Background: Tibetans have a genetic signature in the coding region of their PHD2 gene.
Results: Tibetan PHD2 variant displays markedly impaired binding to the HSP90 cochaperone p23.
Conclusion: Because p23 couples PHD2 to HIF-α hydroxylation, Tibetans possess a loss of function PHD2 allele.
Significance: This study uncovers a mechanism for Tibetan adaptation to high altitude.
Keywords: Evolution, Hypoxia, Hypoxia-inducible Factor, Hypoxia-inducible Factor (HIF), Mutant, EGLN1, PHD2
Abstract
The Tibetan population has adapted to the chronic hypoxia of high altitude. Tibetans bear a genetic signature in the prolyl hydroxylase domain protein 2 (PHD2/EGLN1) gene, which encodes for the central oxygen sensor of the hypoxia-inducible factor (HIF) pathway. Recent studies have focused attention on two nonsynonymous coding region substitutions, D4E and C127S, both of which are markedly enriched in the Tibetan population. These amino acids reside in a region of PHD2 that harbors a zinc finger, which we have previously discovered binds to a Pro-Xaa-Leu-Glu (PXLE) motif in the HSP90 cochaperone p23, thereby recruiting PHD2 to the HSP90 pathway to facilitate HIF-α hydroxylation. We herein report that the Tibetan PHD2 haplotype (D4E/C127S) strikingly diminishes the interaction of PHD2 with p23, resulting in impaired PHD2 down-regulation of the HIF pathway. The defective binding to p23 depends on both the D4E and C127S substitutions. We also identify a PXLE motif in HSP90 itself that can mediate binding to PHD2 but find that this interaction is maintained with the D4E/C127S PHD2 haplotype. We propose that the Tibetan PHD2 variant is a loss of function (hypomorphic) allele, leading to augmented HIF activation to facilitate adaptation to high altitude.
Introduction
Approximately 25,000 years ago, humans colonized the Tibetan plateau, which has an altitude of ∼14,000 feet. Tibetan adaptation to the extreme conditions of this plateau, particularly the marked hypoxia at this altitude (oxygen concentration is 60% of that at sea level), has provided one of the most striking examples of human adaptation (1, 2). Compared with Andeans, who also reside at high altitude, Tibetans display relatively low red cell mass and pulmonary arterial pressure, and relatively high ventilation and exhaled nitric oxide (2). A series of genome-wide investigations of the Tibetan population have recently provided convincing evidence for genetic adaptation (3–9). These studies consistently identified genetic signatures in the EGLN1 (also known as prolyl hydroxylase domain protein 2, or PHD2)2 and EPAS1 (also known as hypoxia-inducible factor-2α, or HIF2A) genes in the Tibetan population (10).
PHD2 and HIF2A are compelling candidate genes in this population because of the key positions that they occupy in the HIF pathway, which is the central pathway for transducing changes in oxygen tension to changes in gene expression (11–13). Under normoxic conditions, PHD2 site-specifically prolyl hydroxylates the α subunit of HIF (of which there are three paralogues: HIF1-α, HIF-2α, and HIF-3α). In HIF-1α, the primary site of hydroxylation is Pro-564; in HIF-2α, it is Pro-531 (14). This post-translational modification allows recognition by the von Hippel-Lindau tumor suppressor protein (VHL), a component of an E3 ubiquitin ligase complex that then targets HIF-α for degradation by the ubiquitin-proteasome pathway. Under hypoxic conditions, prolyl hydroxylation is attenuated, leading to the stabilization of HIF-α, its dimerization with HIF-β, and activation of HIF target genes. These genes participate in a broad range of processes that promote systemic and cellular adaptation to hypoxia. HIF-1α and HIF-2α have overlapping as well as distinct gene targets. For example, HIF-1α activates many of the genes that encode for glycolytic enzymes, whereas HIF-2α is the principal regulator of the ERYTHROPOIETIN (EPO) gene (13, 14).
The current challenge is to identify the molecular basis for how Tibetan-associated haplotypes in the PHD2 and HIF2A genes promote adaptation to high altitude. With regard to the PHD2 gene, resequencing of this gene in 46 Tibetan individuals ruled out a classic de novo mutation sweep (15). Instead, this study reported on selection from standing variation. In particular, there is significant enrichment in the Tibetan population of two nonsynonymous coding region single-nucleotide polymorphisms, originally identified by others (16), that reside in exon 1 of the PHD2 gene: rs186996510 and rs12097901. The single-nucleotide polymorphisms are predicted to produce D4E and C127S amino acid substitutions, respectively, in the PHD2 protein.
Amino acid residues that are essential for the catalytic activity of PHD2 are encoded for largely by exons 2–5 of the PHD2 gene. Exon 1 (amino acids 1–297), in contrast, is most notable for a zinc finger of the MYND (myloid, Nervy, DEAF-1) type, which encompasses amino acids 21–58. This zinc finger shows strong phylogenetic conservation (17, 18) and in fact is present in the PHD2 orthologue in the simplest metazoan, Trichoplax adhaerens (17). We have recently discovered that the function of this zinc finger is to couple PHD2 to the HSP90 pathway (19). Specifically, it binds to a Pro-Xaa-Leu-Glu (PXLE) motif that is present in select HSP90 cochaperones such as p23, and this allows recruitment of PHD2 to HSP90 to facilitate prolyl hydroxylation of HIF-1α, which itself is an HSP90 client protein (20–22).
The presence of the Tibetan-associated rs186996510 (D4E) and rs12097901 (C127S) single-nucleotide polymorphisms in exon 1 of the PHD2 gene led us to explore the possibility that this haplotype might affect PHD2 function, potentially through its zinc finger. Here, we provide evidence that the Tibetan D4E/C127S haplotype impairs PHD2 function and that mechanistically, it leads to a dramatically decreased ability of the PHD2 zinc finger to associate with the HSP90 cochaperone p23. Remarkably, this impairment depends on both the D4E and C127S amino acid substitutions. On the basis of these results, we conclude that the Tibetan PHD2 D4E/C127S haplotype leads to a loss of function (hypomorphic) PHD2 allele, and we propose that this is a key mechanism by which the HIF pathway has been reconfigured for adaptation to chronic hypoxia in this population.
EXPERIMENTAL PROCEDURES
Plasmids
pcDNA3-HA-PHD2 (1–131) was constructed by subcloning the 0.4-kb BamHI/NotI (partial digest) fragment encoding residues 1–131 of pcDNA3-HA-PHD2 (1–196) (19) into the BamHI/NotI site of pcDNA3-HA. pcDNA3-HA-PHD2 (1–173) was constructed by digesting pcDNA3-HA-PHD2 (1–196) with BlpI/XhoI, blunting the ends with the Klenow fragment of Escherichia coli DNA polymerase I, and then religating.
pcDNA3-HA-PHD2 (1–131) D4E/C127S was constructed by amplifying by PCR a 0.4-kb band using pcDNA3-HA-PHD2 (19) as a template and the following primers: PHD2 D4E 5′, 5′-AAG GGG ATC CGA ATT CTT AAG CTC GAC ATG GCC AAT GAG AGC GGC GGT CCC GGC GGG CCG AGC CCG AGC GAG-3′; and PHD2 C127S 3′, 5′-GCC GGC GGC CGC TCT GGA GGG GCT AGC AGC CGC CGC TGG GTC GGC CGG-3′. The product was digested with BamHI and NotI and subcloned into the BamHI/NotI site of pcDNA3-HA to give pcDNA3-HA-PHD2 (1–131) D4E/C127S. pcDNA3-HA-PHD2 (1–196) D4E/C127S was constructed by subcloning the 0.23-kb NotI fragment pcDNA3-HA-PHD2 (1–196) into the NotI site of pcDNA3-HA-PHD2 (1–131) D4E/C127S. pcDNA3-HA-PHD2 (1–173) D4E/C127S was constructed by digesting pcDNA3-HA-PHD2 (1–196) D4E/C127S with BlpI/XhoI, blunting the ends with the Klenow fragment of E. coli DNA polymerase I, and then religating. pcDNA3-HA-PHD2 D4E/C127S was constructed by subcloning the 0.6-kb BamHI/XhoI fragment from pcDNA3-HA-PHD2 (1–196) D4E/C127S into the BamHI/XhoI site of pcDNA3-HA-PHD2 (130–426) (19). pcDNA5/FRT/TO-FlagPHD2 D4E/C127S was constructed by subcloning the 0.6-kb BamHI/XhoI fragment of pcDNA3-HA-PHD2 (1–196) D4E/C127S into the BamHI/XhoI site of pcDNA5/FRT/TO-FlagPHD2 (19). pcDNA3-FlagPHD2 D4E/C127S was constructed by subcloning the 0.6 kb BamHI/XhoI fragment of pcDNA3-HA-PHD2 (1–196) D4E/C127S into the BamHI/XhoI site of pcDNA3-FlagPHD2 (23).
pcDNA3-HA-PHD2 (1–196) D4E was constructed by amplifying by PCR a 0.6-kb band using pcDNA3-HA-PHD2 (1–196) as a template and the following primers, PHD2 D4E 5′ (see above) and BGH rev: 5′-TAG AAG GCA CAG TCG AGG-3′. The product was digested with BamHI and XhoI and subcloned into the BamHI/XhoI site of pcDNA3-HA to give pcDNA3-HA-PHD2 (1–196) D4E. pcDNA3-HA-PHD2 D4E was constructed by subcloning the 0.6-kb BamHI/XhoI fragment from pcDNA3-HA-PHD2 (1–196) D4E into the BamHI/XhoI site of pcDNA3-PHD2 (130–426).
pcDNA3-HA-PHD2 (1–131) C127S was constructed by amplifying by PCR a 0.4-kb band using pcDNA3-HA-PHD2 as a template and the following primers: CMV 5′, 5′-GCG TGT ACG GTG GGA GGT C-3′; and PHD2 C127S 3′ (see above). The product was digested with BamHI and NotI and subcloned into the BamHI/NotI site of pcDNA3-HA to give pcDNA3-HA-PHD2 (1–131) C127S. pcDNA3-HA-PHD2 (1–196) C127S was prepared by subcloning the 0.23-kb NotI fragment pcDNA3-HA-PHD2 (1–196) into the NotI site of pcDNA3-HA-PHD2 (1–131) C127S. pcDNA3-HA-PHD2 C127S was constructed by subcloning the 0.6-kb BamHI/XhoI fragment from pcDNA3-HA-PHD2 (1–196) C127S into the BamHI/XhoI site of pcDNA3-PHD2 (130–426). pFastBac-HT-FlagPHD2 D4E/C127S was prepared by subcloning the 0.4-kb BamHI/NotI fragment of pcDNA3-HA-PHD2 (1–196) D4E/C127S into the BamHI/NotI site of pFastBac-HT-FlagPHD2 (23).
pcDNA3-Flag-p23 W106A was constructed by overlapping PCR. In the first round of PCR, we performed two PCRs. We amplified a 0.32-kb band using pcDNA5/FRT/TO-3×Flag-p23 as a template and the following primers: CMV 5′, 5′-GCG TGT ACG GTG GGA GGT C-3′; and P23 W106A 3′, 5′-GAA TCA TCT TCC CAG TCT TTC GCA TTA TTG AAA TCG ACA CTA AGC CAA TTA AG-3′. We amplified a 0.2-kb band using pcDNA5/FRT/TO-3×Flag-p23 as a template and the following primers: P23 W106A 5′, 5′-CTT AAT TGG CTT AGT GTC GAT TTC AAT AAT GCG AAA GAC TGG GAA GAT GAT TC-3′; and BGH rev, 5′-TAG AAG GCA CAG TCG AGG-3′. The two PCR products were mixed and employed as template in a second round of PCR using the CMV 5′ and BGH rev primers. The product was digested with BamHI and XhoI and subcloned into the BamHI/XhoI site of pcDNA5/FRT/TO-3×Flag.
The plasmid Hsp90 HA (β isoform) (24) was obtained from Addgene (plasmid 22487). pFastBac-HT-HA-HSP90β was constructed by subcloning the 2.2-kb NotI/XbaI fragment of Hsp90 HA into the NotI/XbaI site of pFastBacHTc. pFastBac-HT-HA-HSP90β P709A/L711A/E712A was constructed by standard recombinant DNA methods.
pFastBac-HT-FKBP38 (FK506-binding protein 38) was constructed by subcloning the 1.2-kb BamHI/NotI fragment of pcDNA5/FRT/TO-3×Flag-FKBP38 (19) into the BamHI/NotI site of pFastBacHTc. pFastBac-HT-FlagHIF-1α was prepared by subcloning the 3.2-kb BamHI/XbaI fragment of pcDNA3-FlagHIF-1α (25) into the BamHI/XbaI site of pFastBacHTb. pFastBac-HT-HA-VHL was prepared by subcloning the 0.7-kb NcoI/XhoI fragment of pcDNA3-HA-VHL (23) into the NcoI/XhoI site of pFastBacHTc. pFastBac-HT-p23 was constructed by subcloning the 0.5-kb BamHI/XhoI fragment of pcDNA5/FRT/TO-3×Flag-p23 (19) into the BamHI/XhoI site of pFastBac-HTc. pFastBac-HT-HA-p23 was constructed by subcloning the 0.5-kb BamHI/XhoI fragment of pcDNA5/FRT/TO-3×Flag-p23 into the BamHI/XhoI site of pFastBac-HT-HA-VHL. The sources of pGEX-HIF-1α (531–575), pGEX-HIF-2α (516–549), and all other plasmids have been described (19, 26).
Baculoviruses
Baculoviruses for HT-FlagPHD2 D4E/C127S, HT-p23, HT-HA-p23, HT-HA-HSP90β, HT-HA-HSP90β P709A/L711A/E712A, HT-FKBP38, HT-FlagHIF-1α, and HT-HA-VHL were prepared using the appropriate pFastBac vectors and the Bac-To-Bac system (Invitrogen).
Proteins
GST, GST-HIF-1α (531–575), and GST-HIF-2α (516–549) were purified as described (19). His6-tagged (HT)-FlagPHD2, HT-FlagPHD2 D4E/C127S, HT-p23, HT-HA-p23, HT-HA-HSP90β, HT-HA-HSP90β P709A/L711A/E712A, HT-FKBP38, and HT-FlagHIF-1α were purified from Sf9 cells infected with the appropriate baculovirus using nickel-nitrilotriacetic agarose (Qiagen). HT-HA-VHL was purified as a complex with ElonginB and ElonginC following coinfection of Sf9 cells with a baculovirus for HT-HA-VHL and one for ElonginB/ElonginC (23).
Mass Spectrometry
WT and D4E/C127S HT-FlagPHD2 purified from baculovirus-infected Sf9 cells were subjected to filter-aided sample preparation (27). Mass spectrometry was then performed as described (19) with the following modifications: fragmentation spectra (MS2s) were selected from within a reduced parent mass scan range (600–800 m/z) and using a targeted parent mass list designed to selectively acquire spectra for the indicated peptides. Masses selected for fragmentation included both fully and semitryptic peptides in addition to mass shifts associated with zero, one, or two serine phosphorylation events (+79.966 Da). The data were analyzed using the MaxQuant analytical package (28) version 1.2.2.5 and included N-terminal acetylation, oxidation of methionine, and phosphorylation of serine (with potential accompanying neutral losses) as variable modifications and carbamidomethylation of cysteine as a fixed modification.
Prolyl Hydroxylase Assays
Recombinant WT or D4E/C127S HT-FlagPHD2 was incubated with substrate in a buffer consisting of 20 mm Hepes, pH 8.0, 100 mm KCl, 10 μm FeCl2, 1 mm 2-oxoglutarate, 5 mm ascorbate, 1 mm DTT, and 1 μm ZnCl2 at 37 °C. For hypoxic conditions, the reactions were conducted in either a Ruskinn In Vivo 200 hypoxia work station or a Billups Rothenberg modular incubator perfused with a gas mixture containing the appropriate oxygen concentration. The products were subjected to SDS-PAGE, transferred to Immobilon membranes, blocked with PBS containing 0.5% Tween 20, and 1% bovine serum albumin for 30 min, and then incubated with recombinant HT-HA-VHL (in complex with ElonginB and ElonginC) for 1 h in the same buffer at 4 °C. Following washing, HT-HA-VHL was visualized using anti-HA antibodies conjugated to alkaline phosphatase.
Peptide Binding Assays
Peptides corresponding to HSP90β (703–712) and HSP90α (711–720) were synthesized by Genscript. These peptides, as well as one corresponding to p23 (151–160) possessed an N-terminal tyrosine residue to allow spectrophotometric quantitation (19) as well as an N-terminal biotin.
Peptides (0.5 μg) were prebound to 15-μl aliquots of streptavidin-agarose (Sigma). The resins were incubated with Sf9 lysates containing baculovirus-expressed HT-FlagPHD2 (23) for 1 h with rocking at 4 °C in buffer A (20 mm Tris, pH 7.6, 150 mm NaCl, 10% glycerol, 1% Triton X-100). The resins were washed four times with buffer A and eluted, and the eluates were subjected to SDS-PAGE and Western blotting.
Cell Culture
HEK293 FT cells were maintained and transfected with plasmids as described (19). Luciferase assays were performed as described (19). Hypoxia experiments were performed in an In Vivo 200 hypoxia work station (Ruskinn Technologies).
Mouse Embryonic Fibroblast (MEF) Generation
Phd2+/− mice (29) were crossed, and MEFs were obtained from embryonic day 11.5 Phd2−/− embryos by the primary explant technique (30). MEFs were immortalized by transfection with pSG5-T, which contains the coding sequence for the SV40 large T antigen (31), and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml of streptomycin. Wild type MEFs that were immortalized in the same manner were a gift from Mr. Scott Sutherland.
Stable transfectants were generated by transfection of Phd2−/− MEFs with pcDNA3-Flag, pcDNA3-FlagPHD2, or pcDNA3-FlagPHD2 D4E/C127S, followed by selection using 200 μg/ml G418. Individual G418-resistant clones were screened for PHD2 expression level by Western blotting.
Immunoprecipitations
For purified recombinant proteins, proteins were mixed in buffer B (50 mm Tris, pH 7.5, 100 mm NaCl, 0.5% Triton X-100, and 1 μm ZnCl2) supplemented with 1% bovine serum albumin for 1 h at 4 °C. In some cases lysates from 293 FT cells were included. The reactions were then added to 15-μl aliquots of anti-Flag (M2)-agarose and rocked for 1 h at 4 °C. The resins were washed three times with buffer B and eluted, and the eluates were subjected to SDS-PAGE and Western blotting.
For proteins expressed by transient transfection, cells were lysed in buffer B. The lysates were clarified by centrifugation at 15,800 × g for 10 min at 4 °C. The lysates were then added to 15-μl aliquots of anti-Flag (M2)-agarose and rocked for 1 h at 4 °C. The resins were washed three times with buffer B and eluted, and the eluates were subjected to SDS-PAGE and Western blotting. In the experiments in which FlagPHD2 was immunoprecipitated from stably transfected MEFs, the lysates were precleared by incubation with Sepharose CL-4B (Sigma) prior to incubation with anti-Flag agarose. The results presented are representative of two to four independent experiments.
Western Blotting
The sources of antibodies to p23, FKBP38, Flag tag, HA tag, and mouse Hif-1α, as well as the procedure for Western blotting, have been described (19, 29). Rabbit mAb D31E11 to PHD2 was obtained from Cell Signaling Technology. The E7 monoclonal antibody against β-tubulin developed by Dr. Michael Klymkowsky was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, National Institutes of Health and maintained by the University of Iowa Department of Biology (Iowa City, IA). Protein concentrations of extracts were determined by the Bio-Rad DC protein assay.
Statistical Analysis
Unpaired Student's t test was employed for statistical analysis. Differences were considered significant when p < 0.05.
RESULTS
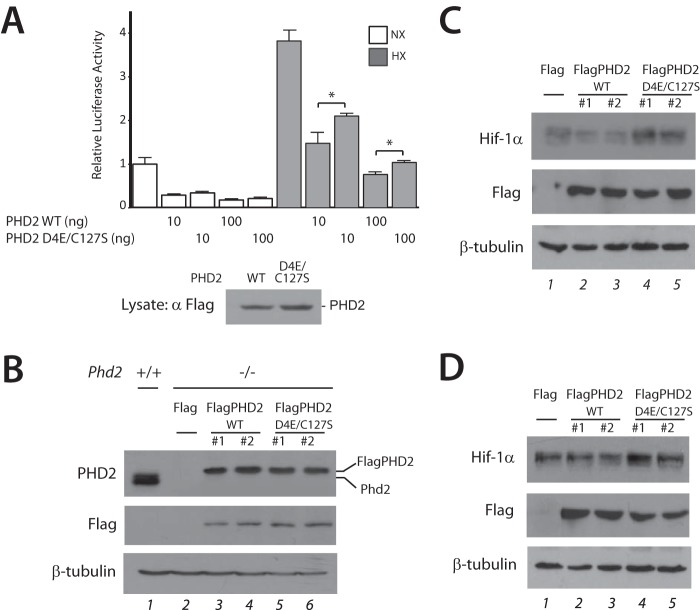
We first transfected HEK293 FT cells with constructs for WT or D4E/C127S PHD2 along with a luciferase reporter gene driven by three copies of a hypoxia response element and maintained cells under normoxia or hypoxia (1% O2). As shown in Fig. 1A, we find that hypoxia activates the reporter gene and that overexpression of WT PHD2 inhibits this activation (seventh and ninth columns). Importantly, we observe that D4E/C127S PHD2 is weaker in its ability to inhibit hypoxia response element activity (eighth and tenth columns).
FIGURE 1.
The D4E/C127S PHD2 haplotype impairs PHD2 regulation of hypoxia response element reporter gene activity and HIF-1α. A, HEK293 FT cells were transfected with 100 ng of (eHRE)3-Luc, 50 ng of pRL-TK, and the indicated doses of pcDNA5/FRT/TO-FlagPHD2 or pcDNA5/FRT/TO-FlagPHD2 D4E/C127S. The total DNA dose was adjusted to 250 ng using pcDNA3. Cells were then exposed to normoxia (NX) or hypoxia (HX, 1% O2) for 16 h, and then firefly luciferase activities were measured and normalized to that of Renilla luciferase. Standard deviations are indicated by the bars (n = 3). *, p < 0.05. Also shown is a Western blot for FlagPHD2. B, MEFs with the indicated genotypes were examined by Western blotting using equal protein amounts for each lane. WT and D4E/C127S indicate that Phd2−/− MEFs were stably transfected with expression vectors for WT and D4E/C127S FlagPHD2, respectively. In lane 2, the Phd2−/− MEFs were stably transfected with pcDNA3-Flag vector. The difference in mobility of bands in the top panel reflects a difference in the molecular masses of murine Phd2 (lane 1, predicted molecular mass = 43 kDa) and Flag-tagged human PHD2 (lanes 3–6, predicted molecular mass = 48 kDa). C and D, Phd2−/− MEFs stably transfected as indicated were subjected to normoxia (C) or hypoxia (D, 5% O2 for 5 h). Equal protein amounts of each cell line were examined by Western blotting.
We generated Phd2−/− MEFs, then stably transfected them with constructs for WT or D4E/C127S PHD2, and then selected two independent clones for each with comparable PHD2 expression levels (Fig. 1B). When exposed to either normoxia or hypoxia (Fig. 1, C and D, respectively), we observe that Hif-1α levels are higher in the D4E/C127S PHD2 expressing MEFs as compared with those expressing WT PHD2 (Fig. 1, C and D top panel, compare lanes 2 and 3 with lanes 4 and 5).
These experiments raise the possibility that D4E/C127S PHD2 might have intrinsically weaker catalytic activity than WT PHD2. To examine this, we prepared recombinant proteins and conducted in vitro prolyl hydroxylase assays. We employed a VHL probe-based far Western analysis, because VHL binds with extremely high specificity to the prolyl hydroxylated form of HIF-α (32, 33). Under a range of oxygen concentrations, including 1, 3, 5, and 21%, we have not seen any consistent differences between WT and D4E/C127S PHD2 in their capacity to hydroxylate GST-HIF-1α (531–575), GST-HIF-2α (516–549), or full-length HIF-1α (data not shown).
Both Asp-4 and Cys-127 reside in exon 1, the most prominent feature of which is a MYND-type zinc finger. This zinc finger binds to a PXLE motif that is found in select HSP90 cochaperones, including p23 and FKBP38 (19). We therefore examined WT and D4E/C127S PHD2 for their capacity to interact with each protein. We first incubated recombinant PHD2 with recombinant p23 or FKBP38, immunoprecipitated the PHD2, and examined the immunoprecipitates. We find that WT PHD2 interacts directly with both p23 and FKBP38; however, we do not observe any significant difference between WT and D4E/C127S PHD2 in their ability to interact with either protein (data not shown).
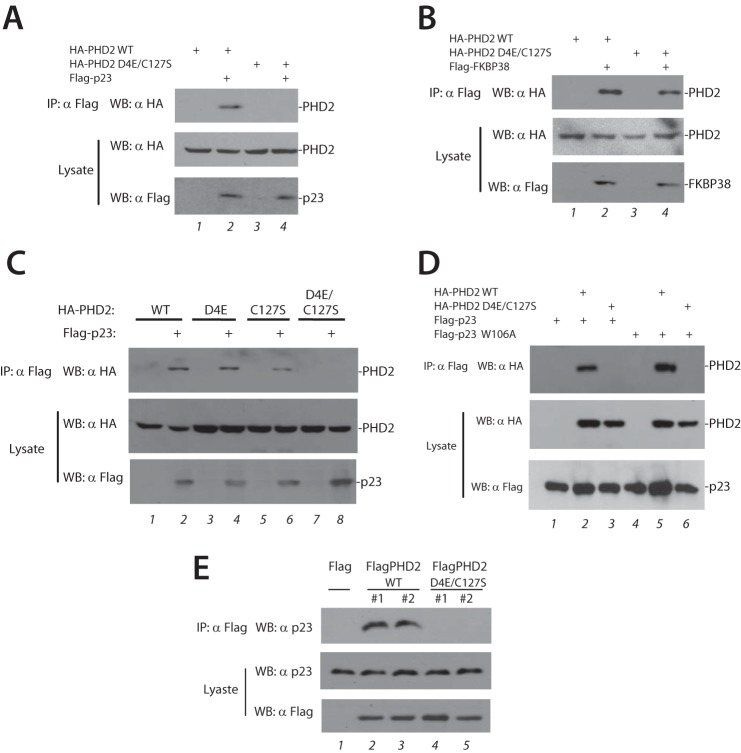
Recognizing that the interaction between purified proteins may not recapitulate their interactions within the cell, we next performed a series of transfection and coimmunoprecipitation studies. We coexpressed either HA-tagged WT or D4E/C127S PHD2 with Flag-p23, immunoprecipitated the latter, and examined the immunoprecipitates for the absence or presence of PHD2. In striking contrast to the results with purified recombinant proteins, we find that D4E/C127S PHD2 is defective in its ability to coimmunoprecipitate with p23 (Fig. 2A, top panel, compare lanes 2 and 4).
FIGURE 2.
The D4E/C127S PHD2 haplotype abolishes interaction with p23. A–D, HEK293 FT cells were transfected with expression plasmids for the indicated proteins. The cells were lysed and subjected to immunoprecipitation with anti-Flag antibodies, and then the immunoprecipitates and aliquots of lysate were examined by Western blotting as indicated. E, Phd2−/− MEFs stably transfected as indicated were lysed and subjected to immunoprecipitation with anti-Flag antibodies, and then the immunoprecipitates and aliquots of lysate were examined by Western blotting as indicated. IP, immunoprecipitation; WB, Western blotting.
When parallel transfection experiments were performed with FKBP38, we find that D4E/C127S PHD2 is only marginally decreased in its interaction as compared with WT (Fig. 2B). We examined the D4E and C127S substitutions individually. Remarkably, we find that both of the individual substitutions preserve the ability of PHD2 to coimmunoprecipitate with p23 (Fig. 2C, top panel, lanes 2, 4, and 6) and that only the combined D4E/C127S substitution abolishes it (Fig. 2C, top panel, lane 8).
It is conceivable that the difference in the results of Fig. 2A and those using purified recombinant proteins may be due to the fact that p23 can exist in a complex with HSP90 within the cell and that the p23·HSP90 complex might interact differently with WT as compared with D4E/C127S PHD2. Previous studies showed that a W106A mutant of p23 is defective in its interaction with HSP90 (34). We therefore tested the role of HSP90-mediated binding by examining the ability of W106A p23 to coimmunoprecipitate PHD2. We find that this mutant p23 can coimmunoprecipitate PHD2 in a manner comparable with WT p23 (Fig. 2D, top panel, lanes 2 and 5). Significantly, the interaction is abolished by the D4E/C127S substitution (Fig. 2D, top panel, lanes 3 and 6). Therefore, the differential interaction of WT versus D4E/C127S PHD2 with p23 is not dependent on the association of p23 with HSP90.
We also examined the aforementioned stably transfected MEFs. WT PHD2 stably expressed in MEFs can coimmunoprecipitate endogenous murine p23 (Fig. 2E, top panel, lanes 2 and 3), and we observe that stably expressed D4E/C127S PHD2 is defective in this capacity (Fig. 2E, top panel, lanes 4 and 5). These results further support the notion that the D4E/C127S substitution impairs function of the zinc finger of PHD2.
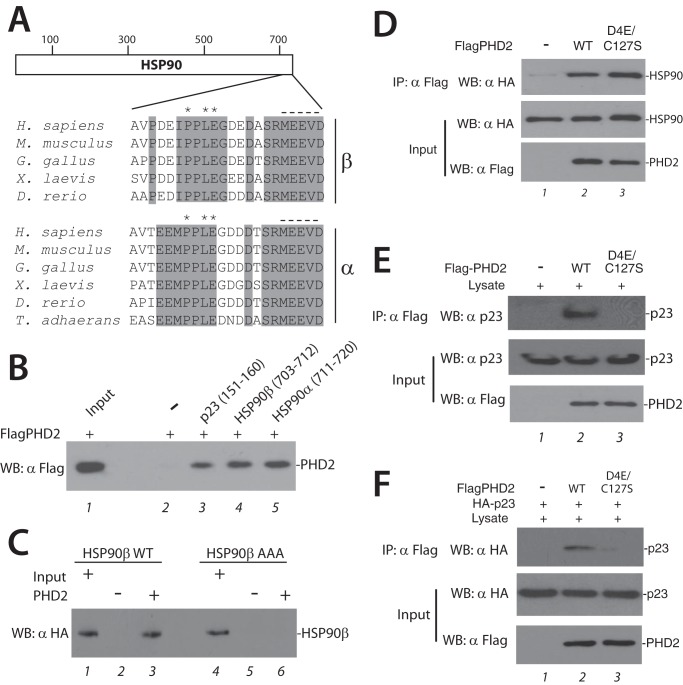
We considered whether the D4E/C127S substitution might also affect interaction with a different PXLE-containing protein. Toward this end, we searched for such a motif and identified one in HSP90 itself (both α and β isoforms; Fig. 3A). As in p23 and FKBP38 (19), the HSP90 PXLE motif is immediately preceded by a hydrophobic residue and is in close vicinity to acidic amino acids that reside N-terminal to the motif. The PXLE motif is conserved in HSP90 from mammalian, bird, amphibian, and fish species, as well as that from the simple metazoan T. adhaerens. In a structure for human HSP90α (293–732) determined by x-ray crystallography, the C-terminal 33 amino acids were not visualized, likely because of high flexibility in this region (35). The last five amino acids of HSP90, MEEVD, are well known to bind to tetratricopeptide repeat domain containing cochaparones such as FKBP51, FKBP52, and FKBP38 (36). To our knowledge, a function for the PXLE motif immediately N-terminal to this MEEVD motif has not yet been assigned.
FIGURE 3.
The D4E/C127S PHD2 haplotype maintains interaction with HSP90. A, schematic diagram of HSP90. The numbers at the top indicate residue numbers. Below are the sequences of the last 22 amino acids from HSP90β and HSP90α from the indicated species. Asterisks indicate P, L, and E of the PXLE motif. The dashed line indicates the MEEVD motif. Shading denotes conservation in the indicated species. B, Sf9 lysates containing HT-Flag-PHD2 were incubated with the indicated biotinylated peptides immobilized on streptavidin-agarose and washed, and the eluates were examined for the presence of PHD2 using anti-Flag antibodies. Input, 1% of total. C, recombinant WT or P709A/L711A/E712A (AAA) HT-HA-HSP90β (2 μg) was incubated with or without HT-FlagPHD2 (2 μg), and the latter was then immunoprecipitated using anti-Flag antibodies. The immunoprecipitates were examined for the presence of HSP90 as shown. Input represents 2% of total. D, recombinant HT-HA-HSP90 (1 μg) was incubated without or with WT or D4E/C127S HT-FlagPHD2 (1 μg), and the latter was then immunoprecipitated using anti-Flag antibodies. The immunoprecipitates were examined for the presence of HSP90 as shown. E, HEK293 FT lysates were incubated without or with recombinant WT or D4E/C127S HT-FlagPHD2 (1 μg), and the latter was then immunoprecipitated using anti-Flag antibodies. The immunoprecipitates and aliquots of lysate were subjected to Western blotting as indicated. F, recombinant HT-HA-p23 (1 μg) was incubated without or with either WT or D4E/C127S HT-FlagPHD2 (1 μg) in the presence of HEK293 FT lysate (100 μg), and the HT-FlagPHD2 was then immunoprecipitated using anti-Flag antibodies. The immunoprecipitates and aliquots of lysate were examined for the presence of HA-p23 as shown. IP, immunoprecipitation; WB, Western blotting.
To investigate the possibility that this motif might bind PHD2, we first immobilized biotinylated peptides with sequences corresponding to HSP90β (703–712), HSP90α (711–720), and, as a positive control, p23 (151–160), on streptavidin-agarose. We incubated recombinant PHD2 with each resin, washed the resins, and then assessed for the absence or presence of PHD2. As shown in Fig. 3B, under conditions where PHD2 binds to the PXLE-containing p23 peptide (lane 3), we also find that it binds PXLE-containing HSP90β and HSP90α peptides (lanes 4 and 5, respectively).
To determine whether this motif in HSP90β (hereafter referred to as HSP90) can mediate binding to PHD2 in the context of full-length HSP90, we prepared recombinant HSP90 from baculovirus-infected Sf9 cells and incubated it with recombinant FlagPHD2. The latter was then immunoprecipitated with anti-Flag antibodies, and the eluates were examined. As shown in Fig. 3C, we observe evidence for a specific interaction between the proteins (compare lanes 2 and 3). Importantly, a mutation of the PXLE motif to AXAA abolishes it (lane 6). We then incubated recombinant PHD2 with recombinant HSP90, immunoprecipitated the PHD2, and examined the immunoprecipitates. As expected, WT PHD2 interacts directly with HSP90 (Fig. 3D, top panel, lane 2). However, we do not observe any appreciable difference between WT and D4E/C127S PHD2 in their ability to interact with HSP90 (lane 3).
We next incubated recombinant Flag-tagged WT or D4E/C127S with HEK293 FT cell lysates, immunoprecipitated the PHD2 with anti-Flag antibodies, and then examined the immunoprecipitates for the absence or presence of endogenous p23 from the lysates. Interestingly, recombinant D4E/C127S PHD2 is substantially weakened in its ability to coimmunoprecipitate p23 (Fig. 3E, top panel, compare lane 2 and 3). We extended these studies by incubating purified Flag-tagged WT or D4E/C127S PHD2 with purified HA-tagged p23 in the presence of HEK293 FT cell lysates, immunoprecipitating the PHD2 with anti-Flag antibodies, and then examining the immunoprecipitates for the recombinant HA-tagged p23. We find that the PHD2-p23 interaction is recapitulated, and importantly, this interaction is diminished by the D4E/C127S haplotype (Fig. 3F, top panel, lanes 2 and 3). Thus, the defective interaction between recombinant D4E/C127S PHD2 and recombinant p23 is observed in the presence of cellular lysate (Fig. 3F), but not in its absence (data not shown).
The aforementioned studies also provide an opportunity to reexamine whether the D4E/C127S substitution affects the ability of PHD2 to interact with the PXLE motif of HSP90. We incubated purified Flag-tagged WT or D4E/C127S PHD2 with purified HA-tagged HSP90 in the presence of HEK293 FT cell lysates, immunoprecipitated the PHD2 with anti-Flag antibodies, and then examined the immunoprecipitates for recombinant HSP90. We observed evidence for the PHD2-HSP90 interaction but, in contrast to results with p23, this interaction is not appreciably affected by the D4E/C127S haplotype (data not shown). From these experiments, we conclude that the D4E/C127S PHD2 haplotype produces a selectively detrimental defect in p23 interaction.
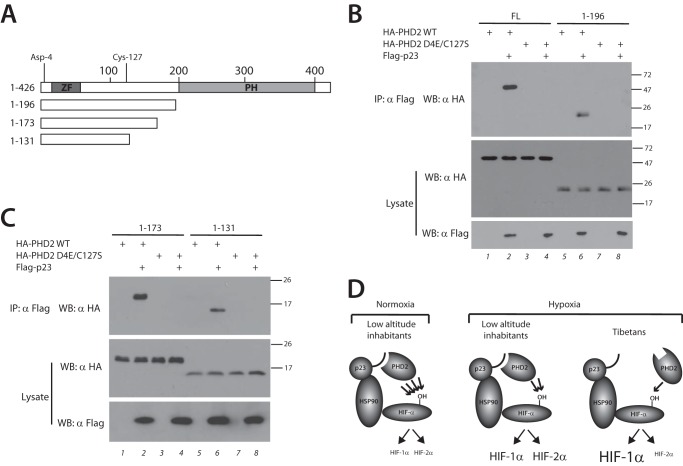
Using transient transfection assays, we next examined whether C-terminal truncations of PHD2 (Fig. 4A) can coimmunoprecipitate with p23. Consistent with our previous study (19), we find that both full-length PHD2 (1–426) and PHD2 (1–196) coimmunoprecipitate with p23 (Fig. 4B, top panel, lanes 2 and 6). We now show here that even smaller fragments of PHD2 that retain the zinc finger, namely PHD2 (1–173) and PHD2 (1–131), also interact with p23 (Fig. 4C, top panel, lanes 2 and 6). Strikingly, the D4E/C127S substitution in each of these constructs abolishes its ability to coimmunoprecipitate with p23 (Fig. 4, B, top panel, lanes 4 and 8, and C, top panel, lanes 4 and 8). These findings highlight the dramatic impact of the D4E/C127S substitution on p23 binding.
FIGURE 4.
Defective p23 binding localizes to the N-terminal domain of PHD2. A, schematic diagram of PHD2. Zinc finger (ZF) and prolyl hydroxylase domain (PH) are indicated. The numbers at the top indicate residue number, with Asp-4 and Cys-127 as shown. Deletions are as indicated. B and C, HEK293 FT cells were transfected with expression plasmids for the indicated proteins. The cells were lysed and subjected to immunoprecipitation with anti-Flag antibodies, and then the immunoprecipitates and aliquots of lysate were examined by Western blotting as indicated. Positions of molecular mass markers (in kDa) are shown to the right. FL, full length; IP, immunoprecipitation; WB, Western blotting. D, model for Tibetan PHD2 variant. Left panel, under normoxic conditions, WT PHD2 in low altitude inhabitants efficiently prolyl hydroxylates HIF-α (indicated by four arrows), leading to low levels of both HIF-1α and HIF-2α. Middle panel, in response to the chronic hypoxia of high altitude, WT PHD2 less efficiently modifies HIF-α (indicated by two arrows), leading to elevated levels of both HIF-1α and HIF-2α. Right panel, the Tibetan PHD2 is impaired relative to WT PHD2 in its ability to hydroxylate HIF-α (indicated by one arrow), leading to an exaggerated HIF-1α response. However, the Tibetan HIF2A allele likely leads to a blunted HIF-2α response, thereby leading to differential activation of HIF-1α versus HIF-2α.
DISCUSSION
We have previously shown that p23 knockdown augments hypoxia-induced HIF-α protein levels and HIF target genes (19), indicating that p23 recruitment to the HSP90 pathway facilitates HIF-α hydroxylation. We now report that the Tibetan PHD2 haplotype disrupts this linkage to p23, leading directly to a model in which the Tibetan variant is impaired in its ability to prolyl hydroxylate HIF-α and hence is a loss of function (hypomorphic) allele (Fig. 4D). This might seem counterintuitive. Low altitude inhabitants who ascend to high altitude (Fig. 4D, middle panel) are predisposed to developing both pulmonary hypertension and erythrocytosis (Monge's disease). A loss of function PHD2 allele would be expected to potentiate HIF-α signaling and thereby exacerbate both.
However, genetic adaptation can result from selection at more than one loci, and multiple studies indicate that Tibetan genetic selection has occurred not only at the PHD2 locus, but also at the HIF2A locus (3, 5–9). Importantly, two of these studies (3, 5) demonstrated both a strong association between the Tibetan HIF2A haplotype and low hemoglobin concentration (which is controlled by the HIF-2α target gene, EPO (37–40)), thereby providing compelling evidence that the HIF2A allele is a loss of function allele. Because HIF-2α also plays a central role in pulmonary hypertension (41–44), the loss of function HIF2A allele would be predicted to blunt the erythrocytosis and pulmonary hypertension that might otherwise result from loss of PHD2 function (Fig. 4D, right panel).
Our model could also allow for potentially beneficial effects of augmented HIF-1α activation, such as increased respiration and nitric oxide production, because HIF-1α plays a role in promoting both (45–48). We therefore propose that the Tibetan population has enriched specific haplotypes of the PHD2 and HIF2A genes to reconfigure the HIF pathway to facilitate high altitude adaptation.
We find a dramatic effect of the D4E/C127S haplotype on PHD2 interaction with p23. Although it was far more significant than the effect seen on PHD2 interaction with either FKBP38 or HSP90, we cannot at present rule out the possibility that subtle alterations in these other interactions may also contribute to the phenotype. Interestingly, the impaired interaction with p23 is seen in the presence of cellular lysates, whether the proteins are transiently expressed, stably expressed, or prepared as recombinant proteins (Figs. 2; 3, E and F; and 4). The impaired interaction is not seen in the absence of lysate (data not shown). This suggests that there may be a conformational change in D4E/C127S versus WT PHD2 in the presence of lysate, a cofactor(s) in the lysate that modulates the PHD2-p23 interaction, or a post-translational modification of PHD2 that is differentially present in D4E/C127S versus WT PHD2. With regard to the latter, the C127S substitution provides a potential phosphoacceptor residue; however, we have thus far not observed phosphorylation at this residue by mass spectrometry (data not shown). The various possibilities mentioned above will require further investigation, as will the possibility that p23 may play a role in the maturation of HIF-α itself. Regardless, the present studies provide genetic evidence for the functional importance of the PHD2-p23 interaction. Moreover, they raise the possibility that therapeutic targeting of this interaction may recapitulate key aspects of adaptation to chronic hypoxia, which is a central feature of common diseases that include ischemic heart, cerebrovascular, and chronic obstructive pulmonary disease.
Acknowledgments
We thank Dr. Melanie Percy, Dr. Terence Lappin, and Dr. Gerard Schellenberg for valuable discussions; Scott Sutherland for the wild type MEFs; and Dr. James Alwine for the gift of the pSG5-T plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grants R21-HL120751 (to F. S. L.), R01-CA153347 (to F. S. L.), and R01-GM090301 (to F. S. L. and S. R. M.).
- PHD
- prolyl hydroxylase domain protein
- HIF
- hypoxia-inducible factor
- HSP
- heat shock protein
- MEF
- mouse embryonic fibroblast
- VHL
- von Hippel-Lindau protein
- HT
- His6-tagged.
REFERENCES
- 1. Scheinfeldt L. B., Tishkoff S. A. (2013) Recent human adaptation: genomic approaches, interpretation and insights. Nat. Rev. Genet. 14, 692–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beall C. M. (2013) Human adaptability studies at high altitude: Research designs and major concepts during fifty years of discovery. Am. J. Hum. Biol. 25, 141–147 [DOI] [PubMed] [Google Scholar]
- 3. Beall C. M., Cavalleri G. L., Deng L., Elston R. C., Gao Y., Knight J., Li C., Li J. C., Liang Y., McCormack M., Montgomery H. E., Pan H., Robbins P. A., Shianna K. V., Tam S. C., Tsering N., Veeramah K. R., Wang W., Wangdui P., Weale M. E., Xu Y., Xu Z., Yang L., Zaman M. J., Zeng C., Zhang L., Zhang X., Zhaxi P., Zheng Y. T. (2010) Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. U.S.A. 107, 11459–11464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simonson T. S., Yang Y., Huff C. D., Yun H., Qin G., Witherspoon D. J., Bai Z., Lorenzo F. R., Xing J., Jorde L. B., Prchal J. T., Ge R. (2010) Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75 [DOI] [PubMed] [Google Scholar]
- 5. Yi X., Liang Y., Huerta-Sanchez E., Jin X., Cuo Z. X., Pool J. E., Xu X., Jiang H., Vinckenbosch N., Korneliussen T. S., Zheng H., Liu T., He W., Li K., Luo R., Nie X., Wu H., Zhao M., Cao H., Zou J., Shan Y., Li S., Yang Q., Asan, Ni P., Tian G., Xu J., Liu X., Jiang T., Wu R., Zhou G., Tang M., Qin J., Wang T., Feng S., Li G., Huasang, Luosang J., Wang W., Chen F., Wang Y., Zheng X., Li Z., Bianba Z., Yang G., Wang X., Tang S., Gao G., Chen Y., Luo Z., Gusang L., Cao Z., Zhang Q., Ouyang W., Ren X., Liang H., Huang Y., Li J., Bolund L., Kristiansen K., Li Y., Zhang Y., Zhang X., Li R., Yang H., Nielsen R., Wang J. (2010) Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bigham A., Bauchet M., Pinto D., Mao X., Akey J. M., Mei R., Scherer S. W., Julian C. G., Wilson M. J., López Herráez D., Brutsaert T., Parra E. J., Moore L. G., Shriver M. D. (2010) Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 6, e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peng Y., Yang Z., Zhang H., Cui C., Qi X., Luo X., Tao X., Wu T., Ouzhuluobu, Basang, Ciwangsangbu, Danzengduojie, Chen H., Shi H., Su B. (2011) Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol. Biol. Evol. 28, 1075–1081 [DOI] [PubMed] [Google Scholar]
- 8. Wang B., Zhang Y. B., Zhang F., Lin H., Wang X., Wan N., Ye Z., Weng H., Zhang L., Li X., Yan J., Wang P., Wu T., Cheng L., Wang J., Wang D. M., Ma X., Yu J. (2011) On the origin of Tibetans and their genetic basis in adapting high-altitude environments. PLoS One 6, e17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu S., Li S., Yang Y., Tan J., Lou H., Jin W., Yang L., Pan X., Wang J., Shen Y., Wu B., Wang H., Jin L. (2011) A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol. Biol. Evol. 28, 1003–1011 [DOI] [PubMed] [Google Scholar]
- 10. Simonson T. S., McClain D. A., Jorde L. B., Prchal J. T. (2012) Genetic determinants of Tibetan high-altitude adaptation. Hum. Genet. 131, 527–533 [DOI] [PubMed] [Google Scholar]
- 11. Semenza G. L. (2012) Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kaelin W. G., Jr., Ratcliffe P. J. (2008) Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 13. Majmundar A. J., Wong W. J., Simon M. C. (2010) Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 40, 294–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee F. S., Percy M. J. (2011) The HIF pathway and erythrocytosis. Annu. Rev. Pathol. 6, 165–192 [DOI] [PubMed] [Google Scholar]
- 15. Xiang K., Ouzhuluobu, Peng Y., Yang Z., Zhang X., Cui C., Zhang H., Li M., Zhang Y., Bianba, Gonggalanzi, Basang, Ciwangsangbu, Wu T., Chen H., Shi H., Qi X., Su B. (2013) Identification of a Tibetan-specific mutation in the hypoxic gene EGLN1 and its contribution to high-altitude adaptation. Mol. Biol. Evol. 30, 1889–1898 [DOI] [PubMed] [Google Scholar]
- 16. Lorenzo F. R., Simonson T. S., Yang Y., Ge R., Prchal J. T. (2010) American Society of Hematology Annual Meeting, Dec 4–7, 2010, Orlando, FL (The American Society of Hematology, Washington, D. C.), Abstract 2602 [Google Scholar]
- 17. Loenarz C., Coleman M. L., Boleininger A., Schierwater B., Holland P. W., Ratcliffe P. J., Schofield C. J. (2011) The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO Rep. 12, 63–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rytkönen K. T., Williams T. A., Renshaw G. M., Primmer C. R., Nikinmaa M. (2011) Molecular evolution of the metazoan PHD-HIF oxygen-sensing system. Mol. Biol. Evol. 28, 1913–1926 [DOI] [PubMed] [Google Scholar]
- 19. Song D., Li L. S., Heaton-Johnson K. J., Arsenault P. R., Master S. R., Lee F. S. (2013) Prolyl hydroxylase domain protein 2 (PHD2) binds a Pro-Xaa-Leu-Glu motif, linking it to the heat shock protein 90 pathway. J. Biol. Chem. 288, 9662–9674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Isaacs J. S., Jung Y. J., Mimnaugh E. G., Martinez A., Cuttitta F., Neckers L. M. (2002) Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1α-degradative pathway. J. Biol. Chem. 277, 29936–29944 [DOI] [PubMed] [Google Scholar]
- 21. Katschinski D. M., Le L., Heinrich D., Wagner K. F., Hofer T., Schindler S. G., Wenger R. H. (2002) Heat induction of the unphosphorylated form of hypoxia-inducible factor-1α is dependent on heat shock protein-90 activity. J. Biol. Chem. 277, 9262–9267 [DOI] [PubMed] [Google Scholar]
- 22. Minet E., Mottet D., Michel G., Roland I., Raes M., Remacle J., Michiels C. (1999) Hypoxia-induced activation of HIF-1: role of HIF-1α-Hsp90 interaction. FEBS Lett. 460, 251–256 [DOI] [PubMed] [Google Scholar]
- 23. Huang J., Zhao Q., Mooney S. M., Lee F. S. (2002) Sequence determinants in hypoxia-inducible factor-1α for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3. J. Biol. Chem. 277, 39792–39800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. García-Cardeña G., Fan R., Shah V., Sorrentino R., Cirino G., Papapetropoulos A., Sessa W. C. (1998) Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature 392, 821–824 [DOI] [PubMed] [Google Scholar]
- 25. Yu F., White S. B., Zhao Q., Lee F. S. (2001) Dynamic, site-specific interaction of hypoxia-inducible factor-1α with the von Hippel-Lindau tumor suppressor protein. Cancer Res. 61, 4136–4142 [PubMed] [Google Scholar]
- 26. Percy M. J., Furlow P. W., Beer P. A., Lappin T. R., McMullin M. F., Lee F. S. (2007) A novel erythrocytosis-associated PHD2 mutation suggests the location of a HIF binding groove. Blood 110, 2193–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wiśniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 28. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 29. Arsenault P. R., Pei F., Lee R., Kerestes H., Percy M. J., Keith B., Simon M. C., Lappin T. R., Khurana T. S., Lee F. S. (2013) A knock-in mouse model of human PHD2 gene-associated erythrocytosis establishes a haploinsufficiency mechanism. J. Biol. Chem. 288, 33571–33584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu J. (2005) Preparation, culture, and immortalization of mouse embryonic fibroblasts. In Current Protocols in Molecular Biology (Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., eds) Chapter 28, Unit 28.21, John Wiley & Sons, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 31. Yu Y., Alwine J. C. (2002) Human cytomegalovirus major immediate-early proteins and simian virus 40 large T antigen can inhibit apoptosis through activation of the phosphatidylinositide 3′-OH kinase pathway and the cellular kinase Akt. J. Virol. 76, 3731–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hon W. C., Wilson M. I., Harlos K., Claridge T. D., Schofield C. J., Pugh C. W., Maxwell P. H., Ratcliffe P. J., Stuart D. I., Jones E. Y. (2002) Structural basis for the recognition of hydroxyproline in HIF-1α by pVHL. Nature 417, 975–978 [DOI] [PubMed] [Google Scholar]
- 33. Min J. H., Yang H., Ivan M., Gertler F., Kaelin W. G., Jr., Pavletich N. P. (2002) Structure of an HIF-1α-pVHL complex: hydroxyproline recognition in signaling. Science 296, 1886–1889 [DOI] [PubMed] [Google Scholar]
- 34. Wochnik G. M., Young J. C., Schmidt U., Holsboer F., Hartl F. U., Rein T. (2004) Inhibition of GR-mediated transcription by p23 requires interaction with Hsp90. FEBS Lett. 560, 35–38 [DOI] [PubMed] [Google Scholar]
- 35. Lee C. C., Lin T. W., Ko T. P., Wang A. H. (2011) The hexameric structures of human heat shock protein 90. PLoS One 6, e19961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taipale M., Jarosz D. F., Lindquist S. (2010) HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528 [DOI] [PubMed] [Google Scholar]
- 37. Scortegagna M., Ding K., Zhang Q., Oktay Y., Bennett M. J., Bennett M., Shelton J. M., Richardson J. A., Moe O., Garcia J. A. (2005) HIF-2α regulates murine hematopoietic development in an erythropoietin-dependent manner. Blood 105, 3133–3140 [DOI] [PubMed] [Google Scholar]
- 38. Gruber M., Hu C. J., Johnson R. S., Brown E. J., Keith B., Simon M. C. (2007) Acute postnatal ablation of Hif-2α results in anemia. Proc. Natl. Acad. Sci. U.S.A. 104, 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kapitsinou P. P., Liu Q., Unger T. L., Rha J., Davidoff O., Keith B., Epstein J. A., Moores S. L., Erickson-Miller C. L., Haase V. H. (2010) Hepatic HIF-2 regulates erythropoietic responses to hypoxia in renal anemia. Blood 116, 3039–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Percy M. J., Furlow P. W., Lucas G. S., Li X., Lappin T. R., McMullin M. F., Lee F. S. (2008) A gain-of-function mutation in the HIF2A gene in familial erythrocytosis. N. Engl. J. Med. 358, 162–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brusselmans K., Compernolle V., Tjwa M., Wiesener M. S., Maxwell P. H., Collen D., Carmeliet P. (2003) Heterozygous deficiency of hypoxia-inducible factor-2α protects mice against pulmonary hypertension and right ventricular dysfunction during prolonged hypoxia. J. Clin. Invest. 111, 1519–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hickey M. M., Richardson T., Wang T., Mosqueira M., Arguiri E., Yu H., Yu Q. C., Solomides C. C., Morrisey E. E., Khurana T. S., Christofidou-Solomidou M., Simon M. C. (2010) The von Hippel-Lindau Chuvash mutation promotes pulmonary hypertension and fibrosis in mice. J. Clin. Invest. 120, 827–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gale D. P., Harten S. K., Reid C. D., Tuddenham E. G., Maxwell P. H. (2008) Autosomal dominant erythrocytosis and pulmonary arterial hypertension associated with an activating HIF2α mutation. Blood 112, 919–921 [DOI] [PubMed] [Google Scholar]
- 44. Tan Q., Kerestes H., Percy M. J., Pietrofesa R., Chen L., Khurana T. S., Christofidou-Solomidou M., Lappin T. R., Lee F. S. (2013) Erythrocytosis and pulmonary hypertension in a mouse model of human HIF2A gain of function mutation. J. Biol. Chem. 288, 17134–17144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kline D. D., Peng Y. J., Manalo D. J., Semenza G. L., Prabhakar N. R. (2002) Defective carotid body function and impaired ventilatory responses to chronic hypoxia in mice partially deficient for hypoxia-inducible factor 1α. Proc. Natl. Acad. Sci. U.S.A. 99, 821–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Melillo G., Musso T., Sica A., Taylor L. S., Cox G. W., Varesio L. (1995) A hypoxia-responsive element mediates a novel pathway of activation of the inducible nitric oxide synthase promoter. J. Exp. Med. 182, 1683–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Palmer L. A., Semenza G. L., Stoler M. H., Johns R. A. (1998) Hypoxia induces type II NOS gene expression in pulmonary artery endothelial cells via HIF-1. Am. J. Physiol. 274, L212–L219 [DOI] [PubMed] [Google Scholar]
- 48. Boutin A. T., Weidemann A., Fu Z., Mesropian L., Gradin K., Jamora C., Wiesener M., Eckardt K. U., Koch C. J., Ellies L. G., Haddad G., Haase V. H., Simon M. C., Poellinger L., Powell F. L., Johnson R. S. (2008) Epidermal sensing of oxygen is essential for systemic hypoxic response. Cell 133, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]