Abstract
The periodontal pathogen Porphyromonas gingivalis has two different lipopolysaccharide (LPS) molecules, O-LPS and A-LPS. We have recently shown that P. gingivalis strain HG66 lacks A-LPS. Here, we found that introduction of a wild-type wbpB gene into strain HG66 restored formation of A-LPS. Sequencing of the wbpB gene from strain HG66 revealed the presence of a nonsense mutation in the gene. The wbpB gene product is a member of the Wbp pathway, which plays a role in the synthesis of UDP-ManNAc(3NAc)A in Pseudomonas aeruginosa; UDP-ManNAc(3NAc)A is sequentially synthesized by the WbpA, WbpB, WbpE, WbpD and WbpI proteins. We then determined the effect of the PGN_0002 gene, a wbpD homolog, on the biosynthesis of A-LPS. A PGN_0002-deficient mutant demonstrated an A-LPS biosynthesis deficiency. Taken together with previous studies, the present results suggest that the final product synthesized by the Wbp pathway is one of the sugar substrates necessary for the biosynthesis of A-LPS.
The periodontal pathogen Porphyromonas gingivalis is a Gram-negative obligate anaerobe. As P. gingivalis cannot utilize saccharides, the bacterium expresses many proteases on the cell surface to utilize peptides as carbon and nitrogen sources. Specifically, cysteine proteases such as Arg-gingipain (Rgp) and Lys-gingipain (Kgp) are considered important P. gingivalis virulence factors1.
P. gingivalis displays black pigmentation on blood agar plates. The black pigmentation is the result of storage of the μ-oxo-dimeric form of heme (iron protoporphyrin IX) on the cell surface2. Kgp can degrade the hemoglobin protein, which holds heme molecules, and it has been shown that the kgp mutant possesses a pigment-less phenotype3. Previous studies using transposon mutagenesis have revealed that porR4 and porT5 are involved in the pigmentation phenotype in P. gingivalis. The porR-type mutation demonstrates significantly reduced gingipain activities on the cell surface but retains these activities in the culture supernatant, suggesting that the PorR-type system plays a role in anchoring the gingipains on the cell surface. Conversely, the porT-type mutation results in no gingipain activities on the cell surface or in the culture supernatant, suggesting that the PorT-type system is involved in the secretion of gingipains onto the cell surface. In P. gingivalis, studies have shown that the PorT-type system is composed of 11 proteins, named the Por secretion system (PorSS)/type IX secretion system (T9SS); this system is widely distributed in the Bacteroidetes phylum except for Bacteroides fragilis and Bacteroides thetaiotaomicron6,7,8.
P. gingivalis has two different lipopolysaccharide (LPS) molecules, O-LPS and A-LPS. O-LPS has the conventional O-antigen of P. gingivalis strain W50 consists of a tetrasaccharide repeat unit composed of -3)- α-D-Galp-(1-6)-α-D-Glcp-(1-4)- α-L-Rhap-(1-3)-β-D-GalNAcp-(1-9. A-LPS has a different O-antigen from P. gingivalis strain W50 and consists of an anionic polysaccharide (APS) repeat unit10,11. Curtis et al.12 obtained a monoclonal antibody (mAb 1B5) that was originally raised against the catalytic domain of the RgpA protease and was later found to cross-react with A-LPS by recognizing a phosphorylated branched mannan in the APS repeat unit10. Recently, we have shown that another monoclonal antibody (mAb TDC-5-2-1) recognizes the O-antigen of O-LPS present in almost all wild-type cells; however, the glycan epitope recognized by this monoclonal antibody has not been identified13.
The porR mutant has been shown to be immunoreactive to mAb TDC-5-2-1, but not to mAb 1B5, indicating that the porR mutant possesses O-LPS, but lacks A-LPS13. We have previously reported that the porR gene encodes a putative transaminase4. However, the exact substrate of the PorR protein has not been identified. As A-LPS is assumed to be a P. gingivalis virulence factor, genes involved in A-LPS biosynthesis have been recently identified. To date, vimA, vimE, vimF14,15, wbpB16, waaL11 ugdA, rfa17, wzy9, gtfB18, PGN_0242, PGN_066319 wbaP, wzx, wzzP13 and porR have been demonstrated to be involved in A-LPS biosynthesis. The PorR, WbpB and UgdA proteins are predicted to participate in the initial synthesis of structural sugar(s) in the APS. The Rfa protein is involved in the synthesis of the core oligosaccharide of LPS molecules. WbaP (initial phosphoryl glycosyl transferase onto undecaprenyl monophosphate), Wzx (O-antigen flippase), Wzy (O-antigen polymerase), WzzP (O-antigen chain length regulator) and WaaL (O-antigen ligase) are involved in the biosynthesis of both O-antigen and A-antigen. The VimA, VimE and VimF proteins are acetyltransferase, hypothetical protein and galactosyltransferase proteins, respectively20,21,22.
P. gingivalis has approximately 30 proteins (called CTD proteins) that contain a conserved C-terminal domain23. Some of these proteins are secreted onto the cell surface via the PorSS/T9SS system and are then bound to A-LPS or secreted in the culture supernatant6,7,19,24. RgpB and HBP35 are used as model proteins to analyze the PorSS/T9SS secretion system in P. gingivalis because these proteins exhibit diffuse bands on a gel, which is indicative of the A-LPS bound form19,25,26.
Whereas wild-type P. gingivalis strains possess membrane associated gingipain activities on the cell surface, P. gingivalis strain HG66 has significantly reduced gingipain activities on the cell surface but still retains the activities in the culture supernatant27. Some CTD proteins, such as RgpB28, peptidyl arginine deaminase29, periodontain30 and CPG7031, have been purified from the culture supernatant of strain HG66. We have recently shown that the latter strain possesses O-LPS, but lacks A-LPS, similar to the porR mutant13. One aim of this study was to determine the reason for the A-LPS deficiency in strain HG66.
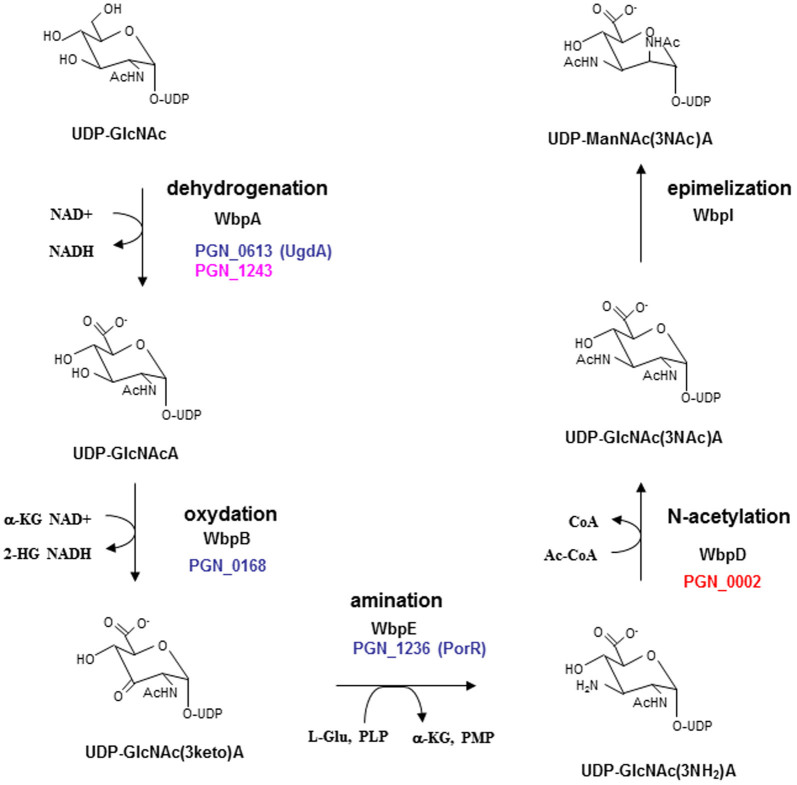
To investigate the lack of A-LPS in HG66, we focused on the relationships between the ugdA, wbpB and porR genes in A-LPS biosynthesis. WbpB belongs to the Wbp pathway, which participates in the synthesis of UDP-2,3-diacetamido-2,3-dideoxy-D-mannuronic acid [UDP- ManNAc(3NAc)A], a precursor of the ManNAc(3NAc)A residue in the B-band O-antigen of Pseudomonas aeruginosa. For UDP-ManNAc(3NAc)A synthesis, WbpA, WbpB, WbpE, WbpD and WbpI react sequentially on the precursor UDP-N-acetyl-glucosamine (UDP-GlcNAc) molecule32,33,34. The UgdA and PorR proteins have high similarity to the WbpA and WbpE proteins, respectively. Therefore, we hypothesized that P. gingivalis may have a similar Wbp pathway. Genomic analysis revealed that ugdA (PGN_0613) and PGN_1243 were wbpA homologs, wbpB (PGN_0168) was a wbpB homolog, porR (PGN_1236) was a wbpE homolog, and PGN_0002 was a wbpD homolog; no wbpI homologs were found.
Our study revealed that the A-LPS deficiency of strain HG66 was the result of a nonsense mutation in the wbpB gene, and likewise, Wbp pathway gene mutants were A-LPS deficient.
Results
Identification of the gene mutation responsible for A-LPS deficiency in strain HG66
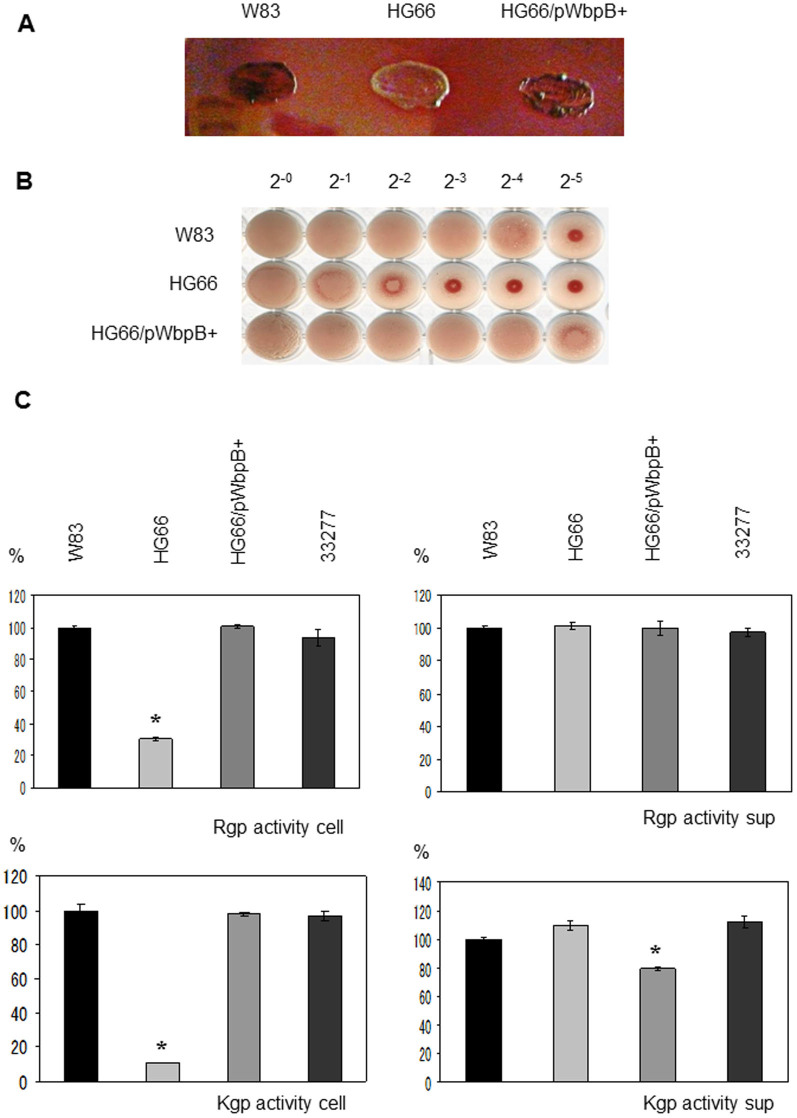
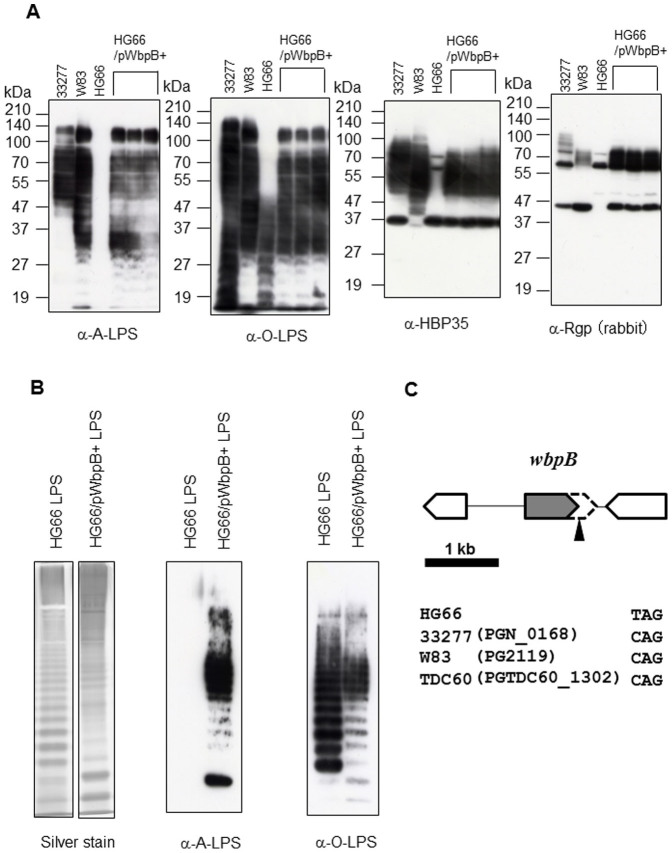
We have recently shown that HG66 possesses O-LPS, but not A-LPS13. When expressed in a wild-type background, porR (PGN_1236), vimA (PGN_1056), vimE (PGN_1055), wbpB (PGN_0168), ugdA (PGN_0613), and wbaP (PGN_1896) mutants retain O-LPS but lose A-LPS13,14,17. We thus hypothesized that mutation of one or more of these genes was responsible for the A-LPS deficient phenotype in strain HG66. Among the above genes, we found that wbpB from strain ATCC 33277 conferred the pigmentation phenotype on strain HG66 (Figure 1A). HG66 cells expressing the wbpB gene from ATCC 33277 showed greater hemagglutination and gingipain activities on the cell surface compared with strain HG66 (Figure 1B, C). We have recently shown that cell lysates from strain HG66 exhibit no diffuse HBP35 and Rgp bands on a gel due to a lack of A-LPS13. Immunoblot analyses revealed that HG66 cell lysates expressing wbpB from ATCC 33277 contained A-LPS when probed with an anti-A-LPS antibody. The lysates also exhibited the diffuse HBP35 and Rgp bands that are also observed in the wild-type ATCC 33277 or W83 strains (Figure 2A). Interestingly, an anti-O-LPS antibody recognized higher molecular mass immunoreactive products in the HG66 cells expressing wbpB from ATCC 33277 compared with strain HG66 (Figure 2A). We next confirmed that purified LPS from HG66 cells expressing wbpB from ATCC 33277 contained both A-LPS and O-LPS by immunoblot analyses (Figure 2B). These results suggested that strain HG66 might encode a non-functional wbpB gene. Therefore, the coding sequence of the wbpB gene from strain HG66 was amplified by PCR and sequenced. We found that a nonsense mutation occurred at amino acid Q240 of the WbpB protein (Figure 2C, Supplemental Text 2). WbpB is an oxidoreductase that forms UDP-GlcNAc(3 keto)A from UDP-GlcNAcA (Figure 3). Additionally, WbpB is a component of the Wbp pathway, which synthesizes UDP-ManNAc(3NAc)A, a precursor of the ManNAc(3NAc)A residue in the B-band O-antigen of P. aeruginosa33,34. The P. gingivalis WbpB protein had high similarity to P. aeruginosa WbpB and Bordetella pertussis WlbA (Supplemental Figure 1). A truncated WbpB protein lacking C-terminal 83 amino acids is likely to be non-functional, although we did not confirm expression at the mRNA or protein levels.
Figure 1. Pigmentation and hemagglutination and gingipain activities of Porphyromonas gingivalis.
(A) Colony pigmentation of Porphyromonas gingivalis cells on blood agar plates. (B) The hemagglutination activities of P. gingivalis strains were measured. (C) The Kgp and Rgp activities of the cell lysates (cell) and vesicle-containing culture supernatants (sup) of W83, HG66, HG66/WbpB+ and ATCC 33277 were measured. The mean of each protease activity of W83 was regarded as 100%. Asterisks indicate significant differences in enzymatic activity between W83 and various strains (P < 0.01). The bars are expressed as the means ± standard deviation for triplicate samples from one of two independent experiments.
Figure 2. Immunoblot and LPS analyses of Porphyromonas gingivalis strain HG66.
(A) Immunoblot analyses of cell lysates from various P. gingivalis strains were performed with anti-A-LPS (mAb 1B5), anti-O-LPS (mAb TDC-5-2-1), anti-HBP35 or anti-Rgp antibodies. Three sets of HG66/WbpB+ strains were obtained from each clone. (B) The LPS fraction from P. gingivalis HG66 or HG66/WbpB+ strain was stained by silver staining. The cropped silver stained gels were run under a same SDS-PAGE gel. Immunoblot analyses were performed with anti-A-LPS (mAb 1B5) or anti-O-LPS (mAb TDC-5-2-1) antibodies. (C) Physical map of the area around the wbpB gene in P. gingivalis. The arrowhead indicates the nonsense mutation in HG66.
Figure 3. Wbp pathway for the biosynthesis of UDP-ManNAc(3NAc)A.
Abbreviations were seen in Supplemental Text 3. Gene homologs of the Wbp pathway in Porphyromonas gingivalis are represented in colored PGN_ numbers.
Genes in the Wbp pathway are involved in the biosynthesis of A-LPS
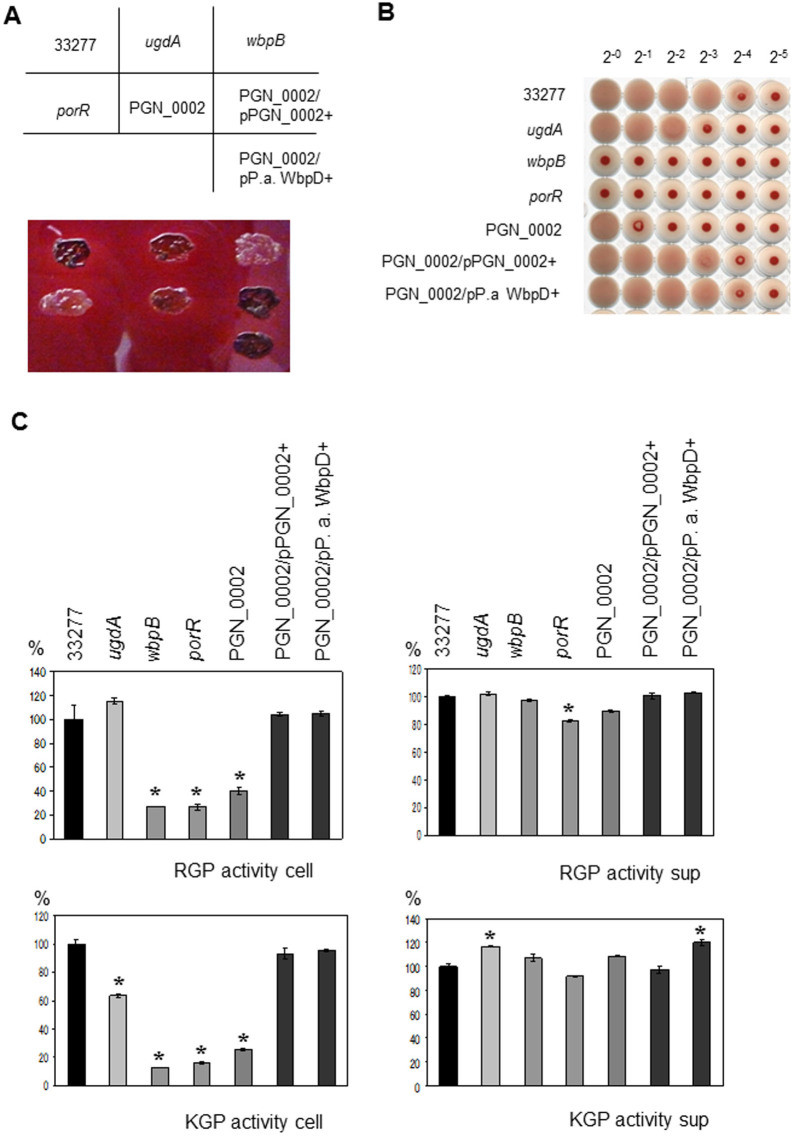
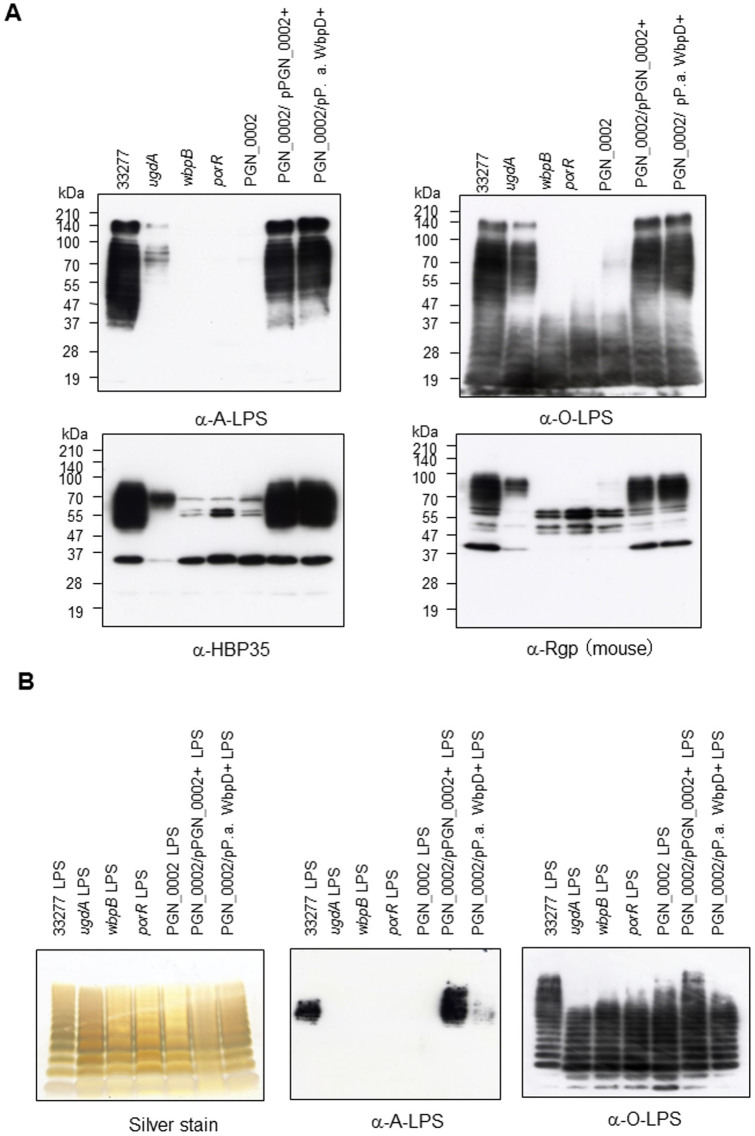
As WbpB belongs to the Wbp pathway, we next hypothesized that the nucleotide-activated sugar formed by the Wbp pathway in P. gingivalis was recognized by an anti-A-LPS antibody. Genomic analysis revealed that PGN_1243 and PGN_0631 (ugdA) were WbpA homologs, PGN_0168 (wbpB) was a WbpB homolog, PGN_1236 (porR) was a WbpE homolog, and PGN_0002 was a WbpD homolog; however, no WbpI homologs were found (Supplemental Figures 1, 2, 3, 4). It has been shown that the ugdA, wbpB and porR gene products are involved in the biosynthesis of A-LPS4,16,17. We focused on the PGN_0002 protein because it may be the last enzyme of the Wbp pathway in P. gingivalis, and it has not yet been characterized. PGN_0002 showed high similarity to P. aeruginosa WbpD and Bordetella pertussis WlbB, and both of these proteins possessed acetyltransferase activity, allowing the formation of UDP-GlcNAc(3NAc)A from UDP-GlcNAc(3NH2)A (Supplemental Figure 4). We constructed full PGN_1243 and PGN_0002 gene deletions; we were able to generate a PGN_0002 mutant, but not a PGN_1243 mutant. The PGN_0002 mutant exhibited a pigment-less phenotype and reduced hemagglutination and gingipain activities on the cell surface compared with wild-type cells (Figure 4A, B, C). Complementation of the PGN_0002 gene or expression of P. aeruginosa wbpD restored the pigmentation phenotype and, hemagglutination and gingipain activities on the cell surface (Figure 4A, B, C). Immunoblot analyses revealed that the wbpB, porR and PGN_0002 mutants were not immunoreactive to anti-A-LPS, and no HBP35 and Rgp diffuse bands were observed (Figure 5A). Interestingly, anti-O-LPS immunoreactive bands in the wbpB, porR and PGN_0002 mutants exhibited a lower molecular mass than those of the wild-type cells. The ugdA mutant showed decreased immunoreactivity to anti-A-LPS and reduced HBP35 and Rgp diffuse bands. The PGN_1243 gene products may compensate for the WbpA-like activity in the ugdA mutant. We also confirmed that purified LPSs from various P. gingivalis mutants in the Wbp pathway possessed O-LPS, but lacked A-LPS (Figure 5B).
Figure 4. Pigmentation and hemagglutination and gingipain activities of various Porphyromonas gingivalis strains.
(A) Colony pigmentation of Porphyromonas gingivalis cells on blood agar plates. (B) The hemagglutination activities of P. gingivalis strains were measured. (C) The Kgp and Rgp activities of the cell lysates (cell) and vesicle-containing culture supernatants (sup) of ATCC 33277 (wild-type), ugdA, wbpB, porR, PGN_0002, PGN_0002/PGN_0002+ and PGN_0002/Pseudomonas aeruginosa (P. a) WbpD+ were measured. The mean of each protease activity of ATCC 33277 was regarded as 100%. Asterisks indicate significant differences in enzymatic activity between ATCC 33277 and various strains (P < 0.01). The bars are expressed as the means ± standard deviation for triplicate samples from one of two independent experiments.
Figure 5. Immunoblot and LPS analyses of various Porphyromonas gingivalis strains.
(A) Immunoblot analyses of cell lysates from various P. gingivalis strains were performed with various antibodies. (B) The LPS fraction from various P. gingivalis strains was stained by silver staining. Immunoblot analyses were performed with antibodies for LPS.
Discussion
Our previous study revealed that CTD proteins, which contain a conserved C-terminal domain, are transported to the cell surface via the type IX secretion system6. P. gingivalis has approximately 30 CTD proteins. Of these, 19 have been shown to be LPS-modified, suggesting that they bind to the A-LPS35. Specifically, RgpB, TapA and HBP35 have been experimentally shown to be anchored to the cell surface by binding to A-LPS25,26,36. As described above, P. gingivalis possesses two different LPS molecules, A-LPS and O-LPS. Recent studies have shown that 15 gene products play a role in the biosynthesis of A-LPS. Of these, the wzy, waaL, gtfB and wzzP gene products are involved in the biosynthesis of both A-LPS and O-LPS; mutations in these genes result in rough or semi-rough LPS9,13,18. Conversely, porR (PGN_1236), vimA (PGN_1056), vimE (PGN_1055), wbpB (PGN_0168), ugdA (PGN_0613) and wbaP (PGN_1896) mutants express O-LPS, but lack A-LPS, suggesting that some of these genes may be involved in A-LPS specific glycan synthesis. A-LPS consists of a lipid A-core and anionic polysaccharide (APS) repeating units containing phosphorylated branched mannan10,11. Paramonov et al.10 has demonstrated that the structure of APS is relatively similar to that of yeast mannan and is predicted to be synthesized by α1→6 and α1→2 mannosyltransferases. However, the genes encoding the mannosyltransferases have not been identified. Recently, the vimF gene product, which is one of proteins necessary for A-LPS biosynthesis, has been demonstrated to possess galactosyltransferase activity22. In addition, not all of the mannosidases are involved in A-LPS synthesis37. Therefore, the postulated structure of APS may not yet be fully understood.
In this study, we found that a wbpB gene mutation was responsible for the non-pigmented phenotype of strain HG66, and we further demonstrated that genes in the Wbp pathway were involved in the biosynthesis of A-LPS. The existence of ManNAc(3NAc)A was first reported in P. aeruginosa LPS more than 30 years ago38. It has been recently shown that the ManNAc(3NAc)A in P. aeruginosa is enzymatically synthesized by WbpA, WbpB, WbpE, WbpD and WbpI, proteins that together have been designated the Wbp pathway33,34,39. Bordetella pertussis has a similar Wbp pathway and possesses two wbpA gene homologs. These homologs are present separately and apart from the other gene homologs located in the wlb gene cluster40. Almost all of the genes in the Wbp pathway are located within a gene cluster in P. aeruginosa and B. pertussis. Gene homologs of wbpA, wbpB, wbpE and wbpD were located separately in the P. gingivalis genome, and no wbpI gene homologs were found in the genome (Figure 6). ManNAc(3NAc)A, the C2-epimer of GlcNAc(3NAc)A, is present not only in the LPS of P. aeruginosa and B. pertussis, but also in the cell wall polysaccharide of the Gram-positive thermophile, Bacillus stearothermophilus41. GlcNAc(3NAc)A has been reported to be present in the LPS of a number of P. aeruginosa strains, including P1-III and P14, the N-linked glycans of the methanogenic archaea, Methanococcus voltae and Methanococcus maripalidus, and the cell surface polysaccharide of the Gram-negative spirochete, Treponema medium42,43,44,45. A partial genome research survey revealed that Thermus thermophiles (Deinococcus-Thermus), Zobellia galactanivorans (Bacteroidetes), Wolinella succinogenes (Epsilonproteobacteria) and Photorhabdus asymbiotica (Gammaproteobacteria) possess 5 genes in the Wbp pathway. Conversely, P. gingivalis and Odoribacter splanchnicus (Bacteroidetes), Sulfurihydrogenibium azorense (Aquificae) and Xenorhabdus bovienii (Gammaproteobacteria) possess 4 genes in the Wbp pathway and lack the wbpI gene (Figure 6). The presence of 5-Wbp pathway gene homologs suggests that a particular species may utilize ManNAc(3NAc)A as a structural sugar, whereas the presence of 4-Wbp pathway gene homologs (lacking wbpI) suggests that a particular species may utilize GlcNAc(3NAc)A as a structural sugar. As ManNAc(3NAc)A and GlcNAc(3NAc)A have C2- and, C3-acetylated and C6-uronic acid forms, they are called di-acetylated mannuronic acid and glucuronic acid, respectively. Considering the conservation and the distribution of these molecules from archaea to Gram-positive and Gram-negative bacteria, di-acetylated uronic acid(s) may be structurally advantageous to endure a variety of environmental stresses.
Figure 6. Gene context for representative gene homologs of the Wbp pathway in Porphyromonas gingivalis ATCC 33277, Thermus thermophiles HB27, Zobellia galactanivorans, Odoribacter splanchnicus, Wolinella succinogenes, Sulfurihydrogenibium azorense, Xenorhabdus bovienii and Photorhabdus asymbiotica.
The lengths of the genes are not drawn to scale. Each homologous set of sequences is represented by one color.
It has been shown that O-glycosylated proteins are widely distributed in bacteria within the Bacteroidetes phylum46. Coyne et al.46 generated a specific antibody against the sugar portion of the O-glycosylated proteins in Bacteroides fragilis. Although this antibody mainly recognized protein bands from Bacteorides species, it also recognized a few P. gingivalis protein bands. To date, OMP85, Mfa1, PGN_0743 (a probable FKBP PPIase), PGN_0876 (a TPR domain protein), PGN_1513 (a hypothetical protein) and PGN_0729 (an outer membrane protein 41 precursor) have been reported to be glycoproteins in P. gingivalis47,48,49. As these glycoproteins each exhibit discrete bands in SDS-PAGE gels, they may be glycosylated by commonly known O-linked or N-linked glycosylation systems. Conversely, A-LPS-modified CTD proteins are expressed via the T9SS and are A-LPS dependent. However, the exact binding mechanism between CTD proteins and A-LPS is not known.
Based on the previously known molecules involved in the biosynthesis of A-LPS, we identified the gene mutation responsible for the non-pigmented phenotype of strain HG66 and found that genes in the Wbp pathway were involved in the biosynthesis of A-LPS. To date, the presence of di-acetylated glucuronic acid in P. gingivalis LPS molecules has not been documented. The analysis of di-acetylated glucuronic acid will shed light on novel features of the T9SS-dependent glycosylation system.
Methods
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Supplemental Tables 1 and 2, respectively50,51,52.
Media and conditions for bacterial growth
P. gingivalis strains were grown anaerobically (80% N2, 10% CO2, 10% H2) in enriched brain-heart infusion (BHI) broth (Becton Dickinson, Franklin Lakes, NJ) or on enriched tryptic soy (TS) agar plates (Nissui, Tokyo, Japan) supplemented with 5 μg/ml hemin (Sigma, St. Louis, MO) and 0.5 μg/ml menadione (Sigma). For blood agar plates, defibrinated laked sheep blood was added to enriched tryptic soy agar at 5%. Luria-Bertani (LB) broth and LB agar plates were used for growth of E. coli strains. Antibiotics were used at the following concentrations: ampicillin (Ap; 10 μg/ml for P. gingivalis, 100 μg/ml for E. coli), erythromycin (Em; 10 μg/ml for P. gingivalis), gentamycin (Gm; 50 μg/ml for P. gingivalis) and tetracycline (Tc; 0.7 μg/ml for P. gingivalis).
Chemicals
The proteinase inhibitors Nα-p-tosyl-L-lysine chloromethyl ketone (TLCK) and iodoacetamide were purchased from Wako (Japan), and leupeptin was obtained from the Peptide Institute (Japan).
Construction of P. gingivalis strains
The oligonucleotides used in this study are listed in Supplemental Table 3. The general manipulation of DNA, restriction and mapping of plasmids and transformation of E. coli and P. gingivalis have been described in detail elsewhere26. The chromosomal DNA from P. gingivalis ATCC 33277 was used as the template for cloning purposes. The construction of various mutants from P. gingivalis ATCC 33277 or complemented strains from the mutants is described in Supplemental Text 1.
Sequencing of the wbpB gene of P. gingivalis HG66
The coding region of the wbpB gene from strain HG66 was amplified using PGN_0168upFw/PGN_0168upRev primers and cloned into the pGEM-T Easy vector. Sequencing was performed by direct sequencing using the purified PCR product or by the common method using the cloned vector with SP6/T7 primers.
Enzymatic assay
Kgp and Rgp activities were determined using the synthetic substrates benzyloxycarbonyl-l-histidyl-l-glutamyl-l-lysine-4-methyl-coumaryl-7-amide (Z-His-Glu-Lys-MCA) and benzyloxycarbonyl-l-phenyl-l-arginine-4-methyl -coumaryl-7-amide (Z-Phe-Arg-MCA), respectively (both from Peptide Institute, Japan). In brief, appropriate amounts of the bacterial cell as well as the bacterial culture supernatant, were added to the reaction mixture (0.25 ml) containing 5 mM cysteine, 20 mM sodium phosphate buffer, pH 7.5, and 10 μM each fluorogenic substrate. After 10 min incubation at 40°C, the reaction was terminated by adding 100 mM sodium acetate buffer, pH 5.0, containing 10 mM iodoacetic acid (0.25 ml). The released 7-amino-4-methyl-coumarine was measured at 460 nm (excitation at 380 nm) by fluorescence spectrophotometer Beckman Coulter DTX 800 (Brea, CA).
Hemagglutination activity
Overnight cultures of P. gingivalis strains in enriched BHI medium were centrifuged, washed once with phosphate-buffered saline (PBS), and suspended in PBS at an optical density of 1.0 at 595 nm. The bacterial suspensions were then diluted in a two-fold series with PBS. A 100-μl aliquot of each suspension was mixed with an equal volume of defibrinated sheep erythrocyte suspension (1% in PBS) and incubated in a round-bottom microtiter plate at room temperature for 3 h.
Preparation of P. gingivalis LPS
A fully grown 200-ml culture was centrifuged, washed once with distilled water and suspended with 5 ml of 10 mM Tris-HCl (pH8.0) containing 2% SDS. Next, 20 μg/ml of DNase and RNase (Sigma, St. Louis, MO. USA) was added at 37°C for 1 h, and 20 μg/ml of proteinase K (Takara, Japan) was added at 37°C overnight. After the overnight incubation, the samples were mixed with preheated 90% phenol at 68°C and stirred at 68°C for 20 min. The samples were centrifuged at 7,000 rpm for 20 min. The aqueous solutions were dialyzed against milliQ water to remove residual phenol. The dialyzed solutions were centrifuged at 100,000 × g for 3 h, and the precipitated samples were dissolved in milliQ water as the LPS fraction. LPS was visualized by silver staining.
Gel electrophoresis and immunoblot analysis
SDS-PAGE and immunoblot analysis were performed as described previously13,19,26. For visualization of LPS, modified SDS-PAGE containing 4 M urea in the separating gel or Tris-Tricine SDS-PAGE was used.
Preparation of antiserum
An anti-HBP35 rabbit polyclonal antibody53 was used to detect HBP35, mAb 1B5 was used to detect A-LPS12, mAb TDC-5-2-1 was used to detect O-LPS and mouse polyclonal antiserum against the amino acid region (E361-L375) within the catalytic domain of RgpB (PGN_1416) was used to detect Rgp13. To prepare rabbit antiserum against the amino acid region (E361-L375) within the catalytic domain of RgpB (PGN_1416), a rabbit was immunized by EveBioscience Co., Ltd. (Wakayama, Japan), and the antiserum was named anti-Rgp (rabbit) to distinguish it from anti-Rgp (mouse) described above.
Statistical analysis
The significance of all described comparisons was established using two-tailed unpaired t tests on triplicate samples with a significance level of 0.01.
Author Contributions
M.S. and K.N. conceived and designed the experiments. M.S. performed the experiments. M.S. and K.N. analysed the data. K.S., H.Y. and M.N. contributed reagents/materials/analysis tools. M.S. and K.N. wrote the manuscript.
Supplementary Material
Supplemental Information
Acknowledgments
This work was supported by Grants-in-Aid (20249073 and 23792110 to KN and MS, respectively) for scientific research from the Ministry of Education, Science, Sports, Culture, and Technology, Japan. We thank to Drs. E. Anderson and J. S. Lam for giving us the generous gift of P. aeruginosa PAO1 strain.
References
- O'Brien-Simpson N. M. et al. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69, 7527–7534 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley J. W., Silver J., Marsh P. J. & Birss A. J. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the μ -oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem. J. 331, 681–685 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K. et al. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J. Biol. Chem. 273, 21225–21231 (1998). [DOI] [PubMed] [Google Scholar]
- Shoji M. et al. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology 148, 1183–1191 (2002). [DOI] [PubMed] [Google Scholar]
- Sato K. et al. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J. Biol. Chem. 280, 8668–8677 (2005). [DOI] [PubMed] [Google Scholar]
- Sato K. et al. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. U S A. 107, 276–281 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K. et al. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS Microbiol. Lett. 338, 68–76 (2013). [DOI] [PubMed] [Google Scholar]
- McBride M. J. & Zhu Y. Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes. J. Bacteriol. 195, 270–278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramonov N. A., Aduse-Opoku J., Hashim A., Rangarajan M. & Curtis M. A. Structural analysis of the core region of O-lipopolysaccharide of Porphyromonas gingivalis from mutants defective in O-antigen ligase and O-antigen polymerase. J. Bacteriol. 191, 5272–5282 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramonov N. et al. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol. Microbiol. 58, 847–863 (2005). [DOI] [PubMed] [Google Scholar]
- Rangarajan M. et al. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J. Bacteriol. 190, 2920–2932 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M. A. et al. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67, 3816–3823 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M. et al. Identification of an O-antigen chain length regulator, WzzP, in Porphyromonas gingivalis. Microbiologyopen 2, 383–401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanterpool E., Roy F., Sandberg L. & Fletcher H. M. Altered gingipain maturation in vimA- and vimE-defective isogenic mutants of Porphyromonas gingivalis. Infect. Immun. 73, 1357–1366 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanterpool E., Roy F. & Fletcher H. M. Inactivation of vimF, a putative glycosyltransferase gene downstream of vimE, alters glycosylation and activation of the gingipains in Porphyromonas gingivalis W83. Infect. Immun. 73, 3971–3982 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaney J. M., Gallagher A., Aduse-Opoku J., Pell K. & Curtis M. A. Mechanisms of resistance of Porphyromonas gingivalis to killing by serum complement. Infect. Immun. 74, 5352–5361 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K. et al. Lipopolysaccharide biosynthesis-related genes are required for colony pigmentation of Porphyromonas gingivalis. Microbiology 155, 1282–1293 (2009). [DOI] [PubMed] [Google Scholar]
- Yamaguchi M. et al. A Porphyromonas gingivalis mutant defective in a putative glycosyltransferase exhibits defective biosynthesis of the polysaccharide portions of lipopolysaccharide, decreased gingipain activities, strong autoaggregation, and increased biofilm formation. Infect. Immun. 78, 3801–3812 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M. et al. Por secretion system-dependent secretion and glycosylation of Porphyromonas gingivalis hemin-binding protein 35. PLoS One 6, e21372 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanterpool E., Roy F. & Fletcher H. M. The vimE gene downstream of vimA is independently expressed and is involved in modulating proteolytic activity in Porphyromonas gingivalis W83. Infect. Immun. 72, 5555–5564 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruni A. W. et al. VimA-dependent modulation of acetyl coenzyme A levels and lipid A biosynthesis can alter virulence in Porphyromonas gingivalis. Infect. Immun. 80, 550–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthiah A. S. et al. In Porphyromonas gingivalis VimF is involved in gingipain maturation through the transfer of galactose. PLoS One 8, e63367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seers C. A. et al. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J. Bacteriol. 188, 6376–6386 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glew M. D. et al. PG0026 is the C-terminal signal peptidase of a novel secretion system of Porphyromonas gingivalis. J. Biol. Chem. 287, 24605–24617 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen K. A., Travis J. & Potempa J. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-Negative bacteria? J. Bacteriol. 189, 833–843 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M. et al. Characterization of hemin-binding protein 35 (HBP35) in Porphyromonas gingivalis: its cellular distribution, thioredoxin activity and role in heme utilization. BMC Microbiol. 10, e152 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa J., Pike R. & Travis J. The multiple forms of trypsin-like activity present in various strains of Porphyromonas gingivalis are due to the presence of either Arg-gingipain or Lys-gingipain. Infect. Immun. 63, 1176–1182 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa J. et al. Comparative properties of two cysteine proteinases (gingipains R), the products of two related but individual genes of Porphyromonas gingivalis. J. Biol. Chem. 273, 21648–21657 (1998). [DOI] [PubMed] [Google Scholar]
- McGraw W. T., Potempa J., Farley D. & Travis J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect. Immun. 67, 3248–3256 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D., Potempa J., Kordula T. & Travis J. Purification and characterization of a novel cysteine proteinase (periodontain) from Porphyromonas gingivalis. Evidence for a role in the inactivation of human alpha1-proteinase inhibitor. J. Biol. Chem. 274, 12245–12251 (1999). [DOI] [PubMed] [Google Scholar]
- Chen Y. Y. et al. CPG70 is a novel basic metallocarboxypeptidase with C-terminal polycystic kidney disease domains from Porphyromonas gingivalis. J. Biol. Chem. 277, 23433–23440 (2002). [DOI] [PubMed] [Google Scholar]
- Wenzel C. Q., Daniels C., Keates R. A., Brewer D. & Lam J. S. Evidence that WbpD is an N-acetyltransferase belonging to the hexapeptide acyltransferase superfamily and an important protein for O-antigen biosynthesis in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 57, 1288–1303 (2005). [DOI] [PubMed] [Google Scholar]
- Westman E. L. et al. Characterization of WbpB, WbpE, and WbpD and reconstitution of a pathway for the biosynthesis of UDP-2,3-diacetamido-2,3-dideoxy-D-mannuronic acid in Pseudomonas aeruginosa. J. Biol. Chem. 284, 11854–11862 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A. & Imperiali B. Biosynthesis of UDP-GlcNAc(3NAc)A by WbpB, WbpE, and WbpD: enzymes in the Wbp pathway responsible for O-antigen assembly in Pseudomonas aeruginosa PAO1. Biochemistry 48, 5446–5455 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith P. D. et al. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J. Proteome Res. 12, 4449–4461 (2013). [DOI] [PubMed] [Google Scholar]
- Kondo Y. et al. Tetratricopeptide repeat protein-associated proteins contribute to the virulence of Porphyromonas gingivalis. Infect. Immun. 78, 2846–2856 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan M., Aduse-Opoku J., Hashim A., Paramonov N. & Curtis M. A. Characterization of the α- and β-mannosidases of Porphyromonas gingivalis. J. Bacteriol. 195, 5297–5307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knirel Y. A., Kocharova N. A., Shashkov A. S., Dmitriev B. A. & Kochetkov N. K. 2,3-Diacetamido-2,3-dideoxy-D-glucuronic acid: a new acidic amino sugar from Pseudomonas aeruginosa type O6 lipopolysaccharide. Carbohydr. Res. 93, C12–13 (1981). [DOI] [PubMed] [Google Scholar]
- Westman E. L. et al. Identification and biochemical characterization of two novel UDP-2,3-diacetamido-2,3-dideoxy-alpha-D-glucuronic acid 2-epimerases from respiratory pathogens. Biochem. J. 405, 123–130 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westman E. L., Preston A., Field R. A. & Lam J. S. Biosynthesis of a rare di-N-acetylated sugar in the lipopolysaccharides of both Pseudomonas aeruginosa and Bordetella pertussis occurs via an identical scheme despite different gene clusters. J. Bacteriol. 190, 6060–6069 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffer C. et al. The diacetamidodideoxyuronic-acid-containing glycan chain of Bacillus stearothermophilus NRS 2004/3a represents the secondary cell-wall polymer of wild-type B. stearothermophilus strains. Microbiology 145, 1575–1583 (1999). [DOI] [PubMed] [Google Scholar]
- Hashimoto M. et al. Structural elucidation of polysaccharide part of glycoconjugate from Treponema medium ATCC 700293. Eur. J. Biochem. 270, 2671–2679 (2003). [DOI] [PubMed] [Google Scholar]
- Okuda S., Murata S. & Suzuki N. Isolation and identification of uridine(5′)-diphospho(1)-2,3-diacetamido-2,3-dideoxy-alpha-d-glucopyranuronic acid from Pseudomonas aeruginosa P1-III. Biochem. J. 239, 733–738 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisin S. et al. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae. J. Biol. Chem. 280, 16586–16593 (2005). [DOI] [PubMed] [Google Scholar]
- Kelly J., Logan S. M., Jarrell K. F., VanDyke D. J. & Vinogradov E. A novel N-linked flagellar glycan from Methanococcus maripaludis. Carbohydr. Res. 344, 648–653 (2009). [DOI] [PubMed] [Google Scholar]
- Coyne M. J. et al. Phylum-wide general protein O-glycosylation system of the Bacteroidetes. Mol. Microbiol. 88, 772–783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao R. et al. Glycosylation of the OMP85 homolog of Porphyromonas gingivalis and its involvement in biofilm formation. Biochem Biophys Res Commun. 365, 784–789 (2008). [DOI] [PubMed] [Google Scholar]
- Zeituni A. E., McCaig W., Scisci E., Thanassi D. G. & Cutler C. W. The native 67-kilodalton minor fimbria of Porphyromonas gingivalis is a novel glycoprotein with DC-SIGN-targeting motifs. J. Bacteriol. 192, 4103–4110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi M. et al. Identification and characterization of novel glycoproteins involved in growth and biofilm formation by Porphyromonas gingivalis. Mol. Oral Microbiol. 27, 458–470 (2012). [DOI] [PubMed] [Google Scholar]
- Shi Y. et al. Genetic analyses of proteolysis, hemoglogin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274, 17955–17960 (1999). [DOI] [PubMed] [Google Scholar]
- Nagano K. et al. Characterization of RagA and RagB in Porphyromonas gingivalis: study using gene-deletion mutants. J. Med. Microbiol. 56, 1536–1548 (2007). [DOI] [PubMed] [Google Scholar]
- Simon R., Priefer U. & Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1, 784–791 (1983). [Google Scholar]
- Abiko Y. et al. Cloning of a Bacteroides gingivalis outer membrane protein gene in Escherichia coli. Arch. Oral Biol. 35, 689–695 (1990). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information