Abstract
Disruption of the histone modification patterns is one of the most common features of human tumors. However, few genetic alterations in the histone modifier genes have been described in tumorigenesis. Herein we show that the histone methyltransferase SETDB1 undergoes gene amplification in non-small and small lung cancer cell lines and primary tumors. The existence of additional copies of the SETDB1 gene in these transformed cells is associated with higher levels of the corresponding mRNA and protein. From a functional standpoint, the depletion of SETDB1 expression in amplified cells reduces cancer growth in cell culture and nude mice models, whereas its overexpression increases the tumor invasiveness. The increased gene dosage of SETDB1 is also associated with enhanced sensitivity to the growth inhibitory effect mediated by the SETDB1-interfering drug mithramycin. Overall, the findings identify SETDB1 as a bona fide oncogene undergoing gene amplification-associated activation in lung cancer and suggest its potential for new therapeutic strategies.
Keywords: SETDB1, gene amplification, overexpression, lung cancer, histone methyltransferase, chemotherapy
Introduction
Disruption of the epigenetic landscape is a common event in cancer cells, leading to significant changes in chromatin structure and gene expression.1, 2 Although information about DNA methylation profiles is widespread, we know much less about the patterns of histone modification disruption in human tumors.3 In this latter setting, it is recognized that there are particular combinations of histone marks for a given promoter associated with the transcriptional silencing of tumor suppressor genes, such as deacetylation of histones H3 and H4, loss of H3K4 trimethylation, and gain of H3-K9 methylation and Lys27 of histone H3 (H3K27) trimethylation.1, 2 At the global level, cancer cells show reduced monoacetylated and trimethylated lysines 16 and 20 of histone H4,4 respectively, and histone acetylation and dimethylation changes in histones H3 and H4, respectively, might have prognostic value.5 Recent efforts to study human cancer genomics have highlighted that point mutations, translocations, deletions and gene amplification events occur in histone modifier enzymes such as histone acetyltransferases, deacetylases, methyltransferases and demethyltransferases.6, 7, 8, 9
One of the best examples of a histone methyltransferase involved in human cancer is the enhancer of zeste homolog 2, a component of the Polycomb repressive complex 2, which represses gene transcription via trimethylation of H3K27 that targets tumor suppressor genes.10, 11, 12, 13 The enhancer of zeste homolog 2 has the hallmarks of an oncogene, particularly in prostate and breast cancer, where elevated levels are found in the more advanced forms of the disease.14, 15, 16 Recently, the enhancer of zeste homolog 2 gain-of-function mutations have also been found in lymphomas.17 However, the list of histone methyltransferases with a role in tumorigenesis is rapidly growing, and includes such examples as the mixed lineage leukemia 1 (ALL-1/HRX),18 hDOT1L,19 which is targeted by chromosomal translocations, and NSD1 that undergoes DNA methylation-associated silencing in solid tumors.20
Among the different histone methyltransferases, SETDB1 has been of increasing interest owing to its involvement in the development of melanoma, where it resides in a recurrently amplified chromosome 1q21 interval and accelerates melanoma onset in a zebrafish model,21 and also because the described 1q21.3 region includes a plausible human melanoma susceptibility gene. SETDB1 is a likely candidate for this.22 Also known as ESET or KMT1E, SETDB1 is a histone H3 lysine 9-specific methyltransferase involved in the transcriptional silencing of euchromatic genes and retroelements.23, 24, 25, 26 Recent reports also show that SETDB1 is essential for proviral silencing and for controlling developmental regulators and chimeric transcripts that maintain the fate of embryonic stem cells.26, 27, 28 In addition to the discoveries in melanoma,21, 22 the role of SETDB1 in cancer was also suggested by its association with the DNA methylation machinery,25 the promyelocytic nuclear leukemia-nuclear bodies29 and by its increased expression in transformed broncoepithelial cells.30
These findings prompted us to investigate the possible presence of SETDB1 gene amplification and its associated overexpression in lung cancer cells and primary tumors. Extra copies of an oncogene may give tumor cells a growth advantage as well as being a mechanism associated with different sensitivity to therapies.31 The identification of amplified target genes is of great importance for cancer diagnosis and prognosis, and ultimately for designing targeted therapies, ERBB2/HER-2 in breast cancer being the best example.32 Thus, we examined whether the SETDB1 gene amplification occurs in lung cancer cell lines and primary tumors, and studied its impact on mRNA and protein expression levels, its functional role in lung cancer growth and its potential value as a biomarker for predicting the response to particular chemotherapeutic agents in lung tumors.
Results and Discussion
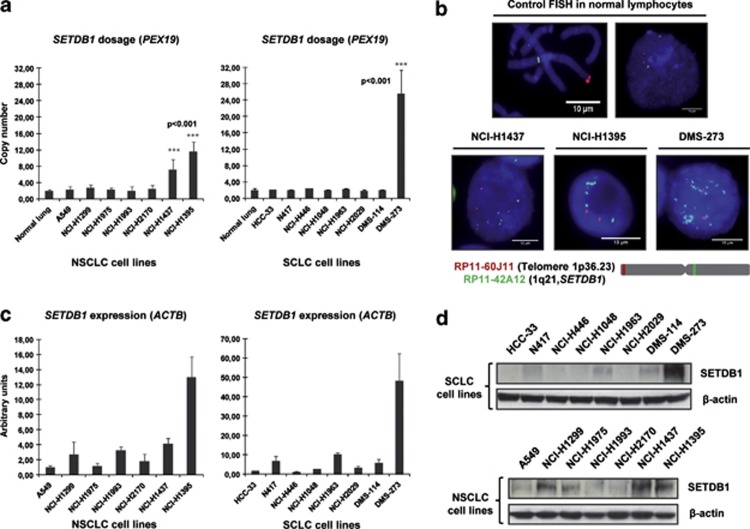
We first screened a collection of 15 human lung cancer cell lines for SETDB1 gene copy-number alterations. These included seven non-small (A549, NCI-H1299, NCI-H1975, NCI-H1993, NCI-H2170, NCI-H1437 and NCI-H1395) and eight small (HCC-33, N417, NCI-H446, NCI-H1048, NCI-H1963, NCI-H2029, DMS-114 and DMS-273) cell lung cancer types. The lung cancer cell lines were purchased from the American Type Culture Collection (Rockville, MD, USA) and were grown and maintained in 10% fetal bovine serum in Roswell Park Memorial Institute medium. Primary normal tissues such as lung epithelium and leukocytes were used as normal SETDB1 copy-number control samples. Using a quantitative genomic PCR approach, we observed that two non-small (NCI-H1437 and NCI-H1395) and one small (DMS-273) cell lung cancer lines had a greater than four-fold change in SETDB1 gene copy number (Figure 1a). This increase was particularly important in the small lung cancer cell line DMS-273 (Figure 1a). The remaining eleven lung cancer cell lines did not present any evident change in SETDB1 gene copy number (that is, amplification, half gene dosage or homozygous deletion). We performed fluorescence in situ hybridization analyses to confirm the presence of SETDB1 gene amplification suggested to occur in the three lung cancer cell lines by the competitive genomic PCR approach (Figure 1b). The fluorescence in situ hybridization technique confirmed the presence of SETDB1 gene amplification in NCI-H1437, NCI-H1395 and DMS-273 (Figure 1b). The applied fluorescence in situ hybridization technique was validated by the observation of the normal copy-number of the SETDB1 gene in lymphocytes (Figure 1b). Using reported copy-number variation data,33 we have determined the size of the amplicon that contains SETDB1 in the three studied lung cancer cell lines: DMS-273 (1 038 210 bp), NCI-H1437 (4 539 342 bp) and NCI-H1395 (4 623 419 bp). SETDB1 is genomically located in the middle of all three amplicons, even in the case of the smallest one (DMS-273; Supplementary Figure S1). In melanoma, the only other tumor type where SETDB1 genetic amplification has been reported,21 all the melanoma cell lines with amplification at this genomic locus (1q21) contain the SETDB1 gene according to the copy-number variation data.33 In addition, in the smallest amplicon detected in the melanoma setting (cell line COLO-679, 2 797 611 bp), SETDB1 is also located right in the middle (Supplementary Figure S1). Thus, SETDB1 is within the smallest identified region of recurrent amplification.
Figure 1.
Determination of SETDB1 gene amplification and its association with RNA and protein overexpression in lung cancer cell lines. (a) Assessment of SETDB1 copy-number by quantitative genomic PCR. Amplification frequency of SETDB1 (evaluated with SYBR Green, Bio-Rad, Hercules, CA, USA) was calculated by the standard curve method using the 7900HT SDS program. To define an internal control gene, we chose chromosome 1p36.23 because it is the least aneuploid region among our cell lines (PEX19 gene). Primers are available upon request. DNA from normal lung was used as the reference standard. Results are reported as n-fold copy-number increase relative to the PEX19 gene. (b) Fluorescence in situ hybridization for the SETDB1 gene. The UCSC genome browser (http://www.genome.ucsc.edu) was used to select the bacterial artificial chromosome (BAC) clone spanning the 1q21 region for the SETDB1 gene: RP11-42A12. A telomeric BAC clone located in the telomeric 1p36.23 region was used as a control. The BACs were obtained from the BACPAC Resource Center at the Children's Hospital Oakland Research Institute (Oakland, CA, USA). SETDB1 and telomeric probes were labeled with Spectrum Green and Red dUTP (Abbott, Wiesbaden, Germany), respectively, using a CGH Nick Translation Reagent Kit (Abbott Molecular Inc., Des Plaines, IL, USA). The samples were counterstained with 4',6-diamidino-2-phenylindole in Vectashield antifade solution (Burlingame, CA, USA). Gene amplification was observed in the interphases of NCI-H1437, NCI-H1395 and DMS-273. Probes were verified to give a single signal on normal commercial lymphocyte metaphase slides (CGH Reagents, Abbott). Quantitative reverse transcription–PCR (c) and western blot (d) demonstrate higher levels of SETDB1 mRNA and protein (ab12317, Abcam, Cambridge, UK), respectively, in amplified cancer cell lines (H1437, NCI-H1395 and DMS-273) than that in unamplified cells. PCR primers are available upon request.
We next considered the possible existence of an association between extra copies of the SETDB1 gene and overexpression of the corresponding RNA transcript and protein using quantitative reverse transcription–PCR and western blot approaches, respectively, (Figures 1c and d). We found that the expression of SETDB1 for both mRNA and protein was enhanced in cancer cell lines harboring the SETDB1 gene amplification event relative to non-amplified cancer cells (Figures 1c and d). In the smallest SETDB1 amplicon (present in DMS-273 cells), thirty-four genes are also co-amplified (Supplementary Figure S1). Using reported microarray expression data, we have observed that, in addition to SETDB1, only 7 of these other 34 genes (21%) are overexpressed in DMS-273, NCI-H1437 and NCI-H1395 in comparison with unamplified lung cancer cells (NCI-H1299 and A549; Supplementary Figure S1). In this regard, we have confirmed by quantitative reverse transcription–PCR that six of these seven genes are overexpressed in the SETDB1-amplified lung cell lines (Supplementary Figure S1).
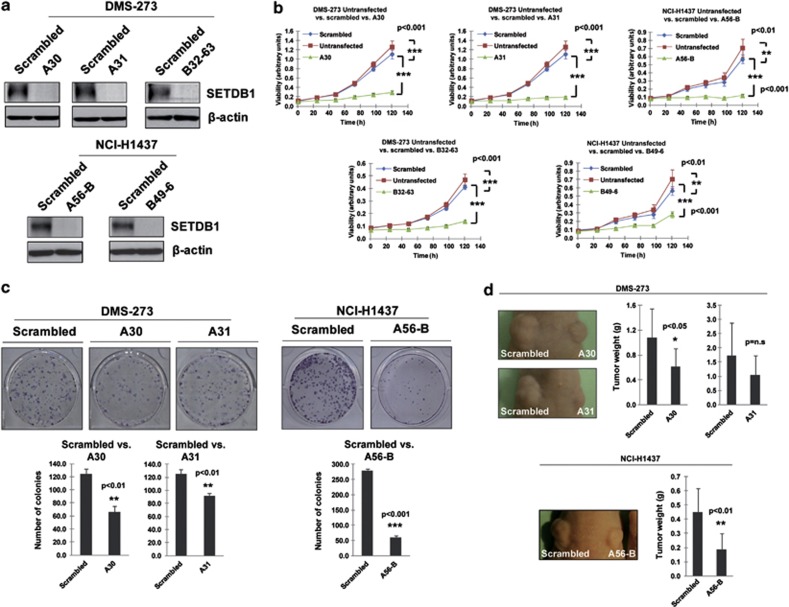
Once we had demonstrated the presence of SETDB1 gene amplification and its associated overexpression in the lung cancer cell lines, we examined its contribution to the tumorigenic phenotype in vitro and in vivo. We first analyzed the effect of SETDB1 depletion in lung cancer cells harboring its gene amplification and its associated overexpression. Supplementary Table S1 includes all the used short hairpin RNA (shRNA) sequences. Three SETDB1 shRNA-depleted clones were established for DMS-273 cells (A30, A31 and B32-63) and two clones for NCI-H1437 (A56-B and B49-6). Experiments for each clone were performed in triplicate. We observed that the reduction of SETDB1 expression in the gene-amplified cells had cancer growth inhibitory features (Figure 2). Upon stable transfection of shRNAs against SETDB1 in the gene-amplified DMS-273 and NCI-H1437 lung cancer cell lines and efficient depletion of the SETDB1 protein (Figure 2a), the cells proved less viable in the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay (Figure 2b and Supplementary Figure S2) and had a markedly reduced percentage colony-formation density in the assay developed on plastic plate (Figure 2c and Supplementary Figure S2). Transfection of the scramble shRNA did not reduce cell viability (Figure 2b and Supplementary Figure S2) and had no impact on the colony formation assay (Figure 2c and Supplementary Figure S2). Confocal microscopy experiments in the NCI-H1437 lung cancer cell line confirmed the expected nuclear staining of the SETDB1 protein (4',6-diamidino-2-phenylindole colocalization; Supplementary Figure S3) and the disappearance of the nuclear signal upon SETDB1 shRNA-mediated depletion (Supplementary Figure S3). SETDB1 knockdown by the shRNA approach in a lung cancer cell line without gene amplification (NCI-H1299) did not cause a significant effect in the cell growth determined by the MTT experiments or the colony formation assay (Supplementary Figure S3), suggesting the SETDB1 dependence for cell growth only in the amplified cancer cells.
Figure 2.
Growth-promoting effects of SETDB1 in lung cancer. (a) Stable downregulation of the SETDB1 gene by short hairpins using two different target sequences for DMS-273 (clones A30/A31 and clone B32-63) and NCI-H1437 (clones A56-B and B46-9). SETDB1 shRNA sequences are available upon request. (b) The short hairpin SETDB1-depleted cells were less viable in the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay than in the untransfected or scrambled shRNA-transfected cells (P-values obtained by the analysis of variance (ANOVA) test). (c) The colony formation assay showed that DMS-273 and NCI-H1437 cells stably transfected with the shRNA against SETDB1 formed significantly fewer colonies than scrambled shRNA-transfected cells (P-values obtained by the ANOVA test). Data shown are means±s.d., n=3. (d) Effect of SETDB1 shRNA-mediated depletion on the growth of DMS-273 and NCI-H1437 xenografts in nude mice. Tumor volume was monitored over time and the tumor was excised and weighed at 30 days. There was a significant decrease in tumor weight in the SETDB1 shRNA-stably transfected cells (P-values obtained by the ANOVA test). Data shown are means±s.d., n=10.
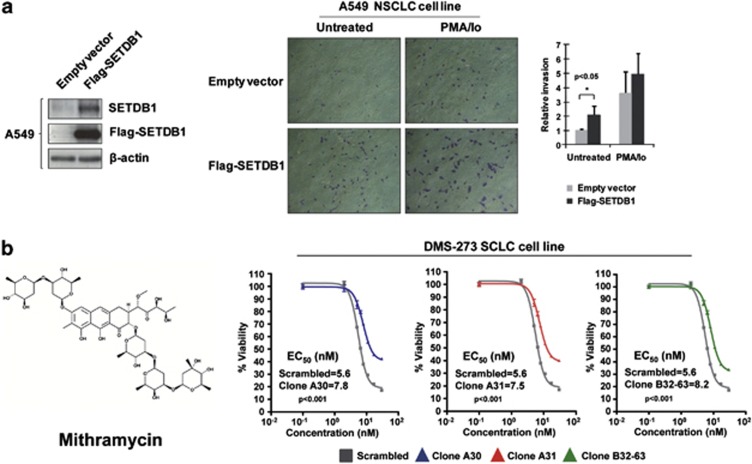
We next tested the ability of SETDB1 shRNA-transfected DMS-273 and -NCI-H1437 cells to form tumors in nude mice compared with scramble shRNA-transfected cells (Figure 2d). DMS-273- and NCI-H1437-scramble shRNA-transfected cells formed tumors rapidly, but cells with shRNA-mediated depletion of SETDB1 had much lower tumorigenicity (Figure 2d). We also studied the oncogenic potential of SETDB1 by evaluating its ability to enhance cell invasion in a lung carcinoma cell line without SETDB1 gene amplification (A549; Figure 3a). To this end, we transfected the plasmids driving the expression of SETDB1 and performed a matrigel invasion assay. We observed a significant increase in the number of invasive cells upon SETDB1 transfection in comparison with empty vector-transfected cells (Figure 3a).
Figure 3.
Impact of SETDB1 on invasiveness and chemosensitivity. (a) Effect of SETDB1 on the invasion potential of A549 cells determined by the matrigel invasion assay. Cells were transfected with 3 μg of Flag-SETDB1 or empty vector in 60 mm dishes. After 24 h, cells were stimulated or not with phorbol myristate acetate (PMA) plus ionomycin (Io) for 30 min. Then cells were trypsinized, and 5 × 104 cells were resuspended in serum-free media and added to the upper compartment of a transwell coated with 1 mg/ml Matrigel (BD Biosciences, Lexington, KY, USA). Media with 10% fetal bovine serum was added in the lower compartment and cells were incubated at 37 °C for 42 h. Invasive cells were fixed with phosphate-buffered saline 4% paraformaldehide, stained with 0.5% violet crystal and visualized and photographed under a × 10 magnification objective with a microscope. Invasive cells were counted using ImageJ 1.45s (Wayne Rasband, National Institutes of Health, Bethesda, MD, USA) and percentage of invasive cells were represented. Results are the mean of at least three experiments by duplicate and the significance was determined using analysis of variance test. *<P=0.05. (b) Cancer cells harboring the SETDB1 gene amplification are sensitive to the decrease in cell viability caused by mithramycin, a SETDB1-interfering drug. 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assays in control-scrambled shRNA DMS-273-transfected cells in comparison with three shRNA-stable downregulated SETDB1 clones (A21, A30 and A31) show enhanced inhibition of viability in cells with SETDB1 gene amplification-mediated overexpression.
We also wondered about gene targets in the amplified lung cancer cells whose expression could be regulated by SETDB1-mediated H3-K9 promoter methylation and that could further explain the above observed impact in cell growth and invasiveness. To find downstream targets of SETDB1 in the amplified lung cancer cell lines DMS-273 and NCI-H1437, we have developed expression microarray analyses (Agilent G4851B 60K, Santa Clara, CA, USA) of both SETDB1 shRNA-depleted cell lines in comparison with their corresponding shRNA-scrambled control cell lines. The microarray expression data obtained are freely available at the Gene Expression Omnibus database: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=tdexdacygiayejy&acc=GSE45175.
Using this approach, we have identified eighteen common genes repressed in both SETDB1-amplified lung cancer cell lines that become upregulated upon SETDB1 shRNA-mediated downregulation (Supplementary Table S2 and Supplementary Figure S4). We have also confirmed the expression changes of the candidate genes by quantitative reverse transcription–PCR and the shift in H3-K9 methylation status in their respective promoters by quantitative chromatin immunoprecitation (Supplementary Figure S4). Related to function, gene ontology analysis of these genes determined ‘regulation of cell proliferation' as the most significantly enriched biological process (false discovery rate=0.0006). Representative examples included the tumor suppressor roles of delta/notch-like epidermal growth factor-related receptor34 and insulin-like growth factor-binding protein 7.35
The observation that the presence of the SETDB1 gene amplification with associated overexpression was critical for the tumorigenesis of these lung cancer cells prompted us to examine whether drugs targeting this pathway might find a therapeutic ‘niche' for their use in the described subset of cases with extra copies of this gene. Similar scenarios have been described for inhibitors of another histone methyltransferase, DOT1L,36 and the BET family of acetyl-lysine-recognizing chromatin ‘adaptor' proteins37, 38, 39 in which hematological malignancies carrying gene-activating events involving targets of these pathways are more sensitive to these drugs. No highly specific inhibitor for SETDB1 has been described in the publicly available literature, but to the best of our knowledge, one agent —mithramycin— could be important in targeting SETDB1.40 Mithramycin is a clinically approved antitumor antibiotic that binds to DNA by interacting with the minor groove and displacing transcriptional activators that bind to GC-rich binding sites.41 Most importantly, it has been shown to suppress basal SETDB1 promoter activity in a dose-dependent manner by inhibiting the binding of Sp transcription factors.40 Thus, we tested whether a putative growth inhibitory effect of this drug in lung cancer cells was dependent on SETDB1 expression. First, we developed western blot analyses for the SETDB1 protein in the SETDB1-amplified lung cancer cell lines DMS-273, NCI-H1437 and NCI-H1395 upon the use of the drug. We found that mithramycin was able to inhibit SETDB1 expression in the three cell lines (Supplementary Figure S5). Using the small lung cancer cell line DMS-273, harboring the previously identified SETDB1 gene amplification, in comparison with three stable short hairpin SETDB1-depleted clones, we observed that the scramble shRNA DMS-273 cells were significantly more sensitive to the growth inhibitory effect mediated by mithramycin than any of the depleted clones (A30, A31 and B32-63; Figure 3 and Supplementary Figure S5). Experiments for each clone were performed in triplicate. Furthermore, we extended the cell viability experiments, using the MTT assay, to the other two SETDB1-amplified lung cancer cell lines (NCI-H1437 and NCI-H1395) and to three SETDB1 non-amplified lung cancer cell lines (DMS-114, A549 and NCI-H1299). The determination of the corresponding EC50 values further confirmed that SETDB1-amplified cell lines (EC50=13.6 nM for NCI-H1437 and EC50=14.7 nM for NCI-H1395) are more sensitive to the drug than the non-amplified cell lines (EC50=347.5 nM for DMS-114, EC50=122 nM for A549 and EC50=32 nM for NCI-H1299; analysis of variance, P<0.001). Thus, at least in vitro, the presence of SETDB1 gene amplification could ‘mark' lung cancer cells that are more sensitive to the inhibition of cell viability associated with the use of mithramycin, for which previous data also suggested SETDB1 as a likely candidate target gene of the drug.40
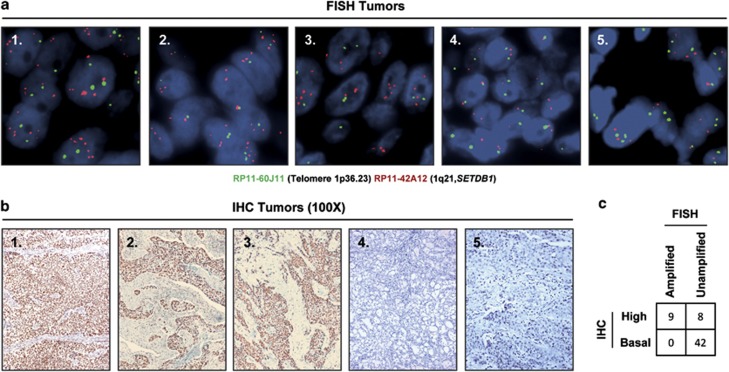
Finally, we sought to demonstrate that the presence of SETDB1 gene amplification was not a specific feature of in vitro grown lung cancer cell lines and that it also occurred in primary tumors of lung cancer patients. In this regard, the 1q chromosome arm undergoes gains (trisomic or tetrasomic) in lung cancer42, 43, 44 that are also associated with overrepresentation of the 1q21 region.45, 46, 47 Recent genomic data48, 49 using single-nucleotide polymorphism microarrays confirm the gain of the SETDB1-1q21 chromosomic region in primary lung tumors. Herein, we performed fluorescence in situ hybridization analyses for the SETDB1 locus using a collection of 59 primary lung tumors, corresponding to 40 non-small cell lung tumors (20 squamous cell carcinoma and 20 adenocarcinomas) and 19 small cell carcinomas (Figure 4a). We identified SETDB1 gene amplification in nine tumors corresponding to 20% (4 of 20), 20% (4 of 20) and 5% (1 of 19) of adenocarcinoma, squamous and small cell lung cancer cases, respectively. Most importantly, we also demonstrated that the presence of extra copies of SETDB1 in these nine primary tumors was associated with overexpression of the SETDB1 protein, as determined by immunohistochemistry in all cases (Figure 4b). Among the remaining fifty unamplified cases, eight (14%) also showed enhanced SETDB1 expression that could be associated with other upstream regulatory events.
Figure 4.
Detection of SETDB1 gene amplification and its associated overexpression in primary tumors from lung cancer patients. (a) Fluorescence in situ hybridization for the SETDB1 gene shows gene amplification in the primary lung tumors 1, 2 and 3. SETDB1 unamplified tumors are shown in the cases 4 and 5. The UCSC genome browser (http://www.genome.ucsc.edu) was used to select the bacterial artificial chromosome (BAC) clone RP11-42A12 spanning the 1q21 region of SETDB1 gene. A telomeric BAC clone located in the telomeric 1p36.23 region was used as a control. (b) Immunohistochemistry for SETDB1 (HPA018142, Sigma-Aldrich, St Louis, MO, USA) shows overexpression of the protein in the above shown three primary lung tumors harboring SETDB1 gene amplification. Minimal expression is detected in the unamplified cases (4 and 5). Magnification × 100. (c) Association between SETDB1 gene amplification and overexpression in the studied fifty-nine cases is shown. Fisher's test, two-tailed P-value<0.0001.
Overall, our results indicate that the histone methyltransferase SETDB1 undergoes gene amplification in the natural history of lung tumorigenesis in non-small and small cell lung cancers. The copy-number gain for SETDB1 is associated with overexpression of the transcript and protein in lung cancer cell lines and primary tumors. From a functional standpoint, SETDB1 exerts growth enhancing activity in vitro and in vivo, as we have shown by depletion and transfection experiments in cell culture and in the nude mice model. Lung cancer cells carrying a SETDB1 gene amplification event are also more sensitive to the antiproliferative action mediated by the antitumoral antibiotic mithramycin, a proposed inhibitor of SETDB1 activity. Thus, our results suggest an oncogenic role for SETDB1 in lung carcinogenesis and raise the possibility of exploring new targeted therapies for the subset of lung tumor patients harboring the SETDB1 gene amplification event.
Acknowledgments
This work was supported by the European Community's Seventh Framework Programme (FP7/2007-2013) under Grant agreement number HEALTH-F2-2010-258677—CURELUNG project, the Institute of Health Carlos III (ISCIII)—PI10/02992, Ministerio de Educación, Ciencia e Innovación Grant SAF2010-14935, Kutxa-Fundación INBIOMED and the Health and Science Departments of the Catalan Government (Generalitat de Catalunya). ME is an ICREA Research Professor.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell. 2010;19:698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Füllgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391–3403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27:396–402. doi: 10.1016/s0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer. 2006;94:179–183. doi: 10.1038/sj.bjc.6602918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi A, Kurokawa M. Key roles of histone methyltransferase and demethylase in leukemogenesis. J Cell Biochem. 2011;112:415–424. doi: 10.1002/jcb.22972. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- Chen H, Tu SW, Hsieh JT. Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. J Biol Chem. 2005;280:22437–22444. doi: 10.1074/jbc.M501379200. [DOI] [PubMed] [Google Scholar]
- Yu J, Cao Q, Mehra R, Laxman B, Yu J, Tomlins SA, et al. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12:419–431. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Beke L, Nuytten M, Van Eynde A, Beullens M, Bollen M. The gene encoding the prostatic tumor suppressor PSP94 is a target for repression by the Polycomb group protein EZH2. Oncogene. 2007;26:4590–4595. doi: 10.1038/sj.onc.1210248. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Jacinto FV, Villanueva A, Fernandez AF, Yamamoto H, Carmona FJ, et al. Silencing of kruppel-like factor 2 by the histone methyltransferase EZH2 in human cancer. Oncogene. 2012;31:1988–1994. doi: 10.1038/onc.2011.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaani E, Nakamura T, Rozovskaia T, Smith ST, Mori T, Croce CM, et al. ALL-1/MLL1, a homologue of Drosophila TRITHORAX, modifies chromatin and is directly involved in infant acute leukaemia. Br J Cancer. 2004;90:756–760. doi: 10.1038/sj.bjc.6601639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Berdasco M, Ropero S, Setien F, Fraga MF, Lapunzina P, Losson R, et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci USA. 2009;106:21830–21835. doi: 10.1073/pnas.0906831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Montgomery GW, Liu JZ, Zhao ZZ, Henders AK, Stark M, et al. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat Genet. 2011;43:1114–1118. doi: 10.1038/ng.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Xia L, Wu DY, Wang H, Chansky HA, Schubach WH, et al. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene. 2002;21:148–152. doi: 10.1038/sj.onc.1204998. [DOI] [PubMed] [Google Scholar]
- Li H, Rauch T, Chen ZX, Szabó PE, Riggs AD, Pfeifer GP. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem. 2006;281:19489–19500. doi: 10.1074/jbc.M513249200. [DOI] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–2489. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Park JS, Kang YK. Dual functions of histone-lysine N-methyltransferase Setdb1 protein at promyelocytic leukemia-nuclear body (PML-NB): maintaining PML-NB structure and regulating the expression of its associated genes. J Biol Chem. 2011;286:41115–41124. doi: 10.1074/jbc.M111.248534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H, Soejima K, Yasuda H, Kawada I, Nakachi I, Yoda S, et al. Deregulation of histone lysine methyltransferases contributes to oncogenic transformation of human bronchoepithelial cells. Cancer Cell Int. 2008;8:15. doi: 10.1186/1475-2867-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2011;9:16–32. doi: 10.1038/nrclinonc.2011.177. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stansky N, Venkatesan K, Margolin AA, Kim S, et al. The cancer cell line encyclopedia enables predictive modeling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Xia S, Lal B, Eberhart CG, Quinones-Hinojosa A, Maciaczyk J, et al. DNER, an epigenetically modulated gene, regulates glioblastoma-derived neurosphere cell differenciation and tumor propagation. Stem Cells. 2009;27:1473–1486. doi: 10.1002/stem.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Lee J, Hagerty SW, Soh BY, McAlpin SE, Cormier KA, et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington's disease. Proc Natl Acad Sci USA. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DE, Jr, Cui DM, Miller DM. Expression of beta-galactosidase under the control of the human c-myc promoter in transgenic mice is inhibited by mithramycin. Oncogene. 1995;10:2323–2330. [PubMed] [Google Scholar]
- Balsara BR, Sonoda G, du Manoir S, Siegfried JM, Gabrielson E, Testa JR. Comparative genomic hybridization analysis detects frequent, often high-level, overrepresentation of DNA sequences at 3q, 5p, 7p, and 8q in human non-small cell lung carcinomas. Cancer Res. 1997;57:2116–2120. [PubMed] [Google Scholar]
- Luk C, Tsao MS, Bayani J, Shepherd F, Squire JA. Molecular cytogenetic analysis of non-small cell lung carcinoma by spectral karyotyping and comparative genomic hybridization. Cancer Genet Cytogenet. 2001;125:87–99. doi: 10.1016/s0165-4608(00)00363-0. [DOI] [PubMed] [Google Scholar]
- Yakut T, Schulten HJ, Demir A, Frank D, Danner B, Egeli U, et al. Assessment of molecular events in squamous and non-squamous cell lung carcinoma. Lung Cancer. 2006;54:293–301. doi: 10.1016/j.lungcan.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Goeze A, Schlüns K, Wolf G, Thäsler Z, Petersen S, Petersen I. Chromosomal imbalances of primary and metastasic lung adenocarcinomas. J Pathol. 2002;196:8–16. doi: 10.1002/path.1009. [DOI] [PubMed] [Google Scholar]
- Sy SM, Wong N, Lee TW, Tse G, Mok TS, Fan B, et al. Distinct patterns of genetic alterations in adenocarcinoma and squamous cell carcinoma of the lung. Eur J Cancer. 2004;40:1082–1094. doi: 10.1016/j.ejca.2004.01.012. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Kawauchi S, Ueda K, Kaneda Y, Oga A, Furuya T, et al. Genomic alterations detected by comparative genomic hybridization in primary lung adenocarcinomas with special reference to the relationship with DNA ploidy. Oncol Rep. 2005;14:1429–1435. [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network Comprenhensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson MD, Yin X, Walter V, Zhao N, Cabanski CR, Hayward MC, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS One. 2012;7:e36530. doi: 10.1371/journal.pone.0036530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.