Abstract
Inteins are mobile genetic elements capable of self-splicing post-translationally. They exist in all three domains of life including in viruses and bacteriophage, where they have a sporadic distribution even among very closely related species. In this review, we address this anomalous distribution from the point of view of the evolution of the host species as well as the intrinsic features of the inteins that contribute to their genetic mobility. We also discuss the incidence of inteins in functionally important sites of their host proteins. Finally, we describe instances of conditional protein splicing. These latter observations lead us to the hypothesis that some inteins have adapted to become sensors that play regulatory roles within their host protein, to the advantage of the organism in which they reside.
Keywords: Bioinformatics, DNA Enzymes, Invasion, Microbiology, Molecular Evolution, Splicing, Conditional Splicing, Intein Gain and Loss, Intein Localization, Protein Splicing
Protein Splicing
Protein splicing is a naturally occurring biochemical process that mediates the post-translational conversion of a precursor polypeptide into a mature and functional protein through the removal of an internal protein element, called an intein (Fig. 1A). The process is analogous to intron splicing at the RNA level. The protein splicing mechanism involves a series of autocatalytic peptide bond rearrangements, where the intein excises itself from the precursor polypeptide with concurrent ligation of the flanking sequences, called exteins (N- or C-exteins relative to the position of intein) (for review, see Refs. 1–3). As a result of this process, two proteins are produced from a single polypeptide product. The term intein refers to both the genetic element in the DNA or RNA and the protein splicing entity.
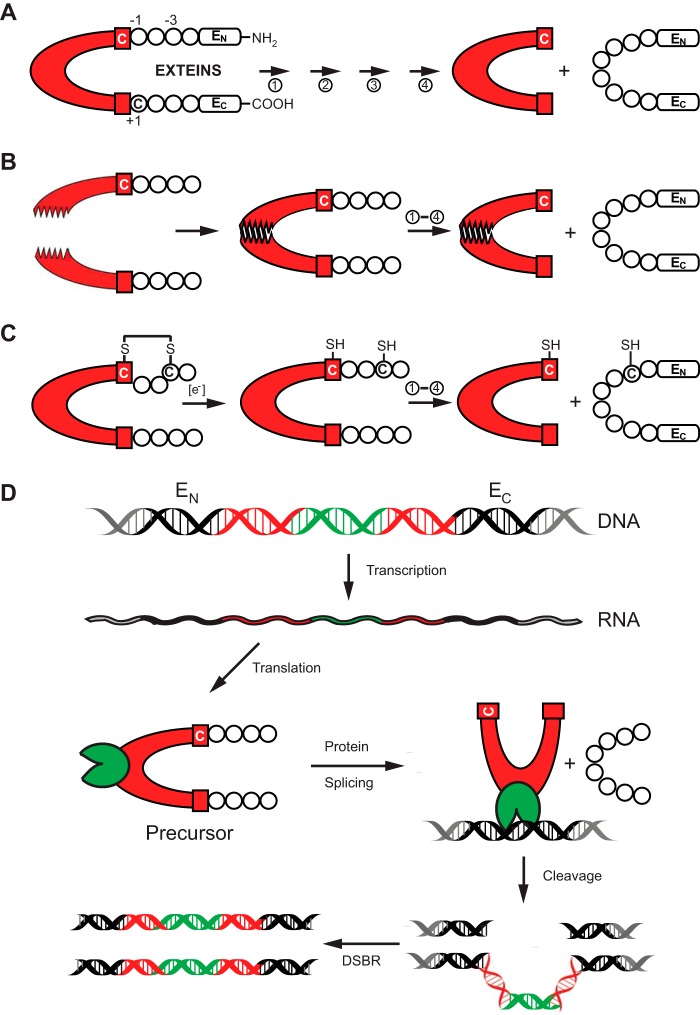
FIGURE 1.
Protein splicing and types of inteins. A, schematic mechanism of protein splicing. The intein is shown in red, flanked by the N-extein (EN) and the C-extein (EC). The four steps of protein splicing are designated by four arrows and are described by Mills et al. (2). The first residue of the intein and that of EC are usually a cysteine, serine, or threonine. They are shown here as Cys1 and Cys+1. B, protein trans-splicing by split inteins. The two halves of the split intein come together via a zipper-like interface. Splicing then proceeds as in A. C, an example of conditional protein splicing. A disulfide bridge, shown between cysteine residues at −3 of EN and at the first residue of the intein (Cys1), prevents protein splicing. In the presence of a reducing reagent, the disulfide bond is broken to release the trapped Cys1 and splicing proceeds. D, HEN-containing inteins are naturally occurring mobile genetic elements. After transcription and translation, precursor undergoes protein splicing with formation of the ligated exteins and intein carrying HEN domain (green). The HEN recognizes and cleaves its cognate intein-less homing site in DNA. The double-strand break is repaired by cellular double-strand break repair (DSBR) machinery using the intein-containing allele as template, resulting in two intron-containing alleles.
Most inteins are expressed within a single polypeptide chain (cis-splicing inteins), but some are split into two polypeptides each containing one extein and an intein fragment (trans-splicing inteins) (4, 5). In the case of split inteins, reassociation of the fragments at a zipper-like interface (6) precedes protein splicing (Fig. 1B). Both cis-splicing and trans-splicing inteins are frequently utilized in various biotechnological applications including protein purification, modification, labeling, and post-translational control of expressed proteins (reviewed in Refs. 7 and 8). The cis-splicing inteins often contain a distinct homing endonuclease (HEN)2 domain (9). HEN-containing inteins are naturally occurring mobile genetic elements. The presence of a HEN provides inteins the ability to transfer their coding elements into homologous alleles at homing sites that lack the intein sequence (Fig. 1D). This HEN-mediated homing process can result in horizontal gene transfer (HGT) of inteins, by invasion of diverse species, followed by vertical transmission of inteins (10, 11). Moreover, HEN-containing inteins are involved in a so-called “homing cycle” that includes two opposing processes, precise intein loss and reinvasion of a newly formed vacant homing site. It is believed that the homing cycle allows the HEN to avoid fixation and functional decay in one locus (12).
Sporadic Distribution of Inteins Relative to the Evolutionary History of Their Host
An intein was initially found in the yeast genome, in the vacuolar ATP synthase catalytic subunit A (VMA) (13, 14). Since then, inteins have been identified in all three domains of life, but none have been discovered in the nuclear genome of a multicellular organism. Notably, the distribution of inteins in different phyla and even among closely related species is sporadic (15, 16). To examine intein distribution, one can survey the intein database, InBase (16), for inteins across the tree of life, including viruses and bacteriophage. To more adequately sample intein occurrence, we also mined inteins from the National Center for Biotechnology Information (NCBI) Gene database (www.ncbi.nlm.nih.gov/gene). Based on the data from the NCBI, we performed a rough assessment of the number of putative intein sequences in sequenced genomes. Additionally, we searched for inteins in Eubacteria and Archaea by filtering the Gene database (Fig. 2; the complete analysis will be published elsewhere). A summary of the intein distribution derived from these databases throughout the tree of life appears in Fig. 2.
FIGURE 2.
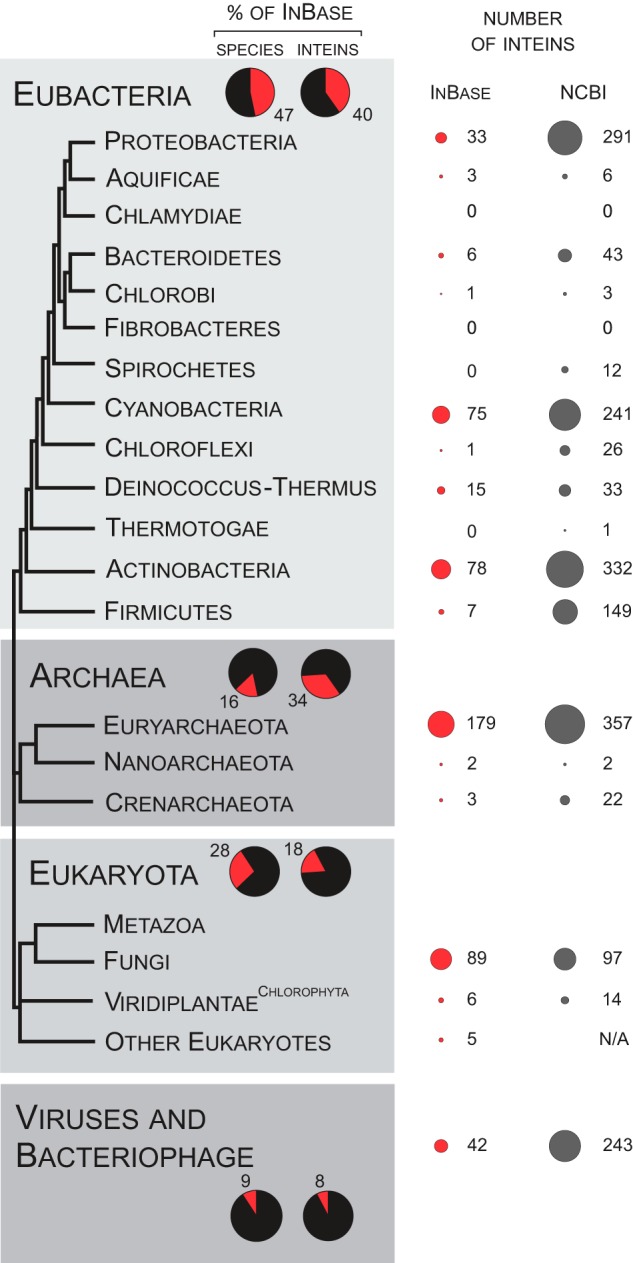
Distribution of inteins in the biosphere. The tree of life is shown after Gribaldo and Brochier-Armanet (93) with minor changes. Thirteen phyla are shown for Eubacteria and three are shown for Archaea. Three kingdoms are indicated for Eukaryota: metazoa or animals, fungi, and viridiplantae or plants (with emphasis on green algae, the Chlorophyta). The additional basal branch is dedicated to all other eukaryotes. Viruses and bacteriophage are not included in the tree and are shown separately at the bottom. In the shaded panels, red sectors show the percentage of Eubacteria, Archaea, Eukaryota, or viruses among all species (left) or inteins within InBase (right), for a total of 100% in each case. On the far right, the total number of inteins submitted to InBase is shown for each group as red circles, with the area of the circles corresponding to the number of inteins. The NCBI column shows results of a preliminary assessment for the intein occurrence in the NCBI Gene database. N/A = not available.
InBase content has some bias in the dataset when compared with the NCBI Gene database because InBase was generated by submissions from individual investigators. There are phyla that are either absent from or underrepresented in InBase (e.g. Spirochaete and Firmicutes), and there are significantly more putative inteins available in the NCBI Gene database than there were submitted into InBase. However, there are many parallels between the two databases, and for both there is clearly a sporadic pattern in intein distribution. The uneven nature of intein occurrence suggests biased intein acquisition, maintenance, and/or loss (Fig. 2).
Intein Flux
Although the dramatic variation in intein occurrence, even among closely related species, suggests that host genomes as a whole and inteins in particular are in flux, fully explaining this phenomenon remains a challenge in evolutionary biology. There are several potential interdependent factors contributing to the observed pattern of intein distribution that should be considered. Undoubtedly, the diverse evolutionary histories of the host genomes contributed to the heterogeneity of inteins at all scales (17, 18). Changes in genome size and frequent HGT should be weighed against gene-specific intein acquisition or loss resulting in the observed intein distribution. Usually, it is assumed that inteins are not under strong selective pressure, either negative or positive, because they splice out efficiently, producing fully functional host proteins. The only evolutionary pressure that is routinely attributed as acting on the inteins is selection for efficient protein splicing (10, 11). However, a number of the genome-reshaping processes might directly affect intein distribution.
Genome streamlining, the selective elimination of unnecessary DNA to decrease the metabolic burden on DNA replication, was proposed as a mechanism for genome reduction in free-living species with very large successful populations (19–21). In contrast, parasitic and symbiotic organisms undergo significant genome size reduction due to relaxation of positive selection, a bias favoring deletions over insertions, and pseudogenization (22–27). Both genome streamlining and adaptation to a symbiotic/parasitic lifestyle would result in intein loss. HGT (not associated with HEN activity and intein mobility), on the other hand, would aid the dissemination of inteins between distantly related species or even across the domains of life. HGT is common among Archaea and Bacteria and is believed to be the essential mechanism for the attainment of genetic diversity (28). Among the most prominent examples of massive and recurrent HGT is the dissemination of antibiotic resistance genes among bacterial pathogens (29–31).
HGT is also an important mechanism in eukaryotic genome evolution, particularly in unicellular organisms (32–35). More and more candidates for horizontally transferred genes are being identified: from Eubacteria to Eukaryota (33); from Eubacteria to Archaea (36); and among Eukaryota (34). Inteins might hitchhike as fragments of the horizontally transferred genes, gene clusters, genome fragments, or even whole chromosomes.
Viruses and bacteriophage represent ideal HGT vectors for intein dissemination (18, 37–39). They can carry intein sequences across cell boundaries as part of their own genomes or transfer cellular genes they acquired as a result of rampant recombination (37). There are many examples of viral and bacteriophage intein-bearing genomes. Among them is a giant virus from a major marine microflagellate grazer Cafeteria roenbergensis, a Mimivirus (40), a virus infecting disparate algae Heterosigma akashiwo (18), and Chrysochromulina ericina (41). There are also inteins in invertebrate-infecting Iridoviridae (42, 43), in the Chlorella virus NY-2A (44), and in coccolithoviruses infecting Emiliania huxleyi (45). Intein-containing haloviruses were described recently in metatranscriptomic analysis of the hypersaline community (46). Bacteriophage that carry inteins are also prevalent and were reported for Bacillus (47, 48), Mycobacteria (49), and Caulobacter (50), among others (16, 51–53).
Distribution of Inteins Is Biased toward Specific Proteins
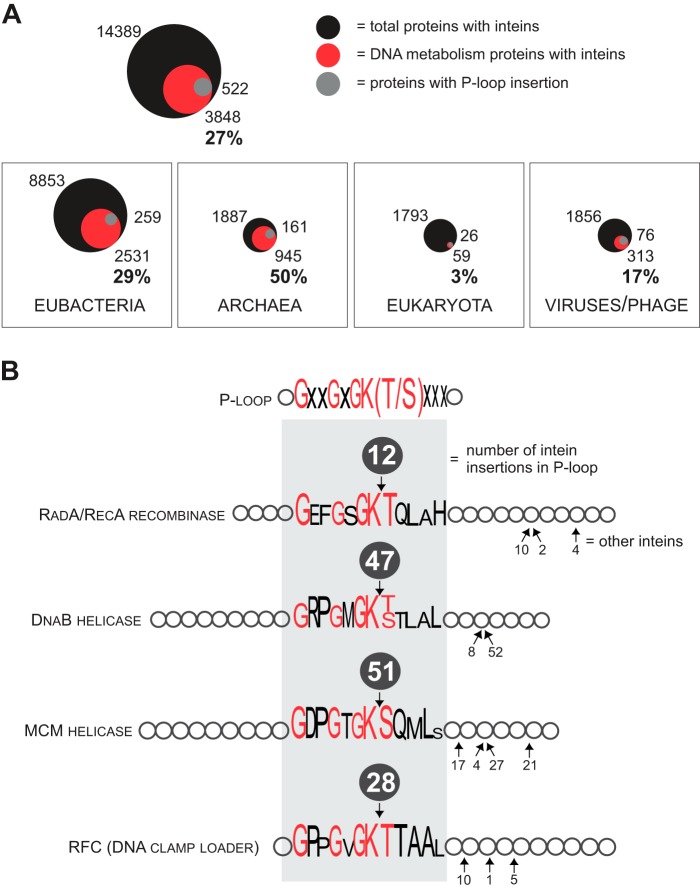
Although inteins occur in proteins with diverse functions, there is a bias for inteins to insert into proteins involved in DNA metabolism, such as polymerases (Pols), helicases, topoisomerases (TOPOs), and ribonucleotide reductases (RNRs) (Table 1; Fig. 3A) (16). Out of 545 inteins reported in InBase, 266 occur either in polymerases, such as PolA, PolB, PolC, DnaE, and RPB1, or in helicases including replicative helicases, DnaB (bacterial), and MCM/Cdc21 (archaeal) (Table 1). A search of the NCBI Protein database revealed that ∼27% of proteins with inteins across all groups corresponded to DNA metabolism proteins with the highest fraction in Archaea (∼50% of all putative intein-containing proteins) and lowest in Eukaryota (only ∼3%; Fig. 3A). The discrepancy between the fractions of intein-containing DNA-related proteins in the two databases is again attributable to different data submission criteria, but in both cases, the preponderance of inteins in DNA metabolism proteins is striking.
TABLE 1.
Some of the most common proteins with intein insertions
| Protein (P-loop insertion: +/−) | Function, process | Intein distribution |
|---|---|---|
| Helicase (DnaB) (+) | Replicative DNA helicase, DNA replication | Eubacteria |
| Mini-chromosome maintenance protein (MCM)/cell division control protein (Cdc21) (+) | Replicative DNA helicase, DNA replication | Archaea |
| Replication factor C (RFC) (+) | DNA clamp loader, DNA replication | Archaea |
| Recombinase (RecA/RadA) (+) | Recombinase, DNA repair | Eubacteria; Archaea |
| SF2 helicase (SWI/SNF2/Rad54) (+) | DNA helicase, DNA repair | Eubacteria |
| SF1 ATP-dependent DNA helicase (UvrD/Rep/PcrA) (+) | DNA helicase, DNA repair | Eubacteria |
| Catalytic α subunit of DNA polymerase III (DnaE) (−) | DNA polymerase subunit, DNA replication | Eubacteria |
| Subunit γ/τ of DNA polymerase III (DnaZX) (−) | DNA polymerase subunit, DNA replication | Eubacteria |
| DNA polymerase I (PolB) (−) | DNA polymerase, DNA replication | Archaea |
| DNA polymerase II, large subunit DP2 (PolC) (−) | DNA polymerase subunit, DNA replication | Archaea |
| Topoisomerase II – DNA gyrase, subunit B (GyrB) (−) | Topoisomerase, DNA replication | Eubacteria; Archaea; Bacteriophage |
| Bacterial DNA polymerase I (PolA) (−) | DNA polymerase, DNA repair | Eubacteria |
| Bacterial DNA polymerase II (PolB) (−) | DNA polymerase, DNA repair | Eubacteria |
| SF6 Holliday junction ATP-dependent DNA helicase (RuvB) (−) | DNA helicase, DNA repair | Eubacteria |
| Phage terminase, large subunit (−) | Terminase, DNA packaging | Bacteriophage |
| Ribonucleoside diphosphate reductase (RNR) (−) | Ribonucleoside diphosphate reductase, DNA synthesis | Eubacteria; Archaea |
| Vacuolar ATP synthase catalytic subunit A (VMA) (−) | Vacuolar H+-pump ATPase | Eukaryota; Archaea |
| Pre-mRNA-processing-splicing factor (PRP8) (−) | Spliceosome factor, splicing | Eukaryota |
FIGURE 3.
Distribution of inteins in proteome. A, intein concentration in proteins of DNA metabolism. Inteins tend to invade Pols, helicases (HELICs), TOPOs, and RNRs. Inteins occur in 27% of Pols, HELICs, TOPOs, and RNRs. The fraction of the inteins found in these proteins is shown for the three domains of life and viruses/bacteriophage. The numbers reflect data available in NCBI Protein database. The total number of the proteins with inteins is indicated on the top, represented by the black circle; the number and proportion (%) of Pols, HELICs, TOPOs, and RNRs with inteins are shown on the bottom, represented by a red circle; the number of proteins with intein inserted into P-loop is indicated on the side, represented by a gray circle. The area of the circles is proportional to the numbers. B, inteins in P-loops of some proteins. The P-loop is a conserved motif commonly found in ATP-binding domains with consensus amino acid sequence GXXGXGK(T/S) (X = any amino acid residue). The loop is a hot-spot for intein insertion in some of the host proteins such as RadA/RecA recombinase, replicative helicases DnaB and MCM, as well as the DNA clamp loader or RFC. The P-loop sequence is shown with the heights of the letters corresponding to the degree of sequence conservation; the most conserved residues are in red. Numbers in the dark gray circles on the top of protein representations correspond to the numbers of species/strains with an intein insertion at that position in the P-loop.
Several hypotheses have been proposed for the biased insertion of inteins into these proteins. First, because the intein HEN is produced simultaneously with its host protein, a possible advantage for an intein could be to ensure its own presence at times of DNA replication and repair. Because intein homing requires the host replication and repair machinery, it is efficient for the intein to be produced in concert with these replication and repair proteins. A second hypothesis suggests that mobility during replication and repair decreases selection against the intein. Indeed, the result of nonspecific activity of the intein HEN can be efficiently repaired during this period, reducing the risk of intein propagation (54). Finally, Pols, helicases, and TOPOs are frequently found in viral genomes (roughly 18% of the total viral proteome available at NCBI) (37). Viruses are efficient vectors for HGT (55–57) and, thus, they might facilitate spreading of inteins across species boundaries (18, 37–39).
Inteins Tend to Be Located at Protein Active Sites
Inteins often occur at sites critical to the function of the proteins, such as catalytic centers and key binding surfaces. Interestingly, a conserved motif containing the Walker A box (58) in a phosphate-binding loop called the P-loop is a hot-spot for intein invasion in some of these proteins. For example, of 33 species/strains with inteins that we found for recombinase RadA/RecA proteins, 12 had insertions in the P-loop, which has been identified in many ATP- and GTP-binding proteins (59) (Fig. 3B). Additionally, P-loop insertions in DnaB and MCM/Cdc21 helicases were found in 47 and 51 species/strains, respectively (Fig. 3B). Although other helicases also had intein insertions in the P-loop, this is not the case for the P-loops of Pols (Table 1) (16). Nevertheless, in line with inteins localizing to the conserved domains that are functionally important, inteins are inserted into either catalytic or ligand-binding sites of RNR, archaeal mini-chromosome maintenance protein (MCM) helicases and archaeal replication factor C (RFC), archaeal/eukaryotic VMA, and eukaryotic RNA polymerases (11, 60, 61).
The rationale behind the localization of inteins to the most critical domains of proteins is still a matter of debate, and might reflect differential targeting, maintenance, or loss. First, targeting of conserved sites might be explained by the specificity of intein HENs. Conserved amino acid residues within functionally important protein motifs limit the range of nucleotide substitutions that can be tolerated at such sites. Thus, the chance to lose the site recognized by the HEN is lower at active centers in comparison with other regions in the protein. Second, purifying selection, which plays an important role in maintaining the long term stability of biological systems by removing deleterious mutations, will preserve the functionally important motifs, facilitating intein invasion into that site by HGT across a wide range of species. Third, inteins could insert themselves into diverse sites throughout a genome, but only insertions at specific sites might become fixed in the population. The retention of inteins in conserved protein motifs would likely be due to the low rate of intein loss through excision. The removal of the intein, if not precise, would disrupt protein function and therefore would be deleterious (11, 60). If the intein were precisely excised from the genome, this new viable intein-less variant would likely be reinvaded if the intein contained an active HEN as a result of the homing cycle (12). A similar argument has been posited for retention of self-splicing introns in functionally important motifs (62, 63).
Finally, the presence of inteins in particular conserved motifs might be explained by an adaptive role of inteins. Mobile elements in general have been shown to evolve diverse roles (64–66). The vast knowledge that has accumulated about genome organization and gene expression during the last two decades has led to a paradigm shift from seeing mobile elements as solely parasitic entities to understanding their dynamic role in evolution of species (66). Despite their importance in biotechnology, inteins remain among the least studied mobile elements in terms of their possible function(s) (67). Nevertheless, the data available, especially on conditional protein splicing, suggest that inteins might be involved in regulation of function of the host protein.
Conditional Protein Splicing and Splicing Regulation
Both native and artificially designed inteins can undergo conditional protein splicing (CPS). CPS depends on the presence of a particular trigger, such as a change in redox state, temperature, small molecules, or light, as has been recently reviewed (7, 8). The existence of stimulus-dependent inteins suggests the possibility that some inteins may adapt to their intracellular niche by becoming post-translational regulatory elements that modulate protein splicing in accord with environmental conditions.
An example of such regulated protein splicing is provided by formation a disulfide bond by cysteine residues involved in the splicing mechanism, thereby trapping the intein inside a precursor protein depending on the redox state. Several inteins were artificially designed where disulfide bond formation controls premature cleavage or splicing reactions (68–70) (Fig. 1C). One of these designs inspired a search for physiologically relevant regulation of protein splicing by a disulfide bond. After determining by mutational analysis that cysteine at the −3 position of the upstream extein can trap the catalytic cysteine (Cys1) at the beginning of the cyanobacterial Ssp DnaE intein (68, 71), data mining revealed several native inteins with cysteine at the −3 position. Examples include the MoaA precursor protein from the thermophilic archaeon Pyrococcus abyssi, a radical S-adenosylmethionine domain protein of sulfate-reducing archaeon Archaeoglobus profundus,and the pyruvate-formate lyase-activating enzyme (PFL-AE) from an uncultured archaeon GZfos13E1 (68).
Strikingly, all three of the above mentioned proteins are predicted to catalyze redox chemistry, and each of the three inteins resides in a conserved CysXXXCysXXCys motif (where X is any amino acid residue). In all cases, the last Cys of the CysXXXCysXXCys motif is Cys1 of the intein, whereas the central Cys corresponds to the Cys−3 that forms a disulfide with Cys1 of the MoaA intein. Although disulfide bond formation and its regulatory role in the radical S-adenosylmethionine domain protein and pyruvate formate-lyase-activating enzyme remain to be elucidated, characterization of the MoaA intein in Escherichia coli revealed that the Cys−3-to-Cys1 disulfide bond can control intein splicing activity depending on the redox state of the host organism (68).
There are other naturally occurring examples of redox-sensitive inteins. Another P. abyssi intein, the PolII intein, can form a disulfide bond between Cys1 and the downstream extein Cys+1 that prevents splicing (72). A second PolII intein, Mma PolII intein from the methanogenic archaeon Methanoculleus marisnigri, can form an internal disulfide bond that modulates splicing activity of the intein depending on the redox state of E. coli or localization to the periplasm or cytoplasm (73).
Protein splicing can also be modulated by temperature. This regulation may have physiological relevance, as it was demonstrated that the activity of various inteins from extreme thermophiles depends on temperature (74–81). In addition to native temperature-dependent inteins, several temperature-sensitive inteins were developed to regulate gene expression using the yeast Sce VMA intein (82, 83). Additionally, some inteins have a modest pH dependence with a preference for low pH, which can be amplified by mutation (84). Although the natural importance of low pH preference for splicing is not yet known, it should be noted that the modulation of the Mtu RecA intein splicing by pH occurs precisely over the range of internal pH (pH 6–8) maintained by Mycobacterium smegmatis and Mycobacterium bovis BCG exposed to high acidity (85).
However, other inteins have been engineered to splice conditionally, and these are worth brief consideration because they may foreshadow the discovery of a similar innate control of splicing. For example, the activity of a trans-splicing intein (Fig. 1B) has been manipulated to depend on the presence of the small molecule, rapamycin, that induces intein reassociation and splicing (86, 87). Another way to control the intein relies on the substitution of the HEN domain by a receptor and utilization of a receptor-ligand binding event to trigger splicing. Examples include estrogen (88–90) and thyroid hormone (89, 91) with their respective receptors, and peroxisome proliferator-activated receptor (92). Although the development of these inteins involves a directed evolution step to tune the intein response to the ligand, the regulation of intein splicing implies some cross-talk between the catalytic center of the intein and extraneous peptide fusions to the intein.
The existence of both native and synthetic CPS anticipates more examples of regulated protein splicing in vivo with the appearance of a functional protein dependent on a specific environmental stimulus. The naturally occurring inteins are thought to have undergone evolution from invasive parasites to persistent mutualists that provide adaptive post-translational regulatory advantage to the host for survival under specific cellular and environmental circumstances. We thus anticipate more examples of CPS of inteins that respond to varied environmental cues in their natural host.
Conclusions
Intein occurrence is sporadic among related species. Inteins frequently reside in proteins involved in DNA metabolism, particularly in their active centers. Consideration of the innate invasiveness of inteins, evolutionary history of intein-containing species, as well as the nature of their protein hosts is necessary for a deeper understanding of intein dynamics and distribution. Although the loss of inteins is likely to be the consequence of genome size reduction in many species, the recurrent invasion of inteins is possible largely due to their intrinsic features as mobile elements and their ability to spread vertically and horizontally among species. An understanding of the evolution and distribution of inteins will shine light on the possible functionality of inteins as unique regulatory elements. Moreover, further studies of inteins could benefit if these genetic elements were viewed as the part of a complex system involving the nature of the split protein, the intein insertion site, the host species, and its environment.
Acknowledgments
We acknowledge Matt Stanger for help with Fig. 1 and Rebecca McCarthy for help with the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM39422 and GM44844 (to M. B.). This is the first article in the Thematic Minireview Series “Inteins.”
- HEN
- homing endonuclease domain
- HGT
- horizontal gene transfer
- CPS
- conditional protein splicing
- VMA
- vacuolar ATP synthase catalytic subunit A
- Pol
- polymerase
- TOPO
- topoisomerase
- RNR
- ribonucleotide reductase
- HELIC
- helicase
- MCM
- mini-chromosome maintenance protein
- RFC
- replication factor C.
REFERENCES
- 1. Paulus H. (2001) Inteins as enzymes. Bioorg. Chem. 29, 119–129 [DOI] [PubMed] [Google Scholar]
- 2. Mills K. V., Johnson M. A., Perler F. B. (2014) Protein splicing: how inteins escape from precursor proteins. J. Biol. Chem. 289, 14498–14505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Volkmann G., Mootz H. D. (2013) Recent progress in intein research: from mechanism to directed evolution and applications. Cell. Mol. Life. Sci. 70, 1185–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Muralidharan V., Muir T. W. (2006) Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat. Methods 3, 429–438 [DOI] [PubMed] [Google Scholar]
- 5. Xu M. Q., Evans T. C., Jr. (2005) Recent advances in protein splicing: manipulating proteins in vitro and in vivo. Curr. Opin. Biotechnol. 16, 440–446 [DOI] [PubMed] [Google Scholar]
- 6. Sorci M., Dassa B., Liu H., Anand G., Dutta A. K., Pietrokovski S., Belfort M., Belfort G. (2013) Oriented covalent immobilization of antibodies for measurement of intermolecular binding forces between zipper-like contact surfaces of split inteins. Anal. Chem. 85, 6080–6088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Topilina N. I., Mills K. V. (2014) Recent advances in in vivo applications of intein-mediated protein splicing. Mob. DNA 5, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shah N. H., Muir T. W. (2014) Inteins: nature's gift to protein chemists. Chem. Sci. 5, 446–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Derbyshire V., Wood D. W., Wu W., Dansereau J. T., Dalgaard J. Z., Belfort M. (1997) Genetic definition of a protein-splicing domain: functional mini-inteins support structure predictions and a model for intein evolution. Proc. Natl. Acad. Sci. U.S.A. 94, 11466–11471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pietrokovski S. (2001) Intein spread and extinction in evolution. Trends Genet. 17, 465–472 [DOI] [PubMed] [Google Scholar]
- 11. Gogarten J. P., Senejani A. G., Zhaxybayeva O., Olendzenski L., Hilario E. (2002) Inteins: structure, function, and evolution. Annu. Rev. Microbiol. 56, 263–287 [DOI] [PubMed] [Google Scholar]
- 12. Burt A., Koufopanou V. (2004) Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr. Opin. Genet. Dev. 14, 609–615 [DOI] [PubMed] [Google Scholar]
- 13. Kane P. M., Yamashiro C. T., Wolczyk D. F., Neff N., Goebl M., Stevens T. H. (1990) Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H+-adenosine triphosphatase. Science 250, 651–657 [DOI] [PubMed] [Google Scholar]
- 14. Hirata R., Ohsumk Y., Nakano A., Kawasaki H., Suzuki K., Anraku Y. (1990) Molecular structure of a gene, VMA1, encoding the catalytic subunit of H+-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 265, 6726–6733 [PubMed] [Google Scholar]
- 15. Perler F. B., Olsen G. J., Adam E. (1997) Compilation and analysis of intein sequences. Nucleic Acids Res. 25, 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perler F. B. (2002) InBase: the Intein Database. Nucleic Acids Res. 30, 383–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Swithers K. S., Soucy S. M., Gogarten J. P. (2012) The role of reticulate evolution in creating innovation and complexity. Int. J. Evol. Biol. 2012, 418964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nagasaki K., Shirai Y., Tomaru Y., Nishida K., Pietrokovski S. (2005) Algal viruses with distinct intraspecies host specificities include identical intein elements. Appl. Environ. Microbiol. 71, 3599–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dufresne A., Garczarek L., Partensky F. (2005) Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 6, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hessen D. O., Jeyasingh P. D., Neiman M., Weider L. J. (2010) Genome streamlining and the elemental costs of growth. Trends Ecol. Evol. 25, 75–80 [DOI] [PubMed] [Google Scholar]
- 21. Swan B. K., Tupper B., Sczyrba A., Lauro F. M., Martinez-Garcia M., González J. M., Luo H., Wright J. J., Landry Z. C., Hanson N. W., Thompson B. P., Poulton N. J., Schwientek P., Acinas S. G., Giovannoni S. J., Moran M. A., Hallam S. J., Cavicchioli R., Woyke T., Stepanauskas R. (2013) Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc. Natl. Acad. Sci. U.S.A. 110, 11463–11468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moran N. A. (2002) Microbial minimalism: genome reduction in bacterial pathogens. Cell 108, 583–586 [DOI] [PubMed] [Google Scholar]
- 23. Moran N. A., Wernegreen J. J. (2000) Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol. Evol. 15, 321–326 [DOI] [PubMed] [Google Scholar]
- 24. Moran N. A. (2007) Symbiosis as an adaptive process and source of phenotypic complexity. Proc. Natl. Acad. Sci. U.S.A. 104, Suppl. 1, 8627–8633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moran N. A., Mira A. (2001) The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2, RESEARCH0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mira A., Ochman H., Moran N. A. (2001) Deletional bias and the evolution of bacterial genomes. Trends Genet. 17, 589–596 [DOI] [PubMed] [Google Scholar]
- 27. Toft C., Andersson S. G. (2010) Evolutionary microbial genomics: insights into bacterial host adaptation. Nat. Rev. Genet. 11, 465–475 [DOI] [PubMed] [Google Scholar]
- 28. Davison J. (1999) Genetic exchange between bacteria in the environment. Plasmid 42, 73–91 [DOI] [PubMed] [Google Scholar]
- 29. Dzidic S., Bedeković V. (2003) Horizontal gene transfer-emerging multidrug resistance in hospital bacteria. Acta Pharmacol. Sin. 24, 519–526 [PubMed] [Google Scholar]
- 30. Ferber D. (2003) Triple-threat microbe gained powers from another bug. Science 302, 1488. [DOI] [PubMed] [Google Scholar]
- 31. Mwangi M. M., Wu S. W., Zhou Y., Sieradzki K., de Lencastre H., Richardson P., Bruce D., Rubin E., Myers E., Siggia E. D., Tomasz A. (2007) Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 104, 9451–9456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andersson J. O. (2005) Lateral gene transfer in eukaryotes. Cell. Mol. Life. Sci. 62, 1182–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dunning Hotopp J. C., Clark M. E., Oliveira D. C., Foster J. M., Fischer P., Muñoz Torres M. C., Giebel J. D., Kumar N., Ishmael N., Wang S., Ingram J., Nene R. V., Shepard J., Tomkins J., Richards S., Spiro D. J., Ghedin E., Slatko B. E., Tettelin H., Werren J. H. (2007) Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756 [DOI] [PubMed] [Google Scholar]
- 34. Keeling P. J. (2009) Functional and ecological impacts of horizontal gene transfer in eukaryotes. Curr. Opin. Genet. Dev. 19, 613–619 [DOI] [PubMed] [Google Scholar]
- 35. Huang J. (2013) Horizontal gene transfer in eukaryotes: the weak-link model. Bioessays 35, 868–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nelson-Sathi S., Dagan T., Landan G., Janssen A., Steel M., McInerney J. O., Deppenmeier U., Martin W. F. (2012) Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of Haloarchaea. Proc. Natl. Acad. Sci. U.S.A. 109, 20537–20542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Filée J., Siguier P., Chandler M. (2007) I am what I eat and I eat what I am: acquisition of bacterial genes by giant viruses. Trends Genet. 23, 10–15 [DOI] [PubMed] [Google Scholar]
- 38. Hambly E., Suttle C. A. (2005) The viriosphere, diversity, and genetic exchange within phage communities. Curr. Opin. Microbiol. 8, 444–450 [DOI] [PubMed] [Google Scholar]
- 39. Culley A. I., Asuncion B. F., Steward G. F. (2009) Detection of inteins among diverse DNA polymerase genes of uncultivated members of the Phycodnaviridae. ISME J. 3, 409–418 [DOI] [PubMed] [Google Scholar]
- 40. Ogata H., Raoult D., Claverie J. M. (2005) A new example of viral intein in Mimivirus. Virol J. 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Larsen J. B., Larsen A., Bratbak G., Sandaa R. A. (2008) Phylogenetic analysis of members of the Phycodnaviridae virus family, using amplified fragments of the major capsid protein gene. Appl. Environ. Microbiol. 74, 3048–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bigot Y., Piégu B., Casteret S., Gavory F., Bideshi D. K., Federici B. A. (2013) Characteristics of inteins in invertebrate iridoviruses and factors controlling insertion in their viral hosts. Mol. Phylogenet. Evol. 67, 246–254 [DOI] [PubMed] [Google Scholar]
- 43. Amitai G., Dassa B., Pietrokovski S. (2004) Protein splicing of inteins with atypical glutamine and aspartate C-terminal residues. J. Biol. Chem. 279, 3121–3131 [DOI] [PubMed] [Google Scholar]
- 44. Fitzgerald L. A., Graves M. V., Li X., Feldblyum T., Nierman W. C., Van Etten J. L. (2007) Sequence and annotation of the 369-kb NY-2A and the 345-kb AR158 viruses that infect Chlorella NC64A. Virology 358, 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Allen M. J., Lanzén A., Bratbak G. (2011) Characterisation of the coccolithovirus intein. Mar. Genomics 4, 1–7 [DOI] [PubMed] [Google Scholar]
- 46. Santos F., Moreno-Paz M., Meseguer I., López C., Rosselló-Mora R., Parro V., Antón J. (2011) Metatranscriptomic analysis of extremely halophilic viral communities. ISME J. 5, 1621–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lazarevic V., Soldo B., Düsterhöft A., Hilbert H., Mauël C., Karamata D. (1998) Introns and intein coding sequence in the ribonucleotide reductase genes of Bacillus subtilis temperate bacteriophage SPβ. Proc. Natl. Acad. Sci. U.S.A. 95, 1692–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lazarevic V. (2001) Ribonucleotide reductase genes of Bacillus prophages: a refuge to introns and intein coding sequences. Nucleic Acids Res. 29, 3212–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hatfull G. F. (2010) Mycobacteriophages: genes and genomes. Annu. Rev. Microbiol. 64, 331–356 [DOI] [PubMed] [Google Scholar]
- 50. Gill J. J., Berry J. D., Russell W. K., Lessor L., Escobar-Garcia D. A., Hernandez D., Kane A., Keene J., Maddox M., Martin R., Mohan S., Thorn A. M., Russell D. H., Young R. (2012) The Caulobacter crescentus phage phiCbK: genomics of a canonical phage. BMC Genomics 13, 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Carvalho C. M., Kropinski A. M., Lingohr E. J., Santos S. B., King J., Azeredo J. (2012) The genome and proteome of a Campylobacter coli bacteriophage vB_CcoM-IBB_35 reveal unusual features. Virol. J. 9, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fouts D. E., Klumpp J., Bishop-Lilly K. A., Rajavel M., Willner K. M., Butani A., Henry M., Biswas B., Li M., Albert M. J., Loessner M. J., Calendar R., Sozhamannan S. (2013) Whole genome sequencing and comparative genomic analyses of two Vibrio cholerae O139 Bengal-specific Podoviruses to other N4-like phages reveal extensive genetic diversity. Virol. J. 10, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dwivedi B., Xue B., Lundin D., Edwards R. A., Breitbart M. (2013) A bioinformatic analysis of ribonucleotide reductase genes in phage genomes and metagenomes. BMC Evol. Biol. 13, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu X. Q. (2000) Protein-splicing intein: genetic mobility, origin, and evolution. Annu. Rev. Genet. 34, 61–76 [DOI] [PubMed] [Google Scholar]
- 55. Canchaya C., Fournous G., Chibani-Chennoufi S., Dillmann M. L., Brüssow H. (2003) Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6, 417–424 [DOI] [PubMed] [Google Scholar]
- 56. Liu H., Fu Y., Jiang D., Li G., Xie J., Cheng J., Peng Y., Ghabrial S. A., Yi X. (2010) Widespread horizontal gene transfer from double-stranded RNA viruses to eukaryotic nuclear genomes. J. Virol. 84, 11876–11887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu H., Fu Y., Li B., Yu X., Xie J., Cheng J., Ghabrial S. A., Li G., Yi X., Jiang D. (2011) Widespread horizontal gene transfer from circular single-stranded DNA viruses to eukaryotic genomes. BMC Evol. Biol. 11, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Saraste M., Sibbald P. R., Wittinghofer A. (1990) The P-loop: a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434 [DOI] [PubMed] [Google Scholar]
- 60. Swithers K. S., Senejani A. G., Fournier G. P., Gogarten J. P. (2009) Conservation of intron and intein insertion sites: implications for life histories of parasitic genetic elements. BMC Evol. Biol. 9, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Goodwin T. J., Butler M. I., Poulter R. T. (2006) Multiple, non-allelic, intein-coding sequences in eukaryotic RNA polymerase genes. BMC Biol. 4, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Goddard M. R., Burt A. (1999) Recurrent invasion and extinction of a selfish gene. Proc. Natl. Acad. Sci. U.S.A. 96, 13880–13885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Belfort M. (1989) Bacteriophage introns: parasites within parasites? Trends Genet. 5, 209–213 [DOI] [PubMed] [Google Scholar]
- 64. Cowley M., Oakey R. J. (2013) Transposable elements re-wire and fine-tune the transcriptome. PLoS Genet. 9, e1003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Emera D., Wagner G. P. (2012) Transposable element recruitments in the mammalian placenta: impacts and mechanisms. Brief. Funct. Genomics 11, 267–276 [DOI] [PubMed] [Google Scholar]
- 66. Faulkner G. J., Carninci P. (2009) Altruistic functions for selfish DNA. Cell Cycle 8, 2895–2900 [DOI] [PubMed] [Google Scholar]
- 67. Wu H., Hu Z., Liu X. Q. (1998) Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U.S.A. 95, 9226–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Callahan B. P., Topilina N. I., Stanger M. J., Van Roey P., Belfort M. (2011) Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications. Nat. Struct. Mol. Biol. 18, 630–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cui C., Zhao W., Chen J., Wang J., Li Q. (2006) Elimination of in vivo cleavage between target protein and intein in the intein-mediated protein purification systems. Protein Expr. Purif. 50, 74–81 [DOI] [PubMed] [Google Scholar]
- 70. Callahan B. P., Stanger M., Belfort M. (2013) A redox trap to augment the intein toolbox. Biotechnol. Bioeng. 110, 1565–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kaneko T., Tanaka A., Sato S., Kotani H., Sazuka T., Miyajima N., Sugiura M., Tabata S. (1995) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 2, 153–166, 191–198 [DOI] [PubMed] [Google Scholar]
- 72. Chen W., Li L., Du Z., Liu J., Reitter J. N., Mills K. V., Linhardt R. J., Wang C. (2012) Intramolecular disulfide bond between catalytic cysteines in an intein precursor. J. Am. Chem. Soc. 134, 2500–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nicastri M. C., Xega K., Li L., Xie J., Wang C., Linhardt R. J., Reitter J. N., Mills K. V. (2013) Internal disulfide bond acts as a switch for intein activity. Biochemistry 52, 5920–5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cambon-Bonavita M. A., Schmitt P., Zieger M., Flaman J. M., Lesongeur F., Raguénès G., Bindel D., Frisch N., Lakkis Z., Dupret D., Barbier G., Quérellou J. (2000) Cloning, expression, and characterization of DNA polymerase I from the hyperthermophilic archaea Thermococcus fumicolans. Extremophiles 4, 215–225 [DOI] [PubMed] [Google Scholar]
- 75. Mills K. V., Dorval D. M., Lewandowski K. T. (2005) Kinetic analysis of the individual steps of protein splicing for the Pyrococcus abyssi PolII intein. J. Biol. Chem. 280, 2714–2720 [DOI] [PubMed] [Google Scholar]
- 76. Mills K. V., Manning J. S., Garcia A. M., Wuerdeman L. A. (2004) Protein splicing of a Pyrococcus abyssi intein with a C-terminal glutamine. J. Biol. Chem. 279, 20685–20691 [DOI] [PubMed] [Google Scholar]
- 77. Shao Y., Paulus H. (1997) Protein splicing: estimation of the rate of O–N and S–N acyl rearrangements, the last step of the splicing process. J. Pept. Res. 50, 193–198 [DOI] [PubMed] [Google Scholar]
- 78. Shao Y., Xu M. Q., Paulus H. (1995) Protein splicing: characterization of the aminosuccinimide residue at the carboxyl terminus of the excised intervening sequence. Biochemistry 34, 10844–10850 [DOI] [PubMed] [Google Scholar]
- 79. Xu M. Q., Comb D. G., Paulus H., Noren C. J., Shao Y., Perler F. B. (1994) Protein splicing: an analysis of the branched intermediate and its resolution by succinimide formation. EMBO J. 13, 5517–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Xu M. Q., Perler F. B. (1996) The mechanism of protein splicing and its modulation by mutation. EMBO J. 15, 5146–5153 [PMC free article] [PubMed] [Google Scholar]
- 81. Xu M. Q., Southworth M. W., Mersha F. B., Hornstra L. J., Perler F. B. (1993) In vitro protein splicing of purified precursor and the identification of a branched intermediate. Cell 75, 1371–1377 [DOI] [PubMed] [Google Scholar]
- 82. Zeidler M. P., Tan C., Bellaiche Y., Cherry S., Häder S., Gayko U., Perrimon N. (2004) Temperature-sensitive control of protein activity by conditionally splicing inteins. Nat. Biotechnol. 22, 871–876 [DOI] [PubMed] [Google Scholar]
- 83. Shih C. K., Wagner R., Feinstein S., Kanik-Ennulat C., Neff N. (1988) A dominant trifluoperazine resistance gene from Saccharomyces cerevisiae has homology with F0F1 ATP synthase and confers calcium-sensitive growth. Mol. Cell Biol. 8, 3094–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wood D. W., Wu W., Belfort G., Derbyshire V., Belfort M. (1999) A genetic system yields self-cleaving inteins for bioseparations. Nat. Biotechnol. 17, 889–892 [DOI] [PubMed] [Google Scholar]
- 85. Rao M., Streur T. L., Aldwell F. E., Cook G. M. (2001) Intracellular pH regulation by Mycobacterium smegmatis and Mycobacterium bovis BCG. Microbiology 147, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 86. Mootz H. D., Blum E. S., Tyszkiewicz A. B., Muir T. W. (2003) Conditional protein splicing: a new tool to control protein structure and function in vitro and in vivo. J. Am. Chem. Soc. 125, 10561–10569 [DOI] [PubMed] [Google Scholar]
- 87. Mootz H. D., Muir T. W. (2002) Protein splicing triggered by a small molecule. J. Am. Chem. Soc. 124, 9044–9045 [DOI] [PubMed] [Google Scholar]
- 88. Buskirk A. R., Ong Y. C., Gartner Z. J., Liu D. R. (2004) Directed evolution of ligand dependence: small-molecule-activated protein splicing. Proc. Natl. Acad. Sci. U.S.A. 101, 10505–10510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Skretas G., Wood D. W. (2005) Regulation of protein activity with small-molecule-controlled inteins. Protein Sci. 14, 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Davis E. O., Sedgwick S. G., Colston M. J. (1991) Novel structure of the recA locus of Mycobacterium tuberculosis implies processing of the gene product. J. Bacteriol. 173, 5653–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gierach I., Li J., Wu W. Y., Grover G. J., Wood D. W. (2012) Bacterial biosensors for screening isoform-selective ligands for human thyroid receptors α-1 and β-1. FEBS Open. Bio. 2, 247–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Li J., Gierach I., Gillies A. R., Warden C. D., Wood D. W. (2011) Engineering and optimization of an allosteric biosensor protein for peroxisome proliferator-activated receptor γ ligands. Biosens. Bioelectron. 29, 132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gribaldo S., Brochier-Armanet C. (2006) The origin and evolution of Archaea: a state of the art. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]