Abstract
Conflicting evolutionary interests between mother and offspring are hypothesized to drive an evolutionary arms race during mammalian pregnancy, and thus, positive selection may cause the rapid divergence of placental proteins that affect maternal or fetal fitness. We investigated the genomic consequences of placental expression in rodents and report that a substantial proportion (20.5%) of genes specifically expressed in the mature placenta are rapidly evolving. Moreover, we found that most rapidly evolving genes belong to just three pregnancy-related gene families: placental cathepsins, prolactins, and placental carcinoembryonic antigens. We then sequenced the most rapidly evolving gene, trophoblast-specific protein alpha (Tpbpa), in nine different Mus species/subspecies and found evidence of positive selection within the Mus lineage, with an excess of nonsynonymous changes clustering near a functionally important interaction site. Together, these results suggest that placental proteins, which mediate interactions between mother and offspring, often may be the targets of evolutionary conflict.
Keywords: adaptive evolution, evolutionary conflict, placenta, positive selection, reproduction
Comparative genomic studies across many diverse taxa consistently have found that those genes related to fertilization and immune responses are among the most rapidly evolving in the genome (e.g., Swanson and Vacquier 2002; Nielsen et al. 2005; Gibbs et al. 2007). Many of these genes are probably involved in evolutionary “arms races,” driven by host–pathogen conflict or male–female sexual conflict, in which they experience strong positive selection to maintain optimal fitness (Dawkins and Krebs 1979; Chapman et al. 2003). Another arena for evolutionary conflict, which is less well studied, is antagonism between mother and offspring during pregnancy. The maternal–fetal conflict hypothesis proposes that there is a selective advantage to offspring who can selfishly maximize their own growth at the expense of the fitness of their mother and siblings, whereas there is an opposing advantage to mothers who can counter these adaptations by maintaining equal investment in all offspring and surviving to breed in future generations (Haig 1993; Zeh and Zeh 2000).
The primary site of physiological exchange between mother and offspring during mammalian pregnancy is the placenta—a temporary organ formed by the fusion of fetal extraembryonic membrane with maternal uterine tissue. At this maternal–fetal interface, a complex interplay of proteins from these two distinct individuals mediates nutrient and waste exchange, immunoregulation, and other pregnancy-related physiological processes. These interactions are likely to result in an evolutionary conflict between maternal and fetal genes (Haig 2008) in which genes under conflict may either experience balancing selection and reach a stable resolution (McVean and Hurst 1997) or experience repeated bouts of positive selection in an arms race to maintain maximal fitness. In this study, we asked whether we could identify placental genes that show strong signatures of continuous positive selection and might therefore be involved in an evolutionary arms race between mother and offspring.
Early comparative genomic screens across mammalian orders (e.g., human vs. mouse) did not identify placental genes to be rapidly evolving as a class (e.g., Clark et al. 2003; Lindblad-Toh et al. 2005), which is unsurprising because many placental genes are lineage-specific and would therefore be missed in comparisons of divergent taxa (Rawn and Cross 2008). As more mammalian genomic data have become available, several reports have revealed signatures of positive selection in placental genes within lineages (e.g., primates and artiodactyls; Hughes et al. 2000; Hou et al. 2009). Here, we investigated placental gene evolution within the rodent lineage, which offers several advantages: 1) rodents have short gestation times and are a widely used experimental model, so we can readily examine subsets of placental genes from each stage of development rather than focusing on the term placenta, which has a distinct expression profile compared with a developing placenta (Knox and Baker 2008), 2) rodents generally have large and multisired litters, which likely intensifies the potential maternal–fetal conflict (Zeh and Zeh 2000), and 3) there are several examples of interfertile monogamous/promiscuous sister species (e.g., Peromyscus maniculatus and P. polionotus, respectively), which allows for the role of mating system, and of particular candidate genes, in maternal–fetal conflict to be tested directly.
For this study, we confined our analysis to placental genes in two rodents, mouse (Mus musculus) and rat (Rattus norvegicus), which share the same type of placenta (Mess 2003) and are the only rodents with well-annotated genomes. Because the timing of expression during development has been demonstrated to correlate with the rate of evolution (e.g., in testes; Good and Nachman 2005), we utilized a comprehensive time course microarray over mouse placental development (Knox and Baker 2008) to group placental genes into “developing” (specifically expressed between E8.5 and 13.5) and “mature” (E13.5 to term) as well as “maternal” (decidua) and “fetal” (fetal chorion) based on the tissue of origin. Finally, we examined whether secreted proteins, which can traverse the maternal–fetal boundary, are more likely to be targets of positive selection.
Our specific goals in examining the molecular evolution of rodent placental genes were to identify the most rapidly evolving genes and to determine whether specific functional classes of genes and/or their developmental timing affect their rate of evolution. By comparing the nonsynonymous substitution rate to the synonymous substitution rate, we estimated the rate of evolution for each gene in Mus and Rattus. High values of ω (ω ≥ 0.5 for pairwise comparisons) indicate elevated rates of amino acid sequence divergence and suggest that some sites are likely to be targets of positive selection (Swanson et al. 2004).
From a total of 786 genes predominantly expressed in the placenta (following Knox and Baker 2008), we aligned 704 coding sequences between mouse and rat based on the Ensembl peptide alignments. We next performed pairwise estimates of ω using maximum likelihood (PAML v4, runmode = −2; Yang 2007); table 1 summarizes these results. When all placental genes are considered together, is significantly higher than the genome-wide average (: 0.25 vs. 0.17, one-way analysis of variance, P < 0.0001), although evolutionary rate is often elevated for genes with tissue-specific expression (Winter et al. 2004). Secreted proteins had a higher than did nonsecreted proteins (table 1) within all gene classes (genome wide and for each placental subclass). When we considered the temporal pattern of expression, we found that genes highly expressed in the mature placenta exhibit significantly faster rates of evolution than genes expressed earlier in placental development (: 0.33 vs. 0.23, P < 0.0001). In fact, secreted proteins from the mature fetal component of the placenta are the most rapidly evolving class of placental genes even compared with other placental secreted genes (N = 30, : 0.56 vs. 0.31, P < 0.005).
Table 1.
Pairwise Comparisons of Placentally Expressed Genes between Mus and Rattus.
| Category | N | Total |
Nonsecreted ϖ (N) | Secreted ϖ (N) | ω ≥ 0.5 (%) | ||
| ϖ | |||||||
| Genome | 18,708 | 0.03 | 0.21 | 0.17 | 0.15 (14,683) | 0.24 (4,025) | 2,438 (13.1) |
| Placental | 704 | 0.08 | 0.24 | 0.25 | 0.20 (469) | 0.35 (235) | 95 (13.5) |
| Developing | 519 | 0.06 | 0.24 | 0.23 | 0.20 (388) | 0.31 (131) | 54 (10.4) |
| Fetal | 342 | 0.06 | 0.24 | 0.22 | 0.19 (290) | 0.36 (52) | 32 (9.4) |
| Maternal | 200 | 0.06 | 0.23 | 0.25 | 0.20 (111) | 0.31 (89) | 29 (14.5) |
| Mature | 191 | 0.09 | 0.26 | 0.33 | 0.23 (85) | 0.41 (106) | 41 (20.5) |
| Fetal | 61 | 0.11 | 0.25 | 0.44 | 0.31 (31) | 0.56 (30) | 21 (34.4) |
| Maternal | 150 | 0.09 | 0.26 | 0.33 | 0.23 (62) | 0.40 (88) | 35 (23.3) |
NOTES.—Categories following Knox and Baker (2008). “Placental genes” exhibit maximal expression in the placenta. Genes classified as “developing” are those overexpressed from E8.5 to 13.5, whereas “mature” genes are overexpressed from E13.5 to term. “Fetal” and “maternal” refer to the tissue of expression and are not mutually exclusive (e.g., a gene may be overexpressed in both fetal and maternal mature placental tissue). N, number of unique Mus/Rattus coding sequence alignments (obtained from Ensembl build 49); , mean nonsynonymous substitution rate; , mean synonymous substitution rate; , mean ω for all genes estimated using PAML v4 (Yang 2007); nonsecreted and secreted, secreted proteins predicted using Phobius, values are significantly different for all secreted/nonsecreted comparisons within a gene class (one-way analysis of variance, P < 0.01); percentage and number of rapidly evolving genes with ω ≥ 0.5 are given.
Table 2 lists the 15 most rapidly evolving genes from our analysis. Notably, we identified rapidly evolving genes from three major rodent placental gene families: carcinoembryonic antigens (CEAs), prolactins, and placentally expressed cathepsins (PECs). All three of these families are comprised of secreted proteins (Zebhauser et al. 2005; Soares et al. 2007; Mason 2008), and the majority of family members represented in our analysis are expressed in the fetal portion of the mature placenta (CEAs: 7 of 8 exhibit fetal expression and 8 of 8 expressed in mature placenta; prolactins: 14 of 16 and 9 of 16; and PECs: 10 of 10 and 8 of 10).
Table 2.
Top 15 Rapidly Evolving Placental Genes Based on Pairwise ω Estimated Using Maximum Likelihood.
| Gene | Chr | dN | dS | ω | Fetal | Maternal | Developing | Mature | Secreted |
| Tpbpa | 13qB2 | 0.30 | 0.20 | 1.50 | X | — | — | X | X |
| Pdgfrl | 8qA4 | 0.09 | 0.06 | 1.44 | — | X | X | — | — |
| 2610036L11Rik | 10qA4 | 0.14 | 0.10 | 1.43 | X | — | X | — | — |
| Tpbpb | 13qB2 | 0.28 | 0.20 | 1.39 | X | X | — | X | X |
| Ctla2a | 13qB2 | 0.17 | 0.14 | 1.22 | X | — | — | X | X |
| Ceacam12 | 7qA2 | 0.27 | 0.24 | 1.12 | X | X | — | X | X |
| Ceacam14 | 7qA2 | 0.26 | 0.24 | 1.07 | X | X | — | X | — |
| Ccl9 | 11qC | 0.41 | 0.40 | 1.05 | — | X | — | X | X |
| Ceacam11 | 7qA2 | 0.25 | 0.24 | 1.04 | X | X | — | X | Xa |
| Pvr | 7qA3 | 0.36 | 0.38 | 0.97 | — | X | — | X | X |
| Glrx | 13qC1 | 0.09 | 0.09 | 0.97 | X | — | X | — | — |
| Spink3 | 18qB3 | 0.29 | 0.30 | 0.96 | X | — | X | — | X |
| Ceacam13 | 7qA2 | 0.22 | 0.24 | 0.95 | X | X | — | X | X |
| Prl3c1 | 13qA3 | 0.21 | 0.23 | 0.93 | — | X | X | — | X |
| Prl7a2 | 13qA3 | 0.18 | 0.19 | 0.92 | X | X | — | X | X |
Chr, chromosomal position in mouse; dN, nonsynonymous substitution rate; dS, synonymous substitution rate; ω, pairwise ω estimated with Paml; fetal, expressed in the fetal portion of the placenta; maternal, expressed in maternal portion; developing, high placental expression during E8.5 to E13.5; mature, high placental expression during E13.5 to term; secreted, predicted signaling molecule (identified with Phobius; Käll et al. 2004). Expression data annotation obtained from Knox and Baker (2008).
From Kataoka et al. (2000).
The CEA family is located on mouse chromosome 7 and comprises a large group of pregnancy-specific glycoproteins (PSGs) and carcinoembryonic antigen-related cell-adhesion molecules (CEACAMs), many of which are rodent specific and exhibit placenta-specific expression (Zebhauser et al. 2005). Although their precise function is unknown, they probably help modulate the maternal immune system (Wynne et al. 2006). In our analysis, the most rapidly evolving placental CEAs are Ceacam11 (ω = 1.04), Ceacam12 (ω = 1.11), Ceacam13 (ω = 0.95), and Ceacam14 (ω = 1.06). In addition, our data set includes CEAs Ceacam18, Psg17, Psg18, and Psg28, which also exhibit elevated rates of evolution (N = 4, = 0.62 ± 0.09).
Placental prolactin hormones are located in a cluster on mouse chromosome 13 and play extremely diverse roles in pregnancy; members of this family are important for placental development, vascularization, stress response, and a variety of other physiological functions (Haig 2008). The most rapidly evolving prolactins we identified were Prl3c1 (ω = 0.93), a maternally expressed prolactin, and Prl7a2 (ω = 0.92), an important stimulator of hematopoiesis in trophoblast (Soares et al. 2007). Additional placental prolactins identified in our analysis also exhibit high rates of evolution (N = 14, = 0.62 ± 0.10).
Finally, the PECs are a rodent-specific family of placental cathepsins that are also located in a cluster on mouse chromosome 13 and include mature proteases (cathepsins 3, 6, 7, 8, M, Q, R) and predicted propeptide inhibitors (Tpbpa, Tpbpb, and Ctla2a). Together, the PECs function to structurally remodel placental tissue during development (Mason 2008). The most rapidly evolving genes from this family are Tpbpa (ω = 1.50), Tpbpb (ω = 1.39), and Ctla2a (ω = 1.22). In addition, the mature proteases as a group are also rapidly evolving (N = 7, = 0.48 ± 0.09), potentially as a consequence of an evolutionary conflict between the placental proteases and their inhibitors.
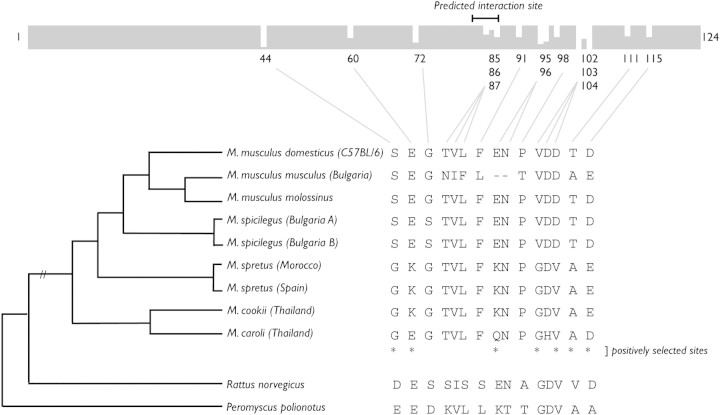
To test for positive selection within a lineage, we sequenced the most rapidly evolving gene, trophoblast-specific protein alpha (Tpbpa), in nine different Mus species/subspecies (fig. 1; Supplementary Material, Supplementary Material online). Under the M8a-M8 model comparison in PAML, we found that Tpbpa was indeed evolving rapidly within the Mus lineage (Likelihood ratio test = 37.5, P < 0.001). The peptide encoded by Tpbpa contains an interaction domain predicted to block to the substrate-binding site of mature cathepsin to prevent premature proteolytic activity (Coulombe et al. 1996) and shows high sequence variation when compared with both placental and nonplacental cathepsins, possibly reflecting different substrate specificities (Deussing et al. 2002). We observed that positively selected sites in Tpbpa clustered significantly near this predicted interaction site (permutation test, P = 0.001); although the sites closest to the interaction site do not appear to be under positive selection, their proximity is suggestive of functional consequences. Furthermore, we observed that 12 of 15 amino acid sites that are variable among Mus species are also variant in either Rattus or deer mice (genus Peromyscus; GenBank accession number EV469066.1) compared with the Mus consensus, which suggests that these may be functionally significant sites and undergo repeated adaptive substitutions.
FIG. 1.
Rapid evolution of Tpbpa in Mus. The conservation plot from the amino acid alignment of Tpbpa from nine Mus species/subspecies in which gray indicates conservation and white shows variation. Divergent amino acid sites are given and displayed next to a species phylogeny (Tucker et al. 2005). Sites marked with “*” are positively selected with P > 99% using the PAML Bayes Empirical Bayes analysis under M8. The corresponding amino acid in Rattus norvegicus and Peromyscus polionotus is shown below each site.
Overall, we show that a subset (∼13.5%) of placental proteins, especially those that are secreted and expressed late in development, probably experience positive selection based on their pattern of sequence divergence. Although a pairwise comparison is a generally low power and conservative test for positive selection, we find supporting evidence that the most rapidly evolving gene Tpbpa is experiencing positive selection in a functionally relevant domain even within the Mus lineage. It is important to note that many genes we identified are part of large gene families in which there may be some functional redundancy; hence, relaxed functional constraint may also contribute to their rapid evolution (Hughes 2007). We avoided the inclusion of pseudogenes by only considering full-length coding sequences that were in-frame and known to be highly expressed in the mouse placenta. Future comparative genomic data from promiscuous and monogamous rodents (e.g., P. maniculatus and P. polionotus, respectively) can test whether the rapidly evolving genes are under positive or relaxed selection because maternal–fetal conflict is hypothesized to be most severe in polyandrous species with multisired litters (Zeh and Zeh 2000).
Several lines of evidence suggest that many of these placental proteins may be involved in an evolutionary conflict between mother and offspring. First, the most rapidly evolving placental genes are expressed in the mature placenta in which the majority of genes are involved in physiological exchange between mother and offspring (Knox and Baker 2008). Second, among genes expressed in the mature placenta, fetal-secreted proteins are the most rapidly evolving class, consistent with their role in directly interacting with maternal physiology or immune response. Although these observations are also consistent with maternal–offspring coadaptation (Wolf and Brodie 1998), we also find evidence for positive selection acting broadly in the placental gene families, CEAs, prolactins, and PECs. These three gene families share characteristics with sexual conflict genes first identified in Drosophila seminal fluid; the most rapidly evolving seminal proteins include secreted proteins such as glycoproteins, proteins with enzymatic functions including proteases and their inhibitors, and proteins involved in the immune response (Wolfner 2002; Ravi Ram and Wolfner 2007). We suggest, then, that evolutionary arms races between mother and offspring may be an important selective force in the evolution of viviparous species, especially those with large and multisired litters.
Supplementary Material
Supplementary file is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org).
Acknowledgments
The authors wish to thank D. Haig, J. Huelsenbeck, L. Turner, P. Vrana, and members of the Hoekstra Lab for helpful discussion. P. Tucker kindly shared all the Mus DNA samples derived from the National Cancer Institute. Thanks also to Daniela Simeonovska-Nikolova and Krastio Dimitrov for their help collecting Bulgarian Mus specimens. This work was supported by an Arnold and Mabel Beckman Young Investigator Award (to H.E.H.) and Putnam Expedition funds from the Museum of Comparative Zoology, Harvard University.
References
- Chapman T, Arnqvist G, Bangham J, Rowe L. Sexual conflict. Trends Ecol Evol. 2003;18:41–47. [Google Scholar]
- Clark A, Glanowski S, Nielsen R, Thomas P, Kejariwal A, Todd M, Tanenbaum D, Civello D, Lu F, Murphy B. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Coulombe R, Grochulski P, Sivaraman J, Ménard R, Mort JS, Cygler M. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996;15(20):5492–5503. [PMC free article] [PubMed] [Google Scholar]
- Dawkins R, Krebs JR. Arms races between and within species. Proc R Soc Lond B Biol Sci. 1979;205:489–511. doi: 10.1098/rspb.1979.0081. [DOI] [PubMed] [Google Scholar]
- Deussing J, Kouadio M, Rehman S, Werber I, Schwinde A, Peters C. Identification and characterization of a dense cluster of placenta-specific cysteine peptidase genes and related genes on mouse chromosome 13. Genomics. 2002;79:225–240. doi: 10.1006/geno.2002.6696. [DOI] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, et al. (176 co-authors) Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Good JM, Nachman MW. Rates of protein evolution are positively correlated with developmental timing of expression during mouse spermatogenesis. Mol Biol Evol. 2005;22:1044–1052. doi: 10.1093/molbev/msi087. [DOI] [PubMed] [Google Scholar]
- Haig D. Genetic conflicts in human pregnancy. Q Rev Biol. 1993;68:495–532. doi: 10.1086/418300. [DOI] [PubMed] [Google Scholar]
- Haig D. Placental growth hormone-related proteins and prolactin-related proteins. Placenta. 2008;29:S36–S41. doi: 10.1016/j.placenta.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Hou Z, Romero R, Uddin M, Than NG, Wildman DE. Adaptive history of single copy genes highly expressed in the term human placenta. Genomics. 2009;93:33–41. doi: 10.1016/j.ygeno.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Green JA, Garbayo JM, Roberts RM. Adaptive diversification within a large family of recently duplicated, placentally expressed genes. Proc Natl Acad Sci U S A. 2000;97:3319–3323. doi: 10.1073/pnas.050002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Takata Y, Nakajima A, Saito S, Huh N. A carcinoembryonic antigen family cDNA from mouse placenta encoding a protein with a rare domain composition. Placenta. 2000;21:610–614. doi: 10.1053/plac.2000.0546. [DOI] [PubMed] [Google Scholar]
- Knox K, Baker JC. Genomic evolution of the placenta using co-option and duplication and divergence. Genome Res. 2008;18:695–705. doi: 10.1101/gr.071407.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade C, Mikkelsen T, et al. (236 co-authors) Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Mason RW. Emerging functions of placental cathepsins. Placenta. 2008;29:385–390. doi: 10.1016/j.placenta.2008.02.006. [DOI] [PubMed] [Google Scholar]
- McVean GT, Hurst LD. Molecular evolution of imprinted genes: no evidence for antagonistic coevolution. Proc R Soc Lond B Biol Sci. 1997;264:739–746. doi: 10.1098/rspb.1997.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mess A. Evolutionary transformations of chorioallantoic placental characters in Rodentia with special reference to hystricognath species. J Exp Zool. 2003;299:78–98. doi: 10.1002/jez.a.10292. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Bustamante C, Clark A, et al. (13 co-authors) A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi Ram K, Wolfner M. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr Comp Biol. 2007;47:427–445. doi: 10.1093/icb/icm046. [DOI] [PubMed] [Google Scholar]
- Rawn SM, Cross JC. The evolution, regulation, and function of placenta-specific genes. Annu Rev Cell Dev Biol. 2008;24:159–181. doi: 10.1146/annurev.cellbio.24.110707.175418. [DOI] [PubMed] [Google Scholar]
- Soares M, Konno T, Alam S. The prolactin family: effectors of pregnancy-dependent adaptations. Trends Endocrinol Metab. 2007;18:114–121. doi: 10.1016/j.tem.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Wong A, Wolfner MF, Aquadro CF. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics. 2004;168:1457–1465. doi: 10.1534/genetics.104.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker PK, Sandstedt SA, Lundrigan BL. Phylogenetic relationships in the subgenus Mus (genus Mus, family Muridae, subfamily Murinae): examining gene trees and species trees. Biol J Linn Soc. 2005;84:653–662. [Google Scholar]
- Winter EE, Goodstadt L, Ponting C. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB. Brodie ED. Coadaptation of parental and offspring characters. Evolution. 1998;52:299–308. doi: 10.1111/j.1558-5646.1998.tb01632.x. [DOI] [PubMed] [Google Scholar]
- Wolfner MF. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity. 2002;88:85–93. doi: 10.1038/sj.hdy.6800017. [DOI] [PubMed] [Google Scholar]
- Wynne F, Ball M, Mclellan A, Dockery P, Zimmermann W, Moore T. Mouse pregnancy-specific glycoproteins: tissue-specific expression and evidence of association with maternal vasculature. Reproduction. 2006;131:721–732. doi: 10.1530/rep.1.00869. [DOI] [PubMed] [Google Scholar]
- Yang Z. Paml 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zebhauser R, Kammerer R, Eisenried A, Mclellan A, Moore T, Zimmermann W. Identification of a novel group of evolutionarily conserved members within the rapidly diverging murine family. Genomics. 2005;86:566–580. doi: 10.1016/j.ygeno.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Zeh DW, Zeh JA. Reproductive mode and speciation: the viviparity-driven conflict hypothesis. Bioessays. 2000;22:938–946. doi: 10.1002/1521-1878(200010)22:10<938::AID-BIES9>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.