Background: Mutations in PARK13 have been implicated in Parkinson disease.
Results: AtPARK13 confers thermotolerance in Arabidopsis and degrades misfolded proteins, including α-synuclein and DJ-1.
Conclusion: AtPARK13 confers thermotolerance through degradation of misfolded proteins.
Significance: Arabidopsis is a complementary model to investigate mechanisms associated with Parkinson disease.
Keywords: Arabidopsis, Parkinson Disease, Protease, Proteomics, Stress, Heat Stress
Abstract
Mutations in HTRA2/Omi/PARK13 have been implicated in Parkinson disease (PD). PARK13 is a neuroprotective serine protease; however, little is known about how PARK13 confers stress protection and which protein targets are directly affected by PARK13. We have reported that Arabidopsis thaliana represents a complementary PD model, and here we demonstrate that AtPARK13, similar to human PARK13 (hPARK13), is a mitochondrial protease. We show that the expression/accumulation of AtPARK13 transcripts are induced by heat stress but not by other stress conditions, including oxidative stress and metals. Our data show that elevated levels of AtPARK13 confer thermotolerance in A. thaliana. Increased temperatures accelerate protein unfolding, and we demonstrate that although AtPARK13 can act on native protein substrates, unfolded proteins represent better AtPARK13 substrates. The results further show that AtPARK13 and hPARK13 can degrade the PD proteins α-synuclein (SNCA) and DJ-1/PARK7 directly, without autophagy involvement, and that misfolded SNCA and DJ-1 represent better substrates than their native counterparts. Comparative proteomic profiling revealed AtPARK13-mediated proteome changes, and we identified four proteins that show altered abundance in response to AtPARK13 overexpression and elevated temperatures. Our study not only suggests that AtPARK13 confers thermotolerance by degrading misfolded protein targets, but it also provides new insight into possible roles of this protease in neurodegeneration.
Introduction
Parkinson disease (PD)2 is the second most common progressive neurodegenerative disorder, characterized neuropathologically by the selective loss of dopaminergic neurons in the substantia nigra pars compacta and Lewy body formation (1). Although most PD cases are sporadic, monogenic PD accounts for 5–10% of all cases (2). A molecular hallmark of PD is the accumulation of misfolded and aggregated α-synuclein (SNCA) within neurons, which has detrimental effects on neuronal survival (3). Autosomal recessive PD forms have been conclusively linked to mutations in PARK2 (4), PINK1 (5), and DJ-1/PARK7 (6), all localizing to mitochondria, ultimately affecting function. Studies also show that mitochondrial dysfunction and oxidative stress pathways are fundamental to PD pathogenesis and that protein misfolding/aggregation, as for SNCA, are central to PD pathology (7).
The HTRA2/Omi/PARK13 mitochondrial serine protease is implicated in PD; however, conflicting reports exist. A cohort of German and Chinese populations showed HTRA2 variants associated with PD (8, 9), and further mutation and polymorphism analysis in Belgian and Taiwanese cohorts show Parkinsonian neurodegeneration (10, 11). However, large genetic association studies also show that there is no overall association of Omi/HtrA2 variants and PD (12). At the molecular level, PARK13 has been shown to exert neuroprotection during stress conditions. Indeed, PARK13 knock-out mice show increased reactive oxygen species (ROS) levels, mitochondrial dysfunction, and Parkinsonian phenotypes (13–15).
The PARK13 protease activity itself also appears to be regulated. PARK13 activity is enhanced by PINK1-mediated phosphorylation, which is important for mitochondrial integrity under stress conditions (13, 16). Although PARK13 has been shown to be a protease, its protein targets remain obscure. To our knowledge, few protein targets affected directly by PARK13 have been reported to date, furthering the challenge of understanding the molecular mechanism of PARK13 in PD pathogenesis.
We previously reported that Arabidopsis thaliana represents a unique complementary PD model. By using Arabidopsis as a multicellular vehicle, we have shown that plant DJ-1 (AtDJ-1a) and human DJ-1DJ-1/PARK7 protect against ROS by interacting with SOD1 (superoxide dismutase 1) and GPX2 (glutathione peroxidase 2), both of which have ROS-quenching properties. Furthermore, we also reported that the PD-associated proteins Parkin, UCHL1, LRRK2, ATP13A2, PINK1, and PARK13 are significantly conserved between humans and Arabidopsis (17, 18).
In this study, we demonstrate that AtPARK13 is a heat stress-induced protease that localizes to mitochondria. We show that increased AtPARK13 levels confer thermotolerance and that unfolded proteins are better AtPARK13 substrates than correctly folded proteins. Furthermore, we show that AtPARK13 and hPARK13 target SNCA and DJ-1/PARK7 for degradation, linking PARK13 directly to crucial PD proteins. Comparative proteomic analysis also identifies AtPARK13-mediated proteome changes, and we identify four proteins showing altered abundance. Combined, our study characterizes the functional role of AtPARK13 in stress protection and provides new insight into possible roles of this protease in neurodegeneration.
EXPERIMENTAL PROCEDURES
Plant Growth and Stress Treatments
Wild type (WT) A. thaliana (ecotype Columbia), mutant lines (SAIL903 and SAIL536), and overexpressing lines were initially grown on half-strength Murashige and Skoog medium (MP Biomedicals) with 1% (w/v) sucrose and transferred to soil after 1 week at 21 °C under long day conditions (16-h light/8-h dark) in plant chambers (Sanyo). For the heat stress treatments, plants were grown at 21, 37, and 42 °C for 1, 2, and 5 h. 10 μm H2O2 (J. T. Baker), 10 μm CuSO4 (Sigma), and 10 μm methyl viologen (Paraquat) (Sigma) were applied to plants for 5 h after 2 weeks at 21 °C for the oxidative stress and metal treatments.
RNA Isolation and RT-PCR
The amino acid sequences of human PARK13, zebrafish PARK13, Drosophila PARK13, and Arabidopsis PARK13 (At5g27660) were obtained from NCBI and aligned using Geneious R6 software (Biomatters Ltd.). Total RNA obtained was using the SurePrep plant/fungi RNA isolation kit (Fisher) from wild type (WT) Arabidopsis seedlings. RNA obtained was treated with 1 unit of DNase I (Thermo Scientific) per μg of RNA in the company-specified buffer for 30 min at 37 °C, followed by a 10-min incubation at 65 °C with 50 mm EDTA. The DNase I-treated RNA was used to synthesize first strand cDNA using the RevertAid first strand cDNA synthesis kit (Thermo Scientific). The cDNA obtained was used to perform preparative RT-PCR using Phusion high fidelity DNA polymerase (Thermo Scientific) (35 cycles) with AtPARK13 forward primer (5′-ATGGCGCGCCATGATGAATTTTCTGAGAAGAGC-3′) (AscI underlined) and reverse primer (5′-ATTTAATTAATCACATGTCTGGATTAGCC-3′) (PacI underlined) (Sigma-Aldrich). The PCR product was subjected to 1% agarose (Fisher) gel electrophoresis. For spatial and stress-induced AtPARK13 transcript analysis, RNA isolation and first strand cDNA synthesis were performed as described. For semiquantitative RT-PCR (25 cycles), TopTaq MasterMix (Qiagen) was used in all cases with AtPARK13 forward and reverse primers. Actin was used as a control in all cases and was amplified using actin forward primer (5′-TGCCAATCTACGAGGGTTTC-3′) and reverse primer (5′-GAACCACCGATCCAGACACT-3′) (Sigma-Aldrich). In all experiments, the PCR products were subjected to 1% agarose gel electrophoresis.
Molecular Cloning
Cloning was carried out using standard techniques. AtPARK13 was cloned into pJET1.2/blunt using the CloneJET PCR cloning kit (Thermo Scientific), binary vectors pBA002 (19) and pER10 (20) at AscI/PacI (Thermo Scientific) restriction sites, YFP fusion vector pWEN18 (19) at the KpnI restriction site, and protein expression vector pET28a(+) (Novagen) at SacI/NotI restriction sites, followed by DNA sequencing (Yale DNA Sequencing Facility). Mature AtPARK13 was designed based on the cleavage sites of hPARK13 and was cloned into protein expression vector pET28a(+) along with mature hPARK13. Primers used for cloning included the following: pBA002-AtPARK13, ATGGCGCGCCATGATGAATTTTCTGAGAAGAGC (AscI underlined) and ATTTAATTAATCACATGTCTGGATTAGCC (PacI underlined); pER10-AtPARK13, ATGGCGCGCCATGATGAATTTTCTGAGAAGAGC (AscI underlined) and ATTTAATTAATCACATGTCTGGATTAGCC (PacI underlined); pWEN18-AtPARK13, ATGGTACCATGATGAATTTTCTGAGAAGAGC (KpnI underlined) and ATGGTACCCATGTCTGGATTAGCCTCC (KpnI underlined); pET28a(+)-AtPARK13, ATGAGCTCATGATGAATTTTCTGAGAAGAGC (SacI underlined) and ATGCGGCCGCCATGTCTGGATTAGCCTCC (NotI underlined); pET28a(+)-mature AtPARK13, ATGAGCTCGCTGCTAGAATTGGACCAGC (SacI underlined) and ATGCGGCCGCTCACATGTCTGGATTAGC (NotI underlined); and pET28a(+)-mature hPARK13, ATGCGGCCGCATGGGCGGGGGTCGGGGTCCTCCG (NotI underlined) and ATCTCGAGTCATTCTGTGACCTCAGGGG (XhoI underlined).
Subcellular Localization Analysis
For subcellular localization studies, pWEN18-AtPARK13-YFP and pCoxIV-ECFP (21) were cotransfected into tobacco leaf cells by particle bombardment and analyzed after 24–48 h using a Nikon A1R confocal laser microscope. pCoxIV-ECFP, harboring the transit sequence of the cytochrome c oxidase IV (coxIV) from yeast fused to ECFP, was used as a marker for mitochondria (21).
Construction of Transgenic Plants and Heat Stress Treatments
pBA002-AtPARK13 and pER10-AtPARK13 were transformed into Agrobacterium tumefaciens (ABI strain) for Arabidopsis transformation by electroporation transformation by the floral dip method (22). Floral dip transformation was performed in the presence of 5% (w/v) sucrose and 0.04% (v/v) Silwet L-77 (Lehle seeds). pBA002 and pER10 transgenic plants were selected on half-strength Murashige and Skoog medium with 0.5% (w/v) BASTA (PlantMedia) and 50 mg/ml kanamycin (Acros), respectively. Selected lines were transferred to half-strength Murashige and Skoog medium without antibiotic and grown for 1 week and then transferred to soil. RNA was isolated from the leaf tissue from the transgenic plants to perform RT-PCR using TopTaq MasterMix with AtPARK13 forward and reverse primers. Two lines with maximum expression of AtPARK13 in pBA002 were selected, seeds were harvested, and the T2 generation of both of the lines was subjected to heat stress at 37 and 42 °C along with controls (both WT and pBA002 vector-only plants) for 12 days and 24 h, respectively. Images were taken at regular intervals to note the phenotypic changes in the AtPARK13-overexpressing plants as compared with the controls.
Protein Expression, Purification, and Enzyme ActivityAssay
pET28a(+)-AtPARK13 full-length, pET28a(+)-mature AtPARK13, and pET28a(+)-mature hPARK13 were purified using standard protocols in Escherichia coli Rosetta (DE3) pLysS (Invitrogen). Purification of mature hPARK13 was performed from the soluble fraction, and full-length and mature AtPARK13 were purified from the insoluble fractions using a His-tagged cobalt-agarose beads affinity column (Goldbio Technology). The purified proteins were analyzed on 12% acrylamide (Bio-World). Full-length AtPARK13 and mature AtPARK13 were refolded by stepwise dialysis from 6 m urea to 0 m urea using Slide-A-Lyzer dialysis cassettes (Pierce), and the concentrations of the proteins were calculated using the Bradford assay (Bio-Rad) and used for the protease assay. The protease assays were performed using the Pierce colorimetric protease assay kit (Pierce) using 83 μm casein as a generic substrate following the manufacturer's protocol. For investigating the effect of ions on the protease activity, 10 mm of Mg2+, Mn2+, Ca2+, Cu2+, Na+, and K+ were used in the assay. Vmax and Km were determined by determining the initial reaction rates (Vo) based on product released at different substrate concentrations as a function of time in a double-reciprocal plot.
Soluble AtPARK13, WT DJ-1, and A107P recombinant proteins were produced in E. coli BL21(DE3) following standard protocols using 0.3 mm isopropyl 1-thio-β-d-galactopyranoside when the A595 reached 0.6–0.7. Cells were resuspended in 50 mm Tris, pH 8.0, 300 mm NaCl, and soluble proteins were extracted using three freeze-thaw cycles. Viscosity was reduced by sonication on ice, and insoluble material was removed by ultracentrifugation at 20,000 rpm in a Sorvall Discovery 90SE Ultracentrifuge, 60 min at 4 °C. The resulting supernatants were purified by nickel-nitrilotriacetic acid metal affinity chromatography using standard methods followed by anion exchange (Q-Sepharose) chromatography. Prior to analysis, proteins were buffer-exchanged using PD-10 spun column chromatography into 50 mm NaHCO3, pH 8.3.
Reaction mixtures for full-length AtPARK13, mature AtPARK13, and mature hPARK13 digestion were made up in a 50-μl final volume with WT SNCA, WT DJ-1, and mutant DJ-1/A107P proteins. Reaction mixtures containing 20 nm SNCA (native or misfolded) or 25 nm DJ-1 proteins (native or misfolded) were initiated by the addition of 4 μm AtPARK13 or 2 μm mature hPARK13. All unfolded substrates were denatured by boiling for 5 min. Reactions were incubated at 42 °C for 30 min and then stopped by the addition of 1× SDS-PAGE loading buffer (43) and boiling for 5 min. Three independent experiments were performed for SNCA, and five independent experiments were performed for DJ-1.
Western Blotting
All samples were subjected to 12% SDS-polyacrylamide gel electrophoresis with running buffer (192 mm glycine, 25 mm Tris base, and 2% (w/v) SDS) at 50 V for 15 min, followed by 75 V for 100 min using a mini-PROTEAN Tetra system (Bio-Rad) and Bio-Rad PowerPac Basic. Gels were transferred to an Immobilon PVDF membrane (Millipore) using transfer buffer (192 mm glycine, 25 mm Tris base, 20% (v/v) methanol, and 0.005% (w/v) SDS) at 200 mm for 2 h. For blocking, the membrane was incubated with 5% (w/v) nonfat dry milk (Bio-Rad) in TBS-T (137 mm NaCl, 15.4 mm Trizma (Tris base) HCl, 0.1% Tween 20, pH 7.6) for 1 h at room temperature. Following blocking, the membranes were incubated with rabbit polyclonal anti-SNCA antibody (Abcam) at a 1:1000 dilution or a mouse monoclonal anti-DJ-1 antibody (ENZO life sciences) at 0.5 μg/ml in 2.5% (w/v) nonfat dry milk overnight at 4 °C. The following day, the membrane was washed with TBS-T three times for 10 min at room temperature, followed by incubation at 1:10,000 dilution with goat anti-rabbit secondary antibody (Pierce) for SNCA and with goat anti-mouse secondary antibody (Pierce) for DJ-1 in 2.5% (w/v) nonfat dry milk for 1 h at room temperature. The membrane was washed with TBS-T three times for 10 min at room temperature and was developed using Pierce ECL Western blot substrate (Thermo Scientific). The signals were recorded using the Molecular Imager Chemi Doc XRS+ imaging system (Bio-Rad).
Two-dimensional Gel Electrophoresis and Mass Spectrometry
Total Arabidopsis protein extracts were subjected to two-dimensional PAGE, and protein spots were analyzed using ImageMaster 2D Platinum version 7 (GE Healthcare) followed by peptide mass fingerprinting and data-dependent sequencing, as described previously (23).
Sample Preparation
Arabidopsis plant seedlings were homogenized with liquid nitrogen in a mortar and pestle and stored at −80 °C until further analysis. Approximately 1 g of the tissue powder was suspended in 3 ml of homogenization buffer (0.05 m phosphate buffer, pH 7.4, containing 0.1 mm phenyl thiourea, 0.1 mm DTT, 0.1 mm PMSF, 1 mm EDTA, and 1× protease/phosphatase inhibitor mixture) and sonicated using a Polytron® PT-2500E homogenizer (KINEMATICA). After sonication, the extract was centrifuged at 4 °C for 15 min at 10,000 × g, and the supernatant was collected for further proteomic analysis.
Protein Determination
The supernatant fractions were concentrated using Amicon® Ultra centrifugal filters (10,000 molecular weight cut-off) by centrifuging at 10,000 × g for 30–60 s, depending on the degree of concentration required, and protein concentration was determined using the Bradford assay. BSA was used as a standard.
Two-dimensional Gel Electrophoresis
Protein samples for separation were precipitated using a standard deoxycholate/trichloroacetic acid/acetone precipitation method (final concentrations: DOC, 0.0175% (w/v); TCA, 6% (w/v)), and the pellets were washed with acetone. These were then mixed with a rehydration solution containing 8 m urea, 2% (w/v) CHAPS, 15 mm DTT, 30 mm thiourea, and 2% IPG buffer, pH 3–10, and traces of bromphenol blue and dissolved on a shaker overnight. The dissolved protein was then applied to ImmobilineTM DryStrip (7 cm, pH 3–10) (GE Healthcare) for overnight rehydration. The isoelectrofocusing was conducted according to conditions recommended by the manufacturer (Bio-Rad). Subsequently, proteins in the gel were reduced with DTT and alkylated with iodoacetamide using standard methods. Electrophoresis in the second dimension was then run on 12% SDS-PAGE at 100 V. Gels were stained overnight with colloidal Coomassie Blue G-250.
Scanning and Analysis of the Images
The stained gels were scanned using EPSON Scan Perfection V750 PRO software (Digital ICE Technologies). The two-dimensional gels were scanned in professional mode at 600 dpi and in 16-bit grayscale. Image analysis software ImageMaster 2D platinum version 7 (GE Healthcare) was used to carry out the spot detection, the background subtraction, and the two-dimensional correction, to determine the differences between protein spots. Spot analysis was carried out by importing all gel images into ImageMasterTM 2D Platinum software version 7.0 (GE Healthcare). In brief, spots were detected automatically with the parameters of smooth = 2, minimal area = 5, and saliency = 10. The patterns of protein spots on each gel were aligned using a landmark spot and matched with the reference gel automatically. Spot normalization was based on relative volume (% volume), which is the volume of each spot divided by the total volume of all spots in the gel. Protein abundance changes were based on the ratio of differences between the vector and overexpressed AtPARK13 as reported by the software.
In-gel Digestion
The differentially expressed protein spots were excised, cut into small pieces, and placed in 0.6-ml Eppendorf tubes. The gel pieces were destained by incubating in 200 μl of 100 mm ammonium bicarbonate/acetonitrile (50:50, v/v) with shaking. When fully destained, the gel fragments were dehydrated with two washes of 100 μl of 100% acetonitrile and were then dried in a vacuum centrifuge (SpeedVac) for 5 min. The protein was then cleaved enzymatically into peptides. For this, trypsin solution (2 μl, 0.02 μg/μl) was added to wet the gel pieces, and incubation was carried out for 4 h at room temperature. Thirty μl of 50 mm ammonium bicarbonate was added to the gel pieces, and they were left overnight at room temperature to allow for diffusion of the peptides from the gel. The digested proteins were stored at −80 °C until further analysis.
Peptide Mass Fingerprinting
After digestion, POROS 20 R2 resin (Applied Biosystems) was added into the digested gel samples with 5% formic acid and 0.2% trifluoroacetic acid for extraction at 4 °C for 4 h on a shaker. Prior to MALDI-MS analysis, the peptide digests were further desalted using ZipTip C18 (Millipore). The ZipTips were conditioned with 10 μl of 0.1% (v/v) TFA twice, 70% (v/v) acetonitrile, 0.1% (v/v) TFA twice and 10 μl of 0.1% (v/v) TFA twice. The sample containing the digests and bead mixture was transferred to the ZipTips and bound to the C18 resin. The loaded tips were then washed with 10 μl of 0.1% (v/v) TFA. The peptide digests were eluted by placing 2 μl of 10 mg/ml α-cyano-4-hydroxycinnamic acid matrix solution in 0.003% (v/v) TFA, 13% (v/v) ethanol, and 84% (v/v) ACN onto the top of the ZipTips and slowly dispensing onto the MALDI plate. Mass spectrometric analysis was performed by a Thermo LTQ XL linear ion trap mass spectrometer (Thermo Scientific) equipped with a vacuum MALDI source after the solvent evaporated at room temperature, and the α-cyano-4-hydroxycinnamic acid matrix was crystallized with peptides on the MALDI plate. A data-dependent acquisition was performed using Xcalibur software, in which the 40 most abundant precursor ions from the survey scan (mass range 700–3500 Da) were chosen, and MS/MS acquisition was triggered to fragment them by collision-induced dissociation. The normalized collision energy was 50%, and the isolation width was 3 Da. The raw files from the LTQ mass spectrometer were analyzed using Mascot Distiller version 2.3.2 (Matrix Science, Boston, MA) for protein identification. Peptide masses were matched against the taxonomy A. thaliana in the National Centre for Biotechnology Information non-redundant (NCBInr) database. One missed trypsin cleavage per peptide was allowed, and an initial mass tolerance of 0.3 Da was used in all searches. Complete carboxyamidation of cysteine sulfhydryls and partial oxidation of methionine were assumed.
Statistical Analysis
ImageJ (National Institutes of Health) and ImageQuant TL version 8.1 software (GE Healthcare) were used for quantification, values are means ± S.E., and Student's t test was utilized. p < 0.05 is denoted as significant.
RESULTS
AtPARK13 Is Similar to Parkinson Disease PARK13/HTRA2 and Localizes to Mitochondria
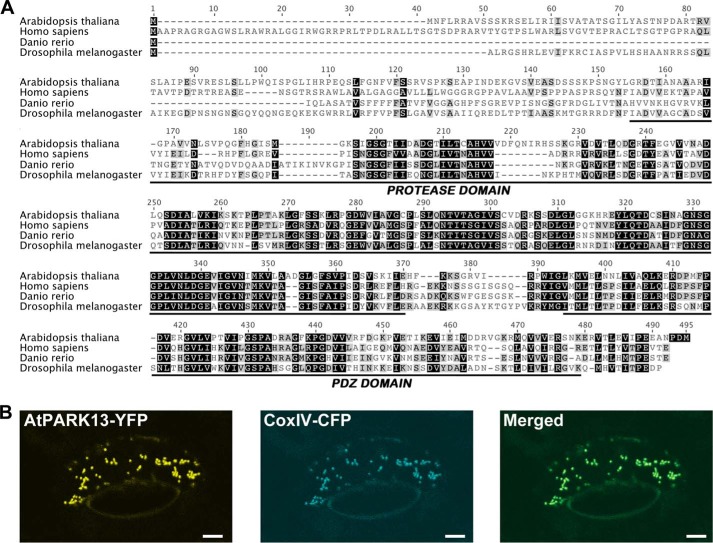
To investigate the conservation of PARK13 across species, we performed prediction analysis suggesting that the Arabidopsis endopeptidase NM122648 (at5g27660) is similar to the human PARK13/HTRA2 (18). Multiple sequence alignment analyses revealed, however, low sequence conservation beyond amino acid residue 411 within the conserved NM122648 PDZ domain. We cloned the at5g27660 cDNA (AtPARK13), revealing an extension of exon 10, by 18 nucleotides, and a shift in the reading frame leading to the conversion of intron 11 into an exonic sequence, effectively fusing exons 11 and 12. The cloned, full-length AtPARK13 is ∼40% similar to PARK13 from humans, Drosophila, and zebrafish, showing high similarity within the protease and PDZ domains (Fig. 1A).
FIGURE 1.
Conservation and subcellular localization of AtPARK13. A, alignment of PARK13 amino acid sequences showing conservation with the protease and PDZ domain from a number of different species. B, confocal microscopy of CaMV35S-AtPARK13-YFP in tobacco leaf cells showing mitochondrial AtPARK13 localization. CoxIV-CFP was used as a mitochondrial marker.
To address the subcellular localization of AtPARK13, we transfected an AtPARK13-YFP transgene into tobacco leaves by particle bombardment. Using confocal laser scanning microscopy analysis, with a CoxIV-CFP transgene as a mitochondrial marker, we found that AtPARK13, like its human counterpart, localizes to mitochondria (Fig. 1B).
AtPARK13 Is Heat Stress-induced but Does Not Respond to Oxidative Stress or Metal Toxicity
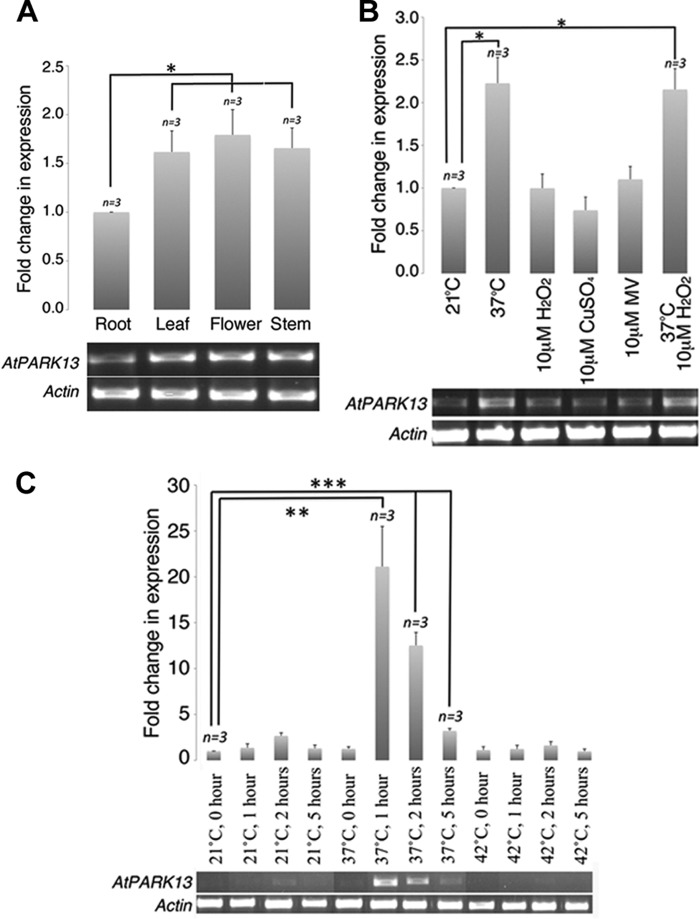
To gain insight into the expression dynamics of AtPARK13, we performed several expression analyses. Spatial expression patterns of AtPARK13 were analyzed by semiquantitative RT-PCR in different plant tissues. We found that AtPARK13 expression levels were significantly higher in aerial parts of the plant, including flowers, stems, and leaves as compared with roots (Fig. 2A). To further analyze possible stress-induced regulation of AtPARK13, we performed semiquantitative RT-PCR on WT plants exposed to elevated temperatures. From this analysis, we found that AtPARK13 transcript levels are significantly up-regulated when plants are shifted from 21 to 37 °C for 5 h (Fig. 2B). To test whether AtPARK13 levels were induced by other stress conditions, we exposed 2-week-old WT plants to 10 μm H2O2, CuSO4, and methyl viologen/Paraquat. No increase in AtPARK13 levels, above baseline, was observed in response to these stress treatments (Fig. 2B). Additionally, combining H2O2 and heat stress (37 °C) had no additive effect on AtPARK13 expression levels (Fig. 2B).
FIGURE 2.
Spatial and heat-induced expression profiles of AtPARK13. A, semiquantitative RT-PCR showing spatial expression patterns of AtPARK13 in A. thaliana. B, semiquantitative RT-PCR showing AtPARK13 expression profiles in response to 37 °C, 10 μm H2O2, CuSO4, methyl viologen/Paraquat (MV), and 37 °C plus 10 μm H2O2 for 5 h. C, semiquantitative RT-PCR showing AtPARK13 expression at 21, 37, and 42 °C for 1, 2, and 5 h. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.E., n = 3.
The kinetics of AtPARK13 expression was also analyzed, showing a rapid ∼20-fold increase in transcript levels after exposure to 37 °C for only 1 h (Fig. 2C). Interestingly, after 2 h at 37 °C, AtPARK13 transcript levels decreased to ∼10-fold above baseline levels with a further reduction after 5 h at 37 °C (Fig. 2C). AtPARK13 transcript levels were not affected by exposure to 42 °C (Fig. 1E).
AtPARK13 Overexpression Confers Thermotolerance
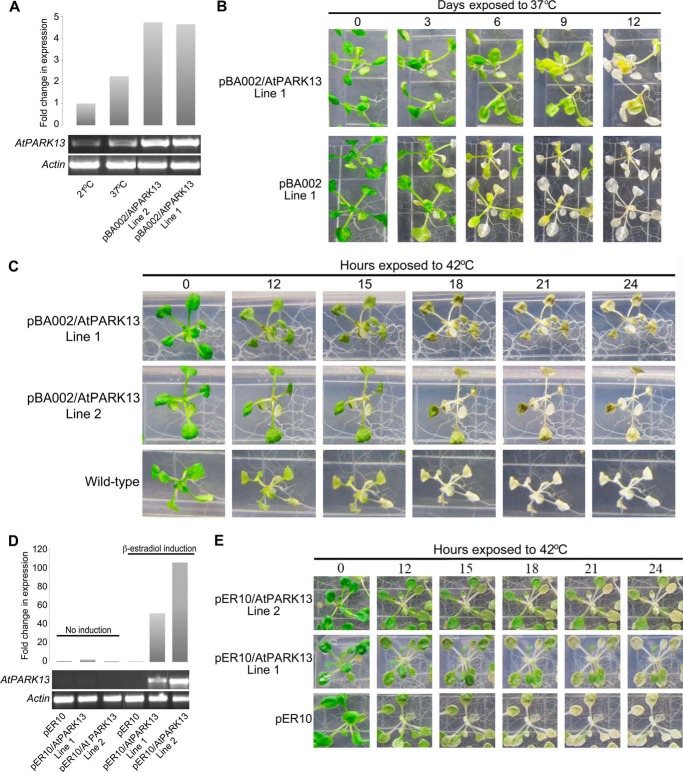
PARK13 knock-out mice show a Parkinsonian phenotype, whereas overexpression confers neuroprotection (24, 25). We initially attempted to characterize two AtPARK13 T-DNA mutant lines (SAIL536 having a T-DNA insertion in the 3′-UTR and SAIL905 having a T-DNA insertion in the coding region); however, no homozygous lines could be recovered (data not shown). We then generated gain-of-function AtPARK13 lines using the CaMV35S promoter (pBA002) (19) and the β-estradiol XVE-inducible promoter (pER10) (20). Two independent lines from each construct were selected, showing high levels of AtPARK13 expression (Fig. 3, A and D). To investigate potential protective roles of AtPARK13, two pBA002-AtPARK13 overexpression lines were subjected to heat stress and compared with transgenic lines containing the empty pBA002 vector and WT plants. We initially tested the effect on survival in AtPARK13-overexpressing plants in response to 37 °C exposure for 12 days. After 6 and 8 days at 37 °C, transgenic plants containing the empty vector (pBA002, line 1) showed clear bleaching, whereas transgenic plants overexpressing AtPARK13 (pBA002/AtPARK13, line 1) showed thermotolerance (Fig. 3B).
FIGURE 3.
AtPARK13 overexpression confers thermotolerance. A, semiquantitative RT-PCR showing AtPARK13 overexpression in two independent pBA002-AtPARK13 transgenic lines as compared with the WT at 21 and 37 °C for 5 h. B, increased resistance of AtPARK13-overexpressing plants as compared with transgenic plants expressing the pBA002 empty vector as a control at 37 °C for 12 days. C, increased resistance to heat stress (42 °C for 24 h) and thermotolerance in AtPARK13 overexpression lines as compared with WT plants. D, semiquantitative RT-PCR showing AtPARK13 overexpression in two independent pER10-AtPARK13 transgenic lines after β-estradiol induction as compared with the uninduced lines. E, increased resistance to heat stress (42 °C for 24 h) and thermotolerance in AtPARK13 overexpression lines with inducible promoter system compared with pER10 empty vector as a control.
We then analyzed the phenotypes of the same transgenic plants in response to 42 °C. After 12 h of exposure to 42 °C, WT plants exhibited bleaching and cell death as compared with plants overexpressing AtPARK13 (Fig. 3C). Further, after 21 and 24 h of exposure to 42 °C, WT plants were completely bleached, whereas AtPARK13-overexpressing plants showed clear cell survival (Fig. 3C).
Because the transgenic pER10-AtPARK13 lines selected exhibited higher AtPARK13 levels (Fig. 3D) compared with the CaMV35S/AtPARK13 transgenic lines analyzed (Fig. 3A), we tested whether the observed thermotolerance was dependent on AtPARK13 levels. As for the pBA002-AtPARK13 transgenic plants (Fig. 3, B and C), we observed clear thermotolerance in the pER10/AtPARK13 transgenic lines as compared with empty vector pER10 plants (Fig. 3E). Moreover, the degree of thermotolerance appeared higher in the pER10/AtPARK13 transgenic plants (Fig. 3E) compared with the pBA002/AtPARK13 plants (Fig. 3C), suggesting a possible dose-dependent effect.
AtPARK13 Is a Protease That Preferentially Acts on Misfolded Proteins
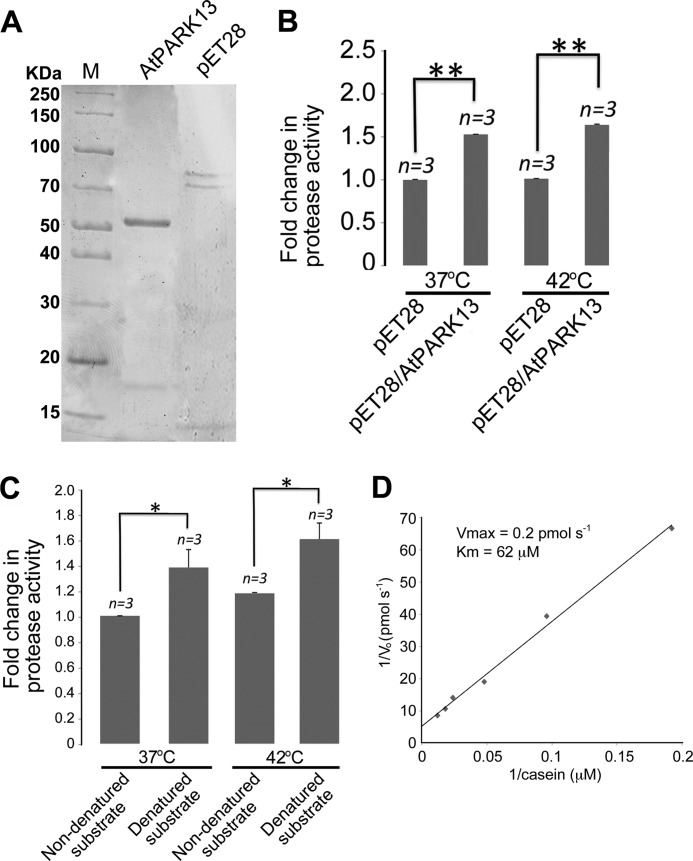
To analyze the AtPARK13 enzymatic activity, we expressed AtPARK13 (pET28a-AtPARK13) in E. coli and purified the protein under denaturing conditions using affinity chromatography (Fig. 4A). Following refolding by stepwise dialysis, protease activity assays, using casein as a generic substrate, demonstrated that AtPARK13 is indeed a protease when compared with fractions purified from E. coli expressing pET28a alone (Fig. 4B). Previous studies have shown that PARK13 activity increases with increasing temperature (26); however, AtPARK13 showed no significant increase in activity at 42 °C as compared with 37 °C (Fig. 4B). Because proteases often recognize unfolded/misfolded proteins, we then addressed whether denatured casein represented a better AtPARK13 substrate as compared with native casein. We found a significant increase in protease activity toward denatured casein as compared with non-denatured casein substrate (Fig. 4C). This suggests that AtPARK13 preferentially acts on misfolded/unassembled proteins. We also analyzed the Vmax and Km in response to misfolded casein was obtained as 0.2 pmol−1 and 62 μm, respectively.
FIGURE 4.
AtPARK13 is a protease that prefers misfolded proteins. A, Coomassie-stained SDS-polyacrylamide gel showing purified AtPARK13 and the purified fraction from the pET28 empty vector control. B, protease activity assays showing that AtPARK13 has significant protease activity toward casein, which is not affected by increased temperature. C, protease activity assays showing that AtPARK13 preferentially degrades misfolded casein. D, Lineweaver-Burk plot showing the Vmax and Km for AtPARK13 in response to misfolded casein. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.E., n = 3.
AtPARK13 and Human PARK13 Can Degrade the PD Proteins SNCA and DJ-1/PARK7
To test whether the AtPARK13 protease could degrade known PD-associated proteins, we exposed purified native SNCA to purified AtPARK13. We found that AtPARK13 could degrade native SNCA, as observed for casein (Fig. 5A). We further observed that misfolded SNCA represents a better target for AtPARK13, showing a ∼2-fold reduction in SNCA levels (Fig. 5A). Because human PARK13 is synthesized as a precursor that becomes processed into a mature cytosolic form (lacking the signal peptide and the trans-membrane domain) in response to apoptotic stimuli (27), we tested whether a mature form of AtPARK13 showed altered protease activity. To this end, we expressed and purified a truncated mature-mimicked version of AtPARK13 (mAtPARK13; amino acids 122–436, lacking the signal peptide and the predicted trans-membrane domain) under denaturing conditions and the mature form of hPARK13 (mhPARK13) under non-denaturing conditions (Fig. 5B). Following refolding by stepwise dialysis, casein degradation assays showed no significant difference in mAtPARK13 activity as compared with full-length AtPARK13 (data not shown). As expected, mhPARK13 was able to degrade casein (data not shown). We then analyzed whether mhPARK13 could degrade SNCA. We observed a ∼2-fold reduction in both folded and unfolded SNCA in response to mhPARK13, similar to the observations with AtPARK13 (Fig. 5, A and C).
FIGURE 5.
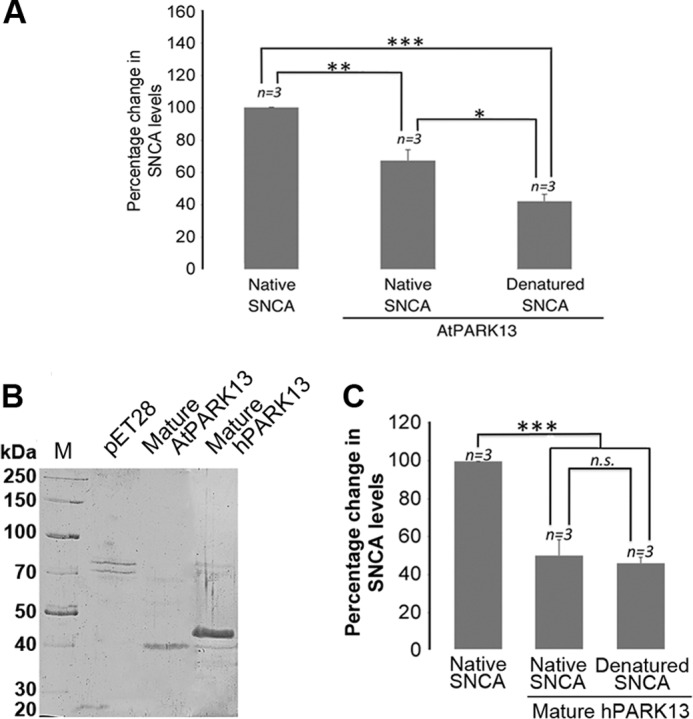
AtPARK13 and hPARK13 degrade SNCA. A, percentage change in native and misfolded SNCA levels in response to AtPARK13. AtPARK13 shows highest activity toward misfolded SNCA. B, Coomassie-stained SDS-polyacrylamide gel showing purified mature AtPARK13 and mature hPARK13 and the purified fraction from the pET28 empty vector control. C, percentage decrease in native and denatured SNCA levels in response to mature hPARK13. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.E., n = 3.
Because the AtPARK13 protein used for this study was purified from the insoluble fraction followed by refolding, we sought to verify our findings using AtPARK13 purified from the soluble fraction in its native state. Soluble AtPARK13 was expressed and purified under native conditions in E. coli alongside an empty vector control showing a molecular weight reminiscent of the processed mature form (Fig. 6A). We next tested whether this mature form of AtPARK13 could act on casein as a generic substrate. We found, as for full-length AtPARK13 (Fig. 4C), that native, processed mAtPARK13 showed clear protease activity against non-denatured and denatured casein (Fig. 6B). We also tested whether the native mAtPARK13 could act on SNCA, and as with full-length AtPARK13 (Fig. 5A), we observed a clear reduction in SNCA (data not shown).
FIGURE 6.
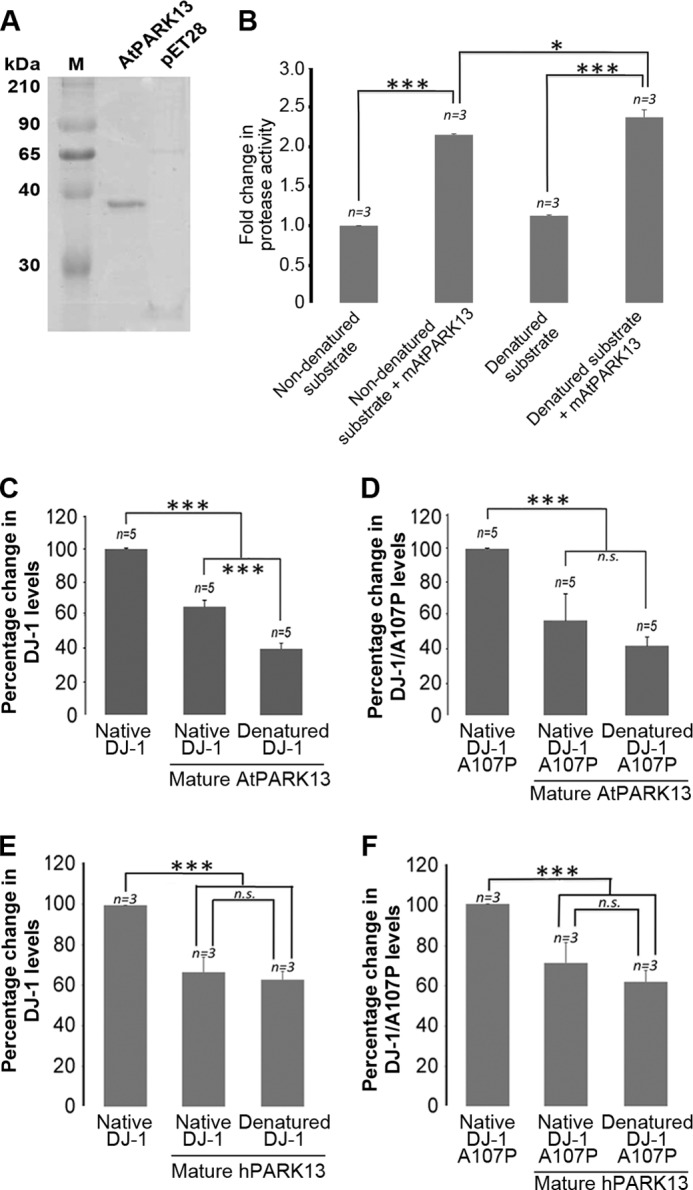
AtPARK13 and hPARK13 degrade DJ-1/PARK7. A, Coomassie-stained SDS-polyacrylamide gel showing purified native AtPARK13 and the purified fraction from the pET28 empty vector control. B, protease activity assays showing that mAtPARK13 degrades folded and misfolded casein. C, percentage decrease in DJ-1/PARK7 levels in response to mAtPARK13. D, percentage decrease in DJ-1/PARK7 A107P levels in response to mAtPARK13. E, percentage decrease in DJ-1/PARK7 levels in response to mhPARK13. F, percentage decrease in DJ-1/PARK7 A107P levels in response to mhPARK13. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.E., n = 3.
We also sought to analyze whether DJ-1/PARK7 could act as a substrate for AtPARK13. To this end, we exposed purified WT DJ-1/PARK7 to purified mAtPARK13. From this analysis, we found that DJ-1/PARK7 levels were reduced by ∼40% when incubated with AtPARK13, suggesting that DJ-1/PARK7 is an AtPARK13 substrate in vitro (Fig. 5C). As with SNCA, denatured DJ-1/PARK7 appears to be a better substrate for mAtPARK13 (Fig. 6C). To further analyze this finding, we also exposed the DJ-1/PARK7 A107P mutated variant to AtPARK13. The DJ-1/PARK7 A107P mutation is presumed to render DJ-1/PARK7 less stable by conformationally altering the three-dimensional structure of the protein. From this analysis, we observed a similar reduction in DJ-1/PARK7 A107P as for WT DJ-1/PARK7 (Fig. 6D). Interestingly, denatured DJ-1/PARK7 A107P did not appear to act as a better substrate for mAtPARK13 (Fig. 6D).
Because the mature form of hPARK13 was able to degrade SNCA, we also tested whether mhPARK13 could also act on WT DJ-1/PARK13 and DJ-1/PARK7 A107P. From these experiments, we found that mhPARK13 could act on both WT DJ-1/PARK7 (Fig. 6E) and on the DJ-1/PARK7 A107P variant (Fig. 6F) but with no significant difference between the native and the denatured forms.
Proteome Changes in Response to AtPARK13 Overexpression and Thermotolerance
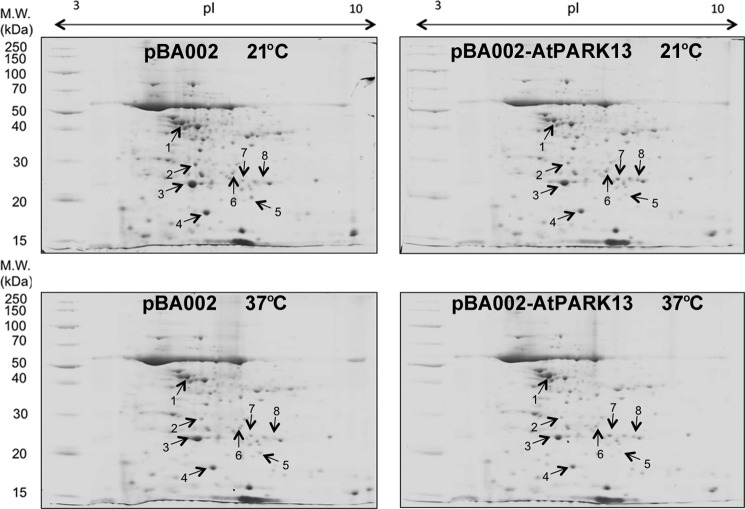
Comparative two-dimensional PAGE was performed on extracts from transgenic plants expressing the vectors pBA002 and pER10 alone and on plants overexpressing pBA002-AtPARK13 (Fig. 3A, Line 1) and pER10-AtPARK13 (Fig. 3B, Line 2) at 21 and 37 °C (Fig. 7). Three biological replicates were used, and proteins showing differential abundance were identified. The three biological replicas comprised extracts from transgenic plants containing the pBA002 and the pER10 vectors. From this analysis, we identified four proteins that showed differential abundance that were then identified by MALDI-linear ion trap peptide mass fingerprinting and tandem mass spectrometry (Table 1). The number of unique peptides and percentage coverage for the four proteins are shown in Table 2 and in supplemental Fig. 1. Three proteins showed decreased abundance in response to AtPARK13 expression, whereas one protein showed increased abundance (Table 1).
FIGURE 7.
Comparative two-dimensional PAGE in response to AtPARK13 overexpression in Arabidopsis at 21 and 37 °C. Protein extracts were separated on a 7-cm pH 3–10 non-linear IPG strip and in the second dimension on a SDS-polyacrylamide gel. Protein spots showing differential expression patterns are numbered.
TABLE 1.
LTQ-MALDI peptide mass fingerprinting of proteins differentially expressed in transgenic Arabidopsis expressing pBA002-AtPARK13 compared with plants expressing pBA002 alone with and without 37 °C heat treatment
| Spot no. | Protein | Swiss-Prot Accession no. | Mr | pI | Average -fold change |
Function | |
|---|---|---|---|---|---|---|---|
| 21 °C | 37 °C | ||||||
| Da | -fold | ||||||
| Down-regulated proteins in pBA002-AtPARK13 | |||||||
| 1 | Photosystem II subunit P-1 | Q42029 | 28,249 | 6.90 | 1.19 ± 0.08a | 0.94 ± 0.21 | Calcium ion binding, poly(U) RNA binding |
| 2 | Peptidyl-prolyl cis-transp isomerase CYP20–3 | P34791 | 28,532 | 8.83 | 1.08 ± 0.18 | 1.23 ± 0.09a | Peptide binding, peptide-prolyl cis-trans isomerase activity |
| 3 | Germin-like protein | P92998 | 23,195 | 6.81 | 2.06 ± 0.84 | 1.824 ± 0.14a | Manganese ion binding, Nutrient reservoir activity |
| Up-regulated protein in pBA002-AtPARK13 | |||||||
| 4 | Glutathione transferase | O80852 | 24,148 | 7.03 | 1.35 ± 0.42 | 1.42 ± 0.01a | Copper ion binding, glutathione binding, glutathione transferase activity, peroxidase activity |
a Significantly (p < 0.05) different proteins at 21 and 37 °C as compared with transgenic plants harboring the empty vector controls.
TABLE 2.
Number of unique peptides, the percentage of coverage, and the score of the proteins shown in Table 1
| Protein no. | No. of unique peptides | Coverage | Score |
|---|---|---|---|
| % | |||
| 1 | 4 | 28 | 500 |
| 2 | 5 | 34 | 512 |
| 3 | 1 | 19 | 138 |
| 4 | 1 | 5 | 41 |
Peptidyl-prolyl cis-trans isomerase CYP20-3 was identified as a moderately down-regulated protein in AtPARK13-overexpressing plants (Table 1, spot 2). AtPARK13 overexpression also resulted in down-regulation of photosystem II subunit P-1 (Table 1, spot 1) and germin-like protein (Table 1, spot 3). In response to AtPARK13 accumulation, we identified glutathione transferase (Table 1, spot 4) as being up-regulated.
DISCUSSION
Our findings show that AtPARK13 in A. thaliana is similar to human PARK13, a protease associated with PD (8, 9, 10, 11, 12). Indeed, AtPARK13 is significantly similar to PARK13 from other species, especially in the protease and the PDZ domain of PARK13 (Fig. 1A). The C-terminal PARK13 terminal PDZ domain generally recognizes and binds hydrophobic sequences of substrates or regulatory peptides and participates in the regulation of PARK13 catalytic activity (28). Indeed, it has been shown that elevated temperatures lead to increased exposure of the protease-PDZ domain interface, increasing proteolytic activity (29). Human PARK13 localizes predominantly to mitochondria (14), and we show that AtPARK13 also localizes to mitochondria in plant cells (Fig. 1B).
AtPARK13 expression is significantly elevated in leaves, flowers, and stems as compared with roots, establishing that AtPARK13 shows higher expression in heat-exposed aerial parts of plants (Fig. 2A). Assuming that aerial plant tissues are exposed to higher temperatures than tissues underground, this is in agreement with microarray data showing elevated AtPARK13 levels in response to heat stress (30). Heat stress may, indeed, uncouple metabolic pathways that ultimately cause accumulation of ROS, in turn triggering a heat stress response (31). Our data further suggest that AtPARK13 expression is specific to heat stress induction and is not affected by ROS or metal toxicity (Fig. 2, B and C).
Plants adapt to changes in temperature rapidly through inhibition of transcription and translation and by increasing expression of heat shock proteins to prevent protein aggregation (32). The AtPARK13 heat stress induction is therefore similar to that observed for heat shock proteins, and with the reported heat-induced PARK13 enzymatic activity (26), AtPARK13 may act in concert with classical heat stress response proteins in response to heat stress.
The retention of chlorophyll in leaves of transgenic plants with increased levels of AtPARK13 plants and better survival after exposure to elevated temperatures (37 and 42 °C), as compared with the control plants, demonstrate that AtPARK13 has a protective role in Arabidopsis conferring thermotolerance (Fig. 3, C and D). Several Arabidopsis genes have been identified as being associated with thermotolerance; however, most of these are also involved in other stress pathways (33). Interestingly, the plastidic FtsH11 protease is involved in thermotolerance in Arabidopsis (34). Plastidic FtsH proteases in Arabidopsis have been shown to degrade unassembled thylakoid membrane proteins (35), photodamaged D1 (36), and light-harvesting complex proteins (37). It is possible that the AtPARK13 protease has a similar function, degrading misfolded/unassembled proteins in response to heat stress. The AtPARK13-associated thermotolerance shown here is in agreement with studies showing PARK13-associated neuroprotection in mice (13).
Although AtPARK13 shows clear protease activity against the generic substrate casein, our results demonstrate that AtPARK13 prefers denatured casein as a substrate (Fig. 4C). This is not surprising because proteases in general prefer unfolded protein substrates, thereby reducing the accumulation of unfolded, non-functional proteins in cells (38). Indeed, bacterial and human HtrA2 proteases have been shown to be involved in the removal of misfolded or damaged proteins (39). The relatively low Vmax and high Km values measured suggest that AtPARK13 has low affinity for casein and converts low amounts of substrate per unit time, possibly suggesting that AtPARK13 harbors substrate specificity. Indeed, misfolded proteins may adopt a variety of structures, only some of which may act as efficient protease substrates.
In plants, organellar proteases preferentially degrade misfolded proteins in response to stress with a high degree of substrate specificity (35, 36). In neurodegenerative diseases, such as PD and Alzheimer disease, pathological hallmarks are represented by protein misfolding and aggregation of SNCA and β-amyloids, respectively (40). A study has shown that PARK13 can facilitate the degradation of the pathogenic A53T SNCA variant but that this is mediated through autophagy (41). We demonstrate that AtPARK13 can degrade SNCA directly, without the involvement of autophagy (Fig. 5A). Furthermore, we show that misfolded SNCA, mimicking transition states in PD pathology, increases the susceptibility of SNCA as a target for AtPARK13-mediated degradation (Fig. 5A). The fact that misfolded SNCA does not appear to represent a better substrate for mhPARK13 (Fig. 5C) may suggest that cytosolic mhPARK13 acts equally on different states of SNCA. This is in agreement with the finding that hPARK13 is processed into a cytosolic mature form in response to apoptotic signals, thereby acting on SNCA as a whole (27). We also show that AtPARK13 can degrade both native WT DJ-1/PARK7 and the A107P mutational variant (Fig. 6, C and D). Interestingly, further unfolding of DJ-1/PARK7 A107P by heat denaturation does not appear to increase its susceptibility as a protease substrate as compared with WT DJ-1/PARK7 (Fig. 6, E and F), suggesting that the A107P mutational variant is sufficiently unfolded to expose additional protease cleavage sites.
It is possible that PARK13 acts as a moderator of native SNCA levels to ensure SNCA homeostasis. Furthermore, our data suggest that hPARK13 may target SNCA as part of a neuroprotective mechanism during neurodegeneration in order to reduce the levels of native and/or misfolded SNCA and subsequent blocking of the proteasome. It is also possible that PARK13 targets DJ-1, thereby modulating appropriate DJ-1 levels, depending on the cellular environment.
Comparative two-dimensional PAGE analysis may give insight into global proteome changes in response to genetic alterations. We found that AtPARK13 overexpression resulted in three proteins showing significant down-regulation, whereas one protein showed up-regulation (Table 1).
Peptidyl-prolyl cis-trans isomerase CYP20-3 was identified as a moderately down-regulated protein in AtPARK13-overexpressing plants, particularly at elevated temperatures (Table 1, spot 2). Peptidyl-prolyl cis-trans isomerases catalyze cis-trans isomerization of imide bonds in proteins and are involved in protein folding (42). Glutathione transferase, involved in ROS protection, was the only protein identified that was up-regulated in response to AtPARK13 (Table 1, spot 4).
In summary, this study demonstrates that AtPARK13, is a mitochondrial protease that confers thermotolerance in A. thaliana. The study suggests that AtPARK13 prefers misfolded protein substrates, suggesting a mode of action in terms of heat stress protection. Importantly, we demonstrate that AtPARK13 and hPARK13 can directly degrade SNCA and DJ-1/PARK7 without autophagy involvement (41). In terms of DJ-1/PARK7, PARK13 may act on mitochondrial DJ-1/PARK7 to maintain protein homeostasis under physiological conditions or to reduce levels of DJ-1/PARK7 mutational variants predominantly targeted to mitochondria. Conversely, during stress conditions, PARK13 may also act on cytosolic DJ-1/PARK7 because it has been shown that PARK13 is released into the cytosol in response to apoptotic stimulus (27). Similarly, we propose that cytosolic PARK13 may act on cytosolic SNCA during stress conditions or apoptotic signals in neurons to reduce its neurotoxicity potential. PD represents a multifaceted and complex neurodegenerative disorder, and our study provides new insight into the role of PARK13 in PD by using A. thaliana as an unconventional, but complementary model system.
Supplementary Material
Acknowledgments
We thank Dr. Lashuel (University of Lausanne, France) for providing purified SNCA. We thank Katherine Moller for language corrections.
This work was supported by grants from the Norwegian Research Council, the Western Norway Regional Health Authority, the Norwegian Centre for Movement Disorders, and the Department of Biological Sciences at St. John's University.

This article contains supplemental Fig. 1.
- PD
- Parkinson disease
- ROS
- reactive oxygen species
- SNCA
- α-synuclein
- mAtPARK13
- mature AtPARK13
- mhPARK13
- mature human PARK13
- TFA
- trifluoroacetic acid
- CFP
- cyan fluorescent protein
- ECFP
- enhanced cyan fluorescent protein
- IPG
- immobilized pH gradient.
REFERENCES
- 1. Hirsch E., Graybiel A. M., Agid Y. A. (1988) Melanized dopaminergic neurons are differentially susceptible to degeneration in Parkinson's disease. Nature 334, 345–348 [DOI] [PubMed] [Google Scholar]
- 2. Martin I., Dawson V. L., Dawson T. M. (2011) Recent advances in the genetics of Parkinson's disease. Annu. Rev. Genomics Hum. Genet. 12, 301–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bartels T., Choi J. G., Selkoe D. J. (2011) α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature 477, 107–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 [DOI] [PubMed] [Google Scholar]
- 5. Valente E. M., Salvi S., Ialongo T., Marongiu R., Elia A. E., Caputo V., Romito L., Albanese A., Dallapiccola B., Bentivoglio A. R. (2004) PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann. Neurol. 56, 336–341 [DOI] [PubMed] [Google Scholar]
- 6. Bonifati V., Rizzu P., Squitieri F., Krieger E., Vanacore N., van Swieten J. C., Brice A., van Duijn C. M., Oostra B., Meco G., Heutink P. (2003) DJ-1 (PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol. Sci. 24, 159–160 [DOI] [PubMed] [Google Scholar]
- 7. Schapira A. H., Gegg M. (2011) Mitochondrial contribution to Parkinson's disease pathogenesis. Parkinsons Dis. 2011, 159160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang C. Y., Xu Q., Weng L., Zhang Q., Zhang H. N., Guo J. F., Tan L. M., Tang J. G., Yan X. X., Tang B. S. (2011) Genetic variations of Omi/HTRA2 in Chinese patients with Parkinson's disease. Brain Res. 1385, 293–297 [DOI] [PubMed] [Google Scholar]
- 9. Strauss K. M., Martins L. M., Plun-Favreau H., Marx F. P., Kautzmann S., Berg D., Gasser T., Wszolek Z., Müller T., Bornemann A., Wolburg H., Downward J., Riess O., Schulz J. B., Krüger R. (2005) Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson's disease. Hum. Mol. Genet. 14, 2099–2111 [DOI] [PubMed] [Google Scholar]
- 10. Lin C. H., Chen M. L., Chen G. S., Tai C. H., Wu R. M. (2011) Novel variant Pro143Ala in HTRA2 contributes to Parkinson's disease by inducing hyperphosphorylation of HTRA2 protein in mitochondria. Hum. Genet. 130, 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bogaerts V., Nuytemans K., Reumers J., Pals P., Engelborghs S., Pickut B., Corsmit E., Peeters K., Schymkowitz J., De Deyn P. P., Cras P., Rousseau F., Theuns J., Van Broeckhoven C. (2008) Genetic variability in the mitochondrial serine protease HTRA2 contributes to risk for Parkinson disease. Hum. Mutat. 29, 832–840 [DOI] [PubMed] [Google Scholar]
- 12. Krüger R., Sharma M., Riess O., Gasser T., Van Broeckhoven C., Theuns J., Aasly J., Annesi G., Bentivoglio A. R., Brice A., Djarmati A., Elbaz A., Farrer M., Ferrarese C., Gibson J. M., Hadjigeorgiou G. M., Hattori N., Ioannidis J. P., Jasinska-Myga B., Klein C., Lambert J. C., Lesage S., Lin J. J., Lynch T., Mellick G. D., de Nigris F., Opala G., Prigione A., Quattrone A., Ross O. A., Satake W., Silburn P. A., Tan E. K., Toda T., Tomiyama H., Wirdefeldt K., Wszolek Z., Xiromerisiou G., Maraganore D. M. (2011) A large-scale genetic association study to evaluate the contribution of Omi/HtrA2 (PARK13) to Parkinson's disease. Neurobiol. Aging 32, 548.e9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martins L. M., Morrison A., Klupsch K., Fedele V., Moisoi N., Teismann P., Abuin A., Grau E., Geppert M., Livi G. P., Creasy C. L., Martin A., Hargreaves I., Heales S. J., Okada H., Brandner S., Schulz J. B., Mak T., Downward J. (2004) Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol. Cell. Biol. 24, 9848–9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Plun-Favreau H., Klupsch K., Moisoi N., Gandhi S., Kjaer S., Frith D., Harvey K., Deas E., Harvey R. J., McDonald N., Wood N. W., Martins L. M., Downward J. (2007) The mitochondrial protease HtrA2 is regulated by Parkinson's disease-associated kinase PINK1. Nat. Cell Biol. 9, 1243–1252 [DOI] [PubMed] [Google Scholar]
- 15. Moisoi N., Klupsch K., Fedele V., East P., Sharma S., Renton A., Plun-Favreau H., Edwards R. E., Teismann P., Esposti M. D., Morrison A. D., Wood N. W., Downward J., Martins L. M. (2009) Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 16, 449–464 [DOI] [PubMed] [Google Scholar]
- 16. Fitzgerald J. C., Camprubi M. D., Dunn L., Wu H. C., Ip N. Y., Kruger R., Martins L. M., Wood N. W., Plun-Favreau H. (2012) Phosphorylation of HtrA2 by cyclin-dependent kinase-5 is important for mitochondrial function. Cell Death Differ. 19, 257–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu X. M., Lin H., Maple J., Björkblom B., Alves G., Larsen J. P., Møller S. G. (2010) The Arabidopsis DJ-1a protein confers stress protection through cytosolic SOD activation. J. Cell Sci. 123, 1644–1651 [DOI] [PubMed] [Google Scholar]
- 18. Xu X. M., Møller S. G. (2011) The value of Arabidopsis research in understanding human disease states. Curr. Opin. Biotechnol. 22, 300–307 [DOI] [PubMed] [Google Scholar]
- 19. Kost B., Spielhofer P, Chua NH. (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16, 393–401 [DOI] [PubMed] [Google Scholar]
- 20. Zuo J., Niu Q. W., Chua N. H. (2000) An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273 [DOI] [PubMed] [Google Scholar]
- 21. Fulda M., Shockey J., Werber M., Wolter F. P., Heinz E. (2002) Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid β-oxidation. Plant J. 32, 93–103 [DOI] [PubMed] [Google Scholar]
- 22. Clough S. J., Bent A. F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 23. Mesquita R. O., de Almeida Soares E., de Barros E. G., Loureiro M. E. (2012) Method optimization for proteomic analysis of soybean leaf: improvements in identification of new and low-abundance proteins. Genet. Mol. Biol. 35, 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu M. J., Liu M. L., Shen Y. F., Kim J. M., Lee B. H., Lee Y. S., Hong S. T. (2007) Transgenic mice with neuron-specific overexpression of HtrA2/Omi suggest a neuroprotective role for HtrA2/Omi. Biochem. Biophys. Res. Commun. 362, 295–300 [DOI] [PubMed] [Google Scholar]
- 25. Jones J. M., Datta P., Srinivasula S. M., Ji W., Gupta S., Zhang Z., Davies E., Hajnóczky G., Saunders T. L., Van Keuren M. L., Fernandes-Alnemri T., Meisler M. H., Alnemri E. S. (2003) Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature 425, 721–727 [DOI] [PubMed] [Google Scholar]
- 26. Zhang X., Chang Z. (2004) Temperature dependent protease activity and structural properties of human HtrA2 protease. Biochemistry 69, 687–692 [DOI] [PubMed] [Google Scholar]
- 27. Suzuki Y., Imai Y., Nakayama H., Takahashi K., Takio K., Takahashi R. (2001) A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell. 8, 613–621 [DOI] [PubMed] [Google Scholar]
- 28. Krojer T., Sawa J., Schäfer E., Saibil H. R., Ehrmann M., Clausen T. (2008) Structural basis for the regulated protease and chaperone function of DegP. Nature 453, 885–890 [DOI] [PubMed] [Google Scholar]
- 29. Zurawa-Janicka D., Jarzab M., Polit A., Skorko-Glonek J., Lesner A., Gitlin A., Gieldon A., Ciarkowski J., Glaza P., Lubomska A., Lipinska B. (2013) Temperature-induced changes of HtrA2(Omi) protease activity and structure. Cell Stress Chaperones 18, 35–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Charng Y. Y., Liu H. C., Liu N. Y., Chi W. T., Wang C. N., Chang S. H., Wang T. T. (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143, 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004) The reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498 [DOI] [PubMed] [Google Scholar]
- 32. Hirt H., Shinozaki K. (eds) (2004) Plant Responses to Abiotic Stress: Topics in Current Genetics, Vol. 4, pp. 73–93, Springer, Heidelberg [Google Scholar]
- 33. Kotak S., Larkindale J., Lee U., von Koskull-Döring P., Vierling E., Scharf K. D. (2007) Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 10, 310–316 [DOI] [PubMed] [Google Scholar]
- 34. Chen J., Burke J. J., Velten J., Xin Z. (2006) FtsH11 protease plays a critical role in Arabidopsis thermotolerance. Plant J. 48, 73–84 [DOI] [PubMed] [Google Scholar]
- 35. Yu F., Park S., Rodermel S. R. (2005) Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol. 138, 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bailey S., Thompson E., Nixon P. J., Horton P., Mullineaux C. W., Robinson C., Mann N. H. (2002) A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277, 2006–2011 [DOI] [PubMed] [Google Scholar]
- 37. Zelisko A., García-Lorenzo M., Jackowski G., Jansson S., Funk C. (2005) AtFtsH6 is involved in the degradation of the light-harvesting complex II during high-light acclimation and senescence. Proc. Natl. Acad. Sci. U.S.A. 102, 13699–13704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Truscott K. N., Bezawork-Geleta A., Dougan D. A. (2011) Unfolded protein responses in bacteria and mitochondria: a central role for the ClpXP machine. IUBMB Life 63, 955–963 [DOI] [PubMed] [Google Scholar]
- 39. Hansen G., Hilgenfeld R. (2013) Architecture and regulation of HtrA-family proteins involved in protein quality control and stress response. Cell Mol. Life Sci. 70, 761–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Selkoe D. J. (2004) Cell biology of protein misfolding: the examples of Alzheimer's and Parkinson's diseases. Nat. Cell Biol. 6, 1054–1061 [DOI] [PubMed] [Google Scholar]
- 41. Li B., Hu Q., Wang H., Man N., Ren H., Wen L., Nukina N., Fei E., Wang G. (2010) Omi/HtrA2 is a positive regulator of autophagy that facilitates the degradation of mutant proteins involved in neurodegenerative diseases. Cell Death Differ. 17, 1773–1784 [DOI] [PubMed] [Google Scholar]
- 42. Pliyev B. K., Gurvits B. Y. (1999) Peptidyl-prolyl cis-trans isomerases: structure and functions. Biochemistry 64, 738–751 [PubMed] [Google Scholar]
- 43. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.