Abstract
Researchers in the field of epigenomics are developing more nuanced understandings of biological complexity, and exploring the multiple pathways that lead to phenotypic expression. The concept of degeneracy—referring to the multiple pathways that a system recruits to achieve functional plasticity—is an important conceptual accompaniment to the growing body of knowledge in epigenomics. Distinct from degradation, redundancy and dilapidation; degeneracy refers to the plasticity of traits whose function overlaps in some environments, but diverges in others. While a redundant system is composed of repeated identical elements performing the same function, a degenerate system is composed of different elements performing similar or overlapping functions. Here, we describe the degenerate structure of gene regulatory systems from the basic genetic code to flexible epigenomic modifications, and discuss how these structural features have contributed to organism complexity, robustness, plasticity and evolvability.
Keywords: epigenetic code, pluripotentiality, robustness, redundancy, DNA methylation, histone modifications, social insect, honey bee
INTRODUCTION
The ability of natural selection to give rise to a large number of nonidentical structures capable of producing similar functions appears to increase both the robustness of biological networks and their adaptability to unforeseen environments by providing them with a large repertoire of alternative functional interactions. Tononi and Edelman [1].
Biological systems can do a lot with a little. Consider, for example, the number of protein-coding genes in various organisms with distinct evolutionary histories. The human genome has to provide developmental cues for generating distinct tissues, organs, hundreds of cell types, and for building and wiring a centralized brain with 100 billion neurons and 100 trillion synaptic connections. And yet the human genome encodes nearly the same number of genes as does the genome of Caenorhabditis elegans, a tiny eutelic nematode with only 959 cells, including just 302 neurons located in dispersed ganglia [2]. Data overwhelmingly indicate there is no simple relationship between gene number, neuron number and evident morphological and behavioural complexities of animals in different phyla [3]. Indeed, organisms with highly advanced brains and complex behaviours, such as primates, have fewer genes than the water flea Daphnia pulex (22 000 and 31 000, respectively). Consequently formulating a genomic interpretation of the development and evolution of new biological forms is proving far more difficult than previously anticipated.
Organisms can employ many different molecular systems to achieve the same end result, and many different designs, such as diverse gastrulation patterns, can be manufactured during embryogenesis to solve the same developmental problem. Frequently, to evolve new morphological or behavioural forms evolution has had to reuse and adapt existing elements by utilizing the inherited genomic sequences in a new context-dependent manner.
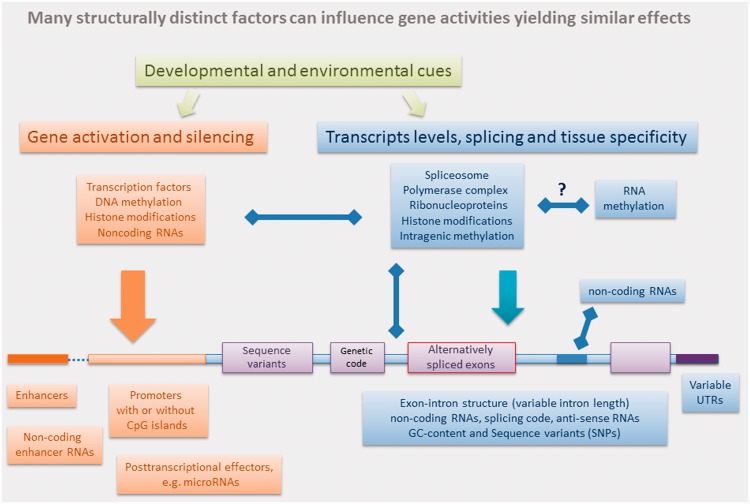
A multi-level regulatory network consisting of such mechanisms as modular utilization of protein domains, alternative splicing and epigenomic modifications of DNA has been the driving force behind the wide radiation, rapid evolution and evolutionary success of eukaryotic organisms. Here, we argue that the degenerate organization of the genome and epigenome is a key feature enabling the evolutionary process to create new forms. A degenerate system contains multiple structurally distinct elements performing similar functions (Box I and II; Figure 1). Ordinarily degeneracy promotes stability in a self-organizing system, but degeneracy also allows elements to functionally diverge by an evolutionary process, and become exapted to a new function without any loss of coherency to the original system.
Box II.
| Structure | Function | Context | |
|---|---|---|---|
| (Unspecified) | Many | Many | Independent |
| Redundancy | One | One | Independent |
| Degeneracy | Many | One | Dependent |
| Pluripotency | One | Many | Dependent |
Redundant components have a structure–function ratio of one-to-one irrespective of context. Degenerate components have a structure-to-function ratio of many-to-one. Pluripotential elements change function according to context.
Key points.
Epigenetic mechanisms operate by utilizing many structurally distinct elements performing similar functions, a principle known as degeneracy.
Degeneracy is a key organizational feature of both the genome and its flexible modifier, the epigenome.
Degeneracy is an important factor in the evolution of new phenotypes; it greatly increases the capacity of a limited and fixed number of genes to generate organismal and behavioural complexities.
Conceptualizing the epigenetic system as degenerate is beneficial for mapping the complex, dynamic, self-organizing pathways of biological activity.
Box I.
| Definitions | |
|---|---|
| Isomorphic | Structures that are identical. For example, the special molds at the Billund factory ensure that each 2 × 2 Lego brick (Design ID 3003) is isomorphic |
| Isofunctional | Performing the identical function. For example, a pen, a pencil and a quill can all perform the same function |
| Heteromorphic | Structurally different elements. For example, a pen and a quill are heteromorphic |
| Degeneracy | The structural variation that underpins functional plasticity. For example, pens and pencils can perform the same function with respect to context. On earth, both pens and pencils can be used to write. Without structural modification, however, only pencils work in space |
Figure 1:
A simplified diagram highlighting the structural complexity of gene regulatory elements in eukaryotic organisms. Left-right arrows indicate bidirectional communications, feed-back and feed-forward loops. For more details, see the main text and selected recent articles [31–36,45,46,54–65].
Degeneracy greatly increases the capacity of a limited and fixed number of genes to generate morphological and behavioural complexities. Here, we review the history of the concept of biological degeneracy, and discuss the importance of degenerate organization for the function and evolution of the genome and epigenome.
BIOLOGICAL DEGENERACY
Degeneracy is an unfortunate word for a useful concept. In contemporary biological science, degeneracy refers to structural variation underlying functional plasticity. Systems with degenerate components have a structure-to-function ratio of many-to-one (Box I and II). Degeneracy is a distributed property of complex adaptive systems that in many circles of science has been hidden in plain sight [4], commonly overlooked because of a reductionist bias [5,6], and ignored because the term itself is misleading [7]. Although degeneracy is known to be a characteristic of genetic codes [8,9], immune systems [10], respiratory network regulation of blood-gas homoeostasis [11], human movement [12,13], cognitive neuroanatomy [14–16], population dynamics [17,18], and as a conceptual tool offered the final solution to the coding problem of DNA [19], the term is still not well comprehended in evolutionary biology.
Commonly, structurally different but functionally similar degenerate components are often mislabelled as redundant, a term that actually refers to identical elements performing the same function (Box I and II). The biological concept of degeneracy suffers from association with a value-laden use outside of the sciences that has gained a host of negative associations [7,20,21].
A living system exhibits degeneracy if it contains multiple different structures that can perform a similar function. The ability to perform the same tasks by different mechanisms prevents unbearable fluctuations and the propagation of cascading failures in a system [22]. A dynamic self-organizing system must also strike a compromise between the over-stabilization of networks and the noise within and between various networks, as too much specificity reduces adaptability [5], whereas some reduced specialization provides a capacity for plasticity and adaptability. It is very common in biological systems for molecules or cells to recognize a range of targets, and for target ranges to partially overlap between different elements. These molecules and cells are called degenerate. Degeneracy operates at multiple levels of complexity. Dissimilar genes can produce the same developmental output; unrelated populations of neurons can subserve the same behavioural task; and different patterns of muscular contraction can yield similar movements. In biological systems, the loss of a component can be compensated by redundant elements (the presence of other identical components) or by degenerate elements (structurally different components that perform the same function with respect to context).
Degeneracy is a phenomenon whereby different structural permutations recurrently lead to similar end results, but with changes in context, the elements of a system might change function in which case they are described as pluripotential with a structure-to-function ratio of one-to-many (Box I and II). Pluripotentiality is the complement of degeneracy [16]. In the immune system, for instance, degeneracy of antigen receptors enables any single epitope to activate many different lymphocyte clones, and simultaneously any single lymphocyte clone is able to recognize many different epitopes [23]. Tieri et al. [24], borrowing a term from Csete and Doyle [25], refer to the overlap between degeneracy and pluripotentiality as a bowtie. Many inputs funnel into a thin knot of interlocking networks and subsequently many corresponding outputs fan out. The prime example of a bowtie is the transcription and translation of DNA to proteins. A large variety of genes produce a few universal polymerase modules—the ‘knot’ of the bowtie—and a large variety of proteins result [26].
Degeneracy is not limited to the internal structures of an organism, but may also occur between internal structures and environmental resources. Deacon [27] gives the example of endogenous ascorbic acid synthesis (vitamin C) existent among some primate lineages and missing in others. All prosimians except Tarsiers synthesize ascorbic acid endogenously but anthropoid primates have lost this function. A shift in diet among anthropoid ancestors has led to a reliance on acquiring ascorbic acid from dietary sources such as fruit. Once food sources containing ascorbic acid were available in reliable and plentiful quantities, the gene responsible for endogenous ascorbic acid synthesis was no longer needed, became selectively neutral, and was free to accumulate mutations without deleterious outcomes for the organism. Mutational variants were no longer eliminated because exogenous ascorbic acid became regularly available. Selection began to operate not simply on genes for ascorbic acid synthesis but also across a distributed network of sensory biases, behavioural inclinations and digestive-metabolic mechanisms that increased the probability of obtaining ascorbic acid from the environment. In this way, within certain degrees of freedom, if there is degeneracy between environmental and genomic factors, then selection can result in an offloading of function from the genome to the environment, or a potential divergence of the environmental and genomic elements leading to the random exploration of adjacent function space.
DEGENERACY IN THE GENOME AND EPIGENOME
Degeneracy is a key organizational feature of our genetic code [8,9]. All but two amino acids are encoded by more than one triplet codon, with each set of codons specific only for one amino acid. In total, there are 64 different codon combinations or ‘ciphers’ in the degenerate genetic code for just 23 amino acids. This evolutionary invention provides several adaptive benefits. For example, bacteria can adapt protein synthesis to a limited availability of certain amino acids, by taking advantage of ‘degeneracy lifting’, a process that allows degenerate systems to display a variety of behaviours, depending on environmental settings. Nutritional perturbations lift the degeneracy of the genetic code by splitting codon families into robust and sensitive synonymous codons that results in 100-fold higher rate of protein synthesis associated with robust codons [9]. However, as the structural complexity of biological systems increases and connectivity of multiple parts becomes non-linear, new molecular strategies are needed to differentially utilize the genome in distinct cell lines of eukaryotic organisms, to coordinate development and cellular identity and to structure and stabilize genome–environment interactions.
Progress in epigenomics has shown that the genome is regulated by a multifaceted and highly degenerate system involving families of transcription factors interacting with various chemical modifications of genomic DNA or its packaging proteins (Figure 1). While these do not alter the underlying genetic code, they do establish regulatory marks along the genome that operate with general transcription factors to coordinate structured gene expression. The epigenetic system is a self-organizing regulatory level of organization that operates above the genome and provides the high level of flexibility needed for coordinated and context-dependent expression of multitudes of genes.
The modern era of epigenetics originated with the seminal works of Conrad Waddington who in 1939 introduced the concept of ‘epigenotype’, a quality whose mode of impact was over the classical genotype [28]. Waddington used this descriptor in the context of developmental canalization to explain how a particular organ is produced by both the genotype and the epigenotype reacting with the external environment. Epigenetic research moved from a genetics-based, to a methylation-based, to a CpG island-based, and more recently to genome-wide methylomics, initiated by the seminal articles of Riggs and associates [29,30]. One often used definition of epigenetics is presented by these authors as ‘the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence’ [30].
One mechanism for epigenetic inheritance is via cytosine methylation whereby gene expression can be modulated by adding or removing methyl groups to the DNA. This process is generally set as part of the normal genetic program, but recent studies have shown that methyl tags can be reset by a variety of factors including viral infection, drug treatment or even simple components of a regular diet [31–36]. Another important epigenetic mechanism operates at the level of chromatin via histone modifications [36,37]. However, at least theoretically, the transmission of epigenetic information might be mediated also by other cellular macromolecules including RNA, proteins and sugars or even lipids and cellular membranes [36]. Experimental evidence suggests that some of these DNA and histone modifications are responsive to both internal and external cues and can be rapidly activated in a context-dependent manner [31–38]. Acting alone or in combination, epigenomic modifications have both short-term and long-term effects on gene regulatory cascades, on DNA replication and chromatin conformation. This vast array of biochemical decorations of genomic sequences acts as a highly flexible epigenetic code that has the capacity to regulate not only individual genes but also entire networks. Adjusting topology of cellular networks by genome-wide reprogramming is a way of re-interpreting genetic hardware to generate contextual epigenetic states. Some of the epigenomic modifications underlying a given epigenetic signature become stable and can be maintained during the life of an organism as a durable phenotype. However, an environmental shift can easily enforce a new round of programming leading to a different more suitable phenotypic outcome [31–38]. In addition to plants and animals, a similar interplay of environmental and epigenetic contexts has also been reported in more elementary pathways, such as the cell autonomous DNA damage response in yeast [39].
Conceptualizing the epigenetic system as degenerate is useful for mapping the complex, dynamic, self-organizing pathways of biological activity. Epigenetic systems exhibit degeneracy since several distinct mechanisms used to control gene regulation can yield similar end results. For example, genes can be turned on and off by the action of transcription factors or by promoter methylation (or chromatin configuration) [40–44]. Alternative splicing is controlled by multifactorial cellular machinery including DNA methylation and histone positioning, acting together or as individual epigenomic modifiers [45–47]. Oxidation of 5-methyl cytosine (5Mc) to 5-hydroxy mC (5hmC) is considered the main pathway in removing the methyl tags from the genome, but in the brain increased levels of 5hmC in gene bodies have been shown to correlate with transcriptional activation [48]. Within the neuronal function-related genes, gain of 5hmC is accompanied by loss of H3K27me3, an important histone mark that is catalysed and maintained by Polycomb Repressive Complex 2 (a histone methyl-transferase, [48]). Although H3K27me3 has been implicated in transcriptional silencing, its localization either in gene bodies or around transcription start sites suggests that its mode of action is degenerate with other epigenetic elements [49]. More evidence suggesting that the multifactorial epigenetic code is degenerate emerges from high-throughput epigenomic data. By integrating genome-wide profiles for a histone mark (H3K36me3) with two sets of transcriptional RNAseq data, Althammer et al. [50] found that epigenetic regulation of gene activities involves multiple intragenic regions, specifically the first exon, first intron and a region downstream of the polyadenylation site. Furthermore, the histone modifications appear to be highly heterogeneous and their regulatory effects depend on the structural features of promoters and genes (GC-content and presence of introns). Their results clearly show that the histone code is highly degenerate with different groups of attributes from different genic regions leading to similar regulatory events.
Another striking example of gene activity regulation via a degenerate epigenomic mechanism is silencing of the ribosomal RNA (rRNA) gene cluster by non-coding RNA [51]. In this model, an RNA molecule, complementary to the rRNA promoter mediates de novo CpG methylation of rRNA genes by interacting with the transcription factor TTF-1 and forming a DNA:RNA triplex, which recruits the DNA methyltransferase DNMT3b. Thus, RNA-dependent DNA methylation acting together with histone modifications might be required to selectively shut down a subset of rRNA genes by forming heterochromatic states that have been shown to be associated with inactive rRNA genes.
At least some of these epigenomic regulatory events appear to be bidirectional. De Almeida et al. [52] have recently shown that gene splicing controlled by trimethylation of histone H3 Lys3 can influence the methylation of H3 Lys3 by recruiting, via an unknown mechanism, methyltransferase HYPB. This process, linked to genes’ structural features (first intron length), not only actively modulates histone H3 Lys 3 modifications but also improves quality of intron splicing. Although introns are demarcated by short sequence motifs, this ‘splicing code’ is poorly conserved and lacks adequate information to recognize correct splicing sites. The emerging view is that a proper choice of splicing events is regulated by a coordinated action of many components including RNA polymerase II, ribonucleoprotein particles, hundreds of auxiliary proteins, chromatin factors, DNA methylation and histone modifications [46,52].
Elaborate manners of regulation, such as interacting DNA-methylation and histone modification systems, are likely to be the hallmarks of the epigenetic code. The combinatorial utilization of flexible epigenomic modifications, together with gene splicing, post-translation protein modification and RNA editing has ample potential to generate functional diversity from a fixed genotype (Figure 1). In contrast to only 64 codon combinations in the genetic code, the number of possible ‘ciphers’ in the epigenetic code or the level of degeneracy could be many orders of magnitude higher [53]. This high level of degeneracy provides virtually unlimited coding potential to ensure both developmental buffering and flexibility to deal with random external factors.
DEGENERACY PROVIDES BOTH STABILITY AND PLASTICITY TO THE EPIGENOME
The idea that epigenetic mechanisms can generate similar phenotypic outcomes by adjusting expression of different genes is not new. Seventy years ago, Waddington argued that epigenetic processes are part of developmental canalization, or buffering of the genotype against external perturbation [28]. He noted that developmental reactions ‘are adjusted to bring about one definite end result regardless of minor conditions, hence the remarkable constancy of the wild type’ [28]. Although Waddington has never used the term degeneracy, his pioneering ideas brought into focus the flexibility of cellular responses to external stimuli and their ability to select ‘a suitable genetically controlled reactivity in the organism’. He illustrated his concept of genotype buffering using the phenomenon of ‘phenocopying’, whereby the phenotypic effect of a mutation can also be created by non-genetic means. For example, in Drosophila, phenocopies for both pigmentation and behaviour of a mutation known as ‘yellow’ can be produced by a number of treatments, including pharmacological inhibitors of tyrosine hydroxylase or by growing larvae on silver nitrate [66,67]. Another notable example of phenotypic buffering against the effect of mutations is the combined action of environmental stress and stochastic expression of protective genes encoding chaperone proteins [68]. Molecular chaperones or heat-shock proteins are considered important drivers of evolutionary change because of their role in folding or assembling macromolecular structures that is highly dependent on receiving specific internal or external signals [69]. In C. elegans, a potentially lethal outcome of a mutation in the transcription factor lin-29 can be significantly weakened after a mild heat shock delivered during early stages of development [69]. This protective effect of stress has been correlated with higher stochastic chaperone expression. However, lower reproductive fitness associated with higher abundance of chaperones suggests that at the whole population level, inter-individual variability in the expression of these proteins provides a better survival strategy in erratic environmental conditions. Encapsulating the heterogeneous construction of phenotypic buffering, the concept of degeneracy highlights the array of pathways that can lead to a similar phenotypic endpoint.
DEGENERACY OF THE EPIGENOME, PHENOTYPIC POLYMORPHISM AND EVOLVABILITY
Permutations of structure that recurrently lead to the same end-point provide a selective repertoire of flexible mechanisms with variable specificities. With these degenerate mechanisms of organization, selection is able to act differentially upon two or more structurally distinct traits that can fulfil the same role within specific contexts. Selective systems have a great number of elements that each respond at varying specificities to incoming signals [70]. In other words, environmental influences can act upon specific elements as well as populations of ‘degenerately responding, possibly less specific, elements’ [71].
Degeneracy means that natural selection acts not only in evolution but also in ontogenesis by sorting stochastic degenerate interactions at multiple levels of complexity [72]. Mathematical modelling has shown that competition for resources by degenerate repertoires is a mechanism of self-organization that can also explain evolutionary branching [73] and possibly even speciation [18]. Degeneracy can account for the divergence of traits into subpopulations and the spontaneous self-structuring of degenerate repertoires into distributions inconsistent with the available resources in the environment [70,71]. In many ways, the inclusion of degeneracy in biological theory answers the call of West-Eberhard to ‘change the way biologists think about the origins of organic diversity’ [74].
New directions in the evolution of an organism start with a population of variably responsive, developmentally plastic mechanisms [75]. As a mechanism of intraspecific variation, degeneracy contributes to evolvability through processes that are not immediately adaptive. As a result of competition for resources, the interaction of degenerate repertoires with selective constraints gives rise to self-organization [70]. Mathematical modelling has revealed that a population of degenerate systems spontaneously manifest self-organization patterns that do not mirror incoming signals or the distribution of resources [70,71]. In other words, degeneracy offers an alternative to adaptation in explaining the features of an organism. Not every trait is an adaptation to the environment that it currently inhabits, or put another way, not all traits were necessarily selected for their current function. Incorporating degeneracy into the study of biological systems brings attention to the many ways that variation can arise while maintaining organism function.
Degenerate epigenomic regulatory systems can diverge through accumulation of random mutation without compromising the phenotype of the organism. This makes possible the evolution of new genome regulatory pathways and potentially new developmental programs that utilize existing coding sequences in new combinations. Unlikely as it may sound growing evidence suggests that such changes more frequently underlie the evolution of new behavioural or morphological structures than the evolution of ‘new’ coding genes.
A particularly pertinent example is the evolution of the human brain. Humans and chimpanzees have almost identical genomes and their brains are anatomically very similar, except that human brain is about three times larger than that of chimpanzees [76]. This significant volumetric expansion of the hominid brain began only 2 million years ago and posed a major challenge for rapid development of new regulatory circuits. It has been argued that a few genomic regions called human accelerated regions (HARs) contain clues to the enhanced changes associated with the evolution of human brains. HAR1, in particular, has been suggested to be important for brain evolution [76]. HAR1 is highly evolutionarily conserved among mammals, but has diverged rapidly in humans since the last common ancestor with chimpanzees [76]. Although HAR1 is essentially deprived of protein coding genes, it contains the so-called non-coding RNA genes including HAR1F that is expressed in the mammalian neocortex and is now a candidate for being part of a process that brought about innovative structural modifications in the human brain. Another interesting example of an RNA-coding gene implicated in human evolution is mir941; a micro RNA that emerged de novo in the human lineage from an unstable tandem repeat DNA sequence between 6 and 1 million years ago. mir941 has been implicated in reorganizing gene regulatory networks controlling cellular differentiation and neurotransmitter signalling [77].
Interestingly, analogous results are emerging from studies in invertebrates. The brain architecture of social honey bees (Apis mellifera) and solitary flies (e.g. Drosophilidae) is virtually identical, but the bees have approximately 10 times more neurons [78,79] in the cephalic ganglia, a richer behavioural repertoire and a unique symbolic ‘language’. Like in humans, the evolutionary behavioural novelties in the honey bees are not associated with an increased gene number. In comparison with other animal species, insects have a relatively small number of genes, ranging from ∼15 000 to 17 000 [2]. Furthermore, non-coding honey bee genes have also been found to be important for brain functions. The expression of a transcription factor-like RNA-coding gene kakusei is associated with an increased neuronal activity in a subset of neurons in brains of foraging bees communicating the location of food resources via the dance ‘language’ [80]. Another non-coding gene Nb-1 has been implicated in modulating octopamine and juvenile hormone release during a worker bee behavioural transition from nursing to foraging [81]. One implication of these findings is that the existing protein and/or gene networks can be rewired by new epigenetic regulatory circuits to generate new forms of communication and social behaviour without inventing novel protein-coding genes. Instead, fast evolving RNA molecules of various types and sizes appear to control the differential recruitment of a hierarchy of general chromatin and DNA modifying complexes to specific loci during differentiation and development. This multilayer process creates multiple functional versions of the same genome, or epigenomes that have that capacity to interpret the genomic information in a context-dependent manner.
The evolution of new developmental programs through changes in epigenetic regulation has enabled the evolution of the highly complex and morphologically specialized social insects. Diagnostic of the advanced social insects is the occurrence in a colony of different morphs specialized for reproductive roles and non-reproductive colony support roles. In the eusocial hymenoptera (e.g. ants, bees and wasps) these are the queen and worker castes. New evidence is revealing how these different phenotypes result from different patterns of epigenetic regulation of the genome. In this way, the action of selection on degenerate epigenetic systems has yielded a whole new level of biological organization that currently dominates most terrestrial ecosystems.
Two phenotypically distinct female honey bees (A. mellifera), queens and workers, are encoded by one genome whose mode of expression can be conditionally modulated by nutritional input [82–85]. Feeding a complex diet known as royal jelly to a growing female larva inhibits global DNA methylation, increases levels of juvenile hormone and correlates with changes in gene expression, which result in the queen phenotype. In contrast, larvae fed less-nutritious worker jelly develop into functionally sterile short-lived worker bees. However, there are no specific ‘queen’ or ‘worker’ genes in the Apis genome. During the initial critical 96 h of larval growth, multiple sensory and secretory systems are involved in receiving, processing and conveying the nutritional information to multilevel, interlocked signaling pathways. The contrasting phenotypes result from threshold-based processes driven by metabolic fluxes, hormonal changes and differential methylation and expression of many genes [83]. All these components have the capacity to respond to environmental change, but their combined and coordinated action has evolved in honey bees as a powerful mechanism for reprograming the entire developmental trajectory with profound consequences for cellular and organismal phenotypes [82–85].
The implications of the honey bee findings are 2-fold. First, they illustrate a candid point that gene products are mere parameters in networks and it is network fluxes and network equilibria that need to be understood [86–88]. There is unlikely to be a single major hub to which a given complex phenotype can be attributed, and hence why the linear explanations of genome-to-phenotype correlations have largely failed [86,87]. Indeed, network modelling of putative gene interactions based on the occurrence of overrepresented motifs in the upstream control regions and differentially expressed genes (DEGs) shows that worker’s network is more interconnected than queen’s network. This suggests that the worker DEGs share more conserved cis-elements when compared with queen DEGs [85].
Second, a fixed number of genes can be epigenetically programmed to yield more than one organismal outcome, suggesting that epigenomic modifiers have the capacity to relax evolutionary constrains on development. These modifiers operate by recruiting only a subset of an organism’s gene repertoire and reusing it in a combinatorial manner. Differential epigenomic modifications of over 2000 genes lead to remodelling multiple sub-networks [83]. Interestingly, the impetus for this process comes from metabolic flux and is driven by the most conserved organic molecules: the metabolic enzymes. Changes in DNA methylation and post-translational alteration in chromatin complexes, lead to both silencing and activation of genes, and result in altered expression profiles at the transcriptomic, proteomic and metabolite levels. The relative contribution of these changes to alternate developmental trajectories will require additional molecular and cell biological data.
CONCLUSION: DEGENERACY AS A DESIGN PRINCIPLE FOR GENOME FUNCTION
Technological innovation is no longer a limiting factor in biomedical sciences. The speed and decreasing cost of second generation DNA sequencing is producing an unparalleled amount of raw data that is expected to drive major breakthroughs in both basic and applied research. The challenge is no longer in generating more data, but in converting them into knowledge. To avoid being distracted by terabases of sequencing reads and their perceived ‘functionality’ we need new conceptual frameworks to fully comprehend the complexity of biology behind the genomic hardware.
In this article, we have argued that degeneracy is a central organizational feature of genomes and epigenomes. Epigenomic systems in particular consist of multiple non-identical elements with partially overlapping functional features. We are certainly not the first to recognize the importance of degenerate organization in the understanding of structure–function relations. Edelman argued that ‘… degeneracy implies a certain relaxation of constraint during development, opening certain evolutionary possibilities in a given set of phenotypes’ and provides ‘the necessary leeway for evolutionary changes that would otherwise lead to blind ends or lethality’ [89]. Here, we have gathered new and emerging examples from epigenetics that show how degeneracy contributes to the stabilization of phenotypes, and can allow the same functional phenotypic outcome to be reached by very different specific mechanisms. These mechanisms are not limited to purely genomic programs, but can involve interactions between the genome, epigenome and environment.
We have also argued that degeneracy has been a critical and overlooked factor in the evolution of new phenotypes. Elements of a degenerate system are free to functionally diverge over evolutionary time without compromising the existing functional output. New random variants that are not immediately selectively advantageous can be maintained in degenerate systems, and through progressive accumulation of new variations interacting with plastic exploration of existing gene regulatory mechanisms new functional pathways and phenotypes can emerge. We have argued that these processes could be a driving force in the evolution of new forms of physiology and behaviour. Epigenomic influences on global regulatory networks in honeys bees in particular have already brought a fresh perspective to the study of epigenetic regulation of development and behaviour [34,35,90–92], and degeneracy may operate far more broadly in facilitating the evolution of new levels of complexity. Deacon [27], for example, offered a sophisticated epistemological approach to understand the role of global degeneracy in the evolution of communication generally. Such ideas are a reminder that the genetic code is not prescriptive and that biological research has to be context-driven. Complementary organisms such as birds [27] and honey bees offer useful systems in which we can test ecologically valid hypotheses about epigenetic morphodynamic processes. Using honey bees and other social insects to study relaxed selection, global degeneracy and social communication is already revealing how radical new forms of social behaviour and communication have evolved [93–95], and could deliver a fatal sting to the idea that adaptations arise through saltatory events or because they are immediately beneficial to a species.
The concept of degeneracy is an important theoretical tool for unpacking the heterogeneous construction of phenotypes as well as the multifarious variable intersecting pathways that compose biological systems. The reiterations of living systems are developmentally constructed and come into being through internal interactions within organisms, interactions between organisms, and interactions between organisms and their surroundings. Novelty and variation arise from these interactions. The functional redistribution of biological activity onto a group of interacting organisms and their environment effectively offloads a degree of genetic control onto epigenetic processes. Degeneracy introduces a susceptibility to developmental influences and enables epigenetic adaptation to the environment. In other words, degeneracy opens a developing organism up to variable causal factors, environmental modification and historical contingency. The availability and recruitment of functional extrasomatic resources creates the possibility for developmental adaptation to the environment without severely compromising the integrity of the system [75]. In this situation, developmental information is distributed throughout an array of internal and external components that each fractionally influences the ongoing dynamics of the system. Degeneracy is not a reductionist account of self-organizing, dynamical and living systems. By understanding biological systems as degenerate, researchers can develop a framework aimed at modelling the coevolutionary and codevelopmental relationship between organisms and their environment.
Acknowledgements
We thank Sylvain Foret for his remarks on an early version of the manuscript and two anonymous reviewers for their constructive comments.
Biographies
Ryszard Maleszka is a professor in the Research School of Biology, ANU. He is spearheading a research theme called ‘From Molecules to Behaviour’ that uses invertebrate model systems to study the genotype to phenotype link.
Paul H. Mason has published in cultural anthropology and biological science. With fieldwork experience in Indonesia, Brazil and India, he is currently investigating the spread of Tuberculosis in developing countries.
Andrew B. Barron is a lecturer in the School of Biological Sciences, Macquarie University. His research examines the mechanisms and evolution of animal cognition.
FUNDING
R.M. is supported by grants from the Australian Research Council (DP1092706) and National Health and Medical Research Council (APP1050593). R.M. and A.B.B. are supported by the ARC grant DP12010180.
References
- 1.Tononi G, Edelman GM. A Universe of Consciousness: How Matter Becomes Imagination. New York: Basic Books; 2000. [Google Scholar]
- 2.Maleszka R. Elucidating the path from genotype to behaviour in honey bees: insights from epigenomics. In: Eisenhardt D, Galizia G, Giurfa M, editors. Honeybee Neurobiology and Behavior: A Tribute to Randolf Menzel. Amsterdam: Springer Publisher; 2012. pp. 373–86. [Google Scholar]
- 3.Miklos GL, Maleszka R. Deus ex genomix. Nat Neurosci. 2000;3:424–5. doi: 10.1038/74786. [DOI] [PubMed] [Google Scholar]
- 4.Edelman GM, Gally JA. Degeneracy and complexity in biological systems. Proc Natl Acad Sci USA. 2001;98:13763–8. doi: 10.1073/pnas.231499798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atamas SP. Les affinités electives. Pour Sci. 2005;46:39–43. [Google Scholar]
- 6.Whitacre J. Degeneracy: a link between evolvability, robustness complexity in biological systems. Theor Biol Med Model. 2010;7:6. doi: 10.1186/1742-4682-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason PH. Degeneracy at multiple levels of complexity. Biol Theory: Integr Dev Evol Cogn. 2010;5:277–88. [Google Scholar]
- 8.Luo LF. The degeneracy rule of genetic code. Orig Life Evol Biosph. 1988;18:65–70. doi: 10.1007/BF01808781. [DOI] [PubMed] [Google Scholar]
- 9.Subramaniam AR, Pan T, Cluzel P. Environmental perturbations lift the degeneracy of the genetic code to regulate protein levels in bacteria. Proc Natl Acad Sci USA. 2013;110:2419–24. doi: 10.1073/pnas.1211077110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohn M. Degeneracy, mimicry and crossreactivity in immune recognition. Mol Immunol. 2005;42:651–5. doi: 10.1016/j.molimm.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Mellen NM. Degeneracy as a substrate for respiratory regulation. Respir Physiol Neurobiol. 2010;172:1–7. doi: 10.1016/j.resp.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayer-Kress G, Yeou-Teh L, Newell KM. Complex systems and human movement. Complexity. 2006;12:40–1. [Google Scholar]
- 13.Barris S, Farrow D, Davids K. Do the kinematics of a baulked take-off in springboard diving differ from those of a completed dive. J Sports Sci. 2012;31:305–13. doi: 10.1080/02640414.2012.733018. [DOI] [PubMed] [Google Scholar]
- 14.Price CJ, Friston KJ. Degeneracy and cognitive anatomy. Trends Cogn Sci. 2002;6:416–21. doi: 10.1016/s1364-6613(02)01976-9. [DOI] [PubMed] [Google Scholar]
- 15.Friston K, Price CJ. Degeneracy and redundancy in cognitive anatomy. Trends Cogn Sci. 2003;7:151–2. doi: 10.1016/s1364-6613(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 16.Noppeney U, Friston KJ, Price CJ. Degenerate neuronal systems sustaining cognitive functions. J Anat. 2004;205:433–42. doi: 10.1111/j.0021-8782.2004.00343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitacre JM, Atamas SP. Degeneracy allows for both apparent homogeneity and diversification in populations. BioSystems. 2012;110:34–42. doi: 10.1016/j.biosystems.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atamas N, Atamas M, Atamas F, et al. Non-local competition drives both rapid divergence and prolonged stasis in a model of speciation in populations with degenerate resource consumption. Theor Biol Med Model. 2012;9:1–23. doi: 10.1186/1742-4682-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crick FHC. On Degenerate Templates and the Adaptor Hypothesis. Medical Research Council Unit for the Study of the Molecular Structure of Biological Systems. Cambridge: England: Cavendish Laboratory; 1955. [Google Scholar]
- 20.Lawrence C. Degeneration under the Microscope at the fin se siécle. Ann Sci. 2009;66:455–71. doi: 10.1080/00033790903005426. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence C. Historical keyword: degeneration. Lancet. 2010;375:975. doi: 10.1016/S0140-6736(10)60425-4. [DOI] [PubMed] [Google Scholar]
- 22.Csermely P. Strong links are important, but weak links stabilize them. Trends Biochem Sci. 2004;29:331–4. doi: 10.1016/j.tibs.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Cohen IR, Hershberg U, Solomon C. Antigen-receptor degeneracy and immunological paradigms. Mol Immunol. 2004;40:993–6. doi: 10.1016/j.molimm.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 24.Tieri P, Castellani GC, Remondini D, et al. Capturing degeneracy of the immune system. In: Flower D, Timmis J, editors. Silico Immunology. New York: Springer Verlag; 2007. pp. 109–18. [Google Scholar]
- 25.Csete M, Doyle J. Bow ties, metabolism and disease. Trends Biotech. 2004;22:446–50. doi: 10.1016/j.tibtech.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Tieri P, Grignolio A, Zaikin A, et al. Network, degeneracy and bow tie. Integrating paradigms and architectures to grasp the complexity of the immune system. Theor Biol Med Model. 2010;7:1–16. doi: 10.1186/1742-4682-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deacon TW. A role for relaxed selection in the evolution of the language capacity. Proc Natl Acad Sci USA. 2010;107:9000–6. doi: 10.1073/pnas.0914624107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waddington CH. An Introduction to Modern Genetics. New York: The Macmillan Company; 1939. [Google Scholar]
- 29.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 30.Russo VEA, Martienssen RA, Riggs AD. Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 31.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 32.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabor Miklos GL, Maleszka R. Epigenomic communication systems in humans and honey bees: from molecules to behaviour. Horm Behav. 2011;59:399–406. doi: 10.1016/j.yhbeh.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 34.Lyko F, Foret S, Kucharski R, et al. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PloS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyko F, Maleszka R. Insects as innovative models for functional studies on DNA methylation. Trends Genet. 2011;27:127–31. doi: 10.1016/j.tig.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Chahwan R, Wontakal SN, Roa S. The multidimensional nature of epigenetic information and its role in disease. Discov Med. 2011;11:233–43. [PubMed] [Google Scholar]
- 37.Dickman MJ, Kucharski R, Maleszka R, Hurd PJ. Extensive histone post-translational modification in honey bees. Insect Biochem Mol Biol. 2013;43:125–37. doi: 10.1016/j.ibmb.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–7. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aslam A, Logie C. Histone H3 serine 57 and lysine 56 interplay in transcription elongation and recovery from S-phase stress. PLoS One. 2010;5:e10851. doi: 10.1371/journal.pone.0010851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Floer M, Bryant GO, Ptashne M. HSP90/70 chaperones are required for rapid nucleosome removal upon induction of the GAL genes of yeast. Proc Natl Acad Sci USA. 2013;110:7101–3. doi: 10.1073/pnas.0800053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mapp AK, Ansari AZ, Ptashne M, et al. Activation of gene expression by small molecule transcription factors. Proc Natl Acad Sci USA. 2000;97:3930–5. doi: 10.1073/pnas.97.8.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kangaspeska S, Stride B, Métivier R, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–5. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 43.Klug M, Heinz S, Gebhard C, et al. Active DNA demethylation in human postmitotic cells correlates with activating histone modifications, but not transcription levels. Genome Biol. 2010;11:R63. doi: 10.1186/gb-2010-11-6-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones MJ, Fejes AP, Kobor MS. DNA methylation, genotype and gene expression: who is driving and who is along for the ride? Genome Biol. 2013;14:126. doi: 10.1186/gb-2013-14-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shukla S, Kavak E, Gregory M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–9. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luco RF, Allo M, Schor IE, et al. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kornblihtt AR, Schor IE, Allo M, et al. When chromatin meets splicing. Nat Struct Mol Biol. 2009;16:902–3. doi: 10.1038/nsmb0909-902. [DOI] [PubMed] [Google Scholar]
- 48.Hahn MA, Qiu R, Wu X, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young MD, Tracy A, Wakefield MJ. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res. 2011;39:7415–27. doi: 10.1093/nar/gkr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Althammer S, Pagès A, Eyras E. Predictive models of gene regulation from high-throughput epigenomics data. Comp Funct Genomics. 2012;2012:284786. doi: 10.1155/2012/284786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmitz KM, Mayer C, Postepska A, et al. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–9. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Almeida SF, Grosso AR, Koch F, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011;18:977–83. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 53.Turner BM. Defining an epigenetic code. Nat Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- 54.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011;2:611–31. doi: 10.1002/wrna.79. [DOI] [PubMed] [Google Scholar]
- 55.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11:426–37. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 56.de Almeida SF, Carmo-Fonseca M. Design principles of interconnections between chromatin and pre-mRNA splicing. Trends Biochem Sci. 2012;37:248–53. doi: 10.1016/j.tibs.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 57.Buratti E, Baralle M, Baralle FE. From single splicing events to thousands: the ambiguous step forward in splicing research. Brief Funct Genomics. 2013;12:3–12. doi: 10.1093/bfgp/els048. [DOI] [PubMed] [Google Scholar]
- 58.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 59.Delatte B, Fuks F. TET proteins: on the frenetic hunt for new cytosine modifications. Brief Funct Genomics. 2013;12:191–204. doi: 10.1093/bfgp/elt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259–66. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 61.Janga SC. From specific to global analysis of posttranscriptional regulation in eukaryotes: posttranscriptional regulatory networks. Brief Funct Genomics. 2012;11:505–21. doi: 10.1093/bfgp/els046. [DOI] [PubMed] [Google Scholar]
- 62.Deal RB, Henikoff S. Chromatin dynamics and the regulation of genome function. Genome Biol. 2010;11:218. doi: 10.1186/gb-2010-11-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harmston N, Lenhard B. Chromatin and epigenetic features of long-range gene regulation. Nucleic Acids Res. 2013;41:7185–99. doi: 10.1093/nar/gkt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clark MB, Choudhary A, Smith MA, et al. The dark matter rises: the expanding world of regulatory RNAs. Essays Biochem. 2013;54:1–16. doi: 10.1042/bse0540001. [DOI] [PubMed] [Google Scholar]
- 65.Amaral PP, Dinger ME, Mattick JS. Non-coding RNAs in homeostasis, disease and stress responses: an evolutionary perspective. Brief Funct Genomics. 2013;12:254–78. doi: 10.1093/bfgp/elt016. [DOI] [PubMed] [Google Scholar]
- 66.Newcomb RD, Lambert DM. The sensitive period for yellow phenocopy induction in Drosophila melanogaster. Experientia. 1988;44:618–21. doi: 10.1007/BF01953317. [DOI] [PubMed] [Google Scholar]
- 67.Di Stefano HS. Effects of silver nitrate on the pigmentation of Drosophila. Am Nat. 1943;77:94–6. [Google Scholar]
- 68.Lindquist S. Protein folding sculpting evolutionary change. Cold Spring Harb Symp Quant Biol. 2009;74:103–8. doi: 10.1101/sqb.2009.74.043. [DOI] [PubMed] [Google Scholar]
- 69.Casanueva MO, Burga A, Lehner B. Fitness trade-offs and environmentally induced mutation buffering in isogenic C. elegans. Science. 2012;335:82–5. doi: 10.1126/science.1213491. [DOI] [PubMed] [Google Scholar]
- 70.Atamas SP. Self-organization in computer stimulated selective systems. Biosystems. 1966;39:143–51. doi: 10.1016/0303-2647(96)01612-7. [DOI] [PubMed] [Google Scholar]
- 71.Atamas SP, Bell J. Degeneracy-driven self-structuring dynamics in selective repertoires. Bull Math Biol. 2009;71:1349–65. doi: 10.1007/s11538-009-9404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kupiec JJ. On the lack of specificity of proteins and its consequences for a theory of biological organisation. Prog Biophys Mol Biol. 2010;102:45–52. doi: 10.1016/j.pbiomolbio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 73.Génieys S, Volpert V, Auger P. Adaptive dynamics: modelling Darwin's divergence principle. C R Biol. 2006;329:876–9. doi: 10.1016/j.crvi.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 74.West-Eberhard MJ. Alternative adaptations, speciation, and phylogeny. Proc Natl Acad Sci USA. 1986;83:1388–92. doi: 10.1073/pnas.83.5.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.West-Eberhard MJ. Developmental Plasticity and Evolution. New York: Oxford University Press; 2003. [Google Scholar]
- 76.Pollard KS, Salama SR, Lambert N, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–72. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 77.Hu HY, He L, Fominykh K, et al. Evolution of the human-specific microRNA miR-94. Nat Commun. 2012;3:1145. doi: 10.1038/ncomms2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Menzel R. The honeybee as a model for understanding the basis of cognition. Nat Rev Neurosci. 2012;13:758–68. doi: 10.1038/nrn3357. [DOI] [PubMed] [Google Scholar]
- 79.Maleszka J, Barron AB, Helliwell PG, et al. Effect of age, behaviour and social environment on honey bee brain plasticity. J Comp Physiol A. 2009;195:733–40. doi: 10.1007/s00359-009-0449-0. [DOI] [PubMed] [Google Scholar]
- 80.Kiya T, Ugajin A, Kunieda T, et al. Identification of kakusei, a nuclear non-coding RNA, as an immediate early gene from the honeybee, and its application for neuroethological study. Int J Mol Sci. 2012;13:15496–509. doi: 10.3390/ijms131215496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tadano H, Yamazaki Y, Takeuchi H, et al. Age- and division-of-labour-dependent differential expression of a novel non-coding RNA, Nb-1, in the brain of worker honeybees, Apis mellifera L. Insect Mol Biol. 2009;18:715–26. doi: 10.1111/j.1365-2583.2009.00911.x. [DOI] [PubMed] [Google Scholar]
- 82.Maleszka R. Epigenetic integration of environmental and genomic signals in honey bees: the critical interplay of nutritional, brain and reproductive networks. Epigenetics. 2008;3:188–92. doi: 10.4161/epi.3.4.6697. [DOI] [PubMed] [Google Scholar]
- 83.Foret S, Kucharski R, Pellegrini M, et al. DNA methylation dynamics, metabolic fluxes, gene splicing and alternative phenotypes in honey bees. Proc Natl Acad Sci USA. 2012;109:4968–73. doi: 10.1073/pnas.1202392109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kucharski R, Maleszka J, Foret S, et al. Nutritional control of reproductive status in honey bees via DNA methylation. Science. 2008;319:1827–30. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 85.Barchuk AR, dos Santos Cristino A, Kucharski R, et al. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev Biol. 2007;7:70. doi: 10.1186/1471-213X-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miklos GLG, Maleszka R. Microarray reality checks in the context of a complex disease. Nat Biotechnol. 2004;22:615–21. doi: 10.1038/nbt965. [DOI] [PubMed] [Google Scholar]
- 87.Miklos GL, Maleszka R. Protein functions and biological contexts. Proteomics. 2001;1:169–78. doi: 10.1002/1615-9861(200102)1:2<169::AID-PROT169>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 88.Foret S, Kucharski R, Pittelkow Y, et al. Epigenetic regulation of the honey bee transcriptome: unravelling the nature of methylated genes. BMC Genomics. 2009;10:472. doi: 10.1186/1471-2164-10-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Edelman GM. Topobiology. New York: Basic Books; 1988. [Google Scholar]
- 90.Lockett GA, Wilkes F, Maleszka R. Brain plasticity, memory and neurological disorders; an epigenetic perspective. NeuroReport. 2010;21:909–13. doi: 10.1097/WNR.0b013e32833e9288. [DOI] [PubMed] [Google Scholar]
- 91.Lockett GA, Helliwell P, Maleszka R. Involvement of DNA methylation in memory processing in the honey bee. NeuroReport. 2010;21:812–6. doi: 10.1097/WNR.0b013e32833ce5be. [DOI] [PubMed] [Google Scholar]
- 92.Lockett GA, Kucharski R, Maleszka R. DNA methylation changes elicited by social stimuli in the brains of worker honey bees. Genes Brain Behav. 2012;11:235–42. doi: 10.1111/j.1601-183X.2011.00751.x. [DOI] [PubMed] [Google Scholar]
- 93.Barron AB, Maleszka R, Helliwell PG, et al. Effects of cocaine on honey bee dance behaviour. J Exp Biol. 2009;211:163–8. doi: 10.1242/jeb.025361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hunt BG, Ometto L, Wurm Y, et al. Relaxed selection is a precursor to the evolution of phenotypic plasticity. Proc Natl Acad Sci USA. 2011;108:15936–41. doi: 10.1073/pnas.1104825108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dornhaus A, Chittka L. Why do honey bees dance? Behav Ecol Sociobiol. 2004;55:395–401. [Google Scholar]