Background: The mechanism of FtsW-mediated Lipid II transport across the bacterial cytoplasmic membrane is unknown.
Results: Transmembrane segment 4 and particularly two charged residues are required for the transport of Lipid II as well as a maximal size of the substrate.
Conclusion: Lipid II is specifically transported possibly through a porelike structure.
Significance: Elucidating how FtsW acts is crucial for understanding how lipid flippases function in general.
Keywords: Antibiotics, Cell Wall, Lipid Transport, Membrane Biogenesis, Phospholipid
Abstract
Synthesis of biogenic membranes requires transbilayer movement of lipid-linked sugar molecules. This biological process, which is fundamental in prokaryotic cells, remains as yet not clearly understood. In order to obtain insights into the molecular basis of its mode of action, we analyzed the structure-function relationship between Lipid II, the important building block of the bacterial cell wall, and its inner membrane-localized transporter FtsW. Here, we show that the predicted transmembrane helix 4 of Escherichia coli FtsW (this protein consists of 10 predicted transmembrane segments) is required for the transport activity of the protein. We have identified two charged residues (Arg145 and Lys153) within this segment that are specifically involved in the flipping of Lipid II. Mutating these two amino acids to uncharged ones affected the transport activity of FtsW. This was consistent with loss of in vivo activity of the mutants, as manifested by their inability to complement a temperature-sensitive strain of FtsW. The transport activity of FtsW could be inhibited with a Lipid II variant having an additional size of 420 Da. Reducing the size of this analog by about 274 Da resulted in the resumption of the transport activity of FtsW. This suggests that the integral membrane protein FtsW forms a size-restricted porelike structure, which accommodates Lipid II during transport across the bacterial cytoplasmic membrane.
Introduction
To successfully produce two identical daughter cells, most bacteria have to undergo at least two fundamental processes during their growth: cell wall synthesis and cell division. The cell wall or peptidoglycan layer is unique to bacteria, and therefore each step in its synthesis is a potentially important target for antibiotic development. In rod-shaped bacteria, cell wall synthesis comprises three main steps: (i) the synthesis of soluble precursors in the cytoplasm, (ii) assembly and translocation of the finale cell wall precursor Lipid II across the membrane, and (iii) the transglycosylation and transpeptidation of the lipid-linked precursor on the exterior side of the membrane (for a recent review, see Ref. 1). The study of the translocation step has been the subject of several endeavors. In this respect, one of the proteins, the cell division protein FtsW, responsible for the translocation of the building block (i.e. Lipid II) across the cytoplasmic membrane was recently identified (2). In view of this advance, the interesting outstanding question regarding the mode of action of this transporter arises. FtsW is essential, is conserved among almost all eubacteria that synthesize a peptidoglycan cell wall, and has no counterpart in humans. Therefore, this protein not only represents a potential new target for antibiotics, but it may be considered as an important model for understanding how energy-independent flippases (membrane proteins that facilitate energy-independent bidirectional transmembrane movement of lipids) interact with and transport their substrates.
FtsW is an integral membrane protein predicted to have 10 transmembrane (TM)2 domains (3). To date, no structural evidence has been reported with regard to the exact mechanism of FtsW function. However, a functional analysis of this protein in the model organism Escherichia coli performed by Pastoret et al. (4), revealed invaluable information about the possible role of various segments of the protein. These authors have shown that the periplasmic loop between TM9 and TM10 is required for the septal localization of penicillin-binding protein 3 (PBP3). A direct interaction between FtsW and PBP3 in vivo and in vitro was recently established (5). In addition, TM9 and TM10 appeared to be involved in the interaction between FtsW and PBP1B. The amino acids in segment Glu240–Ala249 of the periplasmic loop between TM7 and TM8 are indispensable for the participation of FtsW in the septal peptidoglycan assembly, whereas the first fragment (from amino acid residue 1 to 75) is involved in the interaction with FtsQ (TM3 and TM4) (6). Although it is important to gain a better understanding of the potential function of different regions, the function of the protein at the biochemical level, in particular how it accomplishes its role as a transporter, remains obscure. In this study, a structure-function analysis was undertaken in order to obtain insights into the molecular and cellular details of the transport process of Lipid II. We focused on important TMs of FtsW and residues therein and addressed the substrate specificity of FtsW with respect to the size of the substrate and whether phospholipids are also substrates for transport activity of this protein.
Using FtsW mutants, a detailed exploration of which protein segments and amino acid residues contribute to Lipid II transport was performed. As assayed by the ability of each mutant to transport a fluorescent Lipid II analog in the reconstituted system described previously (2), it was demonstrated that the predicted TM4 is important for the transport activity of FtsW. Importantly, this segment contains the charged residues Arg145 and Lys153, which were found to be specifically needed for FtsW function as a transporter. Analyses of the importance of the size of the Lipid II headgroup suggests that the transport by FtsW possibly occurs through a porelike structure with a limited size that is able to accommodate Lipid II during passage across the cytoplasmic membrane.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
The strains and plasmids used in this study are listed in Table 1. All Escherichia coli strains were grown at various temperatures (28, 37, or 42 °C) in LB medium containing 10 g of bactotryptone, 5 g of yeast extract, and 10 g of NaCl per liter or TY (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl per liter, pH 7.0). Where appropriate, kanamycin or ampicillin was added to media at a final concentration of 50 or 100 μg/ml, respectively. Absorbance was measured at 600 nm with a Helios Epsilon spectrophotometer (Thermo Scientific) or with a Biochrom Libra S70 spectrophotometer (Biochrom).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Relevant genotype | Source/Reference |
|---|---|---|
| E. coli strains | ||
| JLB17 | F− thr tr phis thy ara lac gal xyl mtl rspL tonA ftsW (Ts) | Ishino et al. (24) |
| C43(DE3) | BL21 (DE3)-derivative | Avidis (France) |
| LMC500 | (MC4100 LysA) F− araD139 Δ(argF-lac)U169 deoC1 flbB5301 ptsF25rbsR relA1 rpsL150 lysA1 | Taschner et al. (25) |
| Plasmids | ||
| pDML2400 | pET28a, His-ftsW | Pastoret et al. (4) |
| pDSW311 | pDSW209-ftsW | Mercer and Weiss (7) |
| pTrc99A | Expression vector, Ampr | Gift from A. Bouhss |
Generation of Transmembrane Segment ftsW Mutants
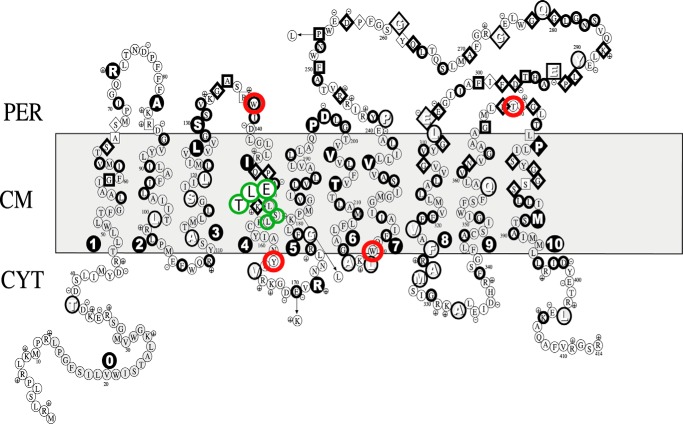
Several constructs have been designed where FtsW systematically lacks various TM segments. Mutants of FtsW were generated that carry a single mutation where the respective amino acid codon is changed in a TAG stop codon (see Fig. 1). For this, plasmid pDML2400 (described by Pastoret et al. (4)) bearing the wild-type ftsW gene was used as a template for site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer's recommendations. The primers for each specific site are listed in Table 2. The mutated genes were confirmed by DNA sequencing.
FIGURE 1.
Putative membrane topology model of E. coli FtsW (reprinted with permission from Ref. 3). The generation of FtsW mutants (used in this study) lacking various TM segments was based on this predicted membrane topology model. Red circles, sites where the stop codons were introduced to create mutants lacking TM10, TM7–TM10, TM5–TM10, or TM4–TM10. Green circles, triple alanine substitution mutants in TM4.
TABLE 2.
Primers used to generate different TAG mutants resulting in truncated forms of FtsW lacking the specified transmembrane segments
| Amino acid | Primers | Mutated protein |
|---|---|---|
| Trp138 (g413a) | 5′-aaaggggcatcgcgttagatcgatctcggtttg-3′ | FtsWΔTM4–10 |
| 5′-caaaccgagatcgatctaacgcgatgccccttt-3′ | ||
| Tyr163 (t489g) | 5′-gctgttttgctatatcgccaactagctggtgcgtaaag-3′ | FtsWΔTM5–10 |
| 5′-ctttacgcaccagctagttggcgatatagcaaaacagc-3′ | ||
| Trp220 (g659a) | 5′-cctggcgggagcgaaattgtagcagttcattg-3′ | FtsWΔTM7–10 |
| 5′-caatgaactgctacaatttcgctcccgccagg-3′ | ||
| Thr369 (a1105t,c1106a,c1107g) | 5′-cggcggggatgttaccgtagaaaggtctgacattgcc-3′ | FtsWΔTM10 |
| 5′-ggcaatgtcagacctttctacggtaacatccccgccg-3′ |
Amino acids substitutions in TM4 were made following the same procedure for site-directed mutagenesis as described above. To construct the mutant R145A, where arginine was changed in alanine, the primer pair 5′-GATCGATCTCGGTTTGCTGGCTATCCAGCCTGCGGAGGCTG-3′ and 5′-CAGCTCCGCAGGCTGGATAGCCAGCAAACCGAGATCGATC-3′ was used. For the mutant K153N where lysine was mutated to asparagine, the primers 5′-GCGGAGCTGACAAATCTGTCGCTGTTTTG-3′ and 5′-CAAAACAGCGACAGATTTGTCAGCTCCGC-3′ were employed (mutated bases are in boldface type). The triple alanine substitutions in TM4a were generated with the primers 5′-AAACAGCGACAGTTTAGCGGCCGCCGCAGGCTGGATACG-3′ and 5′-GAGACCACATGGTCCTTCTTGAG-3′. To create three alanine substitutions in TM4b, the primer pair 5′-GGCGATATAGCAAAAAGCGGCCGCTTTTGTCAGCTCCGC-3′ and 5′-GAGACCACATGGTCCTTCTTGAG-3′ was used. For these two triple alanine mutants, pDSW311 (7) was used as template.
Generation of GFP-FtsW Fusions
To study localization of FtsW, GFP fusions to this protein and its derivatives were needed (because an antibody against FtsW was not available). Wild-type ftsW and substitution mutants with restriction sites for EcoRI and HindIII were obtained by PCR using the primers 5′-CACGAATTCAACAACAACCGTTTATCTCTCCCTCGCCTGAAAATGC-3′ and 5′-GCCGCAAGCTTATCATCGTGAACCTC-3′ (sites underlined) and the plasmid pDML2400 as template. The PCR products were digested with EcoRI and HindIII and ligated into the same sites of the gfp fusion vector pDSW311 (a gift from David Weiss). In this vector, GFP is fused to the N terminus of FtsW. The resulting construct allowed expression of this gfp-ftsW fusion under control of a weak IPTG-regulated trc promoter (7).
The localization of the GFP-FtsW fusions was assessed in the wild-type E. coli strain LMC500. To this end, transformants were cultured overnight at 28 °C in TY supplemented with 100 μg/ml ampicillin and 0.5% glucose (to suppress the expression of the plasmids). Thereafter, the cells were diluted 1:500 in the same medium without glucose and cultured to an A600 of 0.15. To one part of the culture, 10 μm IPTG was then added. After 1.5 h of induction, the cells were fixed in 2.8% formaldehyde and 0.04% glutaraldehyde in growth medium (with agitation) for 15 min at room temperature (8).
Complementation Assay
The complementation assay was performed as described before (4) using E. coli JLB17. This temperature-sensitive strain of ftsW was characterized previously by Pastoret et al. (4). Constructs containing the wild type or substitution mutants of FtsW (the mutants lacking various segments of FtsW could not be tested in vivo because they are involved in several interactions with other proteins as described above) were cloned in pTrc99A vector and transformed to JLB17. The transformants were grown overnight at 28 °C in TY without NaCl supplemented with 0.5% glucose and 20 mg of thymine/liter. The culture was then diluted 1:50 in medium without glucose and grown to A600 of 0.3. Next, the culture was diluted 1:25 in prewarmed medium at 28 and 42 °C and grown at this temperature for an additional 2.5 h.
Immunofluorescence Microscopy
For GFP fluorescence, the cells were first immobilized on 1% agarose following the procedure described by Koppelman et al. (9). Images in the phase-contrast mode or with the GFP filter (U-MNB filter cube with a 470–490-nm band pass excitation filter and a 515-nm long pass emission filter) were obtained using an Olympus BX-60 fluorescence microscope, equipped with a CoolSnap fx (Photometrics) CCD camera through an UPLANFL ×100/1.3 numerical aperture oil objective (Japan). Both images (in the phase-contrast mode and fluorescence) were hyperstacked, and the length and diameter of the cells were determined from the phase-contrast images, and the localization and intensity of the fluorescence signal was analyzed in the fluorescence images of the bacteria using the public domain program Coli-Inspector written by Norbert Vischer (University of Amsterdam), which is based on ImageJ by Wayne Rasband (National Institutes of Health). The map of fluorescence profiles shows all of the measured cells sorted according to length and, for each bacterium, the distribution of the fluorescence along its length axis (10).
Protein Purification
The expression and purification of different His-tagged proteins were performed as described before (2). The constructs for expression of wild type and mutant (different domains of FtsW or substitution mutants) FtsW proteins were transformed to C43 (DE3) cells and purified by nickel affinity chromatography by previously established procedures (2).
Dithionite Reduction Assay
Phospholipids and Bacterial Cell Wall Precursors
Phospholipids and analogues thereof labeled with 7-nitro-2,1,3-benzoxadiazol-4-yl (NBD)-chloride (NBD-PL) were purchased from Avanti Polar Lipids (Alabaster, AL). Stock solutions were prepared in chloroform/methanol (1:1, v/v) and stored under nitrogen at −20 °C. Phospholipids concentration was determined with a lipid phosphorus assay according to Rouser et al. (11). The synthesis and purification of the cell wall precursor NBD-labeled Lipid II was performed as described before (12, 13). All other chemicals were obtained from Sigma.
General Procedure for the Synthesis of NBD-Amino Acid-Alkyne (Compounds I–III)
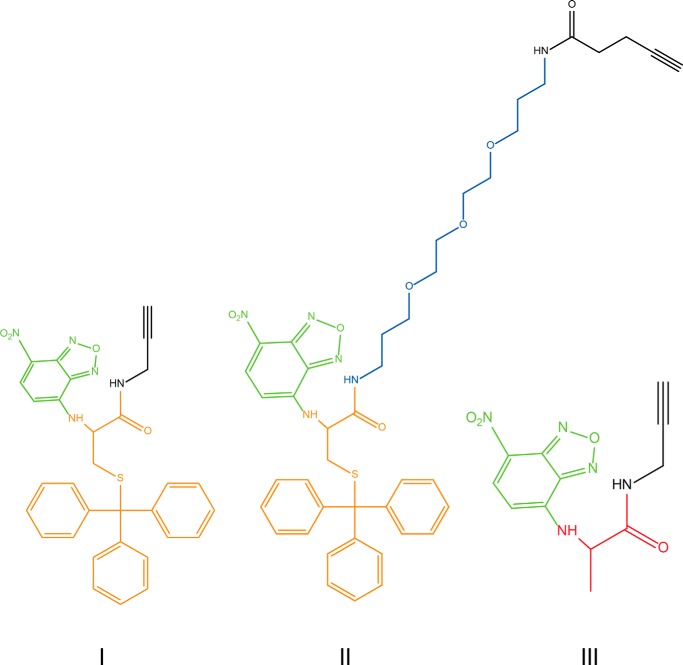
First, 1.4 mmol of the respective amino acid (500 mg of H-Cys(trityl)-OH or 124 mg of alanine) was dissolved in 40 ml of MeOH/water (1:1, v/v). This was followed by the addition of 3 mmol of NBD-Cl in small portions, and finally 3.5 ml of a 1 m NaOH solution was added. The reaction was stirred for 3 h at room temperature, after which 200 ml of CH2Cl2 was added as well as 7 ml of 1 m HCl, and the mixture was washed five times with brine. The CH2Cl2 was evaporated off, and the resulting residue was used without further purification in the next step (yield, ∼50–60%).
The residue was dissolved in 40 ml of CH2Cl2, and 1 mmol of (benzotriazol-1-yloxy)tris(dimethylamino)phosphonium hexafluorophosphate, 1.2 mmol of diisopropylethylamine, and 1.2 mmol of propargyl amine or N-(3-(2-(2-(3-aminopropoxy)ethoxy)ethoxy)propyl)pent-4-ynamide were added, and the reaction mixture was stirred overnight at room temperature. The CH2Cl2 was removed in vacuo, and EtOAc was added (75 ml). The solution was washed with 1 m KHSO4 (2 × 50 ml), saturated NaHCO3 (2 × 50 ml), and brine (2 × 50 ml) and dried over Na2SO4(s). The solvent was removed in vacuo, and the residue was purified using flash chromatography on silica with solvents 4:6 EtOAc/hexane or EtOAc for the reaction with propargylamine or with the alkyne-polyethylene glycol (PEG)-amine spacer, respectively. Calculated compound I C31H25N5O4S: 563.16. Found ESI-MS m/z: 562.2 [M − H]−. Calculated compound II C43H48N6O8S: 808.33. Found ESI-MS m/z: 807.3 [M − H]−. Calculated compound III C12H11N5O4: 289.08. Found ESI-MS m/z: 288.2 [M − H]−.
The alkyne-modified PEG amine linker (in analog 2) was prepared as described previously (14). Briefly, mono-Boc protection of 4,7,10-trioxa-1,13-tridecanediamine was carried out as detailed before (15), after which 4-pentynoic acid was coupled (14). Removal of the Boc group was accomplished using standard conditions (1:1 trifluoroacetic acid/dichloromethane, 1 h) to provide the TFA salt of the desired product as a light yellow oil that was used directly.
Conversion of the Lysine Form of Lipid II into the Azidolysine Form
Lipid II (lysine form, 1 μmol) purified as described earlier (12) was dissolved in 2 ml of water containing 1 mm CuSO4 and 0.5% Triton X-100 (w/v). Subsequently, 10 μmol of imidazole-1-sulfonyl azide hydrochloride (20) was added, followed by 20 μmol of diisopropylethylamine. Complete conversion could be observed after 4 h, upon which the reaction mixture was extracted with 4 ml of butanol, 6 m pyridine acetate, pH 4.2; the butanol phase was applied to a DEAE-cellulose column (1 × 2.5 cm, height/diameter); and the azido-Lipid II was purified as described (12) and stored at −20 °C in chloroform/methanol (1:1, v/v) until use.
General Procedure for Labeling Azido-Lipid II via Click Chemistry
Azido-Lipid II (0.5 μmol) was dissolved in either 2 ml of 10 mm Tris-HCl, pH 7.5, containing 0.1% Triton X-100 or 2 ml of methanol, depending on the solubility of the alkyne-containing compound to be coupled (alkyne label). This was followed by the addition of 2 mm CuSO4 (final concentration) and 1.2 eq of alkyne label. The reaction was started by the addition of 1 mm sodium ascorbate, and the reaction was stirred for 4 h or until completion of the reaction (this can be confirmed by the absence of azido-Lipid II, m/z 950 [M − 2H]2− in a mass spectrum). Each hour, an additional amount (again 1 mm) of sodium ascorbate was added to the reaction. When buffer was used, the reaction mixture was then extracted with 4 ml of butanol, 6 m pyridine acetate, pH 4.2, and the labeled Lipid II was purified as described above. When methanol was used, this was first evaporated in vacuo, followed by the addition of 2 ml of water, and then extracted and purified as outlined above.
Preparation of Proteoliposomes
Large unilamellar vesicles (LUVs) containing fluorescently labeled Lipid II/phospholipids were prepared. This was achieved by mixing 60 mol % 1,2-dioleoyl-sn-glycero-3-phosphocholine, 25 mol % 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 15 mol % 1,2-dioleoyl-sn-glycero-3-phosphoglycerol from stock solutions. For the incorporation of Lipid II/labeled phospholipid, 0.1 mol % NBD-Lipid II/NBD-PL was added to the mixture. Lipids were dried by nitrogen stream, followed by vacuum desiccation for 2 h. The lipid films were hydrated with a buffer containing 10 mm Hepes-KOH, pH 8.0, 100 mm NaCl, 5 mm KCl, and 1 mm MgSO4 to a lipid phosphate concentration of ∼5 mm. The vesicle's suspension was then subjected to 10 cycles of freezing and thawing. Subsequently, unilamellar vesicles were formed by manually extruding the suspension 10 times through 200-nm membrane filters (Anotop 10, Whatman, Maidstone, UK).
Reconstitution of Proteins into Proteoliposomes
For reconstitution experiments, 350 μl of LUVs (5 mm lipid phosphate) were first solubilized with 8 mm Triton X-100, followed by the addition of a purified protein. The final concentration of the proteins was adjusted to a protein/phospholipid molar ratio of ∼1:20,000. This was followed by incubation with gentle agitation for 1 h at 4 °C. Samples were then supplemented with 100 mg/ml Bio-Beads SM-2 adsorbent (Bio-Rad) to remove the detergent. After 2 h of incubation at 4 °C, another 100 mg/ml fresh Bio-Beads suspension was added to the micelle solution, and a second incubation overnight at 4 °C was undertaken. Thereafter, a third incubation with 100 mg/ml fresh Bio-Beads was performed for 2 h. Subsequently, the vesicles were collected by ultracentrifugation at 435,000 × g for 30 min and resuspended in 200 μl of 10 mm Hepes-KOH, pH 8, 100 mm NaCl, 5 mm KCl, and 1 mm MgSO4.
Fluorescence Measurement
The in-to-out translocation of NBD-Lipid II/phospholipid in LUVs and proteoliposomes was measured by determining the percentage of NBD fluorescence that is not available for reduction by 8 mm sodium dithionite, added from a 1 m stock solution in 1 m Tris, pH 11. After quenching, 10 μl of a 10% (w/v) solution of Triton X-100 in water was added to permeabilize the membrane and make all remaining NBD label available for dithionite reduction. Measurements were performed at 20 °C in 1 ml of buffer in a quartz cuvette using an SLM Aminco SPF 500C fluorimeter. The excitation and emission wavelengths were adjusted to 481 and 534 nm, respectively.
RESULTS
TM4 Is Required for the Transport Activity of FtsW
To determine which segments of the transporter are important for the flipping of Lipid II across the bacterial membrane, mutants lacking TM segments of E. coli FtsW were generated (segments were chosen based on the findings published in Ref. 4) and are described above; they were shown to be important for interaction with proteins involved in cell division or cell wall synthesis. The TM mutants were obtained by incorporation of TAG stop codons at specific sites (Fig. 1) using site-directed mutagenesis. The mutants were then assessed for their ability to facilitate the transbilayer movement of Lipid II. To this end, truncated forms of FtsW were purified and reconstituted into LUVs containing NBD-Lipid II. Transport of NBD-Lipid II from the inner to the outer leaflet of the vesicles was monitored using the validated dithionite reduction assay (2). This assay is based on specific quenching of the fluorescence of NBD-Lipid II by the membrane-impermeable reductant dithionite. As depicted in Fig. 2A, the addition of dithionite resulted in quenching of about 50% of the fluorescent signal in LUVs without any protein or containing the control integral membrane protein KcsA. This corresponds to the NBD-Lipid II present in the outer leaflet of the vesicles (NBD-Lipid II in the inner leaflet is protected from quenching by dithionite). Thus, the addition of dithionite results in the rapid conversion of the NBD-Lipid II present in the outer leaflet to the non-fluorescent ABD-Lipid II. This generates a concentration gradient, which can be the driving force for transport. In the presence of wild type FtsW, this quenching increased to almost 70%, reflecting that only 30% of the NBD-Lipid II remained inaccessible to dithionite and thus suggesting transbilayer movement of NBD-Lipid II, reproducing our earlier results (2). Vesicles containing FtsW derived from the mutant lacking TM10, TM7–TM10, or TM5–TM10 showed about 65% quenching of fluorescence upon reduction by dithionite, indicating the translocation of NBD-Lipid II from the inner to the outer leaflet of the bilayer. This implies that the transmembrane segments 5–10 are not essential for the transport activity of FtsW in our assay.
FIGURE 2.
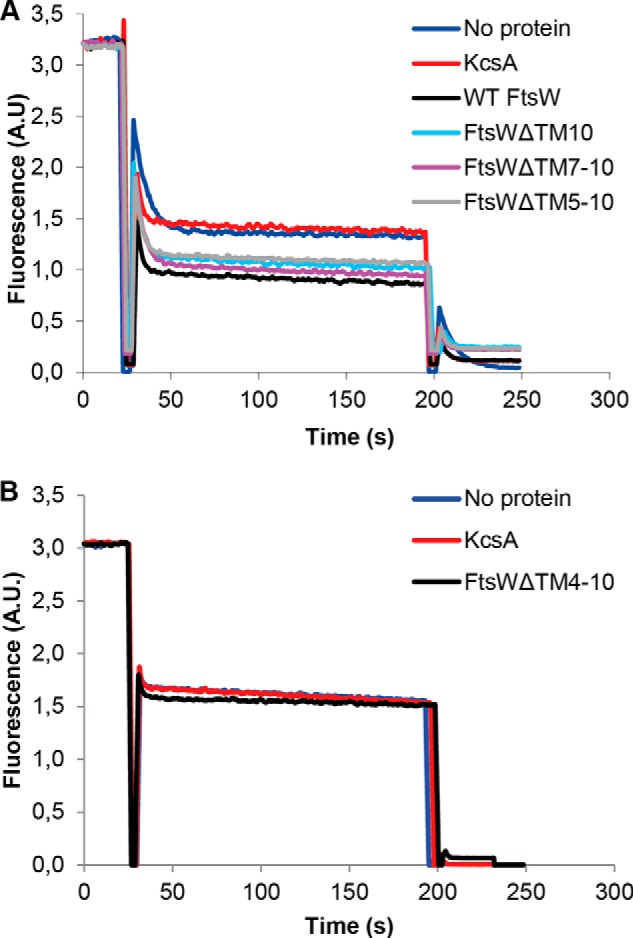
The transport activity of FtsW requires TM4. A, TM5–TM10 are not essential for the transport of NBD-Lipid II. Mutants lacking various segments of FtsW were assessed for transport activity using the dithionite reduction assay. As described under “Materials and Methods,” this was achieved by reconstitution of the purified proteins into LUVs containing NBD-Lipid II symmetrically distributed between inner and outer leaflets of the bilayer. After solubilization with Triton X-100, LUVs were reconstituted with no protein, the control protein KcsA, or FtsW. After the addition of dithionite (1), a reduction of almost 50% of the fluorescence signal is observed in protein-free vesicles and proteoliposomes containing KcsA. On the contrary, about 70% quenching is detectable when vesicles containing wild type FtsW or FtsW lacking TM10 or TM7–TM10 or TM5–TM10 were assayed. When 0.1% Triton X-100 was added (2) to permeabilize the model membranes, a complete quenching of all of the fluorescence is attained. B, TM4 is involved in the transport of NBD-Lipid II. FtsW lacking TM4–TM10 displayed the same effect as the controls (LUVs with no protein or with KcsA) with regard to the extent of quenching by dithionite. All measurements were carried out at 20 °C and are representative of at least three independent experiments. A.U., arbitrary units.
When the mutant lacking TM4–TM10 was tested, the extent of quenching by dithionite was similar (about 50% reduction in fluorescence of NBD-Lipid II) to that of the control LUVs without any protein or reconstituted with KcsA (Fig. 2B). Thus, the transbilayer movement of NBD-Lipid II did not take place, which suggests that TM4 is necessary for the flipping activity of FtsW.
An important observation regarding the TM4 is that it contains charged amino acids. These may be important for the interaction with Lipid II (given its negative charge) and hence the transport activity of FtsW.
Charged Residues within TM4, Arg145 and Lys153, Are Crucial for Flippase Function of FtsW
To assess whether the charged amino acids in TM4 are required for the flippase function of FtsW, the residues Arg145 and Lys153 (the latter is conserved among most bacteria; see Fig. 3) were changed to neutral or uncharged amino acids, and the resulting mutants were assessed for their ability to facilitate transport of Lipid II. Purified FtsW from the mutant where Arg145 was mutated to and the mutant where Lys153 was replaced by asparagine was reconstituted in vesicles with NBD-Lipid II and assayed in the dithionite reduction test. Alteration of the charge via substitutions R145A and K153N resulted in loss of transport activity of FtsW (Fig. 4A), demonstrating the requirement of these residues for the flippase function of the protein. Moreover, the assay was performed with two additional mutants, where three consecutive alanine substitutions in the middle of TM4 were made (Fig. 1). In TM4a, triple alanine substitutions of the amino acids Glu150, Leu151, and Thr152 were made. In TM4b, the amino acids Leu154, Ser155, and Leu156 were substituted. The ability of these FtsW mutants to transport Lipid II was not affected (Fig. 4B). These uncharged amino acids in addition to the negatively charged Glu150 seem therefore not to be required for the translocation of Lipid II.
FIGURE 3.

Sequence alignment of the residues Arg145 and Lys153 in the predicted TM4 of FtsW in E. coli and other bacteria using the Clustal version 2.1 multiple sequence alignment program. The residues Arg145 and Lys153 and their homologues are highlighted in green and red, respectively.
FIGURE 4.
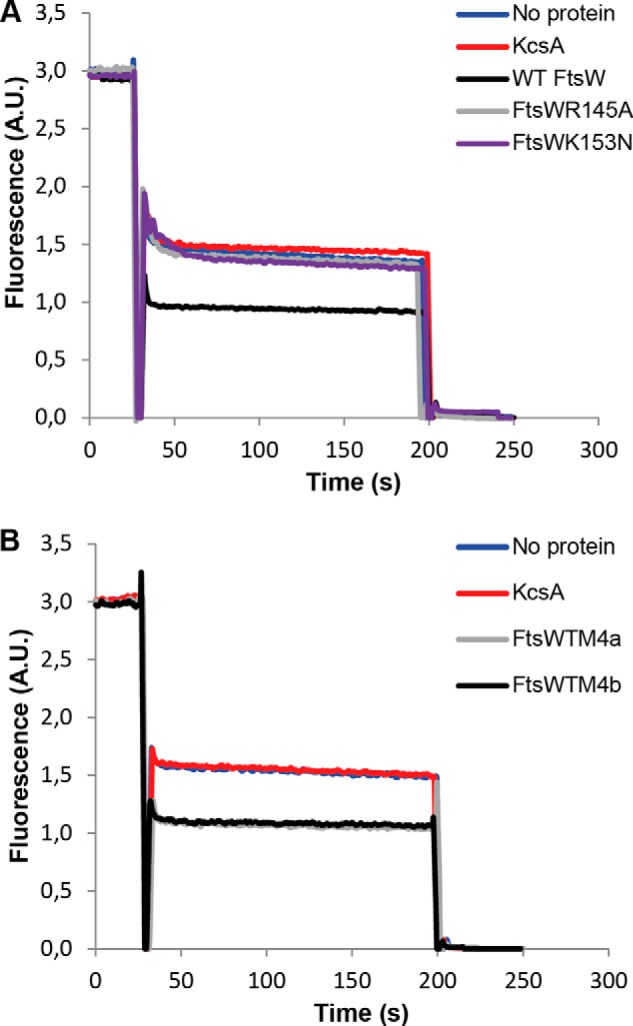
The residues Arg145 and Lys153 within TM4 are essential for the transport activity of FtsW. A, substitution of arginine 145 by alanine abolished the transport activity of FtsW protein. Mutating lysine 153 to asparagine resulted in loss of transport activity of FtsW as well. B, triple alanine substitutions of the amino acids glutamic acid 150, leucine 151, and threonine 152 (TM4a) and the residues leucine 154, serine 155, and leucine 156 (TM4b) (see also Fig. 1 for the putative topology model) did not alter the transport activity of FtsW. The assay was carried out as detailed in the legend to Fig. 2. All measurements were performed at 20 °C and are representative of at least three independent experiments. A.U., arbitrary units.
Changing the Residue Arg145 to Ala or Substitution of Lys153 with Asn Affected the in Vivo Activity of the FtsW Protein
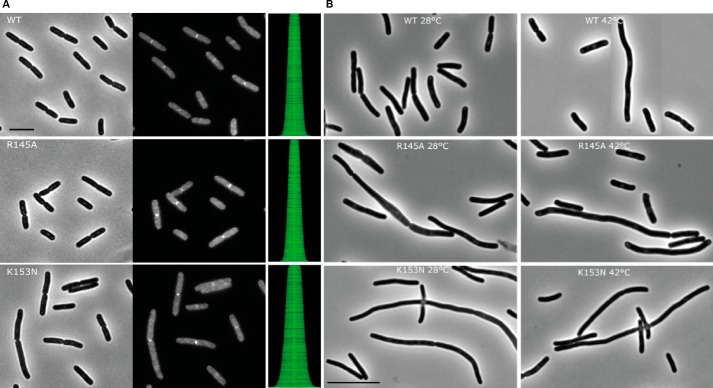
FtsW has been reported to localize to the septum (16). The effect of the charge substitution in FtsW mutants on the arrangement of the protein at the division site was assessed using the wild-type strain E. coli LMC500. FtsW and its derivatives were fused to the C-terminal end of the GFP and placed under the control of a weakened IPTG-inducible trc promoter. Cellular localization of FtsW in LMC500 transformants expressing the GFP-FtsW WT, GFP-FtsWR145A, and GFP-FtsWK153N grown at 28 °C in TY medium was examined by immunofluorescence microscopy. All of these GFP protein fusions were shown to target the division site (Fig. 5A). The map of fluorescence profiles also shows the presence of fluorescence signal at the septum. Thus, the mutations did not affect the localization of the protein at mid-cell, suggesting that (at least) those parts of FtsW that are involved in interactions with other proteins within the divisome are correctly folded and inserted in the membrane.
FIGURE 5.
A, localization of GFP-FtsW fusions in wild-type E. coli LMC500. The constructs containing wild type ftsW and the substitution mutants fused to the C terminus of mut2gfp were transformed to LMC500 and grown at 28 °C in TY medium as described under “Materials and Methods.” The expression of the proteins was induced with 10 μm IPTG for 90 min. Each panel consists of (from left to right) a phase-contrast image, a GFP fluorescence image, and a map of fluorescence profiles of 500- 1000 cells. Scale bar, 5 μm. B, complementation assay of wild-type FtsW and substitution mutants in strain JLB17. The plasmids containing wild type ftsW and the substitution mutants were transformed to JLB17FtsW(Ts) and grown at 28 or 42 °C in TY medium without salt supplemented with thymine as described under “Materials and Methods.” The cells were imaged by phase-contrast microscopy. The image of the FtsW(Ts) cells expressing wild-type FtsW at 42 °C is a composite image to show that in this culture also some filaments were present. Note that the JLB17FtsW(Ts) cells expressing FtsWK153N are thinner than the cells expressing WT FtsW or FtsWR145A. Scale bar, 10 μm.
To assess the mutants for complementation of the E. coli JLB17 (this FtsW temperature-sensitive (Ts) strain produces FtsW G311D that is not functional at the restrictive temperature of 42 °C (4), the constructs were first cloned in the pTrc99A vector (to ensure low expression to closely reflect the wild-type situation) and then transformed to the Ts strain. E. coli JLB17 cells producing FtsW R145A have proven very difficult to obtain; attempts to transform these cells with the target construct often failed or the cells reverted to the wild type genotype or contained other (suppressor) mutations. When the transformation was successful, the cells displayed normal rod-shaped morphology (some elongated cells were also visible) as the wild type at 28 °C but were not able to complement the ftsW(Ts) at 42 °C (Fig. 5B). This same result was attained in the presence of 10 μm IPTG (overproduction of FtsW and its derivatives). Elevated production of wild type FtsW or its derivatives resulted in inhibition of cell growth, which is consistent with the observations reported before (17). The cells expressing FtsW K153N displayed a dominant negative effect because they exhibited filamentous growth at the permissive temperature, denoting that the protein was able to compete with the Ts mutation for localization and prevented it from being functional at 28 °C. At 42 °C, this mutant showed the same morphology, indicating that it cannot complement ftsW(Ts) at this temperature. Altogether, these two substitution mutations did not affect the localization activity of FtsW but altered another essential function, which is consistent with the impairment of the Lipid II transport activity and subsequently the synthesis of the cell wall. To further explore the specificity of FtsW for its substrate Lipid II, the effect of both the wild type and these mutants was assessed on the transport of NBD-PL.
Arg145 and Lys153 Are Not Important for the Transport of NBD-PL
The E. coli membranes contain about 70–80% phosphatidylethanolamine (PE), 20–25% phosphatidylglycerol (PG), and 5% cardiolipin. The transport assay was performed with vesicles containing NBD-PE or NBD-PG, where the NBD fluorophore was linked to the fatty acid chain of the phospholipids. Interestingly, wild-type FtsW was shown to allow the equilibration of NBD-PE and NBD-PG between the two leaflets of the model membrane bilayer (Fig. 6). This capability of FtsW supports the previous postulation that cell wall synthesis depends on ongoing phospholipid synthesis (18).
FIGURE 6.
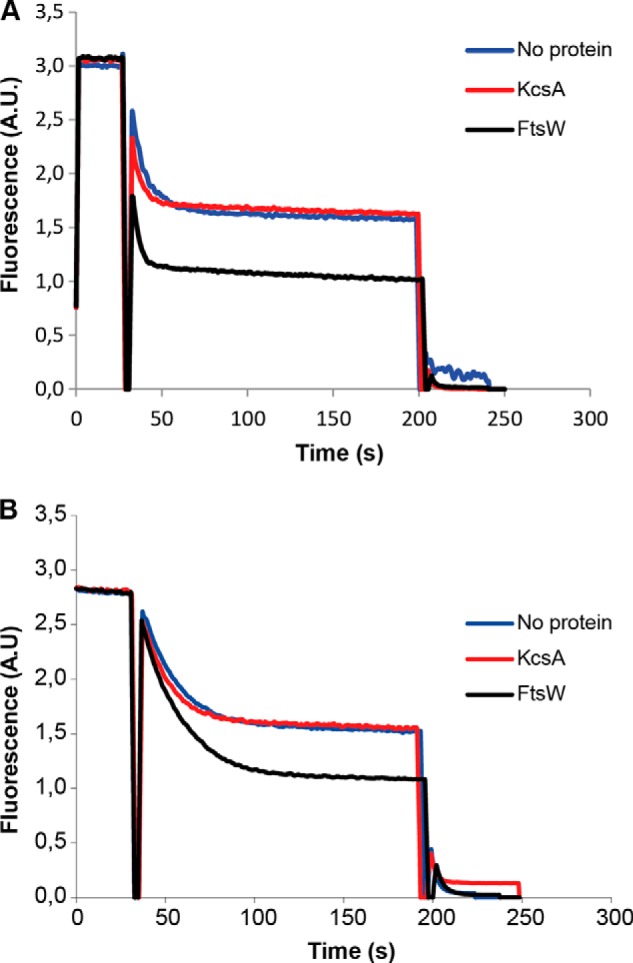
FtsW is able to transport NBD-PE and NBD-PG. LUVs containing NBD-PE (A) or NBD-PG (B) were reconstituted with no protein, KcsA, or FtsW as described before (see details in the legend to Fig. 2).
This phospholipid transport activity of FtsW was not affected by the substitutions R145A and K153N (Fig. 7, A and B). To establish whether there is any headgroup specificity, the effect of these mutants was additionally assessed on the transport of NBD-phosphatidylcholine (PC) (PC is not present in the E. coli membranes). With these substitutions, FtsW was still able to transport NBD-PC (Fig. 7C). This means that these residues are not required for the transport of NBD-PL but are specifically involved in the flipping activity of NBD-Lipid II. This suggests that phospholipids are transported by a different mechanism than Lipid II.
FIGURE 7.
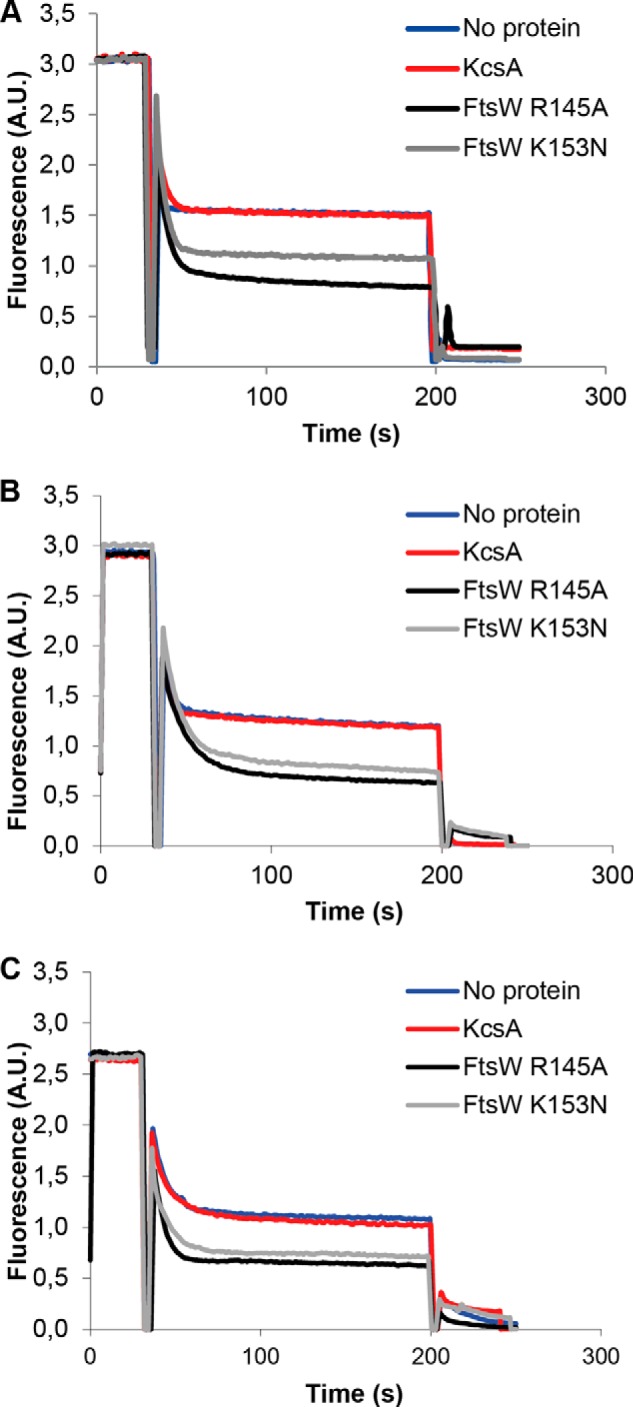
The residues Arg145 and Lys153 of FtsW are not important for the transport of NBD-PL. Substitution mutants R145A and K153N did not affect the ability of FtsW to transport NBD-PE (A) or NBD-PG (B) in model membranes. The flippase activity of FtsW R145A and FtsW K153N was shown to be independent of the headgroup of the phospholipid because these mutants were able to facilitate the transbilayer movement of NBD-PC (C). The assay was carried out as detailed in the legend to Fig. 2.
Transport of Lipid II by FtsW Seems to Occur through a Pore Mechanism
The transport process was demonstrated to occur rapidly and independent of any metabolic energy (2, 13), pointing to a facilitated diffusion mechanism where FtsW facilitates the movement of Lipid II between the two leaflets of the bacterial cytoplasmic membrane. In view of this and the present observations, a pore-mediated mechanism of transport is likely. To test this hypothesis, Lipid II analogues with different sizes were designed to determine the size of the putative pore that could allow the transport of Lipid II.
To ascertain larger flexibility in obtaining Lipid II analogues with variable sizes, it was first tested whether Lipid II was amenable to click chemistry-based approaches. For this, conversion of the ϵ-amino group of the lysine of Lipid II to an azide using the diazotransfer reagent imidazole-1-sulfonyl azide (19) was intended. A 10-fold excess of this reagent could fully convert the amine form of Lipid II into the azide form in 1 h, which caused a shift of Lipid II on thin layer chromatography (TLC) to a higher Rf value (data not shown). This azido form of Lipid II can be readily used in (copper-catalyzed) click reaction in buffers or methanol.
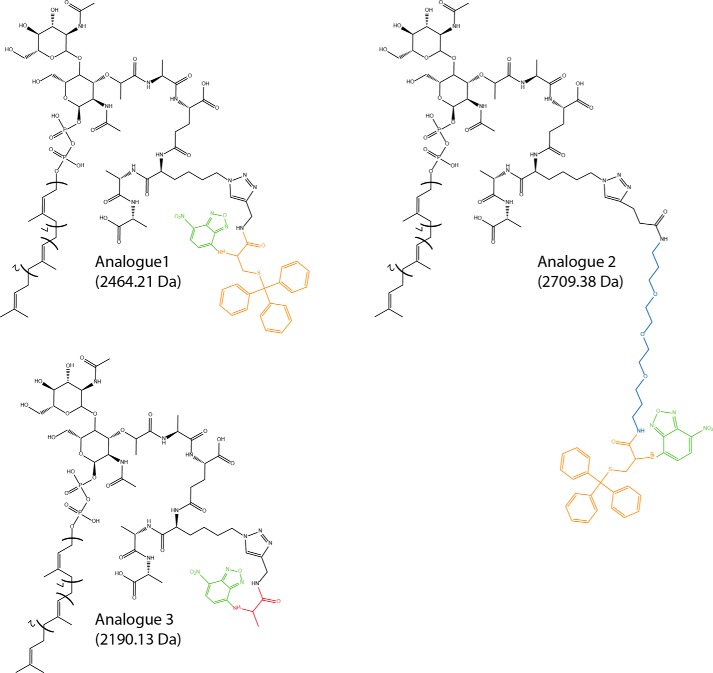
Initially, we attempted to determine the “pore size” of FtsW by following a strategy analogous to that reported on by Bonardi et al. (20), where rigid spherical molecules, linked to the precursor pro-OmpA via maleimide chemistry, were used to probe the size of the SecYEG protein channel (20). A similar approach using the same rigid molecules would require a thiol group on Lipid II. Therefore, a trityl-protected cysteine carrying an NBD group as well as an alkyne group (Fig. 8, compound I) was synthesized that can be coupled to the azido-Lipid II. This would allow in a later stage (after removal of the trityl group) the linking of the rigid spherical molecules via maleimide chemistry. This group was “clicked” to Lipid II and purified. FtsW was then tested for its ability to transport this Lipid II analog (analog 1; Fig. 9). Noticeably, FtsW was not able to facilitate the transbilayer movement of analog 1 (Fig. 10A), suggesting that the attached group was too large to be transported. Next, to confer more flexibility to this variant, a PEG linker between the stem peptide and the NBD-labeled protected cysteine was introduced (analog 2; Fig. 9). Interestingly, analog 2 could be transported by FtsW (Fig. 10B). This signifies that FtsW has a restricted “pore size” through which Lipid II can be transported, whereas flexible attachments, such as a pentaglycine, are tolerated. To further probe the pore size of FtsW, a third analog that was synthesized from an alanine as basal structure was generated. This analog lacks the trityl-protected thiol group (274-Da difference, analog 3; Fig. 9). FtsW allowed the passage of analog 3 (Fig. 10C). Together, these results indicate that accommodation of structures by FtsW requires a maximal size (and a defined shape), which is suggestive of a porelike model of transport.
FIGURE 8.
Structure of compounds I, II, and III used to synthesize Lipid II analogues 1, 2, and 3, respectively. These NBD-amino acid-alkyne compounds were designed and labeled as described under “Materials and Methods.” The NBD group is drawn in green. The cysteine (trityl) backbone is highlighted in orange. The PEG linker in compound II is highlighted in blue, and the Ala backbone in compound III is marked in red.
FIGURE 9.
Structures of Lipid II analogues used in this study. The analogues were synthesized as described under “Materials and Methods.” In these analogues, the NBD fluorophore is highlighted in green. The cysteine (trityl) backbone in analog 1 and 2 is marked in orange. The PEG linker in analog 2 is highlighted in blue, and the Ala backbone in analog 3 is marked in red.
FIGURE 10.
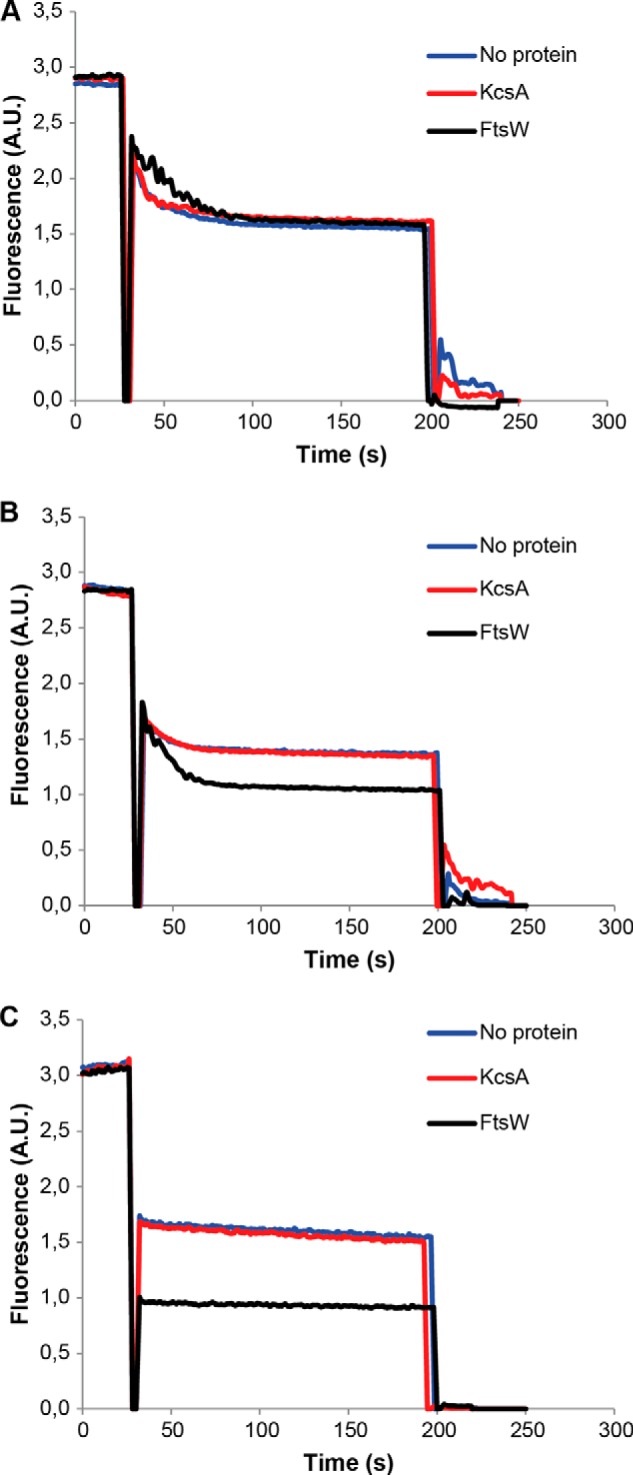
Inhibition of Lipid II analog 1 transport activity of FtsW. In LUVs containing analog 1, the transport was blocked (A), whereas FtsW retained its transport activity in the presence of analog 2 (B). When the size of analog 1 was reduced by about 274 Da (resulting in analog 3), FtsW allowed the passage of this latter (C). The assay was performed as detailed in the legend to Fig. 2.
DISCUSSION
To date, structural information to dissect the function of the FtsW protein as a flippase is lacking. Using our (previously described) reconstituted system, we were able to explore important questions in the transport process of the cell wall precursor Lipid II across the bacterial cytoplasmic membrane; we aimed to (i) identify residues that are required for the transport activity of FtsW and (ii) obtain insights into the nature of the “flipping device” that allows transbilayer movement of Lipid II.
To address the first question and to establish the potential functional importance of various segments of FtsW, different mutants (lacking defined transmembrane segments) were generated, and their effect on the transport of NBD-Lipid II was assessed. Using biochemical reconstitution, we demonstrated that the predicted TM4 is essential for the transport activity of FtsW. This segment was shown to be specifically required for the translocation of Lipid II because the truncated form lacking this domain of the protein was not affected in its ability to facilitate the transbilayer movement of NBD-phospholipids (see below). The essentiality of TM4 in the transport of Lipid II is not completely surprising because this domain contains positively charged residues that could be important for the interaction with the highly negatively charged Lipid II. Within TM4, we identified two charged residues, Arg145 and Lys153, that are specifically involved in the translocation of Lipid II. Changing these charged amino acids to uncharged ones resulted in abrogation of the Lipid II transport activity of FtsW. Interestingly, these substitution mutants were altered in their in vivo activity. Although FtsWR145A and FtsWK153N were able to localize at the division site like the wild type FtsW, the mutants were unable to complement a temperature-sensitive FtsW mutant strain. This indicates that (i) the mutants are correctly folded and inserted into the membrane, and (ii) the mutant proteins are impaired in cell wall assembly, consistent with their loss of transport activity.
During membrane biogenesis, newly synthesized phospholipids at the cytoplasmic side of the membrane must be transported to the exterior side to equilibrate between the two leaflets. This rapid process, essential to maintain bacterial growth, is expected to be mediated by a dedicated membrane protein(s). To assess whether FtsW promotes this route and to further explore the specificity of this protein in flipping Lipid II, we assayed its effect on the transport of NBD-PL. FtsW was capable of facilitating the transmembrane movement of phospholipids (NBD-PE, NBD-PG, and NBD-PC) without displaying any headgroup specificity. The ability of FtsW to transport phospholipids is in agreement with a previous report where the dependence of E. coli peptidoglycan cell wall assembly on ongoing phospholipid synthesis has been proposed (16). The specific requirement of the aforementioned two residues of FtsW in the transport of Lipid II and the lack of (headgroup) specificity in the flipping of phospholipids could point to the occurrence of a binding interaction during the translocation of the much more negatively charged Lipid II, whereas this binding interaction is lacking during the exchange of phospholipids between the leaflets of the membrane. Hence, it is conceivable that FtsW is a specific transporter of Lipid II and that the transport of phospholipids occurs via a different mechanism not necessarily involving TM4. How FtsW mediates the transport of phospholipids remains as yet unclear. It is possible that this process occurs according to the model proposed hitherto by Kol et al. (21). In this model, the highly dynamic nature of the TMs of the membrane protein could disturb the structure of the bilayer at the protein-lipid interface, enabling phospholipids to traverse from one leaflet of the bilayer to the other (21). In this respect, FtsW could resemble rhodopsin, which was recently reported to facilitate bidirectional transport of phospholipids in disc membranes (22, 23).
To understand how FtsW could transport Lipid II across the bacterial cytoplasmic membrane, NBD-Lipid II analogues of different sizes were tested. Although the transmembrane transport of analog 3 was allowed, a 274-Da-bigger variant (analog 1) could not be transported by FtsW. Although the size and the structural differences between these two analogues are not substantial, their different effect would suggest that the transporter possesses a porelike structure of limited size through which the passage of Lipid II is achieved. Interestingly, analog 2, which was even 245 Da larger in molecular mass than analog 1, was transported by FtsW. This is probably due to the flexible linker between the Lipid II headgroup and the NBD-labeled and trityl-protected cysteine moiety that would allow sequential passage through FtsW (see also Fig. 11). Thus, flexibility is important, which would explain why the pentaglycine bridging peptide found in the cell wall of S. aureus (303 Da) is also transported by FtsW.
FIGURE 11.
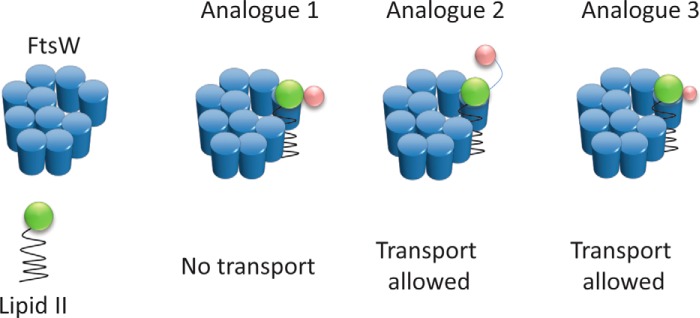
Schematic illustrating that Lipid II transport by FtsW requires restricted size and flexibility of the substrate. FtsW allowed the transport of analog 3, whereas analog 1 that is 247 Da (in molecular mass) bigger than analog 3 could not be transported. This suggests that transport by FtsW possibly occurs through a porelike structure with a limited size. The ability of FtsW to transport analog 2, which is 245 Da larger than analog 1, demonstrates that flexibility of the substrate is important; the presence of the PEG linker in analog 2 (see also Fig. 9) makes it more flexible to move between the leaflets of the membrane, whereas analog 1 (deprived of the linker) is more compact. The additional groups in Lipid II analogues are illustrated by pink spheres.
Altogether, the present findings imply that FtsW could form a porelike structure through which Lipid II is translocated across the cytoplasmic membrane. How binding and subsequent flipping of Lipid II through this structure may occur is still unclear. Solving the crystal structure of FtsW may shed light on this.
Given the essentiality of Lipid II transport across the bacterial cytoplasmic membrane in cell wall synthesis, compounds that can interfere with this step, by inhibiting FtsW, may prove valuable as antibiotics. Furthermore, FtsW is essential for survival, conserved among almost all eubacteria that synthesizes a cell wall, and has no counterpart in humans. These features, therefore, make FtsW an excellent target for exploration as a potential novel target for broad spectrum antibiotics.
Acknowledgments
We thank David Weiss and Kyle Williams for the generous gift of the plasmids with triple alanine substitutions and Ahmed Bouhss for pTrc99A.
This work was supported by European Commission EUR-INTAFAR Grant KSHM-CT-2004-512138, ZonMW Project 20500001, and EU Project FP7-Health-2007-B-223431.
- TM
- transmembrane
- LUV
- large unilamellar vesicle
- NBD
- 7-nitro-2,1,3-benzoxadiazol-4-yl
- NBD-PL
- 7-nitro-2,1,3-benzoxadiazol-4-yl-chloride-labeled phospholipids
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- PC
- phosphatidylcholine
- PG
- phosphatidylglycerol
- PE
- phosphatidylethanolamine
- ESI
- electrospray ionization.
REFERENCES
- 1. Egan A. J., Vollmer W. (2013) The physiology of bacterial cell division. Ann. N.Y. Acad. Sci. 1277, 8–28 [DOI] [PubMed] [Google Scholar]
- 2. Mohammadi T., van Dam V., Sijbrandi R., Vernet T., Zapun A., Bouhss A., Diepeveen-de Bruin M., Nguyen-Distèche M., de Kruijff B., Breukink E. (2011) Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 30, 1425–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lara B., Ayala J. A. (2002) Topological characterization of the essential Escherichia coli cell division protein FtsW. FEMS Microbiol. Lett. 216, 23–32 [DOI] [PubMed] [Google Scholar]
- 4. Pastoret S., Fraipont C., den Blaauwen T., Wolf B., Aarsman M. E., Piette A., Thomas A., Brasseur R., Nguyen-Distèche M. (2004) Functional analysis of the cell division protein FtsW of Escherichia coli. J. Bacteriol. 186, 8370–8379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fraipont C., Alexeeva S., Wolf B., van der Ploeg R., Schloesser M., den Blaauwen T., Nguyen-Distèche M. (2011) The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a sbucomplex in Escherichia coli. Microbiology 157, 251–259 [DOI] [PubMed] [Google Scholar]
- 6. D'Ulisse V., Fagioli M., Ghelardini P., Paolozzi L. (2007) Three functional subdomains of the Escherichia coli FtsQ protein are involved in its interaction with the other division proteins. Microbiology 153, 124–138 [DOI] [PubMed] [Google Scholar]
- 7. Mercer K. L., Weiss D. S. (2002) The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184, 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Den Blaauwen T., Aarsman M. E., Vischer N. O., Nanninga N. (2003) Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol. Microbiol. 47, 539–547 [DOI] [PubMed] [Google Scholar]
- 9. Koppelman C. M., Aarsman M. E., Postmus J., Pas E., Muijsers A. O., Scheffers D. J., Nanninga N., den Blaauwen T. (2004) R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling and is essential for cell division. Mol. Microbiol. 51, 645–657 [DOI] [PubMed] [Google Scholar]
- 10. van der Ploeg R., Verheul J., Vischer N. O., Alexeeva S., Hoogendoorn E., Postma M., Banzhaf M., Vollmer W., den Blaauwen T. (2013) Colocalization and interaction between elongasome and divisome during a preparative cell division phase in Escherichia coli. Mol. Microbiol. 87, 1074–1087 [DOI] [PubMed] [Google Scholar]
- 11. Rouser G., Fkeischer S., Yamamoto A. (1970) Two dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 5, 494–496 [DOI] [PubMed] [Google Scholar]
- 12. Breukink E., van Heusden H. E., Vollmerhaus P. J., Swiezewska E., Brunner L., Walker S., Heck A. J., de Kruijff B. (2003) Lipid II is an intrinsic component of the pore induced by nisin in bacterial membranes. J. Biol. Chem. 278, 19898–19903 [DOI] [PubMed] [Google Scholar]
- 13. van Dam V., Sijbrandi R., Kol M., Swiezewska E., de Kruijff B., Breukink E. (2007) Transmembrane transport of peptidoglycan precursors across model and bacterial membranes. Mol. Microbiol. 64, 1105–1114 [DOI] [PubMed] [Google Scholar]
- 14. Pera N. P., Kouki A., Haataja S., Branderhorst H. M., Liskamp R. M., Visser G. M., Finne J., Pieters R. J. (2010) Detection of pathogenic Streptococcus suis bacteria using magnetic glycoparticles. Org. Biomol. Chem. 8, 2425–2429 [DOI] [PubMed] [Google Scholar]
- 15. Zhang L., Wu Y., Brunsveld L. (2007) A synthetic supramolecular construct modulating protein assembly in cells. Angew. Chem. Int. Ed. Engl. 46, 1798–1802 [DOI] [PubMed] [Google Scholar]
- 16. Ehlert K., Höltje J. V. (1996) Role of precursor translocation in coordination of murein and phospholipid synthesis in Escherichia coli. J. Bacteriol. 178, 6766–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khattar M. M., Begg K. J., Donachie W. D. (1994) Identification of FtsW and characterization of a new ftsW division mutant of Escherichia coli. J. Bacteriol. 176, 7140–7147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang L., Khattar M. K., Donachie W. D., Lutkenhaus J. (1998) FtsI and FtsW are localized to the septum in Escherichia coli. J. Bacteriol. 180, 2810–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goddard-Borger E. D., Stick R. V. (2007) An efficient, inexpensive, and shelf-stable diazotransfer reagent: imidazole-1-sulfonyl azide hydrochloride. Org. Lett. 9, 3797–3800 [DOI] [PubMed] [Google Scholar]
- 20. Bonardi F., Halza E., Walko M., De Plessis F., Nouwen N., Feringa B. L., Driessen A. J. M. (2011) Probing the SecYEG translocation pore size with preproteins conjugated with sizable rigid spherical molecules. Proc. Nat. Acad. Sci. U.S.A. 108, 7775–7780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kol M. A., de Kroon A. I., Killian J. A., de Kruijff B. (2004) Transbilayer movement of phospholipids in biogenic membranes. Biochemistry 43, 2673–2681 [DOI] [PubMed] [Google Scholar]
- 22. Williamson P. (2011) Phospholipid transport: sighting a new face of an old friend. Curr. Biol. 21, R168–9 [DOI] [PubMed] [Google Scholar]
- 23. Menon I., Huber T., Sanyal S., Banerjee S., Barré P., Canis S., Warren J. D., Hwa J., Sakmar T. P., Menon A. K. (2011) Opsin is a phospholipid flippase. Curr. Biol. 21, 149–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ishino F., Jung H. K., Ikeda M., Doi M., Wachi M., Matsuhashi M. (1989) New mutations fts-36, Its-3, and ftsW clustered in the mra region of the Escherichia coli chromosome induce thermosensitive cell growth and division. J. Bacteriol. 171, 5523–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Taschner P. E., Huls P. G., Pas E., Woldringh C. L. (1988) Division behavior and shape changes in isogenic fstZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J. Bacteriol. 170, 1533–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]