Background: Intracellular lipid-binding proteins stimulate lipid-induced gene expression.
Results: Fatty acid-binding protein 5 (FABP5) uses a molecular switch that controls nuclear import when complexed with activating fatty acids.
Conclusion: FABP5 is tuned to selectively stimulate peroxisome proliferation-activated receptor β/δ transactivation in response to specific fatty acids based on their structural features.
Significance: FABPs provide a second level of regulatory control of nuclear receptor-mediated lipid signaling.
Keywords: Fatty Acid, Fatty Acid-binding Protein, Lipids, Nuclear Receptors, Polyunsaturated Fatty Acids, Structural Biology
Abstract
Fatty acid-binding proteins (FABPs) are a widely expressed group of calycins that play a well established role in solubilizing cellular fatty acids. Recent studies, however, have recast FABPs as active participants in vital lipid-signaling pathways. FABP5, like its family members, displays a promiscuous ligand binding profile, capable of interacting with numerous long chain fatty acids of varying degrees of saturation. Certain “activating” fatty acids induce the protein's cytoplasmic to nuclear translocation, stimulating PPARβ/δ transactivation; however, the rules that govern this process remain unknown. Using a range of structural and biochemical techniques, we show that both linoleic and arachidonic acid elicit FABP5's translocation by permitting allosteric communication between the ligand-sensing β2 loop and a tertiary nuclear localization signal within the α-helical cap of the protein. Furthermore, we show that more saturated, nonactivating fatty acids inhibit nuclear localization signal formation by destabilizing this activation loop, thus implicating FABP5 specifically in cis-bonded, polyunsaturated fatty acid signaling.
Introduction
LCFAs,3 in addition to serving as a structural component and energy source of the cell, participate in cellular signaling by modulating the activity of a group of nuclear receptors known as the peroxisome proliferation-activated receptors (PPARs) (1–6). Because of the large number of target genes affected by this family of ligand-regulated transcription factors, LCFAs play a critical role in a variety of cellular processes and their related pathophysiologies, ranging from metabolic defects to cell differentiation and cancer progression (7, 8). However, the relative insolubility of these molecules makes them reliant upon a class of transport proteins, the FABPs, to exert their signaling effects (9–11).
There are nine known FABP members in mammals, each ∼14–15 kDa in size with orthologs found throughout the animal kingdom (12). Although they exhibit a wide range of sequence identity (∼20–70%), all form a twisted β-barrel, composed of 10 anti-parallel β-strands arranged into two orthogonal β-sheets, with a helix-turn-helix lid covering the ligand-binding site (10–12). As members of the intracellular lipid-binding protein (iLBP) family, they have traditionally been thought to be involved in the solubilization/protection of their various hydrophobic cargoes, facilitating ligand movement via passive diffusion between the various compartments of the cell (13, 14). Increasingly, however, FABPs are emerging as specific mediators of precise signaling pathways. For instance, FABP1 facilitates the polyunsaturated fatty acid and fibrate-induced transactivation of PPARα, via direct interaction with the nuclear receptor's ligand binding domain (15–17). A similar role has been observed with FABP4, whereby its ligand-mediated dimerization state governs nuclear import and subsequent ligand delivery to PPARγ (18, 19). Recent findings have even revealed FABPs to be the once enigmatic N-acylethanolamine “transporter,” responsible for endocannabinoid cellular uptake, hydrolysis, and PPARα activation (20, 21).
FABP5 (E-FABP, KFABP, and mal1), first characterized almost 20 years ago in keratinocytes, is one of the most ubiquitously expressed proteins in its class, and it can be found across a broad spectrum of tissue/cell types such as the epidermis, adipose, macrophages, mammary glands, brain, kidney, liver, lung, heart, skeletal muscle, and testis (10, 12, 22). A member of the iLBP subfamily IV, FABP5 binds a wide array of ligands in a 1:1 ratio, including fatty acids and fatty acid metabolites spanning 10–22 carbons in length with various saturation states, as well as the vitamin A metabolite all-trans-retinoic acid and numerous synthetic drugs and probes (10, 23–25). It has also been found to be involved in a range of pathologies, including the metabolic syndrome (26, 27), atherosclerosis (28), cancer (29–33), and potentially certain neurodegenerative diseases (34).
Work conducted by Tan et al. demonstrated the ability of FABP5 to specifically enhance the transactivation of PPARβ/δ, whose known gene targets are involved in cellular glucose and lipid homeostasis (35–37), differentiation (38, 39), and resistance to apoptosis (39, 40). Despite FABP5's promiscuous binding profile, only a subset of fatty acids and other ligands have been shown to result in the protein's nuclear translocation, where it is thought to engage PPARβ/δ, allowing for the channeling of ligand into the nuclear receptor's binding pocket (18). Although previous structural studies have shed considerable light on the role of FABP4 in PPARγ signaling (19, 41), the mechanism underlying select lipid activation (e.g. nuclear translocation) of FABP5 remains unknown. Using a combination of x-ray crystallography, hydrogen deuterium exchange (HDX)-mass spectroscopy, and biochemical and cellular approaches, we have established the presence of a ligand-sensitive tertiary nuclear localization signal (NLS) located on the α1 and α2 helices of FABP5. Furthermore, we show that interaction of a bound ligand with FABP5's β2 loop relays conformational information to the NLS, thereby serving as the driving force for fatty acid-specific nuclear translocation.
EXPERIMENTAL PROCEDURES
Reagents
Chemicals were purchased from Sigma, Fisher, Polysciences, or Cayman, Inc. The pMCSG7-His plasmid was a gift from John Sondek (University of North Carolina, Chapel Hill), whereas pEGFP-N3 was graciously given by Anita Corbett (Emory University, Atlanta, GA).
Cloning and Mutagenesis
Full-length, codon-optimized wild-type human FABP5 (residues 1–135) was subcloned into pMCSG7-His, pCMV-Tag2B, and pEGFP-N3 expression vectors. The NLS-deficient mutant (hFABP5NLSm: K24A, K34A, and R33A) and “double-switch” mutant (hFABP5DSm: M35A and L60A) were generated in the pMCSG7-His pCMV-Tag2B. hFABP5NLSm, hFABP5DSm, and a nuclear export signal mutant (hFABP5NESm: L69A, L94A, and F89A) were generated in pEGFP-N3. All mutagenesis was performed using QuikChange II XL (Stratagene).
Protein Expression and Purification
Full-length human FABP5 in the pMCSG7 vector was transformed into Escherichia coli strain BL21(DE3) cells and expressed as a His6 fusion containing a tobacco etch virus protease cleavage site to facilitate tag removal. Cultures (1.3 liters in TB) were grown to an A600 of ∼0.8 and induced with 1 mm isopropyl β-d-1-thiogalactopyranoside at 37 °C for 4 h. Cell mass was collected by centrifugation at 5000 rpm for 15 min, lysed, and purified by nickel affinity chromatography. The His tag was cleaved by tobacco etch virus protease at 4 °C overnight with simultaneous dialysis into a buffer containing 300 mm NaCl, 50 mm K2HPO4 (pH 7.4), and 5% glycerol and purified to homogeneity by nickel affinity followed by gel filtration chromatography. To generate apo-FABP5, pure protein was delipidated via Bligh and Dyer (42) chloroform/methanol extraction. Denatured protein was then solubilized in buffer composed of 50 mm Tris-HCl (pH 8.0), 6 m guanidinium chloride, and 2 mm DTT and refolded by fast dilution at 4 °C in 20 mm Tris-HCl (pH 8.5), 1.7 m urea, 4% glycerol, and 2 mm DTT. After adjusting the final concentration of urea to 2.0 m, refolded protein was concentrated and dialyzed against PBS at 4 °C overnight before being purified via gel filtration chromatography.
Crystallization, Data Collection, Structural Refinement
Pure FABP5 was concentrated to 15 mg ml−1 in PBS buffer, and crystals of the apoprotein were grown over 2 weeks via hanging drop vapor diffusion at 18 °C from solutions containing 2 μl of FABP5 solution and 1 μl of mother liquor (2 m ammonium sulfate, 300 mm sodium/potassium tartrate, 100 mm sodium citrate (pH 5.6)). Crystals were cryoprotected by immersion in mother liquor containing 15% glycerol and flash-cooled in liquid nitrogen. Data to a resolution of 1.67 Å were collected at 100 K and a wavelength of 1.00 Å at the South East Regional Collaborative Access Team (SER-CAT) beamline (Advanced Photon Source, Argonne, IL) and processed using the HKL-2000 software (43). The structure was solved by molecular replacement using a previously determined structure (PDB 1B56) in PHASER (44). To obtain crystals of the FABP5-linoleic acid complex, apo-FABP5 was exposed to LA at a 1:5 protein/ligand molar ratio in PBS, before being concentrated to 15 mg ml−1. Crystals formed overnight at 4 °C via hanging drop vapor diffusion, using a crystallant consisting of 2.4 m ammonium sulfate, 200 mm sodium/potassium tartrate, and 100 mm sodium citrate (pH 5.6). Crystals were cryoprotected with a 20% glycerol crystallant solution, and data to a resolution of 2.60 Å were collected at Emory University using a Rigaku MicroMax 007 HF generator with a copper anode and a Saturn CCD detector, at a temperature of 100 K. Data indexing and phasing were carried out as described for apo-FABP5. Model building and refinement for both structures were performed using COOT (45) and phenix.refine (46), respectively. Electrostatic surface potential maps of FABP5-LA were calculated by the PDB2PQR Server (47) and the Adaptive Poisson-Boltzmann Solver (48), whereas protein interior volumes were obtained with CASTp using a probe radius of 1.4 Å (49). Figures were generated using the PyMOL Molecular Graphics System (Schrodinger, LLC). Structure validation was performed with MolProbity, showing excellent overall model geometry as the apo- and LA-bound structures received scores in the 99th and 100th percentile, respectively (50). Final coordinates for apo-FABP5 and FABP5-LA have been deposited into the PDB, under accession codes 4LKP and 4LKT.
Cell Localization Assay
COS-7 cells were grown on 10-cm plates in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS) at 37 °C and 5% CO2. At ∼60% confluency, polyethyleneimine (Polysciences) was used to transfect cells with 5 μg of pEGFP-N3 vector harboring full-length hFABP5WT, hFABP5NLSm, hFABP5NESm, or hFABP5DSm, with the DNA-PEI complex being removed 6–8 h after exposure. The following day, cells were checked for fluorescent protein expression and then transferred to Lab-Tek II Chamber Slides (Thomas Scientific catalog no. 154526) in DMEM buffer containing 5% charcoal/dextran-stripped FBS. Twenty four hours post-transfer, cells were exposed to 10 μm fatty acid ligand solubilized in 0.1% EtOH for 30 min at 37 °C, washed three times with ice-cold PBS, fixed with 4% paraformaldehyde, and stained with DAPI. Slides were imaged using a Zeiss LSM510 META Upright confocal microscope (×40/1.3 Oil differential interference contrast objectives) employing Zeiss Zen2009 acquisition software, with both nuclear focusing and enhanced green fluorescent protein (EGFP) imaging conducted at an optical slice of 0.9 μm. Nuclear and cytoplasmic EGFP fusion protein fluorescence intensities were quantified using ImageJ, and the calculated nuclear/cytoplasmic ratios were plotted in Prism 5 (GraphPad Inc., La Jolla, CA). Statistical significance was determined by one-factor ANOVA, with individual comparisons made with Tukey's honestly significant difference (HSD) post hoc tests.
Ligand Binding Assays
Ligand binding was measured via competition of 1-anilinonaphthalene-8-sulfonic acid (1,8-ANS), which displays increased fluorescence when exposed to a hydrophobic environment (51). In brief, both wild-type and mutant hFABP5 were expressed and purified to homogeneity as described above and dialyzed in PBS (pH 8.2). Binding affinity (KD) was derived by monitoring maximal fluorescence intensity of a constant concentration of 500 nm 1,8-ANS with increasing protein concentrations ranging from 20 nm to 424 μm. Blank measurements obtained from protein-only samples were subtracted at each protein concentration tested to obtain the final values. Competition assays were then performed in which the protein was held at a constant concentration of 500 nm (1 μm for the FABP5NLSm palmitic acid competition), with 1,8-ANS also being held constant at either 5 μm (for hFABP5WT and hFABP5DSm) or 10 μm (for hFABP5NLSm) in the presence of increasing fatty acid concentrations from 10 nm to 200 μm. Blanks consisting of 1,8-ANS and fatty acid in the absence of protein were subtracted at each ligand concentration tested. The resulting fluorescence values were used to calculate a Ki value for the fatty acid of interest. Data were collected at 30 °C on a BioTek Synergy plate reader using an excitation filter of 380/20 nm and an emission filter of 460/40 nm and processed in GraphPad Prism 5. Statistical significance was determined by one-factor ANOVA, and individual comparisons were made with Tukey's HSD post hoc tests.
In-cell Activation Assays
MCF-7 cells were transferred to 96-well plates, where they were grown and maintained in high glucose DMEM containing l-glutamine, sodium pyruvate, and phenol red (Invitrogen), supplemented with 10% charcoal/dextran-stripped FBS (Invitrogen), and 1% penicillin/streptomycin (Culture Buffer). One hundred ng well−1 pSG5 vector harboring full-length mouse PPARβ/δ receptor, 100 ng well−1 PPAR-response element-driven firefly luciferase reporter (PPAR-response element X3-TK-luc), and 20 ng well−1 constitutive Renilla luciferase reporter (phRLtk) in the presence or absence of 25 ng well−1 wild-type or mutant variant human FABP5 cloned into the pCMV-Tag2B vector was added to FuGENE HD in Opti-MEM (Invitrogen). This solution was diluted with Culture Buffer (−antibiotic) to a final concentration of 2.2–2.45 ng μl−1 total DNA. 100 μl well−1 of this solution was used to transfect 70–90% confluent cells overnight. Cells were then treated in sextuplicate with 1–100 μm fatty acid ligand or vehicle (ethanol) in high glucose DMEM containing only l-glutamine and 1% penicillin/streptomycin for 24 h (final working ethanol concentration 0.1%), and assayed with Dual-Glo luciferase substrate (Promega). Firefly activity was divided by Renilla activity to account for cell number, viability, and transfection efficiency, and graphs were generated in GraphPad Prism 5. Statistical significance was determined by either one- or two-factor ANOVA, and individual comparisons were made with Tukey HSD or Bonferroni post hoc tests.
Protein Unfolding Assay
Pure hFABP5WT and hFABP5NESm (1 μm, PBS) was exposed to increasing concentrations of guanidinium hydrochloride, and the resulting shift in peak intrinsic fluorescence intensity was measured using a Shimadzu RF-5301PC spectrofluorophotometer at an excitation wavelength of 280 nm with a 5-nm spectral bandwidth. Values were fitted using a four-parameter logistic equation, and the calculated fluorescence shift midpoints were compared via unpaired t test with Welch's correction for unequal variances in GraphPad Prism 5.
HDX
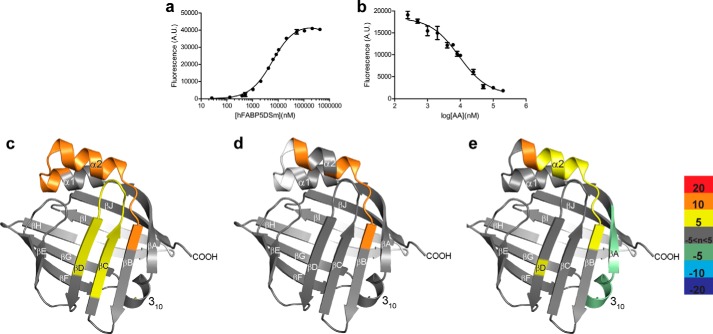
Solution-phase amide HDX was carried out with a fully automated system as described previously (52). Briefly, 4 μl of sample consisting of 10 μm protein and 100 μm ligand in PBS (pH 7.4) was diluted to 20 μl with D2O-containing HDX buffer and incubated at 25 °C for 10, 30, 60, 900, or 3600 s. Following on exchange, back exchange was minimized, and the protein was denatured by dilution to 50 μl in a low pH and low temperature buffer containing 0.1% (v/v) TFA in 5 m urea (held at 1 °C). Samples were then passed across an immobilized pepsin column (prepared in house) at 50 μl min−1 (0.1% v/v TFA, 15 °C); the resulting peptides were trapped on a C8 trap cartridge (Hypersil Gold, Thermo Fisher). Peptides were then gradient-eluted 4–40% (w/v) CH3CN, 0.3% (w/v) formic acid over 5 min at 2 °C across a 1 × 50-mm C18 HPLC column (Hypersil Gold, Thermo Fisher), and electrosprayed directly into an Orbitrap mass spectrometer (LTQ Orbitrap with ETD, Thermo Fisher). Peptide ion signals with a MASCOT score of >20 were used if they had no ambiguous hits using a decoy (reverse) sequence in a separate experiment using a 60-min gradient. The intensity weighted average m/z value (centroid) of each peptide's isotopic envelope was calculated with in-house developed software (53). Each envelope was corrected for back-exchange assuming 70% recovery and accounting for the known deuterium content of the on-exchange buffer. To quantify the difference in exchange rates, we calculated the average percent deuterium uptake for wild-type (WT) FABP5/PoA following 10, 30, 60, 900, and 3600 s of on-exchange by averaging percent deuterium incorporation across all time points. From this value, we subtracted the average percent deuterium uptake measured at the same time points for the matching peptides of the FABP5WT-AA complex. Positive perturbation values indicate exchange rates are faster for these regions within FABP5WT in complex with PoA. To quantify the effects of the DSm we subtracted the percent deuterium uptake for FABP5WT-AA from FABP5DSm-AA, FABP5WT-PoA from FABP5DSm-PoA, and FABP5DSm-AA from FABP5DSm-PoA. The resulting differences in exchange were mapped on the structure of FABP5 and visualized with PyMOL (Schrödinger, LLC).
RESULTS
Overall Structure and Oligomerization Status of Apo- and Holo-FABP5
To elucidate the molecular mechanisms driving ligand-specific FABP5 activation, we determined crystal structures of apo-FABP5 and FABP5 in complex with LA, an ω-6 polyunsaturated pan-PPAR fatty acid agonist that has been shown to trigger FABP5's nuclear translocation (18). Because recombinant FABP5 co-purifies with E. coli LCFAs, delipidation/denaturation of the protein followed by refolding was performed prior to LA exposure and subsequent crystallization (42, 54). The structure of apo-FABP5 was solved in the P43212 space group at high resolution (1.67 Å), with the asymmetric unit comprised of a FABP5 monomer adopting the canonical iLBP fold (Fig. 1a) (54, 55). Interestingly, crystals of the FABP5-LA complex grew only in the P3221 space group, with the resulting 2.6-Å structure revealing four copies of protein in the asymmetric unit (Table 1 and Fig. 1b). Because modulation of dimer interface is thought to account for the ligand-specific nuclear translocation of FABP4 (41), we tested whether fatty acid binding affects the oligomerization status of FABP5. Size exclusion chromatography of both apoprotein (Bligh and Dyer delipidated and refolded) and FABP5 purified in the presence of saturating amounts of LA reveals that FABP5 is monomeric in both the liganded and unliganded state (Fig. 1c). Additionally, dynamic light scattering conducted on untreated recombinant protein indicates that FABP5 exists solely in a monomeric population (data not shown), despite the presence of various bacterial LCFAs, suggesting that fatty acids do not alter the oligomerization state of the protein.
FIGURE 1.
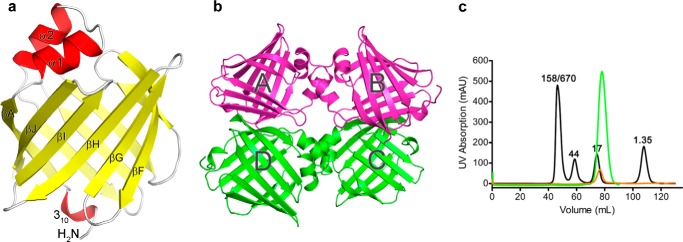
Structural overview of apo- versus LA-bound FABP5. a, tertiary structure of apo-FABP5. Unbound protein adopts the familiar β-barrel fold capped by an α-helical lid, with a 310 helix at the N terminus. b, asymmetric unit of the FABP5-LA crystal, composed of four copies of ligand-bound protein. c, overlay of size exclusion chromatographs for apo- (orange) and LA-complexed (green) FABP5. Standard consisting of 1.35-, 17-, 44-, 158-, and 670-kDa markers is in black.
TABLE 1.
Data collection and refinement statistics (molecular replacement)
| Apo-FABP5 | FABP5-LA | |
|---|---|---|
| Data collection | ||
| Space group | P43212 | P3221 |
| Cell dimensions | ||
| a, b, c | 63.0, 63.0, 74.5 Å | 145.3, 145.3, 81.8 Å |
| a, b, γ | 90.0, 90.0, 90.0° | 90.0, 90.0, 120.0° |
| Resolution | 1.67 Å (1.73 to 1.67 Å)a | 2.60 Å (2.69 to 2.60 Å)a |
| Rsym or Rmerge | 4.2 (38.9) | 12.0 (56.0) |
| I/σI | 42.86 (4.45) | 20.15 (3.03) |
| Completeness | 99.9% (99.4%) | 99.9% (99.9%) |
| Redundancy | 9.1 (6.8) | 9.3 (7.5) |
| Refinement | ||
| Resolution | 1.67 Å | 2.60 Å |
| No. reflections | 18,008 | 31,542 |
| Rwork/Rfree | 18.1/21.5 | 20.5/25.9 |
| No. of atoms | ||
| Protein | 1095 | 4243 |
| Ligand/ion | 15 | 196 |
| Water | 111 | 69 |
| B-factors | ||
| Protein | 19.8 | 46.7 |
| Ligand/ion | 28.3 | 63.8 |
| Water | 31.2 | 43.6 |
| Root mean square deviations | ||
| Bond lengths | 0.015 Å | 0.003 Å |
| Bond angles | 1.825° | 0.624° |
a Data were collected from a single crystal; values in parentheses are for highest resolution shell.
Linoleic Acid Binds FABP5 in Two Distinct Conformations
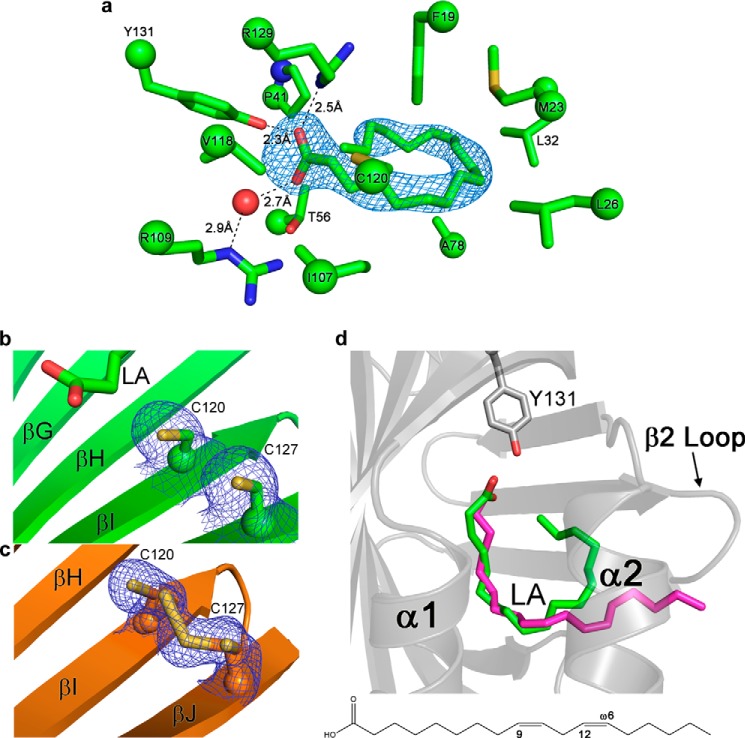
The FABP5-LA interactions are in general very similar to those previously described by Hohoff et al. (54) in their analysis of FABP5 complexed to an E. coli fatty acid. The carboxylic headgroup of LA forms a salt bridge with Arg-129, as well as a hydrogen bond with the hydroxyl moiety of Tyr-131. An additional hydrogen bond interaction is observed between LA and Arg-109 via an ordered water molecule (Fig. 2a). The alkyl tail of the fatty acid is largely stabilized by van der Waals interactions made with the hydrophobic side chains of multiple amino acids that line the binding pocket, including Cys-120. Intriguingly, whereas all previous structural studies of FABP5 have shown this amino acid to participate in disulfide bond formation with Cys-127, electron density reveals the unequivocal presence of both amino acids in their sulfhydryl forms within all four monomers of LA-bound FABP5 (Fig. 2b) (54, 55). In contrast, the apoprotein contains a mixture of cysteine-cystine forms (Fig. 2c). Because the E. coli fatty acid in the Hohoff et al. (54) structure could only be modeled at 50% occupancy, we conclude that the absence of a disulfide bridge within FABP5-LA is the result of the ligand being fully bound, and it likely helps to accommodate the ∼191 Å3 increase (averaged across all four FABP5-LA monomers) in ligand pocket volume as compared with apoprotein (49).
FIGURE 2.
Analysis of FABP5 ligand binding pocket and LA's bound conformations. a, LA is held within FABP5 via a salt bridge and hydrogen bonding with its carboxylic headgroup and hydrophobic interactions with its alkyl tail. b and c, Cys-120 and Cys-127 are unequivocally in their sulfhydryl forms in the presence of LA (b), yet are able to adopt either free or disulfide-bond states within apo-FABP5 (c). The simulated annealing Fo − Fc omit map of electron density was contoured at 2.5σ for a, and 2Fo − Fc electron density maps were modeled at 1σ for b and c. d, conformation of LA when bound to monomers A and B (purple) versus monomers C and D (green) in the FABP5-LA crystal structure (see Fig. 1b).
Visualization of the water network within the binding pocket of the apo- and LA holo-proteins reveals a much higher number of ordered water molecules located within the top half of the β-barrel nearer the α helix lid than in the bottom half closer to the proteins' termini. Although the presence of ligand is responsible for partial rearrangement of this network, waters 2, 7, 15, 31, 38, and 82 in apo-FABP5 remain virtually unaltered between the two structures, suggesting their importance in maintaining binding pocket architecture. Conservation of such a relatively large number of FABP5's ordered waters likely reflects the protein's heavy reliance upon enthalpic versus entropic contributions in binding fatty acids, a property common to the protein class (56).
Although monomers C and D bind LA in the traditional U-conformation, most commonly seen for fatty acids within the binding pockets of iLBP subfamily IV members (FABP3–5 and -7–9), LA adopts a bent or “L” conformation within the pockets of monomers A and B (10, 12, 23). Overlay of the two ligand configurations from monomers B and C reveals a high degree of positional similarity between the acidic headgroup and carbons 1–10 of the aliphatic chain. However, although the cis-9 and -12 double bonds of U-conformation LA provide the two turns necessary for keeping the entire ligand inside the binding cavity, the L-conformation only features the first turn, resulting in the protrusion of the fatty acid tail from the β2 portal loop of the protein (amino acids 58–62) into solvent (Fig. 2d). Although electron density and isotropic B-factors indicate that the U-conformation is more highly ordered, LA is likely able to switch between the two binding states within FABP5 in solution.
Conformation of the Bound Fatty Acid Dictates Activation of FABP5
Studies conducted with FABP4 show that fatty acids that are presumably unable to cause its nuclear translocation bind in a manner that disrupts the protein's β2 portal loop, similar to LA's L-conformation (41, 57). Thus, it was reasoned that LA's U-conformation correlates to its FABP5-activating form, whereas the L-conformation represents a nonactivating binding mode. Based on this rationale, FABP5-activating fatty acids could be predicted based on their propensity to adopt a similar U- versus L-configuration when bound to the protein. To test this hypothesis, four lipids in addition to LA were selected for functional analysis as follows: arachidonic acid (AA), a 20-carbon polyunsaturated ω-6 fatty acid known to activate PPARβ/δ (4); PoA, a 16-carbon monounsaturated ω-7 fatty acid; sapienic acid (SpA), a 16-carbon monounsaturated ω-10 fatty acid; and the fully saturated 16-carbon palmitic acid (PA) (Table 2). Assuming that their ability to favor an activating binding mode within FABP5 correlates to their natural degree of conformational curvature, AA was predicted to be an FABP5-activating fatty acid, similar to LA, although PoA, SpA, and PA were predicted nonactivators.
TABLE 2.
Proposed fatty acid activators and nonactivators of FABP5
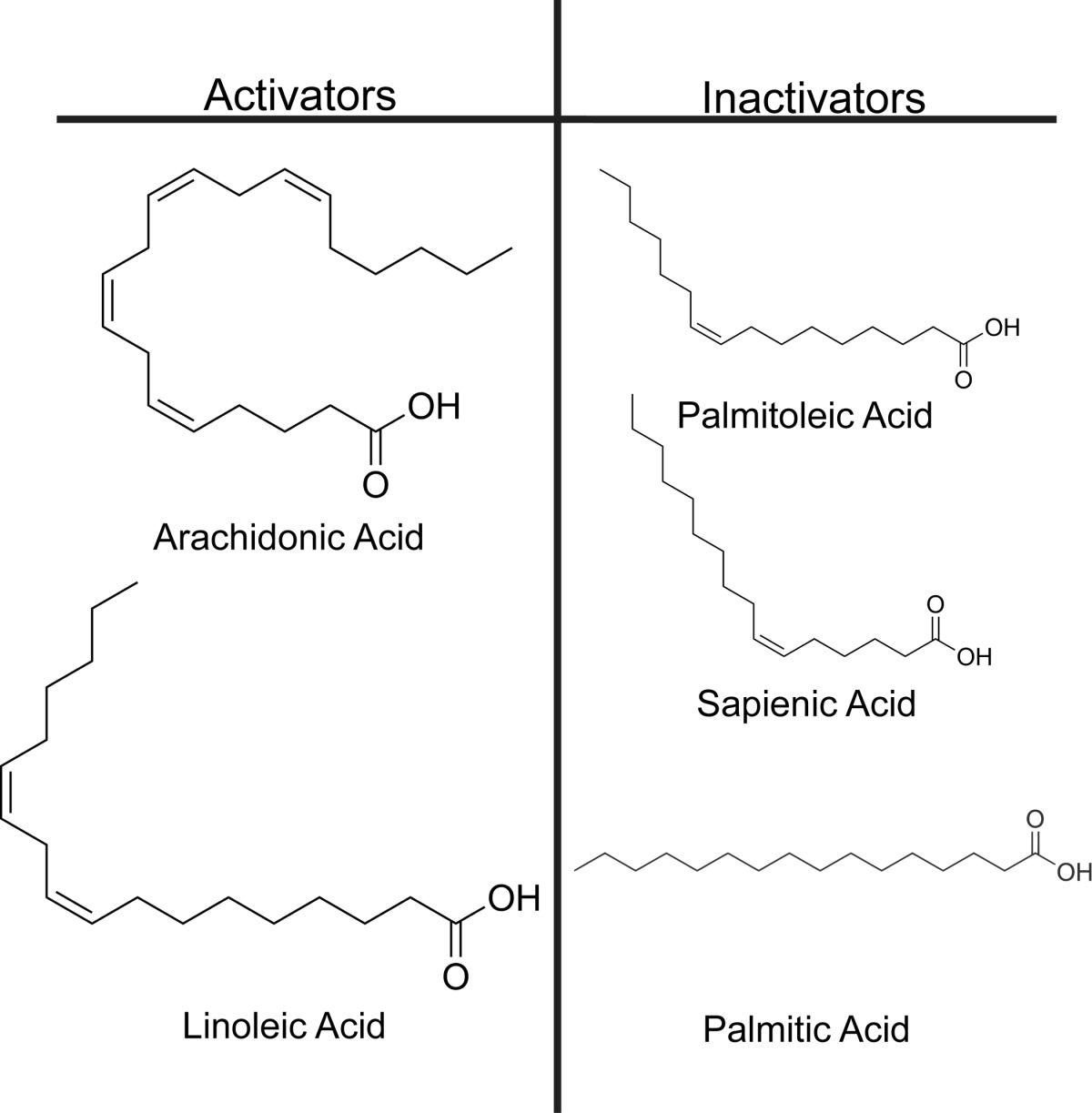
To gauge an appropriate range of ligand concentrations needed for FABP5 activation assays, we determined the affinity of FABP5 for the five fatty acid candidates by testing their ability to displace the fluorophore 1,8-ANS from the lipid binding pocket, as described previously (51). The binding constants obtained for AA, LA, PoA, and PA (Table 3) are somewhat higher than those calculated previously using a similar technique, falling within the range of affinities measured via the Lipidx method (24, 54). Additionally, our results indicate that FABP5 binds AA significantly worse than the other candidates, while exhibiting a relatively high affinity for PA (Fig. 3a). Thus, in our hands, the binding preference of FABP5 approximately correlates with fatty acid aqueous solubility, a phenomenon known as the “solubility hypothesis” that has been used to characterize the ligand affinity trends of proteins throughout the group (58, 59).
TABLE 3.
Binding constants for wild type and mutant FABP5
| FABP5 | Ligand | Binding constant | 95% confidence interval |
|---|---|---|---|
| μm | μm | ||
| WT | ANS | 6.57 (KD) | 5.68–7.47 |
| WT | AA | 2.59 (Ki) | 1.77–3.77 |
| WT | LA | 1.26 (Ki) | 0.84–1.88 |
| WT | PoA | 1.36 (Ki) | 1.12–1.66 |
| WT | SpA | 1.10 (Ki) | 0.89–1.36 |
| WT | PA | 0.67 (Ki) | 0.52–0.87 |
| NLSm | ANS | 12.64 (KD) | 11.50–13.78 |
| NLSm | AA | 9.37 (Ki) | 7.40–11.86 |
| NLSm | LA | 2.17 (Ki) | 1.40–3.37 |
| NLSm | PoA | 1.64 (Ki) | 0.99–2.71 |
| NLSm | SpA | 1.24 (Ki) | 0.66–2.31 |
| NLSm | PA | 3.06 (Ki) | 1.74–5.36 |
| DSm | ANS | 6.10 (KD) | 5.53–6.68 |
| DSm | AA | 5.01 (Ki) | 3.79–6.62 |
| DSm | LA | 3.47 (Ki) | 1.80–6.69 |
| DSm | PoA | 1.78 (Ki) | 0.92–3.45 |
| DSm | SpA | 0.70 (Ki) | 0.31–1.61 |
| DSm | PA | 0.69 (Ki) | 0.54–0.88 |
FIGURE 3.
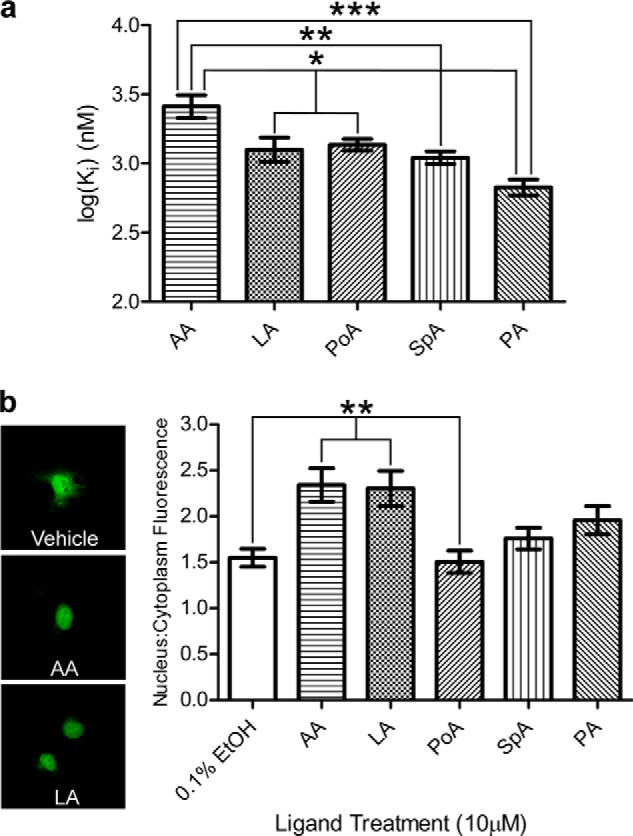
Fatty acid binding and induced nuclear localization of FABP5. a, binding of FABP5 to each fatty acid candidate was measured via 1,8-ANS displacement assay (n = 6), revealing a significantly lower affinity for AA. b, confocal images such as those represented on the left of EGFP-FABP5 expressing COS-7 cells exposed to 10 μm fatty acid or 0.1% ethanol vehicle were quantified using ImageJ (right, n = 30), with LA and AA exposure resulting in higher nuclear/cytoplasmic fluorescence intensity. The mean ± S.E. is shown. Statistical analyses were performed using one-factor ANOVA, with Tukey HSD post hoc tests used for individual comparisons. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
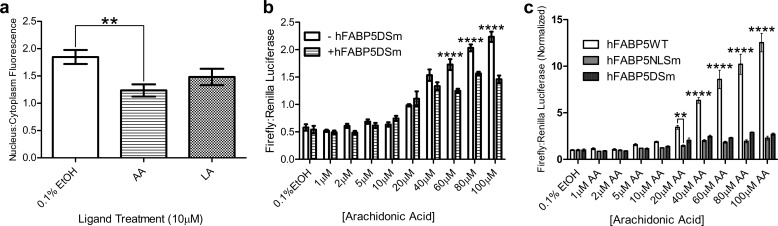
We then examined the proclivity of these fatty acids to drive the FABP5 nuclear localization. Endogenous FABP5 localizes primarily to the cytoplasm; however, overexpression of the protein combined with the innate ability of EGFP to partially localize to the nucleus results in the presence of FABP5-EGFP construct throughout the cell, even in the absence of ligand (Fig. 3b, top panel) (25, 60). Therefore, to obtain a robust measurement of ligand-induced nuclear localization, we quantified the ratio of nuclear fluorescence to cytoplasmic fluorescence, and we averaged this value for 30 cells per condition. LA treatment resulted in a statistically significant increase in nuclear localization compared with 0.1% ethanol (vehicle) treatment only, thus confirming LA as an activating ligand of FABP5 (Fig. 3b). Additionally, AA exposure led to the highest increase in nuclear/cytoplasmic average fluorescence, whereas PA, SpA, and especially PoA, had no significant effect. These results designate AA as a newly discovered FABP5 activator.
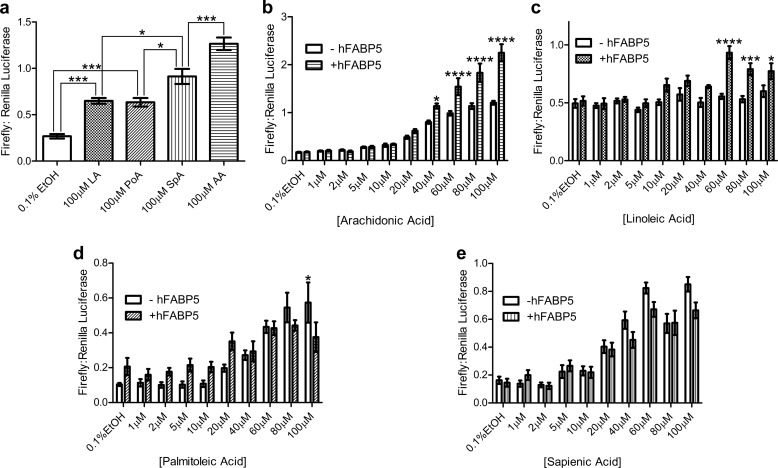
As FABP5 has been shown to participate in a direct signaling pathway with PPARβ/δ (18), we investigated the protein's ability to enhance AA- and LA-induced PPARβ/δ activation. To reduce background, assays were carried out in MCF-7 cells, which produce very low levels of endogenous FABP5. Verification of PPARβ/δ activation as a suitable metric for fatty acid signaling was first conducted in cells overexpressing receptor only. Exposure of cells to high concentrations (100 μm) of either LA or AA resulted in an increase in PPAR promoter-driven luciferase expression (2.4- and 4.7-fold, respectively), corroborating previous findings that both ligands are able to bind (6) and agonize (4) the receptor (Fig. 4a). Surprisingly, the other unsaturated fatty acid candidates were able to activate PPARβ/δ at levels comparable with or greater than LA, allowing their signaling ability to also be measured (Fig. 4a). FABP5 overexpression augmented AA agonism of PPARβ/δ starting at 40 μm, with the effect becoming more pronounced at higher concentrations (Fig. 4b). PPARβ/δ activation in the presence of LA was also enhanced by overexpression of FABP5, the effect being first observed at 60 μm fatty acid and gradually diminishing at 80 and 100 μm (Fig. 4c). In contrast, FABP5 did not enhance PoA- nor SpA-induced receptor activation, even leading to a dampening of response at 100 μm PoA (Fig. 4, d and e).
FIGURE 4.
PPARβ/δ activation by fatty acids in the presence and absence of FABP5 overexpression. a, all unsaturated fatty acid candidates' exposure to MCF-7 cells in the absence of overexpressed FABP5 resulted in PPARβ/δ activation, albeit with varying efficacy, as measured by luciferase reporter assay (n = 6). b and c, overexpression of FABP5 enhanced both AA- and LA-induced PPARβ/δ activation at ligand concentrations of 40–100 and 60–100 μm, respectively (n = 6). d and e, presence of overexpressed FABP5 was unable to enhance activation of PPARβ/δ by the fatty acids PoA and SpA, with an opposing effect seen at the highest concentration of PoA tested (n = 6). Statistical analysis was performed using either one-factor (a) or two-factor (b–e) ANOVA, with Tukey HSD (a) or Bonferroni (b–e) post hoc tests used for individual comparisons. *, p ≤ 0.05; ***, p ≤ 0.001; ****, p ≤ 0.0001. The mean ± S.E. is shown for all data points.
FABP5 Contains a Ligand-sensitive NLS within Its α-Helical Lid
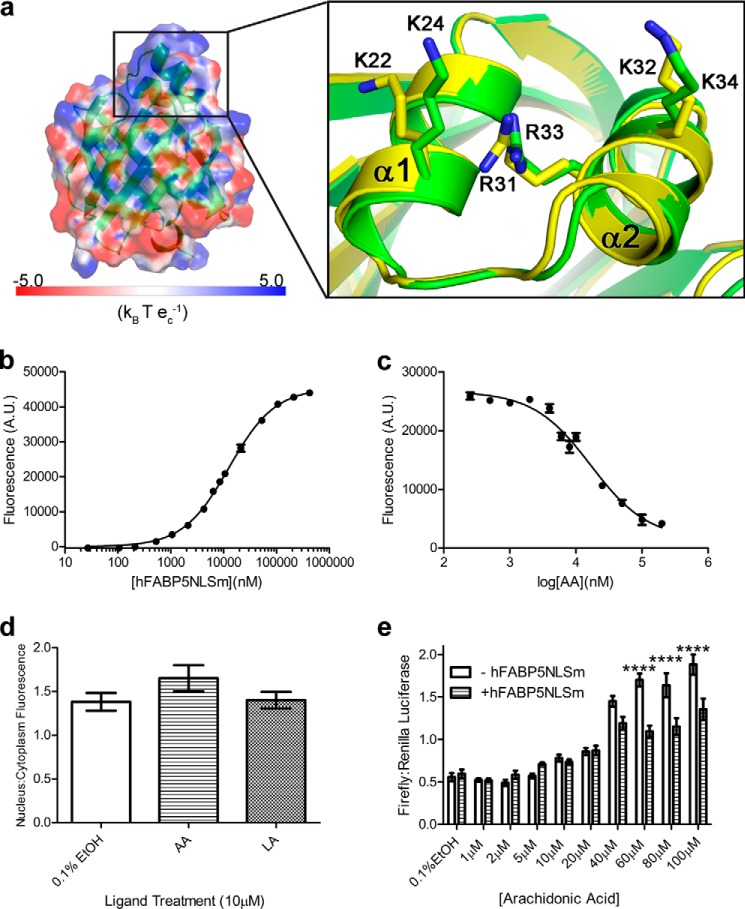
We next set out to determine the structural mechanism responsible for the ligand-specific nuclear translocation of FABP5. FABP5 does not harbor a classical NLS within its primary sequence. However, studies conducted on the iLBPs cellular retinoic acid-binding protein 2 (CRABP-II) (61) and FABP4 (19) prompted us to search for a tertiary, or cryptic, NLS within the three-dimensional structure. As the vast majority of all known NLS motifs are highly basic in nature (62), we created an electrostatic potential map of FABP5, which clearly displays a large patch of positive charge located on the surface of the protein's α-helical cap. Within this region, we identified two well ordered, solvent-exposed lysines (Lys-24 and Lys-34) and an arginine (Arg-33), homologous to the NLS residues of CRABP-II and FABP4 (shown in overlay with monomer C of FABP5-LA) (Fig. 5a).
FIGURE 5.
Tertiary nuclear localization signal of FABP5. a, surface potential of FABP5 bound to LA in the active “U” conformation. Units are in multiples of 26.7 mV (kB, Boltzmann's constant; T, temperature; ec, electron charge). Overlay of activated FABP5-LA (green) with FABP4 bound to 1,8-ANS (yellow, Protein Data Bank code 2ANS) reveals FABP5's homologous tertiary NLS residues as follows: Lys-24, Lys-34, and Arg-33. b and c, FABP5NLSm was able to bind 1,8-ANS (b), which could be displaced from the binding pocket by all fatty acid candidates tested (only AA is shown (c)) (n = 6). d, neither LA nor AA exposure (10 μm) resulted in significant nuclear translocation of EGFP-FABP5NLSm (n = 30), as determined by one-factor ANOVA. e, expression of FABP5NLSm resulted in subdued PPARβ/δ activation at AA concentrations of 60, 80, and 100 μm (n = 6). Statistical analysis was performed using two-factor ANOVA, with Bonferroni post hoc tests used for individual comparisons. ****, p ≤ 0.0001. The mean ± S.E. is shown for all data points.
To test the role of these residues in nuclear import, we created FABP5NLSm, in which Lys-24, Lys-34, and Arg-33 were substituted with alanines. The substitutions had no impact on expression, purification, or stability (data not shown). All fatty acid candidates were able to successfully compete 1,8-ANS from the protein's binding pocket, although the NLS mutant exhibited reduced binding affinity for AA and PA (Fig. 5, b and c, and Table 3). Cell localization assays reveal that FABP5NLSm is unable to translocate to the nucleus in the presence of the activators AA or LA (Fig. 5d). Similarly, FABP5NLSm was unable to enhance AA-induced activation of PPARβ/δ at any of the selected ligand concentrations, instead suppressing PPARβ/δ transactivation at exposure levels of 60, 80, and 100 μm (Fig. 5e). As these concentrations are 6.4-, 8.5-, and 10.7-fold, respectively, over the ligand's Ki value, it is extremely unlikely that such effects are attributable to the mutant's altered AA binding ability. Collectively, these data indicate that a cryptic NLS, located on the α-helical cap of the fatty acid binding pocket, is required for ligand-dependent activation.
NES Equivalent Residues of FABP5 Are Necessary for Protein Stability
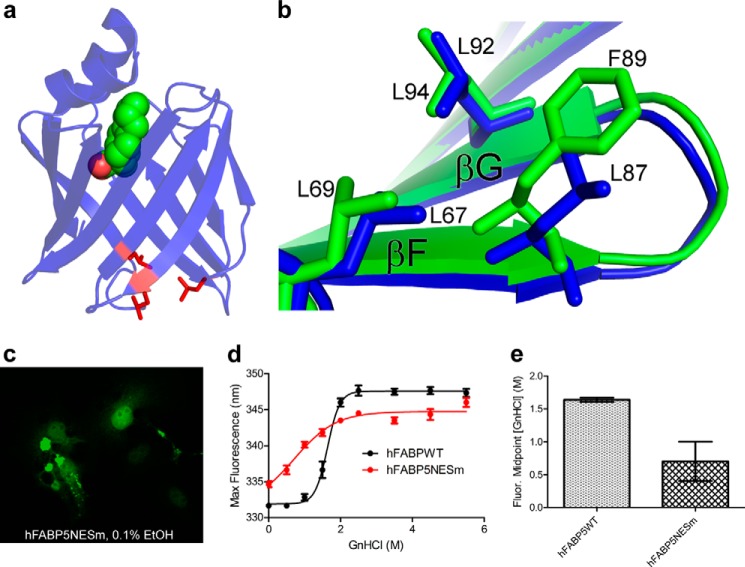
FABP4 has been shown to possess a tertiary NES composed of three leucine residues (Leu-67, Leu-87, and Leu-92) located at the edge of its β-barrel farthest from its α helix lid (Fig. 6a) (19). Given their structural similarity, we reasoned that FABP5 might also have a tertiary NES that is formed from residues equivalent to those that belong to the NES of FABP4. A structural overlay of the two proteins reveals the conservation of two of the three leucines (residues 69 and 94) with the third being Phe-89 in FABP5 (Fig. 6b), a less common but still acceptable NES amino acid substitution because of its ability to preserve the overall hydrophobic character of the signal (63). However, expression of EGFP-tagged FABP5NESm, in which the three residues had been mutated to alanines, resulted in the presence of fluorescing puncta located within both the nuclei and cytoplasm of COS-7 cells (Fig. 6c), suggesting that the mutations may affect structural integrity.
FIGURE 6.
Putative nuclear export signal of FABP5. a, crystal structure of FABP4 bound to LA (Protein Data Bank code 2Q9S), with the leucine residues composing the tertiary NES shown in red. b, overlay reveals strong similarity between the NES of FABP4 (blue) and the potential NES of FABP5 (green). c, representative confocal image of untreated COS-7 cells expressing EGFP-FABP5NESm, displaying aggregation of mutant protein. d, unfolding curves, as measured by protein maximum fluorescence shift, for purified FABP5WT versus FABP5NESm in the absence of ligand addition (n = 6). The mean ± S.E. is shown for all data points. e, comparison of the calculated fluorescence intensity shift midpoint for both proteins reveals that wild-type FABP5 is significantly more resistant to chemical denaturing than the NESm construct. Statistical analysis was performed using an unpaired t test with Welch's correction. *, p ≤ 0.05.
To address this concern, we carried out guanidinium hydrochloride unfolding of both FABP5NESm and wild-type protein, using maximal intrinsic fluorescence intensity wavelength as an indicator of tertiary structure. The unfolding curve of unaltered FABP5 is sigmoidal in nature with the protein exhibiting an average maximum fluorescence wavelength of 331 nm in the absence of denaturant. In contrast, FABP5NESm's curve appears substantially more linear, with an average maximum fluorescence wavelength of nearly 335 nm at 0 m guanidinium hydrochloride, indicating a partially denatured resting state of the mutant protein (Fig. 6d). The concentration of guanidinium hydrochloride necessary to induce the midpoint in maximal intensity wavelength shift varied significantly between proteins, at 1.64 ± 0.03 m for wild-type FABP5 and 0.70 ± 0.30 m for FABP5NESm (Fig. 6e). Therefore, because mutating residues 69, 94, and 98 reduced protein stability, we were unable to positively confirm the presence or identity of the NES.
Ligand-specific Dynamics between β2 Loop and α2 Helix Drives Tertiary NLS Formation
Having identified the cryptic NLS within FABP5, we finally directed our efforts to elucidating a possible driving force responsible for ligand-specific NLS formation. The βC-D, or β2 loop, along with the βE-F loop and α2 helix together constitute the portal domain (64), a feature common throughout the FABPs that is hypothesized to gate ligand access to and from the binding pocket (56) and that has been shown to display structural mobility within FABP5 (55). Analysis of the backbone temperature values for apo-FABP5 lends support for this domain's relatively more labile nature, with the average B-factors of the α2 helix (20.95 Å2) and especially the β2 loop (28.42 Å2) considerably higher than that for the entire protein (16.3 Å2) (Fig. 7a). Furthermore, overlay of FABP5-LA in its proposed activated (monomer C) versus inactivated state (monomer B) reveals that, to avoid a collision with L-conformation LA, the backbone of residues 59–61 within the β2 loop must shift away from the rest of the protein body, assuming a conformation in which there is less contact with α helix 2 (Fig. 7b).
FIGURE 7.
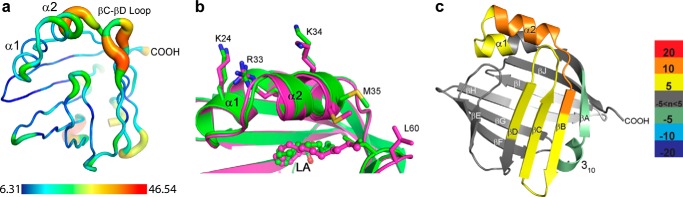
Structural mechanism driving FABP5 activation. a, putty representation of the backbone of apo-FABP5 with B-factor color coordination displaying elevated thermal movement within α2 helix and βC-βD loop. Scale is in units of Å2. b, portal loop of both monomers of FABP5-LA is ordered, yet because of the “L” conformation of LA, it assumes a more open position farther from the α2 helix in its presumed inactivated (purple) versus activated (green) state. c, HDX analysis difference map of FABP5WT-PoA minus FABP5WT-AA average percent deuterium uptake values shows loss of protection from deuterium exchange across the entire portal region of FABP5, including the βC-βD loop and α-helical cap in the presence of PoA versus AA. Portions of the protein colored in white indicate a lack of peptide data coverage. Color bar displays cutoff values for average percent deuterium uptake differences.
To probe the dynamics of FABP5 bound to a nonactivator versus activator, we utilized HDX, which provides an unbiased assessment of backbone motion in solution. The difference in percent deuterium uptake revealed weaker protection of residues that compose the α2 helix and β2 loop within FABP5-PoA relative to FABP5-AA, indicating decreased stabilization of these elements (Fig. 7c). Taken together with our crystallographic data, we hypothesize that the interaction between the α2 helix and the β2 loop via residues Met-35 and Leu-60 determines the activation state of the protein (Fig. 7, b and c). When bound to a fatty acid with a solvent-exposed alkyl tail, the loop must remain open. This breaks contact between Met-35 and Leu-60, destabilizing the α2 helix and thereby rendering FABP5 inactive. Conversely, binding to a more compact, sterically constrained fatty acid such as AA allows for loop closure, providing the additional hydrophobic contacts required to stabilize the α2 helix. This results in a more coalesced formation of the NLS and likely drives protein activation.
To examine the potential role of Met-35 and Leu-60 as ligand conformation sensing “activation switches,” we reduced their hydrophobic interaction via mutation of both residues to alanines, creating a double-switch mutant of FABP5 (FABP5DSm). These mutations had no major impact on ligand binding in vitro (Fig. 8, a and b, and Table 3), yet HDX analysis reveals that they decrease the stability of the α2 helix-β2 loop interface within the mutant protein bound to AA, relative to the wild-type protein AA complex (Fig. 8c). Thus, FABP5DSm does not possess the allosteric coordination that senses and relays information from AA to the NLS. Additionally, comparison of mutant with wild-type protein when both are bound to the nonactivating PoA displays far fewer differences in subsequent deuterium exchange, indicative of largely equal states of loop/helix disruption (Fig. 8d). This effect can also be observed in the HDX analysis of FABP5DSm-PoA relative to FABP5DSm-AA, which, when compared with Fig. 7c, clearly illustrates the mutant's severely hampered ability to structurally distinguish activating from nonactivating ligand (Fig. 8e).
FIGURE 8.
Structural determination of FABP5's activation switch residues. a and b, FABP5DSm was able to bind both the fluorophore 1,8-ANS (a) as well as all fatty acid candidates tested (only AA is shown (b)) (n = 6). c–e, HDX analysis difference map displaying the average percent deuterium uptake values of FABP5DSm-AA minus FABP5WT-AA (c), FABP5DSm-PoA minus FABP5WT-PoA (d), and FABP5DSm-PoA minus FABP5DSm-AA (e) confirms that the mutation of residues Met-35 and Leu-60 results in perturbed βC-βD loop/α2 helix dynamics, resulting in a mutant protein less sensitive to the structural changes brought about by AA versus PoA binding. Portions of the protein colored in white indicate a lack of peptide data coverage. Color bar displays cutoff values for average percent deuterium uptake differences.
Finally, we conducted cellular assays with the double-switch mutant protein to provide biological verification of the results obtained from HDX. Although LA exposure was unable to induce additional FABP5DSm nuclear localization, AA treatment actually resulted in increased cytoplasmic localization (Fig. 9a). Reporter gene assays confirmed a loss in signaling ability of FABP5DSm, with expression of the protein resulting in diminished AA-induced PPARβ/δ activation at 60–100 μm AA (Fig. 9b). These conclusions are further strengthened by the more striking direct comparison of normalized AA dose responses in the presence of wild-type and mutant FABP5 (Fig. 9c). Although overexpression of FABP5WT causes a continual increase in nuclear receptor activation with greater concentrations of AA, expression of either mutant results in a plateau of response beginning at 40 μm ligand. Such a similarity in effect between mutants underscores both the importance of the NLS as well as the switch residues Met-30 and Leu-60 in FABP5 activation.
FIGURE 9.
Biological verification of FABP5's activation switch residues. a, neither LA nor AA exposure induced nuclear translocation of FABP5DSm, with the presence of AA leading to significantly reduced levels of “switch mutant” protein within the nuclei versus cytoplasm of COS-7 cells (n = 30). b, FABP5DSm expression significantly suppressed AA-induced PPARβ/δ activation at concentrations of 60, 80, and 100 μm (n = 6). c, direct comparison of AA dose responses of PPARβ/δ in the presence of FABP5WT, FABP5NLSm, or FABP5DSm. Data sets were taken from Figs. 4b, 5e, and 9b and were normalized internally by dividing all replicate values by the average value obtained from vehicle treatment only. Statistical analyses were performed using either one-factor (a) or two-factor (b and c) ANOVA, with Tukey HSD (a) or Bonferroni (b and c) post hoc tests used for individual comparisons. **, p ≤ 0.01; ****, p ≤ 0.0001. The mean ± S.E. is shown for all data points.
DISCUSSION
Since the discovery of the first FABP's by Ockner et al. (65) over 40 years ago, a wealth of data have steadily accumulated regarding the structure and function of this class of proteins (9). Although the vast majority of structural studies have focused on the determinants of stability and ligand binding, almost no attention outside of FABP1 and -4 has been given to the physical mechanisms driving signal propagation and protein-protein interaction (19, 41, 66–68). We have expanded understanding of FABP signaling by identifying the molecular switch that dictates fatty acid-specific activation, whereby the conformation of bound LCFA relays information from what we now term the “activation loop” (βC-D) of the portal region to the protein's tertiary NLS, consisting of Lys-24, Arg-33, and Lys-34.
In this way, FABP5 shares key mechanistic elements from both FABP4 and CRABP-II, yet ultimately undergoes a method of activation different from either. Like FABP4, only certain ligands cause nuclear localization of the protein (18, 41). However, instead of dimer rearrangement driving the cytosolic exposure of the NLS, FABP5, like CRABP-II, remains monomeric, with binding of activating ligand resulting in stabilization of the NLS that is necessary for nuclear import (41, 61). Interestingly, although this process can occur in as little as 30–60 min for all three proteins, the ensuing enhancement of nuclear receptor-driven gene transcription is most frequently tested 24 h after ligand introduction (19, 69, 70). This time difference could explain, at least in part, why 10 μm AA and LA are sufficient for our localization assays, but not for FABP5-enhanced transactivation of PPARβ/δ, as ligand degradation and metabolism become more relevant over time.
Sequence alignment in Clustal Omega (71) of all nine human FABP members with other iLBPs known to participate in ligand-mediated signaling reveals that FABP8, a major protein constituent of the peripheral nervous system myelin (72), contains residues homologous to the cryptic NLS present in FABP4 and -5 and CRABP-II, as well as to the pair of bulky/hydrophobic amino acids that constitute the ligand-dependent activation switch (Fig. 10). This raises the possibility that myelin FABP could also undergo directed nuclear localization; however, the same NLS homology is also found within the α helices of the iLBP cellular retinol-binding protein I (CRBP-I), where it governs the protein's retinol-dependent interaction with the transmembrane receptor stimulated by retinoic acid 6 (STRA6) (73). Therefore, the potential of FABP8 to engage in a ligand-driven signaling pathway other than nuclear translocation cannot be discounted. Based on its predicted amino acid sequence (data not shown), the same directed inquiries can also be made for the newly discovered FABP12, although its presence within cells has not been documented beyond the mRNA level (74).
FIGURE 10.
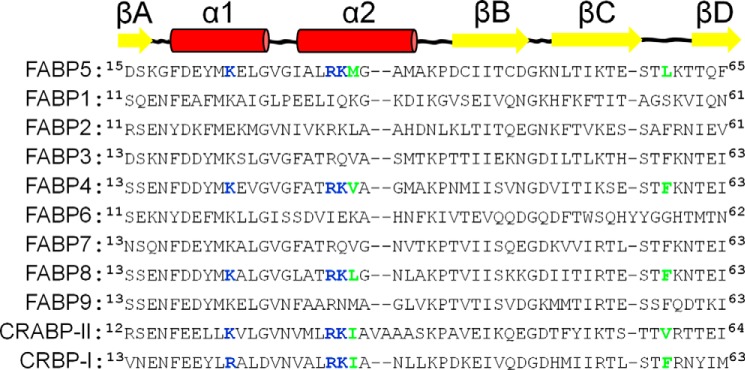
Alignment of FABP1–9 with CRABP-II and CRBP-I. Sequence alignment of the region in FABP5 shown to be most affected by ligand- induced activation with that of the other human FABPs as well as CRABP-II and CRBP-I. FABP8 is the only protein in its class that harbors all three homologous NLS residues (bold, blue) and appropriately bulky/hydrophobic “switch” residues (bold, green), yet is currently untested for ligand-driven nuclear translocation.
Additionally, we have shown that a single fatty acid can adopt at least two unique conformations within the binding pocket of FABP5. Although FABP-bound fatty acid tail mobility has been noted previously via x-ray crystallography (75), our delineation of an active U-conformation versus inactive L-conformer opens up exciting new possibilities for structure-based drug design. The overexpression of FABP5 has been linked to insulin resistance (26), and its signaling is related to cancer cell survival (25), proliferation (29, 32), and metastasis (31, 33), making the protein an ideal candidate for antagonist development. Theoretically, such compounds could exert their influence via one of several mechanisms of action. The first would be to bind and disrupt the portal region, forcing the activation loop into its inactive state. The second would be to bind completely within the binding pocket, allowing closure of the activation loop and subsequent nuclear translocation of the protein, although the compound itself would be unable to bind PPARβ/δ. The third, and likely most potent, would be to improve the nuclear accumulation of current PPARβ/δ antagonists by optimizing their ability to bind and activate FABP5.
Conversely, our fatty acid binding model can be used for the prediction of additional FABP5 activators. We have demonstrated that the state of unsaturation is one of the major determinants of a fatty acid's activation potential, presumably due to its affect on U-conformation preference within the binding pocket. Judging from the configurations seen within our structures as well as that published by Hohoff et al. (54), the first 11–13 carbons share a remarkably close alignment regardless of fatty acid type, thereby placing a greater degree of importance for activator differentiation on the cis-double bonds located more distal to the carboxylate headgroup. As both LA and AA were found to be activators, yet PoA (an ω-7 FA) was not, this suggests an intriguing role for FABP5 as a specific mediator for ω-6 and possibly ω-3 fatty acid signaling. Because all unsaturated fatty acids tested, including oleic acid (data not shown), were able to significantly activate PPARβ/δ in the absence of FABP5, the presence of such a secondary control measure likely serves to ensure preferential activation of the nuclear receptor by this or a similar subset of fatty acids.
Acknowledgments
We gratefully acknowledge Alexa Mattheyses, Jason Fritz, and Debby Martinson (Integrated Cellular Imaging Core, Emory University) and Shuiliang Yu (Case Western Reserve University) for training in microscopy methods, and Katie Doud (Case Western Reserve University) for generous assistance with the PPARβ/δ activation assays. Data for 4LKP were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-BM beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 DK060684 and Grant P30CA138292 from the Emory University Integrated Cellular Imaging Microscopy Core of the Winship Cancer Institute comprehensive cancer center. This work was also supported by start-up funds from Emory University (to E. A. O.).
The atomic coordinates and structure factors (codes 4LKP and 4LKT) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- LCFA
- long chain fatty acid
- FABP
- fatty acid-binding protein
- AA
- arachidonic acid
- NLS
- nuclear localization signal
- PPAR
- peroxisome proliferator-activated receptor
- LA
- linoleic acid
- HSD
- honestly significant difference
- ANOVA
- analysis of variance
- PoA
- palmitoleic acid
- iLBP
- intracellular lipid-binding protein
- DSm
- double-switch mutant
- SpA
- sapienic acid
- PA
- palmitic acid
- NES
- nuclear export signal
- PDB
- Protein Data Bank
- HDX
- hydrogen deuterium exchange
- EGFP
- enhanced green fluorescent protein
- h
- human
- 1,8-ANS
- 1-anilinonaphthalene-8-sulfonic acid
- CRBP-I
- cellular retinol-binding protein I
- CRABP-II
- cellular retinoic acid-binding protein 2.
REFERENCES
- 1. Göttlicher M., Widmark E., Li Q., Gustafsson J. A. (1992) Fatty acids activate a chimera of the clofibric acid-activated receptor and the glucocorticoid receptor. Proc. Natl. Acad. Sci. U.S.A. 89, 4653–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Banner C. D., Göttlicher M., Widmark E., Sjövall J., Rafter J. J., Gustafsson J. A. (1993) A systematic analytical chemistry/cell assay approach to isolate activators of orphan nuclear receptors from biological extracts: characterization of peroxisome proliferator-activated receptor activators in plasma. J. Lipid Res. 34, 1583–1591 [PubMed] [Google Scholar]
- 3. Amri E. Z., Bonino F., Ailhaud G., Abumrad N. A., Grimaldi P. A. (1995) Cloning of a protein that mediates transcriptional effects of fatty acids in preadipocytes. Homology to peroxisome proliferator-activated receptors. J. Biol. Chem. 270, 2367–2371 [DOI] [PubMed] [Google Scholar]
- 4. Forman B. M., Chen J., Evans R. M. (1997) Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc. Natl. Acad. Sci. U.S.A. 94, 4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kliewer S. A., Sundseth S. S., Jones S. A., Brown P. J., Wisely G. B., Koble C. S., Devchand P., Wahli W., Willson T. M., Lenhard J. M., Lehmann J. M. (1997) Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc. Natl. Acad. Sci. U.S.A. 94, 4318–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu H. E., Lambert M. H., Montana V. G., Parks D. J., Blanchard S. G., Brown P. J., Sternbach D. D., Lehmann J. M., Wisely G. B., Willson T. M., Kliewer S. A., Milburn M. V. (1999) Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol. Cell 3, 397–403 [DOI] [PubMed] [Google Scholar]
- 7. Kersten S., Desvergne B., Wahli W. (2000) Roles of PPARs in health and disease. Nature 405, 421–424 [DOI] [PubMed] [Google Scholar]
- 8. Willson T. M., Brown P. J., Sternbach D. D., Henke B. R. (2000) The PPARs: from orphan receptors to drug discovery. J. Med. Chem. 43, 527–550 [DOI] [PubMed] [Google Scholar]
- 9. Furuhashi M., Hotamisligil G. S. (2008) Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat. Rev. Drug. Discov. 7, 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smathers R. L., Petersen D. R. (2011) The human fatty acid-binding protein family: evolutionary divergences and functions. Hum. Genomics 5, 170–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Storch J., McDermott L. (2009) Structural and functional analysis of fatty acid-binding proteins. J. Lipid Res. 50, S126–S131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chmurzyńska A. (2006) The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. J. Appl. Genet. 47, 39–48 [DOI] [PubMed] [Google Scholar]
- 13. Ong D. E., Newcomer M. E., Chytil F. (1994) in The Retinoids: Biology, Chemistry, and Medicine (Sporn M. B., Roberts A. B., Goodman D. S., eds) pp. 288–317, Raven Press, Ltd., New York [Google Scholar]
- 14. Storch J., Thumser A. E. (2000) The fatty acid transport function of fatty acid-binding proteins. Biochim. Biophys. Acta 1486, 28–44 [DOI] [PubMed] [Google Scholar]
- 15. Petrescu A. D., Huang H., Martin G. G., McIntosh A. L., Storey S. M., Landrock D., Kier A. B., Schroeder F. (2013) Impact of L-FABP and glucose on polyunsaturated fatty acid induction of PPARα-regulated β-oxidative enzymes. Am. J. Physiol. Gastrointest. Liver Physiol. 304, G241–G256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Petrescu A. D., McIntosh A. L., Storey S. M., Huang H., Martin G. G., Landrock D., Kier A. B., Schroeder F. (2013) High glucose potentiates L-FABP mediated fibrate induction of PPARα in mouse hepatocytes. Biochim. Biophys. Acta 1831, 1412–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hostetler H. A., McIntosh A. L., Atshaves B. P., Storey S. M., Payne H. R., Kier A. B., Schroeder F. (2009) L-FABP directly interacts with PPARα in cultured primary hepatocytes. J. Lipid Res. 50, 1663–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tan N. S., Shaw N. S., Vinckenbosch N., Liu P., Yasmin R., Desvergne B., Wahli W., Noy N. (2002) Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol. Cell. Biol. 22, 5114–5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ayers S. D., Nedrow K. L., Gillilan R. E., Noy N. (2007) Continuous nucleocytoplasmic shuttling underlies transcriptional activation of PPARγ by FABP4. Biochemistry 46, 6744–6752 [DOI] [PubMed] [Google Scholar]
- 20. Kaczocha M., Glaser S. T., Deutsch D. G. (2009) Identification of intracellular carriers for the endocannabinoid anandamide. Proc. Natl. Acad. Sci. U.S.A. 106, 6375–6380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaczocha M., Vivieca S., Sun J., Glaser S. T., Deutsch D. G. (2012) Fatty acid-binding proteins transport N-acylethanolamines to nuclear receptors and are targets of endocannabinoid transport inhibitors. J. Biol. Chem. 287, 3415–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Siegenthaler G., Hotz R., Chatellard-Gruaz D., Didierjean L., Hellman U., Saurat J. H. (1994) Purification and characterization of the human epidermal fatty acid-binding protein: localization during epidermal cell differentiation in vivo and in vitro. Biochem. J. 302, 363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marcelino A. M., Smock R. G., Gierasch L. M. (2006) Evolutionary coupling of structural and functional sequence information in the intracellular lipid-binding protein family. Proteins 63, 373–384 [DOI] [PubMed] [Google Scholar]
- 24. Simpson M. A., LiCata V. J., Ribarik Coe N., Bernlohr D. A. (1999) Biochemical and biophysical analysis of the intracellular lipid binding proteins of adipocytes. Mol. Cell. Biochem. 192, 33–40 [PubMed] [Google Scholar]
- 25. Schug T. T., Berry D. C., Shaw N. S., Travis S. N., Noy N. (2007) Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell 129, 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maeda K., Uysal K. T., Makowski L., Görgün C. Z., Atsumi G., Parker R. A., Brüning J., Hertzel A. V., Bernlohr D. A., Hotamisligil G. S. (2003) Role of the fatty acid binding protein mal1 in obesity and insulin resistance. Diabetes 52, 300–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hong J., Gu W., Zhang Y., Yan Q., Dai M., Shi J., Zhai Y., Wang W., Li X., Ning G. (2011) Different association of circulating levels of adipocyte and epidermal fatty acid-binding proteins with metabolic syndrome and coronary atherosclerosis in Chinese adults. Atherosclerosis 217, 194–200 [DOI] [PubMed] [Google Scholar]
- 28. Yeung D. C., Wang Y., Xu A., Cheung S. C., Wat N. M., Fong D. Y., Fong C. H., Chau M. T., Sham P. C., Lam K. S. (2008) Epidermal fatty-acid-binding protein: a new circulating biomarker associated with cardio-metabolic risk factors and carotid atherosclerosis. Eur. Heart J. 29, 2156–2163 [DOI] [PubMed] [Google Scholar]
- 29. Schug T. T., Berry D. C., Toshkov I. A., Cheng L., Nikitin A. Y., Noy N. (2008) Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARβ/δ to RAR. Proc. Natl. Acad. Sci. U.S.A. 105, 7546–7551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morgan E., Kannan-Thulasiraman P., Noy N. (2010) Involvement of fatty acid binding protein 5 and PPARβ/δ in prostate cancer cell growth. PPAR Res. 2010 pii: 234629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jeong C. Y., Hah Y. S., Cho B. I., Lee S. M., Joo Y. T., Jung E. J., Jeong S. H., Lee Y. J., Choi S. K., Ha W. S., Park S. T., Hong S. C. (2012) Fatty acid-binding protein 5 promotes cell proliferation and invasion in human intrahepatic cholangiocarcinoma. Oncol. Rep. 28, 1283–1292 [DOI] [PubMed] [Google Scholar]
- 32. Kannan-Thulasiraman P., Seachrist D. D., Mahabeleshwar G. H., Jain M. K., Noy N. (2010) Fatty acid-binding protein 5 and PPARβ/δ are critical mediators of epidermal growth factor receptor-induced carcinoma cell growth. J. Biol. Chem. 285, 19106–19115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levi L., Lobo G., Doud M. K., von Lintig J., Seachrist D., Tochtrop G. P., Noy N. (2013) Genetic ablation of the fatty acid-binding protein FABP5 suppresses HER2-induced mammary tumorigenesis. Cancer Res. 73, 4770–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cologna S. M., Jiang X. S., Backlund P. S., Cluzeau C. V., Dail M. K., Yanjanin N. M., Siebel S., Toth C. L., Jun H. S., Wassif C. A., Yergey A. L., Porter F. D. (2012) Quantitative proteomic analysis of Niemann-Pick disease, type C1 cerebellum identifies protein biomarkers and provides pathological insight. PLoS One 7, e47845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barak Y., Liao D., He W., Ong E. S., Nelson M. C., Olefsky J. M., Boland R., Evans R. M. (2002) Effects of peroxisome proliferator-activated receptor δ on placentation, adiposity, and colorectal cancer. Proc. Natl. Acad. Sci. U.S.A. 99, 303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akiyama T. E., Lambert G., Nicol C. J., Matsusue K., Peters J. M., Brewer H. B., Jr., Gonzalez F. J. (2004) Peroxisome proliferator-activated receptor β/δ regulates very low density lipoprotein production and catabolism in mice on a Western diet. J. Biol. Chem. 279, 20874–20881 [DOI] [PubMed] [Google Scholar]
- 37. Lee C. H., Olson P., Hevener A., Mehl I., Chong L. W., Olefsky J. M., Gonzalez F. J., Ham J., Kang H., Peters J. M., Evans R. M. (2006) PPARδ regulates glucose metabolism and insulin sensitivity. Proc. Natl. Acad. Sci. U.S.A. 103, 3444–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nadra K., Anghel S. I., Joye E., Tan N. S., Basu-Modak S., Trono D., Wahli W., Desvergne B. (2006) Differentiation of trophoblast giant cells and their metabolic functions are dependent on peroxisome proliferator-activated receptor β/δ. Mol. Cell. Biol. 26, 3266–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tan N. S., Michalik L., Noy N., Yasmin R., Pacot C., Heim M., Flühmann B., Desvergne B., Wahli W. (2001) Critical roles of PPARβ/δ in keratinocyte response to inflammation. Genes Dev. 15, 3263–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Di-Poï N., Tan N. S., Michalik L., Wahli W., Desvergne B. (2002) Antiapoptotic role of PPARβ in keratinocytes via transcriptional control of the Akt1 signaling pathway. Mol. Cell 10, 721–733 [DOI] [PubMed] [Google Scholar]
- 41. Gillilan R. E., Ayers S. D., Noy N. (2007) Structural basis for activation of fatty acid-binding protein 4. J. Mol. Biol. 372, 1246–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 43. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 44. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 46. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34, W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Mol Probity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kane C. D., Bernlohr D. A. (1996) A simple assay for intracellular lipid-binding proteins using displacement of 1-anilinonaphthalene 8-sulfonic acid. Anal. Biochem. 233, 197–204 [DOI] [PubMed] [Google Scholar]
- 52. Chalmers M. J., Busby S. A., Pascal B. D., He Y., Hendrickson C. L., Marshall A. G., Griffin P. R. (2006) Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 78, 1005–1014 [DOI] [PubMed] [Google Scholar]
- 53. Pascal B. D., Willis S., Lauer J. L., Landgraf R. R., West G. M., Marciano D., Novick S., Goswami D., Chalmers M. J., Griffin P. R. (2012) HDX workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 23, 1512–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hohoff C., Börchers T., Rüstow B., Spener F., van Tilbeurgh H. (1999) Expression, purification, and crystal structure determination of recombinant human epidermal-type fatty acid binding protein. Biochemistry 38, 12229–12239 [DOI] [PubMed] [Google Scholar]
- 55. Gutiérrez-González L. H., Ludwig C., Hohoff C., Rademacher M., Hanhoff T., Rüterjans H., Spener F., Lücke C. (2002) Solution structure and backbone dynamics of human epidermal-type fatty acid-binding protein (E-FABP). Biochem. J. 364, 725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hanhoff T., Lücke C., Spener F. (2002) Insights into binding of fatty acids by fatty acid binding proteins. Mol. Cell. Biochem. 239, 45–54 [PubMed] [Google Scholar]
- 57. Xu Z., Bernlohr D. A., Banaszak L. J. (1993) The adipocyte lipid-binding protein at 1.6-Å resolution. Crystal structures of the apoprotein and with bound saturated and unsaturated fatty acids. J. Biol. Chem. 268, 7874–7884 [PubMed] [Google Scholar]
- 58. Richieri G. V., Ogata R. T., Kleinfeld A. M. (1994) Equilibrium constants for the binding of fatty acids with fatty acid-binding proteins from adipocyte, intestine, heart, and liver measured with the fluorescent probe ADIFAB. J. Biol. Chem. 269, 23918–23930 [PubMed] [Google Scholar]
- 59. Richieri G. V., Ogata R. T., Zimmerman A. W., Veerkamp J. H., Kleinfeld A. M. (2000) Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interactions. Biochemistry 39, 7197–7204 [DOI] [PubMed] [Google Scholar]
- 60. Seibel N. M., Eljouni J., Nalaskowski M. M., Hampe W. (2007) Nuclear localization of enhanced green fluorescent protein homomultimers. Anal. Biochem. 368, 95–99 [DOI] [PubMed] [Google Scholar]
- 61. Sessler R. J., Noy N. (2005) A ligand-activated nuclear localization signal in cellular retinoic acid binding protein-II. Mol. Cell 18, 343–353 [DOI] [PubMed] [Google Scholar]
- 62. McLane L. M., Corbett A. H. (2009) Nuclear localization signals and human disease. IUBMB Life 61, 697–706 [DOI] [PubMed] [Google Scholar]
- 63. Kutay U., Güttinger S. (2005) Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 15, 121–124 [DOI] [PubMed] [Google Scholar]
- 64. Sacchettini J. C., Gordon J. I., Banaszak L. J. (1989) Refined apoprotein structure of rat intestinal fatty acid binding protein produced in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 86, 7736–7740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ockner R. K., Manning J. A., Poppenhausen R. B., Ho W. K. (1972) A binding protein for fatty acids in cytosol of intestinal mucosa, liver, myocardium, and other tissues. Science 177, 56–58 [DOI] [PubMed] [Google Scholar]
- 66. Hertzel A. V., Hellberg K., Reynolds J. M., Kruse A. C., Juhlmann B. E., Smith A. J., Sanders M. A., Ohlendorf D. H., Suttles J., Bernlohr D. A. (2009) Identification and characterization of a small molecule inhibitor of fatty acid binding proteins. J. Med. Chem. 52, 6024–6031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hostetler H. A., Balanarasimha M., Huang H., Kelzer M. S., Kaliappan A., Kier A. B., Schroeder F. (2010) Glucose regulates fatty acid binding protein interaction with lipids and peroxisome proliferator-activated receptor α. J. Lipid Res. 51, 3103–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Velkov T. (2013) Interactions between human liver fatty acid binding protein and peroxisome proliferator activated receptor selective drugs. PPAR Res. 2013, 938401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Budhu A. S., Noy N. (2002) Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol. Cell. Biol. 22, 2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Majumdar A., Petrescu A. D., Xiong Y., Noy N. (2011) Nuclear translocation of cellular retinoic acid-binding protein II is regulated by retinoic acid-controlled SUMOylation. J. Biol. Chem. 286, 42749–42757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Greenfield S., Brostoff S., Eylar E. H., Morell P. (1973) Protein composition of myelin of the peripheral nervous system. J. Neurochem. 20, 1207–1216 [DOI] [PubMed] [Google Scholar]
- 73. Berry D. C., O'Byrne S. M., Vreeland A. C., Blaner W. S., Noy N. (2012) Cross talk between signaling and vitamin A transport by the retinol-binding protein receptor STRA6. Mol. Cell. Biol. 32, 3164–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu R. Z., Li X., Godbout R. (2008) A novel fatty acid-binding protein (FABP) gene resulting from tandem gene duplication in mammals: transcription in rat retina and testis. Genomics 92, 436–445 [DOI] [PubMed] [Google Scholar]
- 75. Angelucci F., Johnson K. A., Baiocco P., Miele A. E., Brunori M., Valle C., Vigorosi F., Troiani A. R., Liberti P., Cioli D., Klinkert M. Q., Bellelli A. (2004) Schistosoma mansoni fatty acid binding protein: specificity and functional control as revealed by crystallographic structure. Biochemistry 43, 13000–13011 [DOI] [PubMed] [Google Scholar]