Abstract
Objective
To identify a genetic cause for migrating partial seizures in infancy (MPSI).
Methods
We characterized a consanguineous pedigree with MPSI and obtained DNA from affected and unaffected family members. We analyzed single nucleotide polymorphism (SNP) 500K data to identify regions with evidence for linkage. We performed whole exome sequencing and analyzed homozygous variants in regions of linkage to identify a candidate gene and performed functional studies of the candidate gene SLC25A22.
Results
In a consanguineous pedigree with two individuals with MPSI, we identified two regions of linkage, chromosome 4p16.1-p16.3 and chromosome 11p15.4-pter. Using whole exome sequencing, we identified 8 novel homozygous variants in genes in these regions. Only one variant, SLC25A22 c.G328C, results in a change of a highly conserved amino acid (p.G110R) and was not present in control samples. SLC25A22 encodes a glutamate transporter with strong expression in the developing brain. We show that the specific G110R mutation, located in a transmembraine domain of the protein, disrupts mitochondrial glutamate transport.
Interpretation
We have shown that MPSI can be inherited and have identified a novel homozygous mutation in SLC25A22 in the affected individuals. Our data strongly suggest that SLC25A22 is responsible for MPSI, a severe condition with few known etiologies. We have demonstrated that a combination of linkage analysis and whole exome sequencing can be used for disease gene discovery. Finally, as SLC25A22 had been implicated in the distinct syndrome neonatal epilepsy with suppression bursts on EEG, we have expanded the phenotypic spectrum associated with SLC25A22.
Introduction
Migrating partial seizures in infancy (MPSI) is a severe epileptic encephalopathy with onset in early infancy. Infants with MPSI have medically intractable epilepsy, nearly continuous and migrating partial seizures on electroencephalogram (EEG), and poor developmental prognosis. MPSI, first described (and called malignant migrating partial seizures in infancy) in 19951, is not usually associated with an underlying etiology identifiable by examination, neuroimaging, or most clinically available genetic testing. There is a rare association of MPSI with hippocampal sclerosis,1 and one case has been reported with both hippocampal sclerosis and developmental malformation of the temporal lobe.2
There are no prior reports of multiple individuals affected with MPSI within a family and, until recently, MPSI was not widely considered to have a genetic basis.3,4 More recently, however, MPSI has been associated with genetic mechanisms involving three genes: a missense mutation and a deletion containing the gene SCN1A, previously has associated with Severe Myoclonic Epilepsy of Infancy (Dravet syndrome),5,6 an inherited homozygous deletion of PLCB1, previously associated with a distinct early onset epilepsy,7,8 and de novo mutations in the potassium channel-encoding KCNT1.9 In addition, a single case of MPSI has recently been reported in association with duplication 16p11.2.10 In order to find a novel gene for MPSI, we mapped a consanguineous, autosomal recessive pedigree and used whole exome sequencing to identify a gene associated with a severe syndrome that until recently was not recognized as having a genetic basis. Functional studies confirmed the pathogenicity of the mutation we identified.
Subjects and Methods
Subjects with MPSI and their families were ascertained from Saudi Arabia, the USA, Australia, Sweden, and New Zealand. Detailed history, neurological examination, EEG, and neuroimaging were performed clinically. Informed consent was obtained in accordance with the Institutional Review Boards (IRBs) of Boston Children’s Hospital and Beth Israel Deaconess Medical Center in Boston as well as the local IRBs of the referring institutions. Work on de-identified samples at the Duke Center for Human Genome Variation (CHGV) was approved by the IRB of Duke University School of Medicine. Clinical data were reviewed by two neurologists (MAS, AP); neuroimaging was reviewed by three neurologists and one neuroradiologist (MAS, AP, CAW, AJB).
Blood samples were collected from consenting subjects, including affected individuals and their parents and unaffected siblings. DNA was extracted using standard methods from peripheral blood lymphocytes. DNA was digested, amplified, and hybridized to an Affymetrix 500K single nucleotide polymorphism (SNP) array at the Broad Institute (Cambridge, MA).
Assuming an autosomal recessive mode of inheritance with 100 percent penetrance and a susceptibility allele frequency of 0.1 percent, we analyzed SNP data from the pedigree described in this report to evaluate for evidence of genetic linkage using Allegro software.11 Copy number was assessed using dChip software (Affymetrix, Santa Clara, CA).
DNA from one affected individual (EP-201) was used for whole exome sequencing at the Duke CHGV using the Illumina Genome Analyzer IIx massively parallel sequencing system (Illumina, Inc., San Diego, CA). Alignment to the human genome (reference build hg18) was conducted using BWA version 0.5.5.12 SAMtools version 0.1.712 was used to make consensus and variant calls. Annotation, filtering for quality, filtering to remove potential variants present in dbSNP129 or in 220 individuals from a group non-enriched for neuropsychiatric phenotypes, and prediction of functional effects of potential mutations were performed using Sequent Variant Analyzer (SVA) (http://people.genome.duke.edu). After an initial list of variants was generated in this manner, we limited the list of candidate mutations to include only those variants present in the regions for which there was evidence for linkage and only those variants in those regions that were homozygous or that represented possible compound heterozygous mutations. We further eliminated variants present in dbSNP130 after inspection of each variant prior to elimination to ensure that there was evidence that the variant was a true polymorphism. We prioritized variants that were predicted to produce a premature truncation, nonsynonymous coding change, or disruption of a canonical splice site. Further analysis of functional homozygous variants in the regions of linkage was performed using the UCSC Genome Browser (www.genome.ucsc.edu) and Sorting Intolerant from Tolerant (SIFT) mutation prediction software (http://blocks.fhcrc.org/sift/SIFT.html).
Primers were designed using Primer 3 software13 to confirm potentially pathogenic mutations by Sanger sequencing in EP-201 and establish the genotype in the other family members. Sanger sequencing and Sequenom genotyping (San Diego, CA) were used to evaluate for the presence of potential mutations in three additional families and 14 sporadic cases of MPSI, 144 Middle Eastern unaffected individuals, and 576 neurologically normal Caucasian control individuals from the National Institute of Neurological Disorders and Stroke (NINDS) through the Coriell Institute for Medical Research (Camden, NJ).
We overexpressed wild type (WT) SLC25A22 using a commercially available plasmid (Origene, Rockville, MD) tagged with green fluorescent protein (GFP) in COS-7 cells in culture to determine the pattern of cellular localization. We then used site-directed mutagenesis to create a mutant construct containing the c.G328C variant using the QuikChange II mutagenesis system (Stratagene Cloning Systems, La Jolla, CA). We confirmed the wild-type and mutant sequences with polymerase chain reaction (PCR). We overpressed the GFP-tagged mutant construct into COS-7 cells. Transfections were performed using Fugene 6 transfection reagents (Roche, Indianapolis, IN). We stained for GFP as well as a nuclear counterstain, 4',6-diamidino-2-phenylindole (DAPI) and a mitochondrial marker, MitoTracker Red CMXRos.
To conduct functional studies of wild type vs G110R SLC25A22 using a bacterial system, plasmids were constructed containing the coding sequence for wild-type SLC25A22 amplified by PCR from human brain cDNA. The Gly110Arg mutation was introduced into the wild-type SLC25A22 cDNA by overlap-extension PCR. Constructs were transformed into E. coli TOP 10 cells. Overproduction of wild-type and G110R mutant SLC25A22 as inclusion bodies in the cytosol of E. coli was accomplished according to standard methods. The same amount of each recombinant protein was used for in vitro reconstitution of SLC25A22 into liposomes. Then the [14C]glutamate/glutamate exchange in proteoliposomes reconstituted with the recombinant wild-type or the G110R mutant SLC25A22 (also known as glutamate carrier 1, or GC114) was measured after 1-minute incubation (in the initial linear range of uptake to measure the specific activity) and after 60-min incubation (when radioactive uptake by proteoliposomes had approached equilibrium), as described previously.15–18 Full details are provided in the Supplemental Methods section.
Results
MPSI in a Consanguineous Pedigree
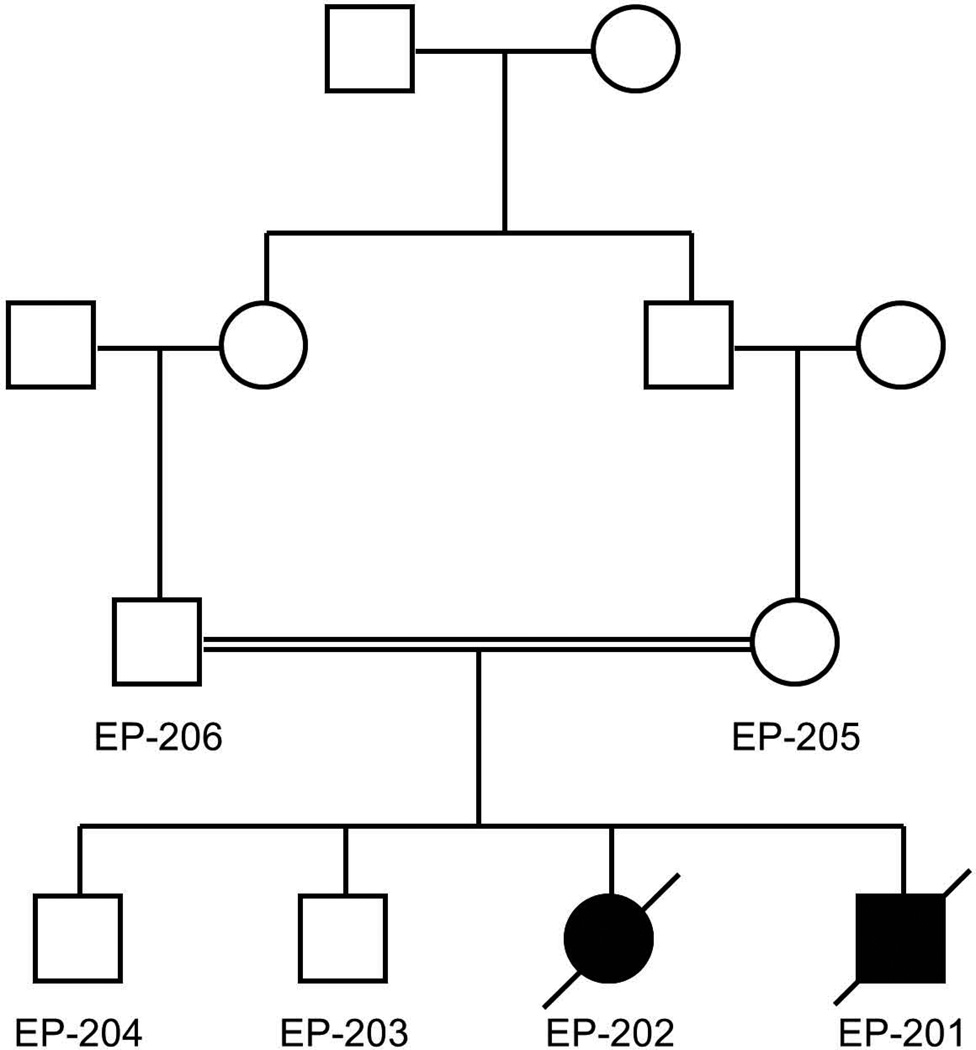
The pedigree shown in Figure 1 depicts two affected children (EP-201 and EP-202) with MPSI, one male and one female. Their parents were first cousins from Saudi Arabia with Sudanese ancestry; there were two unaffected brothers. DNA was obtained from these 6 individuals.
Figure 1.
The affected boy (EP-201) and girl (EP-202) are shown as a black square and circle. The mother and father of the affected children (EP-205 and EP-206) were first cousins from Saudi Arabia. The unaffected brothers (EP-203 and EP-204) are shown as well.
The clinical features of both affected siblings are summarized in Table 1. EP-201, the affected boy, presented with seizures at 1 week of age. The initial seizures appeared as hemi-convulsions on the right side of the body that migrated to the left side within 3 days; accompanying the convulsive activity, there was flushing of the face, staring, and eventually bilateral eyelid blinking. The general examination was unremarkable, and the neurological examination revealed hypotonia and brisk deep tendon reflexes (DTRs). EEG performed at 1 month of age showed a delta brush pattern in the left mid-temporal region (T7), positive spikes in the right mid-temporal region (T8), and high-voltage focal spikes in the right and left frontal regions. Magnetic resonance imaging (MRI), also performed at 1 month of age, was normal. An extensive neuro-metabolic screen included evaluation of serum ammonia, lactate, venous blood gas, and analysis of blood by tandem mass spectrometry as well as evaluation of urine for sulphocystine and xanthine and by gas chromatography and mass spectrometry. The results were all normal. Brainstem auditory evoked responses (BAER), visual evoked potentials (VEP), and electroretinogram (ERG) were all normal. Seizures were only partially responsive to carbamazepine and diazepam treatment. There was delay and subsequent arrest of development, and the child died at 14 months of age.
Table 1.
Clinical Features of Two Siblings with MMPEI
| Patient | Sex | Seizure types | Age of seizure onset |
Examination | Development | Age of death |
|---|---|---|---|---|---|---|
| EP-201 | male | Hemi-convulsive with color change and staring | 1 week | Hypotonic Brisk DTRs | Delay, arrest | 14 months |
| EP-202 | female | Hemi-convulsive | 2 weeks | Hypotonic Brisk DTRs | Delay, arrest | 47 months |
EP-202, the affected girl, presented with seizures at 2 weeks of age. The seizures were described as bilateral and hemi-clonic convulsions of the right side more than the left, accompanied by bilateral eyelid fluttering. The general examination was unremarkable, and the neurological examination revealed hypotonia and brisk DTRs. EEG performed at 6 months of age showed independent focal spikes in a multifocal distribution and partial seizures of multifocal origin. Brain MRI, performed at 3 months of age and again at 2 years of age, revealed a delayed myelination pattern and diffuse thinning of the corpus callosum. This child, in addition to the evaluation undertaken for her brother, had evaluation of creatine kinase, very long chain fatty acids, high-resolution karyotype, and a muscle biopsy (histology and histochemistry to evaluate for mitochondrial dysfunction), all of which were normal. BAER and ERG were normal, but VEP suggested bilateral optic pathway involvement. Seizures were refractory to treatment with phenobarbital, diazepam, pyridoxine, topiramate, and carbamazepine. Development was delayed and subsequently arrested. She died at 47 months of age.
Linkage Analysis and Copy Number Results
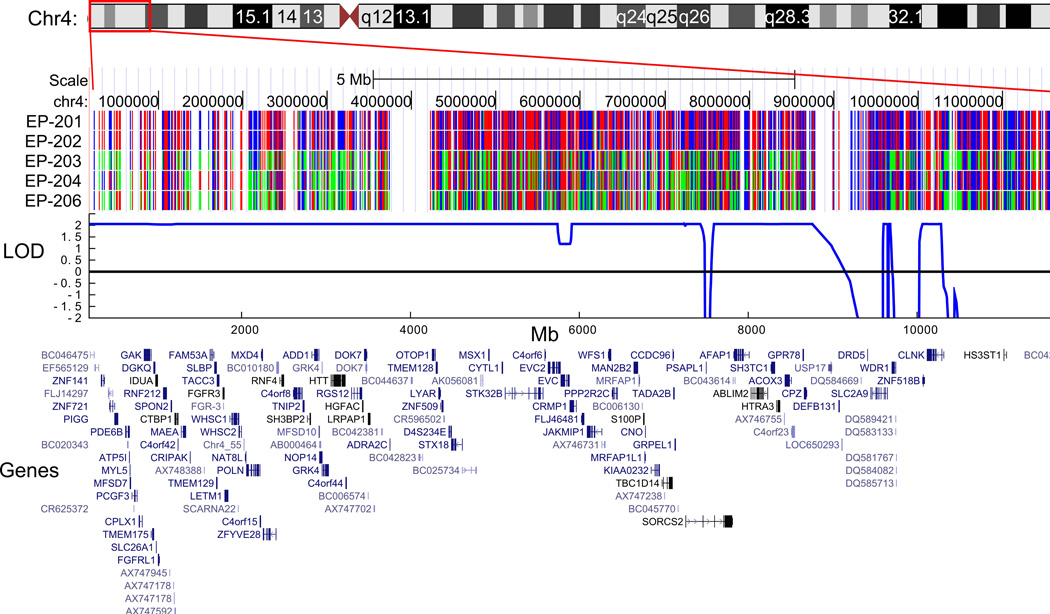
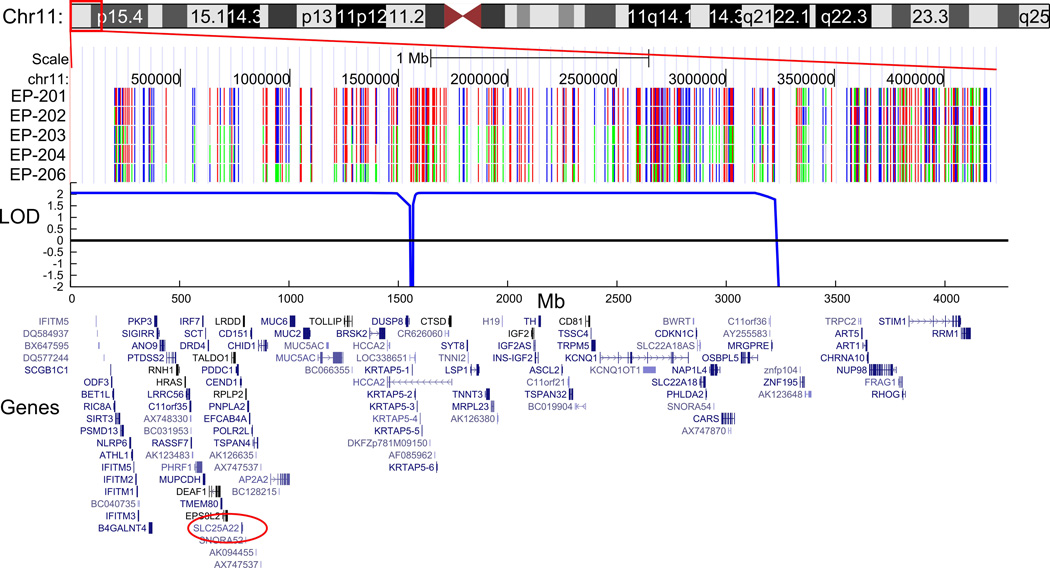
We obtained Affymetrix 500K SNP data from individuals EP-201, EP-202, EP-203, EP-204, and EP-206. In our genome-wide analysis of these data, we found only one region spanning a genetic distance of greater than 2cM with apparent identity by descent at chromosome 4p16.1-p16.3, flanked by SNPs rs10001263 and rs6826371 (base pair position chr4:74,508–10,760,828, hg18), in which SNPs were homozygous in the affected individuals but heterozygous in the unaffected individuals. Linkage analysis of this 9.6 Mb region of homozygosity resulted in a LOD (logarithm of odds) score of 2.06, the maximum expected LOD score for a pedigree of this size and structure.19 A smaller region of 3.2 Mb at chromosome 11p15.4-pter, from the telomere to SNP_A-1791882 (base pair position chr11:1–3,164,265, hg18), was also associated with a maximum LOD score of 2.06. A representation of the SNP genotypes, a plot of the LOD scores, and a display of the genes in the regions are shown in Figure 2. Together these regions contained at least 204 annotated genes. Copy number evaluation of the SNP-derived data array revealed no shared deletions or duplications in the affected individuals.
Figure 2.
The SNP genotype, plot of the LOD score, and schematic display of the genes in the regions are shown for the two regions with maximal LOD score in the EP-200 family. Part (a) depicts the region at chromosome 4p16.1-p16.3, and part (b) depicts the region at chromosome 11p15.4-pter. SLC25A22 is circled in red. The SNPs displayed as red and blue represent homozygous genotypes, whereas those shown in green represent heterozygous genotypes. (The ideograms for chromosomes 4 and 11 are derived from the UCSC Genome Bioinformatics website, www.genome.ucsc.edu.)
Whole Exome Sequencing Results
Initial analysis from whole exome sequencing resulted in over 25,000 single nucleotide variants (SNVs) and small insertions or deletions (“indels”) across the genome. We narrowed our analysis to variants that could be considered “functional” or potentially pathogenic variants on the basis of predicted function (non-synonymous amino acid substitutions, premature stop codons, or interruption of canonical splice sites) and excluded variants that were present in dbSNP129 and dbSNP130 and variants that were homozygous in the whole exomes of any of 220 individuals from a group non-enriched for neuropsychiatric phenotypes and were also run at the Duke Center for Human Genome Variation. Given the autosomal recessive pattern of inheritance, we were interested chiefly in homozygous variants, which consisted of 163 SNVs and 46 indels. In the two regions for which we had evidence for linkage, 4p16.1-p16.3 and 11p15.4-pter, there were 8 SNVs predicted to produce amino acid substitutions (Table 2) and no indels. Details of exon coverage in these two regions are provided in Supplemental Figure 1. There were 113 amplicons in the two regions of homozygosity on chromosomes 4 and 11 that were not covered with more than one read in the exome data. Thus, we performed Sanger sequencing of these amplicons. After three attempts at Sanger sequencing, 86 amplicons were successfully sequenced, and there were no novel homozygous functional variants identified.
Table 2.
Eight novel homozygous single nucleotide variants (SNVs) in the regions of linkage in EP-201, an individual with MMPEI, are all non-synonymous coding variants.
| Gene Symbol |
Location of SNV | Observed SNV |
Amino acid variant |
Strong conservation |
Present in controls |
|---|---|---|---|---|---|
| SLC25A22 | chr11: 782,954 | c.G328C | p.G110R | yes | no* |
| GPR78 | chr4: 8,639,830 | c.G932A | p.R311Q | yes | yes* |
| LETM1 | chr4: 1,813,122 | c.G344A | p.R115H | no | no |
| TRPM5 | chr11: 2,398,165 | c.G512A | p.G171D | no | no |
| SLC37A2 | chr11: 124,452,606 | c.A286G | p.I96V | no | no |
| RNH1 | chr11: 485,007 | c.G1174A | p.A392T | no | no |
| PIWIL4 | chr11: 93,966,413 | c.G1108C | p.A370P | no | no |
| KRTAP5-5 | chr11:1,608,131 | c.C485A | p.P162H | no | no |
For these two genes, the presence of the variants was assessed not only in 220 non-neurologically affected individuals but also in 144 Middle Eastern unaffected individuals and 576 neurologically normal Caucasian control individuals.
Further Analysis of Variants Identified by Whole Exome Sequencing
Of the 8 homozygous SNVs in the regions with evidence for linkage in our MPSI pedigree, there was strong conservation across species and predicted disruption of the protein only for GPR78 and SLC25A22. Sanger sequencing confirmed that both EP-201 and EP-202 were homozygous for the observed SNVs, and the parents and siblings were all heterozygous.
GPR78 is a poorly characterized orphan G-protein coupled receptor. Sanger sequencing of 96 controls from the Middle East unrelated to the EP-200 family for the observed SNV in GPR78 revealed 2/96 individuals to be heterozygous for the R>Q variant. Based on this observation, if the homozygous p.R311Q variant were pathogenic, we would predict that MPSI caused by this mutation alone would be present in at least ~1/10,000 Middle Eastern children (1/4 × 1/48 × 1/48). Since MPSI appears to be far more rare than 1/10,000, we concluded that this variant was an as yet unannotated polymorphism without pathogenic significance.
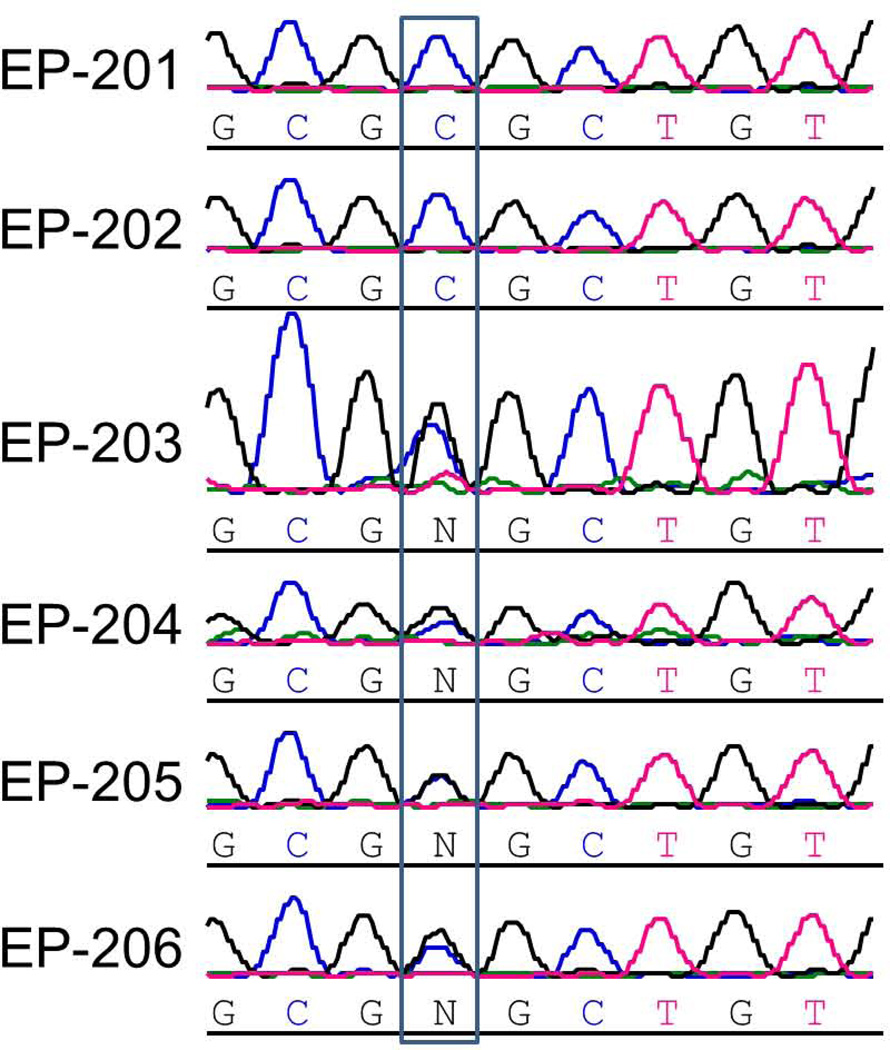
Using Sequenom genotyping, we evaluated the same 96 plus an additional 48 Middle Eastern controls as well as an additional 576 neurologically normal controls from NINDS for the SLC25A22 c.G328C, p.G110R variant. No control individuals possess this variant. We also evaluated SNP data from the NHLBI Exome Sequencing Project, in which 5400 exomes have been made publicly available, and our variant has not reported in this database [Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/), last accessed January 2013]. The SLC25A22 p.G110R change is predicted to affect a glycine residue that is conserved from humans to zebrafish (Fig 3). Mutating the glycine at position 110 is predicted to have a damaging effect using SIFT prediction software.
Figure 3.
(a) The chromatograms from Sanger sequencing confirmation of the homozygous c.G328C mutation are shown for EP-201 and EP-202. The unaffected siblings (EP-203 and E-204) and parents (EP-205 and EP-206) are all heterozygous, as seen by the two differently colored peaks and the denotation of “N”. (b) Amino acid conservation of two cerebral mitochondrial glutamate transporters, SLC25A22 and SLC25A18 (also known as GC1 and GC222), in humans and of SLC25A22 in several other species in the region of a novel SLC25A22 mutation associated with MPSI. The amino acid corresponding to the location of the p.G110R mutation is in bold.
We screened with Sanger sequencing all exons and exon-intron junctions of SLC25A22 in two siblings from another consanguineous pedigree with MPSI from Saudi Arabia that did not have evidence for linkage in the same regions as for EP-201 and EP-202 (linkage data not shown). We also screened two affected siblings from a non-consanguineous family from New Zealand, one of two affected siblings from a non-consanguineous family from Sweden, 12 sporadic cases of MPSI from Australia, and two sporadic cases from the U.S.; we did not find additional cases with a mutation in SLC25A22.
Localization of Wild-type and Mutant SLC25A22 in Cell Culture
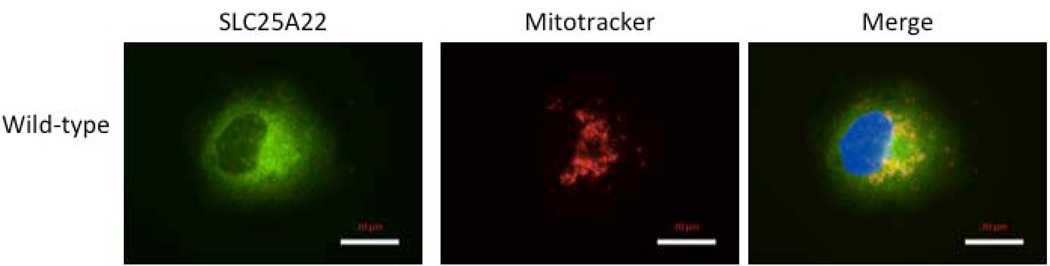
Overexpression of GFP-tagged WT SLC25A22 resulted in a punctate pattern of protein, consistent with mitochondrial localization evident by co-staining with a mitochondrial marker, as well as some widespread signal throughout the cytoplasm (Fig 4a). Overexpression of GFP-tagged mutant SLC25A22 containing the c.G328C variant resulted in a similar punctate pattern with a similar distribution. These data suggest that the mutation does not substantially alter the subcellular localization of the SLC25A22 protein.
Figure 4.
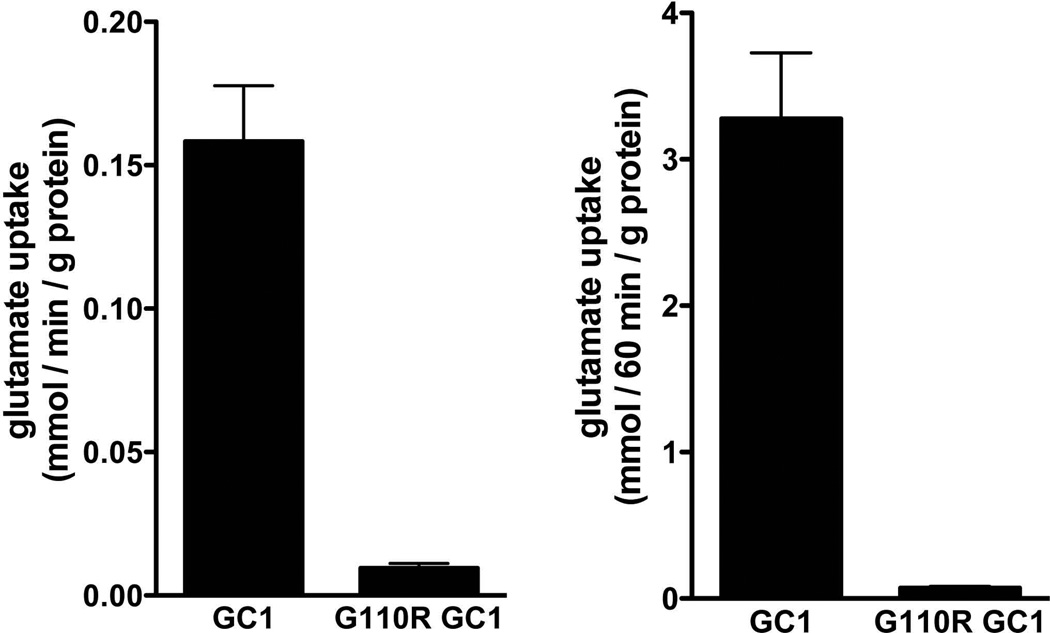
(a) Overexpression of GFP-tagged WT SLC25A22 in COS-7 cells resulted in a punctate pattern of GFP, consistent with mitochondrial localization (green=GFP, blue=DAPI, red=mitochondrial marker MitoTracker Red CMXRos). There is widespread signal throughout the cytoplasm, outlining the boundaries of the cell, seen in the SLC25A22 panel. The middle panel shows labeling of mitochondria, and some colocalization of SLC25A22 and mitochondria is evident as yellow in the third panel, labeled Merge. (b) Transport assays of wild-type and mutant SLC25A22. At time zero, 1 mM [14C]glutamate was added to liposomes reconstituted with WT SLC25A22 or the G110R mutant SLC25A22 and containing 10 mM glutamate. After 1 and 60 minutes’ incubation the uptake of the labeled substrate was terminated by addition of 30 mM pyridoxal 5′-phosphate and 10 mM bathophenanthroline. The data represent the means ± S.D. of four independent experiments performed in duplicate.
Functional analysis of the SLC25A22 G110R mutant
The conservation of the altered glycine residue across species suggests that its mutation might disrupt function of the protein. To assess the pathogenic potential of the c.G328C mutation, wild-type and G110R mutant SLC25A22 were overexpressed in E. coli, purified, and reconstituted into phospholipid vesicles (liposomes). The wild-type protein efficiently catalyzed glutamate/glutamate exchange (Fig 4b). In contrast, despite normal insertion of the mutant protein in the liposomal membrane, negligible transport activity was detected with the G110R mutant even after a long period of incubation (60 minutes). The percentage of [14C]glutamate transported by the G110R mutant GC1 into liposomes was 5.6 ± 1.1 and 2.2 ± 0.3 as compared to that transported by wild-type SLC25A22 after 1 and 60 minutes incubation, respectively, in four experiments. These findings are analogous to what has been shown to be the case with two previously reported epilepsy-related SLC25A22 mutations.20,21
Discussion
We identified a novel homozygous variant in the gene SLC25A22 (also known as GC1), which encodes a mitochondrial glutamate carrier14,22, in a family with two children with the severe epilepsy syndrome MPSI. We began with a classical Mendelian genetics approach to identify regions with evidence for linkage in which there were a large number of candidate genes. We progressed from the stage of linkage analysis to gene discovery using developments in massively parallel sequencing in the age of a well-characterized human genome with excellent annotation of observed variants and conservation across species. A growing list of gene discoveries has unfolded in this fashion23–26 and through exome sequencing of trios with severe epilepsy,27 and we expect the landscape of severe childhood epilepsies to be heavily influenced by this accelerating pace of gene discovery.
Until now, MPSI has been considered a sporadic disorder without familial occurrence.3 Our observation that MPSI may recur in families has important genetic counseling implications, especially as we are now beginning to identify a genetic differential diagnosis for this disorder. Only a few cases of MPSI have had an identifiable etiology, and the testing of SLC25A22 provides a starting point for potential genetic diagnosis in future, especially in cases likely to have a recessive basis due to consanguinity. While such early gene discoveries are not immediately available clinically, whole exome (or genome) sequencing is not far from clinical practice, particularly for patients with severe diagnoses such as intractable infantile onset epilepsy.
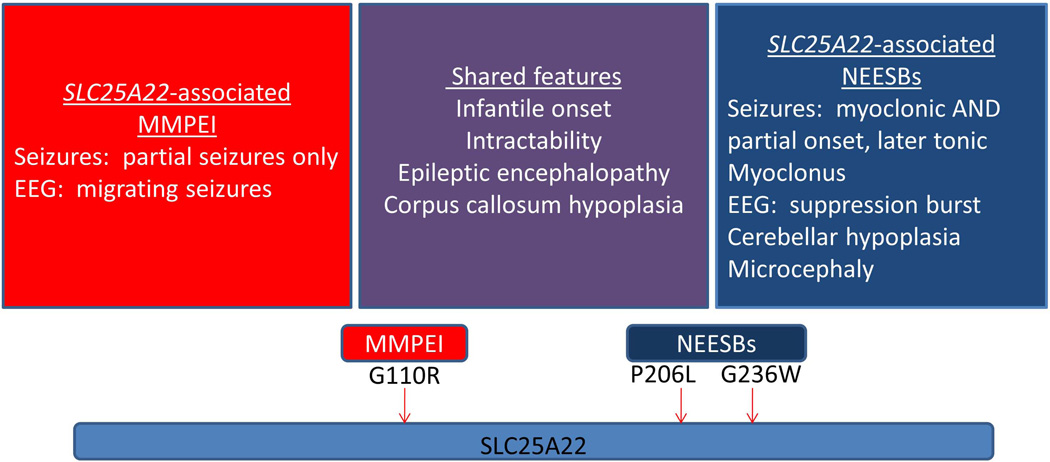
Though there are no prior reports of MPSI associated with mutations in SLC25A22, two distinct mutations in this gene have been associated with neonatal epileptic encephalopathy with suppression bursts (NEESBs) on electroencephalogram (EEG)20,21, a different form of early onset catastrophic epilepsy. Molinari and colleagues demonstrated that the mitochondrial glutamate transporter encoded by this gene is expressed in the developing human nervous system, notably in the hippocampus, cortex, and brainstem by the 15-week fetal stage.20 The homozygous missense mutations in SLC25A22 reported in associated with NEESBs result in the amino acid substitutions p.P206L and p.G236W, both of which are shown to alter the function of SLC25A22.20,21 The mutation we describe in association with MPSI results in the amino acid substitution p.G110R, affecting a different but also highly conserved portion of the mitochondrial glutamate + H+ carrier (Fig 5). Notably, all three of these mutated amino acids are highly conserved not only in the SLC25A22 (GC1) protein but also in the other important, paralogous mitochondrial glutamate transporter SLC25A18 (GC2) (Fig 3). The p.G110R residue is located in the third transmembrane helix of SLC25A22 in a highly conserved glycine-rich region that has been suggested to be involved in opening and closing the mitochondrial carriers on the cytosolic side14,28; the incorporation of a positive charge into the hydrophobic membrane environment would be predicted to be deleterious for the function of SLC25A22. Indeed, our functional studies provide strong evidence that the G110R mutation essentially abrogates the function of the glutamate transporter. Thus, though a few genes in the regions of linkage were not fully covered by our whole exome and Sanger sequencing, and we can not completely rule out the possibility that a synonymous variant affecting splicing was not recognized as such by the current methods of filtering variants, the SLC25A22 mutation provides an excellent explanation for MPSI in our pedigree.
Figure 5.
The chief clinical features of MPSI and NEESBs are shown. A schematic representation of the SLC25A22 glutamate transporter protein, with a single reported protein domain, is shown with the MPSI-associated p.G110R mutation and the two NEESBs-associated mutations, p.P206L and p.G236W20,21 indicated with arrows.
Although both MPSI and NEESBs are devastating disorders with refractory seizures and profound developmental outcome, there are fundamental semiological and electrophysiological differences that distinguish the two epilepsy syndromes, including age range of onset, seizure types, and EEG signature (Fig 5). Specifically, comparing the cases of SLC25A22-associated MPSI and NEESBs, they share corpus callosum hypoplasia, but we do not observe the cerebellar hypoplasia observed in the NEESBs cases in our MPSI cases.20,21 This is, of course, not the first time a gene initially reported to be associated with a well-defined epilepsy syndrome is eventually found to be associated with new and distinct forms of epilepsy. The classic example, mentioned in the Introduction, is that of SCN1A associated with both Dravet syndrome as well as Genetic Epilepsy with Febrile Seizures Plus (GEFS+) and recently with MPSI. Mutations in STXBP1 were described first as a cause of Ohtahara syndrome29 and have since been demonstrated to result in West syndrome and other early onset epileptic encephalopathies.30 Another recent example is that of TBC1D24, a gene associated with both focal epilepsy and intellectual disability31 and familial infantile myoclonic epilepsy.32 The precise phenotypic specificity used to identify families and cases with well-defined clinical syndromes is critical to initial gene discovery. That said, given the clinical heterogeneity already observed with mutations in SLC25A22 and many other genes, we must be able to place each individual observation in a broader context when appropriate. We would predict that mutations in SLC25A22 cause severe phenotypes, but as large cohorts of patients with epileptic encephalopathies and other epilepsy syndromes are sequenced in the coming years, we will be able to better delineate the extent of its phenotypic spectrum. Ultimately, the identification of the full range of phenotypes associated with SLC25A22 and other epilepsy genes may guide further studies of the precise role of this mitochondrial glutamate transporter in epileptogenesis and provide potential avenues of targeted treatment for epileptic encephalopathy more generally.
In summary, we have shown that MPSI can be inherited with an autosomal recessive pattern. Demonstrating the strength of a combined approach coupling linkage analysis in a single pedigree with MPSI with whole exome sequencing and a targeted analysis approach in the era of a well-annotated genome, we identified a novel homozygous mutation in the mitochondrial glutamate transporter SLC25A22. Multiple lines of evidence suggest strongly that SLC25A22 is the gene responsible for MPSI in our pedigree: (1) the high probability of linkage to our candidate loci (reflected by a maximal LOD score of 2.06 for the family), (2) the presence of the homozygous variant in SLC25A22 as the only homozygous novel variant in a gene in these loci that confers an amino acid change predicted to be damaging and that is not present in control individuals, (3) the association of other SLC25A22 mutations with a distinct but also severe infantile epileptic encephalopathy, and (4) functional evidence that the G110R mutation disrupts the normal function of the SLC25A22 protein. Additional evidence would be the presence of the same homozygous mutation in an additional pedigree, but given the rarity and heterogeneity of MPSI, such evidence may be unattainable for some time.
Our finding represents one of the early gene identifications for a neurological disease through the combination of traditional Mendelian genetics and massively parallel sequencing. As only one of the families and cases we studied had MPSI associated with SLC25A22, our work illustrates the etiological heterogeneity of this devastating disease. Furthermore, we have broadened the phenotype of malignant early onset epilepsies associated with SLC25A22. Future studies on additional cohorts of patients with early onset epileptic encephalopathies will be required to determine the full phenotypic range associated with this gene.
Supplementary Material
Acknowledgments
We thank the families of the children with MPSI who participated in this research study. Annapurna Poduri was supported by the NINDS (K23NS069784). Additional funding was provided by the Comitato Telethon Fondazione Onlus No. GGP11139 and the Italian Human ProteomeNet No. RBRN07BMCT_009 (Ferdinando Palmieri). The authors would like to extend their appreciation to the Deanship of Scientific Research at King Saud University, Riyadh, Saudi Arabia for funding of this research through the Research Group Project no 301 (Mustafa A. Salih). Christopher A. Walsh is an Investigator of the Howard Hughes Medical Institute and was also supported by the NINDS (R01NS035129, R01NS035129S), the NIMH (NIMH1RC2MH089952), the Manton Center for Orphan Disease Research, the Dubai Harvard Foundation for Medical Research, the Simons Foundation, and the Nancy Lurie Marks Family Foundation.
Additional funding was provided by Bryan ADRC NIA P30 AG028377.
DNA panels from the NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds) were used in this study. The submitters that contributed samples are acknowledged in detailed descriptions of each panel: NDPT019, NDPT022, NDPT023, NDPT024, NDPT079, and NDPT084.
We acknowledge the following colleagues for supplying control samples: J. Burke, L. Cirulli, V. Dixon, J. Hoover-Fong, C. Hulette, D. Lancet, K. Linney, W. Lowe, P. Lugar, D. Marchuk, J. McEvoy, J. Milner, A. Need, S. Palmer, K. Pelak, E. Pras, E. Ruzzo, V. Sashi, N. Sobriera, D. Valle, and Kathleen Welsh-Bohmer.
We thank Dr. Christian Schubert and Dr. David Goldstein for helpful discussions that shaped the project.
References
- 1.Coppola G, Plouin P, Chiron C, Robain O, Dulac O. Migrating partial seizures in infancy: a malignant disorder with developmental arrest. Epilepsia. 1995 Oct;36(10):1017–1024. doi: 10.1111/j.1528-1157.1995.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 2.Coppola G, Operto FF, Auricchio G, D'Amico A, Fortunato D, Pascotto A. Temporal lobe dual pathology in malignant migrating partial seizures in infancy. Epileptic Disord. 2007 Jun;9(2):145–148. doi: 10.1684/epd.2007.0106. [DOI] [PubMed] [Google Scholar]
- 3.Coppola G. Malignant migrating partial seizures in infancy: an epilepsy syndrome of unknown etiology. Epilepsia. 2009 May;50(Suppl 5):49–51. doi: 10.1111/j.1528-1167.2009.02121.x. [DOI] [PubMed] [Google Scholar]
- 4.Coppola G, Veggiotti P, Del Giudice EM, et al. Mutational scanning of potassium, sodium and chloride ion channels in malignant migrating partial seizures in infancy. Brain & development. 2006 Mar;28(2):76–79. doi: 10.1016/j.braindev.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Freilich ER, Jones JM, Gaillard WD, et al. Novel SCN1A mutation in a proband with malignant migrating partial seizures of infancy. Arch Neurol. 2011 May;68(5):665–671. doi: 10.1001/archneurol.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carranza Rojo D, Hamiwka L, McMahon JM, et al. De novo SCN1A mutations in migrating partial seizures of infancy. Neurology. 2011 Jul 26;77(4):380–383. doi: 10.1212/WNL.0b013e318227046d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poduri A, Chopra SS, Neilan EG, et al. Homozygous PLCB1 deletion associated with malignant migrating partial seizures in infancy. Epilepsia. 2012 Jun 12; doi: 10.1111/j.1528-1167.2012.03538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurian MA, Meyer E, Vassallo G, et al. Phospholipase C beta 1 deficiency is associated with early-onset epileptic encephalopathy. Brain. 2010 Oct;133(10):2964–2970. doi: 10.1093/brain/awq238. [DOI] [PubMed] [Google Scholar]
- 9.Barcia G, Fleming MR, Deligniere A, et al. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat Genet. 2012 Nov;44(11):1255–1259. doi: 10.1038/ng.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bedoyan JK, Kumar RA, Sudi J, et al. Duplication 16p11.2 in a child with infantile seizure disorder. Am J Med Genet A. 2010 Jun;152A(6):1567–1574. doi: 10.1002/ajmg.a.33415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gudbjartsson DF, Thorvaldsson T, Kong A, Gunnarsson G, Ingolfsdottir A. Allegro version 2. Nat Genet. 2005 Oct;37(10):1015–1016. doi: 10.1038/ng1005-1015. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009 Aug 15;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 14.Palmieri F. The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Aspects Med. 2013 Apr-Jun;34(2–3):465–484. doi: 10.1016/j.mam.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Fiermonte G, Paradies E, Todisco S, Marobbio CM, Palmieri F. A novel member of solute carrier family 25 (SLC25A42) is a transporter of coenzyme A and adenosine 3',5'-diphosphate in human mitochondria. J Biol Chem. 2009 Jul 3;284(27):18152–18159. doi: 10.1074/jbc.M109.014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Floyd S, Favre C, Lasorsa FM, et al. The insulin-like growth factor-I-mTOR signaling pathway induces the mitochondrial pyrimidine nucleotide carrier to promote cell growth. Mol Biol Cell. 2007 Sep;18(9):3545–3555. doi: 10.1091/mbc.E06-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmieri F, Klingenberg M. Direct methods for measuring metabolite transport and distribution in mitochondria. Methods Enzymol. 1979;56:279–301. doi: 10.1016/0076-6879(79)56029-7. [DOI] [PubMed] [Google Scholar]
- 18.Vozza A, Blanco E, Palmieri L, Palmieri F. Identification of the mitochondrial GTP/GDP transporter in Saccharomyces cerevisiae. J Biol Chem. 2004 May 14;279(20):20850–20857. doi: 10.1074/jbc.M313610200. [DOI] [PubMed] [Google Scholar]
- 19.Ott JB. Analysis of Human Genetic Linkage. 3rd ed. Baltimore, MD: Johns Hopkins University Press; 1999. [Google Scholar]
- 20.Molinari F, Raas-Rothschild A, Rio M, et al. Impaired mitochondrial glutamate transport in autosomal recessive neonatal myoclonic epilepsy. Am J Hum Genet. 2005 Feb;76(2):334–339. doi: 10.1086/427564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molinari F, Kaminska A, Fiermonte G, et al. Mutations in the mitochondrial glutamate carrier SLC25A22 in neonatal epileptic encephalopathy with suppression bursts. Clin Genet. 2009 Aug;76(2):188–194. doi: 10.1111/j.1399-0004.2009.01236.x. [DOI] [PubMed] [Google Scholar]
- 22.Fiermonte G, Palmieri L, Todisco S, Agrimi G, Palmieri F, Walker JE. Identification of the mitochondrial glutamate transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution of two human isoforms. J Biol Chem. 2002 May 31;277(22):19289–19294. doi: 10.1074/jbc.M201572200. [DOI] [PubMed] [Google Scholar]
- 23.Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010 Jan;42(1):30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholas AK, Khurshid M, Desir J, et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet. 2010 Nov;42(11):1010–1014. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu TW, Mochida GH, Tischfield DJ, et al. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet. 2010 Nov;42(11):1015–1020. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bilguvar K, Ozturk AK, Louvi A, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010 Sep 9;467(7312):207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Investigators EKaE. De novo mutation in the classic epileptic encephalopathies. Nature. 2013 (in press) [Google Scholar]
- 28.Palmieri F, Pierri CL. Structure and function of mitochondrial carriers - role of the transmembrane helix P and G residues in the gating and transport mechanism. FEBS Lett. 2010 May 3;584(9):1931–1939. doi: 10.1016/j.febslet.2009.10.063. [DOI] [PubMed] [Google Scholar]
- 29.Saitsu H, Kato M, Mizuguchi T, et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat Genet. 2008 Jun;40(6):782–788. doi: 10.1038/ng.150. [DOI] [PubMed] [Google Scholar]
- 30.Deprez L, Weckhuysen S, Holmgren P, et al. Clinical spectrum of early-onset epileptic encephalopathies associated with STXBP1 mutations. Neurology. 2010 Sep 28;75(13):1159–1165. doi: 10.1212/WNL.0b013e3181f4d7bf. [DOI] [PubMed] [Google Scholar]
- 31.Corbett MA, Bahlo M, Jolly L, et al. A focal epilepsy and intellectual disability syndrome is due to a mutation in TBC1D24. Am J Hum Genet. 2010 Sep 10;87(3):371–375. doi: 10.1016/j.ajhg.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falace A, Filipello F, La Padula V, et al. TBC1D24, an ARF6-interacting protein, is mutated in familial infantile myoclonic epilepsy. Am J Hum Genet. 2010 Sep 10;87(3):365–370. doi: 10.1016/j.ajhg.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.