Background: The Wnt1 pathway is recognized to play a major role in cancer progression.
Results: The promoter region of the WNT1 gene can form G-quadruplex structures, which regulate WNT1 expression and its downstream signaling pathways.
Conclusion: The Wnt1-mediated migration and invasion activities of cancer cells are inhibited by G-quadruplex stabilizers.
Significance: A pathway-specific strategy is identified to repress cancer metastasis using G-quadruplex stabilizers.
Keywords: Anticancer Drug, Cell Migration, DNA Structure, Promoters, Wnt Pathway, G-quadruplex
Abstract
WNT1 encodes a multifunctional signaling glycoprotein that is highly expressed in several malignant tumors. Patients with Wnt1-positive cancer are usually related to advanced metastasis. Here, we found that a stretch of G-rich sequences located at the WNT1 promoter region is capable of forming G-quadruplex structures. The addition of G-quadruplex structure stabilizers, BMVC and BMVC4, raises the melting temperature of the oligonucleotide formed by the WNT1 promoter G-rich sequences. Significantly, the expression of WNT1 was repressed by BMVC or BMVC4 in a G-quadruplex-dependent manner, suggesting that they can be used to modulate WNT1 expression. The role of G-quadruplex stabilizers on Wnt1-mediated cancer migration and invasion was further analyzed. The protein levels of β-catenin, a mediator of the Wnt-mediated signaling pathway, and the downstream targets MMP7 and survivin were down-regulated upon BMVC or BMVC4 treatments. Moreover, the migration and invasion activities of cancer cells were inhibited by BMVC and BMVC4, and the inhibitory effects can be reversed by WNT1-overexpression. Thus the Wnt1 expression and its downstream signaling pathways can be regulated through the G-quadruplex sequences located at its promoter region. These findings provide a novel approach for future drug development to inhibit migration and invasion of cancer cells.
Introduction
The Wnt-mediated signal pathway controls various cellular functions, including proliferation, survival, migration, and development by trigger downstream signaling cascades (1). The canonical Wnt-mediated signaling pathway is activated by the binding of Wnts to Frizzled receptors and LRP5/6 co-receptors in the plasma membrane. The LRP5/6 is phosphorylated and Dishevelled (Dsh) is recruited to the plasma membrane to interact with Frizzled and inactivate the destruction complex to protect β-catenin from degradation. The accumulation of cytoplasmic β-catenin then translocates to the nucleus where it interacts with transcription factor T-cell factor (Tcf)/lymphoid enhancer factor (Lef) and stimulates the expression of target genes (2, 3). Many of the Wnt-targeted genes are involved in proliferation (myc and cyclin D1), angiogenesis (WISP1), migration (Wrch1), or invasion (MMP7) (4–7). Thus, aberrant activation of Wnt signaling is observed in a variety of tumors. For example, high levels of WNT1 expression in patients are associated with advanced metastasis (8–10). Moreover, Wnt1-positive cancers show higher proliferation capacities, and the overall survival is lower in patients with Wnt1-positive cancer. Thus, developing the Wnt pathway inhibitors has been considered as a therapeutic approach for the treatment of patients with cancers and other Wnt-related diseases (11).
The regulation mechanism of WNT1 expression has been analyzed. Functional analysis of human WNT1 proximal promoter using reporter assays revealed that the 277-bp upstream sequence of WNT1 is sufficient for the control of developmentally regulated expression (12, 13). Pax3, for example, stimulates the transcription of WNT1 through its association with a conserved binding site located at position −130 to −136 within the WNT1 promoter (14). Sequence analysis of this 277-bp upstream sequence also identified two TATA boxes and a stretch of extremely G-rich sequence. Interestingly, the G-rich sequence of the WNT1 promoter contains four runs of at least three contiguous guanines that may form G-quadruplex structures under physiological conditions. Formation of G-quadruplex structures by the G-rich sequences were also found in the promoter regions of other oncogenes including c-myc, VEGF, bcl-2, and c-kit (15–18). Moreover, these G-quadruplex structures were shown to repress the expression of these genes in the presence of G-quadruplex stabilizers (15). Hence, it is possible that the G-rich sequence within the WNT1 promoter might form specific G-quadruplex structure that could be a drug target for regulating WNT1 expression.
Previously, we have developed two carbazole derivatives, 3,6-bis(1-methyl-4-vinylpyridinium) carbazole diiodide (BMVC)4 and 3,6-bis(4-methyl-2-vinylpyrazinium) carbazole diiodide (BMVC4), that bind to G-quadruplex structure formed by telomeric DNA sequences, inhibit telomerase activity, cause telomere shortening, and induce cancer cells into senescence (19, 20). The binding of BMVC and BMVC4 to G-quadruplex structures, however, is not limited to telomeres as they also target a quadruplex forming sequence located at the promoter region of c-myc to repress its expression. Here, we test the possibility of G-quadruplex structure formation by the G-rich sequences of WNT1 promoter and evaluate the potential of repressing WNT1 expression using our developed G-quadruplex stabilizers. We find that the G-rich sequence of WNT1 promoter is capable of forming G-quadruplex structure in the presence of potassium ion. Both BMVC and BMVC4 repressed the expression of WNT1 in a G-quadruplex structure-dependent manner. The Wnt1-mediated signaling pathway was repressed upon the addition of BMVC and BMVC4 in cancer cells. Consequently, the migration and invasion activities of cancer cells were also decreased. Thus, our results provide a novel mechanism of suppressing tumor metastasis through stabilizing the G-quadruplex forming sequence located at the WNT1 promoter.
EXPERIMENTAL PROCEDURES
DNA Preparation
All oligonucleotides were purchased from Bio Basic (Ontario, Canada) and used without further purification. DNA samples were prepared by dissolving oligonucleotides in 10 mm Tris-HCl (pH 7.5). DNA concentrations were determined using a UV-visible absorption nanophotometer (Implen). An appropriate amount of KCl was added to the DNA samples 70 min before experiments. All DNA samples were prepared as described unless specified. For ligand treatments, the DNA samples were incubated with individual ligands for 30 min before experiments. The annealing experiments were conducted by dissolving oligonucleotides in 10 mm Tris-HCl (pH 7.5) containing 5 or 150 mm KCl followed by heat denaturation at 95 °C for 10 min and slow cooling to room temperature (1 min/°C).
Circular Dichroism (CD) Spectra and Melting Temperature Measurement
The CD spectra were recorded using a spectropolarimeter (J-715, Jasco, Japan) with a bandwidth of 2 nm at a scan speed of 50 nm/min and a step resolution of 0.2 nm under N2 over the spectral range of 210–350 nm to monitor the G4 structures. Thermal melting curves were recorded by a Peltier thermal coupler chamber (PFD-425S/15, Jasco) and the molar ellipticity was monitored at 265 nm between 10 and 100 °C with a temperature ramping rate of 1 °C/min rate. The melting temperature (Tm) was measured from the first differential of the melting curve. The induced CD were recorded at the range of 360–600 nm after mixing with 1:1, 1:2, and 1:3 molar ratio of DNA to ligands for 30 min before experiments.
1H Imino Proton NMR Spectra
Experiments were performed on a Bruker AVIII 800 MHz spectrometer equipped with a cryoprobe at 25 °C. The one-dimensional imino proton NMR spectra were measured in H2O/D2O (90%/10%) using jump and return sequences for solvent suppression. The DNA samples were prepared at a strand concentration of 100 μm with an internal reference of DSS (0.1 mm sodium 4,4-dimethyl-4-silapentane-1-sulfonate).
Quantification of WNT1, MMP7, and Survivin RNA
The real-time quantitative RT-PCR was also used to determine the mRNAs of WNT1, MMP7, and survivin. Cells were treated with 10 μm BMVC or BMVC4 for 4 days and then subjected to RT-PCR analysis. Total RNA was isolated using TRIzol reagent (Sigma) and reverse-transcribed by random hexamers using cDNA reverse transcription kit (Applied Biosystem). The reverse-transcribed products were then analyzed using Cyber Green I system (FastStart Universal SYBR Green Master, Roche Applied Science). Primers used for PCR reactions were: WNT1 (forward primer 5′-CTGTCCTGCCTCCTCATC-3′ and reverse primer 5′-GGACCCAGCACAATAAATAGTT-3′), MMP7 (forward primer 5′-CCTCCACTCACTATGTAGA-3′, and reverse primer 5′-ATTCTTATCTCCAACTTCCAA-3′), survivin (forward primer 5′-CCTGGCTCCTCTACTGTT-3′ and reverse primer 5′-ACTCTATTCTGTCTCCTCATCC-3′), GAPDH (forward primer 5′-TAACTCTGGTAAAGTGGATA-3′ and reverse primer 5′-AAGATGGTGATGGGATTT-3′). The parameter Ct is defined as the fractional cycle number at which the fluorescence is generated by the Cyber green I system. Quantifications were first determined by normalizing the Ct values of the tested mRNA with the Ct of GAPDH. The relative values were then obtained using no drug treatment as 100%. Quantification results were obtained from three independent experiments.
Immunoblotting Analysis
The BMVC- or BMVC4-treated H1299 cells were washed twice with phosphate-buffered saline (PBS) and lysed using radioimmune precipitation assay buffer (10 mm Tris-HCl (pH 7.4), 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 100 μm phenylmethylsulfonyl fluoride, 1 mg/ml leupeptin, 1 mg/ml aprotinin, and 25 mm dithiothreitol). Equal amount of total proteins were separated on SDS-PAGE gels and transferred to nitrocellulose membranes. Antibodies against Wnt1 (1:1000 dilution, Spring, REF E3960), MMP7 (1:1000 dilution, GeneTex, GTX104658), β-catenin (1:1000 dilution, GeneTex, GTX61089), and GAPDH (1:5000 dilution, Millipore, MAB374) were used as the primary antibodies. Horseradish peroxidase-conjugated anti-mouse IgG or anti-rabbit IgG (Amersham Biosciences) were used as the secondary antibodies. Blots were visualized using enhanced chemiluminescence system (PerkinElmer Life Sciences).
Construction of WNT1 Reporter Plasmid and Site-directed Mutagenesis
The WNT1 promoter ranging from −600 to 201 relative to the transcription starting site was PCR-amplified from H1299 genomic DNA and cloned upstream to a secreted alkaline phosphatase (SEAP) reporter gene to generate pWNT1-SEAP. To construct a mutant that fails to form G-quadruplex structure, plasmid pWNT1-SEAP was used as the template for mutagenesis using Pfu DNA polymerase (Stratagene). The resulting mutations were verified by DNA sequencing of the plasmids.
SEAP Assay (21)
Secreted alkaline phosphatase was used as the reporter system to monitor the transcriptional activity of WNT1. Here, about 1 × 104 each of cells were grown in 96-well plates and incubated at 37 °C for 24 h and then changed with fresh media. Varying amounts of drugs were added, and cells were incubated for another 24 h. Culture media were collected and heated at 65 °C for 10 min to inactivate heat-labile phosphatases. An equal amount of SEAP buffer (2 m diethanolamine, 1 mm MgCl2, and 20 mm l-homoarginine) was added to the media, and p-nitrophenylphosphate was added to a final concentration of 12 mm. Absorptions at 405 nm were taken, and the rate of absorption increase was determined.
Chromatin Immunoprecipitation Assay (ChIP)
H1299 cells were treated with 10 μm BMVC and cross-linked by 1% formaldehyde at room temperature for 20 min. Cells were harvested by centrifugation, washed by PBS, and lysed by incubating on ice using a buffer containing 50 mm Tris pH 8.0, 1% SDS, 10 mm EDTA, 0.5 mm DTT, and protease inhibitor mixture (Calbiochem). The lysed cells were then sonicated, and the chromatins were harvested by centrifugation. The resulting chromatins were suspended in ChIP buffer (60 mm Tris pH 8.0, 12 mm EDTA, 0.01% SDS, and 0.9% Triton X-100) and immunoprecipitated by anti-nucleolin antibody (GeneTex, GTX30908). The immunoprecipitates were resuspended in elusion buffer (0.2 m NaHCO3, 0.012 m NaCl, and 1% SDS) and then heated at 65 °C for 4 h to remove cross-links. The reactions were then adjusted to 64 mm Tris (pH 6.5) and 16 mm EDTA by adding Tris and EDTA, respectively. Proteinase K was added to the reactions and incubated at 45 °C for 1 h to remove proteins. The resulting DNA samples were extracted by phenol/chloroform and ethanol-precipitated. Quantitative real-time PCR analyses were conducted using primers targeting GAPDH, QFSL (forward 5′-ACACAGTCCAGACACTCTGC-3′ and reversed 5′-ACGTGCAGAAACTCCTTGTTC-3′) and QFSR (forward 5′-CCAGCGCCGCAACTATAAGA-3′ and reversed 5′-GGCGACTTTGGTTGTTGCCC-3′).
Immunofluorescence
H1299 cells were treated with BMVC or BMVC4, fixed, and incubated with anti-β-catenin antibody (1:250 dilution, GeneTex, GTX61089). Visualization of β-catenin was achieved by the addition of rhodamine-conjugated anti-mouse IgG Jackson ImmunoResearch Laboratories, rhodamine RedTM-X-conjugated) and observed under fluorescence microscopy (OLYMPUS BX50). The fluorescence signals were quantified using MetaMorph imaging system software using the value of mock as 1.
Scratch Assay
Cells were treated with 3, 5, or 10 μm BMVC or BMVC4 for 4 days and subjected to scratch assay. The drug-treated cells were plated and scratched with a pipette tip to generate a cell-free zone. Cells were then incubated at 1% serum culture medium to avoid further cell proliferation. Migration of cells toward the cell-free zone was monitored after 24 h using a Leica DFC 420c camera (Leica Microsystems), and the migration rates were determined. The relative migration activities were obtained using the value of solvent control as 1.
Invasion assay
Cells were incubated with 10 μm BMVC or BMVC4 for 4 days, and the invasion assays were then performed using the QCM ECMatrix Cell Invasion assay kit (Millipore, ECM554). Briefly, ∼2 × 104 of the drug-treated cells were grown in the upper chamber in serum-free medium with 20% serum medium in the bottom chamber. Invasive migration of cells through the ECM layer was then determined by crystal violet staining of the bottom of upper chamber. The relative invasion activities were obtained by measurement of A560 and using the value of solvent control as 1.
RESULTS
The G-rich Sequence Located at the WNT1 Promoter Is Capable of Forming G-quadruplex Structure in Vitro
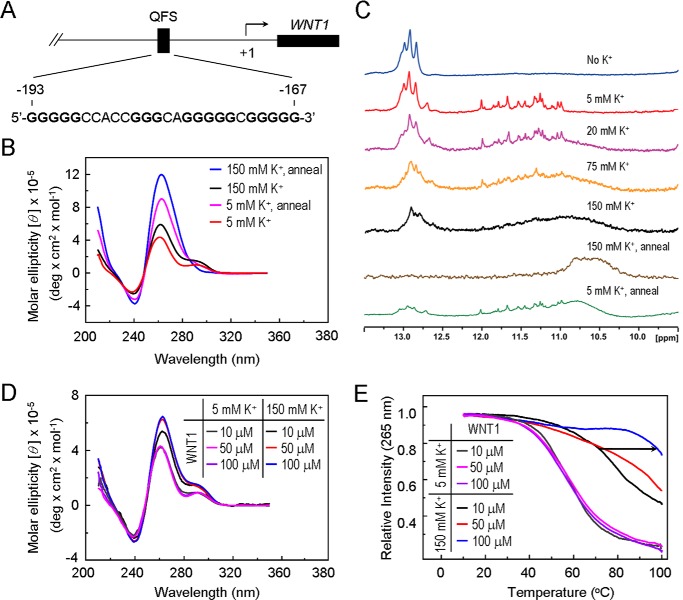
A stretch of G-rich sequence was identified at the −193 to −167 region of the WNT1 promoter (Fig. 1A). To test the potential of this sequence to form G-quadruplex, the CD spectrum analysis was first conducted. CD provides spectral information for the existence of G-quadruplex structures and the determination of DNA strand orientation for a given G-quadruplex structure. Signature CD spectra have previously been determined for several novel G-quadruplex structures in the presence of monovalent cations (22–24). The antiparallel-type G-quadruplexes containing alternating syn- and anti-glycosyl bond conformations have been shown to have a CD spectrum characterized by a positive CD band at ∼295 nm, whereas parallel-type G-quadruplexes composed of all anti-glycosyl bond conformations have a positive CD band at ∼265 nm and a negative CD band at ∼240 nm. An oligonucleotide carrying the G-quadruplex-forming sequence of WNT1 promoter was synthesized here and analyzed by CD to evaluate the formation of G-quadruplex structure. The CD spectrum of WNT1 oligonucleotide displayed a positive CD band at 264 nm and a negative CD band at 240 nm together with a relative weak positive CD band at 292 nm in the presence of 5 mm K+ (Fig. 1B). An increase of the 264-nm intensity was observed in 150 mm K+ solution. This result implies that WNT1 oligonucleotide forms a mixed parallel/antiparallel folding pattern with at least two different G-quadruplex conformations.
FIGURE 1.
The G-rich sequence located at the WNT1 promoter is capable of forming a quadruplex structures in vitro. A, the location of G-rich sequence at the human WNT1 promoter. B, CD spectra of the WNT1 G-rich sequence. The CD spectra of 100 μm WNT1 (5′-GGGGGCCACCGGGCAGGGGGCGGGGG-3′) oligonucleotide were recorded over the spectral range of 210–350 nm. The annealing samples were preheated at 95 °C for 10 min and slowly cooled to room temperature (1 min/°C) in the presence of 5 mm KCl or 150 mm KCl. C, one-dimensional imino proton NMR spectra of WNT1 oligonucleotide. NMR spectra were taken using a Bruker 800 MHz spectrometer. WNT1 oligonucleotide at 100 μm concentration was prepared in the absence of KCl and with the addition of 5, 20, 75, and 150 mm KCl. The NMR spectra of the annealed samples at 100 μm WNT1 oligonucleotide in the presence of 5 mm KCl or 150 mm KCl were also obtained. Concentration-dependent CD spectra (D) and melting curves (E) of 10, 50, and 100 μm WNT1 oligonucleotide were at 5 mm KCl or 150 mm KCl.
Formation of the G-quadruplex structures by the WNT1 G-rich sequence was also examined using NMR spectroscopy. In contrast to the Watson-Crick base pairing that exhibits chemical shift of 13–14 ppm for guanine imino protons (H1) (25), the typical chemical shifts for the guanine imino proton involved G-G pairing were within the range of 10–12 ppm (26, 27). In the absence of potassium ion, the guanine imino protons of WNT1 DNA showed several typical chemical shifts at around 13 ppm (Fig. 1C). The addition of 5 mm K+ caused a major change of chemical shifts, for which a signature chemical shift for G-quadruplex structure was observed centered at 11–12 ppm. With increasing K+ concentrations, the individual guanine peaks became less distinct and were replaced by a less-defined broad band. The results suggest that WNT1 oligonucleotide at low K+ concentrations predominately formed a conformationally defined G-quadruplex structure. Increasing K+ concentration resulted in the formation of less defined high order G-quadruplex structures. Because the position of G-G base pairings within the oligonucleotide could vary at different K+ concentration, the results also suggest that multiple G-quadruplex conformations are formed. Nevertheless, both spectral analyses support that the G-rich sequences within the WNT1 promoter is capable of forming G-quadruplex structure.
To further define the structure of WNT1 oligonucleotide at different K+ concentrations, we conducted annealing experiments using both 5 and 150 mm K+ concentrations. Upon heat-denaturing and slow-cooling, the interactions between bases were maximized. Indeed, CD spectra analysis showed a further enhancement of the 264-nm intensities were apparent at both 5 and 150 mm K+ solution when we annealed the WNT1 oligonucleotide (Fig. 1B). At 150 mm K+ concentration, the NMR spectrum was similar to that observed without the annealing step (Fig. 1C). Significantly, distinct chemical shifts at 11–12 ppm were apparent at 5 mm K+ concentration, indicating that the WNT1 oligonucleotide at 5 mm K+ concentration prefers to form a conformationally defined G-quadruplex structure. High K+ concentrations appear to favor maximization of G-G pairing to form high order G-quadruplex structures.
We next evaluated whether the structure formed by WNT1 oligonucleotide is intramolecular G-quadruplex. Because the formation of intermolecular G-quadruplex is dependent on the concentration of oligonucleotide, whereas intramolecular G-quadruplex is not, it is anticipated that the CD spectra observed in Fig. 1B should not be affected by decreasing the WNT1 oligonucleotide concentration. Indeed, identical spectra of WNT1 oligonucleotide at 5 mm K+ were observed when the DNA concentrations were decreased (Fig. 1D). In contrast, a decrease of the 265-nm CD intensity was observed when the DNA concentration was decreased to 10 μm at 150 mm K+ concentration. Our results also predict that the melting temperature of intramolecular G-quadruplex structure should not be affected by DNA concentration, whereas the intermolecular high order structure should be. The CD melting curves as a function of the DNA concentrations were also conducted. Almost identical melting curves were observed at 5 mm K+ concentration for three different concentrations of DNA (Fig. 1E). The melting temperatures at 5 mm K+ concentration were ∼58 °C for all three DNA concentrations. In contrast, the melting temperature increases at 150 mm K+ solution in a DNA concentration-dependent manner. This striking difference can be explained by the formation of additional intermolecular G-quadruplexes when DNA concentrations were increased at 150 mm K+ solution. Accordingly, we conclude that WNT1 oligonucleotide predominately forms the intramolecular G-quadruplex at low K+ concentrations and sufficient WNT1 oligonucleotides favor the formation of intermolecular G-quadruplexes at high K+ concentrations in vitro.
BMVC and BMVC4 Repressed Wnt1 Expression through Stabilizing the G-quadruplex Structure Formed at Its Promoter
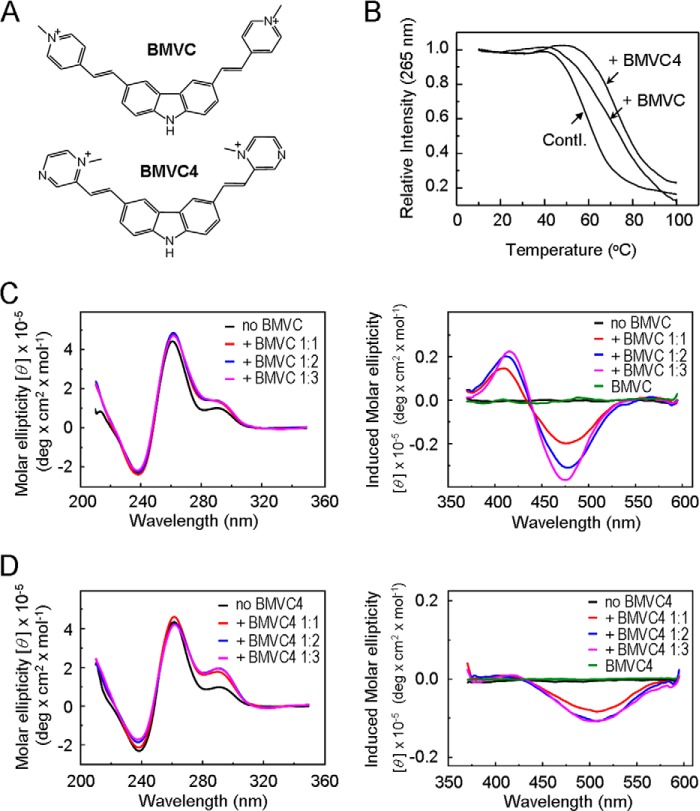
To evaluate the function of the G-rich sequence at the WNT1 promoter, the effects of G-quadruplex structure formation on Wnt1 expression were first analyzed. Previously, we characterized two carbazole derivatives, BMVC and BMVC4 (Fig. 2A), that bound and stabilized the G-quadruplex structures formed by both human telomeric DNA sequences and the quadruplex-forming sequence located at the c-myc promoter (19, 20). These two compounds were employed here to determine whether they could bind and stabilize the G-quadruplex structure formed by the G-rich sequence of WNT1 promoter. The CD at 265 nm was measured as a function of temperature to determine the Tm of WNT1 oligonucleotide in the presence of these compounds. As shown in Fig. 2B, the addition of BMVC or BMVC4 greatly raised the melting temperature of WNT1. The Tm of WNT1 oligonucleotide was increased by 12 and 14 °C upon the addition of BMVC and BMVC4, respectively.
FIGURE 2.
Stabilizing WNT1 G-quadruplex structure by BMVC and BMVC4. A, structures of G-quadruplex stabilizers BMVC and BMVC4. B, BMVC and BMVC4 enhance the T1a of WNT1 G-quadruplex. Temperature-dependent CD signals at 265 nm of the WNT1 quadruplex and upon interaction with BMVC and BMVC4 in 5 mm K+ solution at 8 μm DNA concentration with 16 μm BMVC or BMVC4. C, CD and induced CD spectra of 100 μm Wnt1 oligonucleotide in 5 mm K+ buffer solution with the addition of BMVC from 1:1 to 1:3 molar ratio. D, CD and induced CD spectra of 100 μm Wnt1 oligonucleotide in 5 mm K+ buffer solution with the addition of BMVC4 from 1:1 to 1:3 molar ratio.
We have also performed the CD titration analysis of WNT1 oligonucleotide with different BMVC and BMVC4 concentrations. BMVC did not appear to affect the overall spectra of WNT1 oligonucleotide, suggesting that BMVC did not greatly affect the structure of WNT1 oligonucleotide. Interestingly, the addition of BMVC4 significantly enhanced the CD band at 292 nm (Fig. 2, C and D). Although the G-quadruplex structure formed by the WNT1 promoter is yet to be determined, the results suggest that BMVC4 might induce a conformational change of the G-quadruplex structure. Indeed, differently induced CD spectra were apparent between BMVC and BMVC4 upon interaction with the WNT1 oligonucleotide (Fig. 2, C and D). Together, these results indicate that both BMVC and BMVC4 could bind with and thermally stabilize the G-quadruplex structure formed by the WNT1 oligonucleotide.
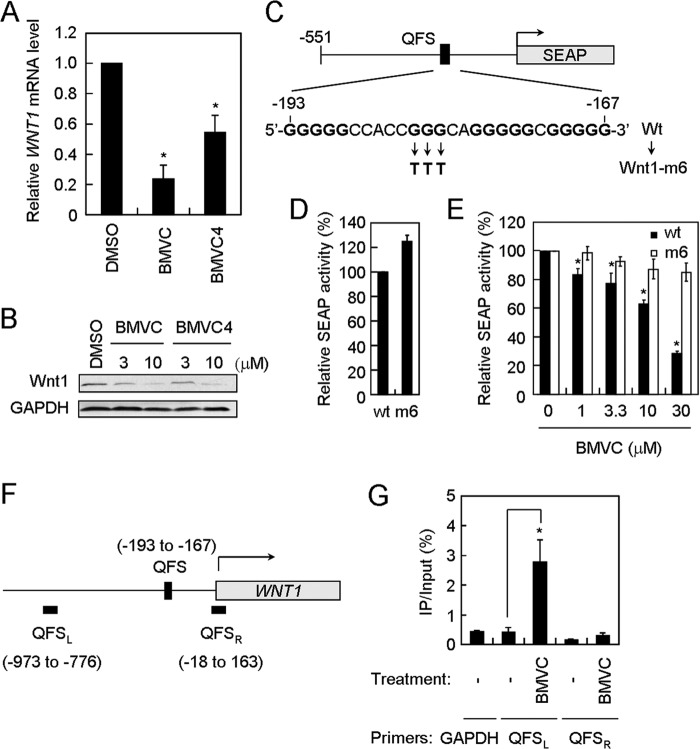
Because the G-quadruplex-forming sequence at the WNT1 promoter could be a target for G-quadruplex stabilizers, the effect of BMVC and BMVC4 on WNT1 expression was next analyzed. Real time RT-PCR analysis shows that the WNT1 mRNA level was significantly decreased in cells that were treated with BMVC or BMVC4 (Fig. 3A). Moreover, immunoblotting analysis of the BMVC- or BMVC4-treated lung cancer H1299 cells shows that the Wnt1 protein levels were decreased by BMVC or BMVC4 treatments in a dose-dependent manner (Fig. 3B). Under the BMVC or BMVC4 conditions, no apparent cytotoxic effect was observed for up to 6 days (19, 20). These results indicate that G-quadruplex stabilizers could reduce the expression of WNT1.
FIGURE 3.
BMVC- and BMVC4-repressed WNT1 expression through stabilizing the G-quadruplex structure formed at its promoter. A, suppression of WNT1 expression by BMVC or BMVC4 at the transcription level. The H1299 cells was incubated with 3 μm BMVC or BMVC4 for 4 days and then subjected to quantitative RT-PCR analysis. The GAPDH expression level was used as an internal control. The mRNA level of WNT1 expression in untreated H1299 cells was defined as 1. Results were obtained from the average of three independent experiments. Asterisks indicate p < 0.05 (p = 4 × 10−4 and 3 × 10−2 for BMVC and BMVC4, respectively). B, BMVC and BMVC4 reduced the protein level of Wnt1. H1299 cells were incubated with 3 or 10 μm BMVC or BMVC4 for 4 days. Total cell extracts were prepared, and immunoblotting analysis was conducted using antibodies against Wnt1 or GAPDH. C, schematic diagrams showed the mutation sites of WNT1 in reporter assays. The G-quadruplex-forming sequences of WNT1 or WNT1-m6 (Wnt1 mutation) were indicated. D, WNT1-m6 mutation did not affect the basal expression level of WNT1. The wild-type and WNT1-m6 reporter plasmids were transfected into H1299 cells and analyzed for their basal expression activity. The relative phosphatase activity of wild-type and WNT1-m6 mutant was presented using the wild-type level as 100%. E, G-quadruplex structure formation was required for suppression of WNT1 expression by BMVC. The WNT1 reporter plasmids were transfected into H1299 cells and then incubated with the indicated concentrations of BMVC for 2 days. The phosphatase activities were then analyzed using the activity of DMSO-treated cells as 100% (right panel). Asterisks indicate p < 0.05 (p = 0.021, 0.046, 0.025, and 0.0026 for BMVC at 1, 3.3, 10, and 30 μm, respectively). F, schematic diagram showed the G-QFS and the PCR sites in chromatin immunoprecipitation assays. The analyzed PCR products and position located both 5′ (QFSL) and 3′ (QFSR) to the QFS are indicated. G, nucleolin binding to WNT1 promoter in BMVC-treated cells. H1299 cells were treated with 10 μm BMVC, cross-linked by 1% formaldehyde, sonicated, and immunoprecipitated by the anti-nucleolin antibody. The immunoprecipitates were then heated at 65 °C for 4 h to remove cross-links and treated with proteinase K, and quantitative real-time PCR analyses using primers targeting GAPDH, QFSL, and QFSR were conducted. The resulting immunoprecipitation values were quantified, divided by the Input controls, and presented as % of immunoprecipitate/input. The average values of three independent experiments are plotted. The asterisk indicates p < 0.05 (p = 0.024).
To show that the repressing effect of WNT1 expression was indeed mediated by stabilizing the G-quadruplex forming sequence by BMVC, a reporter construct with a SEAP gene fused downstream to the WNT1 promoter was generated (Fig. 3C). A mutant construct of three G to T mutations on the G-rich sequence was also generated to disrupt the G-quadruplex structure formation (Wnt1-m6). The expression of SEAP was used as the criterion for the measurement of wild-type and mutant WNT1 expression efficiency. Plasmid carrying wild-type or the mutant was first transfected into H1299 cells, and analyzed for phosphatase activity. Similar luciferase activities were observed for both wild-type and the Wnt1-m6 promoters, suggesting that the mutation did not affect the general transcriptional activity of the WNT1 promoter (Fig. 3D). Upon treatment with BMVC, the expressions of SEAP from the wild-type promoter were reduced (Fig. 3E). In contrast, the expression efficiency of mutant promoter did not respond to BMVC treatments. Thus, formation of G-quadruplex structure is required for suppressing WNT1 expression by BMVC.
Nucleolin has been shown to bind to G-quadruplex structure to activate or repress gene expression (28–30). The binding of nucleolin to the WNT1 promoter was also tested to check whether the repression of expression by BMVC is due to a change in protein binding. Using antibody against nucleolin in a chromatin immunoprecipitation assay, we found nucleolin did not preferentially bind to the G-quadruplex-forming sequences on the WNT1 promoter (Fig. 3, F and G). Interestingly, BMVC treatment increased the binding of nucleolin to the WNT1 promoter, suggesting that the G-quadruplex structure was formed at the WNT1 promoter upon BMVC treatment. These results also indicate that the repressed WNT1 expression might be due to the binding of nucleolin to its promoter. Together our results indicate that BMVC and BMVC4 repressed WNT1 expression through stabilizing the G-quadruplex structure formed at its promoter.
The Wnt1-mediated Signaling Pathway Was Suppressed by G-quadruplex Stabilizers
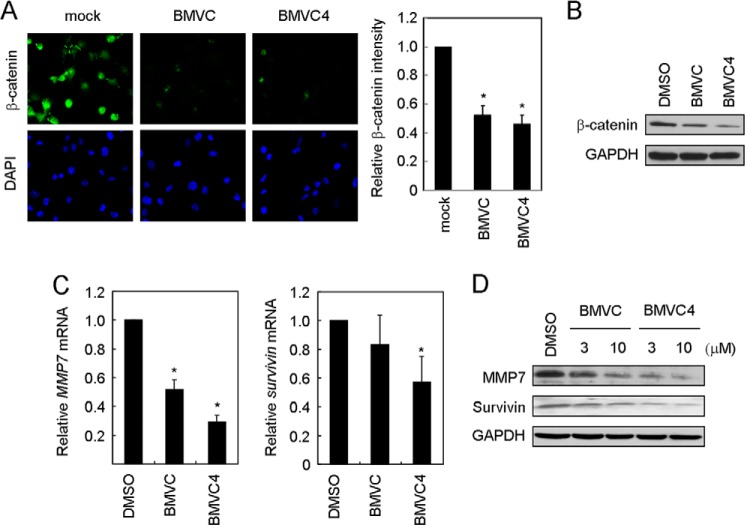
In the canonical Wnt1-mediated signaling pathway, binding of Wnt1 to its receptor prevents β-catenin from degradation. The accumulated β-catenin then enters the nucleus to drive expression of its downstream genes. Here the downstream effect of Wnt1-mediated signaling pathway by G-quadruplex stabilizers was also analyzed. Immunofluorescence analysis of β-catenin showed that both the total and nuclear signals of β-catenin were decreased upon BMVC or BMVC4 treatments (Fig. 4A). Immunoblotting analysis further supported the observation that the β-catenin levels were decreased by BMVC and BMVC4 (Fig. 4B). The expression of two downstream target genes, MMP7 and survivin, was also examined. Both the mRNA and protein levels of MMP7 and survivin were reduced in BMVC-treated or BMVC4-treated cells (Fig. 4, C and D). Because the promoter regions of both MMP7 and survivin do not have G-rich sequences, the inhibitory effects of BMVC and BMVC4 are not likely to be caused by direct repression of MMP7 and survivin by these two compounds. On the other hand, c-myc is also a downstream target gene of the Wnt/β-catenin pathway. Previously we demonstrated that BMVC4 repressed the c-myc expression (19, 20). Because a G-quadruplex forming sequence was also identified at the promoter region of c-myc, it is likely that the repressing effect of c-myc by BMVC4 might be through its own G-quadruplex forming sequence directly and through repressing WNT1 expressing indirectly. Nevertheless, our results indicate that the Wnt1-mediated signaling pathway was suppressed by G-quadruplex stabilizers.
FIGURE 4.
Both BMVC and BMVC4 reduced the Wnt1 downstream mediator β-catenin and the expression of target genes. A, the β-catenin level was decreased in H1299 cells. H1299 cells were treated with BMVC or BMVC4 (10 μm) for 4 days and stained with antibody against β-catenin. Staining of DNA by DAPI was employed to locate the positions of nuclei. The images were taken (left panels), and the relative β-catenin fluorescent levels were measured (right panel). The histograms represent the average of three independent experiments. Asterisks indicate p < 0.05 (p = 1 × 10−3 and 1 × 10−4 for BMVC and BMVC4, respectively). B, immunoblotting analysis of β-catenin in BMVC or BMVC4-treated H1299 cells. Cell extracts were prepared after treating with 10 μm BMVC or BMVC4 for 4 days and then analyzed by immunoblotting assays using antibodies against β-catenin or GAPDH. C, repressing the mRNA of MMP7 and survivin by BMVC and BMVC4. Quantitative RT-PCR was conducted after H1299 cells were treated with 10 μm BMVC or BMVC4. The GAPDH was used as the internal control, and the value of DMSO-treated cells was defined as 1. Asterisks indicate p < 0.05 (MMP7, p = 8 × 10−4 and 8 × 10−3 for BMVC and BMVC4, respectively; survivin, p = 8 × 10−4 and 8 × 10−3 for BMVC and BMVC4, respectively). D, both MMP7 and surviving protein levels were repressed by BMVC and BMVC4. H1299 cells were incubated with 3 or 10 μm carbazole derivatives. The total cell extracts were prepared, and immunoblotting was conducted by antibodies against MMP7, survivin, or GAPDH.
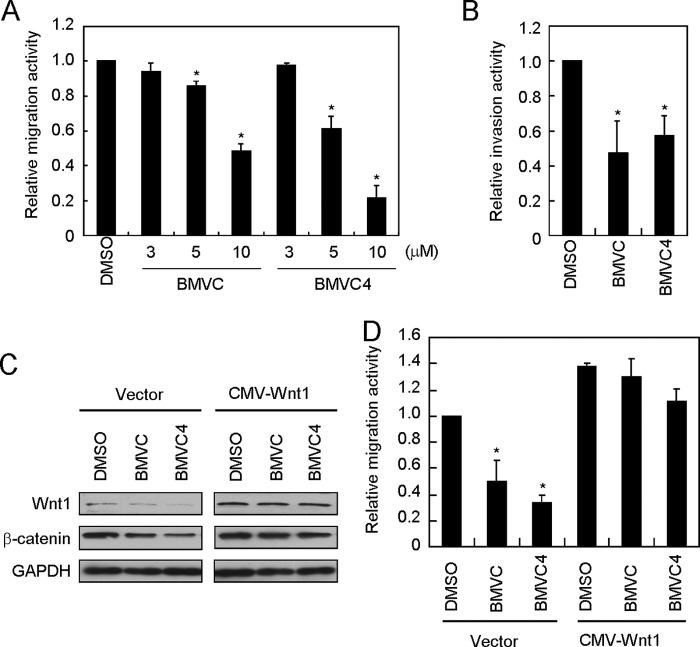
The Wnt1-mediated signaling pathway was shown to have an important role in cancer migration and invasion (1). The cellular effects of suppressing WNT1 expression by G-quadruplex stabilizers were analyzed. Both BMVC and BMVC4 reduced the migration activity of H1299 cells in a dose-dependent manner (Fig. 5A). Similarly, the activity of invasive migration through the extracellular matrix was also decreased by these two compounds (Fig. 5B). These results indicate that BMVC and BMVC4 suppressed the expression of WNT1 and subsequently inhibited the downstream cellular effects.
FIGURE 5.
BMVC and BMVC4 reduce the migration and invasion activities of H1299 cells. A, the migration ability of H1299 cells was suppressed by BMVC and BMVC4. H1299 cells were incubated with 10 μm DMSO or 3, 5, or 10 μm BMVC or BMVC4 for 4 days, and then the treated cells were subjected for scratch assays. Relative migration rates of H1299 cells were determined by migration distance over the time. The value of migration rate in DMSO-treated cells was defined as 1. Asterisks indicate p < 0.05 (p = 0.006 and 0.003 for BMVC at 5 and 10 μm; p = 0.020 and 0.002 for BMVC4 at 5 and 10 μm, respectively). B, reduction of invasion activity by BMVC or BMVC4. H1299 cells were incubated with 10 μm BMVC or BMVC4 for 4 days then analyzed by invasion assays. The cells were stained with crystal violet, dissolved in acetic acid, and quantified for absorption at A560 using a spectrophotometer. The results were expressed as relative values using DMSO-treated cells as 1. Asterisks indicate p < 0.05 (p = 1 × 10−2 and 1 × 10−2 for BMVC and BMVC4, respectively). C, immunoblotting analysis of Wnt1 and β-catenin in WNT1-overexpressing cells. H1299 cells were transfected with plasmid carrying CMV promoter-driven WNT1 or vector plasmid. The cells were incubated with 10 μm BMVC or BMVC4 for 4 days and then examined by immunoblotting analysis using antibodies against Wnt1, β-catenin, or GAPDH. D, WNT1 overexpression reversed the migration inhibitory activity of BMVC and BMVC4. The relative migration rates were determined in 10 μm BMVC- or BMVC4-treated cells that overexpressing WNT1. The relative migration rate in DMSO-treated H1299 cells was defined as 1. Asterisks indicate p < 0.05 (p = 6 × 10−3 for BMVC and 1 × 10−4 for BMVC4 in the vector control group, respectively).
To show that the anti-metastatic effect of BMVC and BMVC4 was through suppression of Wnt1-mediated pathway, we overexpressed WNT1 in BMVC- or BMVC4-treated cells. We expected that expressing of excess amount of Wnt1 should reverse the anti-metastatic effect by BMVC or BMVC4. Plasmid construct harboring WNT1 under the control of a CMV promoter was transfected into H1299 cells. Immunoblotting analysis indicated that the protein levels of Wnt1 and its downstream mediator β-catenin were higher in WNT1-overexpressing cells (Fig. 5C). Although the levels of Wnt1 and β-catenin were suppressed upon BMVC or BMVC4 treatments, neither was significantly reduced when WNT1 was overexpressed. Consistent with the results from immunoblotting analysis, the migration analysis also indicated that the inhibitory effects of BMVC and BMVC4 were reversed in WNT1-overexpressing cells (Fig. 5D). The results indicate that the migration inhibitory effects of BMVC and BMVC4 were mediated through suppressing the Wnt1-mediated signaling pathway.
Suppression of Wnt1-mediated Cell Migration by BMVC and BMVC4 Was Not Limited to H1299 Cells
To test the effects of WNT1 expression and migration upon BMVC and BMVC4 treatments on other cancer cell lines, the MCF7 (breast cancer) and A549 (human lung adenocarcinoma) cells were also tested. In MCF cells, both BMVC and BMVC4 effectively repressed the expression of WNT1 and β-catenin (Fig. 6A). As a result, the migration activity of MCF cells was reduced by these two compounds (Fig. 6B). It was also noted that the level of Wnt1/β-catenin repression by BMVC and BMVC4 correlated with their reduction in migration activity. Interestingly, although BMVC4 effectively repressed the Wnt1/β-catenin levels and the migration activity, BMVC did not appear to cause an inhibitory effect in A549 cells (Fig. 6). Thus, the cellular context might also have a role in modulating the repressing effects of BMVC and BMVC4. But the reason for the differential cellular responses by these two compounds is unclear to us. Nevertheless, the correlation between Wnt1/β-catenin repressing and migration inhibition by BMVC and BMVC4 provides a strong indication that these two compounds inhibit the metastatic activity of cancer cells through repressing the expression of WNT1.
FIGURE 6.
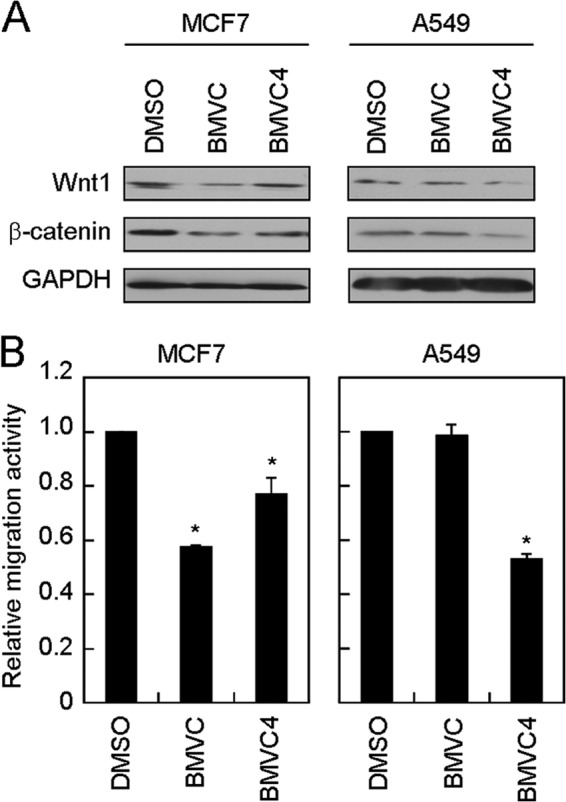
BMVC and BMVC4 suppressed WNT1/β-catenin expression and reduced cell migration in MCF7 and A549 cells. A, BMVC and BMVC4 suppressed Wnt1/β-catenin-signaling pathway in MCF7 and A549 cells. MCF7 or A549 cells were treated with 10 μm BMVC or BMVC4 for 4 days. Total cell extracts were prepared, and immunoblotting analysis was conducted using antibodies against Wnt1, β-catenin, or GAPDH. B, reduction of MCF7 and A549 migration by BMVC or BMVC4. The BMVC or BMVC4-treated MCF7 or A549 cells were subjected to scratch assay. The value of migration rate in DMSO-treated cells was defined as 1. Asterisks indicate p < 0.05 (for MCF7, p = 0.002 and 0.054 for BMVC and BMVC4, respectively; for A549, p = 4 × 10−4 for BMVC4).
DISCUSSION
G-rich sequences can fold into G-quadruplexes, and such structures may play critical roles in biological functions (15–18, 30). It is further suggested that the G-quadruplex-forming sequence might be a potential target for therapeutic intervention (15, 31). However, the G-rich sequences containing four or five consecutive guanines can easily form high order G-quadruplexes or multiple conformations in a K+ solution, especially at sufficient DNA concentrations in vitro (15, 32). Thus, verification of the G-quadruplex structure is crucial not only for its own biological importance but also for its potential usage as a drug target. In this work we found the formation of G-quadruplex structures by the G-rich sequence located at the WNT1 promoter in vitro. We then introduced G-quadruplex stabilizers to elucidate the biological function of WNT1 G-quadruplex in vivo.
The WNT1 G-rich sequence contains three G-tracks with five consecutive guanines. Thus, it is likely that they could form multiple G-quadruplex conformations by varying the position of G-G base pairings within the oligonucleotide. We have provided several lines of evidence to show that the WNT1 oligonucleotide is capable of forming both intramolecular G-quadruplex structure and high order structures through intermolecular interactions in vitro. The conformational exchange is dependent on both the K+ and DNA concentrations. The observed CD and NMR spectral features were similar to the G-rich sequence (bcl2mid) located at the bcl2 promoter (32). It was found that a major intramolecular component of bcl2mid exists at low K+ concentrations, and an additional intermolecular component appears at high K+ concentrations. These two components showed distinct gel migration activities and can be separated by an emulsified induced filtration method (33). The slow migration band in the gel assay was the intermolecular conformers of bcl2mid. It showed a broad feature in the imino proton NMR spectrum as well as a strong positive 265-nm signal and a negative 240-nm signal in the CD spectrum. The fast migration band is bcl2mid monomer that is characterized by several defined chemical shifts in the imino proton NMR spectrum and a positive 295-nm signal in the CD spectrum. Thus, the similarity of CD and NMR spectra between bcl2mid and WNT1 oligonucleotides suggests that the WNT1 oligonucleotide mainly forms intramolecular G-quadruplex at low K+ and DNA concentration in vitro.
It has been shown that the G-quadruplex ligands can repress the expression of c-myc oncogene by stabilizing the G-quadruplex structures (15). Here we show that two G-quadruplex stabilizers, BMVC and BMVC4, increase the Tm of the WNT1 oligonucleotide. Moreover, the CD spectra show no discernible difference upon interaction with BMVC but some appreciable difference upon interaction with BMVC4. The increase of the 292-nm CD signal suggests that BMVC4 can induce non-parallel G-quadruplex conformation. Although they show different binding behaviors, it is of great interest to examine the molecular details of how these two G-quadruplex stabilizers perturb the structure of WNT1 G-quadruplex.
The Wnt1/β-catenin pathway has been well recognized as playing a major role in cancer progression (2). Several of the downstream target genes induced by this pathway are involved in promoting tumor malignancies. For example, cyclin D1 (34), c-myc (4), WISP1 (35), MMP7 (6), and PAI1 (36) are involved in cancer cell proliferation, invasion, and metastasis. Indeed, the aberrant activation of WNT1 has been observed in prostate, lung, breast, liver, and colon tumors (4, 9, 37, 38). Moreover, the Wnt1 level is significantly increased at the invasive front of human prostate carcinoma tissues and the tumor metastasized to lymph nodes and bone (9). Considering the important role of Wnt pathway in cancer development, inhibitors have been designed and developed to block the Wnt pathway (11, 39). Small molecular compounds have been designed to target several mediators of Wnt pathway (40–44). These compounds affect the levels of β-catenin, interfere with the interaction between β-catenin and T-cell factor, increase the levels of axin to destabilize β-catenin, inhibit the kinase activity of CK1 to prevent the phosphorylation of β-catenin, or block the Porc activity to promote acylation of Wnt. Agents targeting Wnt1 directly were also developed. The anti-Wnt1 antibody was used to block the stimulation of Wnt1 downstream signaling pathway. Treatment of anti-Wnt1 antibody was shown to reduce the growth of HCC and colorectal cancer both in vivo and in vitro (37, 38). Here we found that the G-quadruplex structure formed by the G-rich sequence located at the promoter region of WNT1 could be stabilized by BMVC and BMVC4 to repress the expression of WNT1 and the downstream signaling pathway. Significantly, the migration and invasion activities of cancer cells were also inhibited by these two G-quadruplex stabilizers. Our results provide a novel mechanism to repress the expression of WNT1 through stabilizing the G-quadruplex structure located at its promoter. The finding of BMVC and BMVC4 to down-regulate the expression of WNT1 might offer a potent and selective therapeutic strategy for the clinical management of cancer patients with WNT1 overexpression.
Bioinformatic analysis has indicated that about 375,000–377,000 potential intramolecular G-quadruplex-forming sequences (QFSs) could be identified within the human genome (45–47). These QFSs are potential targets for G-quadruplex stabilizers that equally exist in both normal and cancer cells. Thus, an important issue faced by applying G-quadruplex stabilizers as anti-cancer agents is the specificity. We consider that there are at least two levels of specificities that make BMVC and BMVC4 potential cancer-specific targeting agents. First, it was found that there is a bias in the distribution of these putative QFSs as they are preferentially located in proto-oncogenes (48). For example, QFSs within the promoters of c-myc (15), c-kit (49–51), KRAS (52), HIF1 (53), Rb1 (54), VEGF (16, 55), hTERT (56), c-myb (57), PDGFR-β (58), and PDGF-A (59) were found capable of forming G-quadruplex structures, and their expressions were affected by G-quadruplex stabilizers. Thus, although G-quadruplex stabilizers could target multiple places within the genome, the G-quadruplex stabilizers could still preferentially affect oncogenes. Second, it was reported that whereas BMVC is retained in the lysosomes of normal cells, it escapes lysosomal retention and localizes to the nucleus of cancer cells (60). The side-chain property of carbazole derivatives appears to have a critical role in determining their cellular distribution by cancer cells. The hydrogen-bonding capacity and the weak lipophilicity of side chains such as pyridine and pyrazine in BMVC and BMVC4, respectively, direct them to escape from lysosomes and enter the nucleus of cancer cells. These properties make carbazole derivatives promising candidates for anti-cancer chemotherapeutics.
Different G-rich sequences could form various types of G-quadruplex structures and sequences that contain G-tracks of four or five consecutive guanines can form multiple conformations. Although the G-quadruplex structure is not precisely defined in our current analysis, it will be interesting to know what specific structure is responsible for repressing WNT1 expression by BMVC or BMVC4. It is likely that BMVC or BMVC4 might prefer a specific form of G-quadruplex structure for binding. Moreover, because a transcription factor WiF-1 (Wnt1-inducing factor-1) was reported to bind to the G-rich region of WNT1 promoter (13), it will also be interesting to test if BMVC or BMVC4 inhibit the binding of this factor to WNT1 promoter.
Acknowledgments
We thank Dr. Pei-Jen Lou for comments and suggestions. We also thank members of the Lin and Chang groups for helpful discussions.
This work was supported by Academia Sinica AS-98-TP-A04 and AS-102-TP-A07, the National Science Council (100-2311-B-002-016 and 100-2311-B-010-001), and the National Health Research Institute (NHRI-EX101-10050SI).
- BMVC
- 3,6-bis(1-methyl-4-vinylpyridinium) carbazole diiodide
- BMVC4
- 3,6-bis(4-methyl-2-vinylpyrazinium) carbazole diiodide
- SEAP
- secreted alkaline phosphatase
- QFS
- G-quadruplex-forming sequence.
REFERENCES
- 1. Clevers H., Nusse R. (2012) Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 [DOI] [PubMed] [Google Scholar]
- 2. Klaus A., Birchmeier W. (2008) Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer 8, 387–398 [DOI] [PubMed] [Google Scholar]
- 3. Niehrs C. (2012) The complex world of WNT receptor signalling. Nat. Rev. Mol. Cell Biol. 13, 767–779 [DOI] [PubMed] [Google Scholar]
- 4. Wong S. C., Lo S. F., Lee K. C., Yam J. W., Chan J. K., Wendy Hsiao W. L. (2002) Expression of frizzled-related protein and Wnt-signalling molecules in invasive human breast tumours. J. Pathol. 196, 145–153 [DOI] [PubMed] [Google Scholar]
- 5. Tao W., Pennica D., Xu L., Kalejta R. F., Levine A. J. (2001) Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev. 15, 1796–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He W., Tan R. J., Li Y., Wang D., Nie J., Hou F. F., Liu Y. (2012) Matrix metalloproteinase-7 as a surrogate marker predicts renal Wnt/β-catenin activity in CKD. J. Am. Soc. Nephrol. 23, 294–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim Y. C., Clark R. J., Ranheim E. A., Alexander C. M. (2008) Wnt1 expression induces short-range and long-range cell recruitments that modify mammary tumor development and are not induced by a cell-autonomous β-catenin effector. Cancer Res. 68, 10145–10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu X., Sun P. L., Li J. Z., Jheon S., Lee C. T., Chung J. H. (2011) Aberrant Wnt1/β-catenin expression is an independent poor prognostic marker of non-small cell lung cancer after surgery. J. Thorac. Oncol. 6, 716–724 [DOI] [PubMed] [Google Scholar]
- 9. Chen G., Shukeir N., Potti A., Sircar K., Aprikian A., Goltzman D., Rabbani S. A. (2004) Up-regulation of Wnt1 and β-catenin production in patients with advanced metastatic prostate carcinoma: potential pathogenetic and prognostic implications. Cancer 101, 1345–1356 [DOI] [PubMed] [Google Scholar]
- 10. Nakashima T., Liu D., Nakano J., Ishikawa S., Yokomise H., Ueno M., Kadota K., Huang C. L. (2008) Wnt1 overexpression associated with tumor proliferation and a poor prognosis in non-small cell lung cancer patients. Oncol. Rep. 19, 203–209 [PubMed] [Google Scholar]
- 11. Polakis P. (2012) Drugging Wnt signalling in cancer. EMBO J. 31, 2737–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nusse R., Theunissen H., Wagenaar E., Rijsewijk F., Gennissen A., Otte A., Schuuring E., van Ooyen A. (1990) The Wnt1 (int-1) oncogene promoter and its mechanism of activation by insertion of proviral DNA of the mouse mammary tumor virus. Mol. Cell. Biol. 10, 4170–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. St-Arnaud R., Moir J. M. (1993) Wnt1-inducing factor-1: a novel G/C box-binding transcription factor regulating the expression of Wnt1 during neuroectodermal differentiation. Mol. Cell. Biol. 13, 1590–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fenby B. T., Fotaki V., Mason J. O. (2008) Pax3 regulates Wnt1 expression via a conserved binding site in the 5′ proximal promoter. Biochim. Biophys. Acta 1779, 115–121 [DOI] [PubMed] [Google Scholar]
- 15. Siddiqui-Jain A., Grand C. L., Bearss D. J., Hurley L. H. (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-myc transcription. Proc. Natl. Acad. Sci. U.S.A. 99, 11593–11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun D., Guo K., Rusche J. J., Hurley L. H. (2005) Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 33, 6070–6080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dai J., Dexheimer T. S., Chen D., Carver M., Ambrus A., Jones R. A., Yang D. (2006) An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 128, 1096–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fernando H., Reszka A. P., Huppert J., Ladame S., Rankin S., Venkitaraman A. R., Neidle S., Balasubramanian S. (2006) A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry 45, 7854–7860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang F.-C., Chang C.-C., Lou P.-J., Kuo I.-C., Chien C.-W., Chen C.-T., Shieh F.-Y., Chang T.-C., Lin J.-J. (2008) G-quadruplex stabilizer 3,6-bis(1-methyl-4-vinylpyridinium)carbazole diiodide induces accelerated senescence and inhibits tumorigenic properties in cancer cells. Mol. Cancer Res. 6, 955–964 [DOI] [PubMed] [Google Scholar]
- 20. Huang F.-C., Chang C.-C., Wang J.-M., Chang T.-C., Lin J.-J. (2012) Induction of senescence in cancer cells by a G-quadruplex stabilizer BMVC4 is independent of its telomerase inhibitory activity. Br. J. Pharmacol. 167, 393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cullen B. R., Malim M. H. (1992) Secreted placental alkaline phosphatase as a eukaryotic reporter gene. Methods Enzymol. 216, 362–368 [DOI] [PubMed] [Google Scholar]
- 22. Balagurumoorthy P., Brahmachari S. K., Mohanty D., Bansal M., Sasisekharan V. (1992) Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 20, 4061–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kypr J., Kejnovská I., Renciuk D., Vorlícková M. (2009) Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 37, 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Masiero S., Trotta R., Pieraccini S., De Tito S., Perone R., Randazzo A., Spada G. P. (2010) A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org. Biomol. Chem. 8, 2683–2692 [DOI] [PubMed] [Google Scholar]
- 25. Patel D. J., Tonelli A. E. (1974) Assignment of the proton NMR chemical shifts of the T-N3H and G-N1H proton resonances in isolated AT and GC Watson-Crick base pairs in double-stranded deoxyoligonucleotides in aqueous solution. Biopolymers 13, 1943–1964 [DOI] [PubMed] [Google Scholar]
- 26. Feigon J., Koshlap K. M., Smith F. W. (1995) 1H NMR spectroscopy of DNA triplexes and quadruplexes. Methods Enzymol. 261, 225–255 [DOI] [PubMed] [Google Scholar]
- 27. Phan A. T., Patel D. J. (2003) Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 125, 15021–15027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. González V., Hurley L. H. (2010) The C terminus of nucleolin promotes the formation of the c-MYC G-quadruplex and inhibits c-MYC promoter activity. Biochemistry 49, 9706–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Uribe D. J., Guo K., Shin Y. J., Sun D. (2011) Heterogeneous nuclear ribonucleoprotein K and nucleolin as transcriptional activators of the vascular endothelial growth factor promoter through interaction with secondary DNA structures. Biochemistry 50, 3796–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei P.-C., Wang Z.-F., Lo W.-T., Su M.-I., Shew J.-Y., Chang T.-C., Lee W.-H. (2013) A cis-element with mixed G-quadruplex structure of NPGPx promoter is essential for nucleolin-mediated transactivation on non-targeting siRNA stress. Nucleic Acids Res. 41, 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Balasubramanian S., Hurley L. H., Neidle S. (2011) Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 10, 261–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin C.-T., Tseng T.-Y., Wang Z.-F., Chang T.-C. (2011) Structural conversion of intramolecular and intermolecular G-quadruplexes of bcl2mid: the effect of potassium concentration and ion exchange. J. Phys. Chem. B 115, 2360–2370 [DOI] [PubMed] [Google Scholar]
- 33. Tsai Y. L., Wang Z. F., Chen W. W., Chang T.-C. (2011) Emulsified BMVC derivative induced filtration for G-quadruplex DNA structural separation. Nucleic Acids Res. 39, e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rimerman R. A., Gellert-Randleman A., Diehl J. A. (2000) Wnt1 and MEK1 cooperate to promote cyclin D1 accumulation and cellular transformation. J. Biol. Chem. 275, 14736–14742 [DOI] [PubMed] [Google Scholar]
- 35. Xu L., Corcoran R. B., Welsh J. W., Pennica D., Levine A. J. (2000) WISP-1 is a Wnt-1- and β-catenin-responsive oncogene. Genes Dev. 14, 585–595 [PMC free article] [PubMed] [Google Scholar]
- 36. He W., Tan R., Dai C., Li Y., Wang D., Hao S., Kahn M., Liu Y. (2010) Plasminogen activator inhibitor-1 is a transcriptional target of the canonical pathway of Wnt/β-catenin signaling. J. Biol. Chem. 285, 24665–24675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wei W., Chua M. S., Grepper S., So S. K. (2009) Blockade of Wnt-1 signaling leads to anti-tumor effects in hepatocellular carcinoma cells. Mol. Cancer 8, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. He B., Reguart N., You L., Mazieres J., Xu Z., Lee A. Y., Mikami I., McCormick F., Jablons D. M. (2005) Blockade of Wnt-1 signaling induces apoptosis in human colorectal cancer cells containing downstream mutations. Oncogene 24, 3054–3058 [DOI] [PubMed] [Google Scholar]
- 39. Barker N., Clevers H. (2006) Mining the Wnt pathway for cancer therapeutics. Nat. Rev. Drug Discov. 5, 997–1014 [DOI] [PubMed] [Google Scholar]
- 40. Lepourcelet M., Chen Y.-N., France D. S., Wang H., Crews P., Petersen F., Bruseo C., Wood A. W., Shivdasani R. A. (2004) Small-molecule antagonists of the oncogenic Tcf/β-catenin protein complex. Cancer Cell 5, 91–102 [DOI] [PubMed] [Google Scholar]
- 41. Huang S.-M., Mishina Y. M., Liu S., Cheung A., Stegmeier F., Michaud G. A., Charlat O., Wiellette E., Zhang Y., Wiessner S., Hild M., Shi X., Wilson C. J., Mickanin C., Myer V., Fazal A., Tomlinson R., Serluca F., Shao W., Cheng H., Shultz M., Rau C., Schirle M., Schlegl J., Ghidelli S., Fawell S., Lu C., Curtis D., Kirschner M. W., Lengauer C., Finan P. M., Tallarico J. A., Bouwmeester T., Porter J. A., Bauer A., Cong F. (2009) Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 [DOI] [PubMed] [Google Scholar]
- 42. Chen B., Dodge M. E., Tang W., Lu J., Ma Z., Fan C.-W., Wei S., Hao W., Kilgore J., Williams N. S., Roth M. G., Amatruda J. F., Chen C., Lum L. (2009) Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ewan K., Pajak B., Stubbs M., Todd H., Barbeau O., Quevedo C., Botfield H., Young R., Ruddle R., Samuel L., Battersby A., Raynaud F., Allen N., Wilson S., Latinkic B., Workman P., McDonald E., Blagg J., Aherne W., Dale T. (2010) A useful approach to identify novel small-molecule inhibitors of Wnt-dependent transcription. Cancer Res. 70, 5963–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sack U., Walther W., Scudiero D., Selby M., Aumann J., Lemos C., Fichtner I., Schlag P. M., Shoemaker R. H., Stein U. (2011) S100A4-induced cell motility and metastasis is restricted by the Wnt/β-catenin pathway inhibitor calcimycin in colon cancer cells. Mol. Biol. Cell 22, 3344–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Todd A. K., Johnston M., Neidle S. (2005) Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 33, 2901–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huppert J. L., Balasubramanian S. (2005) Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maizels N., Gray L. T. (2013) The G4 genome. PLoS Genet. 9, e1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eddy J., Maizels N. (2006) Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 34, 3887–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Phan A. T., Kuryavyi V., Burge S., Neidle S., Patel D. J. (2007) Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 129, 4386–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hsu S.-T., Varnai P., Bugaut A., Reszka A. P., Neidle S., Balasubramanian S. (2009) A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 131, 13399–13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kuryavyi V., Phan A. T., Patel D. J. (2010) Solution structures of all parallel-stranded monomeric and dimeric G-quadruplex scaffolds of the human c-kit2 promoter. Nucleic Acids Res. 38, 6757–6773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cogoi S., Xodo L. E. (2006) G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 34, 2536–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. De Armond R., Wood S., Sun D., Hurley L. H., Ebbinghaus S. W. (2005) Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1α promoter. Biochemistry 44, 16341–16350 [DOI] [PubMed] [Google Scholar]
- 54. Xu Y., Sugiyama H. (2006) Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb). Nucleic Acids Res. 34, 949–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sun D., Guo K., Shin Y.-J. (2011) Evidence of the formation of G-quadruplex structures in the promoter region of the human vascular endothelial growth factor gene. Nucleic Acids Res. 39, 1256–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Palumbo S. L., Ebbinghaus S. W., Hurley L. H. (2009) Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-Interactive ligands. J. Am. Chem. Soc. 131, 10878–10891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Palumbo S. L., Memmott R. M., Uribe D. J., Krotova-Khan Y., Hurley L. H., Ebbinghaus S. W. (2008) A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res. 36, 1755–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qin Y., Fortin J. S., Tye D., Gleason-Guzman M., Brooks T. A., Hurley L. H. (2010) Molecular cloning of the human platelet-derived growth factor receptor β (PDGFR-β) promoter and drug targeting of the G-quadruplex-forming region to repress PDGFR-β expression. Biochemistry 49, 4208–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Qin Y., Rezler E. M., Gokhale V., Sun D., Hurley L. H. (2007) Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 35, 7698–7713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kang C.-C., Huang W.-C., Kouh C.-W., Wang Z.-F., Cho C.-C., Chang C.-C., Wang C.-L., Chang T.-C., Seemann J., Huang L. J. (2013) Chemical principles for the design of a novel fluorescent probe with high cancer-targeting selectivity and sensitivity. Integr. Biol. 5, 1217–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]