Abstract
The discovery of inteins in the early 1990s opened the door to a wide variety of new technologies. Early engineered inteins from various sources allowed the development of self-cleaving affinity tags and new methods for joining protein segments through expressed protein ligation. Some applications were developed around native and engineered split inteins, which allow protein segments expressed separately to be spliced together in vitro. More recently, these early applications have been expanded and optimized through the discovery of highly efficient trans-splicing and trans-cleaving inteins. These new inteins have enabled a wide variety of applications in metabolic engineering, protein labeling, biomaterials construction, protein cyclization, and protein purification.
Keywords: Protein Chemical Modification, Protein Engineering, Protein Processing, Protein Purification, Protein Self-assembly, Self-cleaving Tag, Intein, Protein Cyclization, Protein Labeling, Protein Splicing
Introduction
The ability of inteins to form and cleave specific peptide bonds in a variety of contexts has enabled the development of powerful new tools in molecular biology. Initial applications focused on self-cleaving affinity tags and protein modification and labeling, and utilized mutations that altered the native splicing reactions described by Perler and co-workers (89) in this series. An important advantage of intein-based methods is that they are generally enzymatic in nature, and therefore exhibit highly specific activities under physiological conditions. Although thiol compounds are used in some applications, the chemistries involved in most intein methods are innocuous to their various target proteins. Intein-based methods have greatly expanded in subsequent years through the discovery of hundreds of additional inteins, and new applications in protein activity regulation and modification have followed. An important underlying theme in this work has been the discovery and development of highly efficient split inteins for advanced applications in vitro and in vivo. These trans-splicing and -cleaving inteins have been employed for applications ranging from the purification of recombinant products to the in vivo control of protein function and labeling.
Protein Purification
One of the first major applications of inteins was the development of self-cleaving affinity tags for the recovery of untagged target proteins in recombinant expression systems (1–3). In these applications, a modified intein is expressed in fusion to an affinity tag and target protein, and once the fusion is affinity-purified, the intein is induced to cleave the target protein from the intein and tag (Fig. 1). The first commercial intein system was released by New England Biolabs in 1997 and employed a modified Sce VMA1 3 intein that is triggered to cleave at its N terminus (IMPACT system), or both N and C termini (IMPACT-CN system), by the addition of thiol compounds (e.g. Refs. 4–7) (Fig. 1A, left). Shortly thereafter, inteins with rapid C-terminal cleaving activity were published. These inteins exhibit suppressed cleavage activity at pH 8.5, allowing purification of the tagged target, and are then induced to cleave by a shift to pH 6.5. The cleavage activity of these inteins is also strongly influenced by temperature, where the most rapid cleaving is observed at 37 °C (8–10) (Fig. 1A, right). The development of these inteins led to a variety of new tag systems and configurations for protein purification, initially including the chitin (3) and maltose-binding proteins (11), as well as non-chromatographic purification tags (12, 13). More recently, small ubiquitin-like modifier (SUMO) and ubiquitin tags have been used to increase expression efficiency and simplify purification (14, 15), and an ELP precipitation tag has been combined with a dockerin-cohesin binding pair to provide optimized expression with a reusable non-chromatographic purification reagent (16).
FIGURE 1.
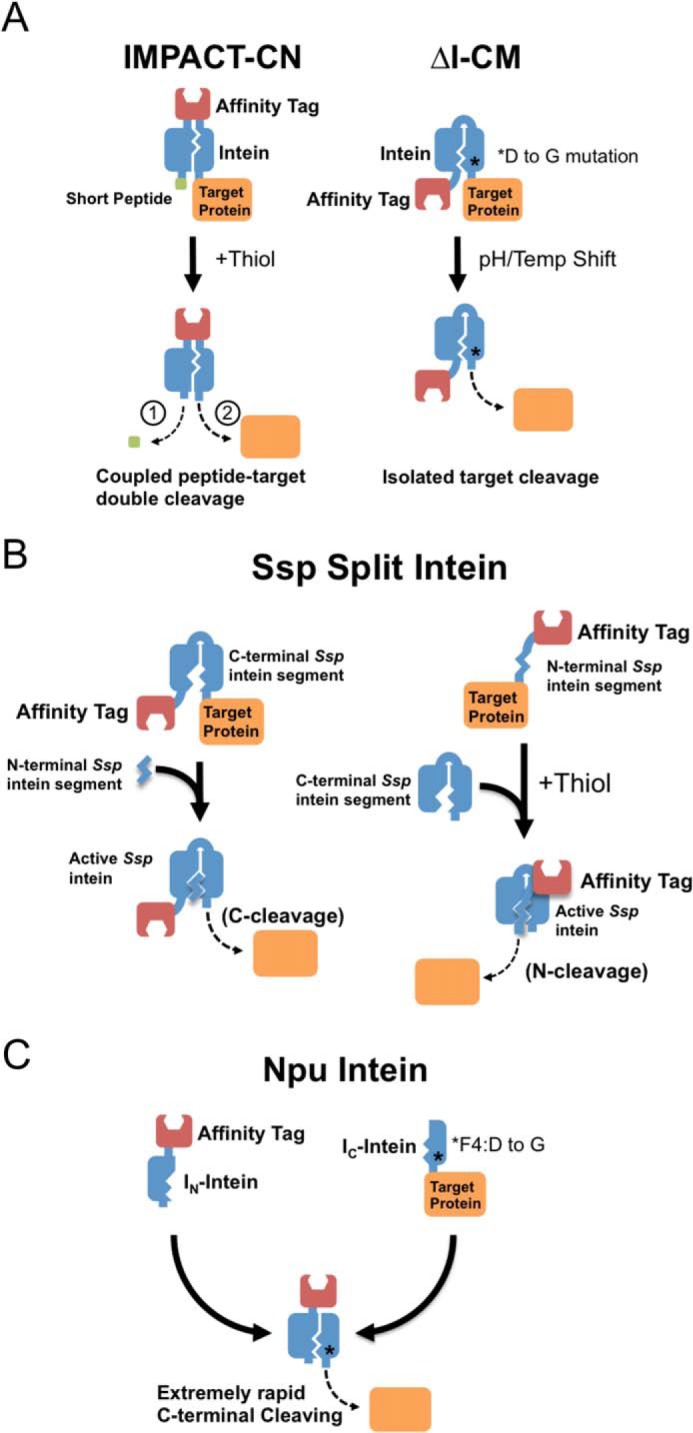
Self-cleaving affinity tags based on inteins. A, initial tags used thiol addition or temperature (Temp) and/or pH changes to induce cleaving in cis-cleaving inteins. The IMPACT-CN system (left) includes an affinity tag within the intein, and C-terminal cleaving is induced via thiol-induced cleavage of a short N-terminal extein peptide (step 1 at bottom). Cleavage of N-extein peptide then leads to rapid C-terminal cleavage (step 2 at bottom), and the N-terminal peptide is subsequently separated from the cleaved target protein by dialysis. The ΔI-CM intein (right) provides C-terminal cleaving in the absence of N-terminal cleavage, where the cleavage reaction is controlled by shifts in pH and/or temperature. In this intein, the cleavage reaction is additionally accelerated through mutation of a conserved aspartic acid to glycine, close to the C terminus of the intein. B, the Ssp DnaB intein has been engineered with an 11-residue deletion at its N terminus to eliminate premature cleaving during expression. C-terminal cleavage of the intein can be induced by the addition of the 11-residue intein segment (left panel), or conversely, N-terminal cleaving from the 11-residue intein segment can be induced by the addition of the remaining intein in the presence of thiol (right panel). C, the Npu DnaE naturally split intein has been engineered with an internal affinity tag to provide extraordinarily rapid cleaving upon reassembly. This intein effectively combines the N-extein removal of the IMPACT-CN system with the aspartic acid to glycine mutation of the ΔI-CM intein, leading to very rapid cleaving that can be controlled by intein reassembly in the presence of zinc.
A key requirement for all intein-based purification systems is the ability to minimize cleavage during protein expression but cleave rapidly once the fusion precursor is purified. Many early intein systems required long incubation times for complete C-terminal cleaving or high concentrations of thiol for efficient N-terminal cleaving. Further, C-cleaving inteins are plagued by premature cleavage during expression, whereas N-cleaving inteins are difficult to use with target proteins containing disulfide bonds. For this reason, recent work has focused on controlling intein function through the reassembly of trans-cleaving inteins, as well as the engineering of disulfide bonds into existing intein systems (17). Early trans-cleaving intein systems were generated by splitting cis-splicing inteins into two parts and inducing cleavage of the target protein by fragment reassembly (18–20). Although this strategy was effective in suppressing premature cleaving during expression, soluble expression and reassembly of the intein fragments were often inefficient (21). The discovery of naturally split inteins had a major impact in this field, however, because the segments of these inteins are capable of efficient expression and reassembly both in vivo and in vitro (22, 23). Recently reported examples of trans-cleaving systems are based on an engineered Ssp DnaB intein and the naturally trans-splicing Npu DnaE intein.
The Ssp DnaB intein has been engineered to suppress premature cleavage via an 11-amino acid deletion at its N terminus (24). This deletion abolishes cleaving activity in the remaining intein, allowing the purification of a C-terminally fused target protein. Cleavage is induced by the addition of the 11-residue peptide (Fig. 1B). Conversely, the 11-residue peptide can also be used as an effective protease target sequence, where N-terminal cleaving from this segment is induced by the addition of the C-terminal segment in the presence of thiol. In both cases, the intein cleaving mechanism releases the target protein from the immobilized tag during purification.
Another split intein purification system is based on the Npu DnaE intein, which naturally exhibits extremely rapid trans-splicing and has been engineered to exhibit rapid C-terminal cleaving upon reassembly (Fig. 1C). In this system, the N-terminal segment of the intein is fused to an affinity tag and immobilized, while the C-terminal segment is fused to the target protein. Association of the intein segments acts as the mechanism for purification, where cleavage during assembly and purification is suppressed by zinc ion. Once purified, rapid cleavage of the target is activated by thiol addition.
Remarkably, this intein can undergo complete cleaving in less than 30 min at room temperature (25, 26) and in only a few hours at 6 °C. This cleavage rate can be attributed to two rational modifications of the Npu intein. The first is the introduction of the previously reported ΔI-CM intein mutation of aspartic acid to glycine in the intein F-motif (9) (Fig. 1A, right), while the second is the repositioning of the affinity tag from the N terminus of the IN segment to the C terminus of the IN segment. Repositioning the affinity tag mimics the N-terminal cleavage of a short extein segment in the IMPACT-CN system, which greatly accelerates C-terminal cleavage (2) (Fig. 1A, left). Thus, these two established cis-splicing intein modifications have been introduced to a naturally fast trans-splicing intein to generate a highly effective C-terminal split intein cleaving system. This system is currently being optimized for expression of proteins in various host cells and is showing particular promise as an effective platform for virtually any expression host (27).
Protein Modification
Inteins are also used to modify proteins through rearrangements of peptide bonds. Applications include the production of proteins with structural modifications, including backbone cyclization, site specific labeling, and proteolysis. In each of these cases, a mutated intein is moved to a non-native context, and the resulting modified splicing reaction is used to generate covalent modifications to the target protein.
Protein Backbone Cyclization
Backbone cyclization of recombinant polypeptides can be accomplished both in vitro and in vivo by either expressed protein ligation (EPL) or protein trans-splicing (PTS) (reviewed in Ref. 28). EPL-based cyclization is accomplished by fusing the target polypeptide N terminus to a peptide leader sequence that ends with a Cys residue, while the C terminus is fused to an engineered intein (Fig. 2A, left). The N-terminal leader sequence can be cleaved in vivo or in vitro by a proteolytic or self-proteolytic event, leaving an N-terminal Cys residue on the target peptide. The Cys residue can then react in an intramolecular fashion with an α-thioester generated by the downstream intein, thus providing a backbone cyclized polypeptide.
FIGURE 2.
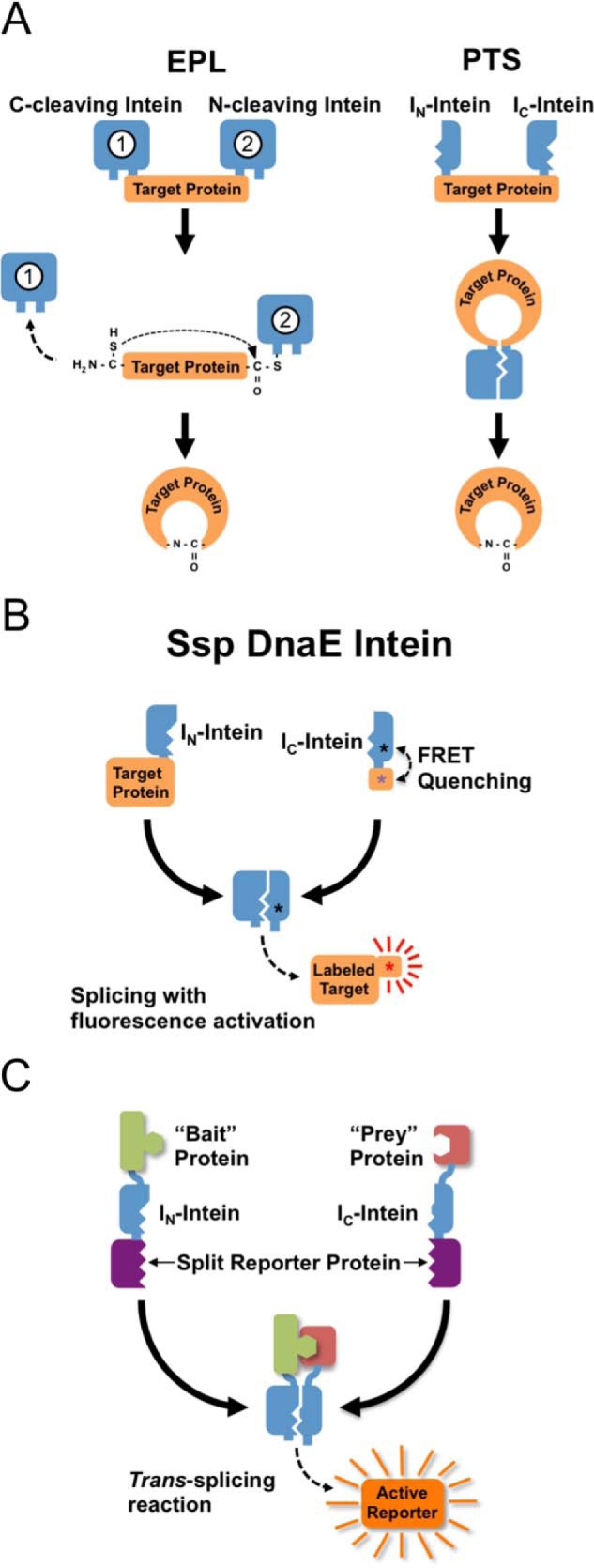
Intein applications involving post-translational modifications of target proteins. A, left panel, EPL methods involve a nucleophilic attack of an N-terminal Cys residue on a thioester formed by a downstream intein. The N-terminal Cys can be generated by a second, upstream intein or by conventional proteolytic cleavage. Right panel, PTS methods produce cyclized proteins through the assembly and splicing of an inverted split intein fused to the N and C terminus of the target protein. B, fluorescent labeling of proteins using a self-quenched intein-based PTS reagent. In this case, PTS simultaneously labels the target protein while releasing the quencher from the dye, thus providing a strong label signal with minimal background from unreacted label. C, protein-protein interactions can be detected by fusing “bait” and “prey” proteins to each half of a weakly interacting trans-splicing intein. Interactions between bait and prey drive assembly and splicing of the intein, resulting in activation of a reporter enzyme.
The use of EPL-mediated backbone cyclization was first reported by Camarero and Muir (29) in 1999, using the N-terminal SH3 domain of the c-Crk protein as a model system. The resulting circular protein domain folded faster and was more stable than the linear counterpart (30). A similar approach has been used by Iwai and Plückthun for the biosynthesis of circular β-lactamase (31) and green fluorescent protein (GFP)(32). The resulting circular proteins were biologically active and found to be more resistant to thermal denaturation (31, 32). The general applicability of this approach has recently been further demonstrated through the cyclization of several disulfide-rich backbone-cyclized peptides, including cyclotide Kalata B1 (33), Bowman-Birk inhibitor SFTI-1 (34), θ-defensins (35), and an artificially cyclized α-defensin (36).
EPL cyclization has also been used inside living cells (37). This technique allows the production and genetic selection of complex libraries of cyclic polypeptides using standard recombinant techniques. These highly stable cyclic scaffolds provide biologists with a powerful molecular tool and have been used to unravel genomic information encoding complex biochemical pathways and protein interaction networks (38, 39). For example, Camarero and co-workers (33, 40) have recently used EPL for in-cell expression of cyclotide-based libraries using standard bacterial host systems. Cyclotides are small globular microproteins with a head-to-tail cyclized backbone, which is further stabilized by three disulfide bonds forming a cysteine knot. This cysteine knot motif (CCK) gives cyclotides exceptional resistance to physical, chemical, and biological degradation, making them ideal scaffolds for the development of novel peptide-based therapeutics (41, 42). The cyclotide libraries were assayed for biological activity and the ability to fold correctly (43). A similar study was also performed by the same group using the naturally occurring cyclic peptide SFTI-1 as molecular scaffold for the generation of genetically encoded libraries (34).
Another approach for the recombinant production of cyclic peptides in vivo, first reported by Benkovic and co-workers (44) using the naturally occurring Ssp DnaE split intein, is PTS (Fig. 2A, right). PTS relies on an intein splicing domain that is split into two fragments (IN and IC). The split intein fragments are not active individually but can bind to each other with high specificity under appropriate conditions to form an active splicing domain in trans. By inverting the order of the intein fragments and fusing them in-frame to the polypeptide to be cyclized, the trans-splicing reaction results in the formation of a backbone-cyclized polypeptide. PTS has been used for the generation of several naturally occurring cyclic peptides, as well as large libraries of small cyclic peptides (45). For example, Benkovic and co-workers (46) have recently used this technology in combination with nonsense codon suppressor tRNA technology to build libraries of cyclic hexapeptides that include non-natural amino acids. These libraries were screened for HIV protease inhibitors using a cell-based lethality assay (46).
PTS has been also successfully used for the generation of larger circular proteins (44, 47). For example, the artificially split Ssp DnaB mini-intein has been used to cyclize TEM-1 β-lactamase in the bacterial periplasm, where the TEM-1 β-lactamase export signal peptide was added to the split intein precursor (47). This group also produced large libraries of small circular peptides using PTS, estimating that ∼ 50% of the combinatorial peptides cyclized efficiently inside the cell. The Kang group (48) recently reported an enhancement of this method, where backbone cyclization through PTS generates an intact c-Myc epitope tag, thus simplifying the detection and purification of the cyclic products.
Although PTS offers a good alternative to EPL for the production of backbone-cyclized polypeptides, it should be noted that most split inteins require specific amino acid residues at the intein-extein junctions for efficient protein splicing (45). The Camarero group (49) has recently shown that the highly efficient Npu DnaE intein is able to tolerate non-native junction sequences, however, making it possible to produce correctly folded cyclotide MCoTI-I containing non-natural fluorescent amino acids in an intracellular bacterial expression system (49). This exciting finding suggests the possibility for high throughput screening of genetically encoded cyclotide libraries for the ability to bind specific bait proteins.
The production of circular polypeptides using standard biological expression systems has also made possible the inexpensive introduction of NMR active isotopes (15N and/or 13C), thus facilitating the use of NMR to study structure-activity relationships of circular polypeptides and their targets (50). The recent development of in-cell NMR using sequential labeling approaches (51) could also be easily interfaced for in-cell screening of genetically encoded libraries of circular polypeptides.
Site-specific Labeling of Proteins
EPL and PTS can also enable the introduction of site-specific modifications to proteins, including glycosylation, biotinylation, ubiquitination, phosphorylation, and segmental isotopic labeling, among others (see Refs. 52 and 53 for recent reviews). These methods can also be applied to the N- and C-terminal labeling of membrane-associated proteins. In this section, we will focus on recent applications where inteins have been used for site-specific labeling of proteins in living cells.
The C terminus of the membrane-associated human transferrin receptor was recently labeled with fluorescein and biotin on the surface of live mammalian cells using the Ssp GyrB split intein (54). Using a similar approach, the N terminus of the monomeric red fluorescent protein (mRFP) was site-specifically labeled with biotin on the surface of CHO cells (55). Mootz and co-workers have also used the Npu DnaE split intein to attach enhanced GFP (eGFP) to trans-membrane and glycosylphosphatidylinositol-anchored membrane proteins using PTS in live cells (90).
PTS has also been used for site-specific labeling of proteins inside live cells. The split inteins used to accomplish this task are usually formed by either short IN fragments or short IC fragments, thereby facilitating their chemical synthesis with chemical probes. In-cell intein-mediated protein labeling has several advantages over other chemoselective ligation techniques. In particular, PTS-mediated labeling uniquely provides site-specific covalent modification of the target protein, and more importantly, the splicing reaction relies on a specific recognition event between the corresponding IN and IC fragments. Hence, the use of orthogonal split inteins (i.e. split inteins that do not cross-react with each other) might allow simultaneous multicolor labeling of proteins within living cells (56).
The in vivo use of PTS for site-specific fluorogenic dye labeling of proteins requires that the labeling process be linked to the simultaneous activation of fluorescence, thus minimizing background signal from unreacted label. Camarero and co-workers (56) have recently used Förster resonance energy transfer (FRET) quenched with a DnaE split intein to accomplish this in living cells. The fluorescent label is introduced at the C terminus of the C-extein, while the quencher is introduced on the IC intein segment (Fig. 2B). The PTS reaction ligates the fluorophore to the protein of interest while simultaneously dissociating the dye from the quencher and activating its fluorescence. This approach was used for site-specific in-cell labeling of the DNA-binding domain (DBD) of the transcription factor YY1 in several human cell lines, showing that this method can be used for modifying proteins to control their cellular localization and alter their biological activity (56).
The C-terminal labeling of a pleckstrin homology (PH) domain with quantum dots inside Xenopus embryos has also been accomplished using PTS with the split DnaE intein (57). A more recent publication demonstrates that a split mini-DnaB intein can also be used for N-terminal labeling (58), thereby demonstrating the potential for using orthogonal split inteins for multiprobe labeling.
PTS has also been used for segmental isotopic protein labeling in live cells (59). This is accomplished by sequentially expressing the labeled and unlabeled fragments in labeled and unlabeled medium, respectively, via a compatible dual expression system with different inducible promoters. For example, Züger and Iwai used in-cell PTS to fuse unlabeled immunoglobulin-binding protein G domain B1 (GB1) to the C terminus of the yeast prion protein Sup35 (residues 1–61), resulting in the production of a fusion protein with improved solubility and stability provided by the NMR-invisible tag (60).
Site-specific Proteolysis
Split inteins can also provide a means for efficient site-specific proteolysis in living cells. For example, the non-canonical Ssp DnaB S1 split intein has recently been used as an efficient and highly site-specific cellular protease (61). In this work, the protease recognition sequence is formed by the 11-residue IN fragment and five native N-extein residues. This sequence is recognized by the corresponding IC fragment, which is called the intein-derived protease. When the two fragments are co-expressed, they bind to each other in trans, reconstituting a fully active splicing domain that cleaves the protein of interest through N-terminal cleavage. This approach, which is analogous to the N-terminal tag cleavage reaction shown in Fig. 2B, has been shown to work efficiently in bacterial and eukaryotic cells. In contrast to other less specific proteases, intein-derived proteases are highly specific, presenting extremely low activity toward other cellular proteins not containing the IN recognition sequence.
Intein-based Biosensors
Intein-based biosensors are usually built from independent protein domains, which can be expressed inside living cells. The signal of interest is recognized by a sensing module, which then induces a change in the splicing activity of a fused intein module, resulting in a change in the activity of a reporter module. A common mechanism by which intein-based biosensors function is through conditional protein splicing (see Ref. 62 for a recent review). Conditional protein splicing of the intein domain is activated by an external stimulus such as the binding of small molecule, light, or a change in temperature, pH, or redox state (62). The splicing reaction restores the function of a reporter domain, which then produces an easily detectible signal. Importantly, the intein domain enables a modular design of these sensors, allowing the easy exchange of sensing and reporter domains. This feature has facilitated the design of sensors for protein-protein interactions, small molecules, redox states, protease activity, DNA methylation, and other types of stimuli.
Detecting Protein-Protein Interactions
Biosensors to detect protein-protein interactions make use of artificially or engineered split inteins with reduced affinity between the IN and IC fragments (Fig. 2C). The interacting proteins or protein domains are fused to the C and N termini of paired IN and IC fragments, respectively, which are in turn fused to the segments of a split reporter module. An interaction between the protein partners brings the split intein fragments into close proximity, leading to intein activation and splicing of the reporter protein. Umezawa and co-workers (63–65) used this concept to design several protein-protein interaction biosensors using GFP and luciferase as reporters in single cell organisms and transgenic animals. These approaches have also been used for intracellular detection of interactions between the phosphorylated insulin receptor substrate 1 and the N-terminal SH2 domain of PI3K kinase (66), the interaction between the MyoD and Id proteins (67), and the epidermal growth factor (EGF)-induced interaction between an oncogenic mutant of Ras and Raf-1 (63).
Detecting Protein-DNA Interaction
A similar PTS-mediated biosensor has also been designed for detecting specific changes in DNA methylation. This biosensor was designed around a split version of the Sce VMA intein, where each intein segment is fused to a pentadactyl zinc finger domain and a split firefly luciferase segment. Binding of both zinc finger domains to their specific DNA target sequences induces intein-mediated reconstitution of the luciferase reporter (68). This biosensor was used to detect the loss of epigenetic silencing and increased accessibility of a DNA sequence near the promoter region of L1PA2 upon treatment with a demethylation drug.
Detecting Small Molecules
Wood and co-workers (69, 70) have designed allosteric intein biosensors for the detection of human nuclear hormone receptor ligands. In this case, a non-splicing mini-intein acts as an allosteric signal transducer between the receptor ligand-binding domain (LBD) and the reporter enzyme. The biosensor relies on a rationally designed fusion of the Mtu RecA mini-intein splicing domain with a human nuclear hormone receptor LBD. This module is then fused to the C terminus of the Escherichia coli maltose-binding protein (MBP) and to the N terminus of the T4 thymidylate synthase enzyme. In practice, the activity of the thymidylate synthase reporter is modulated by hormone binding to the LBD, allowing the production of E. coli cell lines that report the presence of hormone-like ligands through a simple growth phenotype assay (69). Initial studies used the human estrogen (ERα) and thyroid hormone (TRβ-1) receptors and showed that this sensor can differentiate between receptor agonists and antagonists (71). Further, the modular design of the sensor has simplified the development of additional biosensors based on estrogen (ERα and ERβ) (69) and thyroid hormone (TRα-1 and TRβ-1) receptors (72), as well as the human peroxisome proliferator-activated receptor γ (PPARγ) receptor (73) and several animal estrogen receptors (74).
Liu and co-workers (75) have also designed an intein-based biosensor for the detection of estrogens. In this work, an estrogen-sensitive intein was created by replacing the endonuclease region of the Sce VMA intein with the human ERα receptor LBD. The DNA encoding this modified intein was inserted into the constitutively expressed chromosomal lacZ gene. The resulting intein was designed to be able to splice in the presence of estrogenic ligands to produce an active β-galactosidase reporter enzyme (76).
Detecting Redox State
The split Ssp DnaE intein has recently been used for the development of a bacterial redox sensor. This sensor was designed by engineering a new disulfide bond that includes the catalytic N-terminal intein Cys residue (17, 77). The activity of the intein is reported by a FRET pair formed by cyan and yellow fluorescent proteins. In the disulfide-bonded state, the intein is inactive, providing high FRET. When the disulfide bond is reduced, however, the intein becomes active, triggering the N-terminal cleavage of the cyan fluorescent protein with a concomitant decrease in the FRET signal. This biosensor has been successfully used to select hyperoxic mutant E. coli strains (17, 77).
Detecting Protease Activity
In-cell protease activity has been also detected using protein trans-splicing (78). In this work, the luciferase reporter protein backbone was cyclized through PTS using the Ssp DnaE intein with a caspase-3 substrate linker. In the cyclized state, the activity of luciferase is greatly reduced due to steric hindrance, but is fully restored after cleavage by caspase-3. This sensor has enabled real-time sensing of caspase activity in living mice (78).
Gene Delivery and Control in Transgenic Organisms
Split inteins in combination with PTS can also be used to control the delivery of heterologous genes into transgenic organisms (see Ref. 79 for a recent review on plants). This approach relies on splitting the target protein into two segments, which can be post-translationally reconstituted in vivo by PTS. This approach minimizes the risk for transferring particular genes that confer desired traits to unwanted hosts, as in the case of herbicide resistance genes. For example, the metabolic enzyme acetolactate synthase has been split and successfully reconstituted by PTS in E. coli (80). In a similar fashion, the bacterial 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) was also reconstituted by PTS in the chloroplasts of Nicotiana tabacum to produce an herbicide-resistant transgenic crop (81, 82). By splitting the gene between nuclear and chloroplast chromosomes, the potential for recombination and transfer to an undesired host is strongly inhibited. Similar PTS strategies have also been used for the reconstitution of fully active heterologous β-glucuronidase in the plant Arabidopsis thaliana (83) as well as in the leaves of barley, soybean, maize, and pea (84).
The thermostable self-splicing bacterial intein from Thermus thermophilus has been recently used to control a thermostable xylanase (XynB) from Dictyoglomus thermophilum in maize (85). By producing the XynB enzyme within the corn plant itself, the authors have created “self-processing” corn, which hydrolyzes its own cellulosic biomass to simple sugars for fermentation. Transgenic maize expressing active XynB exhibits shriveled seeds and low fertility, but maize expressing the conditionally active XynB-intein fusion produces normal seeds and fertility. By using directed evolution, the authors generated mutants of the XynB-intein construct that could splice only at high temperature to restore wild-type xylanase activity. These strains have great potential in simplifying pretreatment of plant cellulosic biomass to release soluble sugars.
PTS has also enabled the delivery and control of oversized transgenes in mammalian cells and mice. For example, the split DnaB mini-intein has been employed in cell culture (86) and in mice (87) for joining the heavy and light chains of the B-domain deleted factor VIII. In this case, the split genes were delivered by two separate viral vectors (86, 87), and in vivo splicing activity was evaluated via measurements of plasma protein concentration and increased coagulation activity. These findings suggest that PTS can be efficiently used for in vivo production of proteins that would otherwise be too large to be delivered by a single viral vector.
The split DnaE intein has also been employed for the production of Cre recombinase in mice (88). By placing the two split intein-Cre constructs under the control of two different promoters, the split Cre system can be used to interrogate the expression patterns of two genes or promoters while allowing the production of proteins under control of the Cre-Lox P system.
Conclusions
Since their initial discovery in the early 1990s, intein applications have become more efficient, and intein methods are now commonly used in laboratories worldwide. Successes include new understandings of cell and animal physiology, as well as highly effective methods for protein purification. Naturally and artificially split inteins have enhanced the effectiveness of early technologies while enabling the development of new applications in metabolic engineering and drug discovery. These inteins have further enabled new biological tools, allowing specific control over the biological functions of proteins in living cells, plants, and whole animals. Perhaps the most exciting aspect of recent intein technology development is its movement out of the laboratory and into real-world applications in energy and medicine. Future intein applications will build on these techniques, providing new classes of therapeutic proteins and new avenues for synthetic biology. Protein purification methods based on self-cleaving intein tags are now commonly used in laboratories worldwide and are expected to provide a significant platform for the production of commercial enzymes and therapeutic proteins. These developing applications suggest that inteins are becoming a mature and critical biological tool, capable of opening new avenues of scientific research, as well as enhanced transgenic plants and new therapeutic strategies.
This work was supported, in whole or in part, by National Institutes of Health Research Grant R01-GM090323 (to J. A. C.). This is the fourth article in the Thematic Minireview Series “Inteins.”
- VMA
- vacuolar ATP synthase catalytic subunit A
- EPL
- expressed protein ligation
- PTS
- protein trans-splicing
- LBD
- ligand-binding domain
- TR
- thyroid hormone receptor
- ER
- estrogen receptor.
REFERENCES
- 1. Southworth M. W., Amaya K., Evans T. C., Xu M. Q., Perler F. B. (1999) Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. Biotechniques 27, 110–114, 116,, 118–120 [DOI] [PubMed] [Google Scholar]
- 2. Chong S., Montello G. E., Zhang A., Cantor E. J., Liao W., Xu M. Q., Benner J. (1998) Utilizing the C-terminal cleavage activity of a protein splicing element to purify recombinant proteins in a single chromatographic step. Nucleic Acids Res. 26, 5109–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chong S., Mersha F. B., Comb D. G., Scott M. E., Landry D., Vence L. M., Perler F. B., Benner J., Kucera R. B., Hirvonen C. A., Pelletier J. J., Paulus H., Xu M.-Q. (1997) Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene 192, 271–281 [DOI] [PubMed] [Google Scholar]
- 4. Szweda P., Pladzyk R., Kotlowski R., Kur J. (2001) Cloning, expression, and purification of the Staphylococcus simulans lysostaphin using the intein-chitin-binding domain (CBD) system. Protein Expr. Purif. 22, 467–471 [DOI] [PubMed] [Google Scholar]
- 5. Hong S., Toyama M., Maret W., Murooka Y. (2001) High yield expression and single step purification of human thionein/metallothionein. Protein Expr. Purif. 21, 243–250 [DOI] [PubMed] [Google Scholar]
- 6. Thomson C. A., Ananthanarayanan V. S. (2001) A method for expression and purification of soluble, active Hsp47, a collagen-specific molecular chaperone. Protein Expr. Purif. 23, 8–13 [DOI] [PubMed] [Google Scholar]
- 7. Wu C., Seitz P. K., Falzon M. (2000) Single-column purification and bio-characterization of recombinant human parathyroid hormone-related protein (1–139). Mol. Cell. Endocrinol. 170, 163–174 [DOI] [PubMed] [Google Scholar]
- 8. Myscofski D. M., Dutton E. K., Cantor E., Zhang A., Hruby D. E. (2001) Cleavage and purification of intein fusion proteins using the Streptococcus gordonii SPEX system. Prep. Biochem. Biotechnol. 31, 275–290 [DOI] [PubMed] [Google Scholar]
- 9. Wood D. W., Wu W., Belfort G., Derbyshire V., Belfort M. (1999) A genetic system yields self-cleaving inteins for bioseparations. Nat. Biotechnol. 17, 889–892 [DOI] [PubMed] [Google Scholar]
- 10. Mathys S., Evans T. C., Chute I. C., Wu H., Chong S., Benner J., Liu X. Q., Xu M. Q. (1999) Characterization of a self-splicing mini-intein and its conversion into autocatalytic N- and C-terminal cleavage elements: facile production of protein building blocks for protein ligation. Gene 231, 1–13 [DOI] [PubMed] [Google Scholar]
- 11. Wood D. W., Derbyshire V., Wu W., Chartrain M., Belfort M., Belfort G. (2000) Optimized single-step affinity purification with a self-cleaving intein applied to human acidic fibroblast growth factor. Biotechnol. Prog. 16, 1055–1063 [DOI] [PubMed] [Google Scholar]
- 12. Banki M. R., Feng L., Wood D. W. (2005) Simple bioseparations using self-cleaving elastin-like polypeptide tags. Nat. Methods 2, 659–661 [DOI] [PubMed] [Google Scholar]
- 13. Banki M. R., Gerngross T. U., Wood D. W. (2005) Novel and economical purification of recombinant proteins: intein-mediated protein purification using in vivo polyhydroxybutyrate (PHB) matrix association. Protein Sci. 14, 1387–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xie Y. G., Luan C., Zhang H. W., Han F. F., Feng J., Choi Y. J., Groleau D., Wang Y. Z. (2013) Effects of thioredoxin: SUMO and intein on soluble fusion expression of an antimicrobial peptide OG2 in Escherichia coli. Protein Pept. Lett. 20, 54–60 [PubMed] [Google Scholar]
- 15. Wang Z., Li N., Wang Y., Wu Y., Mu T., Zheng Y., Huang L., Fang X. (2012) Ubiquitin-intein and SUMO2-intein fusion systems for enhanced protein production and purification. Protein Expr. Purif. 82, 174–178 [DOI] [PubMed] [Google Scholar]
- 16. Liu F., Chen W. (2013) Engineering a recyclable elastin-like polypeptide capturing scaffold for non-chromatographic protein purification. Biotechnol. Prog. 29, 968–971 [DOI] [PubMed] [Google Scholar]
- 17. Callahan B. P., Stanger M., Belfort M. (2013) A redox trap to augment the intein toolbox. Biotechnol. Bioeng. 110, 1565–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu H., Xu M. Q., Liu X. Q. (1998) Protein trans-splicing and functional mini-inteins of a cyanobacterial dnaB intein. Biochim. Biophys. Acta 1387, 422–432 [DOI] [PubMed] [Google Scholar]
- 19. Mills K. V., Lew B. M., Jiang S., Paulus H. (1998) Protein splicing in trans by purified N- and C-terminal fragments of the Mycobacterium tuberculosis RecA intein. Proc. Natl. Acad. Sci. U.S.A. 95, 3543–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shingledecker K., Jiang S. Q., Paulus H. (1998) Molecular dissection of the Mycobacterium tuberculosis RecA intein: design of a minimal intein and of a trans-splicing system involving two intein fragments. Gene 207, 187–195 [DOI] [PubMed] [Google Scholar]
- 21. Shi C., Meng Q., Wood D. W. (2013) A dual ELP-tagged split intein system for non-chromatographic recombinant protein purification. Appl. Microbiol. Biotechnol. 97, 829–835 [DOI] [PubMed] [Google Scholar]
- 22. Martin D. D., Xu M. Q., Evans T. C., Jr. (2001) Characterization of a naturally occurring trans-splicing intein from Synechocystis sp. PCC6803. Biochemistry 40, 1393–1402 [DOI] [PubMed] [Google Scholar]
- 23. Evans T. C., Jr., Martin D., Kolly R., Panne D., Sun L., Ghosh I., Chen L., Benner J., Liu X. Q., Xu M. Q. (2000) Protein trans-splicing and cyclization by a naturally split intein from the dnaE gene of Synechocystis species PCC6803. J. Biol. Chem. 275, 9091–9094 [DOI] [PubMed] [Google Scholar]
- 24. Volkmann G., Sun W., Liu X. Q. (2009) Controllable protein cleavages through intein fragment complementation. Protein Sci. 18, 2393–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramirez M., Valdes N., Guan D., Chen Z. (2013) Engineering split intein DnaE from Nostoc punctiforme for rapid protein purification. Protein Eng. Des. Sel. 26, 215–223 [DOI] [PubMed] [Google Scholar]
- 26. Zettler J., Schütz V., Mootz H. D. (2009) The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 583, 909–914 [DOI] [PubMed] [Google Scholar]
- 27. Guan D., Ramirez M., Chen Z. (2013) Split intein mediated ultra-rapid purification of tagless protein (SIRP). Biotechnol. Bioeng. 110, 2471–2481 [DOI] [PubMed] [Google Scholar]
- 28. Aboye T. L., Camarero J. A. (2012) Biological synthesis of circular polypeptides. J. Biol. Chem. 287, 27026–27032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Camarero J. A., Muir T. W. (1999) Biosynthesis of a head-to-tail cyclized protein with improved biological activity. J. Am. Chem. Soc. 121, 5597–5598 [Google Scholar]
- 30. Camarero J. A., Fushman D., Sato S., Giriat I., Cowburn D., Raleigh D. P., Muir T. W. (2001) Rescuing a destabilized protein fold through backbone cyclization. J. Mol. Biol. 308, 1045–1062 [DOI] [PubMed] [Google Scholar]
- 31. Iwai H., Plückthun A. (1999) Circular β-lactamase: stability enhancement by cyclizing the backbone. FEBS Lett. 459, 166–172 [DOI] [PubMed] [Google Scholar]
- 32. Iwai H., Lingel A., Plückthun A. (2001) Cyclic green fluorescent protein produced in vivo using an artificially split PI-PfuI intein from Pyrococcus furiosus. J. Biol. Chem. 276, 16548–16554 [DOI] [PubMed] [Google Scholar]
- 33. Kimura R. H., Tran A. T., Camarero J. A. (2006) Biosynthesis of the cyclotide Kalata B1 by using protein splicing. Angew. Chem. Int. Ed. Engl. 45, 973–976 [DOI] [PubMed] [Google Scholar]
- 34. Austin J., Kimura R. H., Woo Y. H., Camarero J. A. (2010) In vivo biosynthesis of an Ala-scan library based on the cyclic peptide SFTI-1. Amino Acids 38, 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gould A., Li Y., Majumder S., Garcia A. E., Carlsson P., Shekhtman A., Camarero J. A. (2012) Recombinant production of rhesus Θ-defensin-1 (RTD-1) using a bacterial expression system. Mol. Biosyst. 8, 1359–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Garcia A. E., Tai K. P., Puttamadappa S. S., Shekhtman A., Ouellette A. J., Camarero J. A. (2011) Biosynthesis and antimicrobial evaluation of backbone-cyclized α-defensins. Biochemistry 50, 10508–10519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Camarero J. A., Fushman D., Cowburn D., Muir T. W. (2001) Peptide chemical ligation inside living cells: in vivo generation of a circular protein domain. Bioorg. Med. Chem. 9, 2479–2484 [DOI] [PubMed] [Google Scholar]
- 38. Sancheti H., Camarero J. A. (2009) “Splicing up” drug discovery. Cell-based expression and screening of genetically-encoded libraries of backbone-cyclized polypeptides. Adv. Drug Deliv. Rev. 61, 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gould A., Ji Y., Aboye T. L., Camarero J. A. (2011) Cyclotides, a novel ultrastable polypeptide scaffold for drug discovery. Curr. Pharm. Des. 17, 4294–4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Camarero J. A., Kimura R. H., Woo Y. H., Shekhtman A., Cantor J. (2007) Biosynthesis of a fully functional cyclotide inside living bacterial cells. Chembiochem 8, 1363–1366 [DOI] [PubMed] [Google Scholar]
- 41. Garcia A. E., Camarero J. A. (2010) Biological activities of natural and engineered cyclotides, a novel molecular scaffold for peptide-based therapeutics. Curr. Mol. Pharmacol. 3, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Henriques S. T., Craik D. J. (2010) Cyclotides as templates in drug design. Drug Discov. Today 15, 57–64 [DOI] [PubMed] [Google Scholar]
- 43. Austin J., Wang W., Puttamadappa S., Shekhtman A., Camarero J. A. (2009) Biosynthesis and biological screening of a genetically encoded library based on the cyclotide MCoTI-I. Chembiochem 10, 2663–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scott C. P., Abel-Santos E., Wall M., Wahnon D. C., Benkovic S. J. (1999) Production of cyclic peptides and proteins in vivo. Proc. Natl. Acad. Sci. U.S.A. 96, 13638–13643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tavassoli A., Benkovic S. J. (2007) Split-intein mediated circular ligation used in the synthesis of cyclic peptide libraries in E. coli. Nat. Protoc. 2, 1126–1133 [DOI] [PubMed] [Google Scholar]
- 46. Young T. S., Young D. D., Ahmad I., Louis J. M., Benkovic S. J., Schultz P. G. (2011) Evolution of cyclic peptide protease inhibitors. Proc. Natl. Acad. Sci. U.S.A. 108, 11052–11056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deschuyteneer G., Garcia S., Michiels B., Baudoux B., Degand H., Morsomme P., Soumillion P. (2010) Intein-mediated cyclization of randomized peptides in the periplasm of Escherichia coli and their extracellular secretion. ACS Chem. Biol. 5, 691–700 [DOI] [PubMed] [Google Scholar]
- 48. Sudheer P. D., Pack S. P., Kang T. J. (2013) Cyclization tag for the detection and facile purification of backbone-cyclized proteins. Anal. Biochem. 436, 137–141 [DOI] [PubMed] [Google Scholar]
- 49. Jagadish K., Borra R., Lacey V., Majumder S., Shekhtman A., Wang L., Camarero J. A. (2013) Expression of fluorescent cyclotides using protein trans-splicing for easy monitoring of cyclotide-protein interactions. Angew. Chem. Int. Ed. Engl. 52, 3126–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Puttamadappa S. S., Jagadish K., Shekhtman A., Camarero J. A. (2010) Backbone dynamics of cyclotide MCoTI-I free and complexed with trypsin. Angew. Chem. Int. Ed. Engl. 49, 7030–7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Burz D. S., Dutta K., Cowburn D., Shekhtman A. (2006) Mapping structural interactions using in-cell NMR spectroscopy (STINT-NMR). Nat. Methods 3, 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Berrade L., Camarero J. A. (2009) Expressed protein ligation: a resourceful tool to study protein structure and function. Cell. Mol. Life Sci. 66, 3909–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Volkmann G., Iwaï H. (2010) Protein trans-splicing and its use in structural biology: opportunities and limitations. Mol. Biosyst. 6, 2110–2121 [DOI] [PubMed] [Google Scholar]
- 54. Volkmann G., Liu X. Q. (2009) Protein C-terminal labeling and biotinylation using synthetic peptide and split-intein. PLoS One 4, e8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ando T., Tsukiji S., Tanaka T., Nagamune T. (2007) Construction of a small-molecule-integrated semisynthetic split intein for in vivo protein ligation. Chem. Commun. (Camb) 4995–4997 [DOI] [PubMed] [Google Scholar]
- 56. Borra R., Dong D., Elnagar A. Y., Woldemariam G. A., Camarero J. A. (2012) In-cell fluorescence activation and labeling of proteins mediated by FRET-quenched split inteins. J. Am. Chem. Soc. 134, 6344–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Charalambous A., Andreou M., Skourides P. A. (2009) Intein-mediated site-specific conjugation of Quantum Dots to proteins in vivo. J. Nanobiotechnology 7, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Charalambous A., Andreou M., Antoniades I., Christodoulou N., Skourides P. A. (2012) In vivo, site-specific, covalent conjugation of quantum dots to proteins via split-intein splicing. Methods Mol. Biol. 906, 157–169 [DOI] [PubMed] [Google Scholar]
- 59. Iwai H., Züger S. (2007) Protein ligation: applications in NMR studies of proteins. Biotechnol. Genet. Eng. Rev. 24, 129–145 [DOI] [PubMed] [Google Scholar]
- 60. Züger S., Iwai H. (2005) Intein-based biosynthetic incorporation of unlabeled protein tags into isotopically labeled proteins for NMR studies. Nat. Biotechnol. 23, 736–740 [DOI] [PubMed] [Google Scholar]
- 61. Volkmann G., Volkmann V., Liu X. Q. (2012) Site-specific protein cleavage in vivo by an intein-derived protease. FEBS Lett. 586, 79–84 [DOI] [PubMed] [Google Scholar]
- 62. Mootz H. D. (2009) Split inteins as versatile tools for protein semisynthesis. Chembiochem 10, 2579–2589 [DOI] [PubMed] [Google Scholar]
- 63. Kanno A., Ozawa T., Umezawa Y. (2009) Bioluminescent imaging of MAPK function with intein-mediated reporter gene assay. Methods Mol. Biol. 574, 185–192 [DOI] [PubMed] [Google Scholar]
- 64. Kanno A., Umezawa Y., Ozawa T. (2009) Detection of apoptosis using cyclic luciferase in living mammals. Methods Mol. Biol. 574, 105–114 [DOI] [PubMed] [Google Scholar]
- 65. Kanno A., Ozawa T., Umezawa Y. (2011) Detection of protein-protein interactions in bacteria by GFP-fragment reconstitution. Methods Mol. Biol. 705, 251–258 [DOI] [PubMed] [Google Scholar]
- 66. Ozawa T., Kaihara A., Sato M., Tachihara K., Umezawa Y. (2001) Split luciferase as an optical probe for detecting protein-protein interactions in mammalian cells based on protein splicing. Anal. Chem. 73, 2516–2521 [DOI] [PubMed] [Google Scholar]
- 67. Paulmurugan R., Umezawa Y., Gambhir S. S. (2002) Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc. Natl. Acad. Sci. U.S.A. 99, 15608–15613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Huang X., Narayanaswamy R., Fenn K., Szpakowski S., Sasaki C., Costa J., Blancafort P., Lizardi P. M. (2012) Sequence-specific biosensors report drug-induced changes in epigenetic silencing in living cells. DNA Cell Biol. 31, S2–S10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Skretas G., Wood D. W. (2005) Regulation of protein activity with small-molecule-controlled inteins. Protein Sci. 14, 523–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Skretas G., Wood D. W. (2005) A bacterial biosensor of endocrine modulators. J. Mol. Biol. 349, 464–474 [DOI] [PubMed] [Google Scholar]
- 71. Skretas G., Meligova A. K., Villalonga-Barber C., Mitsiou D. J., Alexis M. N., Micha-Screttas M., Steele B. R., Screttas C. G., Wood D. W. (2007) Engineered chimeric enzymes as tools for drug discovery: generating reliable bacterial screens for the detection, discovery, and assessment of estrogen receptor modulators. J. Am. Chem. Soc. 129, 8443–8457 [DOI] [PubMed] [Google Scholar]
- 72. Gierach I., Li J., Wu W.-Y., Grover G. J., Wood D. W. (2012) Bacterial biosensors for screening isoform-selective ligands for human thyroid receptors α-1 and β-1. FEBS Open Bio. 2, 247–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li J., Gierach I., Gillies A. R., Warden C. D., Wood D. W. (2011) Engineering and optimization of an allosteric biosensor protein for peroxisome proliferator-activated receptor γ ligands. Biosens. Bioelectron. 29, 132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gierach I., Shapero K., Eyster T. W., Wood D. W. (2013) Bacterial biosensors for evaluating potential impacts of estrogenic endocrine disrupting compounds in multiple species. Environ. Toxicol. 28, 179–189 [DOI] [PubMed] [Google Scholar]
- 75. Buskirk A. R., Ong Y. C., Gartner Z. J., Liu D. R. (2004) Directed evolution of ligand dependence: small-molecule-activated protein splicing. Proc. Natl. Acad. Sci. U.S.A. 101, 10505–10510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liang R., Zhou J., Liu J. (2011) Construction of a bacterial assay for estrogen detection based on an estrogen-sensitive intein. Appl. Environ. Microbiol. 77, 2488–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Callahan B. P., Topilina N. I., Stanger M. J., Van Roey P., Belfort M. (2011) Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications. Nat. Struct. Mol. Biol. 18, 630–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kanno A., Yamanaka Y., Hirano H., Umezawa Y., Ozawa T. (2007) Cyclic luciferase for real-time sensing of caspase-3 activities in living mammals. Angew. Chem. Int. Ed. Engl. 46, 7595–7599 [DOI] [PubMed] [Google Scholar]
- 79. Evans T. C., Jr., Xu M. Q., Pradhan S. (2005) Protein splicing elements and plants: from transgene containment to protein purification. Annu. Rev. Plant Biol. 56, 375–392 [DOI] [PubMed] [Google Scholar]
- 80. Sun L., Ghosh I., Paulus H., Xu M. Q. (2001) Protein trans-splicing to produce herbicide-resistant acetolactate synthase. Appl. Environ. Microbiol. 67, 1025–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chin H. G., Kim G. D., Marin I., Mersha F., Evans T. C., Jr., Chen L., Xu M. Q., Pradhan S. (2003) Protein trans-splicing in transgenic plant chloroplast: reconstruction of herbicide resistance from split genes. Proc. Natl. Acad. Sci. U.S.A. 100, 4510–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dun B. Q., Wang X. J., Lu W., Zhao Z. L., Hou S. N., Zhang B. M., Li G. Y., Evans T. C., Jr., Xu M. Q., Lin M. (2007) Reconstitution of glyphosate resistance from a split 5-enolpyruvyl shikimate-3-phosphate synthase gene in Escherichia coli and transgenic tobacco. Appl. Environ. Microbiol. 73, 7997–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yang J., Fox G. C., Jr., Henry-Smith T. V. (2003) Intein-mediated assembly of a functional β-glucuronidase in transgenic plants. Proc. Natl. Acad. Sci. U.S.A. 100, 3513–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yang J., Henry-Smith T. V., Qi M. (2006) Functional analysis of the split Synechocystis DnaE intein in plant tissues by biolistic particle bombardment. Transgenic Res. 15, 583–593 [DOI] [PubMed] [Google Scholar]
- 85. Shen B., Sun X., Zuo X., Shilling T., Apgar J., Ross M., Bougri O., Samoylov V., Parker M., Hancock E., Lucero H., Gray B., Ekborg N. A., Zhang D., Johnson J. C., Lazar G., Raab R. M. (2012) Engineering a thermoregulated intein-modified xylanase into maize for consolidated lignocellulosic biomass processing. Nat. Biotechnol. 30, 1131–1136 [DOI] [PubMed] [Google Scholar]
- 86. Zhu F., Liu Z., Chi X., Qu H. (2010) Protein trans-splicing based dual-vector delivery of the coagulation factor VIII gene. Sci. China Life Sci. 53, 683–689 [DOI] [PubMed] [Google Scholar]
- 87. Zhu F., Liu Z., Wang X., Miao J., Qu H., Chi X. (2013) Inter-chain disulfide bond improved protein trans-splicing increases plasma coagulation activity in C57BL/6 mice following portal vein FVIII gene delivery by dual vectors. Sci. China Life Sci. 56, 262–267 [DOI] [PubMed] [Google Scholar]
- 88. Wang P., Chen T., Sakurai K., Han B. X., He Z., Feng G., Wang F. (2012) Intersectional Cre driver lines generated using split-intein mediated split-Cre reconstitution. Sci. Rep. 2, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mills K V., Johnson M. A., Perler F. B. (2014) Protein splicing: how inteins escape from precursor proteins. J. Biol. Chem. 289, 14498–14505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Dhar T., Mootz H. D. (2011) Modification of transmembrane and GPI-anchored proteins on living cells by efficient protein trans-splicing using the Npu DnaE intein. Chem. Commun. (Camb), 47, 3063–3065 [DOI] [PubMed] [Google Scholar]


