Abstract
Protein splicing is a posttranslational modification where intervening proteins (inteins) cleave themselves from larger precursor proteins and ligate their flanking polypeptides (exteins) through a multistep chemical reaction. First thought to be an anomaly found in only a few organisms, protein splicing by inteins has since been observed in microorganisms from all domains of life. Despite this broad phylogenetic distribution, all inteins share common structural features such as a horseshoe-like pseudo two-fold symmetric fold, several canonical sequence motifs, and similar splicing mechanisms. Intriguingly, the splicing efficiencies and substrate specificity of different inteins vary considerably, reflecting subtle changes in the chemical mechanism of splicing, linked to their local structure and dynamics. As intein chemistry has widespread use in protein chemistry, understanding the structural and dynamical aspects of inteins is crucial for intein engineering and the improvement of intein-based technologies.
Keywords: NMR, Protein Engineering, Protein Folding, Protein Structure, Protein-Protein Interactions, Intein, Protein Splicing
Introduction
Protein splicing is a posttranslational modification, a multistep autoprocessing event, where intervening proteins (inteins) self-excise themselves from the precursors followed by the ligation of external proteins (exteins) (1). Once thought anomalies, inteins have since been found in all domains of life. Beyond their biological context, inteins are particularly intriguing as their capacity to process the polypeptide backbone makes them useful tools for protein engineering. Despite the fact that all inteins share the same fold and have highly conserved sequence motifs in their active sites, inteins have surprisingly different splicing efficiencies, and their extein sequence preferences differ dramatically. Here, we review studies on intein structure and dynamics that shed light on their complex conserved fold and their divergent efficiency and substrate specificity.
The Intein Fold
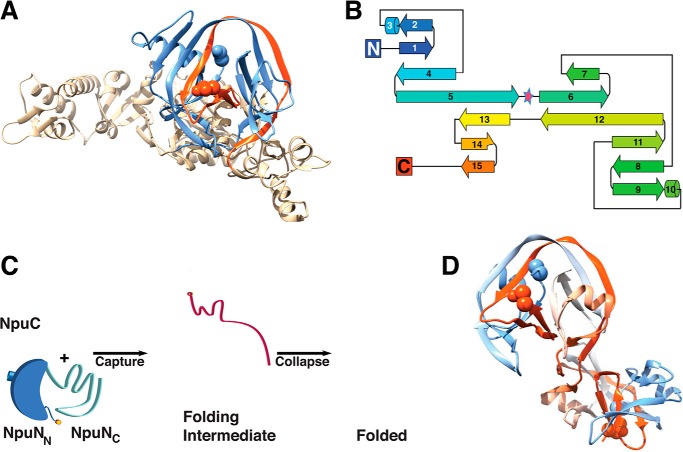
The conserved horseshoe-like fold of inteins (Fig. 1A) has been seen in all intein structures solved by NMR and x-ray crystallography (2–11). The fold comprises primarily β-sheets, loops, and two short helices and has pseudo two-fold symmetry (Fig. 1B). Given this symmetry, it has been proposed that this fold arose due to a gene duplication event of some parent protein (6). The intein fold has three remarkable features: 1) the topology is complex, involving multiple passes of the polypeptide chain back and forth between the symmetry-related halves (Fig. 1B); 2) the extein-bearing termini of the intein are brought in close proximity (<10 Å) for splicing; and 3) multiple protease-like active sites composed of conserved sequence motifs are built around these termini to carry out each of the chemical steps involved in protein splicing. Folding of an intein is coupled to the reaction; the initial structure facilitates the first step of splicing, and thereafter each splicing step causes local conformational changes, affecting the fold of the catalytic apparatus and hence affecting the reaction coordinate and shifting the equilibrium position.
FIGURE 1.
Intein fold. A, T. kodakaraensis Pol-2 intein (Protein Data Bank (PDB) 2CW7) (9). The N-intein (blue) and C-intein (red) are shown. B, secondary structure topology map of Drosophila melanogaster Hog domain, a relative of inteins (PDB 1AT0) (6). The pseudo two-fold symmetry axis is highlighted with a star. C, capture and collapse folding mechanism of the Npu DnaE intein (12). The N-intein is blue (dark blue for NpuNN and light blue for NpuNC), the C-intein is red, and the extein-bearing termini are marked by orange circles. D, three-dimensional domain swapped dimer of a contiguous Npu DnaE intein variant (3).
The splicing motifs are found in two splicing regions, the N-terminal splicing region (N-intein) and C-terminal splicing region (C-intein) (Fig. 2A). In standard contiguous inteins (cis-splicing), these two splicing regions are separated by homing endonuclease or linker sequences, whereas in split inteins (trans-splicing), the splicing regions are translated separately. In a functional intein fold, these two splicing regions, whether in contiguous or in split inteins, are entwined to bring all catalytic residues and the termini together, which is a unique folding/molecular recognition problem preceding protein splicing. Surprisingly, despite the fact that protein splicing is initiated by this folding event, few studies have addressed the folding of inteins (12, 13).
FIGURE 2.
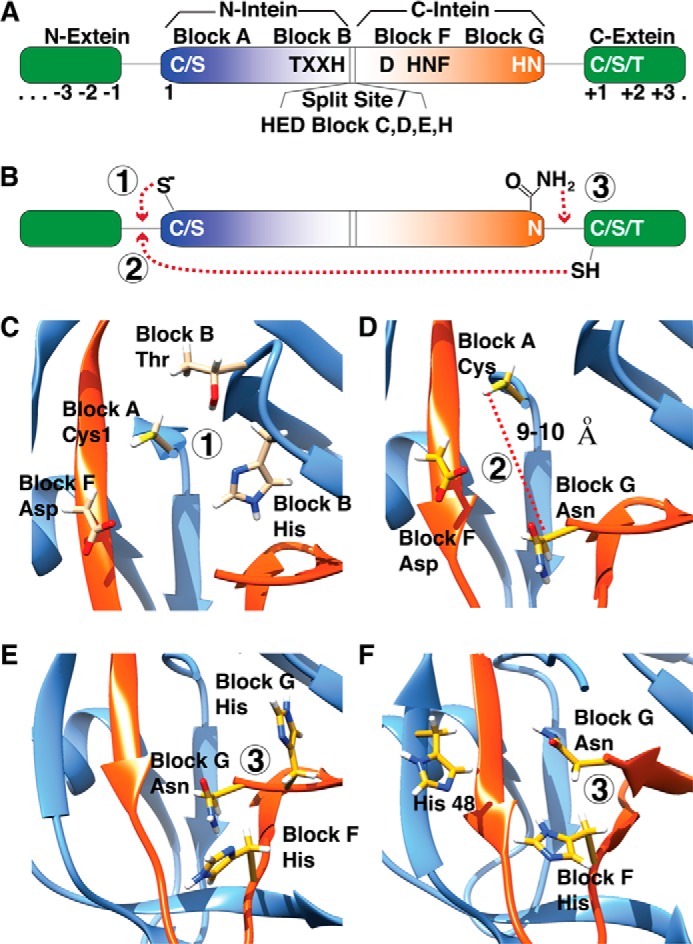
Canonical splicing motifs and mechanism. A, intein is shown as a color gradient from blue to red, and exteins are show in green. The canonical motifs and corresponding blocks are shown. B, a simplified schematic of splicing reaction is shown; the splicing steps 1–3 are shown with circles. C–E, key residues, which take part in each splicing step, steps 1–3 in panel B, are highlighted as yellow sticks on the Mtu RecA intein (PDB 2L8L) (19). F, an alternate mechanism seen in DnaE inteins, lacking penultimate histidine, is shown on the Npu DnaE intein (PDB 2KEQ) (10).
The intertwined fold of fragments requires some degree of disorder in individual partners. In the Npu DnaE split intein, the C-intein (NpuC) is completely disordered, whereas the N-intein is partially disordered at its C-terminal half in isolation (NpuNC) (12) (Fig. 1C). The folding is initiated by electrostatic charge complementarity between the disordered C-intein and the disordered C-terminal half of the N-intein, “capture,” and this intermediate is stabilized further by hydrophobic interactions between the C-intein and the N-terminal half of the N-intein, “collapse” (12) (Fig. 1C). The “capture and collapse” mechanism points toward a general folding pathway of all inteins including contiguous inteins. The initiating electrostatic interactions are unique to split inteins (14), yet it is possible that the “split site” in split inteins could be a point of nucleation in contiguous inteins; in contiguous inteins with homing endonuclease (HE) domains, the folding of the homing endonuclease domain could nucleate intein folding. The intrinsic disorder seen in the Npu DnaE split intein is also seen in the related Ssp DnaE split intein, further supporting the notion that the folding pathway is conserved (13). Surprisingly, the N- and C-terminal splicing regions of different intein molecules can interact to form a domain-swapped dimer (Fig. 1D). As this type of noncovalent interaction between different split inteins creates hybrid active sites and mixed extein combinations, it has been proposed such structures may yield alternative spliced products that could expedite evolution by sampling alternate phenotypes under a constant genetic background (3).
The Structural Basis of Protein Splicing
Protein splicing is a multistep process (Fig. 2B). The first step is N–S/O acyl shift initiated by the N-terminal intein residue, generally a cysteine or serine (15, 16), acting on −1 residue carbonyl carbon (1, 17), resulting in linear (thio)ester intermediate. The next step is the trans-(thio)esterification caused by a nucleophilic attack by C-extein +1 residue, which is a cysteine, serine, or threonine. The resultant branched (thio)ester intermediate is resolved by cyclization of the conserved C-terminal asparagine of intein. Once the succinimide forms, the intein is excised from the exteins. Finally, the linkage between the exteins rearranges from a (thio)ester to an amide in an intein-independent manner, and the C-terminal succinimide of the intein slowly hydrolyzes (18). All these sequential steps require multiple active sites with distinct residues all in close proximity to the intein termini. These active-site residues are conserved among most inteins, but several exceptions exist. Below, we discuss the structural details of each step in protein splicing and discuss the unique ways in which some divergent inteins accommodate mutations in the conserved splicing motifs.
N-to-O/S Acyl Shift
This step is carried out by the N-terminal cysteine or serine of the intein, which attacks the C-terminal extein residue at its carbonyl carbon. The attack is facilitated by the Block B (TXXH) threonine and histidine and the Block F aspartate (Fig. 2C). The Block F aspartate serves as a hydrogen bond donor and stabilizes C1 thiolate (19) and is a strict requirement for N–O/S acyl shift reaction (20). Block B histidine is absent only in the Thermococcus kodakaraensis Tko CDC21-1 intein, and the Tko CDC21-1 intein circumvents the lack of Block B histidine by using an alternate mechanism facilitated by Lys58, which is out of the conserved motifs (21). This first step of splicing, N–O/S acyl shift, requires a substantial challenge as peptide bond cleavage in general has a large kinetic barrier. It is possible that the energy gained from folding into a very stable conformation is coupled directly to overcoming the energy barrier for the initial splicing step. Such a mechanism is observed in the SEA (named after its initial identification in a Sperm protein, in Enterokinase, and in Agrin (50)) domains of human MUC1, where the autoproteolysis is catalyzed by conformational stress (22). In the Ssp DnaE intein structure, the exteins are tightly packed (11), and upon intein assembly, they may create a steric clash at the active site so that the splicing reaction may be the most kinetically accessible escape mechanism from this clash. Also, the structure of the Ssp DnaE intein caught in a redox trap (4), supplemented by further computational and crystallographic evidence (5), shows that the initial rearrangements of residues and the first step, N–O/S acyl shift, are accelerated by localized structural strain on the −1 N-extein residue, mediated by the conserved Block B threonine, which makes bonding interactions with C1 and the −2 N-extein residue. Additionally, some inteins lack a Block A nucleophile; instead they have an alanine or a proline. Several studies have shown a twisted or destabilized N-terminal scissile peptide bond in the precursor protein (5, 23). These inteins directly form a typical branched intermediate upon N-terminal activation (24), or in some cases, they use a Block F cysteine to form a different branched (thio)ester intermediate before the canonical branched intermediate (25). The Mja KlbA intein, with an alanine at N terminus, has a peptide bond in cis-conformation possibly making it unstable or increasing its susceptibility to a nucleophilic attack (7).
trans-(Thio)esterification
This step is the least understood step in protein splicing because it is probably the most difficult step to investigate experimentally (the linear (thio)ester intermediate cannot be isolated). trans-(Thio)esterification requires deprotonation of Cys/Ser/Thr+1 side chain and nucleophilic attack to the linear (thio)ester linkage at Cys/Ser1, which are far apart in high-resolution structures (9–10 Å) (11, 26, 27) (Fig. 2D). In Mtu RecA, Block F aspartate together with C+1A mutation showed no N-cleavage activity, and Block F aspartate mutated to residues that are not hydrogen bond-capable showed similar results, implying that Block F aspartate accomplishes its role in trans-(thio)esterification through hydrogen bond interactions (28), which was also supported by NMR-based pKa studies (19). All these studies indicate that the first two steps of protein splicing are achieved by a complex hydrogen-bonding network of three conserved residues, N-terminal cysteine/serine, Block B histidine, and Block F aspartate (29).
Branched Intermediate Resolution
Branched intermediate (BI)2 resolution is irreversible and achieves the cleavage of the peptide bond between the intein and the C-extein. The side chain nitrogen of C-terminal asparagine performs a nucleophilic attack on the carbonyl carbon of the peptide bond to form a succinimide, subsequently cleaving the intein from BI (30, 31). Indeed, Asn side chain nucleophiles are biochemically rare (also found in N-glycosylation) (32), and succinimide formation in proteins more commonly occurs by nucleophilic attack of a backbone amide nitrogen to an Asn side chain carbonyl carbon, resulting in deamidation of the side chain (33). Both structural and computational studies revealed that Block F and Block G (penultimate) histidines are important in BI resolution; they activate asparagine and protonate the forming amine (18) (Fig. 2E). Interestingly, some inteins lack a penultimate histidine (34). In DnaE split inteins, this histidine is either a serine or an alanine (35). It is not clear how these divergent inteins can circumvent the need for two histidines and carry out splicing reaction. However, there are other proximal residues that may serve as general acids or bases during splicing. Interestingly, although His48 in DnaE inteins has not been implicated in splicing, it is close to the active site and may substitute Block G histidine (Fig. 2F). Although not apparent in crystal structures, it is presumed that each splicing step causes local conformational changes, affecting the reaction coordinate, shifting the equilibrium position. Consistent with this notion, branched intermediate formation in both the Mxe GyrA (18) and the Npu DnaE (36) inteins dramatically enhances Asn cyclization kinetics, indicating that the branched architecture changes the structure and dynamics of the active site. Indeed, NMR data on the precursor GyrA intein and a synthetically trapped branched intermediate indicate that the chemical environment and relaxation dynamics around the scissile peptide bond change upon branch formation.
Extein Dependence on Protein Splicing
It is intriguing that hundreds of different inteins can carry out the same chemistry but with different extein specificity. Different inteins have evolved to splice different proteins in nature, and thus their extein sequence preferences are roughly defined by their native contexts. Deviation from these preferences often results in a profound effect on splicing kinetics and yield, suggesting that the exteins can contribute either chemically or structurally to the intein active site. For example, in the Npu DnaE intein, the splicing is minimally affected by local N-extein sequence, but the variation of the local C-extein sequence dramatically affects splicing efficiency (37–39) by perturbing branched intermediate resolution (40). In the structure of the orthologous Ssp DnaE intein, the native Phe+2 side chain packs against and stabilizes Block F histidine (Fig. 3A). This conserved catalytic histidine lies on a flexible loop and has been implicated as a general base or acid in the BI resolution step in many inteins (11, 18). The flexible loop containing Block F histidine is modulated by the C-extein composition and has been used to engineer Npu into a more extein-tolerant intein. The mutation, D124Y, on this loop stabilizes and orients His125 to a catalytically favorable pose, hence reducing the Phe+2 extein dependence on Npu (36). In a computational study on the Mtu RecA intein, where quantum mechanics/molecular mechanics methods were used and the local extein electronic structure of various +1 residue mutants was investigated, it was found that the mutants have different electron affinities and ionization potentials imposing different energy barriers (41). In the Ssp DnaB mini-intein, analysis of the crystal structure explains why the −1 position can only be occupied by a Gly. Modeling suggests that any bigger residue would abrogate splicing by causing steric clashes (42) (Fig. 3B). Additionally, in the Pho RadA intein, it was shown that the size and charge of the side chain have an impact at position −1 due to electrostatic and packing interactions with the active site; negatively charged amino acids, β-branched amino acids, proline, and amino acids with small side chains have lower splicing efficiency (43) (Fig. 3, C and D).
FIGURE 3.
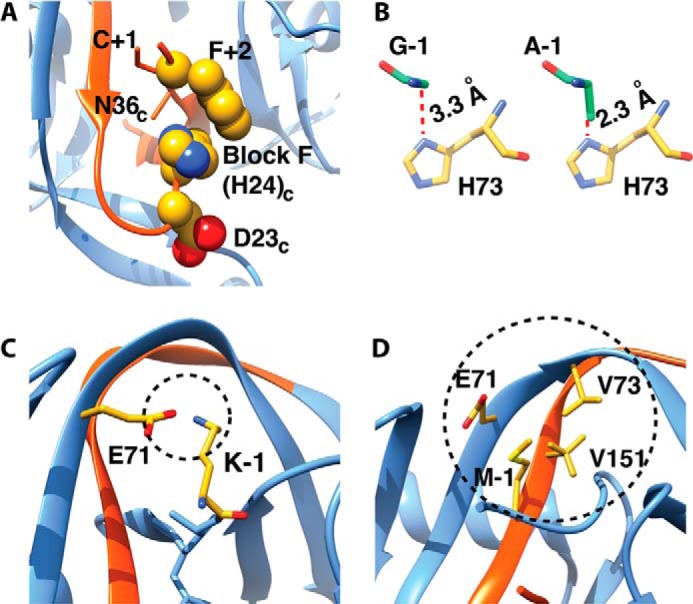
Extein dependence. A, in the DnaE Ssp DnaE intein, Phe+2 packs against Block F histidine, modulating its pose and hence increasing efficiency. B, DnaB Ssp mini-intein can only tolerate Gly at −1 position; any bigger residue (i.e. the alanine shown) creates steric clashes in the active site. C, the native −1 amino acid in the Pho RadA intein (PDB 4E2T) is a lysine that makes charge-charge interaction with Glu71 (Lys−1 was modeled by using CHIMERA (49)). D, −1 residue in Pho RadA mini-intein tightly interacts with Glu71, Val73, and Val151 (PDB 4E2U).
Distal Mutations Affect Protein Splicing
Using mutagenesis strategies, enhancing/activating mutations distal from the active site have been observed, suggesting that the overall structure and dynamics of the intein fold are somewhat plastic (Fig. 4). For example, in DnaE split inteins, several residues that are distant from the active site have profound effects on splicing efficiency; these residues either stabilize the global fold or stabilize and orient the active-site splicing motifs (39). The fast split inteins of this family prefer an aromatic residue at position 56, which is adjacent to the conserved TXXH motif and may facilitate packing interactions and stabilize the TXXH motif. A preferred glutamate at position 89 takes part in an ion cluster shown to be important in stabilizing the intein complex (14), whereas another glutamate is preferred at position 122 close to Block F histidine. Additionally, Glu23 is important for stability and dynamics and has been shown to be an activating point mutation (44, 45). In the minimized Mtu RecA intein, a mutation distal from the active site, V67L, enhances splicing by modifying the active site through improving packing interactions in an internal hydrophobic core (20) and reducing the ensemble distribution (45). This single V67L mutation minimally affects the crystal structure (20) but has profound effects on global dynamics specifically on the dynamics of N and C termini of the intein shown by NMR chemical shift perturbations and H/D exchange (45). Directed evolution studies also show that distal mutations have enhancing effects on splicing (40, 44, 46). In the Ssp DnaB mini-intein, the mutations acquired through directed evolution have additive effects, and the mutant intein becomes more tolerant to the local extein sequences and the constraints these sequences impose on the intein active site (44). Similarly, the Npu DnaE intein, selected against noncanonical C-extein (SGV instead of the native CFN), accumulate mutations distal from the active site (40). These mutations were identified as either a part of or juxtaposed to a spatially contiguous network of amino acids predicted to be energetically coupled to the active site. A directed evolution system, using phage display to generate novel inteins with enhanced activity over a broad pH and temperature range, demonstrated that most mutations that accommodated the stress conditions mapped on the surface of the intein. This again emphasizes that peripheral mutations can cause subtle conformational changes in the active-site environment (47). The mutations proximal to the active site typically orient and/or stabilize active-site residues. The distal mutations appear to either stabilize the intein fold and/or relay perturbations through a contiguous network of amino acids that can alter the active-site structure and dynamics.
FIGURE 4.
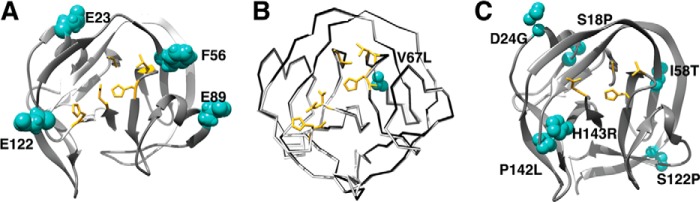
Distal mutations affect catalytic residues and splicing. A, the Npu DnaE intein structure (PDB 2KEQ) highlighting catalytic residues (yellow sticks) and noncatalytic positions (green spheres) that are shown to modulate activity in Ssp DnaE mini-intein (39). B, overlay of the crystal structures of minimized Mtu RecA, ΔΔIhh-V67SM (black, PDB 2IN9), and ΔΔIhh-L67SM (gray, PDB 2IN0) (20). V67L (green spheres) mutation minimally affects the three-dimensional structure but enhances splicing activity. The catalytic residues are shown as yellow sticks. C, the mutations acquired by Ssp DnaE mini-intein after sequential directed evolution by using Kan resistance are shown (green spheres), and the key catalytic residues are highlighted (yellow sticks) (PDB 1MI8) (44).
Conclusions
Thus far, traditional structural biology and biochemistry approaches have elucidated many details of the structural basis for protein splicing. However, a detailed understanding of how the intein fold coordinates the chemistry of its multiple active sites is still lacking, as is a better understanding of the general rules for extein preferences. The fact that intein activity can be modulated by mutations distant from the active site suggests that allosteric networks may play a larger role in determining intein activity than previously thought. Ultimately, a better understanding of the structure and dynamics of inteins is crucial as using this information will be essential to engineer inteins as better biotechnology tools. Currently, some of reactive intermediates in the splicing pathway are not readily experimentally observable by using traditional experimental techniques. However, combinations of synthetic chemistry, directed evolution, and computational modeling (for example, see Ref. 48) are likely to provide insights into these areas, resulting in a better understanding of protein splicing and eventually new, highly efficient inteins for protein engineering.
This work was supported, in whole or in part, by National Institutes of Health Grant GM086868 (to D. C.). This is the third article in the Thematic Minireview Series “Inteins.”
- BI
- branched intermediate.
REFERENCES
- 1. Paulus H. (2000) Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem. 69, 447–496 [DOI] [PubMed] [Google Scholar]
- 2. Aranko A. S., Oeemig J. S., Iwaï H. (2013) Structural basis for protein trans-splicing by a bacterial intein-like domain–protein ligation without nucleophilic side chains. FEBS J. 280, 3256–3269 [DOI] [PubMed] [Google Scholar]
- 3. Aranko A. S., Oeemig J. S., Kajander T., Iwaï H. (2013) Intermolecular domain swapping induces intein-mediated protein alternative splicing. Nat. Chem. Biol. 9, 616–622 [DOI] [PubMed] [Google Scholar]
- 4. Callahan B. P., Topilina N. I., Stanger M. J., Van Roey P., Belfort M. (2011) Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications. Nat. Struct. Mol. Biol. 18, 630–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dearden A. K., Callahan B., Roey P. V., Li Z., Kumar U., Belfort M., Nayak S. K. (2013) A conserved threonine spring-loads precursor for intein splicing. Protein Sci. 22, 557–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hall T. M. T., Porter J. A., Young K. E., Koonin E. V., Beachy P. A., Leahy D. J. (1997) Crystal structure of a Hedgehog autoprocessing domain: homology between Hedgehog and self-splicing proteins. Cell 91, 85–97 [DOI] [PubMed] [Google Scholar]
- 7. Johnson M. A., Southworth M. W., Herrmann T., Brace L., Perler F. B., Wüthrich K. (2007) NMR structure of a KlbA intein precursor from Methanococcus jannaschii. Protein Sci. 16, 1316–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klabunde T., Sharma S., Telenti A., Jacobs W. R., Jr., Sacchettini J. C. (1998) Crystal structure of GyrA intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nat. Struct. Biol. 5, 31–36 [DOI] [PubMed] [Google Scholar]
- 9. Matsumura H., Takahashi H., Inoue T., Yamamoto T., Hashimoto H., Nishioka M., Fujiwara S., Takagi M., Imanaka T., Kai Y. (2006) Crystal structure of intein homing endonuclease II encoded in DNA polymerase gene from hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1. Proteins 63, 711–715 [DOI] [PubMed] [Google Scholar]
- 10. Oeemig J. S., Aranko A. S., Djupsjöbacka J., Heinämäki K., Iwaï H. (2009) Solution structure of DnaE intein from Nostoc punctiforme: structural basis for the design of a new split intein suitable for site-specific chemical modification. FEBS Lett. 583, 1451–1456 [DOI] [PubMed] [Google Scholar]
- 11. Sun P., Ye S., Ferrandon S., Evans T. C., Xu M. Q., Rao Z. (2005) Crystal structures of an intein from the split dnaE gene of Synechocystis sp. PCC6803 reveal the catalytic model without the penultimate histidine and the mechanism of zinc ion inhibition of protein splicing. J. Mol. Biol. 353, 1093–1105 [DOI] [PubMed] [Google Scholar]
- 12. Shah N. H., Eryilmaz E., Cowburn D., Muir T. W. (2013) Naturally split inteins assemble through a “capture and collapse” mechanism. J. Am. Chem. Soc 135, 18673–18681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zheng Y., Wu Q., Wang C., Xu M. Q., Liu Y. (2012) Mutual synergistic protein folding in split intein. Biosci. Repr. 32, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shah N. H., Vila-Perelló M., Muir T. W. (2011) Kinetic control of one-pot trans-splicing reactions by using a wild-type and designed split intein. Angew. Chem. Int. Ed. Engl. 50, 6511–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perler F. B., Olsen G. J., Adam E. (1997) Compilation and analysis of intein sequences. Nucleic Acids Res. 25, 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pietrokovski S. (1994) Conserved sequence features of inteins (protein introns) and their use in identifying new inteins and related proteins. Protein Sci. 3, 2340–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perler F. B. (1998) Protein splicing of inteins and Hedgehog autoproteolysis: structure, function, and evolution. Cell 92, 1–4 [DOI] [PubMed] [Google Scholar]
- 18. Frutos S., Goger M., Giovani B., Cowburn D., Muir T. W. (2010) Branched intermediate formation stimulates peptide bond cleavage in protein splicing. Nat. Chem. Biol. 6, 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Du Z., Zheng Y., Patterson M., Liu Y., Wang C. (2011) pKa coupling at the intein active site: implications for the coordination mechanism of protein splicing with a conserved aspartate. J. Am. Chem. Soc. 133, 10275–10282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Roey P., Pereira B., Li Z., Hiraga K., Belfort M., Derbyshire V. (2007) Crystallographic and mutational studies of Mycobacterium tuberculosis recA mini-inteins suggest a pivotal role for a highly conserved aspartate residue. J. Mol. Biol. 367, 162–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tori K., Cheriyan M., Pedamallu C. S., Contreras M. A., Perler F. B. (2012) The Thermococcus kodakaraensis Tko CDC21-1 intein activates its N-terminal splice junction in the absence of a conserved histidine by a compensatory mechanism. Biochemistry 51, 2496–2505 [DOI] [PubMed] [Google Scholar]
- 22. Macao B., Johansson D. G., Hansson G. C., Härd T. (2006) Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat. Struct. Mol. Biol. 13, 71–76 [DOI] [PubMed] [Google Scholar]
- 23. Romanelli A., Shekhtman A., Cowburn D., Muir T. W. (2004) Semisynthesis of a segmental isotopically labeled protein splicing precursor: NMR evidence for an unusual peptide bond at the N-extein-intein junction. Proc. Natl. Acad. Sci. U.S.A. 101, 6397–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Southworth M. W., Benner J., Perler F. B. (2000) An alternative protein splicing mechanism for inteins lacking an N-terminal nucleophile. EMBO J. 19, 5019–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tori K., Dassa B., Johnson M. A., Southworth M. W., Brace L. E., Ishino Y., Pietrokovski S., Perler F. B. (2010) Splicing of the mycobacteriophage Bethlehem DnaB intein: identification of a new mechanistic class of inteins that contain an obligate Block F nucleophile. J. Biol. Chem. 285, 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ding Y., Xu M. Q., Ghosh I., Chen X., Ferrandon S., Lesage G., Rao Z. (2003) Crystal structure of a mini-intein reveals a conserved catalytic module involved in side chain cyclization of asparagine during protein splicing. J. Biol. Chem. 278, 39133–39142 [DOI] [PubMed] [Google Scholar]
- 27. Poland B. W., Xu M. Q., Quiocho F. A. (2000) Structural insights into the protein splicing mechanism of PI-SceI. J. Biol. Chem. 275, 16408–16413 [DOI] [PubMed] [Google Scholar]
- 28. Pereira B., Shemella P. T., Amitai G., Belfort G., Nayak S. K., Belfort M. (2011) Spontaneous proton transfer to a conserved intein residue determines on-pathway protein splicing. J. Mol. Biol. 406, 430–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Volkmann G., Mootz H. D. (2013) Recent progress in intein research: from mechanism to directed evolution and applications. Cell. Mol. Life Sci. 70, 1185–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shao Y., Xu M. Q., Paulus H. (1995) Protein splicing: characterization of the aminosuccinimide residue at the carboxyl-terminus of the excised intervening sequence. Biochemistry 34, 10844–10850 [DOI] [PubMed] [Google Scholar]
- 31. Xu M. Q., Comb D. G., Paulus H., Noren C. J., Shao Y., Perler F. B. (1994) Protein splicing: an analysis of the branched intermediate and its resolution by succinimide formation. EMBO J. 13, 5517–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwarz F., Aebi M. (2011) Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21, 576–582 [DOI] [PubMed] [Google Scholar]
- 33. Stephenson R. C., Clarke S. (1989) Succinimide formation from aspartyl and asparaginyl peptides as a model for the spontaneous degradation of proteins. J. Biol. Chem. 264, 6164–6170 [PubMed] [Google Scholar]
- 34. Chen L., Benner J., Perler F. B. (2000) Protein splicing in the absence of an intein penultimate histidine. J. Biol. Chem. 275, 20431–20435 [DOI] [PubMed] [Google Scholar]
- 35. Caspi J., Amitai G., Belenkiy O., Pietrokovski S. (2003) Distribution of split DnaE inteins in cyanobacteria. Mol. Microbiol. 50, 1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shah N. H., Eryilmaz E., Cowburn D., Muir T. W. (2013) Extein residues play an intimate role in the rate-limiting step of protein trans-splicing. J. Am. Chem. Soc. 135, 5839–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cheriyan M., Pedamallu C. S., Tori K., Perler F. (2013) Faster protein splicing with the Nostoc punctiforme DnaE intein using non-native extein residues. J. Biol. Chem. 288, 6202–6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iwai H., Züger S., Jin J., Tam P. H. (2006) Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 580, 1853–1858 [DOI] [PubMed] [Google Scholar]
- 39. Shah N. H., Dann G. P., Vila-Perelló M., Liu Z., Muir T. W. (2012) Ultrafast protein splicing is common among cyanobacterial split inteins: implications for protein engineering. J. Am. Chem. Soc. 134, 11338–11341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lockless S. W., Muir T. W. (2009) Traceless protein splicing utilizing evolved split inteins. Proc. Natl. Acad. Sci. U.S.A. 106, 10999–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shemella P. T., Topilina N. I., Soga I., Pereira B., Belfort G., Belfort M., Nayak S. K. (2011) Electronic structure of neighboring extein residue modulates intein C-terminal cleavage activity. Biophys. J. 100, 2217–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwarzer D., Ludwig C., Thiel I. V., Mootz H. D. (2012) Probing intein-catalyzed thioester formation by unnatural amino acid substitutions in the active site. Biochemistry 51, 233–242 [DOI] [PubMed] [Google Scholar]
- 43. Oeemig J. S., Zhou D., Kajander T., Wlodawer A., Iwaï H. (2012) NMR and crystal structures of the Pyrococcus horikoshii RadA intein guide a strategy for engineering a highly efficient and promiscuous intein. J. Mol. Biol. 421, 85–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Appleby-Tagoe J. H., Thiel I. V., Wang Y., Wang Y., Mootz H. D., Liu X. Q. (2011) Highly efficient and more general cis- and trans-splicing inteins through sequential directed evolution. J. Biol. Chem. 286, 34440–34447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Du Z., Liu Y., Ban D., Lopez M. M., Belfort M., Wang C. (2010) Backbone dynamics and global effects of an activating mutation in minimized Mtu RecA inteins. J. Mol. Biol. 400, 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wood D. W., Wu W., Belfort G., Derbyshire V., Belfort M. (1999) A genetic system yields self-cleaving inteins for bioseparations. Nat. Biotechnol, 17, 889–892 [DOI] [PubMed] [Google Scholar]
- 47. Hiraga K., Soga I., Dansereau J. T., Pereira B., Derbyshire V., Du Z., Wang C., Van Roey P., Belfort G., Belfort M. (2009) Selection and structure of hyperactive inteins: peripheral changes relayed to the catalytic center. J. Mol. Biol. 393, 1106–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schwartz S. D., Schramm V. L. (2009) Enzymatic transition states and dynamic motion in barrier crossing. Nat. Chem. Biol. 5, 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 50. Bork P., Patthy L. (1995) The SEA module: a new extracellular domain associated with O-glycosylation. Protein Sci. 4, 1421–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]