Background: Crystal structures of the secondary active MATE multidrug transporter NorM suggest a role for Na+ as a coupling ion.
Results: Biochemical studies in proteoliposomes containing purified NorM demonstrate a dependence of transport on the sodium- and proton-motive force.
Conclusion: Ion/drug antiport by NorM involves both Na+ and H+ cycling.
Significance: Mechanistic information on drug transporters will facilitate the design of new drugs that can poison or circumvent transport activity.
Keywords: Bioenergetics, Enzyme Mechanisms, Membrane Proteins, Membrane Transport, Multidrug Transporters
Abstract
Membrane transporters belonging to the multidrug and toxic compound extrusion family mediate the efflux of unrelated pharmaceuticals from the interior of the cell in organisms ranging from bacteria to human. These proteins are thought to fall into two classes that couple substrate efflux to the influx of either Na+ or H+. We studied the energetics of drug extrusion by NorM from Vibrio cholerae in proteoliposomes in which purified NorM protein was functionally reconstituted in an inside-out orientation. We establish that NorM simultaneously couples to the sodium-motive force and proton-motive force, and biochemically identify protein regions and residues that play important roles in Na+ or H+ binding. As the positions of protons are not available in current medium and high-resolution crystal structures of multidrug and toxic compound extrusion transporters, our findings add a previously unrecognized parameter to mechanistic models based of these structures.
Introduction
Multidrug transporters can confer multidrug resistance on cells by mediating the active extrusion of a broad spectrum of chemically and structurally unrelated antibiotics and toxic compounds out of cells, thus lowering the concentration of these drugs near their targets (1). The phenomenon of multidrug transporters is a worrying threat, because the acquisition of such a single system in a cell can render currently available antibiotics and chemotherapeutic agents ineffective. Bacterial multidrug and toxic compound extrusion (MATE)2 transporters confer clinical resistance to antibiotics such as tigecycline, which was developed to overcome methicillin- and vancomycin-resistant Staphylococcus aureus strains (2–5). Human MATE transporters mediate biliary excretion (via MATE1, also termed SLC47A1) and renal excretion (via MATE1 and MATE2; the latter is also termed SLC47A2) of a diverse array of organic compounds that include the antidiabetic drug metformin (6, 7), antibiotics cephalexin and cephradine (8), antihistamine agent fexofenadine (9), anticancer drug cisplatin (10), and antiviral agent acyclovir (8). Hence, MATE proteins are important determinants of drug pharmacokinetics and bioavailability.
MATE transporters are a relatively new family among MDR transporters and are less well characterized than members of ATP-binding cassette superfamily, major facilitator superfamily, resistance nodulation-cell division family, and small multidrug resistance family (11, 12). Depending on the effect of Na+ on substrate efflux in intact cells, earlier studies on bacterial MATE transporters grouped these proteins into two classes, those that are Na+ dependent, including NorM from Vibrio parahaemeolyticus (13), Vibrio cholerae (14), Neisseria gonorrhoeae (15), and PdrM from Streptococcus pneumonia (16), and those that are not Na+ dependent, including PmpM from Pseudomonas aeruginosa (17), PfMATE from Pyrococcus furiosus (18), and DinF from Bacillus halodurans (19), but that are H+ dependent instead. Evidence for this H+ dependence came from substrate-induced proton transport by PmpM and DinF in inside-out membrane vesicles (17, 19) and PfMATE in spheroplasts (18).
There are three well conserved acidic residues, Asp-32, Glu-251, and Asp-367 in NorM from V. parahaemeolyticus (NorM-VP), and corresponding Asp-36, Glu-255, and Asp-371 in NorM from V. cholera (NorM-VC), which were suggested to be important for transport in functional studies (13) and molecular dynamics simulations (20). A medium resolution crystal structure of NorM-VC was determined in the outward-facing conformation (14) in which these carboxylates are arranged asymmetrically. Glu-255 and Asp-371 are located in C-terminal helical bundle facing the internal cavity, whereas Asp-36 is located in the N-terminal helical bundle, far away from the proposed C-terminal cation-binding site. This asymmetric distribution was confirmed in recent crystal structures of NorM from N. gonorrhoeae (NorM-NG) in outward-facing and drug-bound states (15).
Despite these significant advancements at the structural level, the mechanistic interpretation of MATE structures requires detailed biochemical insights in the antiport reactions that are catalyzed. Here, the energetics of NorM-VC were evaluated in intact cells and synthetic membrane vesicles (proteoliposomes) containing purified and functionally reconstituted protein, in which well defined electrochemical ion gradients were imposed artificially. We demonstrate that NorM-VC simultaneously couples to both the sodium-motive force (SMF) and proton-motive force (PMF), and that catalytic carboxylates have different roles in these coupling reactions.
EXPERIMENTAL PROCEDURES
Expression of NorM-VC in Lactococcal Cells
The norM-VC gene was subcloned from pET19b-NorM-VC (14) into plasmid pHLP5 (a lactococcal plasmid pNZ8048-derivative (21) encoding His-tagged multidrug transporter LmrP (22)) and expressed in Lactococcus lactis strain NZ9000 ΔlmrA ΔlmrCD (23). Cells were grown at 30 °C in M17 medium (Oxoid) supplemented with 0.5% (w/v) glucose and 5 μg/ml of chloramphenicol. Expression of NH2-terminal His10-tagged NorM-VC proteins was induced by the addition of 7.5 ng of nisin A/ml (a 1:1500 dilution of the supernatant of the nisin-producing strain L. lactis NZ9700) at an A660 of 0.3–0.35. Cells were harvested by centrifugation after induction for 1 h. Mutations were introduced into the norM-VC gene in the pET19b vector using the QuikChange method (Agilent Technologies) with mutagenic oligonucleotide primers as listed in Table 1. The mutated genes were sequenced to ensure that only the intended mutations were introduced.
TABLE 1.
Primers used for site-directed mutagenesis
| Mutation | Primersa |
|---|---|
| D36N | GGGCTTTGTCAATACCATCATGGCG |
| CGCCATGATGGTATTGACAAAGCCC | |
| N174A | GGGTTATTGCTCGCTATCCCTCTC |
| GAGAGGGATAGCGAGCAATAACCC | |
| E255Q | GCCGCCCTCTTTTTTCAAGTGACTC |
| GAGTCACTTGAAAAAAGAGGGCGGC | |
| F288A | CTCTTCTTTGGTGGCTATGTTCCC |
| GGGAACATAGCCACCAAAGAAGAG | |
| Y367A | CGGCAATCGCTCAATGTATGGATGC |
| GCATCCATACATTGAGCGATTGCCG | |
| F395A | CCACCGTACAGCTATCTCCTACTGGG |
| CCCAGTAGGAGATAGCTGTACGGTGG | |
| D371N | CAATGTATGAATGCGGTACAAG |
| CTTGTACCGCATTCATACATTG |
a Forward (top) and reverse (bottom) sequences in 5′ → 3′ direction.
Photoaffinity Labeling with Ethidium Monoazide
Membrane vesicles prepared from L. lactis cells (24) (25 mg of total membrane protein in 5 ml) were incubated with 10 μm ethidium monoazide (EMA) bromide in 50 mm potassium Pi buffer (pH 7.4) containing 200 KCl, 5 mm MgSO4 in the presence or absence of 10 mm Na2SO4 at 37 °C for 15 min, and subsequently irradiated with UV light (312 nm) in a Bio-Link BLX photo-reactor (BDH) with 5 UV lamps of 8 watt each. n-Dodecyl-β-d-maltoside (DDM) (Melford and Anatrace) (1% w/v) was added and NorM-VC protein was purified as described under “Ethidium Transport in DNA-loaded Proteoliposomes.” Photolabeled proteins were separated by SDS-PAGE after which the intensity of EMA fluorescence signals was quantified using ImageJ software, version 1.43 (National Institutes of Health).
Ethidium Transport in L. lactis
Ethidium transport assays were conducted as previously described (24, 25) with several modifications. Cells were grown and harvested in mid-exponential phase and washed with 50 mm potassium Pi buffer (pH 7.0) containing 5 mm MgSO4. The harvested cells (A660 of 0.3) were ATP-depleted by incubation in the presence of 0.25 mm 2,4-dinitrophenol for 30 min at 30 °C. Cells were subsequently washed, and resuspended in the potassium Pi buffer to an A660 of 5.0, and kept on ice. Active ethidium efflux was measured in ATP-depleted cells (A660 of 0.5) that were preloaded with 2 μm ethidium bromide for 40 min to a fluorescence level (a.u.) between 250 and 300. The subsequent addition of 25 mm glucose to the cells allowed the generation of metabolic energy. The active efflux of ethidium was then monitored with a PerkinElmer LS 55B fluorimeter using excitation and emission wavelengths of 500 and 580 nm, respectively. To manipulate the composition of the PMF, the ionophores nigericin and/or valinomycin were used at final concentrations of 0.5 μm each, and these were added 3 min before the addition of glucose, together with Na+ or K+ where required.
To measure the rate of facilitated ethidium efflux as a function of the ethidium concentration, ATP-depleted cells were equilibrated with ethidium at concentrations between 0.1 and 30 μm at 30 °C until the saturation level was reached. After equilibration, cells were pelleted by centrifugation at 15,000 × g for 2 min, and resuspended in a volume of 50 μl of the same buffer to an A660 of 20. The assay was then started by the 40-fold dilution of the cell suspension into 2 ml of ethidium-free buffer containing 1 mm Na2SO4, and fluorescence was followed over time. To measure facilitated ethidium efflux in the presence of different concentrations of sodium, cells were preloaded with 2 μm ethidium, pelleted, and diluted 40-fold in the potassium Pi buffer containing concentrations of Na2SO4 ranging from 0.1 to 2 mm. Ethidium efflux rates were calculated over the first 30 s during which efflux was linear. Rates of passive efflux in non-expressing control cells were subtracted from the rates obtained for NorM-expressing cells.
Ethidium Transport in DNA-loaded Proteoliposomes
For reconstitution experiments NorM proteins were expressed from pET19b-based vectors in Escherichia coli Origami DE3 cells, in which the expression level was higher than in L. lactis (about 2 versus 1% of total membrane proteins, respectively, as determined by densitometric analysis on Coomassie-stained SDS-PAGE gels). Cells were grown in LB medium at 30 °C to an A660 of 0.5 in the presence of 50 μg/ml of carbenicillin. NorM-VC expression was induced with 1 mm isopropyl β-d-thiogalactoside for 3 h, and checked on 10% SDS-PAGE. For this purpose, membrane vesicles (10 μg of total membrane protein, see below) were mixed with 30 μl of 3× sample loading buffer containing 6% (w/v) SDS, then mixed with DTT (1 mm final concentration), and subsequently incubated at 20 °C for 10 min before loading on gel (5 μg of protein/lane).
Inside-out membrane vesicles were prepared from washed E. coli cells (26) in 100 mm potassium Pi buffer (pH 7.0) using a Basic Z cell disruptor (Constant Systems) (27). Membrane vesicles were solubilized for 4 h in the presence of 1% (w/v) DDM in 50 mm potassium Pi (pH 8.0), 200 mm K2SO4, 5% glycerol (v/v), 30 mm imidazole and EDTA-free protease inhibitor (Roche Applied Science). The insoluble fraction was removed by ultracentrifugation and NorM-VC protein was purified from the supernatant by Ni2+-nitrilotriacetic acid affinity chromatography. The solubilized His-tagged protein was allowed to bind to the equilibrated resin (1 ml of resin/10 mg of total membrane protein) by mixing for 2 h at 4 °C and the resin was then washed with 15 volumes of wash buffer A (50 mm potassium Pi (pH 8.0) containing 0.2 mm KCl, 10% (v/v) glycerol, 0.05% (w/v) DDM, and 20 mm imidazole (pH 8.0)), then 15 volumes of wash buffer B (50 mm potassium Pi (pH 7.0) containing 0.2 mm KCl, 10% (v/v) glycerol, 0.05% (w/v) DDM, and 20 mm imidazole (pH 8.0)). Purified protein was then eluted with 3–4 volumes of elution buffer (50 mm potassium Pi (pH 7.0) containing 0.2 mm KCl, 5% (v/v) glycerol, 0.05% (w/v) DDM, and 200 mm imidazole (pH 8.0)) using a Bio-spin column (Bio-Rad). The purity of the NorM-VC protein was checked on 10% SDS-PAGE with Coomassie Brilliant Blue staining.
NorM proteins were reconstituted in proteoliposomes prepared from washed E. coli lipids (24) mixed with l-α-phosphatidylcholine (Avanti) at a ratio of 3:1 (w/w) in chloroform. After evaporation of the solvent, the lipids were rehydrated in buffer (pH 6.8) containing 10 mm Tris-Cl, 10 mm potassium Pi, 100 mm Na2SO4, 100 mm NH4SCN, and 1 mg/ml of sonicated calf thymus DNA (Trevigen). The liposome suspension was then extruded 11 times through a 400-nm polycarbonate filter to form unilamellar liposomes of homogenous size. Liposomes were destabilized by the addition of 3 mm Triton X-100, after which purified NorM-VC protein was added at a lipid:protein ratio of 100:1 (w/w). To artificially impose SMF (interior positive and high) and/or PMF (interior positive and acid) or their components, proteoliposomes were diluted 100-fold in pre-cooled buffers (10 °C) as listed in Table 2. After 30 s, 5 μm ethidium was added and the fluorescence of the ethidium-DNA complex formed inside the proteoliposomes was followed over time. In control experiments, proteoliposomes were diluted 100-fold in Buffer 1 in which they were prepared to measure ethidium transport in the absence of the ion gradients. For measurements in the complete absence of Na+ in Fig. 2D, buffers were used in which Na+ was replaced by K+, and all experimental procedures were performed in plastic containers to avoid leaching of Na+ from glass surfaces. Glass fluorimeter cuvettes were rinsed with 0.1 n H2SO4 and ultrapure water before use.
TABLE 2.
Dilution buffers used for artificial imposition of (electro-)chemical ion gradients in proteoliposomes
| Buffera | Forces | Composition of buffer | pH |
|---|---|---|---|
| 1 | No gradient | 10 mm Tris-Cl, 10 mm potassium Pi, 100 mm Na2SO4, 100 mm NH4SCN | 6.8 |
| 2 | SMF plus PMF | 10 mm Tris-Cl, 10 mm potassium Pi | 7.5 |
| 3 | SMF | 10 mm Tris-Cl, 10 mm potassium Pi, 50 mm (NH4)2SO4 | 6.8 |
| 4 | PMF | 10 mm Tris-Cl, 10 mm potassium Pi, 100 mm Na2SO4 | 7.5 |
| 5 | ΔpNa | 10 mm Tris-Cl, 10 mm potassium Pi, 100 mm NH4SCN | 6.8 |
| 6 | Δψ | 10 mm Tris-Cl, 10 mm potassium Pi, 100 mm Na2SO4, 50 mm (NH4)2SO4 | 6.8 |
| 7 | ΔpH | 10 mm Tris-Cl, 10 mm potassium Pi, 100 mm Na2SO4, 100 mm NH4SCN | 7.5 |
a Proteoliposomes were prepared in Buffer 1 and diluted 100-fold in Buffers 1–7 with composition and pH as indicated in the third and fourth columns, respectively, to achieve no gradient, SMF interior positive and high) and/or PMF interior positive and acidic) or their components as indicated in the second column.
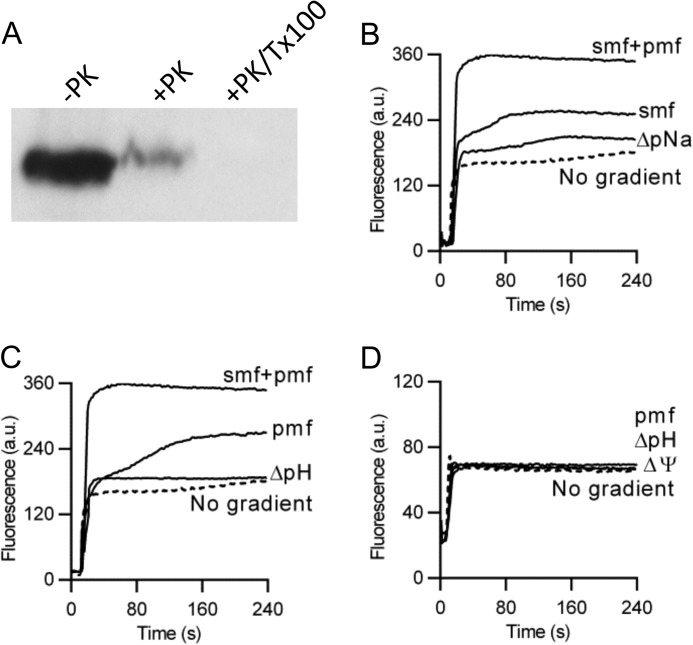
FIGURE 2.
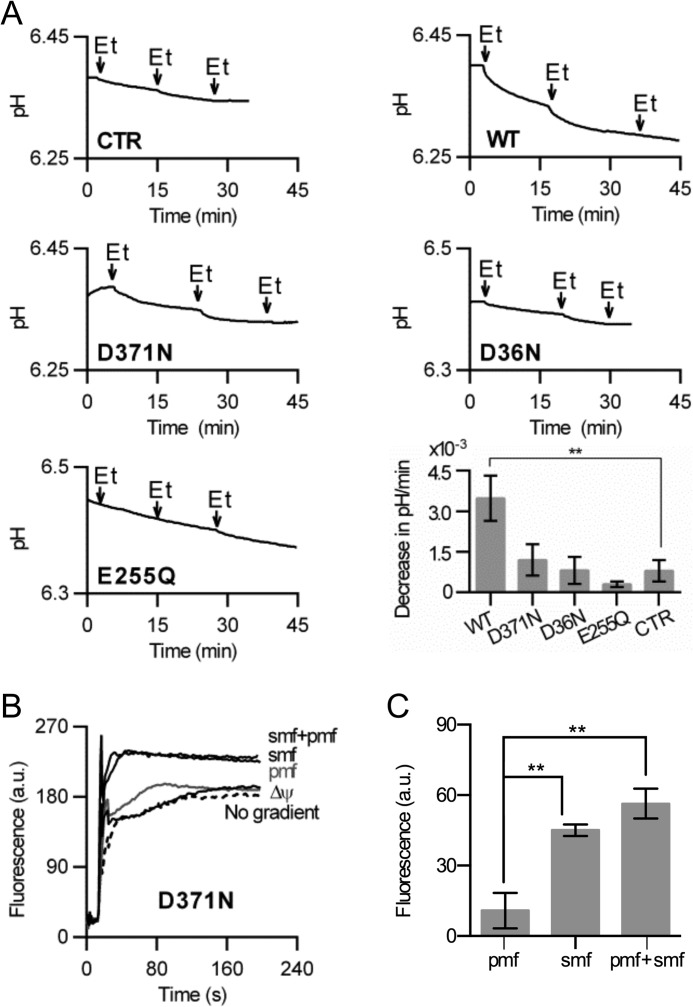
Ethidium transport by NorM-VC in DNA-loaded proteoliposomes. A, availability of the NH2-terminal His10 tag in WT NorM to cleavage by Proteinase K (+PK) at the external side of proteoliposomes compared with control without the protease (−PK). To make the His tag in the liposomal lumen accessible, 1% Triton X-100 was added to the proteoliposomes before proteolysis (+PK/Tx100). Subsequently, the remaining His tag was detected on an immune blot (2.5 μg of protein per lane) probed with anti-His5 antibody. B–D, ethidium transport in proteoliposomes with imposed chemical Na+ gradient (ΔpNa, interior high), sodium-motive force (SMF) (= membrane potential (Δψ)-ZΔpNa, interior positive and high, in which Z ≅ 58 mV at 20 °C), chemical proton gradient (ΔpH, interior acid), proton-motive force (PMF = Δψ-ZΔpH, interior positive and acid), or SMF plus PMF (= Δψ-ZΔpNa-ZΔpH). Experiments with imposed PMF and ΔpH in C were performed in the presence of Na+ ([Na+]in = [Na+]out = 1 mm), whereas those in D were performed in the complete absence of Na+ in plastic containers. The data represent observations in at least three separate experiments using independent batches of purified proteins, cells, and proteoliposomes. Error bars represent the mean ± S.E.
To study the accessibility of the His tag of WT NorM in proteoliposomes to protease digestion in the external buffer, 10 μg of membrane protein in 38.5 μl of 50 mm K-HEPES buffer (pH 7.5) supplemented with 1 mm CaCl2 was incubated in the presence of 2.5 μg/ml of proteinase K (28, 29). Following incubation at 0 °C for 20 min, the reaction was terminated by the addition of 1.5 μl of 200 mm phenylmethanesulfonyl fluoride (PMSF, in ethanol), after which 20 μl of 3× sample-loading buffer and DTT (1 mm final concentration) were added. The samples were incubated at 20 °C for 10 min, after which they were subjected to SDS-PAGE (2.5 μg of protein/lane), and analyzed on immunoblot probed with monoclonal anti-His5 antibody (Sigma). Signals were quantified using ImageJ.
Measurement of Substrate-induced Proton Release
Proton release measurements were conducted as described (30) with several modifications. NorM protein was expressed and purified as described under “Ethidium Transport in DNA-Loaded Proteoliposomes,” and was dialyzed 3 times against 200 volumes of low-buffered proton release solution (10% glycerol, 120 mm NaCl). The first two dialyzes were carried for 1 h, and the last round was carried out overnight at 4 °C in a stirred sealed beaker under a N2 atmosphere. Protein aggregates were removed by centrifugation at 200,000 × g for 30 min. For each pH measurement, purified proteins were diluted to 1 μm in 2 ml of proton-release solution supplemented with 0.05% DDM and 10 mm Na2SO4. The pH was then continuously measured with a pH electrode (VWR) that was connected to a 2-channel data acquisition system ML826 PowerLab (ADInstruments). The Labchart program was used to record the continuous real-time pH changes. The pH was allowed to stabilize for several minutes, after which 5 μl of ethidium was added three times to yield a final concentration of 20 μm.
Curve Fitting and Statistical Analysis
The kinetics of substrate transport was fitted to a hyperbola using the equation y = ax/(b + x), in which y is the transport rate, x is the substrate concentration, a is the maximum rate of the reaction (Vmax), and b is the substrate concentration giving half-Vmax (corresponding to the transport constant, Kt). The kinetic data for the N174A mutant in Fig. 4C were fitted to the Hill equation y = axc/(bc + xc) with similar parameters as above and Hill coefficient denoted as c. All experiments were performed at least three times using independent batches of cells or proteoliposomes. Significance was calculated using the Student's t test and analysis of variance. Statistical significance was calculated at a 95% confidence interval.
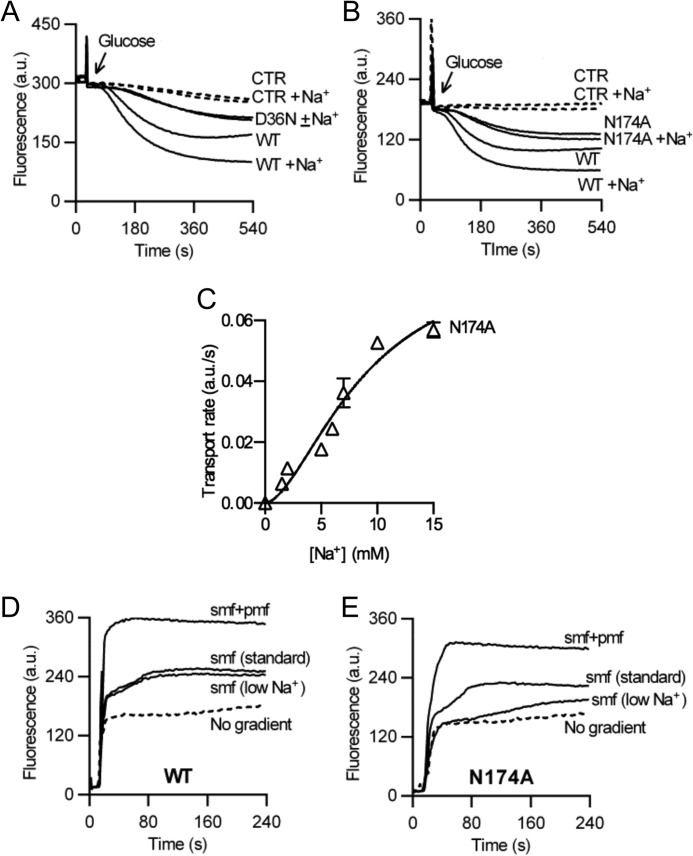
FIGURE 4.
Role of Asp-36 and neighboring residues in Na+ coupling. A and B, in contrast to observations for WT, active ethidium efflux in cells expressing the D36N (A) or N174A (B) mutant was not stimulated by the addition of 1 mm Na+. C, rate of facilitated ethidium efflux as a function of Na+ concentration in ATP-depleted cells containing the N174A mutant demonstrates reduced affinity of the mutant for Na+ compared with WT (compare Fig. 1D). D and E, ethidium transport in DNA-loaded proteoliposomes containing purified WT NorM-VC (D) or N174A protein (E), in which the SMF was imposed artificially. The standard SMF was imposed at [Na+]in/[Na+]out = 100/1 mm, whereas “SMF (low Na+)” was imposed at [Na+]in/[Na+]out = 10/0.1 mm. Traces are based on observations in four separate experiments using independent batches of cells and proteoliposomes. Error bars represent the mean ± S.E.
RESULTS
Ion Coupling in NorM-VC
The x-ray structures of NorM-VC and NorM-NG were captured in a Rb+- or Cs+-bound outward-facing state, which was suggested to represent a Na+-bound state (14, 15), reflecting Na+ dependence in transport. To test whether NorM-VC can utilize the electrochemical Na+ gradient to extrude substrates, NorM-VC was expressed at a level of about 1% of total membrane protein in the drug-hypersensitive L. lactis strain NZ9000 ΔlmrA ΔlmrCD (23) under the control of a nisin A-inducible promoter in the expression plasmid pNZ8048 (Fig. 1A), and we measured the Na+ dependence of ethidium binding and transport (Fig. 1, B–D),
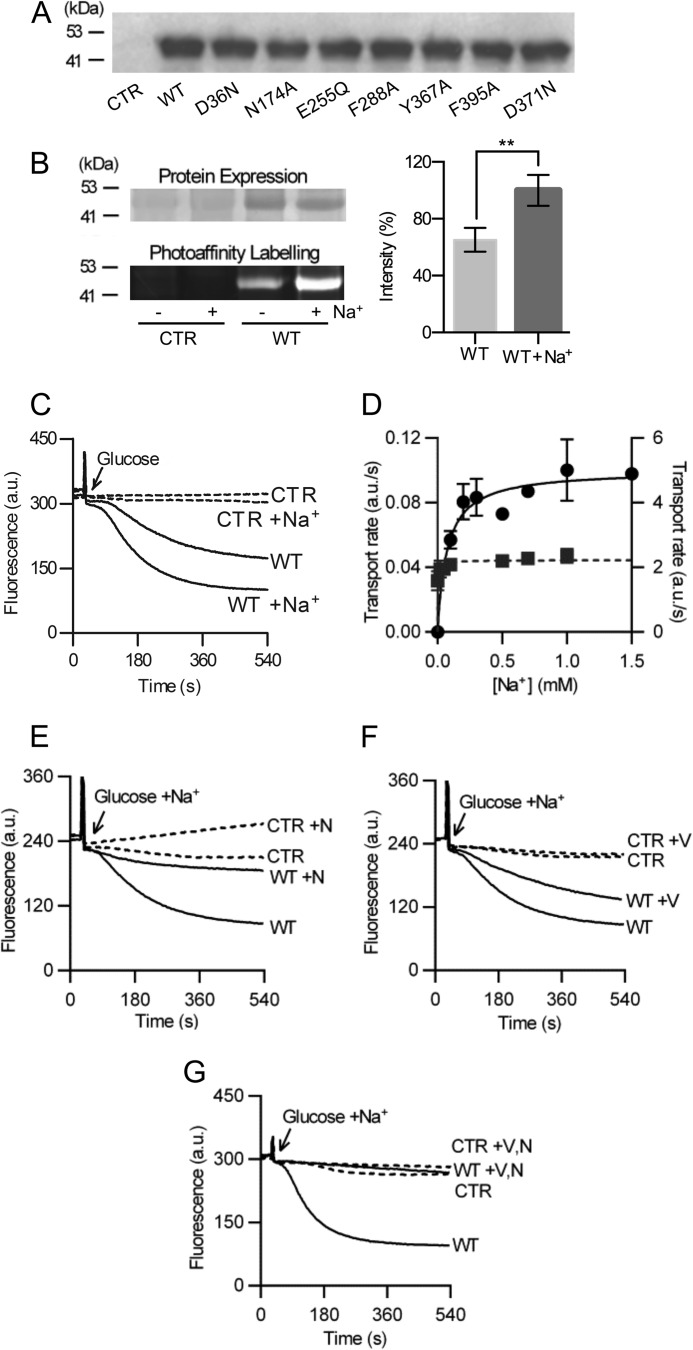
FIGURE 1.
Ethidium binding and transport by NorM-VC in lactococcal membranes. A, Western analysis of total membrane protein in membrane vesicles (5 μg/lane) using anti-His5 antibody shows that wild type (WT) and mutant proteins are expressed at similar levels in the cytoplasmic membrane of L. lactis. B, EMA photolabeling of NorM-VC in the presence or absence of Na+. Lactococcal membrane vesicles (25 mg) containing NorM-VC (WT) or lacking NorM protein (CTR) were incubated with 10 μm EMA in 50 mm Tris-Cl buffer (pH 7.4) containing 10 mm Na2SO4 or K2SO4, and 5 mm MgSO4 at 37 °C for 15 min. Membrane vesicles were exposed to UV irradiation and solubilized total membrane proteins (10 μg) were loaded on a 12% SDS-PAGE gel. Coomassie staining (left panel, top) confirmed equal loading NorM-VC (WT) and lack of the protein in control lanes (CTR). Band intensity of ethidium fluorescence emission of NorM-VC signal (left panel, bottom) was quantified using ImageJ (right panel). **, p < 0.05, significantly different. C, ATP-depleted cells were preloaded with 2 μm ethidium until a saturation level was reached. Active ethidium efflux was initiated by addition of 25 mm glucose as a source of metabolic energy, and 1 mm Na2SO4 (or K2SO4 in control experiments) at the arrow. D, rate of active ethidium efflux (■) (right y axis) as a function of the Na+ concentration in the buffer. For measurements of facilitated ethidium efflux (●) (left y axis), ATP-depleted ethidium-loaded cells were diluted 40-fold in buffer after which the efflux rate was determined as a function of the external Na+ concentration in the absence of glucose. The rate of ethidium efflux in control cells lacking NorM-VC protein was subtracted from the rate observed in NorM-VC expressing cells. E–G, ionophores nigericin (N) or valinomycin (V) or both (0.5 μm each) were added to ethidium-loaded cells 3 min prior to the addition of glucose and 1 mm Na+.
Ethidium binding to NorM-VC was investigated in a photoaffinity labeling approach using EMA (Fig. 1B). Lactococcal plasma membrane vesicles were incubated with 10 μm EMA in the presence or absence of 10 mm Na+ at 37 °C for 15 min and irradiated with 150 joule of UV light. The NorM proteins were then purified and equivalent amounts of proteins were loaded onto an SDS-PAGE gel. Densitometry of signals on Coomassie-stained SDS-PAGE gels (Fig. 1B, left top panel) confirmed equal loading of NorM-VC (98 ± 9% for the sample containing Na+; 100 ± 6% for the sample without Na+) and lack of NorM-VC in control lanes. Measurements of the ethidium fluorescence emission revealed that Na+ significantly enhanced the incorporation of EMA in NorM-VC (Fig. 1B, left bottom panel). Furthermore, cells expressing NorM-VC showed enhanced ethidium efflux activity in the presence of 1 mm Na+ compared with its absence, whereas Na+ had no effect on the changes in fluorescence in control cells (Fig. 1C). These results indicate that Na+ is a coupling cation for NorM-VC. Interestingly, NorM-VC also showed a substantial residual activity in the absence of added Na+ to the buffer (Fig. 1C). Sodium dependence of facilitated ethidium efflux down its chemical gradient in ATP-depleted cells, and active efflux of ethidium in energized cells revealed a Kt = 73 ± 4 μm Na+ and 4.7 ± 0.6 μm Na+, respectively (Fig. 1D) (Table 3). This high affinity for Na+ during active efflux suggests that small traces of Na+ in cell suspensions, chemicals, and desorbed from glass surfaces are sufficient to support Na+-dependent transport activity of NorM-VC. However, external Na+ in μm concentrations does not allow formation of a substantial chemical Na+ gradient across the cytoplasmic membrane, which raises questions about the energetics of NorM-VC-mediated ethidium efflux under these conditions.
TABLE 3.
Kinetic parameters for Na+-dependent ethidium efflux by NorM-VC
| Protein | Ethidium |
Na+ |
||
|---|---|---|---|---|
| Kta | Vmaxa | Ktb | Vmaxb | |
| μm | a.u./s | mm | a.u./s | |
| WT | 2.62 ± 0.17 | 0.27 ± 0.02 | 0.073 ± 0.004 | 0.10 ± 0.01 |
| N174A | 2.71 ± 0.93 | 0.26 ± 0.02 | 8.8 ± 1.7c | 0.08 ± 0.03 |
| F288A | 2.86 ± 0.51 | 0.21 ± 0.01 | 0.37 ± 0.10d | 0.05 ± 0.01 |
| Y367A | 2.58 ± 0.03 | 0.18 ± 0.02 | 0.08 ± 0.03 | 0.05 ± 0.01 |
| F395A | 24.2 ± 10.4e | 0.22 ± 0.05 | 0.10 ± 0.03 | 0.03 ± 0.01 |
| D371N | 2.60 ± 0.24 | 0.04 ± 0.00 | 0.09 ± 0.03 | 0.02 ± 0.00 |
a Apparent Kt and Vmax values were determined in a facilitated ethidium efflux assay in ATP-depleted lactococcal cells at a fixed concentration of 1 mm Na+.
b Apparent Kt and Vmax values were determined in a facilitated ethidium efflux assay in ATP-depleted lactococcal cells at a fixed concentration of 2 μm ethidium.
c Significantly different substrate binding affinity compared to WT, p < 0.01.
d Significantly different substrate binding affinity compared to WT, p < 0.05.
e Significantly different substrate binding affinity compared to WT, p < 0.02.
The energetics of ethidium transport by NorM-VC was initially studied in lactococcal cells using the ionophores valinomycin and nigericin, which can dissipate the membrane potential (Δψ, interior negative) and transmembrane chemical proton gradient (ΔpH, interior alkaline), respectively, of the PMF (= Δψ − ZΔpH, Z ≅ 58 mV at 20 °C). Ionophore(s) at a concentration of 0.5 μm were added 3 min before adding the glucose and 1 mm Na+. Nigericin mediates electroneutral K+/H+ exchange across a membrane resulting in the dissipation of the ΔpH when cells are in high potassium containing buffers, whereas the potassium ionophore valinomycin specifically dissipates the Δψ. The NorM-VC-mediated efflux activity was reduced compared with control in the presence of nigericin (PMF = Δψ) (Fig. 1E) or valinomycin (PMF = −ZΔpH) (Fig. 1F), suggesting that NorM-VC transports ethidium in an electrogenic manner. The addition of both valinomycin and nigericin completely abolished the activity of NorM-VC (Fig. 1G). As the SMF is generated by the activity of sodium/proton antiport activity in L. lactis (31), and dissipation of the PMF will result in the dissipation of the SMF, these data confirm that NorM-VC-mediated efflux is active and dependent on electrochemical ion gradients.
To study the energetics in more detail, ethidium transport was measured in DNA-loaded proteoliposomes containing purified and functionally reconstituted NorM-VC. The orientation of reconstituted NorM-VC in the proteoliposomal membrane was examined through studies on the accessibility of the cytoplasmic NH2-terminal His10 tag to digestion with Proteinase K in the external buffer. Addition of the protease removed 91% of the His tag signal, whereas solubilization of the proteoliposomes with Triton X-100 allowed complete removal of the His tag (Fig. 2A). These data demonstrate that NorM-VC is predominantly (>90%) oriented in an inside-out fashion in the proteoliposomal membrane.
In the proteoliposomes, a Δψ of about 120 mV (interior positive) was imposed through the diffusion of SCN− ([SCN−]in/[SCN−]out, 100/1 mm) from the lumen to the external buffer (Table 2). In addition, a transmembrane chemical Na+ gradient (−ZΔpNa) of about 120 mV (interior high) and/or H+ gradient (−ZΔpH) of about 41 mV (interior acid) were imposed by dilution of Na+-loaded proteoliposomes in buffer ([Na+]in/[Na+]out = 100/1 mm), and by acid shock (pHin/pHout = 6.8/7.5) that was sustained by the outward diffusion of NH3 from proteoliposomes loaded with NH4+. When the ΔpNa was imposed (Fig. 2B), ethidium transport increased compared with the control (no gradient present). The presence of the SMF significantly enhanced ethidium transport in the proteoliposomes compared with the imposition of the ΔpNa only, suggesting that the transport reaction is electrogenic (Fig. 2B). This conclusion is in agreement with the observations in intact cells (Fig. 1, E and F). Interestingly, the imposition of PMF enhanced ethidium transport by NorM-VC in the presence of SMF (Fig. 2B) or Na+ ([Na+]in = [Na+]out = 1 mm) (Fig. 2C), but not in the absence of Na+ (Fig. 2D). The simultaneous imposition of the SMF plus PMF (= Δψ-ZΔpNa-ZΔpH) revealed that the contributions of the PMF and SMF are additive (SMF + PMF = 170 a.u.; SMF = 80 a.u.; PMF = 75 a.u.) (Fig. 2, B and C). Taken together, these data suggest that ethidium transport by NorM-VC is both sodium and proton dependent.
Three well conserved acidic residues were identified in NorM-VC that are important for transport activity (13): Asp-36 (in TM1) in the N-terminal half, and Glu-255 (in TM7) and Asp-371 (in TM10) in a network of aromatic residues in the C-terminal half (Fig. 3). However, the roles of these carboxylates and surrounding residues in the transport reaction have not been examined in detail. Asp-36, Glu-255, and Asp-371 were substituted by their amides, i.e. D36N, D371N, E255Q. In addition, the effect of substitutions of neighboring residues on activity was assessed: Asn-174 (close to Asp-36), and Phe-288, Tyr-367, and Phe-395 (close to Glu-255 and Asp-371) were mutated to alanine. These mutant proteins were expressed at a similar level as WT in the cytoplasmic membrane of L. lactis (Fig. 1A). Except for the F395A mutation (p < 0.02), none of these mutations significantly affected the apparent ethidium binding affinity of WT NorM-VC in efflux assays (Kt = 2.62 ± 0.17 μm ethidium) (Table 3).
FIGURE 3.
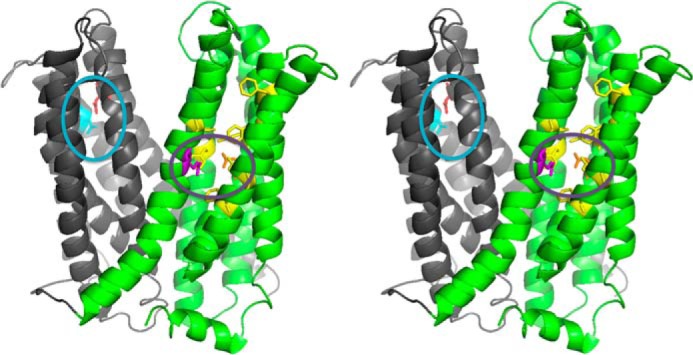
Structure model of outward-facing NorM-VC. Stereoview (PDB 3MKT) showing TM1–6 in gray and TM7–12 in green. Asp-36 (red) and neighboring Asn-174 and Asn-178 residues (light blue) are shown in a blue circle. Glu-255 (purple) and Asp-371 (orange) are shown in a purple circle with neighboring aromatic residues in yellow. Top, extracellular side; bottom, intracellular side.
Studies on Na+ Coupling Reveal a Dominant Role for Residues in Proximity of Asp-36 and Glu-255
We performed ethidium transport assays to compare the activity of the Asp-36 and Asn-174 substitution mutants with that of WT NorM-VC. Cells expressing the D36N protein exhibited a reduction in active efflux activity of ethidium compared with the WT; this residual activity was independent of Na+ and was too low for further studies (Fig. 4A). The ethidium efflux activity of the N174A mutant was reduced compared with WT but was substantially above control, suggesting that Asn-174 is not critical for the transport process (Fig. 4B). Addition of 1 mm Na+ together with the glucose had no effect on the active efflux activity of N174A NorM-VC but significantly enhanced the efflux rate of WT (p < 0.05) (Fig. 4B). Analysis of the sodium dependence of active ethidium efflux by the N174A mutant in cells revealed sigmoidal kinetics with a 1200-fold lower apparent affinity for Na+ (average Kt = 8.8 ± 1.7 mm Na+) compared with WT and Hill number of 1.6 ± 0.7 (Fig. 4C), pointing to a reduction in Na+ binding affinity in the mutant, and to cooperative Na+ binding to two (or more) sites in NorM-VC. Furthermore, the imposition of the standard SMF (with a chemical sodium gradient of [Na+]in/[Na+]out = 100/1 mm) or SMF at reduced Na+ concentrations ([Na+]in/[Na+]out = 10/0.1 mm) was equally effective in driving ethidium transport by WT NorM-VC in proteoliposomes, whereas the SMF at reduced Na+ concentrations was clearly less effective for the N174A mutant (Fig. 4, D and E). These data suggest a reduced binding affinity of the N174A mutant for Na+ compared with WT.
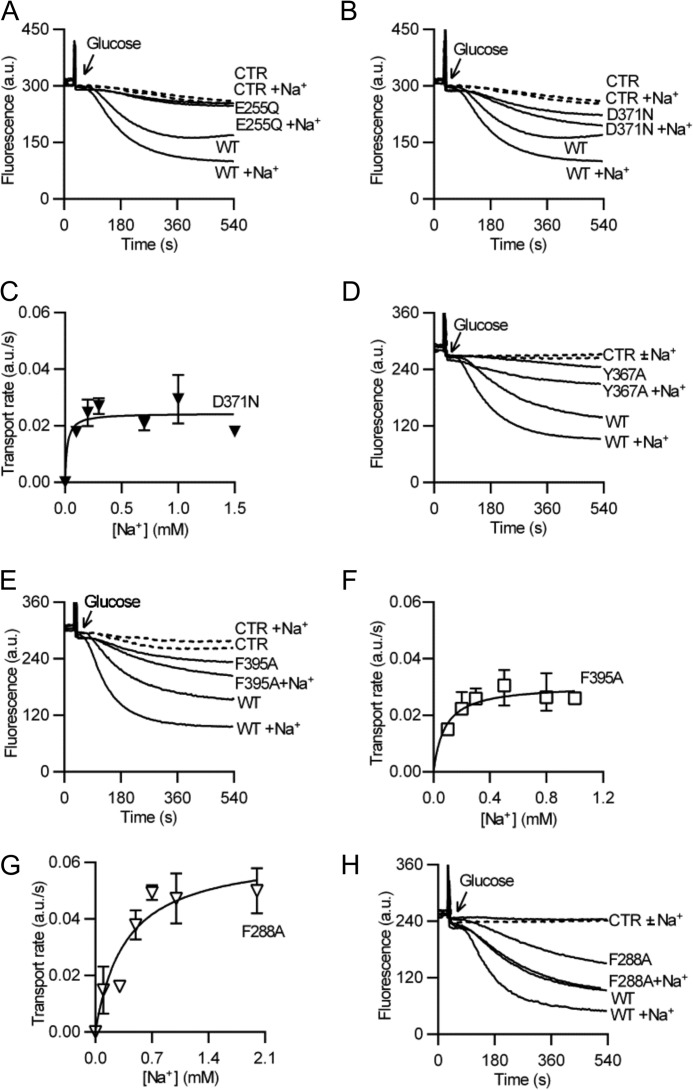
As crystal structures of NorM-VC implicated Glu-255 in the binding of the K+/Na+ analogue Cs+ (14), we tested the effect of the E255Q replacement on the Na+ dependence of transport. The mutant exhibited strongly reduced activity in intact cells (Fig. 5A). We therefore investigated the effect of mutations of residues that are close to Glu-255. Ethidium transport assay in intact cells showed that the D371N mutant, which was reported to inhibit Cs+ binding in soaked NorM-VC crystals (14), effluxed ethidium at a lower rate compared with WT. Similar to WT, however, transport activity was stimulated in the presence of Na+ (Fig. 5B). Furthermore, kinetic analysis of Na+ dependence of facilitated ethidium efflux by the D371N mutant in ATP-depleted cells revealed an unaltered apparent Kt (= 0.09 ± 0.03 mm Na+) compared with WT (Fig. 5C) (Table 3). The findings suggest that the D371N mutation does not affect the Na+ binding affinity of NorM-VC. Comparable results were obtained for Y367A and F395A (Fig. 5, D–F) (apparent Kt = 0.08 ± 0.03 mm Na+ for Y367A and 0.10 ± 0.03 mm Na+ for Phe-395). In contrast, the F288A mutant exhibited a significant 4-fold increase in the apparent Kt for Na+ (0.37 ± 0.10 mm for F288A, p < 0.05 compared with WT) (Fig. 5G). Our observation that ethidium transport by this mutant is stimulated in the presence of 1 mm Na+ (Fig. 5H), which is 2.7-fold above the apparent Kt, suggests that Phe-288 contributes to, but is not critical for Na+ binding. This conclusion is in agreement with crystal structures (14, 15) in which the bound Cs+ in NorM proteins is coordinated by various side chains including Glu-255 and Phe-288 in NorM-VC, and equipositional Glu-261 and Tyr-294 in NorM-NG. Taken together, our findings provide biochemical evidence for a role of regions close to Glu-255 and Asp-36 in Na+-coupled transport by NorM-VC.
FIGURE 5.
Role of Glu-255, Asp-371, and neighboring residues in Na+ coupling. A, B, D, E, and H, active ethidium efflux activity in cells expressing E255Q (A), D371N (B), Y367A (D), F395A (E), or F288A (H) mutant was performed as described in the legend to Fig. 1C. C, F, and G, rates of facilitated ethidium efflux in ATP-depleted cells containing D371N (C), F395A (F), or F288A (G) mutant were measured as a function of the Na+ concentration as described for Fig. 1D. Error bars represent the mean ± S.E. of four independent observations.
Studies on Proton Coupling Suggest a Role for Asp-371
To test ethidium/proton antiport by NorM-VC, the ability of the protein to release protons in response to ethidium binding was tested in direct pH measurements in a low-buffered environment. In the absence of the protein, the addition of ethidium did not significantly affect the pH (Fig. 6A). However, when 1 μm WT NorM-VC was present, the addition of the ethidium caused acidification in a saturable fashion, suggesting that substrate binding triggered the release of proton. To test whether specific carboxyl residues were responsible for proton release, the experiments were repeated with D371N NorM-VC, and the D36N and E255Q mutants. Although the D371N mutant retained a substantial ethidium transport activity compared with WT, the ethidium binding-induced proton release by the mutant was absent compared with WT and in the same range as the minor pH changes observed in solutions without NorM-VC protein (pH decrease in units/min is 3.5 ± 0.8 × 10−3 for WT versus 0.8 ± 0.4 × 10−3 for control, p < 0.01; decrease of 1.2 ± 0.6 × 10−3 for D371N is not significantly different from control) (Fig. 6A). The other two mutants (D36N and E255Q) were too transport-inactive to reveal significant proton release (Fig. 6A), and served as controls showing that proton release from NorM-VC is dependent on a functional transport reaction. Hence, Asp-371 is an important contributor to ethidium binding-induced proton release from NorM-VC.
FIGURE 6.
Studies on H+ coupling. A, effect of saturating amounts of ethidium (Et) on the pH of a low-buffered solution containing purified WT NorM-VC, carboxyl to amide mutants or no protein (CTR). In the histogram, only the decrease in pH/min for WT was significantly elevated compared with CTR (p < 0.005). B, ethidium transport in proteoliposomes containing purified D371N NorM-VC with imposed standard SMF ± PMF (as described in Fig. 2B), or Δψ (interior positive) or PMF (interior positive and acid) in the presence of Na+ ([Na+]in = [Na+]out = 1 mm as described in the legend to Fig. 2C). C, steady-state level in B was determined in four independent experiments. Error bars represent the mean ± S.E. **, p < 0.05, significantly different. The difference between ethidium uptake in the presence of SMF versus SMF + PMF was not significant for the D371A mutant.
The role of Asp-371 in proton coupling was also investigated in proteoliposomes, by comparing the energetics of WT and D371N NorM-VC-mediated ethidium transport. Whereas the imposition of the SMF stimulated the transport of ethidium by the D371N mutant, the imposition of the PMF in the presence of Na+ ([Na+]in = [Na+]out = 1 mm) failed to do so (Fig. 6B). The imposed PMF also failed to significantly enhance SMF-dependent ethidium transport (Fig. 6, B and C). Hence, the D371N mutant lost the PMF dependence of WT protein (Fig. 2, B and C). Furthermore, in contrast to the observations on electrogenic transport by WT, the imposition of the Δψ failed to stimulate ethidium transport by the D371N mutant (Fig. 6B). Taken together, these results indicate a role of Asp-371 in proton coupling in NorM-VC.
DISCUSSION
The use of purified NorM of V. cholerae in artificial phospholipid vesicles (proteoliposomes) in which the composition, magnitude, and orientation of artificial (electro)chemical ion gradients can be carefully manipulated, allowed us to study the coupling mechanism of NorM-VC in detail. We show that ethidium transport by the protein is electrogenic and dependent on the SMF. Surprisingly, these experiments indicate also that the PMF can drive ethidium efflux by NorM-VC in a sodium-dependent fashion (Figs. 1, 4, and 5). To our knowledge, the dependence of NorM-VC on both SMF and PMF is unprecedented for secondary active multidrug transporters.
Recent atomic resolution structures of membrane transporters and channels have shown that binding and transport of specific ions is based on a very precise selectivity for specific ions (32). Carboxyl groups have been implicated in proton binding and dissociation reactions and in interactions with cationic substrates. In recent studies on the proton/drug antiporter LmrP from L. lactis, for example, Asp-142 in the N-terminal half was found to act as a dedicated proton-binding site that facilitates conformational changes underlying the transport cycle. In addition, Asp-235 and Glu-327 in the C-terminal half were found to interact with protons and to facilitate the binding of cationic substrates (12, 25, 27). On the other hand, in the crystallized bacterial sodium-leucine symporter LeuT, one binding site for dehydrated Na+ is formed by a carboxylate moiety and neighboring main chain carbonyl and side chain amide atoms (33). Hence, carboxylates can be part of Na+ binding sites but can also act as proton-binding sites. As the relevance of carboxylates in Na+-dependent MATE transporters has been discussed only in the context of Na+ and cationic substrates (13–16, 20), our novel observations on the SMF and PMF dependence of NorM-VC-mediated transport lead us to re-examine the functional roles of the catalytically important carboxylate Asp-36 (in TM1) in the N-terminal half, and Glu-255 (in TM7) and Asp-371 (in TM10) in a network of aromatic residues in the C-terminal half (Fig. 3). Detailed close-up views on carboxyl-containing regions in NorM-VC are provided by He et al. (14).
The crystal structures of NorM-VC bound to Na+ analogues Rb+ or Cs+ suggested a potential cation-binding site near Asp-371 and Glu-255 (14). Although in our experiments the E255Q replacement could not be tested directly in a reliable fashion in transport experiments due to the severe loss of activity, the residual activity of the D371N mutant is very substantial. Interestingly, the D371N mutation had no significant effect on apparent affinity for Na+ in the ethidium transport reaction (Fig. 5, B and C), making coordination of Na+ by the presumed dyad formed by Glu-255 and Asp-371 in NorM-VC (14, 20) less likely. Indeed, in current structures Asp-371 in NorM-VC and Asp-367 in NorM-NG are located relatively far from the bound Cs+ at a distance of more than 7.2 Å (14, 15). In view of the smaller radius of Na+ compared with Cs+, this distance is expected to be even larger for Na+. The F288A replacement caused a significant 4-fold decrease in the apparent affinity for Na+ (Fig. 5G), consistent with crystal structures of NorM proteins in which Glu-255 and Phe-288 in NorM-VC, and Glu-261 and Tyr-294 in NorM-NG contribute to the coordination of bound Cs+ together with other neighboring aromatic side chains.
In the NorM-VC structures (14), Asp-36 (TM1) is located in the N-terminal helical bundle in proximity to Asn-174 and Asn-178 (TM5). The D36N substitution substantially reduces the activity of NorM-VC, and is associated with a loss of Na+ dependence in residual ethidium efflux activity (Fig. 4A). The substantial transport activity of the N174A mutant allowed a detailed characterization of the transport reactions that are catalyzed. Although the apparent affinity for Na+ in the ethidium efflux reaction by WT protein is high (Kt = 73 ± 4 μm Na+), the N174A mutation decreases this affinity by 1200-fold, pointing to an important role of this region of the N-terminal helical bundle of NorM-VC in Na+ binding (Fig. 4D). The notion that Asp-36 and neighboring residues at the external side of NorM-VC contribute to a second Na+ binding site is consistent with the evidence for two communicating Na+ binding sites in our kinetic experiments with the N174A mutant (Fig. 4D) but is not evident in current x-ray crystal structures (14). High affinity binding of Na+ at the Asp-36 site could facilitate Na+ binding at the Glu-255 site in a transient conformation that precedes the crystallized outward-facing state.
In our studies in proteoliposomes, the D371N was the only replacement that completely abrogated ΔpH and Δψ dependence, but not the ΔpNa dependence, of ethidium transport by NorM-VC (Fig. 6, B and C). Furthermore, whereas ethidium binding causes proton release from WT protein, this phenomenon was not observed for the transport active D371N mutant (Fig. 6A). These results highlight the importance of Asp-371 in proton coupling in NorM-VC, and point to the loss of proton in the antiport reaction catalyzed by the D371N mutant. These data suggest an apparent coupling stoichiometry of electroneutral 1Na+/Et+ for the D371N mutant versus electrogenic (1Na+, 1H+)/Et+ for WT NorM-VC. The observations that ethidium binding to NorM-VC is associated with H+ release (Fig. 6A) but is stimulated by Na+ binding (Fig. 1B) point to different roles of these coupling ions in the ethidium transport reaction. As the D371N mutation does not affect the interaction of NorM-VC with ethidium (Table 3), ethidium binding-induced proton release is most likely based on indirect competition between these cations for binding by NorM-VC. Our data are consistent with recent conclusions that additional factors beyond simple mutual exclusivity of binding of Na+ and drug govern Na+-drug coupling during antiport by NorM (34).
In summary, MATE transporters are thought to fall into two classes that couple substrate efflux to the influx of either Na+ or H+. We establish that NorM-VC simultaneously couples to the sodium-motive force and proton-motive force, and biochemically identify protein regions and residues that play important roles in sodium and proton binding. As the positions of protons are not available in current medium and high-resolution crystal structures of NorM, our findings add a new layer of complexity to mechanistic interpretations of these structures.
Acknowledgments
We thank Zhen Tong for technical assistance, Geoffrey Chang for the generous gift of plasmid pET19b-NorM-VC, and Glenn W. Kaatz and Bryan Schindler for discussions.
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC).
- MATE
- multidrug and toxin extrusion
- DDM
- n-dodecyl-β-d-maltoside
- Δψ
- membrane potential, ΔpH, chemical proton gradient
- ΔpNa
- chemical sodium gradient
- NorM-NG
- NorM from N. gonorrhoeae
- NorM-VC
- NorM from V. cholerae
- NorM-VP
- NorM from V. parahaemolyticus
- PMF
- proton-motive force
- SMF
- sodium-motive force
- TMs
- transmembrane helices
- a.u.
- arbitrary unit
- EMA
- ethidium monoazide.
REFERENCES
- 1. Li X. Z., Nikaido H. (2009) Efflux-mediated drug resistance in bacteria: an update. Drugs 69, 1555–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McAleese F., Petersen P., Ruzin A., Dunman P. M., Murphy E., Projan S. J., Bradford P. A. (2005) A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrob. Agents Chemother. 49, 1865–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaatz G. W., McAleese F., Seo S. M. (2005) Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 49, 1857–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Omote H., Hiasa M., Matsumoto T., Otsuka M., Moriyama Y. (2006) The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 27, 587–593 [DOI] [PubMed] [Google Scholar]
- 5. Kuroda T., Tsuchiya T. (2009) Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta 1794, 763–768 [DOI] [PubMed] [Google Scholar]
- 6. Becker M. L., Visser L. E., van Schaik R. H., Hofman A., Uitterlinden A. G., Stricker B. H. (2010) Interaction between polymorphisms in the OCT1 and MATE1 transporter and metformin response. Pharmacogenet. Genomics 20, 38–44 [DOI] [PubMed] [Google Scholar]
- 7. Tsuda M., Terada T., Mizuno T., Katsura T., Shimakura J., Inui K. (2009) Targeted disruption of the multidrug and toxin extrusion 1 (mate1) gene in mice reduces renal secretion of metformin. Mol. Pharmacol. 75, 1280–1286 [DOI] [PubMed] [Google Scholar]
- 8. Tanihara Y., Masuda S., Sato T., Katsura T., Ogawa O., Inui K. (2007) Substrate specificity of MATE1 and MATE2-K, human multidrug and toxin extrusions/H+-organic cation antiporters. Biochem. Pharmacol. 74, 359–371 [DOI] [PubMed] [Google Scholar]
- 9. Matsushima S., Maeda K., Inoue K., Ohta K. Y., Yuasa H., Kondo T., Nakayama H., Horita S., Kusuhara H., Sugiyama Y. (2009) The inhibition of human multidrug and toxin extrusion 1 is involved in the drug-drug interaction caused by cimetidine. Drug Metab. Dispos. 37, 555–559 [DOI] [PubMed] [Google Scholar]
- 10. Nakamura T., Yonezawa A., Hashimoto S., Katsura T., Inui K. (2010) Disruption of multidrug and toxin extrusion MATE1 potentiates cisplatin-induced nephrotoxicity. Biochem. Pharmacol. 80, 1762–1767 [DOI] [PubMed] [Google Scholar]
- 11. Brown M. H., Paulsen I. T., Skurray R. A. (1999) The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 31, 394–395 [DOI] [PubMed] [Google Scholar]
- 12. van Veen H. W. (2010) Structural biology: last of the multidrug transporters. Nature 467, 926–927 [DOI] [PubMed] [Google Scholar]
- 13. Otsuka M., Yasuda M., Morita Y., Otsuka C., Tsuchiya T., Omote H., Moriyama Y. (2005) Identification of essential amino acid residues of the NorM Na+/multidrug antiporter in Vibrio parahaemolyticus. J. Bacteriol. 187, 1552–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He X., Szewczyk P., Karyakin A., Evin M., Hong W. X., Zhang Q., Chang G. (2010) Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 467, 991–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu M., Symersky J., Radchenko M., Koide A., Guo Y., Nie R., Koide S. (2013) Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl. Acad. Sci. U.S.A. 110, 2099–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hashimoto K., Ogawa W., Nishioka T., Tsuchiya T., Kuroda T. (2013) Functionally cloned pdrM from Streptococcus pneumoniae encodes a Na+ coupled multidrug efflux pump. PLoS One 8, e59525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He G. X., Kuroda T., Mima T., Morita Y., Mizushima T., Tsuchiya T. (2004) An H+-coupled multidrug efflux pump, PmpM, a member of the MATE family of transporters, from Pseudomonas aeruginosa. J. Bacteriol. 186, 262–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tanaka Y., Hipolito C. J., Maturana A. D., Ito K., Kuroda T., Higuchi T., Katoh T., Kato H. E., Hattori M., Kumazaki K., Tsukazaki T., Ishitani R., Suga H., Nureki O. (2013) Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 496, 247–251 [DOI] [PubMed] [Google Scholar]
- 19. Lu M., Radchenko M., Symersky J., Nie R., Guo Y. (2013) Structural insights into H+-coupled multidrug extrusion by a MATE transporter. Nat. Struct. Mol. Biol. 20, 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vanni S., Campomanes P., Marcia M., Rothlisberger U. (2012) Ion binding and internal hydration in the multidrug resistance secondary active transporter NorM investigated by molecular dynamics simulations. Biochemistry 51, 1281–1287 [DOI] [PubMed] [Google Scholar]
- 21. de Ruyter P. G., Kuipers O. P., Beerthuyzen M. M., van Alen-Boerrigter I., de Vos W. M. (1996) Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178, 3434–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Putman M., van Veen H. W., Poolman B., Konings W. N. (1999) Restrictive use of detergents in the functional reconstitution of the secondary multidrug transporter LmrP. Biochemistry 38, 1002–1008 [DOI] [PubMed] [Google Scholar]
- 23. Venter H., Velamakanni S., Balakrishnan L., van Veen H. W. (2008) On the energy-dependence of Hoechst 33342 transport by the ABC transporter LmrA. Biochem. Pharmacol. 75, 866–874 [DOI] [PubMed] [Google Scholar]
- 24. Woebking B., Velamakanni S., Federici L., Seeger M. A., Murakami S., van Veen H. W. (2008) Functional role of transmembrane helix 6 in drug binding and transport by the ABC transporter MsbA. Biochemistry 47, 10904–10914 [DOI] [PubMed] [Google Scholar]
- 25. Schaedler T. A., van Veen H. W. (2010) A flexible cation binding site in the multidrug major facilitator superfamily transporter LmrP is associated with variable proton coupling. FASEB J. 24, 3653–3661 [DOI] [PubMed] [Google Scholar]
- 26. Ambudkar S. V., Zlotnick G. W., Rosen B. P. (1984) Calcium efflux from Escherichia coli: evidence for two systems. J. Biol. Chem. 259, 6142–6146 [PubMed] [Google Scholar]
- 27. Schaedler T. A., Tong Z., van Veen H. W. (2012) The multidrug transporter LmrP protein mediates selective calcium efflux. J. Biol. Chem. 287, 27682–27690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Venter H., Shilling R. A., Velamakanni S., Balakrishnan L., Van Veen H. W. (2003) An ABC transporter with a secondary-active multidrug translocator domain. Nature 426, 866–870 [DOI] [PubMed] [Google Scholar]
- 29. Wang W., van Veen H. W. (2012) Basic residues R260 and K357 affect the conformational dynamics of the major facilitator superfamily multidrug transporter LmrP. PLoS One 7, e38715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fluman N., Ryan C. M., Whitelegge J. P., Bibi E. (2012) Dissection of mechanistic principles of a secondary multidrug efflux protein. Mol. Cell 47, 777–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Makarova K., Slesarev A., Wolf Y., Sorokin A., Mirkin B., Koonin E., Pavlov A., Pavlova N., Karamychev V., Polouchine N., Shakhova V., Grigoriev I., Lou Y., Rohksar D., Lucas S., Huang K., Goodstein D. M., Hawkins T., Plengvidhya V., Welker D., Hughes J., Goh Y., Benson A., Baldwin K., Lee J. H., Díaz-Muñiz I., Dosti B., Smeianov V., Wechter W., Barabote R., Lorca G., Altermann E., Barrangou R., Ganesan B., Xie Y., Rawsthorne H., Tamir D., Parker C., Breidt F., Broadbent J., Hutkins R., O'Sullivan D., Steele J., Unlu G., Saier M., Klaenhammer T., Richardson P., Kozyavkin S., Weimer B., Mills D. (2006) Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U.S.A. 103, 15611–15616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gouaux E., Mackinnon R. (2005) Principles of selective ion transport in channels and pumps. Science 310, 1461–1465 [DOI] [PubMed] [Google Scholar]
- 33. Yamashita A., Singh S. K., Kawate T., Jin Y., Gouaux E. (2005) Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223 [DOI] [PubMed] [Google Scholar]
- 34. Steed P. R., Stein R. A., Mishra S., Goodman M. C., McHaourab H. S. (2013) Na+-substrate coupling in the multidrug antiporter NorM probed with a spin-labeled substrate. Biochemistry 52, 5790–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]