Abstract
The aryl hydrocarbon receptor repressor (AHRR) is a transcriptional repressor of aryl hydrocarbon receptor (AHR) and hypoxia-inducible factor (HIF) and is regulated by an AHR-dependent mechanism. Zebrafish (Danio rerio) possess two AHRR paralogs; AHRRa regulates constitutive AHR signaling during development, whereas AHRRb regulates polyaromatic hydrocarbon-induced gene expression. However, little is known about the endogenous roles and targets of AHRRs. The objective of this study was to elucidate the role of AHRRs during zebrafish development using a loss-of-function approach followed by gene expression analysis. Zebrafish embryos were microinjected with morpholino oligonucleotides against AHRRa or AHRRb to knockdown AHRR protein expression. At 72 h postfertilization (hpf), microarray analysis revealed that the expression of 279 and 116 genes was altered by knockdown of AHRRa and AHRRb, respectively. In AHRRa-morphant embryos, 97 genes were up-regulated and 182 genes were down-regulated. Among the down-regulated genes were several related to photoreceptor function, including cone-specific genes such as several opsins (opn1sw1, opn1sw2, opn1mw1, and opn1lw2), phosphodiesterases (pde6H and pde6C), retinol binding protein (rbp4l), phosducin, and arrestins. Down-regulation was confirmed by RT-PCR and with samples from an independent experiment. The four genes tested (opn1sw1, pde6H, pde6C, and arr3b) were not inducible by 2,3,7,8-tetrachlorodibenzo-p-dioxin. AHRRa knockdown also caused up-regulation of embryonic hemoglobin (hbbe3), suggesting a role for AHRR in regulating hematopoiesis. Knockdown of AHRRb caused up-regulation of 31 genes and down-regulation of 85 genes, without enrichment for any specific biological process. Overall, these results suggest that AHRRs may have important roles in development, in addition to their roles in regulating xenobiotic signaling.
Keywords: microarrays, morpholino oligonucleotides, opsins, cones, development, repressor, zebrafish, aryl hydrocarbon receptor, dioxin, TCDD, AHR, AHRR
The aryl hydrocarbon receptor repressor (AHRR) is a member of the basic helix-loop-helix/Per-AHR nuclear translocator (ARNT)-Sim (bHLH-PAS) protein family and is well-established as a transcriptional repressor of aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor well known for its roles in toxicology, physiology, and development (Haarmann-Stemmann and Abel, 2006; Hahn et al., 2009). AHRR shares a high degree of sequence similarity with AHR in the amino terminus that contains bHLH and PAS-A domains. However, AHRR lacks the PAS-B domain that acts as the ligand-binding region in AHR (Mimura et al., 1999); thus, AHRR cannot bind ligands such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (Karchner et al., 2002) and acts in a ligand-independent fashion. The presence of bHLH and PAS-A domains enables AHRR to heterodimerize with ARNT and bind to AHR response elements (AHREs) in the promoter regions of target genes. AHR regulates AHRR transcription by the binding of AHR-ARNT heterocomplex to AHREs in the promoter regions of the AHRR gene (Mimura et al., 1999) and putative AHREs have been identified in the AHRR promoter regions in a number of species (Haarmann-Stemmann et al., 2007; Karchner et al., 2002).
Competition between AHR and AHRR for binding to AHREs is one mechanism of repression by AHRR (Hahn et al., 2009; Mimura et al., 1999). Recently, other mechanisms of repression by AHRR have been proposed, including small ubiquitin modification (SUMOylation) of AHRR and ARNT (Oshima et al., 2009) and a transrepression mechanism involving protein-protein interactions (Evans et al., 2008). The presence of multiple mechanisms of repression by AHRR suggests that there may be a broader role for this protein in addition to the regulation of AHR signaling.
The specificity of AHRR in suppressing the function of other transcription factors is not fully understood, but recent studies have suggested that transcription factors other than AHR may also be targets of repression by AHRR. Kanno et al. (2008) reported that the repression of estrogen receptor alpha (ERα)-mediated activation of target genes in vitro occurs by direct interaction between AHRR and ERα protein. Similarly, Karchner et al. (2009) demonstrated that AHRR could repress hypoxia-inducible factor 1α (HIF-1α)-mediated transactivation in vitro in cells transfected with a hypoxia response element (HRE)-coupled luciferase construct. In addition, AHRR has been shown to act as a tumor suppressor gene in several types of cancer, in which knockdown of AHRR expression enhances the angiogenic potential of tumors (Zudaire et al., 2008). Furthermore, Mimura et al. (1999) demonstrated that a NF-κB activator, 12-O-tetradecanoylphorbol-13-acetate, induces AHRR transcription. A NF-κB binding site is present in the murine AHRR regulatory region, suggesting the potential for activation of AHRR by other transcription factors. There are also reports linking polymorphisms in AHRR to reproductive abnormalities (Watanabe et al., 2004). AHRR mRNA expression profiles also suggest that AHRR might be involved in physiological processes other than AHR signaling. AHRR mRNA is constitutively expressed in most adult tissues and the fetus in mammals, suggesting multiple biological roles in both adults and developing animals (Bernshausen et al., 2006; Nishihashi et al., 2006; Thackaberry et al., 2005; Yamamoto et al., 2004).
We are using zebrafish embryos as a model system to investigate AHRR function and its possible role in embryonic development and developmental toxicity. Zebrafish possess two AHRR paralogs (AHRRa and AHRRb), thought to be the result of a teleost-specific genome-duplication; phylogenetic analysis and comparative genomics suggests that they are co-orthologs of mammalian AHRR (Evans et al., 2005). We recently demonstrated that knockdown of AHRRa in zebrafish embryos using morpholino oligonucleotides (MOs) causes developmental abnormalities similar to those observed in TCDD-exposed embryos (Jenny et al., 2009). These phenotypes were not observed in embryos injected with AHRRb-MO. Knockdown of AHRRb, but not AHRRa, enhanced the induction of cyp1a, cyp1b, and cyp1c by TCDD, suggesting distinct roles for AHRRa and AHRRb in regulating developmental processes and TCDD-induced embryotoxicity. Thus, the zebrafish embryo model provides a unique opportunity to investigate the functions of these duplicated genes and gain insights into the various roles played by their single mammalian ortholog (Jenny et al., 2009). Based on the above evidence, we hypothesized that AHRRa and AHRRb may regulate distinct sets of genes during embryonic development. Here, we tested this hypothesis by knocking down the individual AHRRs using antisense MOs and profiling the transcriptional changes using microarrays. Our results demonstrate that AHRRa knockdown alters expression profiles of several genes involved in photoreceptor signaling and hematopoiesis.
MATERIALS AND METHODS
Experimental animals
All the experiments were conducted using the Tupfel/Long fin (TL) wild-type strain of zebrafish. The experimental procedures used were approved by the Animal Care and Use Committee of the Woods Hole Oceanographic Institution. Animal maintenance, breeding, and rearing of embryos were carried out as described previously (Jenny et al., 2009; Jonsson et al., 2007).
Morpholino oligonucleotide microinjections
All the MOs used in this study were previously described (Jenny et al., 2009). Fluorescein tagged MOs against AHRRa and AHRRb as well as control MO were obtained from Gene tools, LLC (Philomath, OR). The efficacy of AHRRa and AHRRb MOs in blocking translation was verified in vitro as described earlier (Jenny et al., 2009).
MOs were diluted to 0.18mM in deionized water and approximately 2.1 nl (3.3–3.4 ng) of each MO was injected into the yolk of 1–2 cell embryos using a Narishige IM-300 microinjector (Jenny et al., 2009). Embryos were screened for fluorescence at 3 h postfertilization (hpf) for successful MO incorporation; any embryos that did not have fluorescence were discarded. Three biological replicates of 20 pooled embryos per replicate were set up for each treatment (Ctrl-MO, AHRRa-MO, and AHRRb-MO). Embryos were maintained in 0.3× Danieau's solution at 28.5°C in 14:10 light-dark cycle. At 72 hpf, all replicate samples were collected for each treatment and stored in −80°C for RNA isolation from each set of 20 pooled embryos followed by gene expression analysis using microarrays (two of the replicates) and, subsequently, quantitative, real-time RT-PCR (all three replicates). The microarray analyses were performed using only two replicates because of limitations on the number of arrays available for hybridization at the time these analyses were performed.
TCDD exposure
Two additional experiments were used to obtain an independent assessment of the effects of knocking down AHRRa and AHRRb, as well as to determine the effect of TCDD exposure in MO-injected embryos. (These experiments were part of an earlier study (Jenny et al., 2009) that focused on CYP1 expression after knockdown of AHRRs.) For those experiments, MOs were injected into the zebrafish embryos as described in the above section. At 6 hpf, MO-injected (AHRRa, AHRRb, and Ctrl) or noninjected (control) embryos were divided into three biological replicates of 20 pooled embryos for each treatment (0.1% dimethyl sulfoxide (DMSO) or 2nM TCDD) and exposed for 1 h in glass Petri dishes, as described earlier (Jenny et al., 2009). At the end of the exposure period, embryos were rinsed thoroughly with fresh Danieau solution and maintained at 28.5°C in 14:10 light-dark cycle until sampling at 72 hpf. All treatments were collected at 72 hpf and stored in −80°C for later analysis of gene expression using quantitative, real-time RT-PCR. CYP1 expression data from these experiments was reported previously (Jenny et al., 2009).
RNA isolation
Total RNA was prepared using the RNA STAT60 protocol (Tel-Test Inc., TX). RNA was quantified using a Nanodrop spectrophotometer and assessed for quality using an Agilent 2100 Bioanalyzer Lab-on-chip system. Only samples with RNA integrity number (RIN) between 9.8 and 10 were used for microarray analysis.
Microarray analysis
Whole-genome mRNA expression analysis was performed using Agilent 4×44 K zebrafish microarrays (Agilent Technologies Inc.). Microarray experiments were done in a two-color experimental design by comparing individual samples to a universal reference (UR) pool prepared from pooling equal amounts of total RNA from all the experimental samples.
RNA labeling was done using Agilent Low Input Quick Amp Labeling kit for two-color microarray-based gene expression analysis. Five hundred nanograms of total RNA were reverse-transcribed during which a T7 sequence was introduced into cDNA. T7 RNA polymerase-driven RNA synthesis was used for the preparation and labeling of RNA with fluorescent dyes. UR RNA was labeled with Cy3 and individual sample RNA was labeled with Cy5. The fluorescent cRNA probes were purified using Qiagen RNeasy Mini Kit (Qiagen Inc., Valencia, CA) to remove any excess dye. Equal amount of Cy3 (UR) and Cy5 (samples) labeled cRNA probes were mixed and hybridized to the array. Hybridization was done at 55°C for 20 h in a rotating hybridization oven (20 rpm). The hybridized slides were washed using Agilent microarray wash buffers and scanned using Genepix 4100A scanner (Molecular Devices Corporation, Sunnyvale, CA).
The resulting images were analyzed using Agilent Feature Extraction (AFE) software version 9.5.3. Image analyses included signal and spatial detrending and applying a universal error model. Quality control (QC) reports generated by AFE software were used to assess data quality for each microarray and to identify outliers. The raw data were used in statistical analysis for determining differential gene expression patterns.
Statistical analysis was performed using the Bioconductor package, linear microarray analysis (LIMMA) (Wettenhall and Smyth, 2004). Background-corrected mean signal intensities were used in the analysis. Within-array and between-array normalization was done using intensity-dependent global loess normalization. For statistical analysis, a linear model was fitted and differential gene expression was determined using the empirical Bayes method. This method moderates the standard errors of the log-fold changes and provides statistical power in situations where the sample size is small (Smyth, 2005). Differentially expressed genes were identified using p-values adjusted for multiple testing. Benjamini and Hochberg's method was used to control False Discovery Rate (adjusted p-value < 0.05). In addition, B-statistic – log-odds that the gene is differentially expressed was also taken into consideration for determining differentially expressed genes. All microarray data presented in this manuscript are in accordance with Minimum Information About a Microarray Experiment (MIAME) guidelines and are deposited in the NCBI GEO database (GEO accession number: GSE52229).
Gene Ontology term enrichment and biological network analysis
Differentially expressed genes were functionally annotated with Gene Ontology (GO) terms using DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resource 6.7 (http://david.abcc.ncifcrf.gov/) (Huang da et al., 2009). GO term enrichment for biological process was determined by comparing the list of differentially expressed genes against all the probes on the microarray.
Biological network analysis was conducted to determine the functional association between the proteins encoded by the differentially expressed genes. This was conducted using the String database (www.string-db.org) (Szklarczyk et al., 2011). Default prediction methods were used except text mining and the required confidence score was set as high (0.7). The network obtained was imported into Cytoscape for identification of subnetworks with differentially expressed genes using jActiveModules version 2.23 (Cline et al., 2007; Yeung et al., 2008).
Quantitative real-time PCR
Complementary DNA was synthesized from 2 μg total RNA using random hexamers and the Omniscript cDNA Synthesis Kit (Qiagen). Quantitative PCR was carried out using the iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) in a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad). The PCR conditions used were 95°C for 3 min and 95°C for 15 s/Tm°C for 1 min (40 cycles). The primers used to amplify different transcripts and their annealing temperatures (Tm, °C) are given in Table 1. Each PCR reaction was run in triplicate and a no-template control reaction was run on each PCR plate. Melt curve analysis was performed at the end of each PCR run to ensure that no nonspecific products are amplified. Relative levels of transcript abundance were calculated using the 2−ΔΔCt method (where ΔΔCt = [Ct(GOI) − Ct(β-actin)]TCDD − [Ct(GOI) − Ct(β-actin)]DMSO (GOI, gene of interest; Livak and Schmittgen, 2001). Statistical analysis of qPCR data was conducted using the Prism 4 software package (GraphPad Software Inc., San Diego, CA). Logarithmic transformed relative expression values were used in analysis of variance (ANOVA). One-way ANOVA was used to determine the effect of MOs on gene expression (confirmation of microarray results). Two-way ANOVA was used to determine the effect of MO and TCDD. Bonferroni's post hoc test was used to determine the statistical significance. A p-value of ≤0.05 is considered statistically significant.
TABLE 1. List of qPCR Primers Used in the Study.
| Gene | Accession number | Primers | Tm (°C) | Product size (bp) |
|---|---|---|---|---|
| pde6h | NM_200785.1 | Forward 5′-CAG AAG CTC CAG CAC AGC AC-3′ | 65 | 179 |
| Reverse 5′-GAT GTC TGT GCC GAG ACC CTC-3′ | ||||
| pde6c | NM_200871.1 | Forward 5′-GGA CAT GAC CAA AGA GAA GGA G-3′ | 60 | 170 |
| Reverse 5′-GCA GGA GGT TGG GTA TAA CTT TG-3′ | ||||
| arrestin3b | BC076177.1 | Forward 5′-CTC TGA CAC AGA AAC GTT AGC AG-3′ | 65 | 145 |
| Reverse 5′-CAC CAT CAA CTG AAT CAA CGC-3′ | ||||
| mmp9 | NM_213123.1 | Forward 5′-CAC ATA CAG GAT TTT GAA CTA TTC G-3′ | 59 | 170 |
| Reverse 5′-GAT CAC CGT GAT CTG CTT TCC-3′ | ||||
| opn1sw1 | NM_131319.1 | Forward 5′-CGA TTG CAG GTC TTG TGA CG-3′ | 60 | 196 |
| Reverse 5′-GAC CCT CGG GAA TGT ATC TGC-3′ | ||||
| β-actin | AF057040.1 | Forward 5′-CAA CAG AGA GAA GAT GAC ACA GAT CA-3′ | 65 | 140 |
| Reverse 5′-GTC ACA CCA TCA CCA GAG TCC ATC AC-3′ | ||||
| hbbe3 | NM_001015058.1 | Forward 5′-CGA GAC CTT GAC AAG ATG CTT GGT-3′ | 59 | 175 |
| Reverse 5′-CCT TTT CAA GTC CCT TGA GGA CC-3′ |
RESULTS
Altered Gene Expression Profiles After AHRR Knockdown
To test the hypothesis that AHRRa and AHRRb have distinct roles in regulating gene expression during development, we knocked down each one separately and measured changes in global gene expression by microarray. Altogether, 279 and 116 genes showed significant differential expression in embryos injected with AHRRa-MO or AHRRb-MO, respectively, as compared with embryos injected with the control-MO. Knockdown of AHRRa caused up-regulation of 97 genes and down-regulation of 182 genes, whereas knockdown of AHRRb caused up-regulation of 31 genes and down-regulation of 85 genes in comparison to the control MO-injected group. All differentially expressed genes were hierarchically clustered and the results are shown as a heat map in Figure 1A.
FIG. 1.
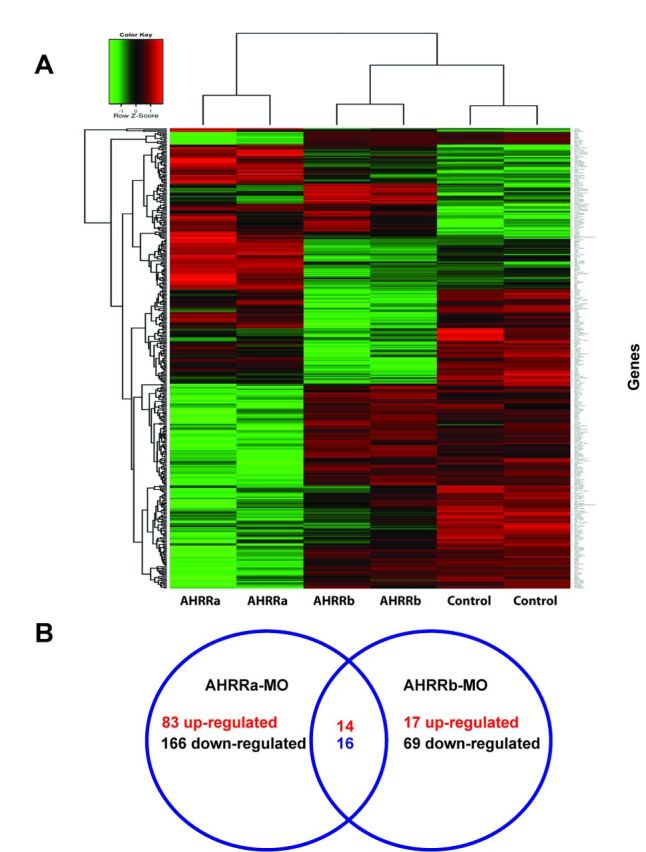
Altered gene expression in response to MO-induced knockdown of AHRR in zebrafish embryos. (A) Heat map representation of gene expression patterns significantly altered by AHRRa and AHRRb knockdown at 72 hpf, in comparison to a control morpholino-injected group. Hierarchial clustering of significant genes was performed. Adjusted p-value less than 0.01 is considered to be statistically significant. (B) Venn diagram showing the total number of up- and down-regulated genes that are uniquely altered by AHRRa and AHRRb MOs at 72 hpf. The set of genes that are differentially expressed by both MOs are shown in the intersection.
The genes with twofold or higher induction with AHRRa knockdown are shown in Table 2 and the list of ≥2-fold down-regulated genes is in Table 3. Genes with a ≥2-fold change in expression after AHRRb knockdown are listed in Table 4 (up-regulated) and Table 5 (down-regulated). Among the genes that were differentially expressed in AHRRa and AHRRb morphants, 30 genes were common to both treatment groups (Fig. 1B; Table 6). Of these, 14 genes were up-regulated and 16 were down-regulated. All but three genes (fibrinogen gamma polypeptide (fgg), cathepsin 1La (ctsl1a), and type IV antifreeze protein precursor (zgc:161979)) were altered in the same direction with both AHRRa and AHRRb knockdown. There was no effect of AHRR knockdowns on mRNA expression of AHR1a, AHR2, HIF-1αb, HIF-2α, HIF-3α, or ARNT2 (other members of these gene subfamilies were not on the array; Supplementary table 1). The full list of all the significantly altered genes is provided in Supplementary table 2 (AHRR-MO differential gene expression.xls).
TABLE 2. List of Genes Up-regulated in AHRRa-MO-injected Zebrafish Embryos.
| Probe name | Description | Accession number | Fold change | Adjusted p-value | B |
|---|---|---|---|---|---|
| A_15_P106583 | Hemoglobin beta embryonic-3 (hbbe3) | NM_001015058 | 16.28 | 0.000 | 14.22 |
| A_15_P107501 | Fibrinogen, gamma polypeptide (fgg) | NM_213054 | 2.43 | 0.000 | 8.07 |
| A_15_P114154 | Matrix metalloproteinase 9 (mmp9) | NM_213123 | 9.59 | 0.000 | 7.09 |
| A_15_P100851 | Cyclin G1 (ccng1) | NM_199481 | 2.27 | 0.000 | 6.41 |
| A_15_P114425 | Uncharacterized protein LOC494049 (zgc:101853) | NM_001008592 | 2.43 | 0.000 | 6.37 |
| A_15_P117758 | Jun B proto-oncogene (junb) | NM_213556 | 2.51 | 0.001 | 5.65 |
| A_15_P106305 | Type IV antifreeze protein precursor (zgc:161979) | NM_001045488 | 2.17 | 0.001 | 5.63 |
| A_15_P102660 | Tumor protein p53 (tp53) | NM_131327 | 2.44 | 0.002 | 4.93 |
| A_15_P111458 | Hemoglobin alpha embryonic-1 (hbae1) | NM_182940 | 2.05 | 0.002 | 4.94 |
| A_15_P118692 | Hypothetical protein LOC550501 (zgc:111983) | NM_001017803 | 2.28 | 0.002 | 4.65 |
| A_15_P103191 | Matrix metalloproteinase 9 (mmp9) | NM_213123 | 3.70 | 0.002 | 4.56 |
| A_15_P120536 | Tumor protein p53 (tp53) | NM_131327 | 2.05 | 0.002 | 4.47 |
| A_15_P111302 | APOEB_BRARE (O42364) apolipoprotein Eb precursor (Apo-Eb), partial (62%)(TC312510) | NM_131098.1 | 2.03 | 0.003 | 4.10 |
| A_15_P118068 | Danio rerio cDNA clone IMAGE: 6965466. | BC160607 | 14.24 | 0.004 | 3.80 |
| A_15_P113920 | CCAAT/enhancer binding protein (C/EBP), beta (cebpb) | NM_131884 | 2.47 | 0.004 | 3.57 |
| A_15_P107566 | Apolipoprotein Eb (apoeb) | NM_131098 | 2.13 | 0.004 | 3.56 |
| A_15_P120997 | Stanniocalcin 1 (stc1) | NM_200539 | 2.33 | 0.006 | 3.15 |
| A_15_P117178 | Fibrinogen, gamma polypeptide (fgg) | NM_213054 | 2.09 | 0.006 | 3.08 |
| A_15_P101751 | Glycine N-methyltransferase (gnmt) | NM_212816 | 2.19 | 0.006 | 3.04 |
| A_15_P102187 | Unknown | 2.04 | 0.008 | 2.64 | |
| A_15_P112685 | Lectin, galactoside-binding, soluble, 1 (galectin 1)-like 1 (lgals1l1) | NM_001005958 | 2.12 | 0.009 | 2.41 |
| A_15_P120578 | Beaded filament structural protein 2, phakinin (bfsp2) (zgc:103750) | NM_001008633 | 2.25 | 0.017 | 1.63 |
| A_15_P116825 | Beaded filament structural protein 2, phakinin (bfsp2) (zgc:103750) | NM_001008633 | 2.31 | 0.022 | 1.15 |
| A_15_P109667 | Crystallin, gamma M2c (crygm2c) | NM_001007783 | 2.18 | 0.023 | 1.12 |
| A_15_P103210 | N-myc downstream regulated gene 1, like (ndrg1l) | NM_200692 | 2.43 | 0.025 | 0.97 |
| A_15_P120206 | Arrestin domain containing 3b (arrdc3b) (zgc:92034) | NM_001004605 | 2.32 | 0.025 | 0.96 |
| A_15_P101920 | Novel protein similar to vertebrate PX domain containing serine/threonine kinase (PXK) | CAK11481.1 | 2.35 | 0.027 | 0.80 |
| A_15_P110005 | Histone H1-like (zgc:110380) | NM_001017660 | 2.15 | 0.041 | 0.07 |
Note. Probe name corresponds to the Agilent probe ID. Fold change values, adjusted p-value and B-statistic are based on the statistical analysis. Only genes that showed ≥2-fold change are shown in the table. The full list of differentially expressed genes is given in Supplementary table 2.
TABLE 3. List of Genes Down-regulated in AHRRa-MO-injected Zebrafish Embryos.
| Probe name | Description | Accession number | Fold change | Adjusted p-value | B |
|---|---|---|---|---|---|
| A_15_P107533 | Phosphodiesterase 6C, cGMP-specific, cone, alpha prime (pde6c) | NM_200871 | −8.09 | 0.000 | 13.30 |
| A_15_P115980 | Opsin 1 (cone pigments), short-wave-sensitive 1 (opn1sw1) | NM_131319 | −7.24 | 0.000 | 13.53 |
| A_15_P114133 | Phosphodiesterase 6H, cGMP-specific, cone, gamma (pde6h) | NM_200785 | −10.09 | 0.000 | 12.54 |
| A_15_P117046 | Opsin 1 (cone pigments), medium-wave-sensitive, 1 (opn1mw1) | NM_131253 | −7.35 | 0.000 | 12.35 |
| A_15_P106879 | Arrestin 3, retinal (X-arrestin) (arr3) | NM_200792 | −4.40 | 0.000 | 11.94 |
| A_15_P120294 | Retinal degradation slow 2 (rds2) | NM_131566 | −5.22 | 0.000 | 11.37 |
| A_15_P111532 | Guanine nucleotide binding protein (G protein), beta polypeptide 3 (gnb3) | NM_213202 | −5.58 | 0.000 | 11.16 |
| A_15_P121399 | Guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 (gnat2) | NM_131869 | −7.08 | 0.000 | 10.98 |
| A_15_P107383 | Phosducin 2 (pdc2) | NM_001025464 | −3.61 | 0.000 | 10.50 |
| A_15_P102875 | Retinal degradation slow 4 (rds4) | NM_131567 | −5.60 | 0.000 | 10.19 |
| A_15_P110693 | Retinal degradation slow 2 (rds2) | NM_131566 | −3.57 | 0.000 | 10.05 |
| A_15_P104722 | Solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3, like (slc25a3l), nuclear gene encoding mitochondrial protein | NM_200715 | −3.25 | 0.000 | 10.05 |
| A_15_P109108 | Villin 1 like (vil1l) | NM_200238 | −3.95 | 0.000 | 9.43 |
| A_15_P119413 | Opsin 1 (cone pigments), short-wave-sensitive 2 (opn1sw2) | NM_131192 | −5.59 | 0.000 | 9.17 |
| A_15_P119429 | Opsin 1 (cone pigments), long-wave-sensitive, 2 (opn1lw2) | NM_001002443 | −2.98 | 0.000 | 8.72 |
| A_15_P111012 | Opsin 1 (cone pigments), long-wave-sensitive, 2 (opn1lw2) | NM_001002443 | −3.37 | 0.000 | 8.54 |
| A_15_P104674 | Retinol binding protein 4, like (rbp4l) | NM_199965 | −3.26 | 0.000 | 8.48 |
| A_15_P104664 | Arrestin 3, retinal (X-arrestin), like (arr3l) | NM_001002405 | −5.21 | 0.000 | 8.32 |
| A_15_P103714 | Guanylate cyclase activator 1C (guca1c) | NM_194393 | −2.79 | 0.000 | 7.59 |
| A_15_P106371 | XM_678537 carboxypeptidase B1 (tissue) isoform 1 (Danio rerio) (exp = −1; wgp = 0; cg = 0), complete | TC311008 | −2.81 | 0.000 | 7.47 |
| A_15_P115516 | Solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3, like (slc25a3l), nuclear gene encoding mitochondrial protein | NM_200715 | −2.82 | 0.000 | 7.11 |
| A_15_P108311 | Chymotrypsinogen B2 precursor (zgc:112160) | NM_001017724 | −4.05 | 0.000 | 6.98 |
| A_15_P116791 | Calcium/calmodulin-dependent protein kinase IG (camk1g) | NM_199966 | −2.26 | 0.000 | 6.56 |
| A_15_P109678 | Calcium binding protein 5b (cabp5b) (zgc:109977) | NM_001020732 | −2.64 | 0.000 | 6.47 |
| A_15_P119059 | Guanylate kinase 1 (guk1) | NM_200724 | −2.52 | 0.000 | 6.42 |
| A_15_P108795 | Retinaldehyde binding protein 1b (rlbp1b) | NM_205690 | −2.99 | 0.000 | 6.34 |
| A_15_P102721 | Chitinase, acidic.2 precursor (zgc:55941) | NM_213249 | −2.53 | 0.000 | 6.21 |
| A_15_P100788 | SH3-domain GRB2-like 2 (sh3gl2) | NM_201116 | −2.08 | 0.001 | 5.83 |
| A_15_P117814 | Trypsin (try) | NM_131708 | −3.13 | 0.001 | 5.30 |
| A_15_P113799 | Chymotrypsinogen B1 (ctrb1) | NM_212618 | −3.05 | 0.002 | 5.05 |
| A_15_P115606 | Q6PBU3_BRARE (Q6PBU3) Stathmin-like 4, complete | NM_213401 | −2.07 | 0.002 | 4.86 |
| A_15_P112264 | Group-specific component (vitamin D binding protein) (zgc:92753) | NM_001002568 | −2.86 | 0.002 | 4.70 |
| A_15_P113364 | Glycogenin, like (gygl) | NM_001002062 | −2.08 | 0.002 | 4.57 |
| A_15_P113940 | Uncharacterized protein LOC445282 precursor (zgc:92041) | NM_001003737 | −4.09 | 0.002 | 4.53 |
| A_15_P107695 | Fatty acid binding protein 10, liver basic (fabp10) | NM_152960 | −2.58 | 0.003 | 4.15 |
| A_15_P110245 | Unknown | −2.64 | 0.004 | 3.81 | |
| A_15_P110192 | Pyrophosphatase (inorganic) (pp) | NM_200733 | −2.00 | 0.004 | 3.83 |
| A_15_P110812 | Trypsin-1-like (zgc:66382) NM_199605 | NM_199605 | −2.98 | 0.004 | 3.78 |
| A_15_P115802 | Cathepsin L, 1 b (ctsl1b) | NM_131198 | −2.79 | 0.004 | 3.69 |
| A_15_P118837 | 3-Oxoacid CoA transferase 1a (oxct1a) | NM_001007291 | −2.28 | 0.004 | 3.66 |
| A_15_P115815 | G-Protein-coupled receptor kinase 7a (grk7a) | NM_001031841 | −2.18 | 0.004 | 3.70 |
| A_15_P112986 | Solute carrier family 31 (copper transporters), member 1 (slc31a1) | NM_205717 | −2.05 | 0.004 | 3.67 |
| A_15_P114820 | Recoverin (rcv1) | NM_199964 | −2.12 | 0.004 | 3.60 |
| A_15_P113750 | Serum/glucocorticoid regulated kinase 1-like (si:ch211–195b13.1) | NM_001077302 | −2.21 | 0.004 | 3.53 |
| A_15_P100752 | Alanine-glyoxylate aminotransferase, like (agxtl) | NM_001002331 | −3.16 | 0.005 | 3.35 |
| A_15_P120962 | ATPase, Na+/K+ transporting, beta 2b polypeptide (atp1b2b) | NM_131838 | −2.07 | 0.005 | 3.36 |
| A_15_P112975 | Transmembrane 4 L six family member 4 (tm4sf4) (zgc:92479) | NM_001003489 | −2.21 | 0.005 | 3.27 |
| A_15_P103143 | Uncharacterized protein LOC327506 (zgc:56085) | NM_199816 | −2.75 | 0.006 | 3.15 |
| A_15_P112053 | Retinal outer segment membrane protein 1b (zgc:56548) | NM_201014 | −2.73 | 0.006 | 3.17 |
| A_15_P120424 | Lysozyme [source: RefSeq_peptide; Acc:NP_631919] | ENSDART00000080549 | −2.04 | 0.006 | 3.15 |
| A_15_P105587 | Prominin 1 b (prom1b) | NM_198071 | −2.28 | 0.006 | 2.98 |
| A_15_P108560 | Internexin neuronal intermediate filament protein, alpha a (inaa) | NM_001144784 | −2.28 | 0.007 | 2.89 |
| A_15_P116896 | Guanine nucleotide binding protein (G protein), beta 5 (gnb5) | NM_200746 | −2.07 | 0.007 | 2.85 |
| A_15_P102378 | Glutamate decarboxylase 1 (gad1) | NM_194419 | −2.50 | 0.007 | 2.80 |
| A_15_P104848 | Tubulin polymerization-promoting protein family member 3 (tppp3) | NM_201335 | −2.35 | 0.007 | 2.80 |
| A_15_P118058 | Unknown | −3.22 | 0.008 | 2.67 | |
| A_15_P103108 | Trypsin-1-like (zgc:66382) NM_199605 | CF595078 | −2.10 | 0.008 | 2.69 |
| A_15_P120202 | SH3-domain GRB2-like 2 (sh3gl2) | NM_201116 | −2.18 | 0.008 | 2.65 |
| A_15_P112659 | Transferrin-a (tfa) | NM_001015057 | −2.17 | 0.009 | 2.45 |
| A_15_P118533 | D. rerio cDNA clone IMAGE: 7395709 | BC092811 | −2.37 | 0.010 | 2.25 |
| A_15_P120388 | Chymotrypsinogen B1 (ctrb1) | NM_212618 | −2.07 | 0.015 | 1.83 |
| A_15_P121355 | Uncharacterized protein LOC393832 (zgc:77748) | NM_200858 | −2.21 | 0.016 | 1.73 |
| A_15_P113063 | Similar to cathepsin L (MGC174155) | NM_001103118 | −2.15 | 0.017 | 1.59 |
| A_15_P114615 | Guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 (ENSDART00000024136) (gngt2a) | XM_001336994 | −2.13 | 0.017 | 1.57 |
| A_15_P103612 | Family with sequence similarity 3, member C (fam3c) | NM_212725 | −2.02 | 0.017 | 1.56 |
| A_15_P118740 | Oxysterol binding protein-like 7 (osbpl7) | NM_001005927 | −2.02 | 0.019 | 1.37 |
| A_15_P101585 | Heat shock protein, alpha-crystallin-related, b11 (hspb11) | NM_001099427 | −2.28 | 0.023 | 1.10 |
| A_15_P110869 | Urate oxidase (uox) | NM_001002332 | −2.19 | 0.026 | 0.87 |
| A_15_P110951 | PREDICTED: D. rerio similar to myosin heavy chain fast skeletal type 2 (LOC100002040) | XM_001339170 | −2.14 | 0.028 | 0.73 |
| A_15_P121427 | Amylase, alpha 2A; pancreatic (amy2a) | NM_213011 | −2.37 | 0.032 | 0.54 |
| A_15_P112046 | Probable protein BRICK1 (zgc:86903) | NP_001002097 | −2.04 | 0.034 | 0.40 |
| A_15_P111425 | Amylase, alpha 2A; pancreatic (amy2a) | NM_213011 | −2.22 | 0.038 | 0.25 |
| A_15_P114488 | Q4SAD3_TETNG (Q4SAD3) chromosome 19 SCAF14691, whole genome shotgun sequence, partial (43%) | TC324157 | −2.00 | 0.039 | 0.17 |
| A_15_P106129 | Ependymin (epd) | NM_131005 | −2.01 | 0.041 | 0.07 |
| A_15_P119722 | Trypsin-1-like (zgc:66382) NM_199605 | CF595078 | −2.23 | 0.048 | −0.15 |
Note. Probe name corresponds to the Agilent probe ID. Fold change values, adjusted p-value and B-statistic are based on the statistical analysis. Only genes that showed ≥2-fold change are shown in the table. The full list of differentially expressed genes is given Supplementary table 2.
TABLE 4. List of Genes Up-regulated in AHRRb-MO-injected Zebrafish Embryos.
| Probe name | Description | Accession number | Fold change | Adjusted p-value | B |
|---|---|---|---|---|---|
| A_15_P114154 | Matrix metalloproteinase 9 (mmp9) | NM_213123 | 3.46 | 0.040 | 1.183 |
| A_15_P103210 | N-myc downstream regulated gene 1, like (ndrg1l) | NM_200692 | 3.08 | 0.017 | 3.093 |
| A_15_P110005 | Histone H1-like (zgc:110380) | NM_001017660 | 2.85 | 0.019 | 2.810 |
| A_15_P110431 | CRCB2_HALSA (Q9HNW1) protein crcB homolog 2, partial (12%) | TC354377 | 2.74 | 0.012 | 5.303 |
| A_15_P120578 | Beaded filament structural protein 2, phakinin (bfsp2) (zgc:103750) | NM_001008633 | 2.30 | 0.027 | 1.883 |
| A_15_P111127 | Danio rerio thioredoxin interacting protein, like (cDNA clone IMAGE:6034364). | BC055213 | 2.28 | 0.016 | 3.342 |
| A_15_P108784 | Protein phosphatase 1, regulatory (inhibitor) subunit 3Ca (ppp1r3ca) (zgc:92069) | NM_001002376 | 2.23 | 0.015 | 3.784 |
| A_15_P109667 | Crystallin, gamma M2c (crygm2c) | NM_001007783 | 2.13 | 0.043 | 0.911 |
| A_15_P113062 | Crystallin, beta A4 (cryba4) | NM_001018125 | 2.10 | 0.032 | 1.656 |
| A_15_P115533 | fw75d04.x1 Gong zebrafish testis D. rerio cDNA clone IMAGE:5616055 3′ sequence | BM571211 | 2.07 | 0.041 | 1.132 |
| A_15_P119407 | Uncharacterized protein LOC561929 (zgc:165347) | NM_001099243 | 2.06 | 0.032 | 1.601 |
TABLE 5. List of Genes Down-regulated in AHRRb-MO-injected Zebrafish Embryos.
| Probe name | Description | Accession number | Fold change | Adjusted p-value | B |
|---|---|---|---|---|---|
| A_15_P113328 | ATP-binding cassette, subfamily F (GCN20), member 2 (abcf2), nuclear gene encoding mitochondrial protein | NM_201315 | −3.54 | 0.015 | 4.422 |
| A_15_P108311 | Chymotrypsinogen B2 precursor (zgc:112160) | NM_001017724 | −2.95 | 0.015 | 4.151 |
| A_15_P120388 | Unknown | −2.86 | 0.012 | 5.176 | |
| A_15_P104868 | Predicted: Danio rerio hypothetical LOC100003999 (LOC100003999) | XM_001343386 | −2.85 | 0.015 | 3.804 |
| A_15_P111425 | Amylase, alpha 2A; pancreatic (amy2a) | NM_213011 | −2.79 | 0.019 | 2.490 |
| A_15_P117308 | Uncharacterized protein LOC692313 precursor (zgc:136461) | NM_001045282 | −2.75 | 0.043 | 0.915 |
| A_15_P121427 | Amylase, alpha 2A; pancreatic (amy2a) | NM_213011 | −2.64 | 0.032 | 1.612 |
| A_15_P110812 | Trypsin-1-like (zgc:66382) NM_199605 | NM_199605 | −2.64 | 0.019 | 2.555 |
| A_15_P100902 | FK506 binding protein 5 (fkbp5) | NM_213149 | −2.54 | 0.004 | 6.970 |
| A_15_P120582 | Purine nucleoside phosphorylase 4b (pnp4b) (zgc:77825) | NM_205643 | −2.43 | 0.045 | 0.818 |
| A_15_P110245 | Unknown | −2.43 | 0.019 | 2.866 | |
| A_15_P110547 | Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (serpina1) | NM_001077758 | −2.38 | 0.019 | 2.665 |
| A_15_P120732 | Q5ERC9_CARAU (Q5ERC9) 14 kDa apolipoprotein, partial (72%) | TC361450 | −2.33 | 0.014 | 4.810 |
| A_15_P106533 | Transferrin-a (tfa) | NM_001015057 | −2.28 | 0.019 | 2.724 |
| A_15_P105980 | Serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 {D. rerio} (exp = −1; wgp = 0; cg = 0), partial (81%) | NM_001013259 | −2.26 | 0.015 | 4.470 |
| A_15_P106963 | Carboxylesterase 2 (intestine, liver) (ces2) (zgc:153863) | NM_001077252 | −2.24 | 0.019 | 2.634 |
| A_15_P110799 | Apolipoprotein A-II (apoa2) zgc:193613 | NM_001130586 | −2.20 | 0.016 | 3.485 |
| A_15_P103108 | Trypsin-1-like (zgc:66382) NM_199605 | CF595078 | −2.20 | 0.017 | 3.198 |
| A_15_P112659 | Transferrin-a (tfa) | NM_001015057 | −2.20 | 0.019 | 2.560 |
| A_15_P116208 | Carboxypeptidase A5 (cpa5) | NM_199271 | −2.19 | 0.022 | 2.201 |
| A_15_P110520 | Growth hormone receptor b (ghrb) | NM_001111081 | −2.18 | 0.043 | 0.998 |
| A_15_P100289 | RRS1 ribosome biogenesis regulator homolog (Saccharomycescerevisiae) (rrs1) | NM_200062 | −2.12 | 0.037 | 1.339 |
| A_15_P113799 | Chymotrypsinogen B1 (ctrb1) | NM_212618 | −2.12 | 0.038 | 1.271 |
| A_15_P107483 | DnaJ (Hsp40) homolog, subfamily C, member 21 (dnajc21) | NM_200044 | −2.11 | 0.015 | 3.981 |
| A_15_P108821 | Eukaryotic translation initiation factor 4A, isoform 1A (eif4a1a) | NM_198366 | −2.10 | 0.015 | 3.864 |
| A_15_P102590 | Heat shock protein 90-alpha 1 (hsp90a.1) | NM_131328 | −2.09 | 0.019 | 2.504 |
| A_15_P117814 | Trypsin (try) | NM_131708 | −2.08 | 0.043 | 1.029 |
| A_15_P100880 | Heat shock protein 47 (hsp47) | NM_131204 | −2.08 | 0.021 | 2.296 |
| A_15_P100921 | Collagen, type X, alpha 1 (col10a1) | NM_001083827 | −2.07 | 0.031 | 1.707 |
| A_15_P118831 | Collagen, type X, alpha 1 (col10a1) | NM_001083827 | −2.06 | 0.016 | 3.451 |
| A_15_P103885 | Methionine adenosyltransferase I, alpha (mat1a) | NM_199871 | −2.06 | 0.019 | 2.820 |
| A_15_P121273 | RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) (rrs1) | NM_200062 | −2.02 | 0.037 | 1.408 |
Note. Probe name corresponds to the Agilent probe ID. Fold change values, adjusted p-value and B-statistic are based on the statistical analysis. Only genes that showed ≥2-fold change are shown in the table. The full list of differentially expressed genes is given in Supplementary table 2.
TABLE 6. List of Genes Differentially Expressed with both AHRRa-MO and AHRRb-MO.
| Agilent probe ID | Gene | AHRRa-MO | AHRRb-MO |
|---|---|---|---|
| A_15_P107501 | Fibrinogen, gamma polypeptide (fgg) | 2.431 | −1.574 |
| A_15_P114154 | Matrix metalloproteinase 9 (mmp9) | 9.588 | 3.458 |
| A_15_P108311 | Chymotrypsinogen B2 precursor (zgc:112160) | −4.052 | −2.946 |
| A_15_P116791 | Calcium/calmodulin-dependent protein kinase IG (camk1g) | −2.262 | −1.800 |
| A_15_P102721 | Chitinase, acidic.2 precursor (zgc:55941) | −2.528 | −1.907 |
| A_15_P117758 | jun B proto-oncogene (junb) | 2.510 | 1.786 |
| A_15_P106305 | Type IV antifreeze protein precursor (zgc:161979) | 2.174 | −1.927 |
| A_15_P117814 | Trypsin (try) | −3.135 | −2.080 |
| A_15_P113799 | Chymotrypsinogen B1 (ctrb1) | −3.050 | −2.124 |
| A_15_P110245 | Unknown | −2.639 | −2.431 |
| A_15_P110812 | Trypsin-1-like (zgc:66382) | −2.981 | −2.636 |
| A_15_P100902 | FK506 binding protein 5 (fkbp5) | −1.892 | −2.542 |
| A_15_P103108 | cDNA clone IMAGE:7015062 | −2.099 | −2.201 |
| A_15_P112659 | Transferrin-a (tfa) | −2.169 | −2.198 |
| A_15_P107950 | si:dkey-42i9.4 (si:dkey-42i9.4) | 1.914 | 1.862 |
| A_15_P108038 | si:dkeyp-113d7.7 (si:dkeyp-113d7.7) | 1.804 | 1.845 |
| A_15_P120388 | Unknown | −2.070 | −2.858 |
| A_15_P120578 | Beaded filament structural protein 2, phakinin (bfsp2) (zgc:103750) | 2.246 | 2.300 |
| A_15_P109667 | Crystallin, gamma M2c (crygm2c) | 2.177 | 2.131 |
| A_15_P103210 | N-myc downstream regulated gene 1, like (ndrg1l) | 2.427 | 3.080 |
| A_15_P110837 | Transmembrane protein 41ab (tmem41ab) | −1.832 | −1.844 |
| A_15_P116208 | Carboxypeptidase A5 (cpa5) | −1.945 | −2.186 |
| A_15_P103675 | Cathepsin L1, a (ctsl1a) | 1.848 | −1.981 |
| A_15_P111425 | Amylase, alpha 2A; pancreatic (amy2a) | −2.216 | −2.793 |
| A_15_P109854 | IMAGE:7278570 5′ | 1.780 | 1.841 |
| A_15_P117166 | Nucleolar complex associated 3 homolog (Saccharomyces cerevisiae) (noc3l) | −1.649 | −1.842 |
| A_15_P110005 | Histone H1-like (zgc:110380) | 2.150 | 2.847 |
| A_15_P116605 | Suppressor of cytokine signaling 3b (socs3b) | 1.706 | 1.967 |
| A_15_P119407 | Uncharacterized protein LOC561929 (zgc:165347) | 1.807 | 2.056 |
| A_15_P121427 | Amylase, alpha 2A; pancreatic (amy2a) | −2.366 | −2.644 |
Note. Fold change values for both the AHRRa and AHRRb MOs are shown. Rows in bold show the genes with opposite responses to AHRRa and AHRRb knockdown.
Functional Annotation of Differentially Expressed Genes
Functional annotation of genes differentially expressed in AHRRa morphants revealed enrichment of genes related to photoreceptor function. Based on GO term analysis (biological process), the majority of the genes are grouped under GO term neurological system process (GO:0050877) and response to stimulus (GO:0009416). The list of genes grouped under each GO term, their enrichment scores, and adjusted p-values are shown in Table 7. Biological network analysis also revealed a network of 17 genes related to cone photoreceptor signaling (Fig. 2). The entire network of all the differentially expressed genes is provided in the supplementary information (Supplementary fig. 1). Similar functional annotation and network analysis for the AHRRb-MO group did not reveal enrichment of genes for any specific biological process.
TABLE 7. Gene Ontology Enrichment Analysis of Genes Differentially Expressed in AHRRa-MO-treated Embryos.
| Gene Ontology term (biological process) | Benjamini Hochberg adjusted p-value |
|---|---|
| NEUROLOGICAL SYSTEM PROCESS (GO:0050877) | 2.1E−4 (Enrichment score: 5.67) |
| ATPase, Na+/K+ transporting, beta 2b polypeptide | |
| Arrestin 3, retinal (X-arrestin) | |
| Arrestin 3, retinal (X-arrestin), like | |
| Glutamate decarboxylase 1 | |
| Guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 | |
| Opsin 1 (cone pigments), long-wave-sensitive, 2 | |
| Opsin 1 (cone pigments), medium-wave-sensitive, 1 | |
| Opsin 1 (cone pigments), short-wave-sensitive 1 | |
| Opsin 1 (cone pigments), short-wave-sensitive 2 | |
| Phosphodiesterase 6C, cGMP-specific, cone, alpha prime | |
| zgc:56548 | |
| zgc:73336 phosphodiesterase 6H, cGMP-specific, cone | |
| RESPONSE TO LIGHT STIMULUS (GO:0009416) | 6.7E−3 (Enrichment score: 3.83) |
| Guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 | |
| Opsin 1 (cone pigments), long-wave-sensitive, 2 | |
| Opsin 1 (cone pigments), medium-wave-sensitive, 1 | |
| Opsin 1 (cone pigments), short-wave-sensitive 1 | |
| Opsin 1 (cone pigments), short-wave-sensitive 2 | |
| Retinaldehyde binding protein 1b | |
| Solute carrier family 31 (copper transporters), member 1 | |
| Tumor protein p53 |
FIG. 2.
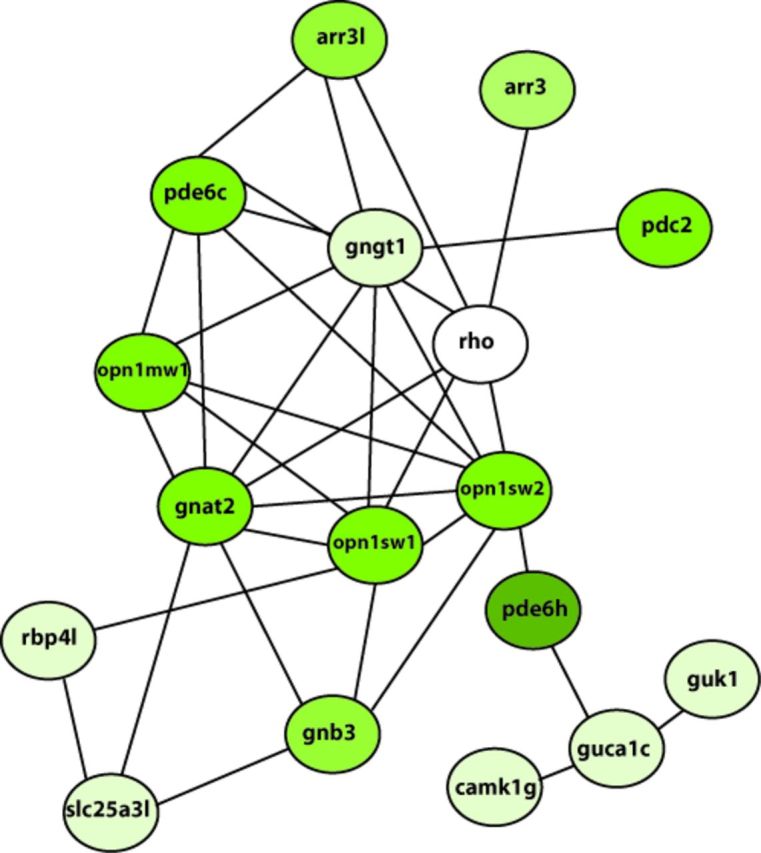
Biological network of genes involved in cone photoreceptor signaling. All the differentially expressed genes from the AHRRa-MO group were analyzed for functional association using the STRING database and statistically significant networks were identified using jActiveModules version 2.23. Only the network of genes involved in cone photoreceptor signaling is shown. The color of each node represents the fold change in gene expression. Different shades of green (light green to dark green) denote the magnitude of down-regulation. Unchanged nodes are colored white.
Confirmation of Microarray Results
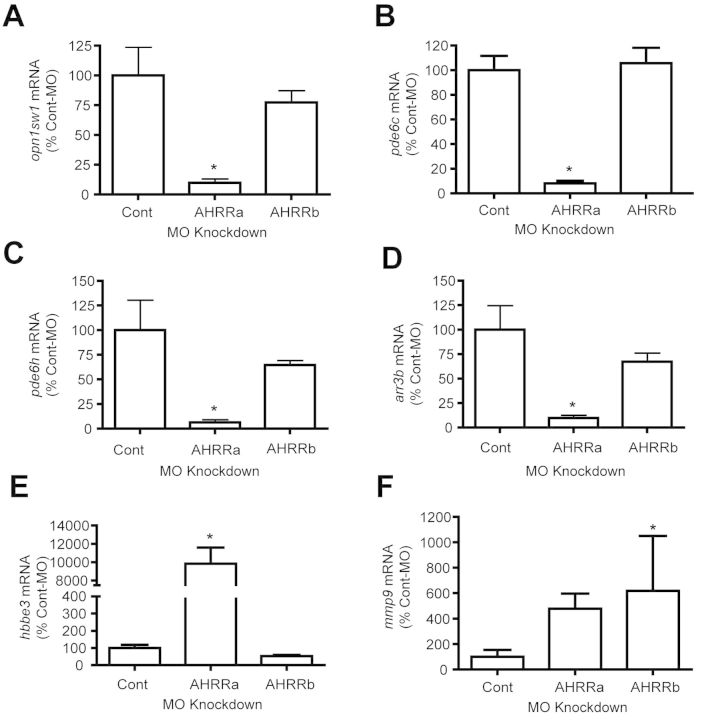
We selected six genes (opn1sw1, pde6c, pde6h, arr3b, hbbe3, and mmp9) that were differentially expressed in AHRRa morphants for confirmation by qRT-PCR (Fig. 3). All the genes except mmp9 showed a change similar to that observed with microarrays. Opn1sw1, pde6c, pde6h, and arr3b showed significant down-regulation after AHRRa knockdown compared with the control-MO and AHRRb-MO groups (Figs. 3A–D). Hbbe3 showed significant up-regulation with AHRRa-MO treatment (Fig. 3E). Mmp9 showed an increasing trend with AHRRa-MO treatment, but it was not significantly different compared with the control group (Fig. 3F). However, mmp9 was significantly up-regulated with AHRRb-MO treatment, as observed with microarrays (Fig. 3F).
FIG. 3.
qRT-PCR confirmation of microarray results. Six genes that were significantly altered by AHRR knockdown as suggested by microarray analysis were selected for qRT-PCR. β-Actin was used as an internal standard. The ΔΔCt method was used to determine the fold change in gene expression. RNA was from larvae sampled at 72 hpf. Values represent mean + SD (one-way ANOVA; n = 3), *p ≤ 0.05.
Effect of TCDD on Gene Expression Patterns in AHRR Knockdown Embryos
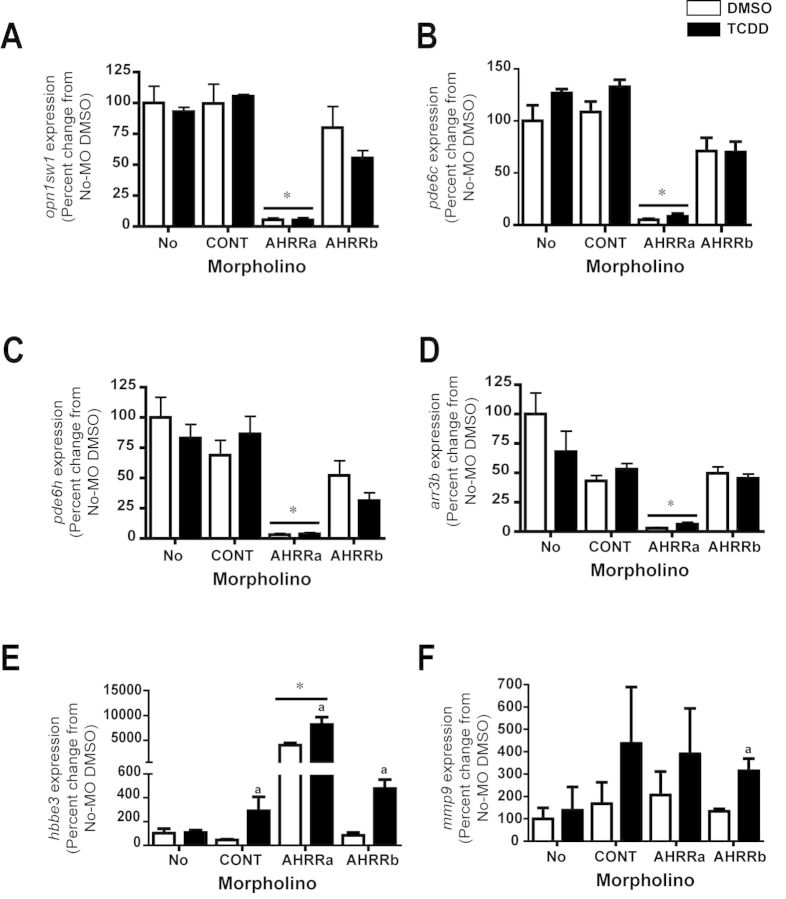
To further examine the effect of AHRR knockdown and to determine whether genes responsive to loss of AHRR are also inducible by the AHR ligand TCDD, we performed qRT-PCR on an independent set of samples from an earlier study that included combinations of AHRR knockdown and TCDD treatment (Jenny et al., 2009). Knockdown of AHRRa caused significant down-regulation of opn1sw1, pde6c, pde6h, and arr3 genes, as observed in the experiment described in the previous section. However, there was no effect of TCDD treatment on the expression of these four genes as compared with vehicle controls, in any of the groups (Figs. 4A–D). The up-regulation of hbbe3 in AHRRa-morphants but not AHRRb-morphants occurred as before. Interestingly, TCDD exposure strongly induced the expression of hbbe3 in control-MO, AHRRa-MO-injected, and AHRRb-MO-injected embryos in comparison to MO-injected, DMSO-exposed embryos (Fig. 4E). In a separate, replicate experiment that also involved injection of AHRR-MOs and exposure to TCDD, the TCDD induction of hbbe3 in control-MO-injected embryos did not occur, but the induction of hbbe3 by TCDD in AHRRa-MO-injected and AHRRb-MO-injected embryos was confirmed (Supplementary fig. 2). TCDD also induced mmp9 gene expression, but only in AHRRb-MO injected embryos (Fig. 4F) and these results were confirmed in the replicate experiment (Supplementary fig. 2).
FIG. 4.
Effect of TCDD on AHRR-MO induced gene expression changes. Injection of AHRR MOs and exposure to TCDD was carried out as described in Materials and Methods section. RNA was from larvae sampled at 72 hpf. β-Actin was used as an internal standard. The ΔΔCt method was used to determine the fold change in gene expression. Values represent mean + SD (two-way ANOVA; n = 3). Bonferroni's post hoc test was used for determining statistical significance. ap ≤ 0.05 TCDD versus DMSO; *p ≤ 0.05 AHRRa-MO group versus no-MO treatment. CYP1 expression data from these experiments was reported previously (Jenny et al., 2009).
DISCUSSION
We have previously demonstrated that the knockdown of AHRRa, but not AHRRb, induces developmental phenotypes similar to those seen with TCDD exposure (Jenny et al., 2009). In order to understand the molecular basis of these phenotypes, transcriptional profiling was carried out using microarrays after knocking down each of the two AHRRs. Comparing the two paralogs, knockdown of AHRRa down-regulated a larger set of genes than did knockdown of AHRRb and there was very little overlap between the two groups, suggesting that they play distinct physiological roles during development.
AHRRa Regulates Genes Involved in Photoreceptor Signaling
Functional annotation of all the differentially expressed genes, especially the large number of down-regulated genes involved in cone photoreceptor signaling, suggests a unique role for AHRRa in eye development. In vertebrates, cone photoreceptors are primarily responsible for color vision and responses to bright light. The cone photoreceptor-specific genes that were down-regulated in AHRRa morphants include those encoding short wave opsins (opn1sw1 and opn1sw2), medium wave opsins (opn1mw1), long wave sensitive opsins (opn1lw2), phosphodiesterase subunits (pde6c and pde6h), cone arrestin (arr3), cone-specific G-protein coupled receptor kinase (grk7a) and transducin α-subunit (gnat2). The down-regulation of several of these (opn1sw1, pde6c, pde6h, and arr3b) was confirmed by qRT-PCR. These genes are critical for vision in zebrafish (Leung and Dowling, 2005; Weger et al., 2011). Opsin is the photopigment found in cone photoreceptors, and based on the sensitivity to different wavelengths of light these receptors can be classified as red (∼565 nm), green (∼520 nm), blue (∼430 nm), and ultraviolet (UV, ∼365 nm). In the zebrafish genome, there are two red (opn1lw1 and opn1lw2), four green (opn1mw1–4), and single blue (opn1sw2) and ultraviolet (opn1sw1) opsin genes (Robinson et al., 1993; Vihtelic et al., 1999). Opsins are membrane-bound G-protein-coupled receptors (GPCR). Photoreceptor signaling begins with the absorption of a photon of light by a GPCR together with 11-cis-retinal, a chromophore. This activates the regulatory G-protein, transducin, and downstream second messenger cGMP by activating cGMP phosphodiesterases (PDE) (Collery and Kennedy, 2009). PDE breaks down cGMP to 5′-GMP, lowering intracellular cGMP concentration and subsequently hyperpolarizing the plasma membrane by closing of the cGMP-gated cationic channels. PDE6 is specific to photoreceptor signaling and pde6c and pde6h are the subunits of PDE specific to cone photoreceptors. Mutations in pde6c cause degeneration of cones in zebrafish (Stearns et al., 2007) and also are responsible for the inherited degenerative disease retinitis pigmentosa in humans (Fadool and Dowling, 2008; Gestri et al., 2012).
Recovery from the photoreceptor response involves phosphorylation of opsin by rhodopsin kinase (grk7a), which prepares the receptor for binding to arrestin (arr3). This prevents transducin-mediated signaling and targets the GPCR for internalization. Recoverin (rcv1), a calcium binding protein, regulates the rate of phosphorylation. Transcript levels of grk7a, arr3, and rcv1 were down-regulated in AHRRa-morphant embryos. Knockdown of grk7a delays the recovery of cone cells from the photoreceptor response in zebrafish (Rinner et al., 2005). In addition to these cone photoreceptor-specific genes, several other eye-related genes were down-regulated with AHRRa-knockdown (Supplementary table 3).
These results suggest that AHRRa has a specific role in cone photoreceptor development. However, it is unclear if all the genes are direct targets of AHRRa or indirectly targeted by yet to be identified mechanisms. Based on the time at which MOs were injected into the embryos (1–2 cell stages) and the time at which the gene expression profiling was done (72 hpf), it is likely that at least some of these changes, and perhaps many, are secondary effects. Measuring the effect of AHRRa knockdown at additional time points during development will help to clarify this. The inability of TCDD to alter the expression of opn1sw1, pde6c, pde6h, or arr3b (Figs. 4A–D) suggests that at least these four genes are not directly regulated by AHR.
One possible explanation for the down-regulation of so many eye-related genes is that AHRRa knockdown impacts early cellular fate specification of progenitor cells by interfering with an essential signaling pathway. These changes could lead to differences in terminal differentiation and altered gene expression patterns in the photoreceptors. In the fruit fly, Drosophila melanogaster, loss of the AHR ortholog spineless disrupts color vision by altering the transcriptional regulation that distinguishes different classes of photoreceptors (Wernet et al., 2006). Even though invertebrate and vertebrate eyes differ morphologically, recent evidence suggests that underlying molecular mechanisms in photoreceptor development are highly conserved (Charlton-Perkins and Cook, 2010). Although there is no direct evidence for a role of AHRR in retinal function in zebrafish, it is possible that AHRRa is expressed in the developing eye, where it could regulate AHR, HIF, or other transcription factors. AHR expression in mammalian eye has been reported (Chevallier et al., 2013; Dwyer et al., 2011; Liu and Piatigorsky, 2011) and we have recently detected expression of AHRRa mRNA in the eyes of adult zebrafish (unpublished results). Future studies will focus on identifying the expression patterns of AHRR and its possible targets in the developing eye.
The toxicological implications of the altered gene expression patterns that we observed in AHRRa-morphant embryos are not clear, but a possible role of AHRRa in the development of the eye suggests a mechanism that could help explain the reported effects of TCDD on visual function in fish (Carvalho and Tillitt, 2004) or ocular defects that appear in AHR-deficient mice (Chevallier et al., 2013; Hu et al., 2013). Expression of AHRR is regulated by AHR and induced by AHR agonists in mammals and fish (Karchner et al., 2002; Mimura et al., 1999). Moreover, AHRR regulates the responses to AHR agonists in adults and embryos (Hosoya et al., 2008; Jenny et al., 2009; Korkalainen et al., 2004, 2005). Thus, induction of AHRR expression by AHR agonists during development or in adult animals could interfere with physiological roles of AHR or other transcription factors involved in the differentiation or function of cells in the eye. In light of our results, a detailed examination of eye development and adult visual function in TCDD-treated animals is warranted.
AHRR Knockdown Induces Stress Signaling
In contrast to the down-regulation of critical players in phototransduction, knockdown of both AHRRa and AHRRb up-regulated two genes that act as chaperone proteins in the eye. Crystallin (crygm2c), a small heat shock protein found exclusively in the lens and cornea of the eye, was up-regulated in AHRRa and AHRRb morphants. Beaded filament structural protein 2 (bfsp2), a lens-specific intermediary filament, was also up-regulated in the morphants. Intermediary filaments act as a scaffold for chaperone proteins and stress-activated kinases (Song et al., 2009). These results suggest that knockdown of AHRRa and AHRRb activated stress signaling in the eye. This is perhaps not surprising considering that AHRR knockdown affected several genes involved in photoreceptor signaling and the induction of chaperone proteins may help to repair any misfolded or damaged proteins. The regulation of crystallin by AHRRa could be associated with AHR signaling, as suggested by the recent finding that AHR regulates crystallin (αb) in mice under normal and TCDD-induced states (Liu and Piatigorsky, 2011).
Role of AHRRa in Erythropoiesis and Hemostasis
We also observed that AHRRa morphants exhibited up-regulation of genes associated with erythropoiesis and hemostasis. The up-regulated genes included embryonic globins involved in oxygen transport (embryonic hemoglobin beta 3 subunit (hbbe3) and alpha subunit (hbae1)) and three genes encoding fibrinogen polypeptides (fga, fgb, and fgg), key players in blood coagulation.
Globins are an important group of proteins involved in oxygen transport. Most vertebrates possess multiple globin genes that vary in their affinity for oxygen and are differentially expressed during development (Brownlie et al., 2003). In zebrafish, hematopoiesis occurs in two distinct waves. The first wave (primitive hematopoiesis) occurs in the first 12–48 hpf, during which progenitor hematopoietic cells with embryonic globin proteins are involved in oxygen transport (Ellett and Lieschke, 2010). In the second wave (definitive erythropoiesis), pluripotent hematopoietic stem cells are formed and adult globin proteins are involved in oxygen transport (Ellett and Lieschke, 2010). This phase extends from 4 to 5 dpf to the adult life.
It remains to be determined whether AHRRa knockdown directly or indirectly derepresses embryonic hemoglobins. We examined that possibility that globins may be AHR-regulated genes by measuring the expression of hbbe3 after exposure of embryos to TCDD. The results provided evidence for induction of this gene by TCDD (Fig. 4E; Supplementary fig. 2), consistent with the possibility that AHRRa may negatively regulate AHR-dependent hbbe3 expression during development. Several transcription factors are involved in the regulation of primitive and definitive phases of erythropoiesis (Paik and Zon, 2010). In light of the emerging understanding of an important role for AHR in hematopoiesis (Boitano et al., 2010; Casado et al., 2010; Lindsey and Papoutsakis, 2012; Singh et al., 2011; Smith et al., 2013), it is conceivable that AHRRa could participate in regulatory interactions involving this function of AHR. One possibility is that knockdown of AHRRa leads to an increase in endogenous AHR activity, disrupting the switch from primitive to definitive erythropoiesis and thereby delaying the appearance of erythrocytes expressing adult globins. Such a disruption of definitive erythropoiesis has been observed in zebrafish embryos in which AHR has been activated exogenously, by exposure to the AHR agonist TCDD (Belair et al., 2001). Similarly, chronic TCDD exposure caused down-regulation of adult hemoglobins in juvenile rainbow trout (Liu et al., 2013). Consistent with the idea that knockdown of AHRRa may lead to enhanced endogenous activity of AHR, the phenotypes observed with AHRRa knockdown are similar to those observed following TCDD treatment (Jenny et al., 2009). Alternatively, or in addition, loss of AHRRa could affect hematopoiesis via altered function of HIF-1α or ARNT (Adelman et al., 1999; Karchner et al., 2009).
Similar to what was seen for embryonic globins, AHRRa knockdown up-regulated fibrinogen polypeptide subunit mRNAs (fga, fgb, and fgg). Compared with hematopoiesis, very little is known about developmental regulation of hemostasis in zebrafish. Fibrinogen is a hexamer consisting of two sets of three polypeptide subunits (alpha, beta, and gamma). Thrombin converts fibrinogen to fibrin during blood coagulation (Fish et al., 2012). High levels of fibrinogen are associated with cardiovascular disease involving the accumulation of fibrinogen, which initiates platelet aggregation and atherogenesis (Tousoulis et al., 2011). Whether the increase in fibrinogen expression causes defects in blood clotting in AHRRa morphant embryos is not yet known. Such an effect could also be associated with increased AHR signaling, as exposure of medaka embryos to TCDD leads to an apparent increase in the formation of blood clots (Kawamura and Yamashita, 2002).
AHRRa May Act as a Tumor Suppressor Gene
Some of our results appear related to recent observations that AHRR acts as a tumor suppressor gene (Zudaire et al., 2008). Knockdown of AHRRa up-regulated two genes (tp53 and ccng1) involved in cell cycle regulation. Tumor protein p53 (tp53) is an important regulator of cell cycle progression and cyclin G1 (ccng1) is involved in the regulation of transition from G1 to S-stage of the cell cycle (Zhao et al., 2003). Previous studies have demonstrated that AHRR acts as a tumor suppressor in several cancer cell lines (Zudaire et al., 2008). Stable transfection of AHRR into ER alpha-positive MCF-7 cells decreased the expression of E2F and cyclin genes important for tumor cell growth when induced with DMBA. In the same study, siRNA-induced down-regulation of AHRR increased the growth of lung carcinoma cell lines and caused them to become more invasive (Kanno et al., 2006). In addition, the region of the chromosome containing the AHRR gene is deleted in several human cancer tissues, suggesting that AHRR plays an important role in tumor suppression by repressing the AHR activity in tumors (Zudaire et al., 2008). Additional studies examining the roles of AHRR in regulation of cell growth are warranted.
One of the off-target effects of MO administration is the activation of tp53 expression (Robu et al., 2007). However, off-target effects involve up-regulation of protein expression without concomitant increase in the mRNA expression (Robu et al., 2007). Our results involving increased tp53 RNA are thus distinct from those associated with off-target effects. In addition, we only observed the induction of tp53 with AHRRa-MO injected embryos and not with AHRRb-MO- and Control-MO-injected embryos, suggesting that the response is specific to loss of AHRRa.
Role of AHR Activation
It is tempting to conclude that the effects occurring upon knockdown of AHRRs result from derepression of AHR signaling. However, because AHRR is known to affect other signaling pathways, such as estrogen and hypoxia signaling (Kanno et al., 2006, 2008; Karchner et al., 2009), it is conceivable that the effects of AHRR knockdown reported here could involve these or other signaling pathways. A limitation of the present study is that MO-based knockdown does not completely eliminate expression of the targeted gene, so there could be functions that are maintained even at the reduced levels of protein expression. Experiments to generate targeted null-mutants of AHRRs are in progress and should provide further insight into AHRR functions and the role of AHR versus other signaling pathways.
CONCLUSIONS
Our results lend further support to our previous suggestion (Jenny et al., 2009) that duplicated AHRR genes (AHRRa and AHRRb) have distinct physiological roles during development in zebrafish. We have identified several genes related to photoreceptor signaling, erythropoiesis, and hemostasis to be differentially expressed in AHRRa knockdown embryos. These results suggest that AHRRa has a role in normal embryonic development. In contrast, AHRRb knockdown affected fewer genes and there was little overlap in the genes differentially expressed in the two AHRR morphants, suggesting that they regulate different processes during development. The results here further demonstrate the value of the zebrafish model and its duplicated AHRRs for obtaining insight into the various functions of AHRR.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Environmental Health Sciences of the U.S. National Institutes of Health (R01ES006272 to M.E.H., R00ES017044 to M.J.J.).
Supplementary Material
Acknowledgments
The authors thank Dr Sibel Karchner for advice on data analysis and presentation, and for reviewing the manuscript prior to submission. We also thank two anonymous reviewers for helpful suggestions that improved the paper.
Disclaimer: The U.S. Government is authorized to produce and distribute reprints for governmental purposes notwithstanding any copyright notation that may appear hereon. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- Adelman D. M., Maltepe E., Simon M. C. Multilineage embryonic hematopoiesis requires hypoxic ARNT activity. Genes Dev. 1999;13:2478–2483. doi: 10.1101/gad.13.19.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belair C. D., Peterson R. E., Heideman W. Disruption of erythropoiesis by dioxin in the zebrafish. Dev. Dyn. 2001;222:581–594. doi: 10.1002/dvdy.1213. [DOI] [PubMed] [Google Scholar]
- Bernshausen T., Jux B., Esser C., Abel J., Fritsche E. Tissue distribution and function of the Aryl hydrocarbon receptor repressor (AhRR) in C57BL/6 and Aryl hydrocarbon receptor deficient mice. Arch. Toxicol. 2006;80:206–211. doi: 10.1007/s00204-005-0025-5. [DOI] [PubMed] [Google Scholar]
- Boitano A. E., Wang J., Romeo R., Bouchez L. C., Parker A. E., Sutton S. E., Walker J. R., Flaveny C. A., Perdew G. H., Denison M. S., et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329:1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie A., Hersey C., Oates A. C., Paw B. H., Falick A. M., Witkowska H. E., Flint J., Higgs D., Jessen J., Bahary N., et al. Characterization of embryonic globin genes of the zebrafish. Dev. Biol. 2003;255:48–61. doi: 10.1016/s0012-1606(02)00041-6. [DOI] [PubMed] [Google Scholar]
- Carvalho P. S., Tillitt D. E. 2,3,7,8-TCDD effects on visual structure and function in swim-up rainbow trout. Environ. Sci. Technol. 2004;38:6300–6306. doi: 10.1021/es034857i. [DOI] [PubMed] [Google Scholar]
- Casado F. L., Singh K. P., Gasiewicz T. A. The aryl hydrocarbon receptor: Regulation of hematopoiesis and involvement in the progression of blood diseases. Blood Cells Mol. Dis. 2010;44:199–206. doi: 10.1016/j.bcmd.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton-Perkins M., Cook T. A. Building a fly eye: Terminal differentiation events of the retina, corneal lens, and pigmented epithelia. Curr. Top. Dev. Biol. 2010;93:129–173. doi: 10.1016/B978-0-12-385044-7.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevallier A., Mialot A., Petit J. M., Fernandez-Salguero P., Barouki R., Coumoul X., Beraneck M. Oculomotor deficits in aryl hydrocarbon receptor null mouse. PLoS One. 2013;8:e53520. doi: 10.1371/journal.pone.0053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collery R. F., Kennedy B. N. Photoreceptor guanylate cyclases and cGMP phosphodiesterases in zebrafish. In: E. Anderson. R., editor. Retinal Degenerative Diseases, Advances in Experimental Medicine and Biology. Vol. 664. New York: Springer; 2009. [DOI] [PubMed] [Google Scholar]
- Dwyer M. A., Kazmin D., Hu P., McDonnell D. P., Malek G. Research resource: Nuclear receptor atlas of human retinal pigment epithelial cells: Potential relevance to age-related macular degeneration. Mol. Endocrinol. 2011;25:360–372. doi: 10.1210/me.2010-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellett F., Lieschke G. J. Zebrafish as a model for vertebrate hematopoiesis. Curr. Opin. Pharmacol. 2010;10:563–570. doi: 10.1016/j.coph.2010.05.004. [DOI] [PubMed] [Google Scholar]
- Evans B. R., Karchner S. I., Allan L. L., Pollenz R. S., Tanguay R. L., Jenny M. J., Sherr D. H., Hahn M. E. Repression of aryl hydrocarbon receptor (AHR) signaling by AHR repressor: Role of DNA binding and competition for AHR nuclear translocator. Mol. Pharmacol. 2008;73:387–398. doi: 10.1124/mol.107.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans B. R., Karchner S. I., Franks D. G., Hahn M. E. Duplicate aryl hydrocarbon receptor repressor genes (ahrr1 and ahrr2) in the zebrafish Danio rerio: Structure, function, evolution, and AHR-dependent regulation in vivo. Arch. Biochem. Biophys. 2005;441:151–167. doi: 10.1016/j.abb.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Fadool J. M., Dowling J. E. Zebrafish: A model system for the study of eye genetics. Prog. Retin. Eye Res. 2008;27:89–110. doi: 10.1016/j.preteyeres.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish R. J., Vorjohann S., Bena F., Fort A., Neerman-Arbez M. Developmental expression and organisation of fibrinogen genes in the zebrafish. Thromb. Haemost. 2012;107:158–166. doi: 10.1160/TH11-04-0221. [DOI] [PubMed] [Google Scholar]
- Gestri G., Link B. A., Neuhauss S. C. The visual system of zebrafish and its use to model human ocular diseases. Dev. Neurobiol. 2012;72:302–327. doi: 10.1002/dneu.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarmann-Stemmann T., Abel J. The arylhydrocarbon receptor repressor (AhRR): Structure, expression, and function. Biol. Chem. 2006;387:1195–1199. doi: 10.1515/BC.2006.147. [DOI] [PubMed] [Google Scholar]
- Haarmann-Stemmann T., Bothe H., Kohli A., Sydlik U., Abel J., Fritsche E. Analysis of the transcriptional regulation and molecular function of the aryl hydrocarbon receptor repressor in human cell lines. Drug Metab. Dispos. 2007;35:2262–2269. doi: 10.1124/dmd.107.016253. [DOI] [PubMed] [Google Scholar]
- Hahn M. E., Allan L. L., Sherr D. H. Regulation of constitutive and inducible AHR signaling: Complex interactions involving the AHR repressor. Biochem. Pharmacol. 2009;77:485–497. doi: 10.1016/j.bcp.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T., Harada N., Mimura J., Motohashi H., Takahashi S., Nakajima O., Morita M., Kawauchi S., Yamamoto M., Fujii-Kuriyama Y. Inducibility of cytochrome P450 1A1 and chemical carcinogenesis by benzo[a]pyrene in AhR repressor-deficient mice. Biochem. Biophys. Res. Commun. 2008;365:562–567. doi: 10.1016/j.bbrc.2007.11.016. [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hu P., Herrmann R., Bednar A., Saloupis P., Dwyer M. A., Yang P., Qi X., Thomas R. S., Jaffe G. J., Boulton M. E., et al. Aryl hydrocarbon receptor deficiency causes dysregulated cellular matrix metabolism and age-related macular degeneration-like pathology. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E4069–E4078. doi: 10.1073/pnas.1307574110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny M. J., Karchner S. I., Franks D. G., Woodin B. R., Stegeman J. J., Hahn M. E. Distinct roles of two zebrafish AHR repressors (AHRRa and AHRRb) in embryonic Development and regulating the response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 2009;110:426–441. doi: 10.1093/toxsci/kfp116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson M. E., Orrego R., Woodin B. R., Goldstone J. V., Stegeman J. J. Basal and 3,3′,4,4’,5-pentachlorobiphenyl-induced expression of cytochrome P450 1A, 1B and 1C genes in zebrafish. Toxicol. Appl. Pharmacol. 2007;221:29–41. doi: 10.1016/j.taap.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y., Takane Y., Izawa T., Nakahama T., Inouye Y. The inhibitory effect of aryl hydrocarbon receptor repressor (AhRR) on the growth of human breast cancer MCF-7 cells. Biol. Pharm. Bull. 2006;29:1254–1257. doi: 10.1248/bpb.29.1254. [DOI] [PubMed] [Google Scholar]
- Kanno Y., Takane Y., Takizawa Y., Inouye Y. Suppressive effect of aryl hydrocarbon receptor repressor on transcriptional activity of estrogen receptor alpha by protein-protein interaction in stably and transiently expressing cell lines. Mol. Cell. Endocrinol. 2008;291:87–94. doi: 10.1016/j.mce.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Karchner S. I., Franks D. G., Powell W. H., Hahn M. E. Regulatory interactions among three members of the vertebrate aryl hydrocarbon receptor family: AHR repressor, AHR1, and AHR2. J. Biol Chem. 2002;277:6949–6959. doi: 10.1074/jbc.M110779200. [DOI] [PubMed] [Google Scholar]
- Karchner S. I., Jenny M. J., Tarrant A. M., Evans B. R., Kang H. J., Bae I., Sherr D. H., Hahn M. E. The active form of human aryl hydrocarbon receptor (AHR) repressor lacks exon 8, and its Pro 185 and Ala 185 variants repress both AHR and hypoxia-inducible factor. Mol. Cell. Biol. 2009;29:3465–3477. doi: 10.1128/MCB.00206-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T., Yamashita I. Aryl hydrocarbon receptor is required for prevention of blood clotting and for the development of vasculature and bone in the embryos of medaka fish, Oryzias latipes. Zoolog. Sci. 2002;19:309–319. doi: 10.2108/zsj.19.309. [DOI] [PubMed] [Google Scholar]
- Korkalainen M., Linden J., Tuomisto J., Pohjanvirta R. Effect of TCDD on mRNA expression of genes encoding bHLH/PAS proteins in rat hypothalamus. Toxicology. 2005;208:1–11. doi: 10.1016/j.tox.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Korkalainen M., Tuomisto J., Pohjanvirta R. Primary structure and inducibility by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) of aryl hydrocarbon receptor repressor in a TCDD-sensitive and a TCDD-resistant rat strain. Biochem. Biophys. Res. Commun. 2004;315:123–131. doi: 10.1016/j.bbrc.2004.01.028. [DOI] [PubMed] [Google Scholar]
- Leung Y. F., Dowling J. E. Gene expression profiling of zebrafish embryonic retina. Zebrafish. 2005;2:269–283. doi: 10.1089/zeb.2005.2.269. [DOI] [PubMed] [Google Scholar]
- Lindsey S., Papoutsakis E. T. The evolving role of the aryl hydrocarbon receptor (AHR) in the normophysiology of hematopoiesis. Stem Cell Rev. 2012;8:1223–1235. doi: 10.1007/s12015-012-9384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. A., Piatigorsky J. Regulation of mouse small heat shock protein alpha b-crystallin gene by aryl hydrocarbon receptor. PLoS One. 2011;6:e17904. doi: 10.1371/journal.pone.0017904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Rise M. L., Spitsbergen J. M., Hori T. S., Mieritz M., Geis S., McGraw J. E., Goetz G., Larson J., Hutz R. J., et al. Gene expression and pathologic alterations in juvenile rainbow trout due to chronic dietary TCDD exposure. Aquat. Toxicol. 2013;140–141:356–368. doi: 10.1016/j.aquatox.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mimura J., Ema M., Sogawa K., Fujii-Kuriyama Y. Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes Dev. 1999;13:20–25. doi: 10.1101/gad.13.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihashi H., Kanno Y., Tomuro K., Nakahama T., Inouye Y. Primary structure and organ-specific expression of the rat aryl hydrocarbon receptor repressor gene. Biol. Pharm. Bull. 2006;29:640–647. doi: 10.1248/bpb.29.640. [DOI] [PubMed] [Google Scholar]
- Oshima M., Mimura J., Sekine H., Okawa H., Fujii-Kuriyama Y. SUMO modification regulates the transcriptional repressor function of aryl hydrocarbon receptor repressor. J. Biol. Chem. 2009;284:11017–11026. doi: 10.1074/jbc.M808694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik E. J., Zon L. I. Hematopoietic development in the zebrafish. Int. J. Dev. Biol. 2010;54:1127–1137. doi: 10.1387/ijdb.093042ep. [DOI] [PubMed] [Google Scholar]
- Rinner O., Makhankov Y. V., Biehlmaier O., Neuhauss S. C. Knockdown of cone-specific kinase GRK7 in larval zebrafish leads to impaired cone response recovery and delayed dark adaptation. Neuron. 2005;47:231–242. doi: 10.1016/j.neuron.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Robinson J., Schmitt E. A., Harosi F. I., Reece R. J., Dowling J. E. Zebrafish ultraviolet visual pigment: Absorption spectrum, sequence, and localization. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6009–6012. doi: 10.1073/pnas.90.13.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu M. E., Larson J. D., Nasevicius A., Beiraghi S., Brenner C., Farber S. A., Ekker S. C. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K. P., Garrett R. W., Casado F. L., Gasiewicz T. A. Aryl hydrocarbon receptor-null allele mice have hematopoietic stem/progenitor cells with abnormal characteristics and functions. Stem Cells Dev. 2011;20:769–784. doi: 10.1089/scd.2010.0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B. W., Rozelle S. S., Leung A., Ubellacker J., Parks A., Nah S. K., French D., Gadue P., Monti S., Chui D. H., et al. The aryl hydrocarbon receptor directs hematopoietic progenitor cell expansion and differentiation. Blood. 2013;122:376–385. doi: 10.1182/blood-2012-11-466722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K. Limma: Linear Models for Microarray Data. New York: Springer; 2005. [Google Scholar]
- Song S., Landsbury A., Dahm R., Liu Y., Zhang Q., Quinlan R. A. Functions of the intermediate filament cytoskeleton in the eye lens. J. Clin. Invest. 2009;119:1837–1848. doi: 10.1172/JCI38277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns G., Evangelista M., Fadool J. M., Brockerhoff S. E. A mutation in the cone-specific pde6 gene causes rapid cone photoreceptor degeneration in zebrafish. J. Neurosci. 2007;27:13866–13874. doi: 10.1523/JNEUROSCI.3136-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackaberry E. A., Jiang Z., Johnson C. D., Ramos K. S., Walker M. K. Toxicogenomic profile of 2,3,7,8-tetrachlorodibenzo-p-dioxin in the murine fetal heart: Modulation of cell cycle and extracellular matrix genes. Toxicol. Sci. 2005;88:231–241. doi: 10.1093/toxsci/kfi301. [DOI] [PubMed] [Google Scholar]
- Tousoulis D., Papageorgiou N., Androulakis E., Briasoulis A., Antoniades C., Stefanadis C. Fibrinogen and cardiovascular disease: Genetics and biomarkers. Blood Rev. 2011;25:239–245. doi: 10.1016/j.blre.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Vihtelic T. S., Doro C. J., Hyde D. R. Cloning and characterization of six zebrafish photoreceptor opsin cDNAs and immunolocalization of their corresponding proteins. Vis. Neurosci. 1999;16:571–585. doi: 10.1017/s0952523899163168. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Sueoka K., Sasagawa I., Nakabayashi A., Yoshimura Y., Ogata T. Association of male infertility with Pro185Ala polymorphism in the aryl hydrocarbon receptor repressor gene: Implication for the susceptibility to dioxins. Fertil. Steril. 2004;82(Suppl. 3):1067–1071. doi: 10.1016/j.fertnstert.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Weger B. D., Sahinbas M., Otto G. W., Mracek P., Armant O., Dolle D., Lahiri K., Vallone D., Ettwiller L., Geisler R., et al. The light responsive transcriptome of the zebrafish: Function and regulation. PLoS One. 2011;6:e17080. doi: 10.1371/journal.pone.0017080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernet M. F., Mazzoni E. O., Celik A., Duncan D. M., Duncan I., Desplan C. Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 2006;440:174–180. doi: 10.1038/nature04615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall J. M., Smyth G. K. limmaGUI: A graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- Yamamoto J., Ihara K., Nakayama H., Hikino S., Satoh K., Kubo N., Iida T., Fujii Y., Hara T. Characteristic expression of aryl hydrocarbon receptor repressor gene in human tissues: Organ-specific distribution and variable induction patterns in mononuclear cells. Life Sci. 2004;74:1039–1049. doi: 10.1016/j.lfs.2003.07.022. [DOI] [PubMed] [Google Scholar]
- Yeung N., Cline M. S., Kuchinsky A., Smoot M. E., Bader G. D. Exploring biological networks with Cytoscape software. Curr. Protoc. Bioinform. 2008 doi: 10.1002/0471250953.bi0813s23. Chapter 8, Unit 8 13. [DOI] [PubMed] [Google Scholar]
- Zhao L., Samuels T., Winckler S., Korgaonkar C., Tompkins V., Horne M. C., Quelle D. E. Cyclin G1 has growth inhibitory activity linked to the ARF-Mdm2-p53 and pRb tumor suppressor pathways. Mol. Cancer Res. 2003;1:195–206. [PubMed] [Google Scholar]
- Zudaire E., Cuesta N., Murty V., Woodson K., Adams L., Gonzalez N., Martinez A., Narayan G., Kirsch I., Franklin W., et al. The aryl hydrocarbon receptor repressor is a putative tumor suppressor gene in multiple human cancers. J. Clin. Invest. 2008;118:640–650. doi: 10.1172/JCI30024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.