure 1:A1). A similar beneficial effect was not observed for C2/C2 recipients (RR 0.93 [0.63–1.37]; P=0.71; Table III, Figure 2: A1). Factors that may contribute to the improved LFS are reductions in leukemic relapse, treatment related mortality (TRM) and GVHD, either singly or in combination. Our analyses demonstrate that reduced incidence of leukemia relapse is the predominant protective effect. No significant correlations were observed with the risks of TRM, acute GVHD or chronic GVHD. Thus, C1/x recipients paired with KIR-Better/Best donors experienced significantly less relapse than C1/x recipients with KIR-Neutral donors (RR 0.70 [0.56–0.87]; P=0.0018; Table III, Figure 1: B1). Five years after transplantation, the frequencies of relapse in C1/x recipients based on donor KIR were 27% vs. 38%, respectively. Although a 4% absolute relapse protection was observed in C2/C2 recipients receiving grafts from KIR-Better/Best vs. KIR-Neutral donors, this trend was not statistically significant (Table III, Figure 2: B1). In all these analyses of the interactions between donor KIR and the Bw4, C1 and C2 epitopes of HLA class I, significant benefits were observed only with C1 epitopes of recipient HLA-C. No significant interactions with donor KIR were demonstrated with recipient C2 and Bw4 or with donor Bw4, C1 and C2.
Table III.
KIR-Better/Best Donors Improve Outcomes for C1/x Recipients
| Outcome | Donor KIR Group | All Transplants | HLA-Matched | HLA-Mismatched | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | RR | P | N | RR | P | N | RR | P | |||
| Recipient HLA C1/x | LFS | KIR-Neutral | 865 | 1.00 | 502 | 1.00 | 363 | 1.00 | |||
| KIR-Better/Best | 354 | 0.78 (0.67–0.91) | 0.0015 | 201 | 0.86 (0.69–1.07) | 0.17 | 153 | 0.70 (0.56–0.88) | 0.003 | ||
| Relapse | KIR-Neutral | 903 | 1.00 | 521 | 1.00 | 382 | 1.00 | ||||
| KIR-Better/Best | 367 | 0.70 (0.56–0.87) | 0.0018 | 211 | 0.79(0.59–1.06) | 0.11 | 156 | 0.61 (0.43–0.88) | 0.008 | ||
| TRM | KIR-Neutral | 893 | 1.00 | 516 | 1.00 | 377 | 1.00 | ||||
| KIR-Better/Best | 363 | 0.89 (0.72–1.09) | 0.26 | 207 | 1.02 (0.75–1.40) | 0.90 | 156 | 0.82 (0.62–1.10) | 0.19 | ||
| Recipient HLA C2/C2 | LFS | KIR-Neutral | 148 | 1.00 | 77 | 1.00 | 71 | 1.00 | |||
| KIR-Better/Best | 75 | 0.93 (0.63–1.37) | 0.71 | 42 | 0.81 (0.45–1.43) | 0.47 | 33 | 1.28 (0.67–2.45) | 0.45 | ||
| Relapse | KIR-Neutral | 152 | 1.00 | 80 | 1.00 | 72 | 1.00 | ||||
| KIR-Better/Best | 80 | 0.80 (0.47–1.36) | 0.41 | 43 | 0.64 (0.29–1.42) | 0.27 | 37 | 1.20 (0.51–2.83) | 0.68 | ||
| TRM | KIR-Neutral | 150 | 1.00 | 79 | 1.00 | 71 | 1.00 | ||||
| KIR-Better/Best | 78 | 0.94 (0.57–1.55) | 0.81 | 42 | 0.89 (0.42–1.89) | 0.76 | 37 | 0.98 (0.45–2.16) | 0.96 | ||
Figure 1. Interactions between KIR-Better/Best donors and recipient HLA-C1 improve LFS and protect against relapse, especially in HLA-mismatched transplants.
Donors were assigned to KIR-Neutral and KIR-Better/Best groups based on KIR genotyping. Probabilities of LFS are provided by Kaplan Meier curves (A) and cumulative incidence probabilities are shown for relapse (B). Each outcome is shown comparing KIR-Neutral donors with KIR-Better/Best donors in HLA-C1/x recipients for all transplants (1), HLA-matched transplants (2) and the HLA-mismatched transplants (3). The estimated rates are presented for LFS and relapse at 5 years. P values were calculated from multivariate analyses comparing relative risks of outcomes for KIR-Neutral and KIR-Better/Best donor groups.
Figure 2. HLA-C2/C2 recipients do not experience enhanced protection from KIR-Better/Best donors.
Donors were assigned to KIR-Neutral and KIR-Better/Best groups based on KIR genotyping and recipients were designated based on their HLA-C allotypes (C1/x and C2/C2). Probabilities of LFS are provided by Kaplan Meier curves (A) and cumulative incidence probabilities are shown for relapse (B). Each outcome is shown comparing KIR-Neutral donors with KIR-Better/Best donors in HLA-C2/C2 recipients for all transplants in all transplants (1), HLA-matched transplants (2) and the HLA-mismatched transplants (3). The estimated rates are presented for LFS and relapse at 5 years. P values were calculated from multivariate analyses comparing relative risks of outcomes for KIR-Neutral and KIR-Better/Best donor groups.
An HLA-C mismatch further reduces relapse for transplants with KIR B donors and C1/x recipients
Our study cohort consisted of similar numbers of HLA-matched (57%) and HLA-mismatched (43%) transplants. This balance enabled a robust evaluation of the effects of HLA mismatch on the interactions of donor KIR with recipient HLA class I. In this analysis, the effects of KIR-Better/Best donors were always compared to those of KIR-Neutral donors (Table III). For HLA-matched transplantation, KIR-Better/Best donors increased LFS and reduced relapse for C1/x recipients compared to C2/C2 recipients (Table III, Figure 1: A2 and B2, Figure 2: A2 and B2) but the difference was not significant. For HLA-mismatched transplants, a stronger, statistically significant effect was observed involving KIR-Better/Best donors and C1/x recipients. Compared to KIR-Neutral donors, LFS was enhanced (RR 0.70 [0.56–0.88]; P=0.003) and relapse was reduced (RR 0.61 [0.43–0.88]; P=0.008; Table III, Figure 1: A3 and B3). Again, C2/C2 recipients derived no significant benefit from a KIR-Better/Best donor (Table III, Figure 2: A3 and B3).
Having demonstrated the beneficial effect of an HLA mismatch on the interaction between donor KIR B and recipient C1, further analyses were performed on the set of 676 HLA-mismatched transplants to determine which HLA genes were involved. We first compared the effects of HLA class I and II mismatch. Improved LFS and relapse protection were observed for C1/x recipients receiving transplants from KIR-Better/Best donors in the subset of 457 HLA-class I mismatched transplants (RR 0.69 [0.54–0.88]; P=0.0029, and RR 0.62 [0.42–0.92]; P=0.019, respectively: Table IV A), but not in the subset of 81 HLA class II mismatched transplants (Table IVA). No differences between HLA class I and class II mismatched transplants were seen in the C2/C2 recipients (data not shown). To identify the specific HLA class I gene responsible for the interaction, we next compared the outcomes for transplants mismatched at HLA-A, or –B (N = 180) or at HLA-C (N=277). The added benefit of an HLA mismatch for a transplant involving a KIR-Better/Best donor and a C1/x recipient was observed only for HLA-C mismatched transplants (RR 0.57 [0.40–0.79]; P=0.001, and RR 0.54 [0.33–0.88]; P=0.013, respectively, Table IV B). Again, no differences were observed in the C2/C2 recipients (data not shown). We next determined whether the benefit of an HLA-C mismatch is the consequence of a KIR-ligand mismatch between transplant donor and recipient. In the circumstance of KIR-ligand mismatch, when the donor expresses C1 or C2 ligand which is lacking in the recipient, donor NK cells can respond alloreactively to the recipient’s cells because they are missing self inhibitory signals. We compared LFS and relapse risk between transplants which included mismatches at HLA-C (n= 460) vs. those with KIR-ligand mismatches based on C1 and C2 (n=60) for the C1/x recipient group. In this small subset, KIR-ligand mismatched transplants were not associated with additional protection (data not shown), demonstrating that KIR-ligand mismatch does not contribute added benefit to other types of HLA-C mismatch. In previous analyses of HLA alone, the degree of HLA-matching correlates with better transplant outcomes(24). Consideration here of the interaction between the HLA and KIR gene systems has shown a benefit for mismatching HLA-C in the particular context of transplantation involving KIR-Better/Best donors and C1/x recipients.
Table IV.
C1/x Recipients Receiving Transplants with Mismatch at HLA-C Benefit from Enhanced Protection with KIR-Better/Best Donors
| A: Transplants with Mismatch at HLA Class I or Class I and Class II vs. Mismatch at HLA Class II | |||||||
|---|---|---|---|---|---|---|---|
| Donor KIR Group | Includes Class I Mismatch | Class II Mismatched Only | |||||
| N | RR | P | N | RR | P | ||
| LFS | KIR-Neutral | 310 | 1.00 | 53 | 1.00 | ||
| KIR-Better/Best | 131 | 0.69 (0.54–0.88) | 0.0029 | 22 | 1.31 (0.57–3.02) | 0.53 | |
| Relapse | KIR-Neutral | 323 | 1.00 | 59 | 1.00 | ||
| KIR-Better/Best | 134 | 0.62 (0.42–0.92) | 0.019 | 22 | 1.45 (0.47–4.50) | 0.52 | |
| B: HLA Class I Mismatch Transplants at HLA-C vs. HLA-A and/or HLA-B | |||||||
|---|---|---|---|---|---|---|---|
| Donor KIR Group | HLA-C Mismatched | HLA-A,B Mismatched | |||||
| N | RR | P | N | RR | P | ||
| LFS | KIR-Neutral | 185 | 1.00 | 125 | 1.00 | ||
| KIR-Better/Best | 83 | 0.57 (0.40–0.79) | 0.001 | 48 | 0.97 (0.63–1.49) | 0.89 | |
| Relapse | KIR-Neutral | 192 | 1.00 | 131 | 1.00 | ||
| KIR-Better/Best | 85 | 0.54 (0.33–0.88) | 0.013 | 49 | 0.68 (0.32–1.45) | 0.32 | |
All KIR B genes contribute to improved clinical outcome associated with KIR B/x donors
Next we examined the extent to which each of the seven KIR B genes individually affected the outcomes associated with the HLA-C1, C2 and Bw4 KIR-ligand status of the recipients. The relative risks for each outcome were determined for 7 groups of KIR B/x donors in comparison to KIR A/A donors. Each of these groups corresponded to the subset of donors carrying one of the 7 KIR genes specific to the KIR-B haplotype. Because most KIR B/x donors have more than one KIR B haplotype-specific gene, each donor is represented in more than one of the 7 groups.
In the full cohort of transplants, C1/x recipients benefited from increases in LFS associated with all 7 KIR B/x donor groups compared to KIR A/A donors (Table V:A). Only KIR2DS1 and KIR3DS1 were associated with about a 20% reduction against relapse (p=0.052 and 0.044 respectively). A striking difference was noted based on the HLA match status of the transplant, as significantly improved LFS and relapse protection were seen only for HLA-mismatched transplants. There was no effect in matched transplants (Table V: B, C). In the HLA-mismatched transplants, multivariate analyses showed that each of the seven KIR B genes contributed clinical benefit of similar magnitude; RR ranged from 0.65–0.80 (P=0.0032–0.055) for LFS and from 0.57–0.70 (P=0.0038–0.036) for relapse (Table V: C). In contrast, for transplantation of C2/C2 recipients, the clinical outcomes were similar for all 7 groups of KIR B/x donors, where KIR B/x donor group has no effect on survival or relapse protection (Table VI). Consequently, no particular donor KIR B haplotype genes interact with recipient C1 to increase LFS or reduce relapse.
Table V.
Impact of Individual Donor KIR B Genes on LFS and Relapse in C1/x Recipients
| A. All Transplants | ||||||
|---|---|---|---|---|---|---|
| Donor KIR Group | n | RR | P | n | RR | P |
|
|
|
|
||||
| LFS | Relapse | |||||
| KIR A/A | 404 | 1.00 | 422 | 1.00 | ||
| KIR B/x with 2DS2+ | 615 | 0.85 (0.73–0.99) | 0.041 | 639 | 0.86 (0.69–1.06) | 0.15 |
| KIR B/x with 2DS5+ | 382 | 0.85 (0.72–1.01) | 0.068 | 397 | 0.83 (0.66–1.05) | 0.12 |
| KIR B/x with 2DL2+ | 610 | 0.86 (0.74–1.00) | 0.056 | 634 | 0.87 (0.70–1.07) | 0.19 |
| KIR B/x with 2DS1+ | 463 | 0.84 (0.71–0.99) | 0.036 | 483 | 0.80 (0.64–1.00) | 0.052 |
| KIR B/x with 3DS1+ | 448 | 0.83 (0.70–0.98) | 0.027 | 469 | 0.79 (0.63–0.99) | 0.044 |
| KIR B/x with 2DS3+ | 306 | 0.81 (0.67–0.97) | 0.025 | 321 | 0.85 (0.66–1.09) | 0.20 |
| KIR B/x with 2DL5+ | 584 | 0.86 (0.74–1.01) | 0.061 | 608 | 0.87 (0.71–1.08) | 0.20 |
| B. HLA Matched | ||||||
|---|---|---|---|---|---|---|
| Donor KIR Group | n | RR | P | n | RR | P |
|
|
|
|
||||
| LFS | Relapse | |||||
| KIR A/A | 233 | 1.00 | 240 | 1.00 | ||
| KIR B/x with 2DS2+ | 344 | 0.96 (0.77–1.19) | 0.71 | 361 | 1.04 (0.79–1.38) | 0.77 |
| KIR B/x with 2DS5+ | 221 | 0.96 (0.76–1.21) | 0.73 | 231 | 1.04 (0.76–1.41) | 0.82 |
| KIR B/x with 2DL2+ | 341 | 0.96 (0.77–1.19) | 0.70 | 358 | 1.04 (0.79–1.37) | 0.79 |
| KIR B/x with 2DS1+ | 274 | 0.96 (0.77–1.20) | 0.71 | 287 | 0.98 (0.73–1.32) | 0.90 |
| KIR B/x with 3DS1+ | 267 | 0.93 (0.74–1.17) | 0.54 | 281 | 0.97 (0.72–1.31) | 0.86 |
| KIR B/x with 2DS3+ | 178 | 0.97 (0.75–1.25) | 0.78 | 189 | 1.07 (0.77–1.50) | 0.67 |
| KIR B/x with 2DL5+ | 341 | 0.99 (0.80–1.23) | 0.95 | 358 | 1.09 (0.82–1.44) | 0.55 |
| C. HLA Mismatched | ||||||
|---|---|---|---|---|---|---|
| Donor KIR Group | n | RR | P | n | RR | P |
|
|
|
|
||||
| LFS | Relapse | |||||
| KIR A/A | 171 | 1.00 | 182 | 1.00 | ||
| KIR B/x with 2DS2+ | 271 | 0.78 (0.62–0.99) | 0.037 | 278 | 0.68 (0.49–0.95) | 0.023 |
| KIR B/x with 2DS5+ | 161 | 0.75 (0.58–0.99) | 0.040 | 166 | 0.59 (0.40–0.88) | 0.0090 |
| KIR B/x with 2DL2+ | 269 | 0.80 (0.63–1.01) | 0.055 | 276 | 0.70 (0.51–0.98) | 0.036 |
| KIR B/x with 2DS1+ | 189 | 0.73 (0.56–0.94) | 0.015 | 196 | 0.59 (0.41–0.85) | 0.0052 |
| KIR B/x with 3DS1+ | 181 | 0.73 (0.57–0.95) | 0.018 | 188 | 0.57 (0.39–0.83) | 0.0038 |
| KIR B/x with 2DS3+ | 128 | 0.65 (0.48–0.86) | 0.0032 | 132 | 0.58 (0.38–0.88) | 0.0098 |
| KIR B/x with 2DL5+ | 243 | 0.75 (0.59–0.96) | 0.020 | 250 | 0.63 (0.44–0.88) | 0.0076 |
Table VI.
Impact of Individual Donor KIR B Genes on LFS and Relapse in C2/C2 Recipients
| A. All Transplants | ||||||
|---|---|---|---|---|---|---|
| Donor KIR Group | n | RR | P | n | RR | P |
| LFS | Relapse | |||||
| KIR A/A | 74 | 1.00 | 77 | 1.00 | ||
| KIR B/x with 2DS2+ | 113 | 1.09 (0.73–1.63) | 0.69 | 119 | 1.17 (0.67–2.07) | 0.58 |
| KIR B/x with 2DS5+ | 75 | 1.22 (0.78–1.92) | 0.38 | 76 | 0.89 (0.48–1.65) | 0.72 |
| KIR B/x with 2DL2+ | 112 | 1.07 (0.71–1.61) | 0.74 | 117 | 1.16 (0.65–2.08) | 0.61 |
| KIR B/x with 2DS1+ | 92 | 1.16 (0.75–1.80) | 0.51 | 96 | 0.99 (0.56–1.77) | 0.97 |
| KIR B/x with 3DS1+ | 90 | 1.10 (0.72–1.69) | 0.65 | 94 | 1.06 (0.60–1.87) | 0.84 |
| KIR B/x with 2DS3+ | 60 | 1.26 (0.75–2.10) | 0.38 | 63 | 1.06 (0.56–2.01) | 0.85 |
| KIR B/x with 2DL5+ | 117 | 1.09 (0.73–1.61) | 0.68 | 121 | 0.98 (0.57–1.69) | 0.93 |
| B. HLA Matched | ||||||
|---|---|---|---|---|---|---|
| Donor KIR Group | n | RR | P | n | RR | P |
| LFS | Relapse | |||||
| KIR A/A | 37 | 1.00 | 39 | 1.00 | ||
| KIR B/x with 2DS2+ | 66 | 1.13 (0.63–2.02) | 0.68 | 68 | 0.94 (0.43–2.05) | 0.88 |
| KIR B/x with 2DS5+ | 36 | 1.26 (0.62–2.54) | 0.52 | 36 | 0.81 (0.31–2.12) | 0.66 |
| KIR B/x with 2DL2+ | 65 | 1.08 (0.60–1.95) | 0.79 | 66 | 0.86 (0.39–1.91) | 0.71 |
| KIR B/x with 2DS1+ | 45 | 1.25 (0.63–2.47) | 0.52 | 46 | 0.90 (0.37–2.21) | 0.82 |
| KIR B/x with 3DS1+ | 44 | 1.15 (0.58–2.28) | 0.69 | 45 | 1.00 (0.41–2.43) | 0.99 |
| KIR B/x with 2DS3+ | 33 | 0.95 (0.43–2.13) | 0.91 | 34 | 0.85 (0.34–2.12) | 0.72 |
| KIR B/x with 2DL5+ | 60 | 1.18 (0.63–2.20) | 0.60 | 61 | 0.82 (0.36–1.84) | 0.63 |
| C. HLA Mismatched | ||||||
|---|---|---|---|---|---|---|
| Donor KIR Group | n | RR | P | n | RR | P |
| LFS | Relapse | |||||
| KIR A/A | 37 | 1.00 | 38 | 1.00 | ||
| KIR B/x with 2DS2+ | 47 | 1.57 (0.71–3.48) | 0.27 | 51 | 1.62 (0.56–4.72) | 0.38 |
| KIR B/x with 2DS5+ | 39 | 1.07 (0.49–2.33) | 0.86 | 40 | 0.92 (0.36–2.36) | 0.86 |
| KIR B/x with 2DL2+ | 47 | 1.53 (0.69–3.40) | 0.30 | 51 | 1.81 (0.59–5.55) | 0.30 |
| KIR B/x with 2DS1+ | 47 | 1.18 (0.56–2.47) | 0.67 | 50 | 1.26 (0.51–3.08) | 0.62 |
| KIR B/x with 3DS1+ | 46 | 1.21 (0.58–2.49) | 0.61 | 49 | 1.27 (0.52–3.08) | 0.60 |
| KIR B/x with 2DS3+ | 27 | 2.33 (0.73–7.47) | 0.15 | 29 | 1.31 (0.42–4.08) | 0.64 |
| KIR B/x with 2DL5+ | 57 | 1.13 (0.58–2.24) | 0.72 | 60 | 1.09 (0.45–2.62) | 0.85 |
Discussion
In the present study, which was designed to determine whether the differential clinical effects of donor centromeric and telomeric encoded KIR could involve interactions with HLA-Bw4, HLA-C1 and HLA-C2, we analyzed a cohort of patients who received HLA-matched and mismatched URD grafts without T-cell depletion following myeloablative preparative regimens. In this large cohort, KIR B donors reduced relapse and improved LFS in both HLA-matched and mismatched transplants. We now demonstrate a significantly protective interaction between donor KIR and recipient C1. In C1/x recipients, KIR-Better/Best donors were associated with improved LFS, attributed to an 11% reduction in relapse rate. The protective effect of this interaction was strongest in the HLA-mismatched transplants, specifically those with class I mismatch for at the C locus. Thus, we have demonstrated that donor KIR B, recipient C1, and an HLA-C mismatch between donor and recipient are all factors that interact to reduce leukemia relapse and increase the LFS after URD HCT as treatment for AML. The correlation of donor KIR B and recipient C1 with protection from relapse raises the strong possibility that interactions between C1 epitopes and the C1-reactive KIR encoded by KIR B haplotypes is a molecular mechanism underlying the improved transplant outcome. Inhibitory KIR2DL2 is the only C1 receptor encoded by KIR B. Moreover, the KIR2DL2 gene, in combination with the KIR2DS2 gene, defines the common Cen-B motif that in homozygous form defines the KIR-Best transplant donors(14). These correlations are consistent with the interaction of C1 with KIR2DL2 being an important contributor to the observed clinical benefits.
While the data support a model in which an interaction between recipient C1 and donor KIR2DL2 enhances NK cell education and improves clinical outcome, we must consider the alternatives. Three KIR genes encode receptors that discriminate HLA-C1 and HLA-C2. These comprise the inhibitory C1 receptor KIR2DL2/3, the inhibitory C2 receptor KIR2DL1 and the activating C2 receptor KIR2DS1. Although KIR2DL2 and KIR2DL3 are both inhibitory receptors that recognize C1, KIR2DL2 is specific to Cen-B haplotypes and KIR2DL3 is specific to Cen-A haplotypes. These receptors differ in four potentially important ways. First, KIR2DL2 has higher avidity for C1 than KIR2DL3, which can affect the education of NK cells mediated by the C1 ligand(25). Second, KIR2DL2 has crossreactivity with C2(25, 26) which can alter NK cell education and produce NK cells that are educated by and responsive to both C1 and C2. The presence of the KIR2DL2 gene causes a major reduction in frequency of NK cells expressing KIR2DL1(27). This mechanism is independent of the presence or absence of the C1 or C2 epitope and is a potential mechanism by which KIR B and Cen-B can mediate beneficial clinical effects in the absence of C1. Fourth, the KIR2DL1 alleles that are in linkage disequilibrium with the KIR2DL2 gene are functionally weaker in ligand-binding affinity or signaling function(28) than those in linkage disequilibrium with KIR2DL3 (23, 28, 29).
The differences between KIR2DL2 and KIR2DL3 may not account for all the beneficial clinical effect associated with KIR B donors. KIR2DS1 is specific to KIR B haplotypes, and the KIR2DL1 allotypes carried by B haplotypes expressed at lower frequencies by NK cells(27). We have previously demonstrated a benefit, albeit less significant, associated with Tel-B in the absence of Cen-B(14). That finding is consistent with this analysis of individual KIR-B specific genes. We have shown that all seven genes contributed equally to the clinical benefit, specifically in C1/x but not C2/C2 recipients. It has previously been reported that C2/C2 homozygous patients with AML had more relapse after HLA-C matched URD HCT (12) and HLA-matched sibling HCT(11).
We also observed significant relapse protection associated with the telomeric KIR2DS1 and KIR3DS1 in the total cohort, consistent with other reported associations with those KIR and improved transplant outcome(30, 31). Venstrom et al have reported that donor KIR2DS1 is associated with reduced relapse and better LFS for AML patients who received an URD HCT that is HLA matched or has a single HLA mismatch(30). Based on a mechanism described by Fauriat et al in which NK cells expressing only KIR2DS1 from healthy C2/C2 donors are hyporesponsive (32), Venstrom et al propose that those cells recognize leukemia and improve LFS via recognition of missing self when transplanted into C1/x recipients.
Because of the compact arrangement of genes in the KIR locus and the extensive linkage disequilibrium between KIR genes, these data should be interpreted with caution(33). For example, KIR2DS2 and KIR2DL2 in Cen-B haplotypes are in almost complete linkage disequilibrium. Several characteristics of the KIR system, including the coordinated transcription of the KIR genes, their variegated expression, and the haplotypic gene-content variation support a model in which the KIR genotype reduces the effect of individual genes. Additionally, KIR gene content analyses alone could be misleading given that the differences between KIR allotypes affect the affinity for HLA class I ligands as well as signaling function(25, 28, 34, 35). Application of high resolution typing of KIR alleles will investigate this possibility. It is also important to emphasize that we can address only the modulation of NK cell education and function by polymorphic KIR and HLA and not by the contributions of the many conserved receptors and ligands that affect these processes(36). Lastly, one must consider the possibility that allogeneic disparity contributes to the graft-versus-leukemia protection mediated by T cells. This could be mediated directly, by an allogeneic response that provides T cell help to NK cells, or indirectly through reciprocal activation of dendritic cells and NK cells that function to bridge the innate and adaptive immune response(37). With those caveats, we propose that the many differences between centromeric and telomeric KIR A and B haplotype receptors result in substantial influences on NK-cell education and repertoire development, which in turn alters NK-cell mediated graft-versus-leukemia reactions following URD HCT for AML.
Independent of the underlying molecular mechanism, there is a general consensus that KIR B/x donors improve outcome for AML patients receiving T-cell containing, myeloablative HCT. We have demonstrated that interactions with HLA-C1 augment the effect of a KIR B/x donor, specifically by enhancing relapse protection, most significantly in transplants mismatched at HLA-C. For the 15% of recipients who are C2/C2, our analysis did not detect additional improvements in survival or reductions in relapse based in interactions with KIR B donors. Larger studies will be needed to test the validity of this result. Understanding the interactions between KIR B/x donors and recipient HLA-C1 is particularly important because a considerable majority (~85%) of US transplant recipients are HLA-C1/x. The findings presented here are being further tested in our ongoing multi-center prospective study incorporating KIR genotyping into URD selection for AML coordinated through the National Marrow Donor Program and Center for International Blood and Marrow Transplant Research (clinicaltrials.gov NCT01288222).
Supplementary Material
Acknowledgments
This work was supported utilizing the Masonic Cancer Center Oncology Medical Informatics and Services shared resource.
Footnotes
This work was supported by National Institutes of Health/NCI grant P01 111412, and in part by NIH P30 CA77598 utilizing the Masonic Cancer Center Oncology Medical Informatics shared resource. The Center for International Blood and Marrow Transplant Research is supported by Public Health Service Grant/Cooperative Agreement U24-CA76518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI), and the National Institute of Allergy and Infectious Diseases (NIAID); a Grant/Cooperative Agreement 5U01HL069294 from NHLBI and NCI; a contract HHSH234200637015C with Health Resources and Services Administration (HRSA/DHHS); 2 grants, N00014-08-1-1207 and N00014-10-1-0204, from the Office of Naval Research; and others. The views expressed in this article do not reflect the official policy or position of the National Institute of Health, the Department of the Navy, the Department of Defense, or any other agency of the US Government.
Authorship Contributions and Disclosure of Conflicts of Interest:
Sarah Cooley, Daniel J. Weisdorf, Lisbeth A. Guethlein, Peter Parham and Jeffrey S. Miller designed this study, analyzed data and wrote the manuscript.
John P. Klein and Tao Wang performed the biostatistical analysis for this study and wrote the manuscript.
Steven G.E. Marsh, Stephen Spellman, and Michael D. Haagenson provided clinical data integration, assisted with data interpretation and assisted in writing the manuscript.
Martha Ladner, Koy Saeteurn and Elizabeth Trachtenberg performed all KIR genotyping, were involved in data analysis and assisted in writing the manuscript.
Conflict of interest statement: We declare that there is no conflict of interest on behalf of the authors.
References
- 1.Shilling HG, Young N, Guethlein LA, Cheng NW, Gardiner CM, Tyan D, Parham P. Genetic control of human NK cell repertoire. J Immunol. 2002;169:239–247. doi: 10.4049/jimmunol.169.1.239. [DOI] [PubMed] [Google Scholar]
- 2.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, Posati S, Rogaia D, Frassoni F, Aversa F, Martelli MF, Velardi A. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 3.Ruggeri L, Capanni M, Casucci M, Volpi I, Tosti A, Perruccio K, Urbani E, Negrin RS, Martelli MF, Velardi A. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999;94:333–339. [PubMed] [Google Scholar]
- 4.Ruggeri L, Mancusi A, Capanni M, Urbani E, Carotti A, Aloisi T, Stern M, Pende D, Perruccio K, Burchielli E, Topini F, Bianchi E, Aversa F, Martelli MF, Velardi A. Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood. 2007;110:433–440. doi: 10.1182/blood-2006-07-038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruggeri L, Mancusi A, Burchielli E, Aversa F, Martelli MF, Velardi A. Natural killer cell alloreactivity in allogeneic hematopoietic transplantation. Curr Opin Oncol. 2007;19:142–147. doi: 10.1097/CCO.0b013e3280148a1a. [DOI] [PubMed] [Google Scholar]
- 6.Hsu KC, Gooley T, Malkki M, Pinto-Agnello C, Dupont B, Bignon JD, Bornhauser M, Christiansen F, Gratwohl A, Morishima Y, Oudshoorn M, Ringden O, van Rood JJ, Petersdorf E. KIR ligands and prediction of relapse after unrelated donor hematopoietic cell transplantation for hematologic malignancy. Biol Blood Marrow Transplant. 2006;12:828–836. doi: 10.1016/j.bbmt.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 7.Malmberg KJ, Schaffer M, Ringden O, Remberger M, Ljunggren HG. KIR-ligand mismatch in allogeneic hematopoietic stem cell transplantation. Mol Immunol. 2005;42:531–534. doi: 10.1016/j.molimm.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 8.Giebel S, Locatelli F, Lamparelli T, Velardi A, Davies S, Frumento G, Maccario R, Bonetti F, Wojnar J, Martinetti M, Frassoni F, Giorgiani G, Bacigalupo A, Holowiecki J. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003;102:814–819. doi: 10.1182/blood-2003-01-0091. [DOI] [PubMed] [Google Scholar]
- 9.Farag SS, Bacigalupo A, Eapen M, Hurley C, Dupont B, Caligiuri MA, Boudreau C, Nelson G, Oudshoorn M, van Rood J, Velardi A, Maiers M, Setterholm M, Confer D, Posch PE, Anasetti C, Kamani N, Miller JS, Weisdorf D, Davies SM. The effect of KIR ligand incompatibility on the outcome of unrelated donor transplantation: a report from the center for international blood and marrow transplant research, the European blood and marrow transplant registry, and the Dutch registry. Biol Blood Marrow Transplant. 2006;12:876–884. doi: 10.1016/j.bbmt.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Leung W. Use of NK cell activity in cure by transplant. Br J Haematol. 2011;155:14–29. doi: 10.1111/j.1365-2141.2011.08823.x. [DOI] [PubMed] [Google Scholar]
- 11.Cook MA, Milligan DW, Fegan CD, Darbyshire PJ, Mahendra P, Craddock CF, Moss PA, Briggs DC. The impact of donor KIR and patient HLA-C genotypes on outcome following HLA-identical sibling hematopoietic stem cell transplantation for myeloid leukemia. Blood. 2004;103:1521–1526. doi: 10.1182/blood-2003-02-0438. [DOI] [PubMed] [Google Scholar]
- 12.Giebel S, Locatelli F, Wojnar J, Velardi A, Mina T, Giorgiani G, Krawczyk-Kulis M, Markiewicz M, Wylezol I, Holowiecki J. Homozygosity for human leucocyte antigen-C ligands of KIR2DL1 is associated with increased risk of relapse after human leucocyte antigen-C-matched unrelated donor haematopoietic stem cell transplantation. Br J Haematol. 2005;131:483–486. doi: 10.1111/j.1365-2141.2005.05797.x. [DOI] [PubMed] [Google Scholar]
- 13.Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT, Marsh SG, Guethlein LA, Parham P, Miller JS, Weisdorf DJ. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood. 2009;113:726–732. doi: 10.1182/blood-2008-07-171926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Le CT, Marsh SG, Geraghty D, Spellman S, Haagenson MD, Ladner M, Trachtenberg E, Parham P, Miller JS. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood. 2010;116:2411–2419. doi: 10.1182/blood-2010-05-283051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houtchens KA, Nichols RJ, Ladner MB, Boal HE, Sollars C, Geraghty DE, Davis LM, Parham P, Trachtenberg EA. High-throughput killer cell immunoglobulin-like receptor genotyping by MALDI-TOF mass spectrometry with discovery of novel alleles. Immunogenetics. 2007;59:525–537. doi: 10.1007/s00251-007-0222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollenbach JA, Ladner MB, Saeteurn K, Taylor KD, Mei L, Haritunians T, McGovern DP, Erlich HA, Rotter JI, Trachtenberg EA. Susceptibility to Crohn’s disease is mediated by KIR2DL2/KIR2DL3 heterozygosity and the HLA-C ligand. Immunogenetics. 2009;61:663–671. doi: 10.1007/s00251-009-0396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol. 2002;38:1007–1021. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]
- 18.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 19.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gumperz JE, Barber LD, Valiante NM, Percival L, Phillips JH, Lanier LL, Parham P. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J Immunol. 1997;158:5237–5241. [PubMed] [Google Scholar]
- 21.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, Easterbrook P, McVicar DW, Maenaka K, Parham P, Carrington M, Dong T, Rowland-Jones S. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 22.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, Rowland-Jones S, Braud VM. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 23.Pyo CW, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, Marsh SG, Miller JS, Parham P, Geraghty DE. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS One. 2010;5:e15115. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersdorf EW. Optimal HLA matching in hematopoietic cell transplantation. Curr Opin Immunol. 2008;20:588–593. doi: 10.1016/j.coi.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 26.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 27.Schonberg K, Sribar M, Enczmann J, Fischer JC, Uhrberg M. Analyses of HLA-C-specific KIR repertoires in donors with group A and B haplotypes suggest a ligand-instructed model of NK cell receptor acquisition. Blood. 2011;117:98–107. doi: 10.1182/blood-2010-03-273656. [DOI] [PubMed] [Google Scholar]
- 28.Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das Gupta N, Holladay M, Rooney B, Leung W. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. 2009;114:5182–5190. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venstrom JM, Pittari G, Gooley TA, Chewning JH, Spellman S, Haagenson M, Gallagher MM, Malkki M, Petersdorf E, Dupont B, Hsu KC. HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med. 2012;367:805–816. doi: 10.1056/NEJMoa1200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venstrom JM, Gooley TA, Spellman S, Pring J, Malkki M, Dupont B, Petersdorf E, Hsu KC. Donor activating KIR3DS1 is associated with decreased acute GVHD in unrelated allogeneic hematopoietic stem cell transplantation. Blood. 2010;115:3162–3165. doi: 10.1182/blood-2009-08-236943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fauriat C, Ivarsson MA, Ljunggren HG, Malmberg KJ, Michaelsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood. 2010;115:1166–1174. doi: 10.1182/blood-2009-09-245746. [DOI] [PubMed] [Google Scholar]
- 33.Gourraud PA, Meenagh A, Cambon-Thomsen A, Middleton D. Linkage disequilibrium organization of the human KIR superlocus: implications for KIR data analyses. Immunogenetics. 2010;62:729–740. doi: 10.1007/s00251-010-0478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frazier WR, Steiner N, Hou L, Dakshanamurthy S, Hurley CK. Allelic variation in KIR2DL3 generates a KIR2DL2-like receptor with increased binding to its HLA-C ligand. J Immunol. 2013;190:6198–6208. doi: 10.4049/jimmunol.1300464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hilton HG, Vago L, Older Aguilar AM, Moesta AK, Graef T, Abi-Rached L, Norman PJ, Guethlein LA, Fleischhauer K, Parham P. Mutation at positively selected positions in the binding site for HLA-C shows that KIR2DL1 is a more refined but less adaptable NK cell receptor than KIR2DL3. J Immunol. 2012;189:1418–1430. doi: 10.4049/jimmunol.1100431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, Bjorklund AT, Flodstrom-Tullberg M, Michaelsson J, Rottenberg ME, Guzman CA, Ljunggren HG, Malmberg KJ. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood. 2010;116:3853–3864. doi: 10.1182/blood-2010-04-281675. [DOI] [PubMed] [Google Scholar]
- 37.Andrews DM, Scalzo AA, Yokoyama WM, Smyth MJ, Degli-Esposti MA. Functional interactions between dendritic cells and NK cells during viral infection. Nat Immunol. 2003;4:175–181. doi: 10.1038/ni880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.