Background: HOP2 protein promotes recombination and is required for meiotic chromosome synapsis.
Results: The N terminus of HOP2 has a winged head DNA binding structure.
Conclusion: The solution structure of the winged head DNA binding domain integrates biochemical and functional aspects of HOP2 recombinational function.
Significance: Determining the three-dimensional structure of HOP2 is crucial to understand the mechanism of HOP2 action.
Keywords: DNA-binding Protein, DNA Repair, Homologous Recombination, NMR, Protein Structure, DNA Binding
Abstract
The HOP2 protein is required for efficient double-strand break repair which ensures the proper synapsis of homologous chromosomes and normal meiotic progression. We previously showed that in vitro HOP2 shows two distinctive activities: when it is incorporated into a HOP2-MND1 heterodimer, it stimulates DMC1 and RAD51 recombination activities, and the purified HOP2 alone is proficient in promoting strand invasion. The structural and biochemical basis of HOP2 action in recombination are poorly understood; therefore, they are the focus of this work. Herein, we present the solution structure of the amino-terminal portion of mouse HOP2, which contains a typical winged helix DNA-binding domain. Together with NMR spectral changes in the presence of double-stranded DNA, protein docking on DNA, and mutation analysis to identify the amino acids involved in DNA coordination, our results on the three-dimensional structure of HOP2 provide key information on the fundamental structural and biochemical requirements directing the interaction of HOP2 with DNA. These results, in combination with mutational experiments showing the role of a coiled-coil structural feature involved in HOP2 self-association, allow us to explain important aspects of the function of HOP2 in recombination.
Introduction
In meiosis, two rounds of chromosome segregation follow only one round of DNA replication. This serves a fundamental function by halving the number of chromosomes that is required for sexual reproduction. The arrangement of maternal and paternal homologous chromosomes into pairs allows them to act as a single unit when microtubules attach and align them on the meiotic spindle. This ensures their orderly segregation to opposite poles of the cell at the first meiotic cell division so that each gamete receives only one copy of each chromosome. In most organisms, homologous chromosomes pairs are stabilized by a physical link provided by the crossovers, the products of homologous recombination, which are viewed cytologically as chiasmata (reviewed in Refs. 1–4). Consequently, mutations that cause loss or misregulation of recombination are invariably associated with increased errors in meiotic chromosome segregation and the generation of aneuploid gametes.
Critical functions in homologous recombination are provided by the ubiquitous RAD51 and the meiotic specific DMC1 recombinases. These enzymes search for homologous DNA sequences to accomplish template repair by promoting the invasion of intact double-stranded DNA (dsDNA)3 by single-stranded DNA ends arising from resection of dsDNA (5). Remarkably, efficient function of the recombinases requires interactions with accessory proteins. Several lines of evidence have demonstrated that HOP2 and MND1 are indispensable for meiosis via their interactions with DMC1 and RAD51 (5–11). A Saccharomyces cerevisiae Hop2 deletion mutant exhibits a profound failure in meiosis due to a uniform arrest at meiosis I with chromosomes engaged in synapsis with non-homologous partners (12). Moreover, Hop2−/− mouse spermatocytes show meiotic arrest and limited chromosome synapsis, consistent with a failure in double-strand break repair (13). Consistent with these data, we and others (6–11) observed that the purified mouse HOP2-MND1 heterodimer physically interacts with DMC1 and RAD51, greatly stimulating the recombinase activities of these proteins. An additional function for HOP2 as a recombinase, independent of DMC1/RAD51, has been proposed as purified HOP2 catalyzes the formation of homologous DNA pairing in meiotic recombination (7, 8, 14). Taken together, these results indicate that HOP2 may have dual role in mammalian meiotic homologous recombination (14).
Here, we present the NMR solution structure of an amino-terminal fragment of HOP2 which reveals a DNA binding domain folded in a typical winged helix conformation, a common motif among the helix-turn-helix DNA binding proteins (15). In addition, we propose a model for the HOP2-DNA complex based on results obtained from chemical shift perturbations in the presence of DNA oligonucleotide, protein docking on DNA, and mutational analysis. Along with revealing the role of a coiled-coil structural feature in HOP2 involved in protein self-association, our results help explain important aspects of the molecular mechanism of recombination mediated by HOP2.
MATERIALS AND METHODS
Protein Expression and Purification
The plasmid pET22b was used as vector for the overexpression of mouse His6-1–84HOP2 in Escherichia coli BL21(DE3). The uniformly 15N-labeled 1–84HOP2 sample was prepared by growing cells at 37 °C to an A600 nm of 0.7. The culture medium (optimal M9 minimal) contained 1 g/liter 15NH4Cl, 2 g/liter mm unlabeled glucose, 50 μg/ml of kanamycin, 1× BME vitamins, 13 μm FeSO4, 0.5 mm MgSO4, and 1× trace elements. Protein expression was stimulated by addition of isopropyl β-d-galactosidase to a final concentration of 1 mm. After 18 h, cells were harvested by centrifugation and lysed, and 1–84HOP2 was purified from the soluble fraction as described previously (11). The final yield of purified 15N-labeled protein was ∼6 mg per liter. The uniformly 13C,15N-labeled 1–84HOP2 sample was produced according to the same procedure, apart from the fact that [13C]glucose was used at a concentration of 2 g/liter. Purification of full-length, truncated versions, and point mutants of HOP2 used in DNA binding and chemical cross-linking experiments were performed as described previously (11).
NMR Spectroscopy
A 450-μl sample of 6 mg/ml 13C,15N-labeled 1–84HOP2 in a buffer of 120 mm NaCl, 10 mm imidazole, 5% glycerol, and 5% D2O was used for NMR experiments. All NMR experiments were performed at 20 °C on a Varian INOVA 600 MHz spectrometer with a Nalorac 5-mm 1H,13C,15N-labeled PFG triple resonance probe and using software VNMRJ with the BioPack suite of pulse programs. 15N-Heteronuclear single quantum coherence (HSQC), HNCO, CO(CA)NH, HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH experiments were performed for protein backbone assignment. 13C-HSQC, H(CCO)NH, and TOCSY-NHSQC, H(CC)H-TOCSY, and C(CO)NH experiments were carried out for side chain assignments. NOESY-NHSQC was obtained for evaluation of structural quality (see below). Data were processed with NMRPipe (16), and peaks were picked using Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). The NMR peaks were first assigned using automatic assigning program PINE (17). The assignments were then verified or corrected manually.
Structure Calculation
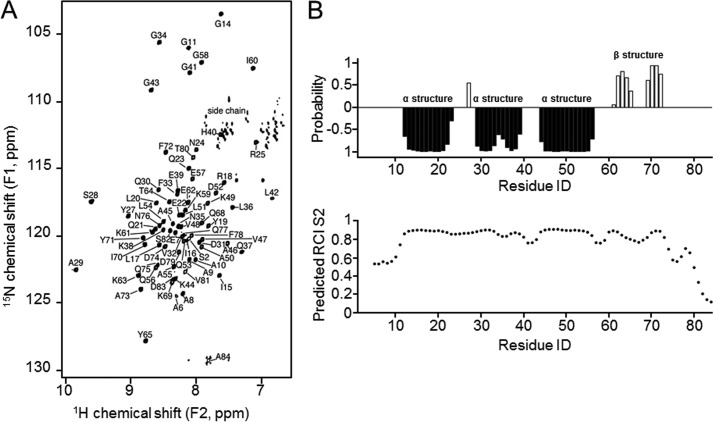
For structure resolution, we first obtained 15N-HSQC spectrum of 1–84HOP2 (Fig. 1A). Peaks in the 15N-HSQC spectra are well dispersed and the proton chemical shifts span a large range (7.8 to 9.8 ppm), indicating a well-folded protein. In addition to the two prolines that do not contribute to 15N-HSQC signal, the first four residues, Ala-12, and Gly-66 were not observed, most likely due to unfavorable dynamics. For the 82 non-proline residues, 74 backbone amide 15N, 1HN (90%), 78 Hα (95%, not double counting the two glycine protons), 76 C′ (93%), 78 Cα (95%), and 72 Cβ (88%) were assigned (Table 1). Furthermore, 48 Cγ, 27 Cδ, 8 Cϵ, 114 Hβ, 62 Hγ, 35 Hδ, and 23 Hϵ were assigned. Random coil index order parameters (S2) (18), secondary element propensity, and backbone dihedral angle restraints were predicted from chemical shifts using program TALOS+ (19).
FIGURE 1.
15N-HSQC NMR spectrum and secondary structure analysis of 1–84HOP2. A, peaks corresponding to chemical shifts were assigned with a set of three-dimensional NMR experiments. A typical 15N-HSQC spectrum is shown. B, secondary structures and order parameters predicted using assigned chemical shifts. Negative probabilities indicate propensity for α-helical structures, and positive probabilities indicate β-strands (top panel). Shown is a chemical shift estimation of protein backbone mobility for 1–84HOP2 (lower panel). RCI, random coil index.
TABLE 1.
Inputs for CS-ROSETTA and structure validation for HOP2 (residues 11–74)
| Chemical shifts used in CS-ROSETTA | |
| Cα | 62 |
| Cβ | 56 |
| C′ | 60 |
| N | 59 |
| HN | 59 |
| Ha (count one for each glycine) | 62 |
| Manually refined NOESY-NHSQC peaks 1,000 lowest energy models (of 40,000) | 1097 |
| CS-ROSETTA scaled energy | −106.1 (2.7) |
| Cα r.m.s.d. | 0.8 (0.3) |
| RPF DP score | 0.744 (0.005) |
| 20 models | |
| CS-ROSETTA scaled energy | −112.7 (2.2) |
| Cα r.m.s.d. | 0.3 (0.1) |
| RPF DP score | 0.745 (0.004) |
| r.m.s.d. from average coordinates (Å) | |
| Backbone atoms | 0.28 (0.08) |
| All atoms | 0.8 (0.1) |
| Ramachandran statisticsa | |
| Most favored regions (%) | 97.2 (1.1) |
| Additional allowed regions (%) | 2.8 (1.1) |
| Generously allowed (%) | 0 (0) |
| Disallowed regions (%) | 0 (0) |
| G-factorsa | |
| φ-ψ | 0.29 (0.03) |
| All dihedrals | 0.42 (0.02) |
| Covalent | 0.68 (0.00) |
| Overall | 0.52 (0.01) |
| Structure Z-scoresb | |
| 2nd generation packing quality | 1.671 (0.216) |
| Ramachandran plot appearance | 0.429 (0.500) |
| χ-1/χ-2 rotamer normality | 8.442 (0.721) |
| Backbone conformation | −4.629 (1.083) |
| RMS Z-scoresb | |
| Bond lengths | 0.454 (0.004) |
| Bond angles | 0.360 (0.001) |
| ω angle restraints | 0.577 (0.038) |
| Side chain planarity | 1.867 (0.005) |
| Improper dihedral distribution | 0.613 (0.002) |
| Inside/outside distribution | 0.817 (0.015) |
a From PROCHECK.
b From WHAT_CHECK.
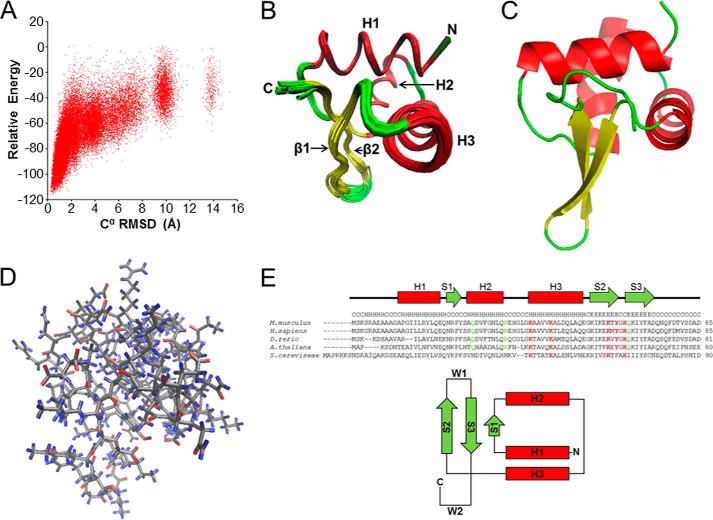
NMR structures were calculated using program CS-ROSETTA hosted on a web server at the Biological Magnetic Resonance Data Bank, utilizing the protein backbone 15N, 1HN, 1Hα, 13Cα, 13Cβ and 13C′ chemical shifts (20, 21). Unlike conventional NMR structure determination methods, CS-ROSETTA does not require any NOE distance restraints, which are often ambiguous and very cumbersome to obtain (22). Experimental chemical shifts are important to narrow down the selection of peptide fragments for building trial structures and to guide the effective sampling of the conformation space (20). CS-ROSETTA has been demonstrated to fold structures accurately for proteins up to 12 kDa using only chemical shifts (20, 22, 23). The side chain packing in structures determined by CS-ROSETTA has been found to be very accurate when compared with crystal structures, even though side chain constraints are not explicitly used in the procedure (20, 23). The side chain accuracy has been attributed to the ROSETTA all-atom force field that includes hydrogen bonding, side chain packing, polar solvation, and backbone and side chain torsional energy (20). The energy r.m.s.d. plot obtained shows convergence toward the lower left corner with many low energy structures clustered with their Cα r.m.s.d. <2 Å from the lowest energy structure (Fig. 2A and Table 1). This indicates that the calculated final structures are highly reliable (20). Next, 1,000 lowest-energy structures were validated for consistence with NOE distances. This was done by calculating recall, precision, and F-measure score (RPF) and DP scores for each of these structures against a manually refined peak list from an unassigned NOESY-NHSQC spectrum (22, 24). The DP scores of all 1,000 structures are found to be >0.73 (Table 1), which confirms that the folds are correct and structures are of high quality. The 20 best structures were chosen to represent the final structural ensemble of the protein by setting the thresholds for a combination of CS-ROSETTA all-atom energy, r.m.s.d., and DP scores (Table 1). The quality of these 20 structures was assessed using PROCHECK (25) and WHAT CHECK (26).
FIGURE 2.
NMR structure of 1–84HOP2. A, NMR structure determination of 1–84HOP2. Shown is a plot of chemical shift-rescored ROSETTA all-atom energy versus Cα r.m.s.d. from the lowest energy structure showing convergence of structure calculation. B, diagram of superimposed 20 refined low-energy structures. C, carboxyl-terminal; N, amino-terminal; H, helix; β, beta. Secondary structures are colored as follows: red, helices, yellow, β-sheets, and green, loops. C, schematic diagram of the lowest energy structure. Secondary structures are colored as described in B. D, backbone atom representation of 1–84HOP2. Gray, carbon, nitrogen, and sulfur; blue, hydrogen; and red, oxygen. E, alignment of the HOP2 amino-terminal primary sequence and topology of a typical winged helix fold. Highlighted are conserved (red) and partially conserved (green) amino acids. The secondary structure prediction for the mouse sequence was obtained using the nnpredict program; H, α-helix. S represents β-strands, and W1 and W2 are loops or wings. M. musculus, Mus musculus; H. sapiens, Homo sapiens; D. rerio, Danio rerio; A. thaliana, Arabidopsis thaliana; S. cereviseae, Saccharomyces cereviseae.
NMR Measurements of Protein-Oligonucleotide Interactions
For protein-DNA interaction studies, dsDNA (supplemental Table S1) was dissolved in the same buffer as the protein, reaching a concentration of 6.8 μg/μl. The control 15N-HSQC NMR spectrum was first acquired for 450 μl of 5 mg/ml 15N-labeled protein sample, followed by titration of the DNA solution at 10-μl increments for a total of 70 μl. The 15N-HSQC NMR spectra were acquired at each titration point, and the scaled chemical shift perturbation was calculated using the equation, δ(i) = ((δNi − δN0)2 + (10 × (δHi − δH0))2)½, where δN0 and δH0 represent 15N and 1H chemical shifts for protein only, and δNi and δHi represent those at the ith titration point.
HOP2-dsDNA Model
To model an interaction between HOP2 and DNA duplex, the apo-HOP2 structure was superimposed upon the BlaI plus DNA NMR structure (Protein Data Bank code 2P7C (27)). The region of the HOP2 main chain path was then modeled on the BlaI structure from the beginning of helix 3 (residue 45) to conform to an optimal path for DNA binding. The joining residues (residues 36–44) between helix 2 of the N-terminal portion and helix 3 were modeled and minimized using the FALC-Loop modeling server (28).
Generation of 1–84HOP2 Point Mutant Proteins
PCR-based site-directed mutagenesis was used to generate single and multiple point mutations in 1–84HOP2 gene. Briefly, primers (oligonucleotides 5–23, supplemental Table S1) were designed to replace the codon of a target amino acid by an alanine; PCR was used to amplify the mutant gene followed by assembly into the plasmid pRSFDuet1 using the Gibson assembly method (29). Transformation was done in XL2-Blue ultra-competent cells (Agilent Technologies). Point mutations were confirmed by DNA sequencing. Single point mutants (Q30A, K38A, K44A, K49A, D52A, E57A, E62A, K67A, Q68A, K69A, and Y71A) and triple point mutants (Q30A/K38A/K44A, Q30A/K44A/K49A, K63A/Y65A/K67A, and Y65A/K67A/Q68A) were developed.
Gel Shift Assay Measurements of Protein-DNA Interactions
The protein (concentration as indicated) was incubated with 720 nm (nucleotides) 32P-labeled 30-bp oligonucleotide in the following buffer: 20 mm Tris-HCl (pH 7.4), 2 mm MgCl2, and 50 mm NaCl in a volume of 20 μl for 10 min at 37 °C. The samples were mixed with 3 μl of loading buffer (30% glycerol, 0.1% bromphenol blue) and analyzed by electrophoresis in 14% polyacrylamide gels in 1× TAE buffer at 5 V/cm for 5 h. The gels were exposed to radiation sensitive films, and the band corresponding to free dsDNA oligonucleotide was quantitated using ImageJ software.
Chemical Cross-linking Experiments
The protein samples (15 μm 1–217HOP2, 1–84HOP2, 1–125HOP2, 126–217HOP2, and 144–217HOP2) were equilibrated for 10 min at room temperature in the presence of reaction buffer (30 mm HEPES (pH 7.4), 50 mm NaCl, and 10% glycerol). The samples were then further incubated with ethylene glycol bis(succinic acid N-hydroxysuccinimide ester) (Sigma) for 20 min at room temperature in a total reaction volume of 20 μl. The cross-linkers were then quenched through the addition of 1 μl of 1 m Tris-HCl (pH 8) followed by incubation for 10 min at room temperature. Finally, the samples were resolved in 5–10 and 15% SDS-PAGE for full-length and HOP2 fragments, respectively.
RESULTS AND DISCUSSION
The HOP2 Amino-terminal DNA-binding Domain Belongs to the Winged Helix Family
The amino-terminal domain (amino acids 1–84) structure of mouse HOP2 was solved using two- and three-dimensional NMR spectroscopy, making use of uniformly 13C,15N-labeled proteins (Fig. 1A). We then used the backbone chemical shifts to estimate secondary structure (TALOS + (19)). As shown in Fig. 1B (top panel), there are three α-helices (amino acids 12–23, 29–39, and 44–56) and two β-strands (amino acids 61–65 and 69–72). In addition, Tyr-27 also has a propensity to be in β-strand conformation. In the determined three-dimensional structure (see below), both Tyr-27 and Ser-28 are in a short β-strand. The chemical shifts also allowed estimation of protein backbone mobility because the random coil chemical shifts are often associated with highly flexible regions (18). The predicted order parameters (S2) indicate that the regions of definite secondary structure are well defined, whereas both ends (Met-1–Ala-10 and Gln-75–Ala-84) show low degree of local order indicating significant sub-nanosecond timescale dynamics (Fig. 1B, lower panel).
The backbone and Cβ chemical shifts were utilized to calculate protein structure using the CS-ROSETTA software (Fig. 2A) (20). With the thresholds for a combination of CS-ROSETTA all-atom energy, Cα r.m.s.d., and DP scores (energy ≤ −110; r.m.s.d. ≤ 0.415 Å, and DP ≥ 0.74) (Table 1), 20 structures were chosen to represent the final structural ensemble of HOP2 (Fig. 2, B–D). The overall quality of these structures was assessed using PROCHECK (25) and WHAT CHECK (26). These structures are found to have a backbone r.m.s.d. value of 0.3 Å and an all atoms r.m.s.d. of 0.8 Å from the average structure. The structural Z-scores from WHAT CHECK procedure are good except for the backbone conformation, but much better RMS Z scores indicate that the local geometry of the structure is reliable (31). Additionally, all the ψ and φ angles fall either in the most favored or additional allowed regions in the Ramachandran plot. The good geometry scores are due to the fact that structures obtained by CS-ROSETTA are built from homology peptide fragments (homology model) augmented with chemical shifts and that these structures are not directly constrained by NOE distances as in conventional NMR methods.
Inspection of the final NMR ensemble (see Table 1 for statistics) reveals a well ordered core region folded in a typical winged head/fork head structure (Fig. 2). This winged helix motif is a compact α/β-structure consisting of two wings (W1 (Tyr-61–Gln-64) and W2 (Ala-69–Asn-72)), three α helices (H1 (Ala-8–Gln-19), H2 (Ala-25–Glu-35)), and H3 (Lys-40–Glu-53) and three β-strands (S1 (Arg-21–Ser-24), S2 (Ile-56–Tyr-61), and S3 (Gln-64–Ala-69)), arranged in order H1-S1-H2-H3-S2-W1-S3-W2 (Fig. 2E). The amino-terminal half of the motif is largely helical, whereas the carboxyl-terminal half is composed of two of the three strands forming the twisted antiparallel β-sheet and the two loops of wings, W1 and W2 (15). Wing W1 connects strand S2 and S3, and wing W2 extends from strand S3 to the carboxyl-terminal of the DNA binding domain. With some variations, the winged helix structure classifies HOP2 as a member of the winged helix protein family, which is a member of the DNA recognition helix-turn-helix superfamily (15). Individual amino acids and the predicted secondary structure of the HOP2 winged helix domain are highly similar among species (Fig. 2E). This indicates that the HOP2 winged helix DNA binding domain mode has been well-conserved throughout evolution.
NMR Analysis of the Interactions of 1–84HOP2 with DNA
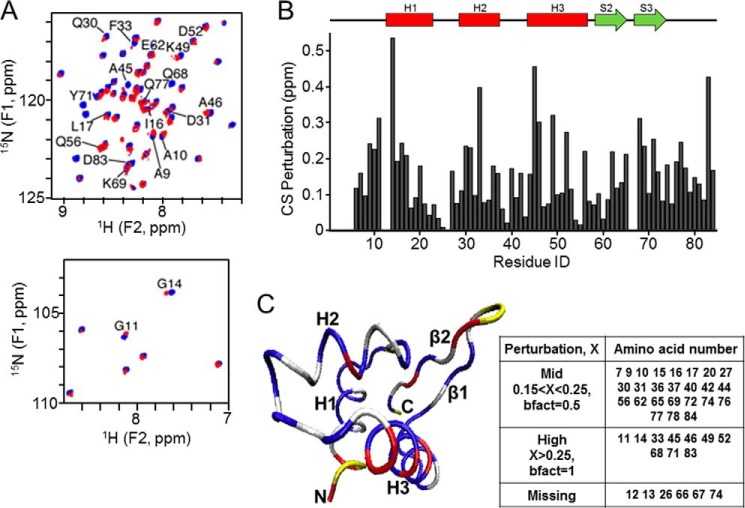
We began analyzing the 1–84HOP2 DNA recognition mode by following changes on the chemical shift of HOP2 amino acids induced by interaction with DNA. Chemical shift perturbation is a sensitive method that can provide DNA binding location. 15N-HSQC spectra of 1–84HOP2 were recorded with increasing amounts of a 30-bp dsDNA oligonucleotide (oligonucleotides 1 and 2, supplemental Table S1). Because the 15N-HSQC spectra of DNA-free 1–84HOP2, and of dsDNA-bound proteins share many similarities (Fig. 3A), the amide 1H and 15N chemical shifts of the amino-terminal domain of DNA-bound 1–84HOP2 were assigned with reference to the DNA-free protein assignments. This global comparison also indicates that 1–84HOP2 does not undergo major structural rearrangements upon binding to dsDNA. The 15N,1H chemical shift changes (perturbation defined in the experimental section) for backbone amide groups show the highest shift or intensity changes in helix H3 (amino acids Ala-45, Ala-46, Lys-49, and Asp-52), and in the β-sheet, especially close to the wing W1 (amino acids Gln-68 and Tyr-71) (Fig. 3, A–C). The residues affected by dsDNA binding (highlighted on the HOP2 structure in Fig. 3C) define a possible DNA-binding surface located on a single side of the HOP2 winged helix domain. Similar regions are typically implicated in coordinating DNA for a large number of the winged helix family members (15). These results suggest the importance of H3 and W1 structure in DNA binding. The titration curves of perturbation versus DNA concentration were analyzed for the 10 most perturbed sites (data not shown), showing that the perturbation values are saturated at the final few titration points. The apparent dissociation constant that gives half-maximum chemical perturbation Kd = 33 ± 10 μm.
FIGURE 3.
Chemical shift mapping of 1–84HOP2 interacting with DNA. A, DNA titration monitored by 15N-HSQC, showing two regions with significant peak displacements. Blue, initial; red, final. B, graph of the chemical shift variations between the DNA free 1–84HOP2 and the 1–84HOP2-DNA complex plotted against residue number. C, chemical shift perturbation color coded on the structure of 11–74HOP2. Blue, δ < 0.15; gray, 0.15 ≤ δ < 0.25; red, δ > 0.25; yellow, missing due to prolines or unfavorable dynamics. The table contains a list of amino acids involved in the chemical shift perturbation by dsDNA. bfact, B factor.
HOP2 Winged Helix Domain DNA Recognition Mode
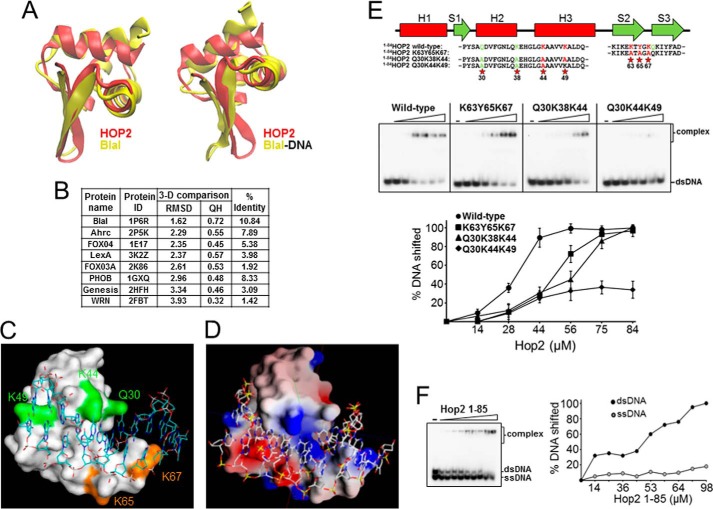
The N-terminal of HOP2 has been shown to be required for efficient formation of the DMC1-mediated synaptic complex (11). Here, we investigate the DNA recognition mode of 1–84HOP2 by comparing the three-dimensional structure of HOP2 with protein-dsDNA complexes reported for other winged helix proteins. We first compared the structure of 1–84HOP2 with other winged helix proteins structures obtained in the absence of DNA using the VMD software package (32). We included in our analysis eight winged helix proteins (BlaI, Ahrc, Fox04, LexA, FOX03a, PhoB, Genesis, and WRN) whose structures in the presence or absence of dsDNA have been studied by x-ray crystallography, NMR, or computational tools (Fig. 4, A and B) (33–40). The most similar three-dimensional structure is that of the BlaI DNA recognition domain (r.m.s.d., 1.62 Å; QH, 0.72; % identity, 10.84). Although no significant sequence homology could be detected by primary sequence similarity searches, BlaI presents a fold very similar to that of 1–84HOP2 (Fig. 4A). Notably, the length of the helices and the loop in the wing regions are similar, and the angles between the three helices are conserved. We propose a model for the interaction between the 1–84HOP2 winged head domain and dsDNA by comparison with the structure of the BlaI-dsDNA complex (Fig. 4, C and D) as described under “Materials and Methods.” Although this method is unable to resolve the precise position of the amino acids and the base pair contacts, it suggests that the HOP2 winged helix domain uses a canonical mode for DNA binding (15). In this mode, helix H3, called the recognition helix, is presented to the major groove of the DNA. The wings (in particular W1) and the amino-terminal part also make contacts with the minor groove of the DNA. This has been observed in the majority of winged helix proteins (reviewed in Ref. 15) and is unlike RFX1, which makes most of the contacts with the DNA major groove via wing W1 with H3 overlying the minor groove (41).
FIGURE 4.
DNA recognition by 1–84HOP2. A, three-dimensional similarity of the NMR structure of 1–84HOP2 and the crystal structure of BlaI obtained in the absence or presence of dsDNA. B, results of three-dimensional (3-D) comparison of 1–84HOP2 and other winged helix proteins. Protein ID corresponds to the protein structure without DNA. C, surface representation of the 1–84HOP2-dsDNA complex model. A 30-bp dsDNA oligonucleotide corresponding to the Bacillus licheniformis BlaI operator op1 (oligonucleotides 3 and 4, supplemental Table S1) was used. Amino acids highlighted in green correspond to helices H2 and H3 and in orange to β1 and wing 1. D, electrostatic potential surface (GRASP) representation of the 1–84 HOP2-dsDNA complex model. Highlighted are amino acids with high potential (5.1 maximum) in blue and low potential (−2.0 minimum) in red. E, DNA binding properties of 1–84HOP2 point mutants. The top panel shows the mutants generated in this study. Analysis of wild-type (1–84 amino acids) and mutant variants of 1–84HOP2 (K63A/Y65A/K67A, Q30A/K38A/K44A, Q30A/K44A/K49A) (0, 14, 28, 44, 56, 75, and 84 μm) for DNA binding. The mean values ± S.D. from three independent experiments were plotted (lower panel). F, 1–84HOP2 show high DNA binding preference for dsDNA. ssDNA, single-stranded DNA.
Effect of Point Mutations on DNA Binding by the 1–84HOP2 Fragment
Results of the chemical shift perturbation in the presence of dsDNA and the protein docking on dsDNA suggest that H3 and W1 structures have functional importance in DNA binding. Prompted by these observations and to dissect the DNA binding sites of the complex, we constructed 1–84HOP2 point mutants to be deficient for DNA binding. 1–84HOP2 shows a marked preference for dsDNA (apparent Kd (dsDNA), 32 μm) (Fig. 4F); therefore, we tested the effect of these mutations on HOP2-dsDNA complex formation. We began by identifying conserved regions within the HOP2 winged head domain enriched in basic amino acids, which may be involved in DNA engagement via ionic interaction, and aromatic residues, predicted to participate in stacking interactions with DNA bases. We replaced the highly conserved Lys-63, Tyr-65, and Lys-67 residues encompassing a β-hairpin at W1 to alanine. The KYK mutation has a significant effect on the DNA binding ability; however, association of 1–84HOP2 to DNA is not abolished (Kd, 52 μm) (Fig. 4E). This is consistent with previous DNA mutational studies showing that a full-length HOP2 mutant carrying three point mutations within the winged head domain had substantially reduced DNA-binding ability (11). Similar results were obtained with a HOP2 mutant in which Tyr-65, Lys-67, and Gln-68 amino acids were replaced by alanine (data not shown). These results indicate that the W1 region participates in DNA binding and that at least one more region of 1–84HOP2 is involved in dsDNA binding. To test the participation of H3 and H2 in DNA binding, we generated two sets of mutants. In one mutant, we changed the highly conserved Lys-44 and Lys-49 (located in the helix H3) and Gln-30 (located in the helix H2) to alanine (Q30A/K44A/K49A mutant). For the second mutant, the amino acids Gln-30, Lys-38, and Lys-44 were changed to alanine (Q30A/K38A/K44A mutant). Q30A/K38A/K44A shows a substantial decrease in dsDNA binding (Kd, 58 μm) and the Q30A/K44A/K49A mutant shows a clear impairment in DNA binding (only 30% of dsDNA substrate bound at 84 μm protein) (Fig. 4E). These results indicate the relative importance of helix H3 with notable participation of the highly conserved Lys-49 amino acid in engaging DNA (Fig. 4C). Single amino acid changes for any of the mutants described above resulted in wild-type levels of DNA binding.
Charged residues distributed over the surface of HOP2 could potentially act as loci for DNA binding. The GRASP program (42) was used to calculate an accessible surface area and a corresponding surface potential map of the 1–84HOP2 structure. The protein area proposed to bind DNA can be superimposed nicely with a charged region of the protein electrostatic surface (Fig. 4D). The residues Lys-44, Lys-49, and Lys-67 overlap with high potential areas within the DNA binding surface. This is additional evidence suggesting the involvement of these amino acids in coordinating DNA. In summary, our results are in good agreement with a canonical winged helix domain DNA interaction mode in which helix H3 and W1 are the major responsible structures coordinating dsDNA binding.
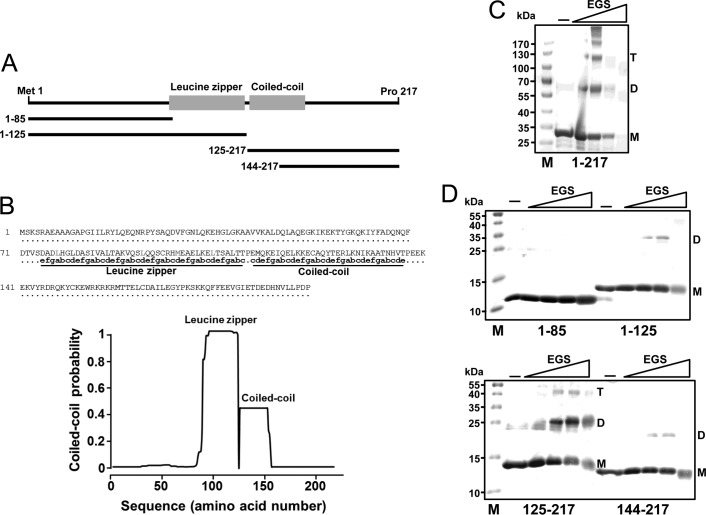
HOP2 Self-association Is Mediated by a Coiled-coil Structure
Amino acid sequence and secondary structure analysis of full-length mouse HOP2 predicts the presence of two coiled-coil structures encompassing amino acids 84–124 and 126–155, respectively (Fig. 5, A and B). The formation and location of these coiled-coil motifs are highly conserved from yeast to human. We previously showed that mutations affecting only amino acids 126–155 but not the N-terminal (amino acids 84–124) coiled-coil structure disrupt the formation of the HOP2-MND1 heterodimer (8). Here, we show that the 126–155 amino acid coiled-coil structure is involved in oligomerization of HOP2. We determined the oligomeric composition of full-length (amino acids 1–217) and truncated versions (amino acids 1–84, 1–125, 126–217, and 144–217 (11)) of HOP2 using chemical cross-linking. We observed that full-length HOP2 forms dimers, tetramers, and a minor amount of higher order oligomers (Fig. 5C). Disruptions within the 133 amino acids from the carboxyl-terminal of HOP2 (1–84HOP2) abolished the formation of dimers and tetramers (Fig. 5D). This indicates that the winged head amino-terminal region of HOP2 does not promote oligomerization alone. The 1–125HOP2 mutant showed a low level of dimer formation only at high concentrations of cross linker, indicating that the 84–124-amino acid coiled-coil structure has a minor role in HOP2 self-association. In contrast, the 126–217HOP2 truncation mutant, which contains the 126–155-amino acid coiled-coil domain, formed a high level of dimers and tetramers even at lower concentrations of cross linker. This characteristic is strongly affected by protein concentration and ionic strength (data not shown). The role of this presumptive coiled-coil structure in HOP2 oligomerization is confirmed by the reduced levels of oligomers observed in the 144–217HOP2 mutant, which removes the first 19 amino acids of the domain.
FIGURE 5.
A coiled-coil structure in the central region of HOP2 mediates protein self-association. A, scheme of wild-type and carboxyl- and amino-terminal mutants HOP2 used in this study. B, sequence of HOP2 putative coiled-coil domain and coiled-coil prediction plot by the Multicoil software. C, products of chemical cross-link of full-length HOP2. D, products of chemical crosslink of 1–84, 1–125, 126–217, and 144–217 HOP2 truncation mutants. M, monomer; D, dimer; T, tetramer. EGS, ethylene glycol bis(succinic acid N-hydroxysuccinimide ester (chemical cross-linker).
CONCLUSIONS
We and others (7–9, 11, 14) previously showed that in vitro, the mouse HOP2 protein show two distinctive activities: 1) when it is incorporated into a HOP2-MND1 heterodimer, it stimulates DMC1 and RAD51 recombination activities, and 2) HOP2 alone is proficient in promoting strand invasion (7, 8, 14). At present, the structural bases supporting these HOP2 biochemical actions is unknown and therefore limits the mechanistic understanding of HOP2 recombination function. In this work, we present the solution structure of the mouse 1–84HOP2 fragment using NMR techniques. Our results reveal that the HOP2 amino-terminal domain displays a compact α/β-structure arranged in a typical winged-head DNA binding domain with order H1-S1-H2-H3-S2-W1-S3-W2. This classifies HOP2 as a member of the winged helix protein family of DNA binding proteins (15).
The winged helix proteins constitute a subfamily within the large ensemble of helix-turn-helix proteins. Since the discovery of the winged helix motif (43), different groups of topologically related proteins with diverse biological functions have been characterized. In the most common (canonical) mode of DNA binding, the DNA interactions, including any specificity determining contacts, map to the recognition helix (H3) within the DNA major groove, with other less important interactions involving the loop W1 and the minor groove of the DNA substrate. By NMR solution structure, biochemical, bioinformatic, and mutagenic analyses, we provide evidence that helix H3 and the loop W1 of HOP2 are involved in DNA recognition.
Interestingly, a large number of winged helix proteins exhibiting the canonical recognition mode show DNA-binding sequence specificity. For example, the BlaI repressor regulates the expression of antibiotic resistance proteins by specific contacts occurring between the base pairs of the TACA motif in the DNA operator sequence and a conserved amino acid residue of the repressor helix H3 (33). Based on the canonical DNA binding mode of HOP2 and high similarity of the HOP2 winged helix structure with sequence specific DNA binding proteins such as BlaI and a number of other transcription factors (Fig. 4B), it is tempting to suggest that HOP2 may also have preference for specific DNA sequences. However, further studies, such as the analysis of HOP2 DNA binding sites at specific genomic locations or the testing of oligonucleotide library sequences for DNA binding will be needed to test this possibility.
DNA recognition by winged helix proteins may be affected by the oligomerization state of the protein. For example, two molecules of the MotA transcription factor bind cooperatively to their DNA consensus sequence (30). In this case, the formation of the active dimer DNA binding form is mediated by the hydrophobic dimer interface of a coiled-coil interaction. Reminiscent of this DNA binding mode, our results show the importance of a carboxyl-terminal coiled-coil structure (amino acids 126–155) but not of the amino-terminal coiled-coil domain in HOP2 self-association (Fig. 5). This is in agreement with our previous results showing that this structure is required for the interaction of HOP2 and MND1 (8). Notably, recent work in the mouse MND1 protein indicated the presence of a highly conserved winged-head domain required for efficient HOP2-MND1 complex binding to DNA (11). Taken together, these results suggest that a coiled-coil structural feature facilitates the formation of an active HOP2-MND1 and HOP2-HOP2 dimer DNA binding form. We speculate that two winged head domains arranged in either a parallel or antiparallel mode cooperate for efficient DNA binding or may regulate HOP2 DNA binding to certain genomic regions.
Previous biochemical studies have shown that, when incorporated into the HOP2-MND1 complex, HOP2 is critical for efficient homologous DNA pairing mediated by the DMC1 and RAD51 recombinases (6–10). This stimulatory function can be explained by HOP2-MND1 acting in two critical steps of recombinase-promoted homologous pairing (9, 10). One, by stabilizing the DMC1 single-stranded DNA nucleoprotein filament and second, by facilitating the conjoining of DNA molecules through the capture of dsDNA by the DMC1 single-stranded DNA nucleoprotein filament. Our results showing the solution structure and biochemical properties of a major DNA binding site for HOP2 provide structural support for a recently proposed model in which the N-terminal DNA binding domain of HOP2, which preferentially binds dsDNA, is positioned at a distance to the DMC1 single-stranded DNA nucleoprotein filament to capture dsDNA (11). These HOP2 functions help assemble synaptic complex and permit the efficient sampling of potential homologous DNA sequences.
Supplementary Material
Acknowledgment
We thank Dr. Margaret Eastman (Oklahoma State-wide Shared NMR Facility) for technical assistance.
This work was supported by NIGMS, National Institutes of Health Award 1P20GM103636 and by Oklahoma Center for the Advancement of Science and Technology Grant HR10-48S (to R. J. P.). This work was also supported by National Institutes of Health Grant GM097713 (to D. H. Z.) and Oklahoma Center for the Advancement of Science and Technology Grant HR12-050 (to D. H. Z.).
The atomic coordinates and structure factors (code 2MH2) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Chemical shift assignments of 1–84HOP2 have been deposited with BioMagResBank (accession no. 19613).

This article contains supplemental Table 1.
- dsDNA
- double-stranded DNA
- HSQC
- heteronuclear single quantum coherence
- r.m.s.d.
- root mean square deviation
- DP
- fold discriminating power.
REFERENCES
- 1. Bascom-Slack C. A., Ross L. O., Dawson D. S. (1997) Chiasmata, crossovers, and meiotic chromosome segregation. Adv. Genet. 35, 253–284 [DOI] [PubMed] [Google Scholar]
- 2. Petronczki M., Siomos M. F., Nasmyth K. (2003) Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112, 423–440 [DOI] [PubMed] [Google Scholar]
- 3. Zickler D., Kleckner N. (1998) The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32, 619–697 [DOI] [PubMed] [Google Scholar]
- 4. Roeder G. S. (1997) Meiotic chromosomes: it takes two to tango. Genes Dev. 11, 2600–2621 [DOI] [PubMed] [Google Scholar]
- 5. Neale M. J., Keeney S. (2006) Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen Y. K., Leng C. H., Olivares H., Lee M. H., Chang Y. C., Kung W. M., Ti S. C., Lo Y. H., Wang A. H., Chang C. S., Bishop D. K., Hsueh Y. P., Wang T. F. (2004) Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl. Acad. Sci. U.S.A. 101, 10572–10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Petukhova G. V., Pezza R. J., Vanevski F., Ploquin M., Masson J. Y., Camerini-Otero R. D. (2005) The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 12, 449–453 [DOI] [PubMed] [Google Scholar]
- 8. Pezza R. J., Petukhova G. V., Ghirlando R., Camerini-Otero R. D. (2006) Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2-Mnd1 complex. J. Biol. Chem. 281, 18426–18434 [DOI] [PubMed] [Google Scholar]
- 9. Pezza R. J., Voloshin O. N., Vanevski F., Camerini-Otero R. D. (2007) Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 21, 1758–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chi P., San Filippo J., Sehorn M. G., Petukhova G. V., Sung P. (2007) Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 21, 1747–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao W., Saro D., Hammel M., Kwon Y., Xu Y., Rambo R. P., Williams G. J., Chi P., Lu L., Pezza R. J., Camerini-Otero R. D., Tainer J. A., Wang H. W., Sung P. (2014) Mechanistic insights into the role of Hop2-Mnd1 in meiotic homologous DNA pairing. Nucleic Acids Res. 42, 906–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leu J. Y., Chua P. R., Roeder G. S. (1998) The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell 94, 375–386 [DOI] [PubMed] [Google Scholar]
- 13. Petukhova G. V., Romanienko P. J., Camerini-Otero R. D. (2003) The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell 5, 927–936 [DOI] [PubMed] [Google Scholar]
- 14. Pezza R. J., Voloshin O. N., Volodin A. A., Boateng K. A., Bellani M. A., Mazin A. V., Camerini-Otero R. D. (2014) The dual role of HOP2 in mammalian meiotic homologous recombination. Nucleic Acids Res. 42, 2346–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gajiwala K. S., Burley S. K. (2000) Winged helix proteins. Curr. Opin. Struct. Biol. 10, 110–116 [DOI] [PubMed] [Google Scholar]
- 16. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 17. Bahrami A., Assadi A. H., Markley J. L., Eghbalnia H. R. (2009) Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput. Biol. 5, e1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Berjanskii M. V., Wishart D. S. (2005) A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 127, 14970–14971 [DOI] [PubMed] [Google Scholar]
- 19. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen Y., Lange O., Delaglio F., Rossi P., Aramini J. M., Liu G., Eletsky A., Wu Y., Singarapu K. K., Lemak A., Ignatchenko A., Arrowsmith C. H., Szyperski T., Montelione G. T., Baker D., Bax A. (2008) Consistent blind protein structure generation from NMR chemical shift data. Proc. Natl. Acad. Sci. U.S.A. 105, 4685–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shen Y., Vernon R., Baker D., Bax A. (2009) De novo protein structure generation from incomplete chemical shift assignments. J. Biomol. NMR 43, 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raman S., Huang Y. J., Mao B., Rossi P., Aramini J. M., Liu G., Montelione G. T., Baker D. (2010) Accurate automated protein NMR structure determination using unassigned NOESY data. J. Am. Chem. Soc. 132, 202–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang Y., Schneider W. M., Shen Y., Raman S., Inouye M., Baker D., Roth M. J., Montelione G. T. (2010) Fully automated high-quality NMR structure determination of small 2H-enriched proteins. J. Struct. Funct. Genomics 11, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang Y. J., Powers R., Montelione G. T. (2005) Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J. Am. Chem. Soc. 127, 1665–1674 [DOI] [PubMed] [Google Scholar]
- 25. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 26. Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Errors in protein structures. Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 27. Boudet J., Duval V., Van Melckebeke H., Blackledge M., Amoroso A., Joris B., Simorre J. P. (2007) Conformational and thermodynamic changes of the repressor/DNA operator complex upon monomerization shed new light on regulation mechanisms of bacterial resistance against β-lactam antibiotics. Nucleic Acids Res. 35, 4384–4395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ko J., Lee D., Park H., Coutsias E. A., Lee J., Seok C. (2011) The FALC-Loop web server for protein loop modeling. Nucleic Acids Res. 39, W210–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A., 3rd, Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 [DOI] [PubMed] [Google Scholar]
- 30. Finnin M. S., Cicero M. P., Davies C., Porter S. J., White S. W., Kreuzer K. N. (1997) The activation domain of the MotA transcription factor from bacteriophage T4. EMBO J. 16, 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Spronk C. A. E. M., Nabuurs S. B., Krieger E., Vriend G., Vuister G. W. (2004) Validation of protein structures derived by NMR spectroscopy. Prog. Nucl. Mag. Res. Sp. 45, 315–337 [Google Scholar]
- 32. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 33. Melckebeke H. V., Vreuls C., Gans P., Filée P., Llabres G., Joris B., Simorre J. P. (2003) Solution structural study of BlaI: implications for the repression of genes involved in β-lactam antibiotic resistance. J. Mol. Biol. 333, 711–720 [DOI] [PubMed] [Google Scholar]
- 34. Tsai K. L., Sun Y. J., Huang C. Y., Yang J. Y., Hung M. C., Hsiao C. D. (2007) Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Res. 35, 6984–6994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kitano K., Kim S. Y., Hakoshima T. (2010) Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure 18, 177–187 [DOI] [PubMed] [Google Scholar]
- 36. Zhang A. P., Pigli Y. Z., Rice P. A. (2010) Structure of the LexA-DNA complex and implications for SOS box measurement. Nature 466, 883–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jin C., Marsden I., Chen X., Liao X. (1999) Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex. J. Mol. Biol. 289, 683–690 [DOI] [PubMed] [Google Scholar]
- 38. Blanco A. G., Sola M., Gomis-Rüth F. X., Coll M. (2002) Tandem DNA recognition by PhoB, a two-component signal transduction transcriptional activator. Structure 10, 701–713 [DOI] [PubMed] [Google Scholar]
- 39. Garnett J. A., Marincs F., Baumberg S., Stockley P. G., Phillips S. E. (2008) Structure and function of the arginine repressor-operator complex from Bacillus subtilis. J. Mol. Biol. 379, 284–298 [DOI] [PubMed] [Google Scholar]
- 40. Boura E., Rezabkova L., Brynda J., Obsilova V., Obsil T. (2010) Structure of the human FOXO4-DBD-DNA complex at 1.9 A resolution reveals new details of FOXO binding to the DNA. Acta Crystallogr. D Biol. Crystallogr. 66, 1351–1357 [DOI] [PubMed] [Google Scholar]
- 41. Gajiwala K. S., Chen H., Cornille F., Roques B. P., Reith W., Mach B., Burley S. K. (2000) Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 403, 916–921 [DOI] [PubMed] [Google Scholar]
- 42. Nicholls A., Sharp K. A., Honig B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 [DOI] [PubMed] [Google Scholar]
- 43. Costa R. H., Grayson D. R., Darnell J. E., Jr. (1989) Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and α1-antitrypsin genes. Mol. Cell. Biol. 9, 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.